Abstract
Recently, a novel variant of mecA known as mecC (mecALGA251) was identified in Staphylococcus aureus isolates from both humans and animals. In this study, we identified a Staphylococcus xylosus isolate that harbors a new allotype of the mecC gene, mecC1. Whole-genome sequencing revealed that mecC1 forms part of a class E mec complex (mecI-mecR1-mecC1-blaZ) located at the orfX locus as part of a likely staphylococcal cassette chromosome mec element (SCCmec) remnant, which also contains a number of other genes present on the type XI SCCmec.
TEXT
Methicillin resistance in staphylococci is encoded by mecA, encoding the penicillin-binding protein 2a (PBP2a), which has a low affinity for beta-lactam antibiotics (1). As a result, the transpeptidase activity of PBP2a is functional at normally inhibitory concentrations of beta-lactam antibiotics, allowing cell wall synthesis to occur (2–4). Recently, a novel variant of mecA was identified in Staphylococcus aureus from cattle (5), humans, and a range of other animal species (6) in Denmark, France, The Netherlands, Ireland, Germany, Belgium, and the United Kingdom (5, 7–11). This subtype was originally designated mecALGA251 but has since been renamed mecC and shares 70% nucleotide identity with the conventional mecA gene. The mecC gene is present with its cognate regulators mecI-mecR1, as part of a class E mec complex that shares structural similarity (mecI-mecR1-mecC-blaZ) with a mec gene complex found in Macrococcus caseolyticus (12). The class E complex is present as part of a larger, 29.4-kb, type XI staphylococcal cassette chromosome mec element (SCCmec) inserted at orfX; this element also includes the recombinase genes ccrAB and arsenic resistance genes. In this work, we describe a highly related mecC homolog present in the orfX locus in a Staphylococcus xylosus.
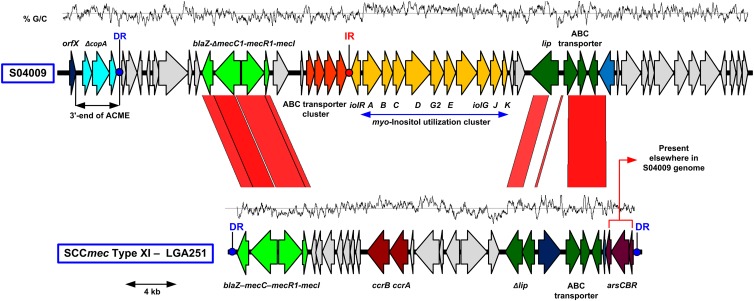
A search of the EMBL nucleotide database identified submission of sequences from S. xylosus strain S04009 (13) with a high degree of similarity (>90%) to mecC (5, 8). However, antimicrobial susceptibility testing of strain S04009 by disk diffusion with oxacillin and cefoxitin showed it to be susceptible to both antibiotics but resistant to penicillin using British Society for Antimicrobial Chemotherapy (BSAC) criteria (version 10.2) (data not shown). Therefore, we submitted the mecC-positive strain S04009 and a mecC-negative S. xylosus isolate, S040010, for whole-genome sequencing to further characterize the mecC-containing region. Illumina library preparation was carried out as described by Quail et al. (14), and HiSeq sequencing was carried out following the manufacturer's standard protocols (Illumina, Inc.). Genome sequencing confirmed the presence of a mecC homolog in S04009 located downstream of the S. xylosus orfX homolog, a region associated with horizontally transferred elements (Fig. 1). Immediately downstream of orfX in S04009 is a 3.3-kb region that shows a high degree of similarity (>95% nucleotide identity) to the 3′ end of the arginine catabolic mobile element (ACME) in the S. aureus USA300 strain FPR3757 (EMBL accession no. CP000255) and contains a truncated version of copA, an ATPase copper transporter (15). Next to this region is the mec complex. The mecC gene in S04009 shares 93.5% nucleotide identity to mecC in S. aureus LGA251 and 69.9% to mecA from S. aureus strain MRSA252. Based on the current guidelines for reporting mecA homologs, the S04009 mecC gene is a new allotype of the LGA251 mecC, herein referred to as mecC1 (16). Sequence analysis of the mecC1 gene identified a frameshift mutation close to the 5′ end of the gene, resulting in a truncated 64-amino-acid (aa) product, providing a molecular basis for the oxacillin and cefoxitin susceptibility of strain S04009. mecC1 is found in a homologous class E mec gene complex (mecI-mecR1-mecC1-blaZ) which has been previously reported in S. aureus (5, 8) The presence of the blaZ gene is likely to account for the observed penicillin resistance of S04009 despite mecC1 being inactivated. mecI, mecR1, and blaZ in S04009 share 91.1%, 90.0%, and 90.9% nucleotide identity, respectively, with their homologs in LGA251. Downstream of the mec gene complex is a hypothetical protein conserved in a number of coagulase-negative Staphylococcus (CoNS) species, followed by a tandem pair of ATP-binding cassette transporters (ABC transporters). After the final ABC transporter gene, there is an imperfect 53-bp inverted repeat (IR), which suggests that this region was once part of a separate mobile element or has undergone deletion mediated by this repeat. Immediately upstream of this is a myo-inositol (MI) utilization cluster, which was previously identified in strain S04009 by subtractive hybridization (13). Downstream from the MI utilization cluster are more genes present in joining region 1 (J1) in the type XI SCCmec in LGA251. The lipase gene, which is present as two truncated pseudogenes (SARLGA251_00420 and SARLGA251_00430) in LGA251, is intact in S04009. Adjacent to this are genes for an ABC transporter permease, an ABC transporter ATPase, and a conserved hypothetical protein (SARLGA251_00470-490) with 96%, 97%, and 98% nucleotide identity, respectively, to those in LGA251. Downstream of the conserved ABC transporter genes in S04009 is a gene for a major facilitator superfamily (MFS) protein that is absent from SCCmec type XI, ending the region of homology. Interestingly, arsR, arsB, and arsC are present in S04009 and share 83%, 88%, and 91% nucleotide identity, respectively, with their homologs in LGA251. However, they are not found proximal to the orfX region but are instead associated with a Tn554-like transposon and are inserted at a different location in the S04009 genome (data not shown).
Fig 1.
Comparison of the orfX region in S. xylosus S04009 (EMBL accession no. HE993884) and the SCCmec type XI in LGA251 (EMBL accession no. FR821779). Red areas are regions conserved between the two sequences, and homologous coding sequences (CDSs) are in bright and dark green. Other CDSs of interest discussed in the text are highlighted in color. Blue and red dots indicate direct repeats (DR) and inverted repeats (IR), respectively. The GC content is shown above each region.
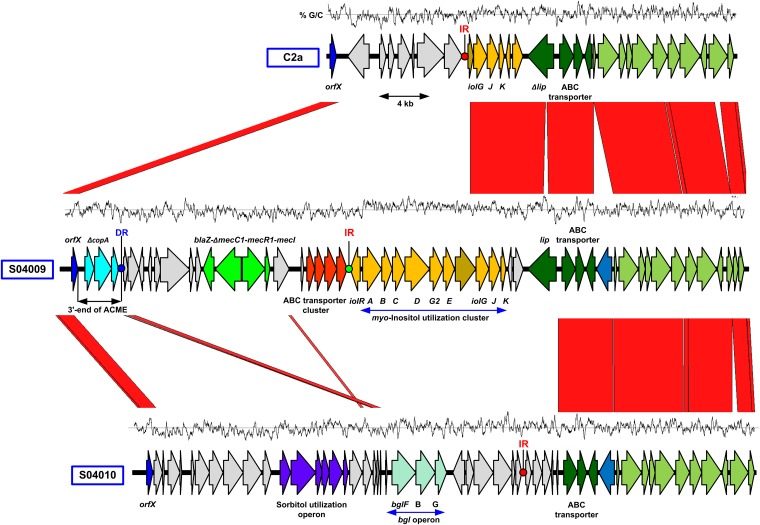
In order to further understand the evolutionary history of the mecC1-containing element in S. xylosus, we compared the orfX locus of strain S. xylosus S04009 with those of two other S. xylosus strains, S040010, and a third S. xylosus strain, C2a (S. Leroy, unpublished data). Immediately downstream of the orfX in C2a is an ∼9-kb region absent from S04009, which shares blocks of homology to Enterococcus faecalis D32 (EMBL accession no. CP003726) at the 5′ end and to Staphylococcus haemolyticus JCSC1435 (EMBL accession no. AP006716) at the 3′ end. This region contains a number of genes associated with mobile elements, including a truncated abortive phage infection protein (AIPR), a type I restriction modification system restriction subunit, and two genes that likely encode an McrBC 5-methylcytosine restriction system. Immediately flanking this region is a truncated copy of the putative Na+/myo-inositol cotransporter, which is interrupted by a 55-bp imperfect inverted repeat (Fig. 2). In S04010, downstream of orfX is an ∼30 kb region which is absent from both S04009 and C2a. This region displays short regions of homology to corresponding regions in E. faecalis, Staphylococcus carnosus subsp. carnosus, S. aureus, and a number of other Gram-positive species. The region proximal to orfX contains a number of hypothetical proteins and, like C2a, a putative restriction modification system. Downstream of this is a sorbitol utilization operon which is found next to a type IV SCCmec in S. aureus strain VRS3a (17) and part of a SCCmecWAMRSA40 composite island (EMBL accession no. JQ746621) which is found on the chromosome in S. carnosus strain TM300 (17). The sorbitol operon is also present in E. faecalis strain D32. Further downstream from this are three genes that make up a bgl (aryl-β,d-glucoside) operon. Downstream from this is an ∼200-bp region that shares 91% nucleotide identity with the IR-containing region in C2a (Fig. 2), the IR itself being identical in 50 of 55 nucleotides. Further small regions of homology exist between S04009 and S04010, consisting of an ∼750 bp region immediately downstream of the ACME DR in S04009 and a region just before the bgl operon in S04010. In order to ascertain the prevalence of mecC1 in S. xylosus strains, we screened a total of 114 S. xylosus isolates from a wide range of sources, though with a deliberate bias toward isolates from bovine milk, as this was the original source of the strain S04009 (Table 1). (Additional information about S. xylosus strains screened for mecC is presented in Table 2.) We screened the strains by PCR using primers for mecC/mecC1, blaZ, and mecA and universal staphylococcal 16S primers (Table 3). Neither mecA, mecC, nor blaZ was detected in any of these isolates.
Fig 2.
Comparison of the orfX regions in S. xylosus strains C2a (EMBL accession no. HE993886), S04009 (EMBL accession no. HE993884), and S04010 (EMBL accession no. HE993885). Red areas show regions conserved between the two sequences, and homologous CDSs are in light and dark green. Other CDSs of interest discussed in the text are highlighted in color. Blue dots indicate direct repeats (DR), and green and red dots show inverted repeats (IR) (the inverted repeats in C2a and S04010 are virtually identical). The GC content of the region is shown above each genome schematic. The eighth gene in the myo-inositol cluster that is truncated in C2a and intact in S04009 is indicated with shading.
Table 1.
Bacterial strains used in this study
Table 2.
Overview of Staphyloccocus xylosus isolates screened for mecC
| No. of isolates screened | Relevant characteristics | Reference |
|---|---|---|
| 15 | The Netherlands, bovine milk; oxacillin MIC ≥ 0.5 μg/ml | 26 |
| 20 | Switzerland, bovine milk; oxacillin MIC ≥ 0.5 μg/ml | This work |
| 5 | France, various sources | 13 |
| 3 | Switzerland, horse skin; 2 isolates with oxacillin MIC ≥ 0.5 μg/ml | 27 |
| 70 | United States, bovine milk and streak canals | 28 and this work |
| 1 | United States, human skin; ATCC 29971 | 29 |
Table 3.
Oligonucleotide primers used in this study
| Primer name | Sequence (5′→3′) | Target | Reference |
|---|---|---|---|
| mecC1 + 2_F | 5′-AAGTTAATCAAAAATGGGTTCAGC-3′ | mecC | This work |
| mecC1 + 2_R | 5′GGTTGTAATGCTGTACCAGATCC-3′ | mecC | This work |
| blaZ_XI_F | 5′-CGTTTTGCWTATGCTTCCAC-3′ | blaZ | This work |
| blaZ_XI_R | 5′-CKGGTTCTTTTCTAGATGGATG-3′ | blaZ | This work |
| MecA1 | GTA GAA ATG ACT GAA CGT CCG ATA A | mecA | 30 |
| MecA2 | CCA ATT CCA CAT TGT TTC GGT CTA A | mecA | 30 |
| 16SF | CCTATAAGACTGGGATAACTTCGGG | 16S rDNA | 31 |
| 16SR | CTTTGAGTTTCAACCTTGCGGTCG | 16S rDNA | 31 |
The finding that multiple components of the type XI SCCmec are present in contiguous blocks in the chromosome of S. xylosus S04009 suggests that this element may represent the remnants of an ancestral SCCmec element. Given the lack of any SCCmec flanking repeats in S04009 and the change in the GC content after the inverted repeat between the MI utilization cluster region and the mecC1-containing region, it is not clear if these two regions represent a single larger element or multiple independent acquisitions by an ancestral strain. The presence of the truncated MI cluster in C2a does suggest that the MI utilization cluster was part of a single contiguous block with the lipase and the ABC transporters in both S04009 and C2a. The finding that the arsenic resistance genes are also present in S04009 in association with a transposon further highlights a potential mechanism for the acquisition of these genes into the type XI SCCmec. Therefore, based on the available evidence, we suggest that the class E mec complex in S. xylosus was part of a larger ancestral SCCmec element which probably included the MI cluster and the lipase and ABC transporters and that this element has undergone gradual deletion and acquisition (of the arsenic resistance genes) to the type XI SCCmec identified in S. aureus LGA251 (5). The fact that we found no other S. xylosus strains harboring mecC or blaZ suggests that mecC1 might be present in only a minor subset of S. xylosus isolates. It is noteworthy that S. xylosus is present in fermented foods such as sausage (18, 19) and cheese (20), highlighting another potential route for the transmission of antibiotic resistance genes from the environment to human flora (21). Given the recent discovery of mecR2, a third regulator of mecA expression, it would also be interesting to see if the expression of mecC1 is positively regulated in the same way by xylR, encoding the xylose operon repressor (present in S. xylosus S04009), which is a close homolog of the mecR2 regulator (22). In S. xylosus, as in other staphylococci, the orfX locus is a site for the integration of multiple SCC-like elements. The strains analyzed in this study have metabolic utilization clusters present at orfX, which may reflect the biological niche occupied by the S. xylosus isolates included in this study. In addition, regions of DNA are present in both C2a and S40010 with close homology to E. faecalis strain D32, an isolate from a pig (23). This indicates that horizontal gene transfer between enterococci and staphylococci is a relatively common occurrence (24, 25), an important observation in relation to the transfer of vancomycin resistance to S. aureus. In conclusion, this study further highlights the fact that CoNS from both humans and animals are an important reservoir of resistance genes that have the potential to be transferred into more pathogenic staphylococcal species.
Nucleotide sequence accession numbers.
The nucleotide sequences determined for the orfX region of S04009, S04010, and C2a have been deposited in the EMBL database under accession numbers HE993884, HE993885, and HE993886, respectively.
ACKNOWLEDGMENTS
This work was supported by a Medical Research Council Partnership grant (G1001787/1) held between the Department of Veterinary Medicine, University of Cambridge (M.A.H.), the School of Clinical Medicine, University of Cambridge (S.J.P.), the Moredun Research Institute (R.N.Z.) and the Wellcome Trust Sanger Institute (J.P. and S.J.P.).
We are grateful to Alexander Tomasz and Hermínia de Lencastre for their comments on the manuscript.
Footnotes
Published ahead of print 28 December 2012
REFERENCES
- 1. Hartman BJ, Tomasz A. 1984. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J. Bacteriol. 158:513–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lim D, Strynadka NC. 2002. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 9:870–876 [DOI] [PubMed] [Google Scholar]
- 3. Ubukata K, Nonoguchi R, Matsuhashi M, Konno M. 1989. Expression and inducibility in Staphylococcus aureus of the mecA gene, which encodes a methicillin-resistant S. aureus-specific penicillin-binding protein. J. Bacteriol. 171:2882–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fuda C, Suvorov M, Vakulenko SB, Mobashery S. 2004. The basis for resistance to beta-lactam antibiotics by penicillin-binding protein 2a of methicillin-resistant Staphylococcus aureus. J. Biol. Chem. 279:40802–40806 [DOI] [PubMed] [Google Scholar]
- 5. Garcia-Alvarez L, Holden MT, Lindsay H, Webb CR, Brown DF, Curran MD, Walpole E, Brooks K, Pickard DJ, Teale C, Parkhill J, Bentley SD, Edwards GF, Girvan EK, Kearns AM, Pichon B, Hill RL, Larsen AR, Skov RL, Peacock SJ, Maskell DJ, Holmes MA. 2011. Methicillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect. Dis. 11:595–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paterson GK, Larsen AR, Robb A, Edwards GE, Pennycott TW, Foster G, Mot D, Hermans K, Baert K, Peacock SJ, Parkhill J, Zadoks RN, Holmes MA. 2012. The newly described mecA homologue, mecALGA251, is present in methicillin-resistant Staphylococcus aureus isolates from a diverse range of host species. J. Antimicrob. Chemother. 67:2809–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cuny C, Layer F, Strommenger B, Witte W. 2011. Rare occurrence of methicillin-resistant Staphylococcus aureus CC130 with a novel mecA homologue in humans in Germany. PLoS One 6:e24360 doi:10.1371/journal.pone.0024360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shore AC, Deasy EC, Slickers P, Brennan G, O'Connell B, Monecke S, Ehricht R, Coleman DC. 2011. Detection of staphylococcal cassette chromosome mec type XI carrying highly divergent mecA, mecI, mecR1, blaZ, and ccr genes in human clinical isolates of clonal complex 130 methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 55:3765–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kriegeskorte A, Ballhausen B, Idelevich EA, Kock R, Friedrich AW, Karch H, Peters G, Becker K. 2012. Human MRSA isolates with novel genetic homolog, Germany. Emerg. Infect. Dis. 18:1016–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sabat AJ, Koksal M, Akkerboom V, Monecke S, Kriegeskorte A, Hendrix R, Ehricht R, Kock R, Becker K, Friedrich AW. 2012. Detection of new methicillin-resistant Staphylococcus aureus that carry novel genetic homologue and important virulence determinants. J. Clin. Microbiol. 50:3374–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stegger M, Andersen PS, Kearns A, Pichon B, Holmes MA, Edwards G, Laurent F, Teale C, Skov R, Larsen AR. 2012. Rapid detection, differentiation and typing of methicillin-resistant Staphylococcus aureus harbouring either mecA or the new mecA homologue mecA(LGA251). Clin. Microbiol. Infect. 18:395–400 [DOI] [PubMed] [Google Scholar]
- 12. Tsubakishita S, Kuwahara-Arai K, Baba T, Hiramatsu K. 2010. Staphylococcal cassette chromosome mec-like element in Macrococcus caseolyticus. Antimicrob. Agents Chemother. 54:1469–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dordet-Frisoni E, Dorchies G, De Araujo C, Talon R, Leroy S. 2007. Genomic diversity in Staphylococcus xylosus. Appl. Environ. Microbiol. 73:7199–7209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. 2008. A large genome center's improvements to the Illumina sequencing system. Nat. Methods 5:1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diep BA, Gill SR, Chang RF, Phan TH, Chen JH, Davidson MG, Lin F, Lin J, Carleton HA, Mongodin EF, Sensabaugh GF, Perdreau-Remington F. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731–739 [DOI] [PubMed] [Google Scholar]
- 16. Ito T, Hiramatsu K, Tomasz A, de Lencastre H, Perreten V, Holden MTG, Coleman DC, Goering R, Giffard PM, Skov RL, Zhang K, Westh H, O'Brien F, Tenover FC, Oliveira DC, Boyle-Vavra S, Laurent F, Kearns AM, Kreiswirth B, Ko KS, Grundmann H, Sollid JE, John JF, Daum R, Soderquist B, Buist G. 2012. Guidelines for reporting novel mecA gene homologues. Antimicrob. Agents Chemother. 56:4997–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kos VN, Desjardins CA, Griggs A, Cerqueira G, Van Tonder A, Holden MTG, Godfrey P, Palmer KL, Bodi K, Mongodin EF, Wortman J, Feldgarden M, Lawley T, Gill SR, Haas BJ, Birren B, Gilmore MS. 2012. Comparative genomics of vancomycin-resistant Staphylococcus aureus strains and their positions within the clade most commonly associated with methicillin-resistant S. aureus hospital-acquired infection in the United States. mBio 3:e00112–12 doi:10.1128/mBio.00112-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Even S, Leroy S, Charlier C, Zakour NB, Chacornac JP, Lebert I, Jamet E, Desmonts MH, Coton E, Pochet S, Donnio PY, Gautier M, Talon R, Le Loir Y. 2010. Low occurrence of safety hazards in coagulase negative staphylococci isolated from fermented foodstuffs. Int. J. Food Microbiol. 139:87–95 [DOI] [PubMed] [Google Scholar]
- 19. Perreten V, Giampa N, Schuler-Schmid U, Teuber M. 1998. Antibiotic resistance genes in coagulase-negative staphylococci isolated from food. Syst. Appl. Microbiol. 21:113–120 [DOI] [PubMed] [Google Scholar]
- 20. Coton E, Desmonts MH, Leroy S, Coton M, Jamet E, Christieans S, Donnio PY, Lebert I, Talon R. 2010. Biodiversity of coagulase-negative Staphylococci in French cheeses, dry fermented sausages, processing environments and clinical samples. Int. J. Food Microbiol. 137:221–229 [DOI] [PubMed] [Google Scholar]
- 21. Resch M, Nagel V, Hertel C. 2008. Antibiotic resistance of coagulase-negative staphylococci associated with food and used in starter cultures. Int. J. Food Microbiol. 127:99–104 [DOI] [PubMed] [Google Scholar]
- 22. Arêde P, Milheiriço C, de Lencastre H, Oliveira DC. 2012. The anti-repressor MecR2 promotes the proteolysis of the mecA repressor and enables optimal expression of β-lactam resistance in MRSA. PLoS Pathog. 8:e1002816 doi:10.1371/journal.ppat.1002816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Larsen J, Schonheyder HC, Singh KV, Lester CH, Olsen SS, Porsbo LJ, Garcia-Migura L, Jensen LB, Bisgaard M, Murray BE, Hammerum AM. 2011. Porcine and human community reservoirs of Enterococcus faecalis, Denmark. Emerg. Infect. Dis. 17:2395–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Noble WC, Virani Z, Cree RG. 1992. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol. Lett. 72:195–198 [DOI] [PubMed] [Google Scholar]
- 25. Weigel LM, Clewell DB, Gill SR, Clark NC, McDougal LK, Flannagan SE, Kolonay JF, Shetty J, Killgore GE, Tenover FC. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569–1571 [DOI] [PubMed] [Google Scholar]
- 26. Sampimon OC, Lam TJ, Mevius DJ, Schukken YH, Zadoks RN. 2011. Antimicrobial susceptibility of coagulase-negative staphylococci isolated from bovine milk samples. Vet. Microbiol. 150:173–179 [DOI] [PubMed] [Google Scholar]
- 27. Schnellmann C, Gerber V, Rossano A, Jaquier V, Panchaud Y, Doherr MG, Thomann A, Straub R, Perreten V. 2006. Presence of new mecA and mph(C) variants conferring antibiotic resistance in Staphylococcus spp. isolated from the skin of horses before and after clinic admission. J. Clin. Microbiol. 44:4444–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Park JY, Fox LK, Seo KS, McGuire MA, Park YH, Rurangirwa FR, Sischo WM, Bohach GA. 2011. Detection of classical and newly described staphylococcal superantigen genes in coagulase-negative staphylococci isolated from bovine intramammary infections. Vet. Microbiol. 147:149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schleifer KH, Meyer SA, Rupprecht M. 1979. Relatedness among coagulase-negative staphylococci: deoxyribonucleic acid reassociation and comparative immunological studies. Arch. Microbiol. 122:93–101 [DOI] [PubMed] [Google Scholar]
- 30. Perez-Roth E, Claverie-Martin F, Villar J, Mendez-Alvarez S. 2001. Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J. Clin. Microbiol. 39:4037–4041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mason WJ, Blevins JS, Beenken K, Wibowo N, Ojha N, Smeltzer MS. 2001. Multiplex PCR protocol for the diagnosis of staphylococcal infection. J. Clin. Microbiol. 39:3332–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]