Abstract
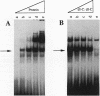
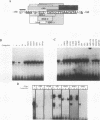
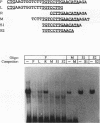
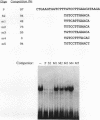
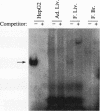
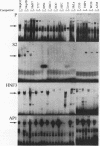
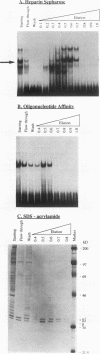
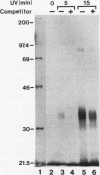
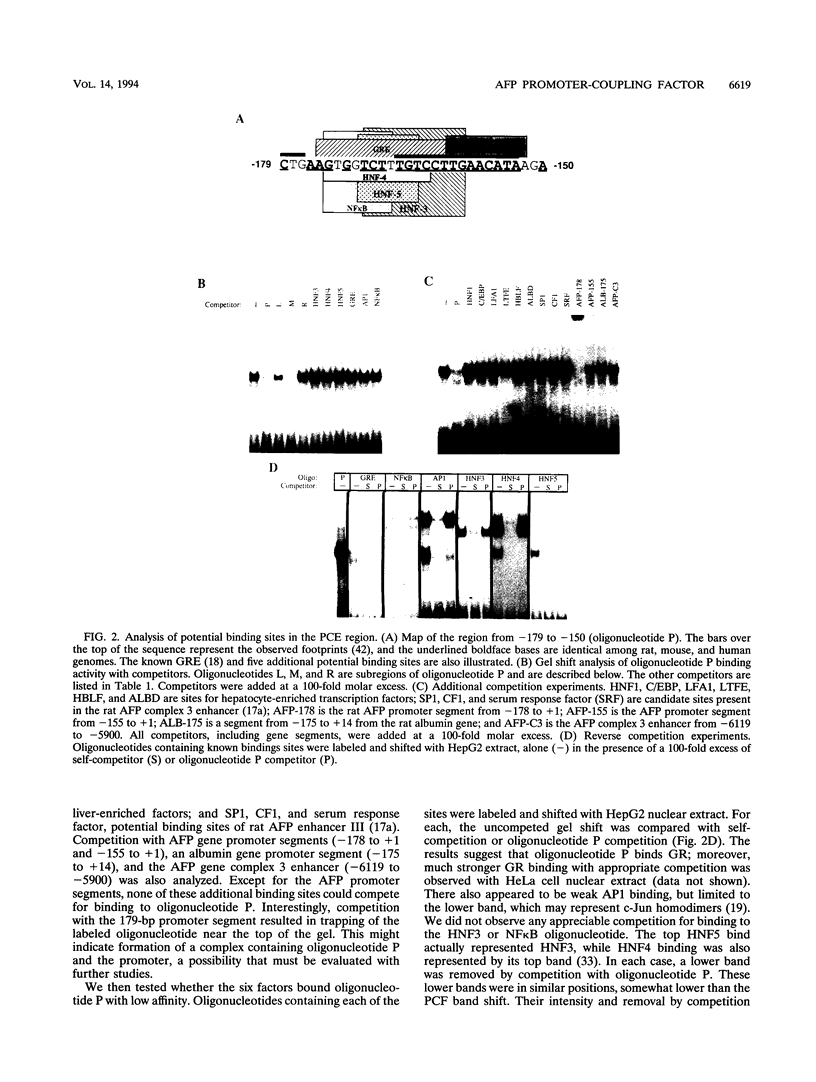
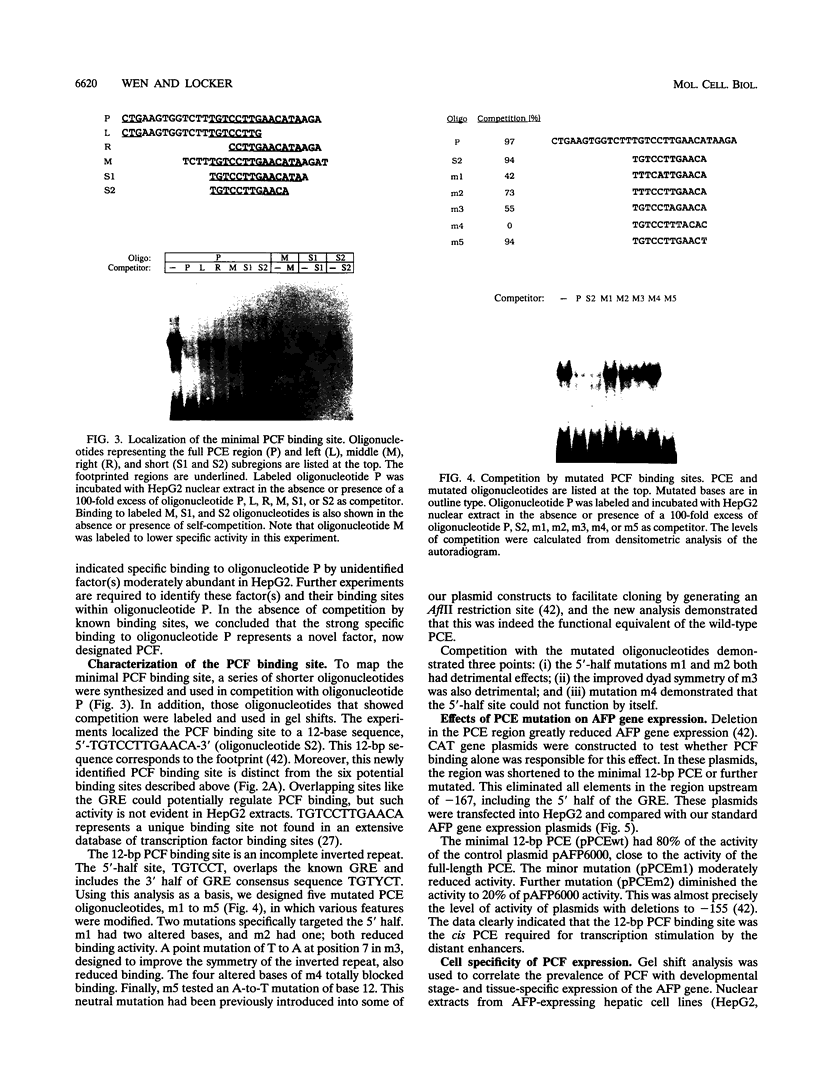
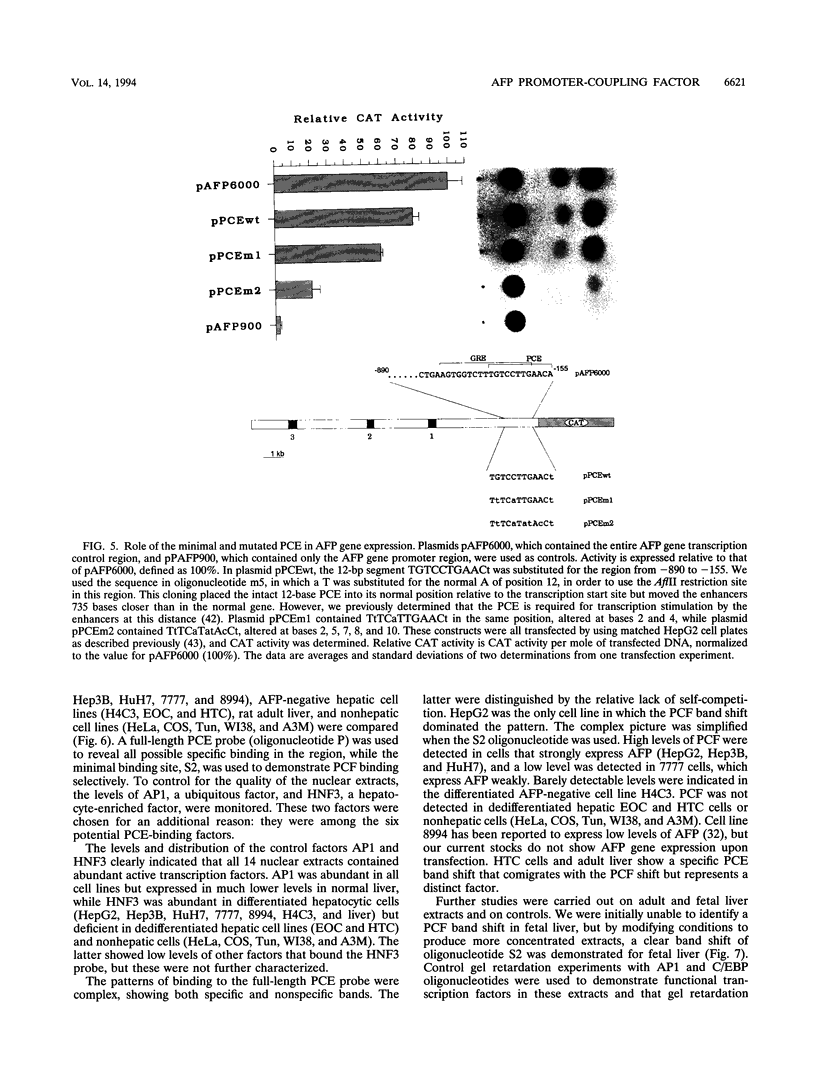
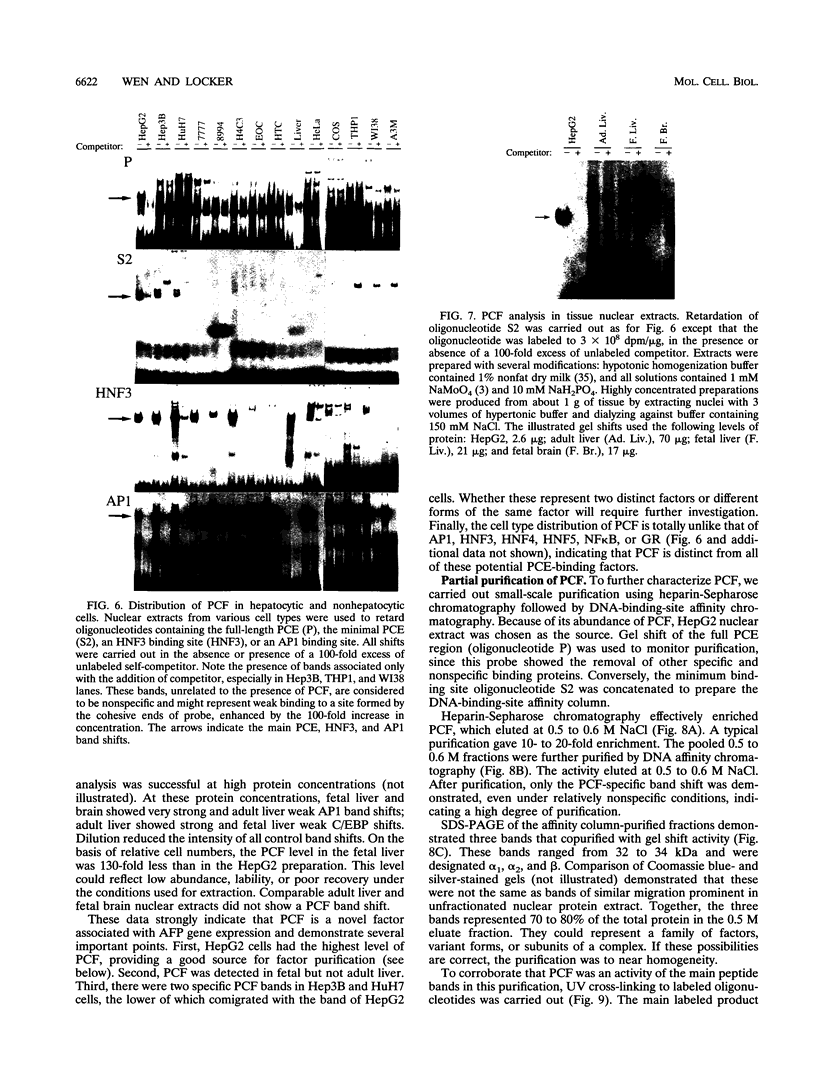
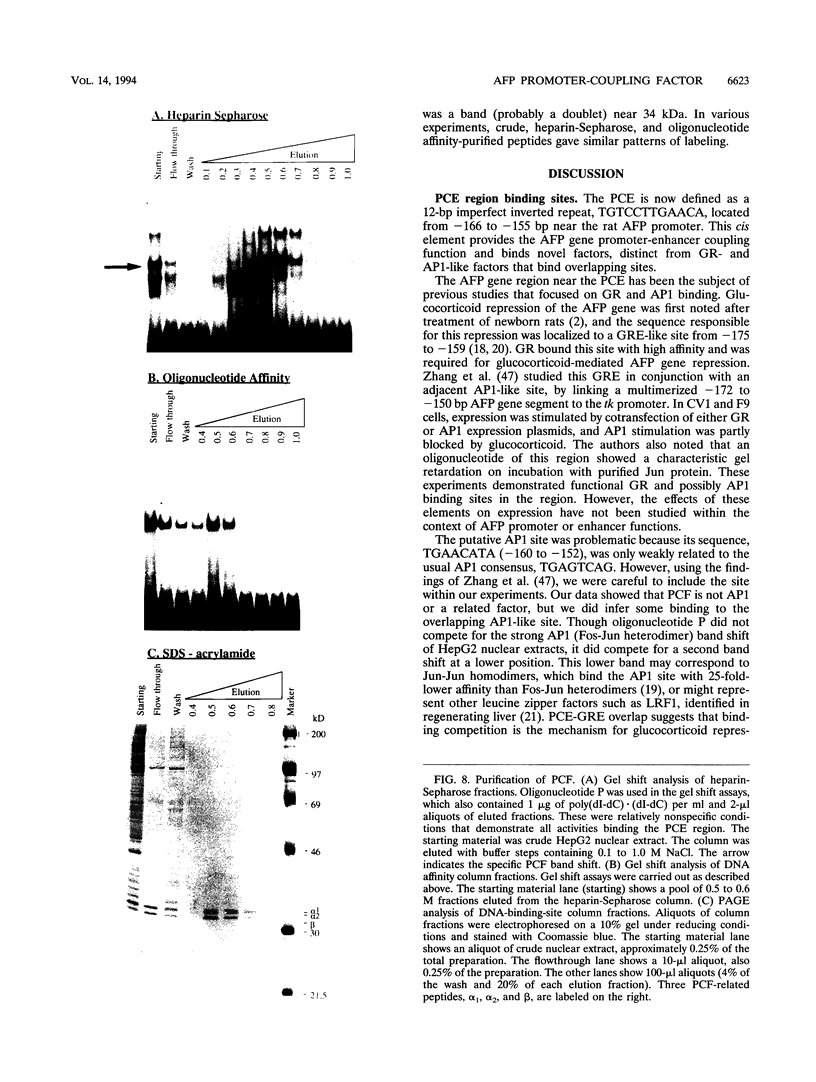
We recently characterized a promoter-linked coupling element (PCE) in the rat alpha-fetoprotein (AFP) gene required for strong transcriptional stimulation by distant enhancers (P. Wen, N. Crawford, and J. Locker, Nucleic Acids Res. 21:1911-1918, 1993). In this study, oligonucleotide gel retardation and competition experiments defined the PCE as a 12-bp binding site, TGTCCTTGAACA, an imperfect inverted repeat from -166 to -155 near the AFP promoter. A factor that bound this site (PCF) was abundant in HepG2 nuclear extracts and detectable in extracts from several other AFP-producing hepatocarcinoma cell lines and fetal liver. Hepatocytic cell lines that did not express AFP, nonhepatocytic cell lines, adult liver, and fetal brain did not show the factor. Experiments excluded the possibility that PCF activity was due to binding of glucocorticoid receptor or an AP1-like factor that bound overlapping sites. Competition experiments with several mutant oligonucleotides determined that the optimum PCF binding site was TGTCCTTGAAC(A/T). Mutations decreased binding or totally abolished binding activity. In expression plasmids, PCE mutations strongly reduced gene expression. UV cross-linking to a PCE probe identified peptide bands near 34 kDa. PCF was purified by heparin-Sepharose chromatography followed by affinity binding to oligomerized PCE DNA. The product resolved as a complex of three peptides (PCF alpha 1, PCF alpha 2, and PCF beta, 32 to 34 kDa) on sodium dodecyl sulfate-acrylamide gels. The peptide sizes and gel patterns are unlike those of any of the well-described hepatic transcription factors, and the binding site has not been previously reported. PCF thus appears to be a novel transcription factor.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bacus S. S., Stancovski I., Huberman E., Chin D., Hurwitz E., Mills G. B., Ullrich A., Sela M., Yarden Y. Tumor-inhibitory monoclonal antibodies to the HER-2/Neu receptor induce differentiation of human breast cancer cells. Cancer Res. 1992 May 1;52(9):2580–2589. [PubMed] [Google Scholar]
- Belanger L., Hamel D., Lachance L., Dufour D., Tremblay M., Gagnon P. M. Hormonal regulation of alpha1 foetoprotein. Nature. 1975 Aug 21;256(5519):657–659. doi: 10.1038/256657a0. [DOI] [PubMed] [Google Scholar]
- Bernier D., Thomassin H., Allard D., Guertin M., Hamel D., Blaquière M., Beauchemin M., LaRue H., Estable-Puig M., Bélanger L. Functional analysis of developmentally regulated chromatin-hypersensitive domains carrying the alpha 1-fetoprotein gene promoter and the albumin/alpha 1-fetoprotein intergenic enhancer. Mol Cell Biol. 1993 Mar;13(3):1619–1633. doi: 10.1128/mcb.13.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Breborowicz J., Tamaoki T. Detection of messenger RNAs of alpha-fetoprotein and albumin in a human hepatoma cell line by in situ hybridization. Cancer Res. 1985 Apr;45(4):1730–1736. [PubMed] [Google Scholar]
- Briggs M. R., Kadonaga J. T., Bell S. P., Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986 Oct 3;234(4772):47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Brunel F., Ochoa A., Schaeffer E., Boissier F., Guillou Y., Cereghini S., Cohen G. N., Zakin M. M. Interactions of DNA-binding proteins with the 5' region of the human transferrin gene. J Biol Chem. 1988 Jul 25;263(21):10180–10185. [PubMed] [Google Scholar]
- Buzard G., Locker J. The transcription control region of the rat alpha-fetoprotein gene. DNA sequence and homology studies. DNA Seq. 1990;1(1):33–48. doi: 10.3109/10425179009041345. [DOI] [PubMed] [Google Scholar]
- Camper S. A., Tilghman S. M. Postnatal repression of the alpha-fetoprotein gene is enhancer independent. Genes Dev. 1989 Apr;3(4):537–546. doi: 10.1101/gad.3.4.537. [DOI] [PubMed] [Google Scholar]
- Chiles T. C., Liu J. L., Rothstein T. L. Cross-linking of surface Ig receptors on murine B lymphocytes stimulates the expression of nuclear tetradecanoyl phorbol acetate-response element-binding proteins. J Immunol. 1991 Mar 15;146(6):1730–1735. [PubMed] [Google Scholar]
- Chodosh L. A., Carthew R. W., Sharp P. A. A single polypeptide possesses the binding and transcription activities of the adenovirus major late transcription factor. Mol Cell Biol. 1986 Dec;6(12):4723–4733. doi: 10.1128/mcb.6.12.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Grayson D. R., Darnell J. E., Jr Multiple hepatocyte-enriched nuclear factors function in the regulation of transthyretin and alpha 1-antitrypsin genes. Mol Cell Biol. 1989 Apr;9(4):1415–1425. doi: 10.1128/mcb.9.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Grayson D. R., Xanthopoulos K. G., Darnell J. E., Jr A liver-specific DNA-binding protein recognizes multiple nucleotide sites in regulatory regions of transthyretin, alpha 1-antitrypsin, albumin, and simian virus 40 genes. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3840–3844. doi: 10.1073/pnas.85.11.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa R. H., Lai E., Grayson D. R., Darnell J. E., Jr The cell-specific enhancer of the mouse transthyretin (prealbumin) gene binds a common factor at one site and a liver-specific factor(s) at two other sites. Mol Cell Biol. 1988 Jan;8(1):81–90. doi: 10.1128/mcb.8.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Martin P. L., Shastry B. S., Roeder R. G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- Godbout R., Ingram R. S., Tilghman S. M. Fine-structure mapping of the three mouse alpha-fetoprotein gene enhancers. Mol Cell Biol. 1988 Mar;8(3):1169–1178. doi: 10.1128/mcb.8.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange T., Roux J., Rigaud G., Pictet R. Cell-type specific activity of two glucocorticoid responsive units of rat tyrosine aminotransferase gene is associated with multiple binding sites for C/EBP and a novel liver-specific nuclear factor. Nucleic Acids Res. 1991 Jan 11;19(1):131–139. doi: 10.1093/nar/19.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin M., LaRue H., Bernier D., Wrange O., Chevrette M., Gingras M. C., Bélanger L. Enhancer and promoter elements directing activation and glucocorticoid repression of the alpha 1-fetoprotein gene in hepatocytes. Mol Cell Biol. 1988 Apr;8(4):1398–1407. doi: 10.1128/mcb.8.4.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis T. D., Georgopoulos K., Greenberg M. E., Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988 Dec 2;55(5):917–924. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- Houart C., Szpirer J., Szpirer C. The alpha-foetoprotein proximal enhancer: localization, cell specificity and modulation by dexamethasone. Nucleic Acids Res. 1990 Nov 11;18(21):6277–6282. doi: 10.1093/nar/18.21.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J. C., Laz T., Mohn K. L., Taub R. Identification of LRF-1, a leucine-zipper protein that is rapidly and highly induced in regenerating liver. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3511–3515. doi: 10.1073/pnas.88.9.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantzen H. M., Strähle U., Gloss B., Stewart F., Schmid W., Boshart M., Miksicek R., Schütz G. Cooperativity of glucocorticoid response elements located far upstream of the tyrosine aminotransferase gene. Cell. 1987 Apr 10;49(1):29–38. doi: 10.1016/0092-8674(87)90752-5. [DOI] [PubMed] [Google Scholar]
- Kakkis E., Riggs K. J., Gillespie W., Calame K. A transcriptional repressor of c-myc. Nature. 1989 Jun 29;339(6227):718–721. doi: 10.1038/339718a0. [DOI] [PubMed] [Google Scholar]
- Knowles B. B., Howe C. C., Aden D. P. Human hepatocellular carcinoma cell lines secrete the major plasma proteins and hepatitis B surface antigen. Science. 1980 Jul 25;209(4455):497–499. doi: 10.1126/science.6248960. [DOI] [PubMed] [Google Scholar]
- Liu J. K., DiPersio C. M., Zaret K. S. Extracellular signals that regulate liver transcription factors during hepatic differentiation in vitro. Mol Cell Biol. 1991 Feb;11(2):773–784. doi: 10.1128/mcb.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muglia L., Locker J. Developmental regulation of albumin and alpha-fetoprotein gene expression in the rat. Nucleic Acids Res. 1984 Sep 11;12(17):6751–6762. doi: 10.1093/nar/12.17.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi H., Hashimoto T., Miyao Y., Tjong K. K., Chan J., Tamaoki T. A position-dependent silencer plays a major role in repressing alpha-fetoprotein expression in human hepatoma. Mol Cell Biol. 1991 Dec;11(12):5885–5893. doi: 10.1128/mcb.11.12.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls M. C., Rippe R. A., Veloz L., Brenner D. A. Transcription factors nuclear factor I and Sp1 interact with the murine collagen alpha 1 (I) promoter. Mol Cell Biol. 1991 Aug;11(8):4065–4073. doi: 10.1128/mcb.11.8.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramji D. P., Tadros M. H., Hardon E. M., Cortese R. The transcription factor LF-A1 interacts with a bipartite recognition sequence in the promoter regions of several liver-specific genes. Nucleic Acids Res. 1991 Mar 11;19(5):1139–1146. doi: 10.1093/nar/19.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz W. A., Crawford N., Locker J. Albumin and alpha-fetoprotein gene expression and DNA methylation in rat hepatoma cell lines. Exp Cell Res. 1988 Feb;174(2):433–447. doi: 10.1016/0014-4827(88)90313-8. [DOI] [PubMed] [Google Scholar]
- Sladek F. M., Zhong W. M., Lai E., Darnell J. E., Jr Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990 Dec;4(12B):2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- Tewari M., Dobrzanski P., Mohn K. L., Cressman D. E., Hsu J. C., Bravo R., Taub R. Rapid induction in regenerating liver of RL/IF-1 (an I kappa B that inhibits NF-kappa B, RelB-p50, and c-Rel-p50) and PHF, a novel kappa B site-binding complex. Mol Cell Biol. 1992 Jun;12(6):2898–2908. doi: 10.1128/mcb.12.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J. M., Schibler U. Tissue-specific expression of the gene encoding hepatocyte nuclear factor 1 may involve hepatocyte nuclear factor 4. Genes Dev. 1991 Dec;5(12A):2225–2234. doi: 10.1101/gad.5.12a.2225. [DOI] [PubMed] [Google Scholar]
- Tilghman S. M., Belayew A. Transcriptional control of the murine albumin/alpha-fetoprotein locus during development. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5254–5257. doi: 10.1073/pnas.79.17.5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman R. Identification and purification of a polypeptide that binds to the c-fos serum response element. EMBO J. 1987 Sep;6(9):2711–2717. doi: 10.1002/j.1460-2075.1987.tb02564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tronche F., Rollier A., Herbomel P., Bach I., Cereghini S., Weiss M., Yaniv M. Anatomy of the rat albumin promoter. Mol Biol Med. 1990 Apr;7(2):173–185. [PubMed] [Google Scholar]
- Trujillo M. A., Letovsky J., Maguire H. F., Lopez-Cabrera M., Siddiqui A. Functional analysis of a liver-specific enhancer of the hepatitis B virus. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3797–3801. doi: 10.1073/pnas.88.9.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacher J., Tilghman S. M. Dominant negative regulation of the mouse alpha-fetoprotein gene in adult liver. Science. 1990 Dec 21;250(4988):1732–1735. doi: 10.1126/science.1702902. [DOI] [PubMed] [Google Scholar]
- Wen P., Crawford N., Locker J. A promoter-linked coupling region required for stimulation of alpha-fetoprotein transcription by distant enhancers. Nucleic Acids Res. 1993 Apr 25;21(8):1911–1918. doi: 10.1093/nar/21.8.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen P., Groupp E. R., Buzard G., Crawford N., Locker J. Enhancer, repressor, and promoter specificities combine to regulate the rat alpha-fetoprotein gene. DNA Cell Biol. 1991 Sep;10(7):525–536. doi: 10.1089/dna.1991.10.525. [DOI] [PubMed] [Google Scholar]
- Xanthopoulos K. G., Prezioso V. R., Chen W. S., Sladek F. M., Cortese R., Darnell J. E., Jr The different tissue transcription patterns of genes for HNF-1, C/EBP, HNF-3, and HNF-4, protein factors that govern liver-specific transcription. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3807–3811. doi: 10.1073/pnas.88.9.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmour J., Little A. M., Schendel D. J., Parham P. The HLA-A,B "negative" mutant cell line C1R expresses a novel HLA-B35 allele, which also has a point mutation in the translation initiation codon. J Immunol. 1992 Mar 15;148(6):1941–1948. [PubMed] [Google Scholar]
- Zhang D. E., Hoyt P. R., Papaconstantinou J. Localization of DNA protein-binding sites in the proximal and distal promoter regions of the mouse alpha-fetoprotein gene. J Biol Chem. 1990 Feb 25;265(6):3382–3391. [PubMed] [Google Scholar]
- Zhang X. K., Dong J. M., Chiu J. F. Regulation of alpha-fetoprotein gene expression by antagonism between AP-1 and the glucocorticoid receptor at their overlapping binding site. J Biol Chem. 1991 May 5;266(13):8248–8254. [PubMed] [Google Scholar]