LETTER
Espedido and coworkers found that up to 38.7% of methicillin-resistant Staphylococcus aureus (MRSA) clonal group ST239-MRSA-III strains in a Sydney hospital carried an arginine catabolic mobile element (ACME) type II variant that inserted between orfX and the staphylococcal cassette chromosome mec element (SCCmec) (1), whereas Ghaznavi-Rad and coworkers detected ACME in 1.1% of ST239-MRSA-III strains isolated from a Kuala Lumpur hospital, but did not specify its insertion point (2). The practical implications of such strains cannot be underestimated, especially in institutions where MRSA screening is predominantly performed using commercial molecular tests, because the majority of these currently detect the presence of sequences in the junction between orfX and SCCmec, and will potentially yield false-negative results if the sequences on the two sides of the junction are separated by a 13-kb ACME insertion.
Given Singapore's geographical proximity to and relations with Australia and Malaysia, we postulated that such ACME-positive strains are likely to be present locally. We had recently assembled a randomly selected sample of 110 clinical ST239-MRSA-III strains isolated between 2000 and 2010 from 3 local hospitals for whole-genome sequencing, which was performed following previously described protocols (3, 4). De novo assembly of the genomes using Velvet v0.7.03 (5) revealed 14 (12.7%) ST239 strains with ACME, and contigs bridging orfX and ACME as well as ACME and SCCmec III were found in 13 of the strains, whereas one strain appeared to have ACME inserted before sasH (data not shown). This was confirmed by orfX-ACME-specific PCR (1), which was positive for the initial 13 strains and negative for the last. None of the ST22-MRSA strains carried ACME.
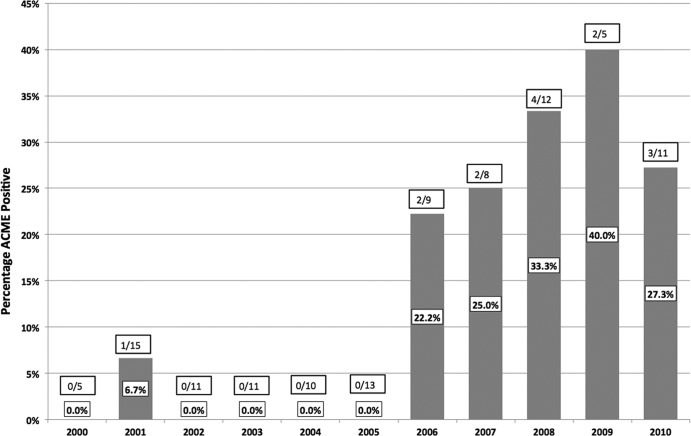
The distribution of ACME-positive ST239 strains—which were present in all 3 hospitals—is shown in Fig. 1. Despite the small number of ST239 strains collected each year, it is striking that ACME tests were positive in at least 20% of ST239-MRSA strains isolated since 2006, and that virtually all (92.9%) strains had ACME inserted into the orfX-SCCmec junction.
Fig 1.
Distribution of ACME-positive ST239 MRSA strains in Singaporean hospitals over time. The sole ACME-positive MRSA strain where the insertion point was outside the orfX-SCCmec junction was isolated in 2008.
As proof of concept, we ran saline suspensions of six of these clinical strains (viable counts ranging from 1.8 to 8.2 × 103 CFU/ml) along with an ACME-negative MRSA strain (ATCC BAA-1026; viable count, 1.6 × 102 CFU/ml) control on the Xpert MRSA assay (Cepheid, Sunnyvale, CA), following the manufacturer's instructions. Only the ATCC MRSA strain and the strain with ACME inserted outside the orfX-SCCmec junction yielded strongly positive results, while all the other strains were negative on the Xpert MRSA system. Parallel aliquots of these suspensions were plated and subsequently demonstrated good growth of MRSA colonies that again tested positive on mecA PCR (6). Although we did not test other commercial orfX-SCCmec molecular MRSA assays, it is extremely unlikely that these would be able to detect such ACME-positive MRSA strains.
Our results suggest that ST239-MRSA-III strains with ACME inserted between orfX and SCCmec are an emerging threat in the region. The inability of current commercial orfX-SCCmec molecular tests to detect such strains will complicate MRSA control efforts wherever such strains are prevalent, and institutions where ST239 is the prevalent MRSA clone are advised to test for the prevalence of such strains prior to investing in current commercial molecular testing platforms for MRSA.
ACKNOWLEDGMENTS
The work was partially supported by the Wellcome Trust (grant 098051) and a National Medical Research Council (Singapore) Research Training Fellowship grant.
Footnotes
Published ahead of print 14 January 2013
REFERENCES
- 1. Espedido BA, Steen JA, Barbagiannakos T, Mercer J, Paterson DL, Grimmond SM, Cooper MA, Gosbell IB, van Hal SJ, Jensen SO. 2012. Carriage of an ACME II variant may have contributed to methicillin-resistant Staphylococcus aureus sequence type 239-like strain replacement in Liverpool Hospital, Sydney, Australia. Antimicrob. Agents Chemother. 56:3380–3383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ghaznavi-Rad E, Nor Shamsudin M, Sekawi Z, Khoon LY, Aziz MN, Hamat RA, Othman N, Chong PP, van Belkum A, Ghasemzadeh-Moghaddam H, Neela V. 2010. Predominance and emergence of clones of hospital-acquired methicillin-resistant Staphylococcus aureus in Malaysia. J. Clin. Microbiol. 48:867–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Harris SR, Feil EJ, Holden MT, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Köser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 366:2267–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zerbino DR. 2010. Using the Velvet de novo assembler for short-read sequencing technologies. Curr. Protoc. Bioinformatics 31:11.5.1–11.5.12 doi:10.1002/0471250953.bi1105s31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oliveira DC, de Lencastre H. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]