Abstract
Helicobacter pylori is a globally important and genetically diverse gastric pathogen that infects most people in developing countries. Eradication efforts are complicated by antibiotic resistance, which varies in frequency geographically. There are very few data on resistance in African strains. Sixty-four Gambian H. pylori strains were tested for antibiotic susceptibility. The role of rdxA in metronidazole (Mtz) susceptibility was tested by DNA transformation and sequencing; RdxA protein variants were interpreted in terms of RdxA structure. Forty-four strains (69%) were resistant to at least 8 μg of Mtz/ml. All six strains from infants, but only 24% of strains from adults, were sensitive (P = 0.0031). Representative Mtz-resistant (Mtzr) strains were rendered Mtz susceptible (Mtzs) by transformation with a functional rdxA gene; conversely, Mtzs strains were rendered Mtzr by rdxA inactivation. Many mutations were found by Gambian H. pylori rdxA sequencing; mutations that probably inactivated rdxA in Mtzr strains were identified and explained using RdxA protein's structure. All of the strains were sensitive to clarithromycin and erythromycin. Amoxicillin and tetracycline resistance was rare. Sequence analysis indicated that most tetracycline resistance, when found, was not due to 16S rRNA gene mutations. These data suggest caution in the use of Mtz-based therapies in The Gambia. The increasing use of macrolides against respiratory infections in The Gambia calls for continued antibiotic susceptibility monitoring. The rich variety of rdxA mutations that we found will be useful in further structure-function studies of RdxA, the enzyme responsible for Mtz susceptibility in this important pathogen.
INTRODUCTION
Helicobacter pylori chronically infects most people in developing countries (1, 2), typically starting in infancy (2–4) and lasting for life. It also remains a significant pathogen in industrialized countries, infecting some 10 to 40% of adults in many societies. Chronic H. pylori infection is a major cause of gastric (stomach) and duodenal ulcers and gastric cancer (5–7). It also increases the risk of infection by other gastrointestinal pathogens, iron deficiency anemia, and infant malnutrition and growth faltering, especially among the very poor (8, 9). These latter conditions are of particular concern in The Gambia, a small developing country on the West Coast of Africa. Fortunately, many H. pylori-associated illnesses can be prevented or cured by the timely eradication of the bacterium, which typically entails a 1 to 2 weeks of treatment variously with metronidazole (Mtz), amoxicillin (Amo), and clarithromycin (Cla), when affordable, and/or tetracycline (Tet), in combination with a proton pump inhibitor such as omeprazole, and/or bismuth, where allowed by local regulations (10). H. pylori transmission tends to be highly localized and preferentially intrafamilial (11, 12) in industrialized societies and often also between households in the local community in developing country settings (13). Given the relatively localized transmission, successful eradication from many members of a household or community might markedly diminish the risk of new infections, especially of newborns, and thereby contribute importantly to public health.
Resistance to useful antimicrobials, especially Mtz and Cla, has been a major problem in some societies, even among people not previously treated for their H. pylori infections. Such resistance is generally attributable to inadvertent H. pylori exposure during treatment for other conditions (14). Mtz itself is an innocuous prodrug that is activated by chemical reduction to hydroxylamine type compounds, which are bactericidal to H. pylori (15). In the strains studied to date, mostly from industrialized societies, a modest level of Mtz resistance (e.g., to 8 or 16 μg of Mtz/ml) was usually associated with inactivation of the gene rdxA, which encodes a nonessential oxygen-insensitive NAPDH nitroreductase that chemically reduces Mtz in vitro (16). Higher-level resistance in rdxA mutant strains, e.g., to 32 μg of Mtz/ml, resulted from the inactivation of frxA, a related but generally less strongly transcribed nitroreductase gene; however, higher-level resistance can result from mutations in any of several additional genes that likely also affect intracellular redox potential (16–18). The hydroxylamine-type derivatives of Mtz that RdxA protein generates are mutagenic, such that exposure to sublethal Mtz concentrations (15) induces as well as selects for mutations to Mtz resistance.
No commonly used anti-H. pylori drugs other than Mtz are known to require activation to render them bactericidal, nor to be so highly mutagenic. In addition, the several resistances to these other drugs identified to date involve specific mutational changes that alter the target's function. In particular, resistance to the related macrolides erythromycin (Ery) and clarithromycin (Cla), which are used in anti-H. pylori therapy, is usually achieved by point mutations at either of two adjacent sites in 23S rRNA (19, 20) that diminish macrolide binding to the ribosome. Cla resistance seems to be rare in many societies but common (more than one-fourth of strains) in others (21, 22). The observed prevalence probably reflects a combination of the very few rRNA sites in which sequence changes can confer resistance and are not too deleterious for the bacterium, a need to incorporate any resistance mutation in both 23S rRNA genes to achieve a resistance phenotype, and the intensity of macrolide use for other infections and thereby inadvertent exposure of resident H. pylori strains.
Tet resistance is much rarer than Mtz or Cla resistance (23, 24), although several bona fide resistant strains have been identified and analyzed. In the best-described case, modest resistance resulted from three contiguous changes in the Tet binding pocket in 16S rRNA (positions 965 to 967) (23, 25). Lower-level resistance was achieved by mutation at one or two of these positions and/or by mutations in genes in other chromosomal locations that have not yet been identified but are suspected to affect bacterial permeability or efflux (23). Amo resistance is also very rare, but where it is found it has been ascribed to mutation in a penicillin-binding protein involved in cell wall metabolism (26–30).
The present study of drug susceptibility and resistance in Gambian H. pylori strains was motivated in part by considering that H. pylori is a genetically very diverse species, with different genotypes predominating in different well-separated geographic regions, even in different parts of Africa (31) and that most studies of drug susceptibility and resistance have focused on strains from Europe, the Americas, or Asia. As with many infection-related topics, there have been far fewer critical studies of antimicrobial resistance and susceptibility of H. pylori strains from Africa, especially those from West Africa—the ancestral home of most people of African ethnicity in the Americas. Given H. pylori transmission preferentially within families and local communities (11–13), West African strains may well have contributed to H. pylori gene pools in the Americas.
Bearing in mind H. pylori's impact on public health worldwide, The Gambia included, and the distinctiveness of African strains, we assessed here the frequencies of resistance to Amo, Cla, Ery, Mtz, and Tet in a set of 64 strains from Gambian citizens. We tested the importance of rdxA status for Mtz susceptibility and resistance by transformation and DNA sequence analysis, and interpreted amino acid sequence differences in RdxA protein in terms of its recently determined structure (32). We also tested by DNA sequencing whether 16S rRNA gene mutations could be responsible for the very few Tet-resistant (Tetr) isolates found as minority components of mixed H. pylori populations from several patient biopsy specimens.
MATERIALS AND METHODS
Patients.
Sixty-four patients were enrolled in the present study: 35 male and 29 female, ranging from 18 months to 70 years, with a mean age of 30 years. These patients were part of a group recruited during a primary study of H. pylori genetic diversity reported previously (33). All subjects were Gambian citizens and provided written informed consent. Children younger than 18 years were enrolled only after parental/guardian written informed consent. The study was approved by the Gambia Government/MRC joint ethics committee and the international Review Board of the U.S. National Institute of Health Division of Microbiology Infectious Diseases.
Bacterial cultures.
H. pylori strains were cultured, generally as pools of bacteria, from gastric biopsy samples on brain heart infusion (BHI) agar (Difco, USA) supplemented with 7% horse blood, 0.4% IsoVitaleX, trimethoprim at 5 μg/liter, vancomycin at 6 μg/liter, and amphotericin at 8 μg/liter. The plates were incubated at 37°C in a microaerobic atmosphere for 3 days as previously described (33). Exponential growth of these strains was obtained by culturing on fresh BHI agar medium for 24 h. Strains were stored at −70°C as suspensions of fresh exponentially growing cells suspended in BHI broth with 20% glycerol.
MIC determined by agar dilution.
Freshly grown bacteria were screened for antibiotic resistance by suspending them in phosphate-buffered saline (pH 7.2) (Invitrogen, USA) at a 105 McFarland standard. A series of sequential 10-fold dilutions was prepared from this suspension, and 10 μl of each dilution was spot inoculated onto BHI medium containing antibiotics appropriate for this experiment as follows: Amo (2 μg/ml), Cla (2 μg/ml), Ery (2 μg/ml), Tet (2 μg/ml), and Mtz (8 μg/ml). When resistance was encountered, susceptibility to higher concentrations of the antibiotic was tested. The plates were incubated at 37°C under microaerobic conditions for 5 days. A strain was considered to be susceptible to a concentration of an antibiotic that caused at least a 10-fold decrease in the efficiency of colony formation by individual cells (efficiency of plating [EOP]) as previously described (16, 21, 23, 34).
DNA extraction.
Total genomic DNA was extracted using a commercial kit (rDNA minikit; Qiagen, United Kingdom) according to the manufacturer's guidelines and stored at −20°C.
RAPD-PCR.
RAPD [random(ly) amplified polymorphic DNA] typing was carried out using two arbitrary primers (1254 [5′-CCGCAGCCAA-3′] and 1283 [5′-GCGATCCCCA-3′]) as previously described (35). The cycling parameters were 45 cycles of 94°C for 1 min, 36°C for 1 min, and 72°C for 2 min. The amplified products were detected by electrophoresis on a 1.5% (vol/vol) agarose gel with ethidium bromide (500 ng/ml), and the bands were visualized by using Gel Doc 2000 (Bio-Rad Laboratories, Milan, Italy).
16S rRNA gene and rdxA gene sequencing.
To detect changes in 16S rRNA sequences associated with Tet resistance, 16S rRNA genes were amplified by PCR with the primers 16S-F (5′-CGGTTACCTTGTTACGACTTCAC-3′) and 16S-R (5′-TATGGAGAGTTTGATCCTGGCTC-3′), and the amplified 16S rRNA genes were sequenced as previously described (23). To detect mutations associated with Mtz resistance, the rdxA genes of 51 strains (33 Mtzr and 18 Mtzs) were amplified by PCR with the primers rdxAF (5′-GTTTCGTTAGGGATTTTATTGTATGCTA-3′) and rdxAR (5′-CACCCCTAAAAGAGCGATTAAAACCATT-3′), the PCR products were sequenced, and the sequences were edited, aligned, and analyzed by using the DNAStar program (version 7; Lasergene, USA) and the Clustalw2 program.
Mutant RdxA protein structure analysis.
The impact on protein structure of mutations found in rdxA genes from Mtzr and Mtzs H pylori strains was assessed by modeling the mutations in the X-ray structure of the RdxA enzyme (PDB ID 3QDL) (32) using Swiss-PdbViewer (36). Distances from mutated residues to flavin mononucleotide (FMN) atoms or to the closest atoms of the associated monomer of the RdxA dimer were also calculated using Swiss-Pdbviewer. Solvent exposure of mutated residues was calculated using the ProtSA server (37). Proteins encoded by genes with insertion, deletion, or nonsense (stop) codon mutations were not modeled. Individual point mutations at the protein surface and far from the FMN group and dimer interface were considered to be neutral and did not affect RdxA function. Mutations resulting in small to large residue replacement at the protein core or leading to the loss of hydrogen bonds between apoprotein and FMN, the burial of polar or charged residues, or the replacement of Gly residues exhibiting dihedral angles not allowed to other residues were regarded as structurally disruptive and thus potentially causing the loss of RdxA function.
Statistical analysis.
Comparisons of antimicrobial resistance in strains from children versus adults, males versus females, and the distribution of rdxA nonsense mutations in different groups of strains were determined using the Fisher exact test; a P value of <0.05 was considered significant.
RESULTS
Of the 64 strains tested for Mtz susceptibility, 20 (31.2%) were sensitive (single cells unable to form colonies) on medium with Mtz at 8 μg/ml, and 44 (68.8%) were resistant. All six strains from infants (18 to 31 months old) were sensitive to this modest level of Mtz, whereas only 14 (24%) of 58 strains from adults were sensitive (P = 0.0031; Table 1). The prevalence of Mtz resistance in males versus females was 66% versus 72%, respectively (P > 0.05; Table 1 and Fig. 1).
Table 1.
MICs for Mtz against H. pylori isolated from males versus females
| Subject group | Sex | No. of subjects |
||||||
|---|---|---|---|---|---|---|---|---|
| Mtz MIC (μg/ml) |
Total | |||||||
| <8 | 8 | 16 | 32 | 64 | 128 | |||
| Adult | Male | 8 | 2 | 8 | 12 | 1 | 0 | 31 |
| Female | 6 | 1 | 5 | 14 | 1 | 0 | 27 | |
| Infant | Male | 4 | 0 | 0 | 0 | 0 | 0 | 4 |
| Female | 2 | 0 | 0 | 0 | 0 | 0 | 2 | |
| Total | 20 | 3 | 13 | 26 | 2 | 0 | 64 | |
Fig 1.
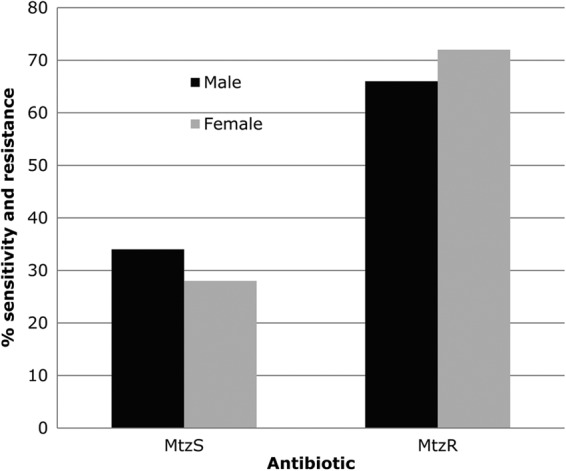
Prevalence of antibiotic-resistant isolates in males and females.
All but 3 of the 44 strains that were resistant to 8 μg of Mtz/ml also grew well on medium with 16 μg of Mtz/ml. In addition, 28 of these strains also grew on medium with 32 μg of Mtz/ml; and 2 of the 28 grew on medium with 64 μg of Mtz/ml. None of our 44 strains grew on medium with 128 μg of Mtz/ml (Fig. 2).
Fig 2.
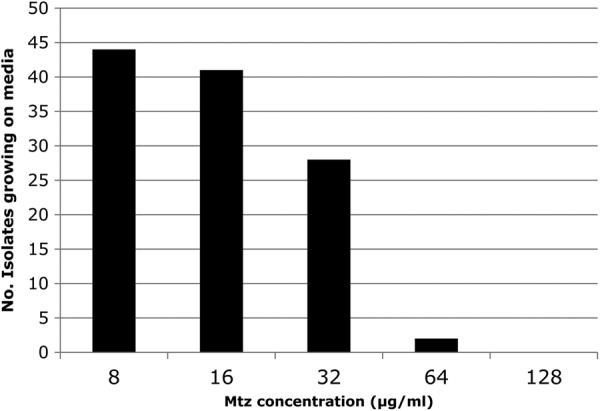
Number of isolates growing on metronidazole medium. The numbers of isolates that formed colonies on different levels of metronidazole medium were determined and graphed.
In further tests of strains that did not grow with 8 μg of Mtz/ml, 2 of the 6 representative strains from infants (18 to 31 months old), and 2 of 14 representative strains from adults grew on medium with only 4 μg of Mtz/ml.
rdxA (nitroreductase) gene analysis.
A transformation test was used to determine whether Mtzs strains were distinct metabolically from most susceptible reference strains in requiring more than just rdxA inactivation to achieve Mtz resistance (16–18). Eleven Mtzs strains were transformed with genomic DNA from a derivative of strain 26695 whose rdxA gene had been replaced with a chloramphenicol-resistant (Camr) cassette (ΔrdxA-cat). Each of the 10 to 20 Camr transformant colonies tested from each of the 11 strain transformations grew well on agar with 8 μg of Mtz/ml. This outcome indicates that most or all Mtzs Gambian H. pylori strains are just one mutational (rdxA inactivation) step away from becoming resistant. In a converse experiment, we tested whether mutation in rdxA was important for the resistance of Mtzr Gambian strains. This entailed transforming 12 representative Mtzr strains with genomic DNA from an H. pylori strain containing a kanamycin-resistant (Kanr) cassette inserted next to a functional rdxA gene. We expected that a fraction of Kanr transformants would acquire the donor strain's rdxA+ (functional) allele (38), even though most might retain the recipient rdxA mutant allele because H. pylori transformation tends to involve mostly short DNA fragments (39). At least 2 of the 20 to 30 Kanr transformants scored from each of 12 Mtzr recipient strains were found to be Mtzs on agar with 8 μg of Mtz/ml, even though most Kanr transformants remained Mtzr. We infer that these few Kanr Mtzs transformants had gained the donor's functional rdxA allele and thereby conclude that rdxA inactivation is needed for most or all Mtzs Gambian H. pylori strains if they are to become Mtzr.
Sequence comparison of rdxA from Mtzr and Mtzs strains.
The rdxA gene was PCR amplified and sequenced from 33 Mtzr (MIC range, 8 to 32 μg/ml) and 18 Mtzs Gambian strains (MIC < 8 μg/ml). The average rdxA sequence diversities were 3.6% in Mtzr strains and 3.4% in Mtzs strains (3.5% overall), which is within the range of diversities among Gambian strain housekeeping genes (range, 1.2 to 4.6%; mean, 2.9%), and whose protein products also act internally in these Gambian strains (O. Secka, unpublished data).
Of the 33 Mtzr strains characterized, 15 (45.5%) contained nonsense (translation stop) codons within the rdxA orf, including 13 of the 19 resistant to 32 μg of Mtz/ml; in contrast, only 2 of the 14 isolates with lower-level resistance (8 to 16 μg/ml) contained nonsense mutations in rdxA (P = 0.004, Table 2). This difference in distribution is in accord with nonsense mutations causing protein truncation and thus a complete loss of RdxA function. Some missense mutations diminish but do not entirely eliminate an encoded protein's activity and thus would confer only leaky phenotypes (lower-level Mtz resistance in the case of rdxA), whereas many others would be well tolerated and have little if any effect on activity of the encoded protein.
Table 2.
rdxA nonsense and frameshift mutationsa
| Strain | Concn (μg/ml) | Frameshift | Mutation description (base position)b | Mutation (codon, base position)c |
|---|---|---|---|---|
| Mtzr | ||||
| 71R | 32 | 0 | Substitution* (523), C→T or G→T | Stop codon (175, 523) |
| 83R | 32 | +1 | Insertion (576) | Stop codon (205, 613) |
| 93R | 32 | 0 | Substitution (415), C→T | Stop codon (139, 415) |
| 100R | 32 | 0 | Substitution* (523), C→T or G→T | Stop codon (175, 523) |
| 114R | 32 | 0 | Substitution (19), G→T | Stop codon (7, 19) |
| 115R | 32 | −1 | Deletion (496) | Stop codon (167, 499) |
| 121R | 16 | +2 | Insertion (6,7) | Stop codon (14, 40) |
| 123R | 32 | +1 | Insertion (23) | Stop codon (23, 67) |
| 205R | 32 | +1 | Insertion (595) | Stop codon (205, 613) |
| 239R | 8 | +1 | Insertion (313) | Stop codon (110, 328) |
| 244R | 32 | −7 | Deletion ×7 (179) | Stop codon (74, 220) |
| 249R | 32 | +5 | Insertion ×5 (178) | Stop codon (64, 190) |
| 263R | 32 | +1 | Insertion (193) | Stop codon (73, 217) |
| 269R | 32 | −1 | Deletion (191) | Stop codon (76, 226) |
| 270R | 32 | +1 | Insertion (193) | Stop codon (74, 220) |
| Mtzs | ||||
| JS114S | <8 | +4 | Insertion ×4 (300) | Stop codon (111, 331) |
| JS124S | <8 | +4 | Insertion ×4 (300) | Stop codon (111, 331) frameshift +4 |
| 101S | <8 | −1 | Deletion (86) | Stop codon (33, 97) |
Not listed in this table are other Mtzr strains that contained a 3-nucleotide deletion at codon 191, a 21-nucleotide (7-codon) deletion starting at codon 18, or the many missense mutations that resulted in amino acid differences in the encoded protein relative to rdxA of reference strain 26695. These amino acid replacements are shown in Fig. S1 in the supplemental material.
*, A total of 17 isolates have a “C” and 14 isolates have a “G” at position 523 (see Fig. S1 in the supplemental material).
The complete RdxA protein is 210 amino acids long.
Three of 33 Mtzr (9.0%) strains had insertions of one or two nucleotides and thereby rdxA frameshift mutations, which would result in new amino acid sequences distal to the mutant site and thereby loss of rdxA function. In addition, two strains contained in-frame deletions of 3 and 21 nucleotides, which do not cause changes in RdxA protein sequences distal to the mutant sites. Thirteen of 33 (39%) Mtzr strains had neither translation stop nor indel mutations in rdxA, but their rdxA alleles differed from those in Mtzs strains by numerous substitutions. Consideration of translation products in terms of H pylori RdxA protein's recently reported tridimensional structure (32) (PDB ID 3QDL) identified nine substitutions found only in Mtzr strains (S43L, P44L, and G163D [each in two strains] and A80I, A80T, C87Y, S158R, G189C, and A206T [each in one strain]) that are likely to have decreased RdxA function due to effects on stability, dimerization, and FMN binding and also indicated that the many other differences might not interfere with these best understood of RdxA protein's functional properties. One substitution found in Mtzr strains, R16H, that should decrease FMN binding affinity was also found in an Mtzs strain.
The basis of Mtz resistance in two strains (127R and 104R, MIC = 32 μg/ml) is not clear from RdxA structure considerations. Their 210-residue RdxA proteins differ at many positions from the RdxA protein whose structure was determined (Hp0954 of reference strain 26695). However, all of the differences present in strain 127R (D59N, R90K, G98S, R131K, V172I, A183V, Q197K, and V204I) and most (Q6H, D59N, R90K, G98S, R131K, E133K, G170S, V172I, E175Q, A183V, E194K, Q197K, and V204I) in 104R were also present in many other resistant and susceptible strains and thus may not perturb function. We interpret that most of these differences simply reflect neutral mutations in accord with H. pylori's great genetic diversity. None of the three mutations that were specific to 104R (E133K, G170S, and E194K) is expected to affect RdxA function because they appeared on the protein surface far from the FMN binding site. Thus, why strains 127R and 104R were resistant is not obvious: possibly their particular combinations of changes in RdxA diminished function (conferred resistance), or these strains might have polar mutations in the upstream DNA that we did not sequence.
Among the 18 rdxA sequences from Mtzs strains, 3 (17%) had internal stop codons (Table 2), and 3 others had point mutations that also might lead to rdxA inactivation, R16H (in one strain) because it should decrease RdxA affinity for FMN′s negatively charged phosphate and A67V (in two strains) because it entails replacement of small alanine by bulky valine in the protein core, although direct tests are needed to learn how severely this replacement affects protein stability and function. If these mutations do indeed cause rdxA inactivation, the Mtzs phenotypes might stem from high-level frxA expression (16, 18); the possibility of nonsense suppressor mutant tRNAs in certain strains also merits consideration. The nonsense mutations between Mtzs and Mtzr strains to 32 μg of Mtz/ml was significant (P = 0.0025, Table 2); however, overall, the nonsense mutations between Mtzs and Mtzr strains was not (P = 0.065, Table 2).
Fifteen substitutions were found only in Mtzr isolates (H25R, S43L, P44L, A80I, A80T, C87Y, H97T, E133K, S158R, G163D, G170S, G189C, E194K D205A, and A206T), which suggests that some of them might decrease RdxA function. Conversely, five were only found in Mtzs isolates (S30G, T31A, A67V, A68V, and Q197R) and thus might be neutral (of these five, only A67V is suspected of decreasing RdxA function, as noted above).
Susceptibility of Gambian strains to other antibiotics.
All 64 of our Gambian H. pylori cultures grown directly from gastric biopsy specimens were found to be highly sensitive to the closely related macrolides Cla and Ery. All cultures were also Tets and Amos (Table 3), although one and four of them contained rare Amor and Tetr cells able to grow on medium with 2 μg of Amo or Tet/ml, respectively (frequencies of 10−3 to 10−4). Further tests of one Tetr colony from each of these unusual Tetr subclone-containing cultures showed that their MICs ranged from 2 to 4 μg of Tet/ml and that each was indistinguishable by RAPD-DNA fingerprinting from the predominant Tets strains from the same biopsy specimen (data not shown). These four sets of strains were not closely related to one another, as expected, since they came from different persons. We found that the PCR-amplified 16S rRNA gene of each Tetr strain was identical in sequence to that of its Tets sibling from the same biopsy specimen. We conclude that in each of these four cases, Tet resistance is due to mutation in a gene distinct from that for 16S rRNA.
Table 3.
Susceptibility of Gambian H. pylori strains to amoxicillin, clarithromycin, erythromycin, and tetracycline
| MIC (μg/ml) | No. of susceptible strainsa |
|||
|---|---|---|---|---|
| Amo | Cla | Ery | Tet | |
| <2 | 64 | 64 | 64 | 64 |
| 2 | 0 | 0 | 0 | 0 |
| 8 | − | − | − | − |
| 16 | − | − | − | − |
| 32 | − | − | − | − |
| 64 | − | − | − | − |
| 128 | − | − | − | − |
−, strains sensitive at 2 µg/ml were not tested at higher concentrations (8 to 128 μg/ml).
DISCUSSION
H. pylori infection contributes importantly to several human diseases in both developing and industrialized countries and directly impacts on health care systems worldwide. Its public health impact is of particular concern in developing countries because the prevalence of infection is so very high (1, 2).
Here, we scored susceptibility and resistance to clinically relevant anti-H. pylori agents using a test of efficiency of colony formation by single cells. This test is especially useful for scoring susceptibility to Mtz because Mtz can be both mutagenic and bactericidal; it both induces and selects for resistance mutations (15, 16, 18). We found that more than two-thirds of Gambian H. pylori strains were Mtzr. This high prevalence can be explained by the relative low cost and easy availability of Mtz in The Gambia, as is typical of developing countries worldwide. Our results are in accord with other reports of many Mtzr strains elsewhere in Africa (Senegal, Nigeria, South Africa Cameroon, and Egypt) (40–44), India, and Latin America (45, 46). Typically, somewhat less than half of H. pylori strains from Europe and North America have been found to be Mtzr (22), likely reflecting the tighter control of Mtz usage in industrialized than in developing countries. Our observation that all six strains from infants (18 to 31 months of age) were sensitive to 8 μg of Mtz/ml, in contrast to 24% of those from older people (≥14 years) (P = 0.003), also merits further examination, especially in light of the possibility that rdxA function might contribute to fitness during the establishment of infection.
Prior studies with other sets of strains had shown that Mtz resistance typically involves inactivation of rdxA, which encodes a nitroreductase that converts Mtz from prodrug to bactericidal agent by chemical reduction. However, H. pylori strains also contain a related gene, frxA, which also confers Mtz susceptibility if highly expressed, independent of rdxA status (16, 43). Our DNA transformation studies indicated that each of 11 Mtzs Gambian strains tested required only rdxA inactivation to gain a Mtzr phenotype and, conversely, that resistance involved rdxA inactivation in each of the 12 Mtzr strains tested.
Sequence analysis of rdxA genes from our strains identified loss-of-function mutations that should cause Mtz resistance similar to those found in previous studies (16, 18, 47). In particular, rdxA nonsense (stop codon) mutations were more common in strains with moderate level resistance (32 μg/ml) than in strains with lower-level resistance (8 to 16 μg/ml), in agreement with other findings (16, 18, 47). However, 3 of 18 Mtzs strains had rdxA-null nonsense mutations, and 3 others had point mutations that also possibly might result in inactive RdxA proteins. These observations are consistent with other reports (16, 18, 48, 49) that rdxA inactivation need not always lead to Mtz resistance and can be explained by postulating higher-level expression of the related frxA nitroreductase gene (16, 18).
We also found that most Mtzr strains had mutations scattered across the rdxA gene that are not likely to contribute to their resistant phenotype. This is in accord with H. pylori's well-known great sequence diversity, seen here in the rdxA gene.
Increasingly frequent Cla resistance, up to one-fourth or more of H. pylori strains, has been reported in Europe and North America (21, 22). High prevalence, where encountered, has been attributed to use of macrolides to treat respiratory infections (22, 50–52). In The Gambia, Ery is now routinely used to treat lower respiratory tract and skin infections. However, we found no resistance to Cla or Ery in the 64 H. pylori isolates we tested.
Tet and Amo are also much used. Tet, especially, is inexpensive and readily available in local drug stores in The Gambia, often without prescription. However, its easy availability did not result in much resistance in Gambian H. pylori strains: Tetr strains were recovered from pooled cultures from only four patients and then only as very rare cells in the population. This outcome is in accord with the rarity of Tet resistance in H. pylori from other parts of the world (22, 53). None of our four Tetr strains had mutational changes in the several allowed positions in the 16S rRNA Tet binding pocket that can result in modest Tet resistance (23, 25). Thus, by default, their resistances are likely to stem from mutations in other loci—a class of mutations previously interpreted as more easily achieved but also likely to diminish H. pylori fitness (23). We propose that the low probability of mutation at just a few specific rRNA sites and the low fitness conferred by Tet resistance mutations in other genes explains the rarity of Tetr H. pylori, despite considerable exposure, in The Gambia. Resistance to Amo was also very rare in Gambian strains, much as has been reported for other geographic regions (22, 53).
Conclusion.
The increased prevalence of resistance to antibiotics used against H. pylori is of great concern, especially in developing countries where the costs of even the least expensive first line drugs are a burden to average citizens. This contributes to the urgency of monitoring antibiotic resistant strain frequencies to help clinicians effectively manage patients and their antibiotic regimens and to effectively deal with treatment failure. The rich repertoire of rdxA mutations found in our many Gambian H. pylori strains should be useful for future studies of RdxA structure and function, of how RdxA and its FrxA homolog make H. pylori susceptible to prodrugs such as Mtz, and of the roles of these two nitroreductase enzymes in H. pylori's central metabolic networks.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by funds from the Medical Research Council Unit (Fajara, The Gambia) and U.S. National Institutes of Health grants RO3-AI061308, R21-AI078237, and R21-AI088337, as well as by fund BFU2010-16297 (Ministerio de Ciencia e Innovación, Spain) and Grupo Protein Target B89 (Diputación General de Aragón, Spain). J.J.G. was funded by the Banco Santander Central Hispano, the Fundación Carolina, and the Universidad de Zaragoza.
We thank the members of the endoscopy team of the Clinical Services and Microbiology Department, Medical Research Council Unit, Fajara, The Gambia, and all of the patients and parents who made this study possible.
O.S., J.E.T., R.A.A., and D.E.B. conceived and designed the experiments. O.S. performed the experiments. O.S., D.E.B., M.A., J.J.G., J.S., R.A.A., and J.E.T. analyzed the data. T.C., M.T., R.W., J.E.T., and V.T. contributed reagents, materials, and analysis tools. O.S., D.E.B., J.J.G., J.S., R.A.A., and J.E. wrote the paper.
Footnotes
Published ahead of print 21 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00517-12.
REFERENCES
- 1. Dunn BE, Cohen H, Blaser MJ. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Frenck RW, Jr, Clemens J. 2003. Helicobacter in the developing world. Microbes Infect. 5:705–713 [DOI] [PubMed] [Google Scholar]
- 3. Thomas JE, Dale A, Harding M, Coward WA, Cole TJ, Weaver LT. 1999. Helicobacter pylori colonization in early life. Pediatr. Res. 45:218–223 [DOI] [PubMed] [Google Scholar]
- 4. Windle HJ, Kelleher D, Crabtree JE. 2007. Childhood Helicobacter pylori infection and growth impairment in developing countries: a vicious cycle? Pediatrics 119:e754–759 [DOI] [PubMed] [Google Scholar]
- 5. Atherton J. 2006. The pathogenesis of Helicobacter pylori-induced gastro-duodenal diseases. Annu. Rev. Pathol. 1:63–96 [DOI] [PubMed] [Google Scholar]
- 6. Crowe S. 2005. Helicobacter infection, chronic inflammation, and the development of malignancy. Curr. Opin. Gastroenterol. 21:32–38 [PubMed] [Google Scholar]
- 7. Wroblewski LE, Peek RM, Jr, Wilson KT. 2010. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin. Microbiol. Rev. 23:713–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Passaro DJ, Taylor DN, Meza R, Cabrera L, Gilman RH, Parsonnet J. 2001. Acute Helicobacter pylori infection is followed by an increase in diarrheal disease among Peruvian children. Pediatrics 8:e87. [DOI] [PubMed] [Google Scholar]
- 9. Thomas JE, Dale A, Bunn JEG, Harding M, Coward WA, Cole TJ, Weaver LT. 2004. Early Helicobacter pylori colonisation: the association with growth faltering in The Gambia. Arch. Dis. Child. 89:1149–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Boer W, Tytgat GNJ. 2000. Treatment of Helicobacter pylori infection. BMJ 320:31–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Raymond J, Thiberge JM, Chevalier C, Kalach N, Bergeret M, Labigne A, Dauga Raymond CJ, Thiberge JM, Chevalier C, Kalach N, Bergeret M, Labigne A, Dauga C. 2004. Genetic and transmission analysis of Helicobacter pylori strains within a family. Emerg. Infect. Dis. 10:1816–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schwarz S, Morelli G, Kusecek B, Manica A, Balloux F, Owen RJ, Graham DY, van der Merwe S, Achtman M, Suerbaum S. 2008. Horizontal versus familial transmission of Helicobacter pylori. PLoS Pathog. 4:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Herrera PM, Mendez M, Velapatiño B, Santivañez L, Balqui J, Finger SA, Sherman J, Zimic M, Cabrera L, Watanabe J, Rodríguez C, Gilman RH, Berg DE. 2008. DNA-level diversity and relatedness of Helicobacter pylori strains in shantytown families in Peru and transmission in a developing-country setting. J. Clin. Microbiol. 46:3912–3918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rimbara E, Fischbach LA, Graham DY. 2011. Optimal therapy for Helicobacter pylori infections. Nat. Rev. Gastroenterol. Hepatol. 8:79–88 [DOI] [PubMed] [Google Scholar]
- 15. Sisson G, Jeong JY, Goodwin A, Bryden L, Rossler N, Lim-Morrison S, Raudonikiene A, Berg DE, Hoffman PS. 2000. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori rdxA+ nitroreductase gene. J. Bacteriol. 182:5091–5096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jeong JY, Mukhopadhyay AK, Dailidiene D, Wang Y, Velapatiño B, Gilman RH, Parkinson AJ, Nair GB, Wong BC, Lam SK, Mistry R, Segal I, Yuan Y, Gao H, Alarcon T, Brea ML, Ito Y, Kersulyte D, Lee HK, Gong Y, Goodwin A, Hoffman PS, Berg DE. 2000. Sequential inactivation of rdxA HP0954 and frxA HP0642 nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J. Bacteriol. 182:5082–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Albert TJ, Dailidiene D, Dailide G, Norton JE, Kalia A, Richmond TA, Molla M, Singh J, Green RD, Berg DE. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods 2:951–953 [DOI] [PubMed] [Google Scholar]
- 18. Jeong JY, Mukhopadhyay AK, Akada JK, Dailidiene D, Hoffman PS, Berg DE. 2001. Roles of FrxA and RdxA nitroreductases of Helicobacter pylori in susceptibility and resistance to metronidazole. J. Bacteriol. 183:5155–5162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raymond J, Burucoa C, Pietrini O, Bergeret M, Decoster A, Wann A, Dupont C, Kalach Raymond NJ, Burucoa C, Pietrini O, Bergeret M, Decoster A, Wann A, Dupont C, Kalach N. 2007. Clarithromycin resistance in H. pylori strains isolated from French children: prevalence of different mutations and coexistence of clones harbouring two different mutations in the same biopsy. Helicobacter 12:157–163 [DOI] [PubMed] [Google Scholar]
- 20. Versalovic J, Shortridge D, Kibler K, Griffy MV, Beyer J, Flamm RK, Tanaka SK, Graham DY, Go Versalovic MFJ, Shortridge D, Kibler K, Griffy MV, Beyer J, Flamm RK, Tanaka SK, Graham DY, Go MF. 1996. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 40:477–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bruce MG, Bruden DL, McMahon BJ, Hennessy TW, Reasonover A, Morris J, Hurlburt DA, Peters H, Sacco F, Martinez P, Swenson M, Berg DE, Parks D, Parkinson AJ. 2006. Alaska sentinel surveillance for antimicrobial resistance in Helicobacter pylori isolates from Alaska native persons, 1999–2003. Helicobacter 11:581–588 [DOI] [PubMed] [Google Scholar]
- 22. Mégraud F. 2004. Recent advances in clinical practice Helicobacter pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut 53:1374–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dailidiene D, Bertoli MT, Miciuleviciene J, Mukhopadhyay AK, Dailide G, Pascasio MA, Kupcinskas L, Berg DE. 2002. Emergence of tetracycline resistance in Helicobacter pylori: multiple mutational changes in 16S ribosomal DNA and other genetic loci. Antimicrob. Agents Chemother. 46:3940–3946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Debets-Ossenkopp YJ, Herscheid AJ, Pot RG, Kuipers EJ, Kusters JG, Vandenbroucke-Grauls CM. 1999. Prevalence of Helicobacter pylori resistance to metronidazole, clarithromycin, amoxicillin, tetracycline and trovafloxacin in The Netherlands. J. Antimicrob. Chemother. 43:511–515 [DOI] [PubMed] [Google Scholar]
- 25. Trieber CA, Taylor DE. 2002. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J. Bacteriol. 184:2131–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dore MP, Graham DY, Sepulveda AR. 1999. Different penicillin-binding protein profiles in amoxicillin-resistant Helicobacter pylori. Helicobacter 4:154–161 [DOI] [PubMed] [Google Scholar]
- 27. Gerrits MM, Schuijffel D, van Zwet AA, Kuipers EJ, Vandenbroucke-Grauls CM, Kusters JG. 2002. Alterations in penicillin-binding protein 1A confer resistance to beta-lactam antibiotics in Helicobacter pylori. Antimicrob. Agents Chemother. 46:2229–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paul R, Postius S, Melchers K, Schäfer KP. 2001. Mutations of the Helicobacter pylori genes rdxA and pbp1 cause resistance against metronidazole and amoxicillin. Antimicrob. Agents Chemother. 45:962–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qureshi NN, Morikis D, Schiller NL. 2011. Contribution of specific amino acid changes in penicillin binding protein 1 to amoxicillin resistance in clinical Helicobacter pylori isolates. Antimicrob. Agents Chemother. 55:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rimbara E, Noguchi N, Kawai T, Sasatsu M. 2008. Mutations in penicillin-binding proteins 1, 2, and 3 are responsible for amoxicillin resistance in Helicobacter pylori. J. Antimicrob. Chemother. 61:995–998 [DOI] [PubMed] [Google Scholar]
- 31. Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, Falush D, Stamer C, Prugnolle F, van der Merwe SW, Yamaoka Y, Graham DY, Perez-Trallero E, Wadstrom T, Suerbaum S, Achtman M. 2007. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445:915–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martínez-Júlvez M, Rojas AL, Olekhnovich I, Angarica VE, Hoffman PS, Sancho J. 2012. Structure of RdxA: an oxygen insensitive nitroreductase essential for metronidazole activation in Helicobacter pylori. FEBS J. 279:4306–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Secka O, Antonio M, Tapgun M, Berg DE, Bottomley C, Thomas V, Walton R, Corrah T, Adegbola RA, Thomas JE. 2011. PCR-based genotyping of Helicobacter pylori of Gambian children and adults directly from biopsy specimens and bacterial cultures. Gut Pathog. 3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nahar S, Mukhopadhyay AK, Khan R, Ahmad MM, Datta S, Chattopadhyay S, Dhar SC, Sarker SA, Engstrand L, Berg DE, Nair GB, Rahman M. 2004. Antimicrobial susceptibility of Helicobacter pylori strains isolated in Bangladesh. J. Clin. Microbiol. 42:4856–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Akopyanz N, Bukanov NO, Westblom TU, Kresovich S, Berg DE. 1992. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 20:5137–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 [DOI] [PubMed] [Google Scholar]
- 37. Estrada J, Bernadó P, Blackledge M, Sancho J. 2009. ProtSA: a web application for calculating sequence specific protein solvent accessibilities in the unfolded ensemble. BMC Bioinformatics 10:104 doi:10.1186/1471-2105-10-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Choi SS, Chivers PT, Berg DE. 2011. Point mutations in Helicobacter pylori's fur regulatory gene that alter resistance to metronidazole, a prodrug activated by chemical reduction. PLoS One 6:e18236 doi:10.1371/journal.pone.0018236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin EA, Zhang XS, Levine SM, Gill SR, Falush D, Lin EA, Zhang XS, Levine SM, Gill SR, Falush D, Blaser MJ. 2009. Natural transformation of Helicobacter pylori involves the integration of short DNA fragments interrupted by gaps of variable size. PLoS Pathog. 5:e1000337 doi:10.1371/journal.ppat.1000337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seck A, Mbengue M, Gassama-Sow A, Diouf L, Ka MM, Boye CS. 2009. Antibiotic susceptibility of Helicobacter pylori isolates in Dakar, Senegal. J. Infect. Dev. Countries 3:137–140 [DOI] [PubMed] [Google Scholar]
- 41. Sherif M, Mohran Z, Fathy H, Rockabrand DM, Rozmajzl PJ, Frenck RW. 2004. Universal high-level primary metronidazole resistance in Helicobacter pylori isolated from children in Egypt. J. Clin. Microbiol. 42:4832–4834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Smith SI, Oyedeji KS, Arigbabu AO, Atimomo C, Coker AO. 2001. High amoxycillin resistance in Helicobacter pylori isolated from gastritis and peptic ulcer patients in Western Nigeria. J. Gastroenterol. 36:67–68 [DOI] [PubMed] [Google Scholar]
- 43. Tanih NF, McMillan M, Naidooc N, Ndipd LM, Weaver LT, Ndip RN. 2010. Prevalence of Helicobacter pylori vacA, cagA, and iceA genotypes in South African patients with upper gastrointestinal diseases. Acta Trop. 116:68–73 [DOI] [PubMed] [Google Scholar]
- 44. Ndip RN, Malange Takang AE, Ojongokpoko JE, Luma HN, Malongue A, Akoachere JF, Ndip LM, MacMillan M, Weaver LT. 2008. Helicobacter pylori isolates recovered from gastric biopsies of patients with gastroduodenal pathologies in Cameroon: current status of antibiogram. Trop. Med. Int. Health 13:848–854 [DOI] [PubMed] [Google Scholar]
- 45. Eisig JN, Silva FM, Barbuti RC, Navarro-Rodriguez T, Moraes-Filho JP, Pedrazzoli J., Jr 2011. Helicobacter pylori antibiotic resistance in Brazil: clarithromycin is still a good option. Arch. Gastroenterol. 48:261–264 [DOI] [PubMed] [Google Scholar]
- 46. Mukhopadhyay AK, Kersulyte D, Jeong JY, Datta S, Ito Y, Chowdhury A, Chowdhury S, Santra A, Bhattacharya SK, Azuma T, Nair GB, Berg DE. 2002. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J. Bacteriol. 182:3219–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten SJ, Berg DE, Hoffman PS. 1998. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene rdxA that encodes an oxygen-insensitive NADPH nitroreductase. Mol. Microbiol. 28:383–393 [DOI] [PubMed] [Google Scholar]
- 48. Chisholm SA, Owen RJ. 2003. Mutations in Helicobacter pylori rdxA gene sequences may not contribute to metronidazole resistance. J. Antimicrob. Chemother. 51:995–999 [DOI] [PubMed] [Google Scholar]
- 49. Kwon DH, Hulten K, Kato M, Kim JJ, Lee M, El-Zaatari FA, Osato MS, Graham DY. 2001. DNA sequence analysis of rdxA and frxA from 12 pairs of metronidazole-sensitive and -resistant clinical Helicobacter pylori isolates. Antimicrob. Agents Chemother. 45:2609–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ben Mansour K, Burucoa C, Zribi M, Masmoudi A, Karoui S, Kallel L, Chouaib S, Matri S, Fekih M, Zarrouk S, Labbene M, Boubaker J, Cheikh I, Ben Hriz M, Siala N, Ayad A, Filali A, Ben Mami N, Najjar A, Maherzi A, Tahar Sfar M, Fendri C. 2010. Primary resistance to clarithromycin, metronidazole and amoxicillin of Helicobacter pylori isolated from Tunisian patients with peptic ulcers and gastritis: a prospective multicentre study. Ann. Clin. Microbiol. Antimicrob. 9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sezgin O, Aslan G, Altintaş E, Tezcan S, Serin MS, Emekdaş G. 2008. Detection of point mutations on 23S rRNA of Helicobacter pylori and resistance to clarithromycin with PCR-RFLP in gastric biopsy specimens in Mersin, Turkey. Turk J. Gastroenterol. 19:163–167 [PubMed] [Google Scholar]
- 52. Tagliabue C, Techasaensiri C, Torres JP, Katz K, Meek C, Kannan TR, Coalson JJ, Esposito S, Principi N, Leff R, Baseman JB, Hardy RD. 2011. Efficacy of increasing dosages of clarithromycin for treatment of experimental Mycoplasma pneumoniae pneumonia. J. Antimicrob. Chemother. 66:2323–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Osato MS, Reddy R, Reddy SG, Penland RL, Malaty HM, Graham DY. 2001. Pattern of primary resistance of Helicobacter pylori to metronidazole or clarithromycin in the United States. Arch. Intern. Med. 161:1217–1220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


