Abstract
The majority of HIV-1 integrase amino acid sites are highly conserved, suggesting that most are necessary to carry out the critical structural and functional roles of integrase. We analyzed the 34 most variable sites in integrase (>10% variability) and showed that prevalent polymorphic amino acids at these positions did not affect susceptibility to the integrase inhibitor dolutegravir (S/GSK1349572), as demonstrated both in vitro (in site-directed mutagenesis studies) and in vivo (in a phase IIa study of dolutegravir monotherapy in HIV-infected individuals). Ongoing clinical trials will provide additional data on the virologic activity of dolutegravir across subject viruses with and without prevalent polymorphic substitutions.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) integrase (IN), which incorporates reverse-transcribed HIV-1 DNA into the host cell, is an essential enzyme for HIV replication (1), making the inhibition of IN an attractive target for new HIV therapies. Prevalent polymorphisms can affect the antiviral activity of some HIV antiretrovirals, as has been shown recently for the maturation inhibitor bevirimat (2) and historically for nonnucleoside reverse transcriptase inhibitors in HIV-2 infection. Thus, an examination of the effects of polymorphic sites of the IN gene on susceptibility to integrase inhibitors (INIs) is important.
Integrase inhibitors are a new class of antiretroviral agents that overall possess relatively complex constellations of primary and secondary resistance mutations and cross-resistance profiles; consequently, different studies have used different lists for defining INI resistance-associated mutations, with different frequency definitions for what constitutes a polymorphic residue in HIV IN (3–7). Lataillade et al. (4) analyzed the prevalence of natural polymorphisms for IN genes sequenced from INI treatment-naive HIV-1 clade B clinical strains. They used a low-stringency polymorphism cutoff: the occurrence of a single mutation in a single sequence out of their 243 IN sequences analyzed. They found that major INI resistance-associated mutations, including those associated with high-level resistance to the previous INIs (i.e., raltegravir [RAL] and elvitegravir [EVG]), occurred infrequently in samples from INI treatment-naive patients. More specifically, the major amino acid substitutions located within the catalytic domain known to contribute to high-level INI resistance were not generally found to occur as natural polymorphisms; however, two unique samples containing N155H and Q148H were identified in samples originating from Australia. Rhee et al. (7) evaluated the prevalence of natural HIV-1 IN polymorphisms from clinical group M (multiple clades) strains from >1,500 INI treatment-naive subjects. In their analyses, the defined INI resistance mutations were overall nonpolymorphic (defined as a cutoff of 0.5% frequency), with the exception of E157Q, which was present at a 1.1% frequency and was considered to be a primary mutation for elvitegravir (4). In the Rhee study, secondary resistance mutations (e.g., L74M, T97A, V151I, G163R, and S230N) possessed polymorphism rates between 0.5% and 2.0%. In another study, Ceccherini-Silberstein et al. (3) carried out a detailed review of RAL and EVG clinical resistance-associated mutations and compared these with polymorphisms in naive IN sequences. In concordance with the Lataillade and Rhee studies, the INI resistance-associated mutations were found to be similarly infrequent in untreated populations. Furthermore, the authors showed that the majority of amino acid positions (sites) in treatment-naive IN (180 of 288 [62.5%]) are highly conserved (<1% variability) and consist of sites involved in critical functions of the protein. Overall, these findings highlight the relative lack of natural polymorphisms in the HIV-1 IN gene that facilitate drug resistance pathways and reinforce the potential advantage of pursuing IN as a target for the treatment of HIV; however, clinical efficacy data are required to further examine the effects of any naturally occurring IN polymorphisms on clinical responses in patients.
There have been multiple analyses looking at the response in the clinic based on natural variation. In the case of RAL, two different analyses have been presented (8, 9). Marcelin et al. (9) concluded that “HIV-1 subtype and baseline integrase polymorphisms do not influence the virologic response to RAL,” whereas Armenia et al. (8) concluded that “among all integrase polymorphisms, only T122I and S119P were associated with poorer virologic response to raltegravir either at 24 or 48 weeks during treatment.” Overall, these studies indicate that the effect of polymorphisms on response and resistance in the INI class is not as clear as has been observed for bevirimat (2) and suggest that additional investigation may be needed.
Dolutegravir (DTG; S/GSK1349572) is being developed for INI-naive patients as a once-daily, unboosted, HIV INI. To date, limited cross-resistance to clinically relevant RAL- and EVG-resistant mutants has been demonstrated in vitro (10). In the in vitro resistance profiling of DTG produced by passage of HIV-1 strain IIIB (from a chronically infected cell line), IN substitutions L101I, T124A, S153F, and S153Y were observed; notably, however, the HIV-1 IIIB viral population contains ∼40% T124A at baseline (11). Interestingly, the resistance profile of DTG generated by in vitro passage of HIV-1 NL432 (starting virus from an infectious clone) demonstrated that the only IN substitutions observed were E92Q and G193E, and neither substitution reduced DTG susceptibility (12, 13).
The antiviral activity of DTG has been evaluated in a phase IIa, multicenter, randomized, parallel, double-blind, placebo-controlled study investigating three doses of DTG (2, 10, and 50 mg) monotherapy compared with placebo for 11 days in HIV-infected subjects (14). All DTG dose groups had significant reductions in plasma HIV-1 RNA from baseline (P < 0.001), with median changes of −1.5 log10, −2.0 log10, and −2.5 log10 for the 2-mg, 10-mg, and 50-mg dose groups, respectively (compared to +0.05 log10 in subjects receiving placebo), with low variability in either pharmacokinetics or virologic response within each dosing group; these results led us to hypothesize that naturally prevalent polymorphisms in IN do not affect virologic response to therapy with DTG.
Accordingly, we aimed to characterize the prevalent polymorphisms in IN gene sequences obtained from HIV-1-infected populations collected before the broad use of INIs and to assess the effect of these natural IN polymorphisms on fold change (FC) to DTG using two methods: resistance determination in vitro and antiviral responses after DTG monotherapy in subjects with and without IN polymorphisms.
(These data were presented in part at the 50th Interscience Conference on Antimicrobial Agents and Chemotherapy, Boston, MA, September 2010.)
MATERIALS AND METHODS
Generation of a master INI treatment-naive reference sequence alignment.
Integrase inhibitor-naive coding sequences were extracted from two major online repositories: the Los Alamos National Laboratory (LANL) HIV Sequence Database (http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html) and the Stanford Resistance Database (SRD) (6, 15). The LANL sequences generated after 2005 (when late-stage clinical development of RAL was initiated) were excluded, whereas INI-naive sequences from SRD were identified as defined by database annotations and were included. Data from both sources were refined by filtering for full-length IN sequences (≥800 bp) and retaining only those that were mapped to a patient ID. Sequences from both databases were subsequently merged, and only nonredundant sequences were retained. A master reference alignment was then generated after sequence alignment using MUSCLE (16).
A cutoff of >10% variation at the amino acid level was used to define a position as polymorphic. This conservative cutoff ensured that an identified polymorphism would be relevant to clinical data and would not be identified on the basis of sequencing error or sporadic mutations, as a public data set was used in this analysis. Percentage conservation for each site in the master reference alignment was calculated with respect to the most prominent residue at that position. An additional bootstrap analysis was also carried out to ensure that the conservation estimates were reproducible and robust with respect to the addition of new data. This was done by randomly sampling 50 subalignments (with replacement) from the master alignment and calculating the error in the sample estimates. To remove the bias associated with patient sampling, each subalignment had 50 randomly sampled sequences and only 1 representative sequence per patient.
For positions with amino acid mixtures, the weights of the corresponding amino acids were adjusted downward. For example, if a certain mixture position was coded for serine or threonine, then serine and threonine would each be given a weight of 0.5. In comparison, an unambiguous codon coding for serine would be given a weight of 1.
Determination of the influence of polymorphisms on in vitro DTG susceptibility using site-directed molecular clones.
A panel of site-directed molecular clones (SDMs) was constructed from pNL432 with single, double, and triple mutants. This panel comprised IN substitutions identified during in vitro passage with DTG as well as IN polymorphic sites. Fold change susceptibility (FC in IC50 relative to wild-type control virus) to DTG and RAL was measured using HeLa CD4 cells (11, 17), and IC50s are defined as the half-maximal effect.
Evaluation of the effect of polymorphisms on HIV-1 RNA response to DTG.
In the phase IIa dose-ranging study of DTG monotherapy, HIV-infected subjects were randomized to receive DTG (2, 10, or 50 mg) as monotherapy or placebo every 24 h for 10 days. The primary efficacy endpoint was the change from baseline in plasma HIV-1 RNA at day 11 as determined by the COBAS Amplicor HIV-1 monitor test, version 1.5, ultrasensitive preparation (Roche Molecular Diagnostics, Branchburg, NJ) (14). Secondary endpoints included the analysis of viral IN genotypes and phenotypes using the GeneSeq and PhenoSense assays for INIs (Monogram Biosciences, Inc., South San Francisco, CA) (18). Results from this testing are reported as FC in IC50 relative to that of the wild-type control virus. The PhenoSense Integrase assay (Monogram Biosciences, Inc.) has biological cutoffs in FC of 2.5 for DTG and 1.5 for RAL.
This study was conducted in accordance with good clinical practice procedures, all applicable regulatory requirements, and the guiding principles of the Declaration of Helsinki. The study protocol was reviewed and approved by the Copernicus Group Institutional Review Board. All patients provided written informed consent before entry in the study.
Change from baseline in log10 HIV-1 RNA was examined at day 11 after adjusting for the effect of dose in a linear analysis of variance model at polymorphic sites, and conserved changes were identified in passage (e.g., S153, G193). Residuals from this model (i.e., the difference between a subject's observed response and the average response at the dose the subject received) were categorized by the presence of a non-wild-type amino acid at each of the codons identified as polymorphic (and for which there was at least 1 nonconsensus amino acid). The effect of polymorphism at each position was assessed for the day 11 response using a multivariate linear model that adjusted for dose and for the presence of nonreference amino acids at each of the identified positions. This model was fitted once for each position. The P value for the effect of each polymorphism at each position was calculated from the linear model's Wald test of the estimated effect of the presence of nonreference amino acids. The P values for these 30 assessments of the effect of polymorphism were not adjusted for multiplicity.
RESULTS
Prevalence of IN variability and polymorphisms.
A total of 2,997 IN sequences were identified and aligned. The major genomic subtypes provided in this data set (n = 1,954) included B (33%), A1 (5.78%), C (24%), and CRF01_AE (5%). Analysis of naive sequences obtained from LANL and SRD revealed a high degree of conservation across the 288 amino acid residues (Fig. 1). The majority of positions (59%) were found to vary by <1% at the amino acid level (i.e., >99% conservation). These highly conserved positions spanned long regions across the IN protein.
Fig 1.
Integrase amino acid conservation of 2,997 sequences from the Los Alamos National Laboratory HIV Sequence Database and the Stanford Resistance Database (http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html) (6, 15).
A total of 34 positions met the criteria for a polymorphic site (>10% amino acid diversity) and were found to be distributed across IN, with some positions directly adjacent to each other (e.g., sites 134, 135, and 136; Fig. 2; see also Table S1 in the supplemental material). A bootstrap analysis indicated that the conservation estimates of these 34 polymorphic sites were highly reproducible, with site 119 recording the highest error rate (75.53% ± 1.05%). Both sites 101 and 124 were found to be highly polymorphic (58.20% ± 0.94% and 52.24 ± 0.94% error rate, respectively). Mutations at site 74 that add or modulate resistance to RAL and EVG (when present with other primary mutations) (11, 19) were found in 11.5% of sequences. Frequencies of each amino acid identified were as follows: L74L, 88.57%; L74I, 8.94%; and L74M, 2.19%. No other position identified as a major INI mutation or INI resistance-associated substitution was found to be polymorphic at ≥10%.
Fig 2.

Integrase amino acid distribution at 34 polymorphic sites (>10%) from the Los Alamos National Laboratory HIV Sequence Database and the Stanford Resistance Database (http://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html) (6, 15). The color of amino acids reflects their chemical properties: polar amino acids, green; basic amino acids, blue; acidic amino acids, red; hydrophobic amino acids, black.
Effect of polymorphisms on INI susceptibility in vitro.
In addition to the polymorphic substitutions identified from the survey of LANL and SRD sequences, this study also investigated previously identified substitutions that arose during in vitro passage (11). The SDMs with polymorphic amino acid changes T124A and L101I had DTG FC values below the assay cutoff for reduced susceptibility. Fold change for SDMs ranged from 0.91 for the L101I/T124A double substitution to 2.5 for the nonpolymorphic substitution S153Y, which arose during in vitro passage of DTG (Table 1). The mean FC for L101I was 1.4 for DTG and 1.2 for RAL, each of which is within the assay's variability. The addition of polymorphisms L101I and T124A to the S153F mutation had an FC within the assay's variability of 2. It is important to note that the DTG trough concentrations were 19-fold above the protein-adjusted inhibitory concentration at 90% (IC90) of 152 nM that was observed at the once-daily, 50-mg dose (20); this is well above the maximum FC for all SDMs tested in this study.
Table 1.
In vitro drug susceptibility for site-directed mutants with polymorphisms and substitutions observed during DTG passagea
| Amino acid substitution(s) in NL432 | Where observed | DTG FC, mean ± SD | RAL FC, mean ± SD |
|---|---|---|---|
| L101I | Polymorphism/passage | 1.40 ± 0.36 | 1.20 ± 0.12 |
| T124A | Polymorphism/passage | 0.95 ± 0.19 | 0.82 ± 0.08 |
| L101I/T124A | Polymorphism | 0.91 | 0.77 |
| S153F | Passage | 1.60 ± 0.20 | 1.30 ± 0.14 |
| S153Y | Passage | 2.50 ± 1.1 | 1.30 ± 0.19 |
| L101I/S153F | Polymorphism/passage | 2.00 ± 0.11 | 1.30 ± 0.54 |
| L101I/T124A/S153F | Polymorphism/passage | 1.90 ± 0.24 | 1.40 ± 0.14 |
| E92Q | Passage | 1.60 ± 0.12 | 3.50 ± 1.4 |
| G193E | Passage | 1.30 | 1.30 |
Assay variability of 2-fold changes. Data were previously presented (11) and are results from 3 to 5 independent experiments.
Effect of polymorphisms on response to DTG.
Of the 34 codons identified as polymorphic, 6 were not represented in the baseline isolates from participants in the phase IIa study. Virus from all 28 subjects receiving DTG had wild-type amino acids at codons 21, 84, 100, 269, and 278 and a non-wild-type amino acid at codon 234.
Differences in the group median FC values were calculated for DTG and RAL at sites 101 and 124 (Table 2). All 101 and 124 genotypes had similar IC50s and overlapping variance ranges for DTG and RAL. The average FC to DTG was 0.79, and the average FC to RAL was 0.89. No polymorphism showed an average shift in susceptibility of >2-fold to either DTG or RAL, and no median FC observed was greater than the FC assay variability (i.e., FC = 2).
Table 2.
Fold change response to DTG and RAL for day 1 isolate samples (n = 35) with variation at amino acids 101 and 124
| Variation at site: |
No. of subjects | DTG FC, median (range) | RAL FC, median (range) | |
|---|---|---|---|---|
| 101 | 124 | |||
| L101L | T124T | 6 | 0.64 (0.47–0.89) | 0.72 (0.5–0.95) |
| L101L | T124A | 5 | 0.97 (0.63–1.17) | 0.91 (0.58–1.05) |
| L101L | T124N | 7 | 0.78 (0.66–0.82) | 0.92 (0.83–1.41) |
| L101L | T124S | 2 | 0.825 (0.79–0.86) | 0.965 (0.96–0.97) |
| L101L | T124T/N | 1 | 0.89 | 0.82 |
| L101L | T124T/N/D/A | 1 | 0.89 | 1.15 |
| L101I | T124T | 5 | 0.79 (0.72–0.88) | 0.86 (0.84–1.16) |
| L101I | T124A | 3 | 0.81 (0.78–0.89) | 0.79 (0.76–0.97) |
| L101I | T124N | 3 | 0.85 (0.46–0.86) | 0.97 (0.89–0.98) |
| L101I | T124S | 1 | 0.74 | 0.64 |
| L101I | T124T/N | 1 | 0.8 | 1.16 |
The residuals (i.e., differences in change from baseline in log10 HIV-1 RNA at day 11 after adjusting for the effect of dose in a linear model) at positions of polymorphic substitution and conserved changes identified in passage (i.e., S153, G193) are shown in Fig. 3. These observed medians were consistent with random variation from a mean of zero and a small variance (<0.3 log10 copies/ml).
Fig 3.
Dose-adjusted differences in viral load (log10) response at day 11 by polymorphisms present at day 1 from a phase IIa study (n = 28). The number within each circle represents the number of genotypes polymorphic at the given position on day 1. For each codon with >1 nonconsensus amino acid, the median and interquartile range of residuals are plotted. Box plots are provided on the right to show the random variation from the mean of zero for the polymorphisms tested.
The effect of substitutions at each position was tested using a multivariate linear model that adjusted for dose and the presence or absence of substitutions at each position (1 position at a time). Figure 4 shows the 30 P values for tests of the effect of substitutions at each of the 30 positions analyzed. The P values ranged from 0.035 to 0.99 (median, 0.56; interquartile range, 0.21 to 0.70). The P values shown in Fig. 4 are not adjusted for multiple comparisons, because any such adjustments rendered even the smallest P value observed (P = 0.035) nonsignificant. When plotted against the corresponding quantiles from a uniform distribution, the distribution of P values shows a close approximation to such a distribution (i.e., numbers randomly and evenly sampled between 0 and 1). For example, the dashed line shows that just under 5% of codons (1 of 30 tested) had a P value of ≤0.05 in a test of the significance of their effect on the day 11 response. The dotted line shows that just over 20% of codons (7 of 30 tested) had a P value of ≤0.2. This pattern of P values is consistent, with no position examined having an effect on the day 11 response to DTG. In summary, the results of the in vivo analysis support the hypothesis that the polymorphisms had no impact on the day 11 response to DTG.
Fig 4.
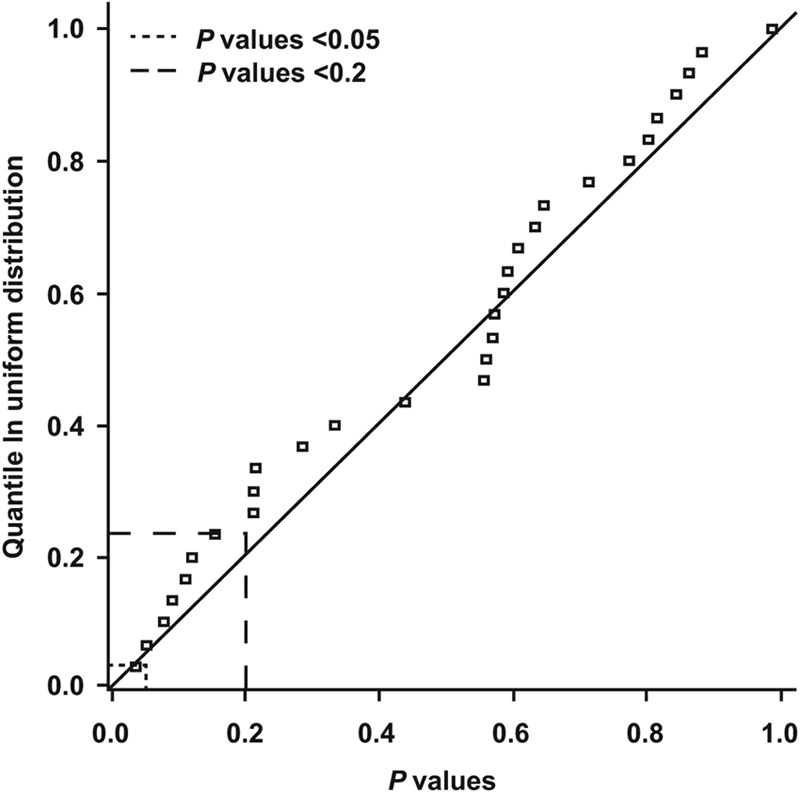
Theoretical quantile distribution of P values for tests of the effect of amino acid substitutions using a multivariate linear model adjusted for dose and the presence or absence of substitutions at each position.
DISCUSSION
Although the majority (59%) of IN amino acids from the large sample data set (∼2,997 sequences) of carefully selected INI-naive sequences were >99% conserved, 34 positions in this data set were polymorphic based on having >10% variation at the amino acid level. This cutoff provided a reasonable number of polymorphic sites for studying DTG susceptibility, and the number of polymorphic sites identified compares well with previous studies that probed smaller numbers of sequences from targeted HIV-1 clinical samples (3, 4, 6). Our analysis identified position 74 as a polymorphic site. The L74M substitution is a recognized INI resistance-associated substitution that does not decrease INI susceptibility alone but that can significantly reduce RAL and EVG susceptibility when present with major INI mutations (11). In this analysis, L74M was detected in our panel of sequences at a frequency of 2.9%; other studies have confirmed this low frequency of L74M in antiretroviral therapy-naive patients (3, 21).
Several groups have considered the role of polymorphisms in terms of implications for resistance to DTG (22–24). The prevalence data showed similar frequencies as previously described (7), and it was concluded that L101I and T124A were present in approximately 50% of subjects examined. Of note, these studies evaluated only the prevalence of polymorphisms L101I and T124A and did not have data on clinical response or in vitro drug susceptibility for their analyses. The Métifiot et al. study (23) examined these two polymorphisms from a structural standpoint and again could find no supporting evidence for their impact on DTG susceptibility.
Variability at polymorphic sites 101 and 124 was significant in our study (>50% for L101I and T124A). However, testing of SDMs at sites 101 and 124 as either single or double mutants demonstrated no effect on DTG susceptibility. Baseline clinical samples taken from participants in our phase IIa study showed no differences in DTG susceptibility in the presence or absence of L101I or T124A (Table 2). Most importantly, subjects receiving 10 days of DTG monotherapy demonstrated no apparent difference in viral load decline with any polymorphism, including those involving sites 101 and 124 (Fig. 3). Notably, virologic responses have also been excellent in the phase IIb SPRING-1 study; 90% of treatment-naive subjects on DTG plus two nucleoside reverse transcriptase inhibitors (NRTIs) had HIV-1 RNA of <50 copies/ml after 48 weeks of therapy, compared with 82% treated with efavirenz plus two NRTIs (20). Given the high prevalence of polymorphisms at these positions, one would expect lower short- or long-term responses if such polymorphisms were associated with response.
Although this analysis focused only on the presence of nonreference amino acids at the 34 polymorphic positions, all subjects had multiple, additional amino acid substitutions compared with the reference. No attempt was made in this analysis to factor out their contributing influence to response. Thus, these results indicate that no single polymorphism led to a substantial increase or decrease in clinical response, a finding confirmed by the nonsignificant effects of the presence of nonreference amino acids in multivariate models of that day 11 response. The high response rates through 48 weeks observed in the phase IIb SPRING-1 study and two large phase III studies (20, 25, 26) further underscore the consistent and potent activity of DTG across INI-naive patients despite polymorphisms, such as those at positions 101 and 124 in HIV-1 IN.
Our data from INI-naive patients suggest that polymorphisms at the most variable positions in wild-type IN do not reduce susceptibility to DTG. Ongoing trials will provide additional data on the virologic activity of DTG across subject viruses with and without prevalent polymorphic substitutions.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by ViiV Healthcare.
All listed authors meet the criteria for authorship set forth by the International Committee for Medical Journal Editors. All authors are employees of GlaxoSmithKline, ViiV Healthcare, or Shionogi & Co., Ltd.
We acknowledge the following individuals for their editorial assistance during the development of the manuscript: Todd Parker, Chris Lawrence, and Christine Levesque.
Footnotes
Published ahead of print 7 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.01791-12.
REFERENCES
- 1. LaFemina RL, Schneider CL, Robbins HL, Callahan PL, LeGrow K, Roth E, Schleif WA, Emini EA. 1992. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J. Virol. 66:7414–7419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lu W, Salzwedel K, Wang D, Chakravarty S, Freed EO, Wild CT, Li F. 2011. A single polymorphism in HIV-1 subtype C SP1 is sufficient to confer natural resistance to the maturation inhibitor bevirimat. Antimicrob. Agents Chemother. 55:3324–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ceccherini-Silberstein F, Malet I, D'Arrigo R, Antinori A, Marcelin AG, Perno CF. 2009. Characterization and structural analysis of HIV-1 integrase conservation. AIDS Rev. 11:17–29 [PubMed] [Google Scholar]
- 4. Lataillade M, Chiarella J, Kozal MJ. 2007. Natural polymorphism of the HIV-1 integrase gene and mutations associated with integrase inhibitor resistance. Antivir. Ther. 12:563–570 [PubMed] [Google Scholar]
- 5. Myers RE, Pillay D. 2008. Analysis of natural sequence variation and covariation in human immunodeficiency virus type 1 integrase. J. Virol. 82:9228–9235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rhee S-Y, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31:298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rhee S-Y, Liu TF, Kiuchi M, Zioni R, Gifford RJ, Holmes SP, Shafer RW. 2008. Natural variation of HIV-1 group M integrase: implications for a new class of antiretroviral inhibitors. Retrovirology 5:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Armenia D, Malet I, Fabeni L, Reigadas S, Guillot V, Dunn D, Kaiser R, Zazzi M, Van Laethem K, Artese A, Garrido C, Masquelier B, Garcia F, Calvez V, Antinori A, Marcelin AG, Perno CF, Geretti AM, Ceccherini-Silberstein F, on behalf of the CORONET Study Group and in collaboration with CHAIN 2011. Impact of baseline HIV-1 integrase polymorphisms on virological outcome in a large European cohort of patients starting a raltegravir-containing regimen. Antivir. Ther. 16(Suppl 1):A69 [Google Scholar]
- 9. Marcelin AG, Delaugerre C, Flandre P, Descamps D, Morand-Joubert L, Amiel C, Ferre V, Izopet J, Masquelier B, Calvez V, the ANRS AC11 Resistance Group 2011. Factors associated with virological response to raltegravir in experienced HIV-infected patients. Antivir. Ther. 16(Suppl 1):A74 [Google Scholar]
- 10. Powderly WG. 2010. Integrase inhibitors in the treatment of HIV-1 infection. J. Antimicrob. Chemother. 65:2485–2488 [DOI] [PubMed] [Google Scholar]
- 11. Kobayashi M, Yoshinaga T, Seki T, Wakasa-Morimoto C, Brown KW, Ferris R, Foster SA, Hazen RJ, Miki S, Suyama-Kagitani A, Kawauchi-Miki S, Taishi T, Kawasuji T, Johns BA, Underwood MR, Garvey EP, Sato A, Fujiwara T. 2011. In vitro antiretroviral properties of S/GSK1349572, a next-generation HIV integrase inhibitor. Antimicrob. Agents Chemother. 55:813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sato A, Seki T, Kobayashi M, Yoshinaga T, Fujiwara T, Underwood M, Garvey E, Johns B. 2009. In vitro passage of drug resistant HIV-1 against a next generation integrase inhibitor (INI), S/GSK1349572. Abstr. 49th Intersci. Conf. Antimicrob. Agents Chemother., abstr H-932 [Google Scholar]
- 13. Seki T, Kobayashi M, Wakasa-Morimoto C, Yoshinaga T, Sato A, Fujiwara T, Underwood M, Garvey E, Johns B. 2010. S/GSK1349572 is a potent next generation HIV integrase inhibitor and demonstrates a superior resistance profile substantiated with 60 integrase mutant molecular clones. Abstr. 17th Conf. Retroviruses Opportunistic Infect., abstr 555 [Google Scholar]
- 14. Min S, Sloan L, DeJesus E, Hawkins T, McCurdy L, Song I, Stroder R, Chen S, Underwood M, Fujiwara T, Piscitelli S, Lalezari J. 2011. Antiviral activity, safety, and pharmacokinetics/pharmacodynamics of dolutegravir as 10-day monotherapy in HIV-1-infected adults. AIDS 25:1737–1745 [DOI] [PubMed] [Google Scholar]
- 15. Shafer RW. 2006. Rationale and uses of a public HIV drug-resistance database. J. Infect. Dis. 194(Suppl 1):S51–S58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nakahara K, Wakasa-Morimoto C, Kobayashi M, Miki S, Noshi T, Seki T, Kanamori-Koyama M, Kawauchi S, Suyama A, Fujishita T, Yoshinaga T, Garvey EP, Johns BA, Foster SA, Underwood MR, Sato A, Fujiwara T. 2009. Secondary mutations in viruses resistant to HIV-1 integrase inhibitors that restore viral infectivity and replication kinetics. Antiviral Res. 81:141–146 [DOI] [PubMed] [Google Scholar]
- 18. Fransen S, Gupta S, Danovich R, Hazuda D, Miller M, Witmer M, Petropoulos CJ, Huang W. 2009. Loss of raltegravir susceptibility by human immunodeficiency virus type 1 is conferred via multiple nonoverlapping genetic pathways. J. Virol. 83:11440–11446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cooper DA, Steigbigel RT, Gatell JM, Rockstroh JK, Katlama C, Yeni P, Lazzarin A, Clotet B, Kumar PN, Eron JE, Schechter M, Markowitz M, Loutfy MR, Lennox JL, Zhao J, Chen J, Ryan DM, Rhodes RR, Killar JA, Gilde LR, Strohmaier KM, Meibohm AR, Miller MD, Hazuda DJ, Nessly ML, DiNubile MJ, Isaacs RD, Teppler H, Nguyen B-Y. 2008. Subgroup and resistance analyses of raltegravir for resistant HIV-1 infection. N. Engl. J. Med. 359:355–365 [DOI] [PubMed] [Google Scholar]
- 20. van Lunzen J, Maggiolo F, Arribas JR, Rakhmanova A, Yeni P, Young B, Rockstroh JK, Almond S, Song I, Brothers C, Min S. 2011. Once daily dolutegravir (S/GSK1349572) in combination therapy in antiretroviral-naive adults with HIV: planned interim 48 week results from SPRING-1, a dose-ranging, randomised, phase 2b trial. Lancet Infect. Dis. 12:111–118 [DOI] [PubMed] [Google Scholar]
- 21. Varghese V, Liu TF, Rhee SY, Libiran P, Trevino C, Fessel WJ, Shafer RW. 2010. HIV-1 integrase sequence variability in antiretroviral naïve patients and in triple-class experienced patients subsequently treated with raltegravir. AIDS Res. Hum. Retroviruses 26:1323–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marcelin A-G, Malet I, Fabeni L, Armenia D, Fourati S, Masquelier B, Katlama C, Perno CF, Calvez V, Ceccherini-Silberstein F. 2010. Resistance-associated mutations to integrase inhibitor S/GSK1349572 in HIV-1 integrase inhibitor-naive and raltegravir-experienced patients. Abstr. 17th Conf. Retroviruses Opportunistic Infect., abstr 554 [Google Scholar]
- 23. Métifiot M, Marchand C, Maddali K, Pommier Y. 2010. Resistance to integrase inhibitors. Viruses 2:1347–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Saladini FP, Meini G, Bianco C, Monno L, Punzi G, Pecorari M, Borghi V, Di Pietro M, Filice G, Gismondo MR, Micheli V, Penco G, Carli T, De Luca A, Zazzi M, Collaborative Group ARCA 2011. Prevalence of mutations related to possible resistance to dolutegravir in raltegravir naïve and pretreated patients. Abstr. 13th European AIDS Conf., abstr PE5.1/4 [DOI] [PubMed] [Google Scholar]
- 25. Raffi F, Rachlis A, Stellbrink Hardy H-JWD, Torti C, Orkin C, Bloch M, Podzamczer D, Pokrovsky V, Almond S, Margolis D, Min S, the SPRING-2 Team 2012. Once-daily dolutegravir (DTG; S/GSK1349572) is noninferior to raltegravir (RAL) in antiretroviral-naive adults: 48 week results from SPRING-2 (ING113086). Abstr. 14th Int. AIDS Conf., abstr THLBB04 [Google Scholar]
- 26. Walmsley S, Antela A, Clumeck N, Duiculescu D, Eberhard A, Gutiérrez F, Hocqueloux L, Maggiolo F, Sandkovsky U, Granier C, Wynne B, Pappa K. 2012. Dolutegravir (DTG; S/GSK1349572) + abacavir/lamivudine once daily statistically superior to tenofovir/emtricitabine/efavirenz: 48-week results—SINGLE (ING114467). Abstr. 52nd Intersci. Conf. Antimicrob. Agents Chemother., abstr H-556b [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.




