Abstract
Upon screening of plant-derived natural products against hepatitis C virus (HCV) in the replicon system, we demonstrate that lucidone, a phytocompound, isolated from the fruits of Lindera erythrocarpa Makino, significantly suppressed HCV RNA levels with 50% effective concentrations of 15 ± 0.5 μM and 20 ± 1.1 μM in HCV replicon and JFH-1 infectious assays, respectively. There was no significant cytotoxicity observed at high concentrations, with a 50% cytotoxic concentration of 620 ± 5 μM. In addition, lucidone significantly induced heme oxygenase-1 (HO-1) production and led to the increase of its product biliverdin for inducing antiviral interferon response and inhibiting HCV NS3/4A protease activity. Conversely, the anti-HCV activity of lucidone was abrogated by blocking HO-1 activity or silencing gene expression of HO-1 or NF-E2-related factor 2 (Nrf2) in the presence of lucidone, indicating that the anti-HCV action of lucidone was due to the stimulation of Nrf-2-mediated HO-1 expression. Moreover, the combination of lucidone and alpha interferon, the protease inhibitor telaprevir, the NS5A inhibitor BMS-790052, or the NS5B polymerase inhibitor PSI-7977, synergistically suppressed HCV RNA replication. These findings suggest that lucidone could be a potential lead or supplement for the development of new anti-HCV agent in the future.
INTRODUCTION
Approximately 170 million people across the world are infected with hepatitis C virus (HCV). The virus causes chronic inflammation of the liver that ultimately leads to severe consequences such as cirrhosis and hepatocellular carcinoma (1). The HCV genome is a 9.6-kb positive single-strand RNA molecule encoding a single polyprotein of 3,000 amino acids that is processed by viral and cellular proteases to produce structural (C, E1, and E2) and nonstructural (NS2, NS3, NS4A, NS4B, NS5A, and NS5B) proteins (2). Pegylated alpha interferon (PEG–IFN-α) combined with ribavirin is the only recommended standard therapy for hepatitis C patients at present. However, the current therapeutic regimens only achieve a 40 to 50% cure rate in genotype 1 HCV-infected patients (3). In addition, the severe side effects of the current treatments, including depression, fatigue, flu-like symptoms, and hemolytic anemia, often lead to treatment discontinuation (4). Despite recent advancement of therapeutics with the approval of NS3/4A protease inhibitors, telaprevir and boceprevir, in combination with PEG–IFN-α and ribavirin, viral resistance and side effect to both inhibitors was still observed in clinical studies (5–7). Therefore, it is essential to develop new anti-HCV agents for therapeutic purposes.
Heme oxygenase-1 (HO-1), the rate-limiting enzyme in the oxidative degradation of heme, protects against oxidative stress and liver inflammation (8). Its reaction products including biliverdin, carbon monoxide, and ferrous iron are regarded as potential cytoprotectants (9). Among the various intracellular molecules capable of inducing HO-1, BTB and CNC homolog 1 (Bach1) and nuclear factor erythroid-derived 2 related factor 2 (Nrf2) play crucial roles in the regulation of HO-1 expression for the maintenance of cellular redox homeostasis (10). Nrf2 is essential for the transcriptional induction of antioxidant response element (ARE)-mediated phase II detoxifying and antioxidant genes via heterodimerization with small Maf (sMaf) proteins. In contrast, Bach1 heterodimerizes with sMaf proteins and interferes with HO-1 expression by competing the binding of the Nrf2-sMAF complex to the ARE of the HO-1 promoter region, which serves as a transcription repressor (11). Recent advances have revealed that the upregulation of HO-1 expression or overproduction of its metabolite is a promising strategy for suppressing HCV replication by activating the HO-1 transcriptional activator Nrf2 or targeting its transcriptional repressor Bach1 (12–14). The HO-1 product biliverdin has been demonstrated to be a major effector against viral replication by increasing the antiviral IFN response and inhibiting HCV NS3/4A protease activity (12, 14). Consequently, the discovery of HO-1-inducible agents may offer an advantage of therapeutic strategy by simultaneously targeting both viral and host factors for future HCV therapies.
By screening many natural products from plant using cell culture-based HCV replicon system, we identified a phytochemical, lucidone (Fig. 1A), isolated from the fruit of Lindera erythrocarpa that specifically inhibited HCV RNA replication. L. erythrocarpa Makino, belonging to the family Lauraceae, is widely cultured in Asian countries such as Taiwan, Japan, Korea, and China, and its fruits are traditionally used in folk medicine because of their extensive pharmacological properties, such as analgesic, digestive, diuretic, antidotal, and antibacterial properties (15). The lucidone derivatives, such as methyllinderone and methyllucidone, also exhibited anti-inflammation activity and anti-farnesyl protein transferase (16, 17). Recent in vitro and animal model studies have demonstrated that lucidone exerts anti-inflammatory activity with significant suppression of iNOS and COX-2 production (16, 18) and antimelanogenic activity (19). In the present study, we characterize the anti-HCV activity of lucidone and evaluate its possible mechanism of action against HCV replication.
Fig 1.
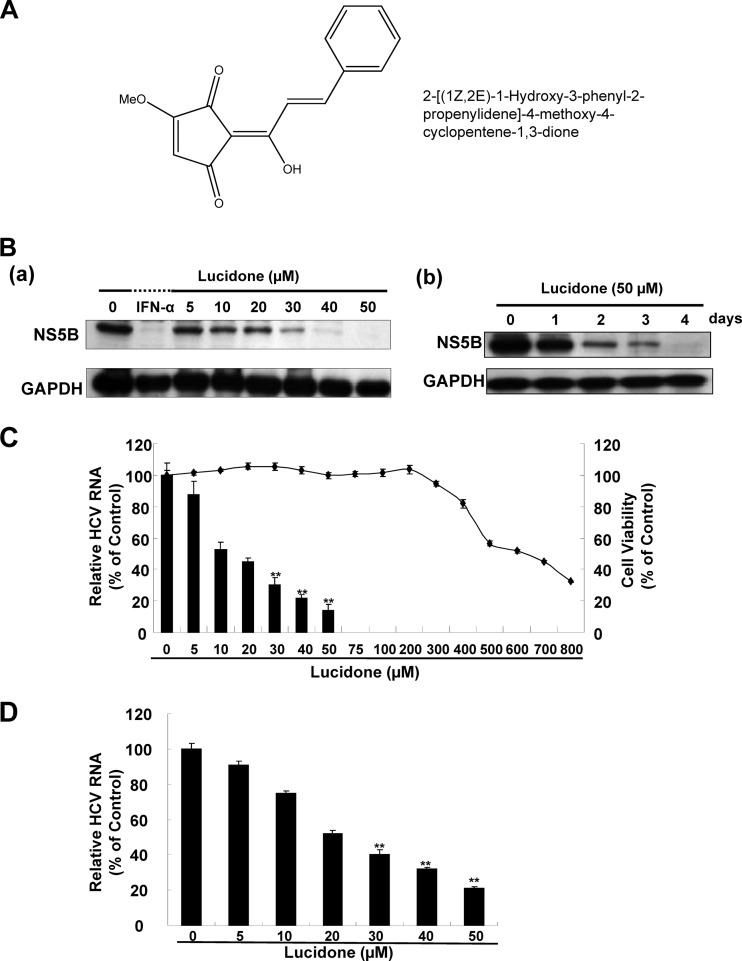
Effect of lucidone on HCV protein expression and RNA replication. (A) Structure of lucidone. The molecular formula is C15H12O4. (B) Inhibitory effects of lucidone on HCV protein synthesis in concentration (a)- and time (b)-dependent analyses. Ava5 cells were exposed to different concentrations (0, 5, 10, 20, 30, 40, and 50 μM) of lucidone for 4 days or different lengths of time (1 to 4 days) at concentrations of 50 μM. Treatment with 100 U of IFN-α/ml served as a positive control. Western blotting was performed with anti-HCV NS5B and anti-GAPDH antibodies. GAPDH (glyceraldehyde-3-phosphate dehydrogenase) protein levels confirmed equal loading of cell lysates. (C and D) Inhibitory effects of various concentrations of lucidone on HCV RNA replication in HCV replicon (C) and HCV JFH-1 (D) infectious systems. The total RNA of lucidone-treated Ava5 cells or JFH-1-infected Huh-7 cells was extracted to quantify HCV RNA levels by qRT-PCR. The relative HCV RNA levels were normalized by cellular gapdh mRNA expression. Cellular toxicity was evaluated by the MTS assay. The results are expressed as means of normalized data ± the SD for triplicate experiments. The error bars denote the SD of the mean. *, P < 0.05; **, P < 0.01.
MATERIALS AND METHODS
Cell culture and reagents.
Ava5 cells (20), a human hepatoma cell line (Huh-7 derivative) harboring HCV subgenomic replicon RNA, were cultured in Dulbecco modified Eagle medium (DMEM) with 10% heat-inactivated fetal bovine serum, 1% antibiotic-antimycotic, 1% nonessential amino acids, and 1 mg of G418/ml. IFN alfa-2a (Roferon-A) was purchased from Roche, Ltd. BMS-790052 and PSI-7977 were purchased from Shanghai Haoyuan Chemexpress Co., Ltd. Telaprevir was purchased from Legend Stat International Co., Ltd. These compounds were stored at a concentration of 10 mM in 100% dimethyl sulfoxide (DMSO). The final concentration of DMSO was maintained at 0.1% for all experiments.
Plasmid construction.
The pHO-1-Luc vector, kindly provided by Anupam Agarwal (University of Alabama), was used to measure the transcription activity of HO-1 (21). p3xARE-Luc, a reporter vector containing triple repeats of the Nrf2-dependent ARE, was used to measure the translocation and transcription activity of Nrf2. The AREs (TGACTCAGC) flanked by XhoI were inserted into the promoterless firefly luciferase vector pMCS-Luc (Stratagene, La Jolla, CA) and are designated as p3xARE-Luc. pISRE-Luc, a reporter vector containing firefly luciferase under the control of an IFN-stimulated response element (ISRE), was used to measure IFN response-dependent transcription activity (Stratagene). HO-1 (NM_002133), Nrf2 (NM_006164), and enhanced green fluorescent protein (EGFP) shRNA (as a negative control) were purchased from the National RNAi Core Facility, Institute of Molecular Biology/Genomic Research Center, Academia Sinica, Taiwan. The cloned DNA fragments were verified by DNA sequencing.
Western blot assay.
A standard procedure was used for Western blotting (22). Membranes were probed with either anti-NS5B (1:5,000; Abcam, Cambridge, MA), anti-GAPDH (1:10,000; GeneTex, CA), anti-HO-1 (1:3,000; GeneTex), anti-BVR (1:1,000; Abcam, Cambridge, MA), or anti-Nrf2 (1:3,000; GeneTex) antibody. Signals were detected using an ECL detection kit (Perkin-Elmer, CT).
Quantification of cellular mRNA and HCV RNA.
Total RNA was extracted from cell lysates using a total RNA miniprep purification kit (GMbiolab Co., Ltd., Taiwan) according to the manufacturer's instructions. Quantitative real-time RT-PCR (qRT-PCR) was performed using an ABI Step One real-time PCR system (ABI, Warrington, United Kingdom). Each sample was normalized by the endogenous glyceraldehyde 3-phosphate dehydrogenase (gapdh) gene expression. The primers used are listed in Table 1.
Table 1.
Oligonucleotides used for real-time RT-PCR
| Oligonucleotidea | Sequence (5′–3′) |
|---|---|
| 5′NS5B | GGAAACCAAGCTGCCCATCA |
| 3′NS5B | CCTCCACGGATAGAAGTTTA |
| 5′GAPDH | GTCTTCACCACCATGGAGAA |
| 3′GAPDH | ATGGCATGGACTGTGGTCAT |
| 5′OAS1 | CAAGCTTAAGAGCCTCATCC |
| 3′OAS1 | TGGGCTGTGTTGAAATGTGT |
| 5′OAS2 | ACAGCTGAAAGCCTTTTGGA |
| 3′OAS2 | GCATTAAAGGCAGGA AGCAC |
| 5′OAS3 | CACTGACATCCCAGACGATG |
| 3′OAS3 | GATCAGGCTCTTCAGCTTGG |
| 5′PKR | ATGATGGAAAGCGAACAAGG |
| 3′PKR | GAGATGATGCCATCCCGTAG |
| 5′IFN-α2 | GCA AGT CAA GCT GCT CTG TG |
| 3′IFN-α2 | GAT GGT TTC AGC CTT TTG GA |
| 5′IFN-α17 | AGG AGT TTG ATG GCA ACC AG |
| 3′IFN-α17 | CAT CAG GGG AGT CTC TTC CA |
GAPDH, glyceraldehyde 3-phosphate dehydrogenase; OAS, 2,5-oligoadenylate synthetase; PKR, protein kinase R; IFN-α, alpha interferon.
HCV JFH-1 infection assay.
The inhibitory effect of compound on HCV infection was assayed as previously described (23). In brief, the Huh-7 cells were seed at density of 4 × 104 cells/well in 24-wells culture plate and infected with 100 μl of HCV JFH-1 particles at a multiplicity of infection of 0.1 for 6 h, followed by incubation with various concentrations of lucidone for an additional 3 days. The total RNAs were then extracted and subjected to qRT-PCR to measure the mRNAs of HCV and GAPDH as described above.
Cytotoxicity assay.
Cells seeded in a 96-well plate at a density of 5 × 103 cells/well were exposed to various concentrations of crude extracts and the pure compounds. The cells were incubated at 37°C in an atmosphere of 5% of CO2 for 4 days. Cell viability was determined by the MTS assay using the CellTiter 96 Aqueous One solution cell proliferation assay system (Promega, WI) according to the manufacturer's instructions. Absorbance was detected at 490 nm using a Bio-Rad 550 plate reader (Bio-Rad, Hertfordshire, United Kingdom).
Transfection and luciferase activity assay.
To evaluate the regulation of HO-1 Nrf2, or IFN response by lucidone, Ava5 cells were transfected with 1 μg of pHO-1-Luc, p3xARE-Luc, or pISRE-Luc using T-Pro reagent (Ji-Feng Biotechnology Co., Ltd., Taiwan) in accordance with the manufacturer's instructions. To evaluate the role of HO-1 and Nrf2 in HCV replication by lucidone, Ava5 cells were transfected with increasing concentrations of the HO-1 or Nrf2 shRNA expression vector (pHO-1-shRNA and pNrf2-shRNA; 0.25 to 2 μg) in the presence of 30 μM lucidone. Each transfection complex contained 0.1 μg of a secreted alkaline phosphatase (SEAP) expression vector (pSEAP), which served as a transfection control. After 3 days of incubation, cell lysates were prepared for luciferase activity assay with the Bright-Glo luciferase assay system (Promega, Madison, WI) in accordance with the manufacturer's instructions and Western blotting with specific antibodies. The supernatants were harvested for SEAP activity assay with a Phospha-Light assay kit (Tropix, Foster City, CA). For each experiment, luciferase activity was normalized to the SEAP activity. The basal level of promoter activity was defined as 1 for comparison of the fold values after normalization of the luciferase activity.
Intracellular bilirubin measurement.
Cells were seeded in six-well plates at a density of 4 × 105 and then treated with lucidone at various concentrations. After 3 days of incubation, the cells were lysed using lysis buffer (25 mM Tris-phosphate [pH 7.8], 2 mM dithiothreitol [DTT], 2 mM CDTA [1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid], 10% glycerol, 1% Triton X-100). The total bilirubin concentration was measured by using a MeDiPro direct bilirubin test kit (Formosa Biomedical Technology Corp., Taiwan) according to the manufacturer's protocol. The assay is calibrated using a CFAS (calibrator for automated systems; Roche Diagnostics, Ltd., USA). The absorbance was detected at 546 and 660 nm using an Epoch microplate spectrophotometer (Bio-Tek Instruments, Inc., United Kingdom).
Cell-based and in vitro transcription and translation (TnT) assay for NS3/4A activity.
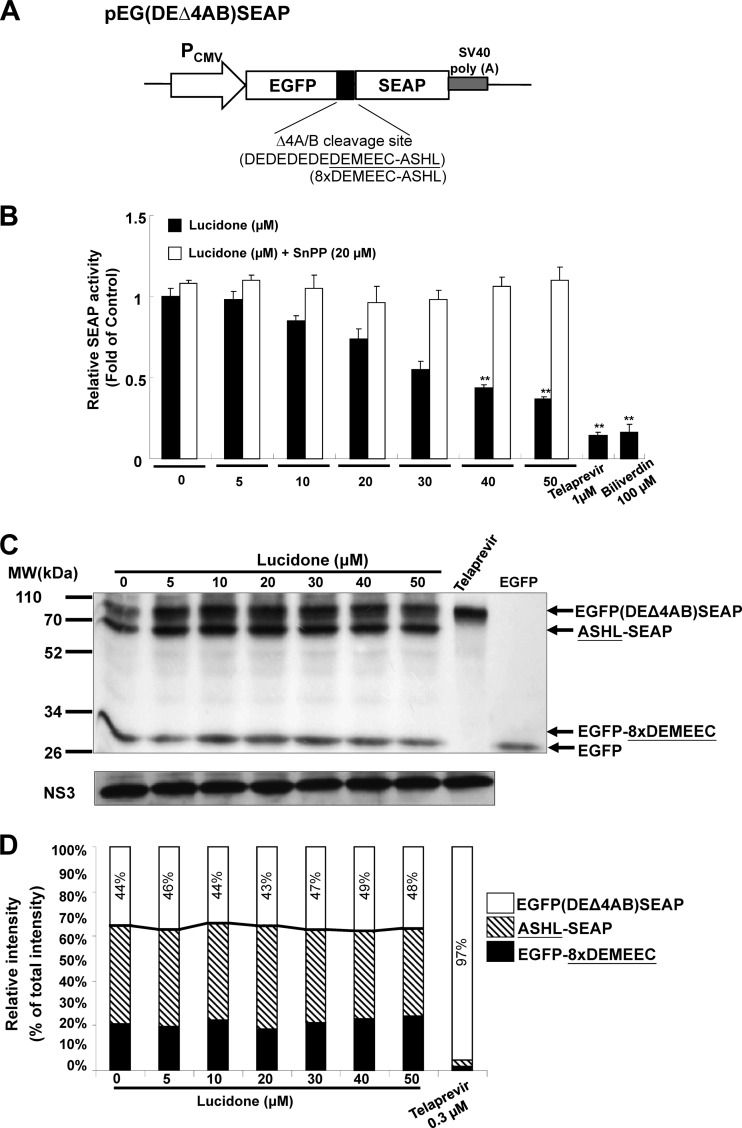
To evaluate the effects of NS3/4A protease activity by lucidone inside cells, the Huh-7 cells were transfected with 1 μg of NS3 response reporter vector pEG(DEΔ4AB)SEAP and 0.5 μg of the NS3/4A expression vector pNS3/4A using the T-Pro reagent (Ji-Feng Biotechnology Co., Ltd., Taiwan). Each transfection complex contained 0.1 μg of a firefly luciferase expression vector (pFLuc), which served as a transfection control. Subsequently, the transfected cells were incubated with different concentrations of lucidone with or without 20 μM HO-1 inhibitor SnPP. The reporter activity assays were performed as described above. An in vitro-coupled TnT reactions for NS3/4A protease activity assay were performed as described previously (24). In brief, the NS3/4A protease and EGFP(DEΔ4AB)SEAP substrate protein were generated by respective incubation of 1 μg of pNS3/4A or pEG(DEΔ4AB)SEAP in the presence of 1 mM methionine or 355 Ci of [35S]methionine (Institute of Isotopes Co., Ltd., Budapest, Hungary)/ml in a total volume of 50 μl of rabbit reticulocyte lysate solution, where incubation of pEGFP alone served as the identity of cleaved product EGFP derived from NS3/4A protease-mediated proteolytic processing of EG(DEΔ4AB)SEAP. Reactions were carried out at 30°C for 2.5 h and then terminated by adding 5 U of Tagetin RNA polymerase inhibitor (Epicentre, Madison, WI). The NS3/4A protease activity assay was performed by coincubation of 10 μl of non-radioisotope-labeled NS3/4A protease with 10 μl of 35S-labeled EGFP(DEΔ4AB)SEAP in the presence of various concentrations of lucidone in a total volume of 25 μl at 30°C for 15 min. Treatment with NS3/4A protease inhibitor telaprevir (0.3 μM) served as a positive control. The reactants were subsequently analyzed by SDS-PAGE and autoradiography. Densitometric quantification was analyzed with Quantity One one-dimensional analysis software (Bio-Rad Laboratories, Inc., USA).
Analysis of drug synergism.
Ava5 cells were treated with serially diluted lucidone (3, 6.125, 12.5, 25, and 50 μM) in combination with serially diluted IFN-α (7.5, 15, 30, and 60 U/ml), the HCV NS3/4A protease telaprevir (0.075, 0.15, 0.3, and 0.6 μM) (25), the HCV NS5A inhibitor BMS-790052 (1, 2, 4, and 8 pM) (26), and the RNA-dependent RNA polymerase nucleoside inhibitor PSI-7977 (10, 20, 40, and 80 nM) (27). Three days later, the total RNA was extracted to quantify HCV RNA levels by qRT-PCR. The relative RNA levels were normalized by cellular gapdh mRNA expression. Combination index (CI) values were analyzed using the CalcuSyn software (Biosoft, Cambridge, United Kingdom), a computer program based on the method of Chou and Talalay (28, 29). The 95% confidence intervals for the dose-response values were used to determine the data statistically. According to the percent inhibition of HCV RNA, the CI value is calculated using the following formula: CI = (Da + Db)/(Dxa + Dxb) + DaDb/DxaDxb. Da and Db are the concentrations of drugs A (for example, lucidone) and B (for example, IFN-α), respectively, required to inhibit X% of HCV RNA as single agents, whereas Dxa and Dxb are the concentrations of A and B, respectively, required to inhibit X% of HCV RNA in combination, for which treatment 0.1% DMSO was considered as a negative control. The effect of multiple drug combinations is presented as antagonism (CI > 1), additivity (CI = 1), or synergism (CI < 1). In addition, traditional isobologram analysis was used to confirm the drug-drug interaction (30).
Preparation of nuclear extract.
Nuclear extracts were prepared using hypotonic and high-salt buffer extraction as previously described (31). Briefly, Ava5 cells were seeded in a 6-cm dish at a density of 4 × 105 cells/dish and then treated with or without lucidone at the indicated dose. After 3 days and the indicated times of incubation, the cells were lysed using the ice-cold hypotonic buffer (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 10% Nonidet P-40 [pH 7.9]). After centrifugation at 7,000 × g for 15 min, the resulting nuclear pellets were extracted with high-salt nuclear extraction buffer (20 mM HEPES, 1.5 mM MgCl2, 0.2 mM EDTA, 0.6 M KCl, 0.2 mM DTT, 0.5 mM DTT [pH 7.9]) at 4°C for 30 min. The protease inhibitors and phosphatase inhibitors were added to hypotonic buffer and high-salt buffer immediately before use. Finally, nuclear proteins were collected after centrifugation at 20,000 × g for 15 min and stored at −80°C until use.
Statistical analysis.
The results were analyzed and are presented as means ± the standard deviations (SD) for at least three independent experiments. The statistical significance was analyzed by using the Student t test.
RESULTS
Lucidone inhibits HCV replication.
To assess the potential of lucidone in inhibiting HCV replication, Ava5 cells, a parent Huh-7-derived cell line harboring an HCV subgenomic RNA replicon (20), were treated with lucidone either at increasing concentrations for 4 days or at the single concentration of 50 μM for various times of incubation (1 to 4 days). The inhibitory effect of lucidone on HCV protein synthesis was then examined by Western blotting. The results indicated that lucidone markedly decreased the HCV NS5B protein levels in a concentration- and time-dependent manner (Fig. 1Ba and b). The inhibitory effect of lucidone on HCV RNA replication was examined by qRT-PCR in HCV replicon cells. As expected, HCV RNA levels were suppressed by lucidone in a concentration-dependent manner (Fig. 1C, left axis). The calculated 50% effective concentration (EC50) of lucidone for reducing HCV RNA levels was 15 ± 0.5 μM in HCV replicon cells. The cell viability assay revealed a 50% cytotoxic concentration (CC50) value of 620 ± 5 μM (Fig. 1C, right axis) for lucidone, which indicated that lucidone is not cytotoxic at effective antiviral concentrations. In addition, we performed HCV JFH-1 infectious assay to confirm the inhibitory effect of lucidone on viral RNA replication, with an EC50 of 20 ± 1.1 μM (Fig. 1D). Because of an acceptable selectivity index (SI; CC50/EC50) of ∼31, we suggested that lucidone could be potential as a promising lead compound for development of new anti-HCV agent.
Lucidone upregulates HO-1 expression in an HCV replicon system.
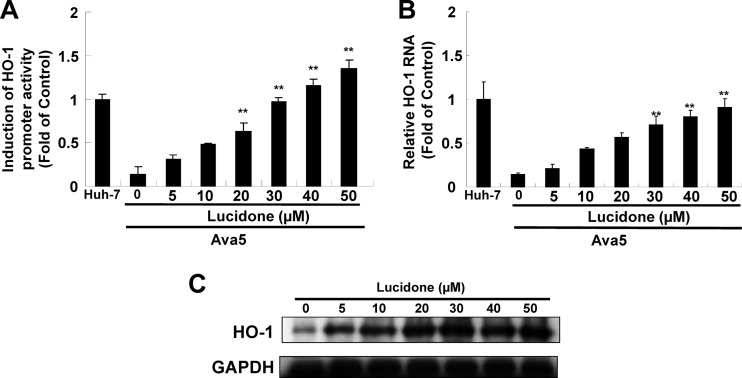
Lucidone has been demonstrated to effectively downregulate lipopolysaccharide (LPS)-induced inflammation by blocking iNOS and COX-2 (16, 18, 32). Because HO-1 is a critical regulator involved in the suppression of iNOS and COX-2 expression in response to inflammation (33, 34), we next examined whether lucidone could modulate HO-1 expression in HCV replicon cells. We first performed a HO-1 promoter-based reporter assay. Ava5 cells were transfected with the reporter plasmid pHO-1-Luc carrying the HO-1 promoter-driven firefly luciferase gene and then incubated with increasing concentrations of lucidone for 3 days, for which pHO-1-Luc-transfected-Huh-7 cells provided the basal level of HO-1 expression. As shown in Fig. 2A, the luminescent signal was markedly diminished in Ava5 cells compared to the signal in Huh-7 cells, indicating that HCV proteins strongly downregulated HO-1 gene transcription. Subsequently, lucidone concentration dependently enhanced the luminescent signal in Ava5 cells compared to that in lucidone-untreated Ava5 cells. Similar to the results obtained in the promoter activity assay, qRT-PCR analysis clearly revealed that lucidone increased HO-1 RNA levels in a concentration-dependent manner (Fig. 2B). Immunoblot analysis was performed to confirm the induction of HO-1 protein synthesis by lucidone in a concentration-dependent manner (Fig. 2C).
Fig 2.
Effect of lucidone on HO-1 expression. (A to C) Concentration-dependent induction of HO-1 promoter activity (A), RNA transcription (B), and protein synthesis by lucidone (C). For the promoter activity assay, Ava5 cells were transiently transfected with 1 μg of the HO-1 promoter reporter vector pHO-1-Luc. Subsequently, the transfected cells were treated with the indicated concentrations (0 to 50 μM) of lucidone for 3 days, and the total cell lysates were analyzed for luciferase activity. The pHO-1-Luc-transfected-Huh-7 cells provided the basal level of HO-1 promoter activity, which was defined as 1. For RNA and protein analysis, Ava5 cells were incubated with the indicated concentrations (0 to 50 μM) of lucidone for 3 days. The total RNA of Huh-7 cells and lucidone-treated Ava5 cells was extracted to quantify HO-1 mRNA levels by qRT-PCR. The HO-1 mRNA levels in Huh-7 cells was defined as 1. Total cell lysates were extracted and analyzed by Western blotting with anti-HO-1 and anti-GAPDH (loading control) antibodies. The results are expressed as the mean fold values of normalized data ± the SD for triplicate experiments. The error bars denote the SD of the mean fold values.*, P < 0.05; **, P < 0.01.
Lucidone augments antiviral IFN response and suppresses HCV NS3/4A protease activity through biliverdin production.
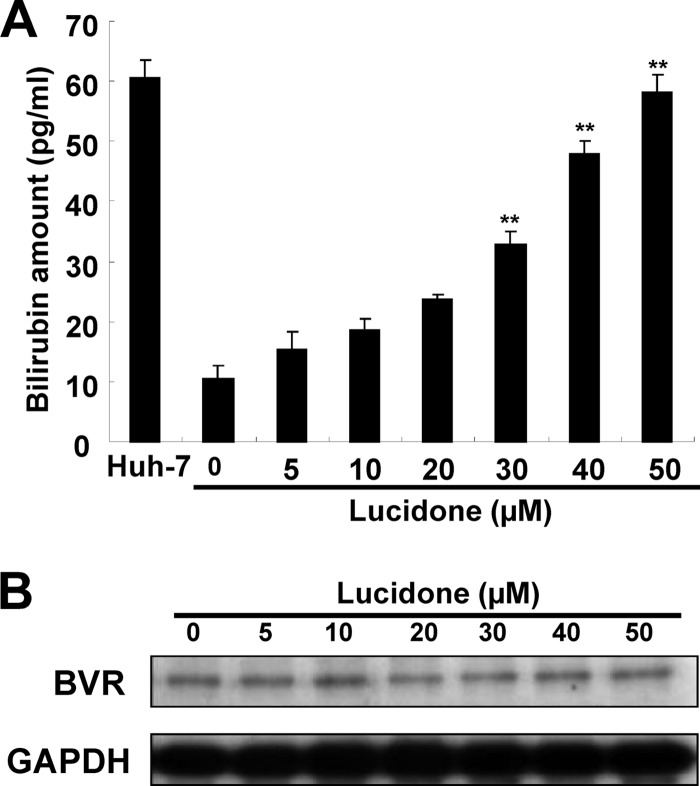
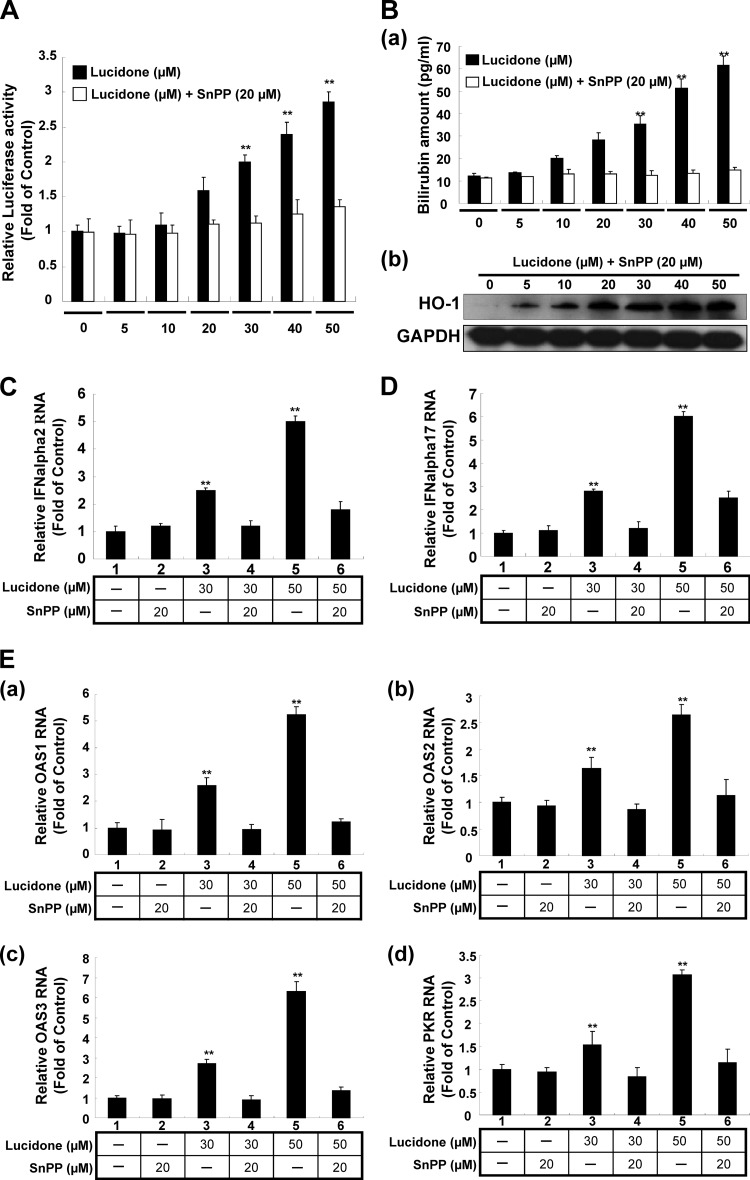
Recently, Lehmann et al. and Zhu et al. have demonstrated that the HO-1 product biliverdin inhibits viral replication by increasing the antiviral interferon response and blocking HCV protease activity (12, 14). To verify whether lucidone could enhance biliverdin production due to the increase of HO-1 production, we first measured the amount of bilirubin, the biliverdin metabolite, to reflect the intracellular biliverdin levels following lucidone treatment. The results revealed that lucidone concentration-dependently increased the bilirubin levels (Fig. 3A). Biliverdin reductase (BVR) is an important catalase for bilirubin formation from biliverdin in HO-1-dependent reaction (35). To rule out the possibility that the accumulation of bilirubin was due to the induction of BVR by lucidone, we further detected the effect of lucidone on BVR expression. As shown in Fig. 3B, the BVR protein levels were not interfered by lucidone under increasing concentrations. Because the conversion of biliverdin to bilirubin is proportional, we concluded that lucidone treatment resulted in the increase of biliverdin, which may lead to anti-HCV activity. To verify whether lucidone inhibited HCV replication via its aforementioned anti-HCV effects, we performed a transient ISRE activity assay using the ISRE-mediated firefly luciferase expression vector. Ava5 cells were transfected with pISRE-Luc reporter plasmids and incubated with increasing concentrations of lucidone for 3 days. As shown in Fig. 4A, lucidone treatment significantly increased ISRE-mediated luciferase activity at effective antiviral concentrations (black columns). Conversely, treatment of the specific competitive HO-1 inhibitor tin protoporphyrin (SnPP) significantly abrogated the transcriptional induction of ISRE by lucidone (white column). Furthermore, our results confirmed that SnPP reversed the inductive effect of lucidone on ISRE-mediated activity due to completely blocking enzymatic activity of HO-1 because the lucidone-enhanced bilirubin levels were dramatically reduced by SnPP at tested concentration (Fig. 4Ba). In contrast, SnPP did not interfere concentration-dependent induction of HO-1 protein levels by lucidone (Fig. 4Bb). In addition, we further measured the expression of IFN-α2 and -α17 in lucidone-treated Ava5 cells (30 and 50 μM) by qRT-PCR analysis. As shown in Fig. 4C and D, lucidone significantly induced the mRNA levels of both IFN genes compared to no lucidone treatment (lanes 1, 3, and 5). Subsequently, we examined the mRNA levels of critical IFN-mediated antiviral genes (36), including protein kinase R (PKR), 2′-5′-oligoadenylate synthetase 1 (OAS1), OAS2, and OAS3 under the same experimental conditions. As expected, lucidone significantly induced the expression of PKR, OAS1, OAS2, and OAS3 compared to their expression in lucidone-untreated cells (Fig. 4Ea to d, lanes 1, 3, and 5). Conversely, treatment of HO-1 inhibitor SnPP significantly abrogated the expression of IFN-α2 and -α17 (Fig. 4C and D, lanes 2, 4, and 6) and the induction of antiviral genes (Fig. 4Ea to d, lanes 2, 4, and 6). Treatment with SnPP alone served as a mock control (lane 2). For the cell-based HCV NS3/4A protease activity assay, Huh-7 cells were cotransfected with the NS3/4A expression vector pNS3/4A-myc and its substrate vector pEG(DEΔ4AB)SEAP (37) (Fig. 5A), which contained a NS3 cleavage site between EGFP and SEAP, in the presence of increasing concentrations of lucidone for 3 days. Treatment with telaprevir (25) or biliverdin (14) served as positive controls. As shown in Fig. 5B, a concentration-dependent reduction of SEAP activity was observed in lucidone-treated cells compared to its activity in lucidone-untreated cells (black columns), reflecting that NS3/4A protease activity was inhibited by lucidone inside cells. Conversely, treatment of SnPP attenuated the inhibitory effect of lucidone on NS3/4A protease activity by detection of SEAP activity (white columns). To examine whether lucidone directly reacted with NS3/4A protease, we performed a cell-free transcription and translation (TnT) of NS3/4A activity assay (24). As shown in Fig. 5C, the cleavage of the EG(DEΔ4AB)SEAP fusion protein by NS3/4A protease was occurred in the absence or presence of increasing concentrations of lucidone compared to telaprevir treatment, a positive control of protease inhibition. Densitometric quantification of the uncleaved- and cleaved-specific bands showed that there was no difference in the residual EG(DEΔ4AB)SEAP levels, remaining from 43 to 49%, from TnT experiments with or without lucidone (Fig. 5D, white squares), indicating that the cleavage efficiency of NS3/4A protease was not interfered by lucidone. Compared to the findings in cell-based reporter assay described above, we indicated that lucidone did not directly target NS3/4A protease. Taken together, these findings are suggestive that lucidone treatment leads to substantial biliverdin production through HO-1 induction, which results in the effective inhibition of HCV replication via the combination of the antiviral interferon response and anti-NS3/4A protease activity. Therefore, despite the enhanced level of IFN-stimulated gene (ISG), the expression of lucidone-treated cells was lower than that of IFN-treated cells at the effective concentration of anti-HCV activity (data not shown), and lucidone was shown to act in an additive manner on anti-HCV replication by both of the antiviral actions described above.
Fig 3.
Effect of lucidone on bilirubin production and biliverdin reductase (BVR) protein expression. (A) Concentration-dependent production of bilirubin by lucidone. Ava5 cells were incubated with the indicated concentrations (0 to 50 μM) of lucidone for 3 days, and the total cell lysates were harvested for quantification of bilirubin concentration using a MeDiPro direct bilirubin test kit and CFAS (calibrator for automated systems). Parental Huh-7 cells provided the basal amount of bilirubin. (B) No significant difference in BVR protein expression in the presence of lucidone. Ava5 cells were exposed to different concentrations (0 to 50 μM) of lucidone for 3 days, and total cell lysates were subjected to Western blotting using anti-BVR and anti-GAPDH antibodies. GAPDH protein levels confirmed equal loading of cell lysates. The results are expressed as the mean values ± the SD for triplicate experiments. The error bars denote SD of the mean. *, P < 0.05; **, P < 0.01.
Fig 4.
Induction of the antiviral IFN responses by lucidone in HCV replicon cells. (A) Concentration-dependent induction of ISRE activity by lucidone. (B) Restoration of lucidone-induced HO-1 activity by HO-1 inhibitor SnPP. (C to E) Concentration-dependent induction of gene expression of IFN-α2 (C) and IFN-α17 (D) and IFN-mediated gene expression (E, panels a to d) by lucidone. For reporter analysis, Ava5 cells were transiently transfected with 1 μg of the IFN response reporter vector pISRE-Luc. Subsequently, the pISRE-Luc-transfected cells were incubated with the indicated concentrations (0 to 50 μM) of lucidone with or without 20 μM HO-1 inhibitor SnPP for 3 days, and total cell lysates were analyzed for luciferase activity. Luciferase activity in lucidone-untreated cells was defined as 1. For the detection of bilirubin production and HO-1 expression, the total cell lysates of lucidone-treated Ava5 cells were harvested for the quantification of bilirubin concentration and Western blot analysis using the MeDiPro direct bilirubin test kit combined with CFAS (calibrator for automated systems) and anti-HO-1 antibody, respectively, under the same assay conditions. GAPDH protein levels confirmed equal loading of cell lysates. For gene expression analysis, the total RNA of lucidone-treated Ava5 cells was extracted to quantify the RNA levels of IFN-α2, IFN-α17, OAS1, OAS2, OAS3, and PKR by qRT-PCR analysis under the same assay conditions. The relative RNA levels were normalized by cellular gapdh mRNA expression. The RNA level in lucidone-untreated Ava5 cells was defined as 1. Each value represents the mean fold of normalized data ± the SD for triplicate experiments. The error bars denote the SD of the mean. *, P < 0.05; **, P < 0.01.
Fig 5.
Effect of lucidone on the HCV NS3/4A protease activity. (A) Schematic representation of the NS3 response reporter vector pEG(DEΔ4AB)SEAP. The decapeptide sequence, named 8×DEMEEC-ASHL, corresponding to the NS4A/B junction was inserted between egfp and seap. (B) Concentration-dependent reduction of NS3/4A protease activity by lucidone in cell-based analysis. Huh-7 cells were transiently cotransfected with 1 μg of the pEG(DEΔ4AB)SEAP vector and 0.5 μg of the NS3/4A expression vector pNS3/4A. Subsequently, the transfected cells were treated with the indicated concentrations (0 to 50 μM) of lucidone with or without 20 μM SnPP. After 3 days, total cell lysates were analyzed for SEAP activity. Treatment with 1 μM telaprevir or 100 μM biliverdin served as the positive controls. (C) No significant inhibition of NS3/4A protease activity by lucidone in cell-free TnT analysis. The reaction mixtures contained non-radioisotope-labeled NS3/4A protein and 35S-labeled EG(DEΔ4AB)SEAP substrate protein generated by a cell-free TnT system (Promega) in the absence or presence of increasing concentrations of lucidone. Treatment with 0.3 μM telaprevir severed as a positive control. TnT product of EGFP alone served as an indictor of protease-mediated proteolytic product from EG(DEΔ4AB)SEAP. After incubation for 15 min at 30°C, reactants were subjected to SDS-PAGE and autoradiography. Western blotting with anti-NS3 antibody was performed to reveal equal amounts of NS3/4A protein in each reaction. (D) Densitometric quantification was performed to present the relative cleavage of EG(DEΔ4AB)SEAP in the absence or presence of lucidone. The arrowheads indicated the expected sizes of EG(DEΔ4AB)SEAP, ASHL-SEAP, EGFP-8×DEMEEC, and EGFP. The relative intensity of EG(DEΔ4AB)SEAP was determined as the ratio of EG(DEΔ4AB)SEAP band to total predominant bands corresponding to the TnT products, expressed as the remaining percentage. Each value represents the mean fold ± the SD of triplicate experiments after normalization of luciferase activities. The error bars denote the SD of the mean. *, P < 0.05; **, P < 0.01.
Anti-HCV activity of lucidone is mediated by HO-1 induction and activity.
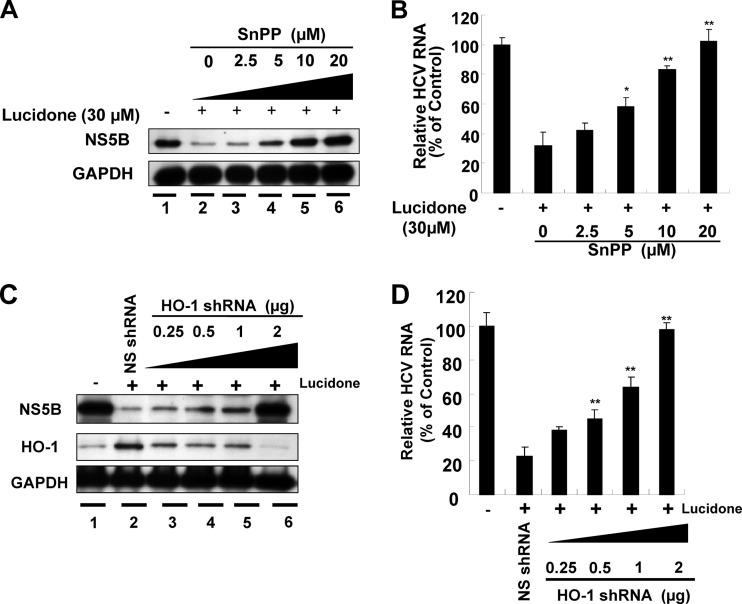
Subsequent experiments were performed to further investigate whether the anti-HCV activity of lucidone is mediated through HO-1 expression. Ava5 cells were incubated with increasing concentrations of the SnPP (2.5, 5, 10, and 20 μM) in the presence of lucidone (30 μM) for 3 days. Western blotting and qRT-PCR were used to determine the inhibitory effect of combinational treatment on HCV protein and RNA expression, respectively. As shown in Fig. 6A, SnPP treatment reversed the inhibitory effect of lucidone on HCV protein synthesis (lanes 3 to 6) in a concentration-dependent manner compared to the effects of no lucidone treatment (lane 1) and lucidone treatment in the absence of SnPP (lane 2). Similarly, HCV RNA levels were gradually restored by increasing the concentrations of SnPP (Fig. 6B). To rule out the possibility of a nonspecific effect of a pharmacological inhibitor, we performed an RNA interference technique to disrupt HO-1 function. Lucidone-treated or untreated Ava5 cells were transfected with either specific HO-1 shRNA (0.25 to 2 μg) or nonspecific control EGFP shRNA vectors. The effect of HO-1 shRNA on HO-1 and HCV protein synthesis was examined by Western blotting. As shown in Fig. 6C, HO-1 shRNA markedly reduced lucidone-mediated HO-1 induction (middle panel, lanes 3 to 6), and simultaneously reversed the inhibitory effect of lucidone on HCV protein synthesis (upper panel, lanes 3 to 6), whereas control shRNA had no effect on HO-1 expression (middle panel, lane 2) and the recovery of HCV protein synthesis in the presence of lucidone (upper panel, lane 2). It is noteworthy that the recovery of HCV protein levels was comparable to that in lucidone-untreated Ava5 cells when HO-1 expression was efficiently silenced (lanes 1 and 6). Likewise, the percent recovery of HCV RNA levels was correlated to the increase in the amount of HO-1 shRNA compared to their recovery in untransfected Ava5 cells in the presence of lucidone (Fig. 6D). Taken together, these results clearly revealed that HO-1 upregulation contributes to the antiviral action of lucidone.
Fig 6.
Restoration of lucidone-suppressed HCV protein synthesis and RNA replication by HO-1 inhibition. Ava5 cells were treated with or without different concentrations (0 to 20 μM) of the HO-1 specific inhibitor SnPP in the presence of 30 μM lucidone. After 3 days, HCV protein synthesis (A) and RNA replication (B) were analyzed by Western blotting and qRT-PCR, respectively. Ava5 cells were transfected with either different amounts (0.25 to 2 μg) of the HO-1-specific shRNA or 2 μg of nonspecific EGFP shRNA vectors. After incubation for 12 h, cells were refreshed with complete medium with or without 30 μM lucidone for an additional 3 days. Protein synthesis (C) and HCV RNA replication (D) were analyzed by Western blotting and qRT-PCR, respectively. Each value represents the mean ± the SD of triplicate experiments. The error bars denote SD of the mean. *P < 0.05; ** P < 0.01.
Lucidone triggers nuclear translocation of Nrf2 and stimulates Nrf2-mediated transcriptional activity for HO-1 induction.
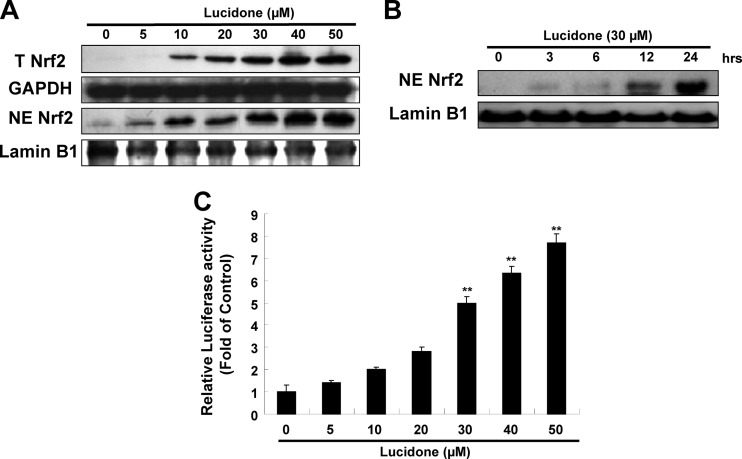
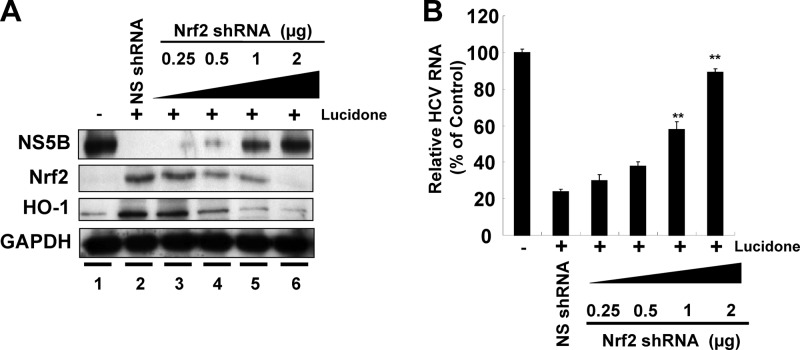
Nrf2 is one of the important nuclear factors that transcriptionally activate HO-1 expression. To examine whether Nrf2 is involved in HO-1 induction by lucidone, we first analyzed the effect of lucidone on Nrf2 expression in Ava5 cells. As shown in Fig. 7A, lucidone increased total Nrf2 protein levels and nuclear Nrf2 accumulation in a concentration-dependent manner. A time-dependent accumulation of nuclear Nrf2 in response to lucidone treatment was also observed (Fig. 7B). According to the qRT-PCR analysis, we found that lucidone caused the elevation of Nrf2 expression at the transcriptional level (data not shown). We next used a p3xARE-Luc luciferase reporter construct to verify the specificity of lucidone in the induction of ARE-mediated HO-1 upregulation in response to Nrf2 binding. p3×ARE-Luc-transfected Ava5 cells were incubated with increasing concentrations of lucidone for 3 days. As expected, lucidone enhanced luciferase activity in a concentration-dependent manner (Fig. 7C). Compared to the findings in the lucidone-untreated cells, an ∼8-fold increase in luciferase activity was observed at a lucidone concentration of 50 μM. We next silenced gene expression of Nrf2 using shRNA expression vector to verify involvement of Nrf2-mediated HO-1 upregulation in anti-HCV activity of lucidone. As shown in Fig. 8A, the increasing gene silence of Nrf2 gradually reduced lucidone-mediated HO-1 induction (second and third panels, lanes 3 to 6) and simultaneously reversed the inhibitory effect of lucidone on HCV protein synthesis (first panel, lanes 3 to 6), whereas control shRNA had no effect on HO-1 expression and the recovery of HCV protein synthesis in the presence of lucidone (lane 2). Likewise, the percentage recovery of HCV RNA levels was correlated to the increase in the amount of Nrf2 shRNA (Fig. 8B). Taken together, these results clearly revealed that Nrf2-mediated HO-1 induction may contribute to the antiviral action of lucidone.
Fig 7.
Effects of lucidone on Nrf2 expression in HCV replicon cells. (A) Concentration-dependent induction of Nrf2 expression by lucidone. Ava5 cells were incubated with the indicated concentrations (0 to 50 μM) of lucidone for 3 days. The cytoplasmic and nuclear fractions were separated from the cell lysate and analyzed by Western blotting with anti-Nrf2, anti-lamin B, and anti-GAPDH (loading control) antibodies. (B) Time-dependent nuclear translocation of Nrf2. Ava5 cells were incubated with 30 μM lucidone. The nuclear fractions were extracted, and Western blotting was performed at the indicated time points. (C) Concentration-dependent induction of Nrf2-mediated ARE transactivation. Ava5 cells were transfected with the reporter plasmid p3xARE-Luc and incubated with various concentrations of lucidone (0 to 50 μM). After 3 days, total cell lysates were harvested for the luciferase activity assay. Luciferase activity in lucidone-untreated cells was defined as 1. Each value represents the mean fold ± the SD of triplicate experiments after normalization of SEAP activities. The error bars denote the SD of the mean fold values. *, P < 0.05; **, P < 0.01.
Fig 8.
Restoration of lucidone-suppressed HCV protein synthesis (A) and RNA replication (B) by Nrf2 gene silencing. Ava5 cells were transfected with either different amounts (0.25 to 2 μg) of the Nrf2-specific shRNA or 2 μg of nonspecific EGFP shRNA vectors. After incubation for 12 h, the cells were refreshed with complete medium with or without 30 μM lucidone for an additional 3 days. Each protein synthesis and HCV RNA replication were analyzed by Western blotting and qRT-PCR, respectively. Each value represents the mean ± the SD of triplicate experiments. The error bars denote the SD of the mean. *, P < 0.05; **, P < 0.01.
Lucidone synergistically inhibits HCV replication in combination with IFN-α or viral enzyme inhibitors.
To evaluate combination treatments, we treated Ava5 cells with lucidone in combination with IFN-α, the NS3/4A protease inhibitor telaprevir (25), which is a drug currently approved by the U.S. Food and Drug Administration, the NS5A inhibitor BMS-790052 (26), or the NS5B polymerase inhibitor PSI-7977 (27), a prodrug of 2′-F-2′-C-methyluridine monophosphate, at various fixed ratios of concentrations as described in Materials and Methods. After 3 days, total RNAs were harvested and quantified by qRT-PCR analysis. The inhibitory effect of the combination treatments on HCV replication was calculated using the isobologram method and CalcuSyn software (28, 30). The results of the combination studies expressed as the mean of CI values at an effective dose of 50% (ED50), 75% (ED75) or 90% (ED90) inhibition. By definition, a CI values of 1 denotes additivity, a CI value of <1 denotes synergism, and a CI value of >1 denotes antagonism. As shown in Table 2, the CI values for the ED50, ED75, and ED90 ranged from 0.21 to 0.78, indicating that the combination treatments synergistically inhibited HCV replication according to the guidelines of the program. We also observed an enhanced induction of ISG expression upon combination treatment with lucidone (50 μM) and PEG–IFN-α (50 U/ml), compared to monodrug treatment (data not shown), which supported the results of synergistic effect for lucidone in combination with IFN-α. A traditional isobologram of each combination was presented in Fig. S1 in the supplemental material. There was no significant cytotoxic effect observed in each combination at the higher concentrations using MTS assay (see Fig. S2 in the supplemental material), which excluded the possibility of synergistic cytotoxicity upon combination treatment.
Table 2.
Effects of a combination of lucidone and various inhibitors on HCV replicationa
| Combination compound | Mean CI ± SD at |
Influence | ||
|---|---|---|---|---|
| ED50 | ED75 | ED90 | ||
| IFN-α | 0.61 ± 0.012 | 0.53 ± 0.034 | 0.47 ± 0.072 | Synergistic |
| Telaprevir | 0.78 ± 0.04 | 0.66 ± 0.047 | 0.56 ± 0.069 | Synergistic |
| BMS-790052 | 0.48 ± 0.095 | 0.42 ± 0.069 | 0.37 ± 0.05 | Synergistic |
| PSI-7977 | 0.45 ± 0.087 | 0.30 ± 0.1 | 0.21 ± 0.104 | Synergistic |
Ava5 cells were treated with combinations of various concentrations of lucidone and IFN-α, telaprevir, BMS-790052, or PSI-7977 for 3 days. The anti-HCV activity was determined by using qRT-PCR to analyze the HCV RNA levels. The combination index (CI) values for the effective dose for 50% (ED50), 75% (ED75), or 90% (ED90) inhibition were calculated using the CalcuSyn computer program. Results are expressed as mean values for three independent experiments. The CI values indicate the degree of interaction of the potential drugs; values of <1, 1, and >1 are indicative of synergistic, additive, and antagonistic effects, respectively.
DISCUSSION
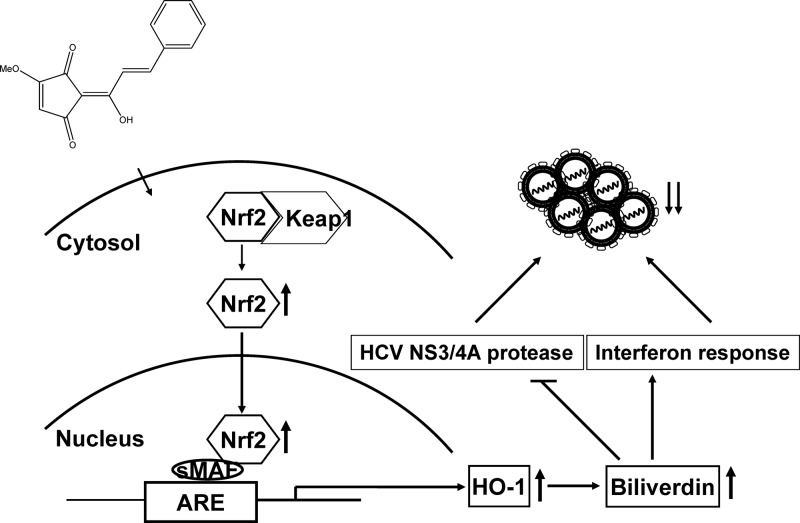
In the present study, we identified a phytochemical lucidone with strong inhibitory effects on HCV replication in both HCV replicon and JFH-1-infected cells (Fig. 1). In addition, we found that lucidone treatment resulted in the elevation of cellular HO-1 expression at the transcriptional level (Fig. 2), leading to accumulation of its product biliverdin for anti-HCV activity (Fig. 3). These results are supported by recent findings of Lehmann et al. and Zhu et al. demonstrating that the HO-1 metabolic product biliverdin inhibited viral replication by targeting IFN signaling pathway and viral NS3/4A protease (12, 14) (Fig. 4 and 5). For comparison, lucidone displayed the efficient inhibition of viral replication by as much as 80% at a concentration of 50 μM in both assay systems (Fig. 1), which was similar to or even better than that of bilividin on anti-HCV replication at concentrations of approximately 50 to 200 μM in those earlier reports. Investigation of the mechanism disclosed that HO-1 inhibition by HO-1 inhibitor or shRNA gradually abrogated the anti-HCV activity of lucidone (Fig. 6). Moreover, we observed high accumulation of cellular and nuclear Nrf2 and subsequently confirmed the specific transactivation of HO-1 expression by Nrf2 using an ARE-luciferase activity assay in lucidone-treated cells (Fig. 7). In contrast, depletion of Nrf2 by shRNA resulted in the abrogation of HO-1 induction and anti-HCV activity of lucidone, clearly indicating that the inhibitory effect of lucidone on HCV replication was because of Nrf2-ARE-dependent HO-1 induction (Fig. 8). The proposed model of anti-HCV action of lucidone was illustrated in Fig. 9. Lucidone enhances Nrf2 expression and triggers its nuclear translocation, leading to an activation of HO-1 expression. Subsequently, an accumulation of its metabolite biliverdin then inhibits HCV replication through increasing antiviral interferon response and blocking HCV NS3/4A protease activity.
Fig 9.
Model for the inhibitory action of lucidone on HCV replication.
A previous report demonstrated that treatment with miRNA-196, which targets Bach1, a transcriptional repressor of HO-1, resulted in HO-1 upregulation and subsequent suppression of HCV replication (13). In addition to suppressing HCV replication, the induction or overexpression of HO-1 has been demonstrated to interfere with the replication of other viruses, such as HIV (38), hepatitis B virus (39), and enterovirus 71 (40). Therefore, targeting HO-1 may be a potential therapeutic approach to inhibit virus infection. The present study is the first time to identify a phytocompound as a protective agent against HCV replication via HO-1 induction. In addition to Nrf2, nuclear factor-κB (NF-κB) and activator protein-1 (AP-1) are also involved in the transactivation of HO-1 expression (10). We will further investigate the effect of lucidone on the NF-κB and AP-1 expression in terms of HO-1 upregulation. In addition, HO-1 gene expression can be transcriptionally mediated by a number of intracellular signaling molecules, such as extracellular signal-regulated protein kinase (ERK) (41), p38 mitogen-activated protein kinase (MAPK), c-Jun N-terminal kinase, protein kinase C, and phosphatidylinositol 3-kinase (8). Among them, the activation of MAPK/ERK kinase-ERK1/2 signaling has been reported to be involved in anti-HCV activity upon oxidative stress (42). Moreover, lucidone was previously reported to exert protective effects against LPS-induced inflammation through NF-κB/MAPK signaling pathways (18). Therefore, it is worthwhile to further investigate additional molecules or signaling pathways involved in HO-1 activation by lucidone to clearly demonstrate the detailed molecular mechanism by which lucidone blocks HCV replication.
A number of new direct-acting antivirals (DAAs) for treating HCV infection are currently under development. Telaprevir, a recently approved NS3/4A protease inhibitor, significantly improves the sustained virological response rate in combination with the standard of care of PEG-IFN and ribavirin among patients with chronic genotype 1 HCV infection (43). Although the antiviral efficacy of protease inhibitor triple therapy appears to be positive, this regimen still has several challenges, such as anemia, a well-recognized side effect of IFN-based regimens, and the emergence of DAA-related resistant variants due to the high replication rate of the virus and the low fidelity of the NS5B polymerase (44). Indeed, targeting host factors required for the viral life cycle has been considered a favorable strategy to overcome the genetic variability of the viral genome because the mutation rate of host genes is lower than that of viral genes (45). Accordingly, combinations of DAAs and host-targeted antivirals may provide alternative regimens to overcome these limitations. In addition to targeting host HO-1 expression, lucidone exhibits synergistic activity against HCV replication in combination with other promising inhibitors against distinct targets of HCV (Table 2). In regard to lucidone as a potentially clinically useful drug or adjuvant, assessment of anti-HCV activity assay in vivo animal mode is required. Recently, Kumar et al. have performed in vivo experiments to demonstrate anti-inflammatory activity of lucidone against lipopolysaccharide (LPS)-induced inflammation in mice (18). After the intraperitoneal injection with 200 mg of lucidone/kg, the concentration of lucidone in plasma was 368.5 μg/ml (1,439.4 μM), which is ∼29-fold higher than the effective concentration of lucidone (50 μM) on reducing HCV RNA levels by 80% an in vitro cell-based anti-HCV activity assay. For the development of new drugs, the proper translation from an mouse animal drug dosage to a human dosage is essential. The body surface area normalization method is more commonly used to safely predicate a suitable starting dose for a human clinical trial from animal toxicology data, which incorporated several physiologic parameters between different mammalian species, such as oxygen utilization, caloric expenditure, basal metabolic rate, blood volume, circulating plasma proteins, and renal function (46). For example, the safe starting dose of lucidone used in the adult human is approximated to be 16.22 mg/kg according to the formula for dose translation by the km factor. In addition, predication of in vivo human metabolic drug clearance from in vitro metabolism data and/or in vivo animal experiments is an important assessment for clinical studies. For predicting the hepatic clearance of drug, several liver models, such as the well-stirred parallel tube and dispersion, have been used to analyze pharmacokinetic parameters using in vitro hepatic microsome and isolated hepatocytes from animals or humans (47, 48). These animal experiments will provide practical information on developing lucidone as an adjuvant in current regimens against HCV in the future.
In summary, we found that lucidone possesses anti-HCV activity. An investigation of the mechanism(s) disclosed that the inhibition of HCV replication by lucidone was because of biliverdin production through Nrf2-mediated HO-1 induction, which indicated the feasibility of using selective HO-1 inducers to improve the efficacy of cellular defense pathways against HCV infection and also inhibit viral protease activity.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Charles Rice (Rockefeller University and Aapth, LCC, USA) for kindly providing the Con1b replicon plasmid, the human hepatoma cell line Huh-7, and the HCV subgenomic replicon-containing cell line Ava5. We also thank T. Wakita (National Institute of Infectious Diseases, Japan) for providing the JFH-1 plasmid and Anupam Agarwal (University of Alabama) for providing the pHOGL3/9.4 plasmid.
This study was funded by grants from the National Science Council of Taiwan (NSC 100-2311-B-037-001 and NSC 101-2311-B-037-002-MY3) and the Kaohsiung Medical University Research Foundation of Taiwan (KMUER014).
Footnotes
Published ahead of print 17 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.02053-12.
REFERENCES
- 1. Jahan S, Ashfaq UA, Qasim M, Khaliq S, Saleem MJ, Afzal N. 2012. Hepatitis C virus to hepatocellular carcinoma. Infect. Agents Cancer 7:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Giannini C, Brechot C. 2003. Hepatitis C virus biology. Cell Death Differ. 10(Suppl 1):S27–S38 [DOI] [PubMed] [Google Scholar]
- 3. Hayashi N, Takehara T. 2006. Antiviral therapy for chronic hepatitis C: past, present, and future. J. Gastroenterol. 41:17–27 [DOI] [PubMed] [Google Scholar]
- 4. Schaefer M, Mauss S. 2008. Hepatitis C treatment in patients with drug addiction: clinical management of interferon-alpha-associated psychiatric side effects. Curr. Drug Abuse Rev. 1:177–187 [DOI] [PubMed] [Google Scholar]
- 5. Susser S, Welsch C, Wang Y, Zettler M, Domingues FS, Karey U, Hughes E, Ralston R, Tong X, Herrmann E, Zeuzem S, Sarrazin C. 2009. Characterization of resistance to the protease inhibitor boceprevir in hepatitis C virus-infected patients. Hepatology 50:1709–1718 [DOI] [PubMed] [Google Scholar]
- 6. Sarrazin C, Zeuzem S. 2010. Resistance to direct antiviral agents in patients with hepatitis C virus infection. Gastroenterology 138:447–462 [DOI] [PubMed] [Google Scholar]
- 7. Sarrazin C, Kieffer TL, Bartels D, Hanzelka B, Muh U, Welker M, Wincheringer D, Zhou Y, Chu HM, Lin C, Weegink C, Reesink H, Zeuzem S, Kwong AD. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767–1777 [DOI] [PubMed] [Google Scholar]
- 8. Farombi EO, Surh YJ. 2006. Heme oxygenase-1 as a potential therapeutic target for hepatoprotection. J. Biochem. Mol. Biol. 39:479–491 [DOI] [PubMed] [Google Scholar]
- 9. Sass G, Seyfried S, Parreira Soares M, Yamashita K, Kaczmarek E, Neuhuber WL, Tiegs G. 2004. Cooperative effect of biliverdin and carbon monoxide on survival of mice in immune-mediated liver injury. Hepatology 40:1128–1135 [DOI] [PubMed] [Google Scholar]
- 10. Paine A, Eiz-Vesper B, Blasczyk R, Immenschuh S. 2010. Signaling to heme oxygenase-1 and its anti-inflammatory therapeutic potential. Biochem. Pharmacol. 80:1895–1903 [DOI] [PubMed] [Google Scholar]
- 11. Dhakshinamoorthy S, Jain AK, Bloom DA, Jaiswal AK. 2005. Bach1 competes with Nrf2 leading to negative regulation of the antioxidant response element (ARE)-mediated NAD(P)H:quinone oxidoreductase 1 gene expression and induction in response to antioxidants. J. Biol. Chem. 280:16891–16900 [DOI] [PubMed] [Google Scholar]
- 12. Lehmann E, El-Tantawy WH, Ocker M, Bartenschlager R, Lohmann V, Hashemolhosseini S, Tiegs G, Sass G. 2010. The heme oxygenase 1 product biliverdin interferes with hepatitis C virus replication by increasing antiviral interferon response. Hepatology 51:398–404 [DOI] [PubMed] [Google Scholar]
- 13. Hou W, Tian Q, Zheng J, Bonkovsky HL. 2010. MicroRNA-196 represses Bach1 protein and hepatitis C virus gene expression in human hepatoma cells expressing hepatitis C viral proteins. Hepatology 51:1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu Z, Wilson AT, Luxon BA, Brown KE, Mathahs MM, Bandyopadhyay S, McCaffrey AP, Schmidt WN. 2010. Biliverdin inhibits hepatitis C virus nonstructural 3/4A protease activity: mechanism for the antiviral effects of heme oxygenase? Hepatology 52:1897–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ichino K, Tanaka H, Ito K, Tanaka T, Mizuno M. 1988. Two new dihydrochalcones from Lindera erythrocarpa. J. Nat. Prod. 51:915–917 [DOI] [PubMed] [Google Scholar]
- 16. Wang SY, Lan XY, Xiao JH, Yang JC, Kao YT, Chang ST. 2008. Anti-inflammatory activity of Lindera erythrocarpa fruits. Phytother. Res. 22:213–216 [DOI] [PubMed] [Google Scholar]
- 17. Oh HM, Choi SK, Lee JM, Lee SK, Kim HY, Han DC, Kim HM, Son KH, Kwon BM. 2005. Cyclopentenediones, inhibitors of farnesyl protein transferase and anti-tumor compounds, isolated from the fruit of Lindera erythrocarpa Makino. Bioorg. Med. Chem. 13:6182–6187 [DOI] [PubMed] [Google Scholar]
- 18. Senthil Kumar KJ, Hsieh HW, Wang SY. 2010. Anti-inflammatory effect of lucidone in mice via inhibition of NF-κB/MAP kinase pathway. Int. Immunopharmacol. 10:385–392 [DOI] [PubMed] [Google Scholar]
- 19. Kumar KJ, Yang JC, Chu FH, Chang ST, Wang SY. 2010. Lucidone, a novel melanin inhibitor from the fruit of Lindera erythrocarpa Makino. Phytother. Res. 24:1158–1165 [DOI] [PubMed] [Google Scholar]
- 20. Blight KJ, Kolykhalov AA, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974 [DOI] [PubMed] [Google Scholar]
- 21. Hill-Kapturczak N, Sikorski E, Voakes C, Garcia J, Nick HS, Agarwal A. 2003. An internal enhancer regulates heme- and cadmium-mediated induction of human heme oxygenase-1. Am. J. Physiol. Renal Physiol. 285:F515–F523 [DOI] [PubMed] [Google Scholar]
- 22. Lee JC, Tseng CK, Chen KJ, Huang KJ, Lin CK, Lin YT. 2010. A cell-based reporter assay for inhibitor screening of hepatitis C virus RNA-dependent RNA polymerase. Anal. Biochem. 403:52–62 [DOI] [PubMed] [Google Scholar]
- 23. Lee JC, Tseng CK, Wu SF, Chang FR, Chiu CC, Wu YC. 2011. San-Huang-Xie-Xin-Tang extract suppresses hepatitis C virus replication and virus-induced cyclooxygenase-2 expression. J. Viral Hepat. 18:e315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee JC, Shih YF, Hsu SP, Chang TY, Chen LH, Hsu JT. 2003. Development of a cell-based assay for monitoring specific hepatitis C virus NS3/4A protease activity in mammalian cells. Anal. Biochem. 316:162–170 [DOI] [PubMed] [Google Scholar]
- 25. Lin K, Perni RB, Kwong AD, Lin C. 2006. VX-950, a novel hepatitis C virus (HCV) NS3-4A protease inhibitor, exhibits potent antiviral activities in HCV replicon cells. Antimicrob. Agents Chemother. 50:1813–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lemm JA, Leet JE, O'Boyle DR, 2nd, Romine JL, Huang XS, Schroeder DR, Alberts J, Cantone JL, Sun JH, Nower PT, Martin SW, Serrano-Wu MH, Meanwell NA, Snyder LB, Gao M. 2011. Discovery of potent hepatitis C virus NS5A inhibitors with dimeric structures. Antimicrob. Agents Chemother. 55:3795–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lam AM, Espiritu C, Bansal S, Micolochick Steuer HM, Niu C, Zennou V, Keilman M, Zhu Y, Lan S, Otto MJ, Furman PA. 2012. Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of hepatitis C virus. Antimicrob. Agents Chemother. 56:3359–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chou TC, Talalay P. 1984. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 22:27–55 [DOI] [PubMed] [Google Scholar]
- 29. Chou TC, Talalay P. 1981. Generalized equations for the analysis of inhibitions of Michaelis-Menten and higher-order kinetic systems with two or more mutually exclusive and nonexclusive inhibitors. Eur. J. Biochem. 115:207–216 [DOI] [PubMed] [Google Scholar]
- 30. Tallarida RJ. 2001. Drug synergism: its detection and applications. J. Pharmacol. Exp. Ther. 298:865–872 [PubMed] [Google Scholar]
- 31. Abmayr SM, Yao T, Parmely T, Workman JL. 2006. Preparation of nuclear and cytoplasmic extracts from mammalian cells. Curr. Protoc. Mol. Biol. Chapter 12:Unit 12-11 [DOI] [PubMed] [Google Scholar]
- 32. Senthil Kumar KJ, Wang SY. 2009. Lucidone inhibits iNOS and COX-2 expression in LPS-induced RAW 264.7 murine macrophage cells via NF-κB and MAPK signaling pathways. Planta Med. 75:494–500 [DOI] [PubMed] [Google Scholar]
- 33. Lee MY, Lee JA, Seo CS, Ha H, Lee H, Son JK, Shin HK. 2011. Anti-inflammatory activity of Angelica dahurica ethanolic extract on RAW264.7 cells via upregulation of heme oxygenase-1. Food Chem. Toxicol. 49:1047–1055 [DOI] [PubMed] [Google Scholar]
- 34. Jun MS, Ha YM, Kim HS, Jang HJ, Kim YM, Lee YS, Kim HJ, Seo HG, Lee JH, Lee SH, Chang KC. 2011. Anti-inflammatory action of methanol extract of Carthamus tinctorius involves in heme oxygenase-1 induction. J. Ethnopharmacol. 133:524–530 [DOI] [PubMed] [Google Scholar]
- 35. Kravets A, Hu Z, Miralem T, Torno MD, Maines MD. 2004. Biliverdin reductase, a novel regulator for induction of activating transcription factor-2 and heme oxygenase-1. J. Biol. Chem. 279:19916–19923 [DOI] [PubMed] [Google Scholar]
- 36. Murray EJ, Burden F, Horscroft N, Smith-Burchnell C, Westby M. 2011. Knockdown of USP18 increases alpha 2a interferon signaling and induction of interferon-stimulating genes but does not increase antiviral activity in Huh7 cells. Antimicrob. Agents Chemother. 55:4311–4319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Iro M, Witteveldt J, Angus AG, Woerz I, Kaul A, Bartenschlager R, Patel AH. 2009. A reporter cell line for rapid and sensitive evaluation of hepatitis C virus infectivity and replication. Antiviral Res. 83:148–155 [DOI] [PubMed] [Google Scholar]
- 38. Devadas K, Dhawan S. 2006. Hemin activation ameliorates HIV-1 infection via heme oxygenase-1 induction. J. Immunol. 176:4252–4257 [DOI] [PubMed] [Google Scholar]
- 39. Protzer U, Seyfried S, Quasdorff M, Sass G, Svorcova M, Webb D, Bohne F, Hosel M, Schirmacher P, Tiegs G. 2007. Antiviral activity and hepatoprotection by heme oxygenase-1 in hepatitis B virus infection. Gastroenterology 133:1156–1165 [DOI] [PubMed] [Google Scholar]
- 40. Tung WH, Hsieh HL, Lee IT, Yang CM. 2011. Enterovirus 71 induces integrin beta1/EGFR-Rac1-dependent oxidative stress in SK-N-SH cells: role of HO-1/CO in viral replication. J. Cell Physiol. 226:3316–3329 [DOI] [PubMed] [Google Scholar]
- 41. Lenz O, Verbinnen T, Lin TI, Vijgen L, Cummings MD, Lindberg J, Berke JM, Dehertogh P, Fransen E, Scholliers A, Vermeiren K, Ivens T, Raboisson P, Edlund M, Storm S, Vrang L, de Kock H, Fanning GC, Simmen KA. 2010. In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435. Antimicrob. Agents Chemother. 54:1878–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yano M, Ikeda M, Abe K, Kawai Y, Kuroki M, Mori K, Dansako H, Ariumi Y, Ohkoshi S, Aoyagi Y, Kato N. 2009. Oxidative stress induces anti-hepatitis C virus status via the activation of extracellular signal-regulated kinase. Hepatology 50:678–688 [DOI] [PubMed] [Google Scholar]
- 43. Saxena V, Terrault N. 2012. Hepatitis C virus treatment and liver transplantation in the era of new antiviral therapies. Curr. Opin. Organ Transplant. 17:216–224 [DOI] [PubMed] [Google Scholar]
- 44. Schaefer EA, Chung RT. 2012. Anti-hepatitis C virus drugs in development. Gastroenterology 142:1340–1350 e1341 [DOI] [PubMed] [Google Scholar]
- 45. Salloum S, Tai AW. 2012. Treating hepatitis C infection by targeting the host. Transl. Res. 159:421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Reagan-Shaw S, Nihal M, Ahmad N. 2008. Dose translation from animal to human studies revisited. FASEB J. 22:659–661 [DOI] [PubMed] [Google Scholar]
- 47. Ito K, Houston JB. 2004. Comparison of the use of liver models for predicting drug clearance using in vitro kinetic data from hepatic microsomes and isolated hepatocytes. Pharm. Res. 21:785–792 [DOI] [PubMed] [Google Scholar]
- 48. Naritomi Y, Terashita S, Kimura S, Suzuki A, Kagayama A, Sugiyama Y. 2001. Prediction of human hepatic clearance from in vivo animal experiments and in vitro metabolic studies with liver microsomes from animals and humans. Drug Metab. Dispos. 29:1316–1324 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.