Abstract
Spreading manure containing antibiotics in agriculture is assumed to stimulate the dissemination of antibiotic resistance in soil bacterial populations. Plant roots influencing the soil environment and its microflora by exudation of growth substrates might considerably increase this effect. In this study, the effects of manure from pigs treated with sulfadiazine (SDZ), here called SDZ manure, on the abundance and transferability of sulfonamide resistance genes sul1 and sul2 in the rhizosphere of maize and grass were compared to the effects in bulk soil in a field experiment. In plots that repeatedly received SDZ manure, a significantly higher abundance of both sul genes was detected compared to that in plots where manure from untreated pigs was applied. Significantly lower abundances of sul genes relative to bacterial ribosomal genes were encountered in the rhizosphere than in bulk soil. However, in contrast to results for bulk soil, the sul gene abundance in the SDZ manure-treated rhizosphere constantly deviated from control treatments over a period of 6 weeks after manuring, suggesting ongoing antibiotic selection over this period. Transferability of sulfonamide resistance was analyzed by capturing resistance plasmids from soil communities into Escherichia coli. Increased rates of plasmid capture were observed in samples from SDZ manure-treated bulk soil and the rhizosphere of maize and grass. More than 97% of the captured plasmids belonged to the LowGC type (having low G+C content), giving further evidence for their important contribution to the environmental spread of antibiotic resistance. In conclusion, differences between bulk soil and rhizosphere need to be considered when assessing the risks associated with the spreading of antibiotic resistance.
INTRODUCTION
Sulfadiazine (SDZ) is an antibiotic compound that has been intensively used since the 1940s for the treatment of infectious diseases in animal husbandry, mainly in pig production (1, 2). More than 96% of the SDZ administered to pigs can be recovered in manure as the parent antibiotic compound or as its metabolites (3). Through field fertilization by manuring, SDZ and its metabolites enter arable soils in bioactive amounts (4, 5). Although the concentration of readily extractable SDZ in soil was reported to decrease rapidly (6, 7), SDZ might still select resistant populations and stimulate horizontal spread of antibiotic resistance genes, leading to an increased abundance of bacterial populations carrying resistance genes (reviewed in reference 8). In bacteria, SDZ inhibits the dihydropteroate synthase (DHPS), a crucial enzyme in the folic acid biosynthesis pathway, by competing with the natural substrate p-amino-benzoic acid (9). Three alternative DHPS genes conferring SDZ resistance are known (sul1, sul2, and sul3), coding for enzymes with a lower affinity for sulfonamides (10). These resistance genes are often located on plasmids (11, 12). Manure was shown to be a hot spot for bacteria carrying antibiotic resistance genes and mobile genetic elements (3, 13–18) and may enhance the horizontal gene transfer of antibiotic resistance genes in soil (19), although it was shown that bacterial populations of manure and soil may be quite distinct (20). Additionally, nutrients provided by manure might stimulate the growth of bacteria in soil, increasing the effect of SDZ, which is active on growing cells (21, 22). In microcosm experiments, synergistic effects of manure and SDZ on resistance levels were demonstrated (23), and an accumulation of sul1 and sul2 genes by repeated application of manure containing SDZ (SDZ manure) to soil was shown (24). LowGC-type plasmids were identified as a major vector of sul2 in manure and manure-treated soils (25). The rhizosphere is known as a hot spot of horizontal gene transfer (26, 27), which appears to be controlled by exudation and root growth affecting the cell density, distribution, and metabolic activity (for a review, see reference 28). Cereals and pasture plants, for example, transfer up to 50% of total assimilated carbon into soil (29). Root exudates include, e.g., sugars, organic acids, and amino acids, but their composition depends on different factors, such as plant species, developmental stage, and stress factors (for a review, see reference 30). A microcosm study revealed that artificial root exudates can increase bacterial community tolerance to SDZ, suggesting a similar effect caused by roots from field plants (31). Recently, it was observed for the field experiment reported here that the dissipation of CaCl2-extractable SDZ was accelerated under field conditions compared to a laboratory experiment and especially in the rhizosphere, a result which might be related to differences in temperature and soil moisture (6) and hence may reduce the exposure of bacteria to SDZ. This assumption was supported by the results of a mesocosm experiment, in which a faster dissipation of SDZ in the rhizosphere of maize than in bulk soil was observed and could be correlated with a lower abundance of sul1 and sul2 relative to 16S rRNA gene abundance (13). Although little is known regarding the influence of plants growing in manured soil on the abundance and transfer of antibiotic resistance genes and selection of resistant bacterial populations under field conditions, this information is crucial for an integrative risk assessment.
In this study, a field experiment was conducted to test our hypothesis that the rhizosphere of plants and repeated application of manure from pigs treated with SDZ increase the transfer and abundance of antibiotic resistance genes in soil. The dynamics of sulfonamide resistance genes in field plots with maize and grass pasture were analyzed because these plants are typically fertilized with large amounts of manure (32, 33). Field plots were treated repeatedly with manure from SDZ-treated or untreated pigs in a randomized block design. Samples were taken over a period of 175 days and analyzed by cultivation-independent methods (quantitative PCR [qPCR] of total community DNA, exogenous plasmid isolation) to quantify the sul1 and sul2 gene abundance and transferability.
MATERIALS AND METHODS
Manure production.
Manure was produced at the experimental farm Frankenforst (University of Bonn), which keeps approximately 100 breeding sows and their descendants for scientific purposes. The fattening pigs (Pietrain × German Landrace, Pietrain × Duroc) used for manure production were treated with iron and vaccinated against enzootic pneumonia at an early age (≤10 days), and no further drugs were administered to them. During the experiments, their feed was composed of wheat (46%), barley (29.5%), soy (2%), soybean meal (20%), and mineral feed (2.5%).
For the SDZ treatment, pig slurry for each application was produced separately by intramuscular administration of the prescribed dose of SDZ (30 mg/kg body weight) on four consecutive days and collection of slurry (SDZ manure) for 10 days. SDZ injection solution (200 mg/ml) was kindly provided by Vetoquinol Biowet (Gorzow Wielkopolski, Poland). For the control treatment, pig slurry was simultaneously collected from untreated pigs in a separate pen. After collection, manure was stored until its application to the soil in plastic barrels at ambient temperature. Basic manure properties varied between the treatments and between the application days (e.g., 2.6 to 8.8% dry matter, 3.5 to 6.2 kg N/m3). Concentrations of SDZ (mean ± standard error [SE] of five laboratory replicates) measured in manure were 30 ± 2, 131 ± 12, and 82 ± 13 mg/kg (fresh weight) for the applications 1, 2, and 3, respectively (6). In addition, the manures contained 19 ± 0 (1st application), 53 ± 2 (2nd application), and 32 ± 2 (3rd application) mg/kg of the SDZ metabolite N-acetyl-SDZ, which is quickly transformed back into the parent compound SDZ in soil. The control manure contained no SDZ.
Field experiment.
The field trial with grassland and maize plots (3 by 6 m; n = 4) arranged in a randomized block design was performed as described in reference 6 from May 2009 until June 2010 near Jülich (Germany) on a Luvisol described earlier (7). Air temperature and precipitation were recorded by a meteorological station and are reported elsewhere (6). An amount of 30 m3/ha of the collected manure was applied manually using modified water cans on 19 May (day 0). The manure was incorporated into a depth of 12 cm on maize plots with a rototiller before the sowing of maize (Zea mays L.) but was not incorporated on the grassland plots. During this and the following applications, the full penetration of manure into the grassland plots took a maximum of 1 h, depending on the previous weather conditions. The penetration depth of the manure was not monitored, and we therefore limited the sampling to the uppermost 4 cm of the grassland plots. Grassland was composed of 47% Lolium perenne L., 20% Festuca pratensis L., 17% Phleum pratense L., 10% Poa pratensis L., and 6% Trifolium repens L. and thus represented a typical permanent pasture mix (51). Grassland plots had been sown about 2 months before the start of the experiment. Due to German regulations limiting the maximum amount of manure that can be applied to agricultural fields per year to an equivalent of 170 kg N ha−1 year−1 (34), the amounts of manure for the second and third applications were reduced. The second manure application (10 m3/ha) between maize rows and on mown grass followed on 7 July (day 49), with a third one (10 m3/ha) on 29 September (day 133), 1 day after the maize was harvested and the grass was mown. After the third application, the harvested maize plots were rototilled again. Before each application, the manure was mixed with the same amount of water to achieve a more uniform distribution on the field plots. Soil samples were collected on days 0, 2, 7, 14, 28, 42, 48, 49, 56, 63, 77, 105, 132, 133, 140, 147, 161, and 175 as previously described (6). Rhizosphere was defined as soil adhering to the roots after gentle shaking. Sufficient amounts of maize rhizosphere could be collected from day 42 until day 132 (harvest) only.
DNA extraction.
To estimate the water content of each sample, 1 g of soil was dried for 48 h at 105°C and the difference in weight was measured. To obtain the rhizosphere samples, 5 g of root material with attached soil was treated in a stomacher blender three times with 15 ml 0.85% sodium chloride solution for 30 s at high speed (Stomacher 400 laboratory blender; Seward, United Kingdom). The suspensions were decanted into 50-ml Falcon tubes. The rhizosphere solutions and 10 ml manure from each day of application were centrifuged (3,780 × g, 15 min; Biofuge Stratos, Heraeus, Germany), and the pellets were homogenized. DNA from 0.5 g rhizosphere pellet, 0.5 g manure pellet, or 0.5 g bulk soil was extracted using the FastDNA SPIN kit for soil (MP Biomedicals, Heidelberg, Germany), followed by a purification step using the Geneclean spin kit (MP Biomedicals, Heidelberg, Germany), according to the manufacturer's instructions.
Exogenous plasmid isolation.
Exogenous plasmid isolation was essentially done as described previously (13, 17) using Escherichia coli K-12 CV601gfp (Rifr mini Tn5::gfp-nptII) as the recipient. One gram of bulk soil or rhizosphere pellet (see previous paragraph) was resuspended in 9 ml of 0.85% sodium chloride solution, supplemented with 5 sterile glass beads (diameter, 4 mm), and incubated in an overhead shaker (GFL 3025; Braunschweiger Labor Bedarf, Braunschweig, Germany) for 2 h, except for rhizosphere samples of day 48, which were not incubated. Four milliliters of the bulk soil or rhizosphere suspension was mixed with recipient cells and further processed as described previously (13, 17). Serial dilutions of resuspended cells from filter matings were spread on selective media supplemented with kanamycin (50 mg/liter), rifampin (50 mg/liter), SDZ (50 mg/liter), and cycloheximide (100 mg/liter) and incubated for two days at 28°C. Transconjugants were confirmed by green fluorescent protein (GFP) fluorescence and transferred to fresh plates of the selective medium. Recipient numbers were determined by applying 20-μl drops in three replicates per serial dilution on plate count agar (PCA) with rifampin and kanamycin (50 mg/liter). The transfer frequencies (TF) were calculated from CFU counts using the following formula:
Detection and quantification of target genes.
The sul1, sul2, and bacterial 16S rRNA genes (rrn) were quantified by real-time PCR 5′-nuclease assays in an CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA) as described previously (23, 25). To adjust for differences in bacterial DNA and amplification efficiency between samples, target numbers of the respective genes were divided by the rrn gene numbers and the results were log transformed. The relative gene abundance obtained was determined for sampling days 2, 7, 28, 42, 48, 56, 77, 106, 132, and 175. The sul3 gene abundance was not determined due to an expected low abundance which was observed previously for the same soil and different manure samples (23). For the PCR replicon typing, plasmids were extracted from transconjugants (see above) using the Qiagen plasmid minikit (Qiagen, Hilden, Germany) as described previously (13) or using phenol extraction (35). A total of 192 and 100 transconjugants were analyzed, uniformly drawn from the different treatments and soil compartments of days 48 and 132, respectively (see above). The rep gene of the LowGC-type plasmid group was detected by amplification of a 912-bp fragment in a standard PCR as previously described (25), using plasmid pHHV216 as a positive control.
Statistical analysis.
Significant differences between transfer frequencies were tested in pairwise comparisons using the Tukey test (P < 0.05; SAS 9.2; SAS Institute Inc., Cary, NC). For calculating the effect of SDZ manure over time (E), the differences in sul1 and sul2 relative abundance between control and SDZ manure treatment were calculated for each time point as follows:
For that purpose, the relative abundance values were ordered for each time point and treatment in a size-dependent manner.
The effect was plotted against the days to the previous manuring, and correlation was tested by linear regression analysis (P < 0.05; SAS 9.2; SAS Institute Inc., Cary, NC).
For the evaluation of the influence of the factors time, treatment, and compartment for the relative abundance of the dependent parameters sul1 and sul2, a three-way analysis of variance (ANOVA) was calculated using the procedure MIXED of SAS 9.2 (SAS Institute Inc., Cary, NC).
RESULTS
sul gene relative abundance influenced by repeated application of manure.
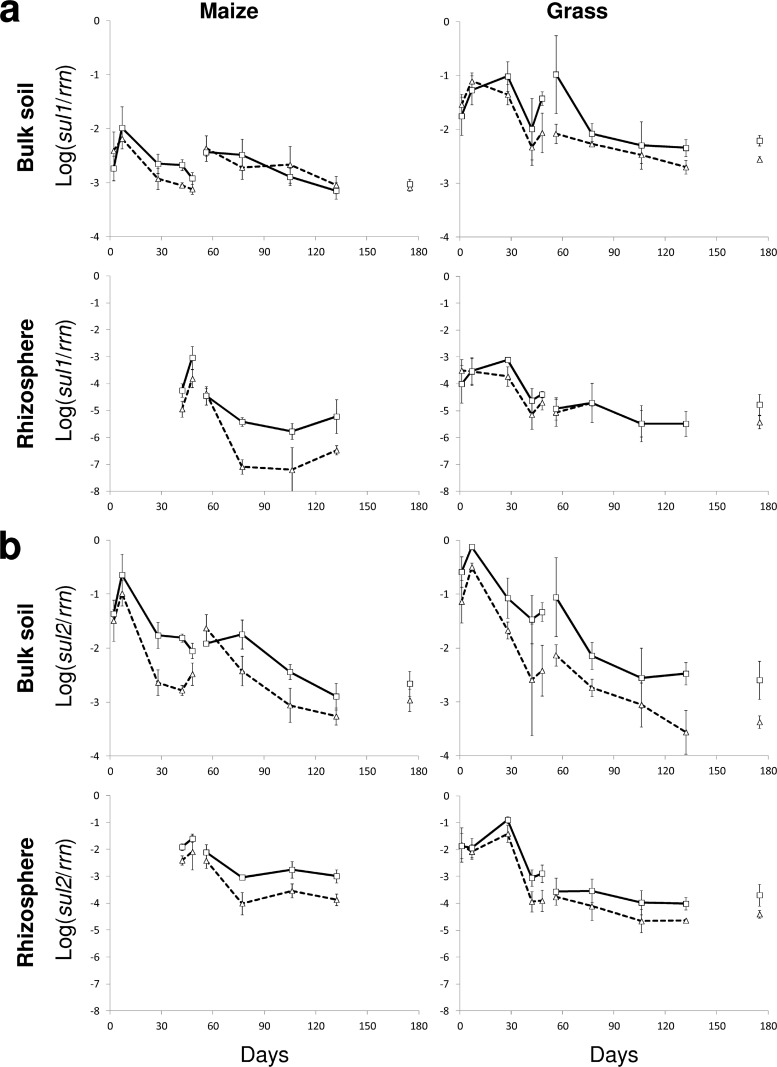
The effect of manure from pigs treated with SDZ or not on the abundance of sul resistance genes in bulk soil and the rhizosphere of grass and maize was analyzed in total community DNA. For that purpose, the relative sul gene abundance related to the number of 16S rRNA genes (rrn) was measured at several time points. In untreated soil (before application of manure, day −1), the relative sul abundances were in the range of −5.9 ± 0.5 log units for sul1 and −4.1 ± 0.2 log units for sul2 (one composite sample, technical replication; n = 4), while in the manures, the relative abundances for sul1 and sul2 were up to 4 and 3 log units higher, respectively, with relative abundances of −1 to −3 log units for sul1 and −1 to −2 log units for sul2 (composite samples). The relative sul2 abundance was on average 0.6 log units higher in the manure from SDZ-treated pigs than in the control manure (t test; P < 0.05), and the same trend was found for sul1 (0.4 log units, P = 0.052). Two and 7 days after the first application of manure from pigs treated with SDZ or not, the relative sul abundance in bulk soil was increased by more than 4 orders of magnitude for sul1 and by more than 3 orders of magnitude for sul2 (Fig. 1) compared to that of untreated soils (see above). In the rhizosphere of grass the values for sul1 and sul2 were about 2 and 1 order of magnitude lower than in the bulk soil, respectively. The sampling of the maize rhizosphere started 42 days after the first application of manure. At that point in time the rhizosphere showed a relative sul abundance similar to that observed in the grass rhizosphere after 7 days. Following the initial increase, a continuous decrease in relative abundance of sul1 and sul2 was observed for both manure treatments in bulk soil and the rhizosphere of grass and maize. This decrease seemed to be faster in the beginning with a slower decrease over time, especially in the rhizosphere, where the relative sul abundance tended to remain at a constant level starting 77 days after the first application of manure. Interestingly, the second and third applications of manure led to a minor increase in relative sul gene abundance, or a slight decrease of up to 1 log unit could be observed. The numbers of rrn/ml manure measured were in the range of 10 to 12 log units for the three applications. Independently from the application of manure, the absolute number of rrn/g soil (dry weight) was rather constant over time and ranged from around 9.5 to 10.5 log units for bulk soil and from around 10 to 11 log units for the rhizosphere.
Fig 1.
Time course of the relative abundance of sulfadiazine (SDZ) resistance genes sul1 (a) and sul2 (b) in bulk soil and rhizosphere of maize and grass. Soils treated with manure from pigs treated with SDZ or not are indicated by boxes connected by solid lines and triangles connected by dashed lines, respectively. Data points are connected by mere convenience, while no connection was drawn between data points taken before and after application of manure on days 0, 49, and 133. Error bars indicate standard deviations of results for 3 or 4 experimental replicates.
Effect of time, treatment, and soil compartment on sul relative abundance.
For the majority of the sampling dates, the relative abundance of sul genes was higher in the SDZ manure treatment than in the control manure treatment (Fig. 1). The statistical evaluation (three-way ANOVA) revealed that the relative abundances of sul1 and sul2 for grass and maize plots were significantly influenced by the factors time, treatment, and soil compartment (Table 1). No significant interaction between these factors was found except for sul1 in maize, where the treatment had a significant effect in the rhizosphere (P < 0.0001) but not in the bulk soil (P = 0.194).
Table 1.
Three-way ANOVA analyzing the influence of time, SDZ treatment, and compartment (bulk soil and rhizosphere) and respective correlations
| Parameter | Resulta |
|||
|---|---|---|---|---|
| Maize |
Grass |
|||
| sul1 | sul2 | sul1 | sul2 | |
| Treatment | 13.57** | 45.40*** | 17.43*** | 68.07*** |
| Time | 8.63*** | 34.97*** | 33.77*** | 61.74*** |
| Compartment | 323.21*** | 19.47*** | 1,561.09*** | 260.89*** |
| Treatment × compartment | 9.02** | 0.94 | 0.12 | 2.48 |
| Time × treatment | 0.63 | 1.70 | 1.86 | 1.18 |
F values are shown, and significance is indicated by asterisks, with P <0.005 and P < 0.0001 indicated by ** and ***, respectively.
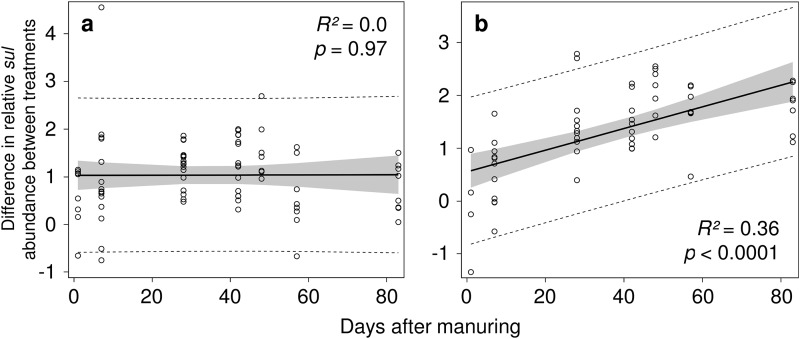
To analyze the time period over which the SDZ manure further increased the level of sul genes compared to that in the control, the differences in relative sul1 and sul2 abundance between control and SDZ treatment were plotted against the days after manuring, and the significance of the increase was tested by a linear regression model (Fig. 2). In bulk soil, the difference in abundance of sul genes between SDZ treatment and control was constant over time, suggesting that the resistance level increased in the first few days after manuring and then remained at that elevated level compared to the control (Fig. 2a). In contrast, in the rhizospheres of maize and grass the difference in abundance of sul genes between SDZ treatment and control significantly and progressively increased for 6 weeks after manuring (Fig. 2b). No further deviation of sul gene abundance between treatment and control was observed from days 42 to 83 after manuring (Fig. 2b). On average, the relative sul abundance in the SDZ treatment compared to that in the control was increased by around 1 order of magnitude in the bulk soil samples, while in the rhizosphere samples the difference between results for SDZ-treated and control samples increased from about one-half to 2 orders of magnitude.
Fig 2.
Effect of SDZ on relative sul1 and sul2 abundance in bulk soil (a) and rhizosphere (b) expressed by plotting the days after the previous application of manure against the differences in log units between results for control and SDZ treatment. Solid lines correspond to a linear regression model, shaded areas to their associated 95% confidence interval, and dotted lines to their 95% prediction limits.
Transferability of sulfonamide resistance and LowGC-type plasmids.
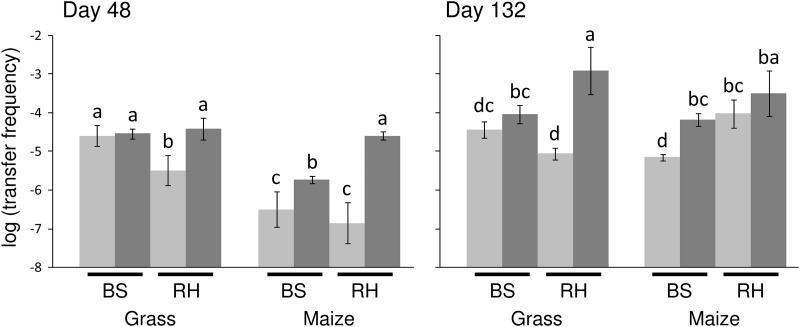
The effect of manure from SDZ-treated pigs on the transfer frequency of SDZ resistance in bulk and rhizosphere soil was analyzed by exogenous plasmid isolation on days 48 and 132. Bulk soil and rhizosphere were differently affected by the SDZ treatment in grass and maize plots (Fig. 3). On day 48 the transfer frequency in the control treatment was about 1 to 2 orders of magnitude higher in the grass (−4.5 to −5.5 log units) than in the maize plots (−6.5 to −7 log units), while on day 132 both were in the same range of −5 to −4 log units. Because the methods of extraction of the microbial fraction from bulk soil and rhizosphere were different on day 48, the transfer frequencies between these compartments could be quantitatively compared only for day 132. On this day, the transfer frequency in the control treatment of the rhizosphere of maize was increased compared to that of bulk soil while no difference was observed in the grass plots (Tukey test; P < 0.05).
Fig 3.
Effect of manure containing sulfadiazine (SDZ) on frequencies of sulfonamide resistance transfer from bulk soil (BS) and rhizosphere (RH) bacteria to E. coli K-12 CV601gfp. Transfer frequencies per recipient cell were determined by filter matings on days 48 and 132 for field plots sown with grass and maize. Differing letters indicate significant differences of means in pairwise comparisons (Tukey test; P < 0.05) for each day. Treatments of soil with control manure and manure containing SDZ are indicated by light and dark gray, respectively. Error bars indicate standard deviations of results for 4 replicates (except maize rhizosphere treatment without SDZ, day 48, where n = 3).
The SDZ treatment generally increased the transfer frequency in the bulk soil of maize, while no increase was observed in the bulk soil of grass. A clear increase could also be observed in the SDZ-treated rhizosphere of both plants compared to that in the control except for the maize rhizosphere on day 132. The highest increase in transfer frequency by about 2 orders of magnitude was observed for the maize rhizosphere on day 48 and for the grass rhizosphere on day 132.
Plasmid DNA was extracted from a total of 292 transconjugants and amplified using rep gene-specific primers for the LowGC-type plasmid group. A 912-bp PCR product characteristic of LowGC plasmids was obtained from 97% (187 of 192 plasmids) and 98% (98 of 100 plasmids) of plasmids captured on days 48 and 132, respectively.
DISCUSSION
Input of sul genes with manure and their selection in soil.
After the first application of manure, the relative abundance of the sul genes in bulk soil and rhizosphere increased by up to 4 orders of magnitude, reaching values observed in the manures. The resistance gene level was only slightly, but significantly, higher in the SDZ manure-treated soil than in the control manure-treated soil, which was comparable to the difference between the manures from SDZ-treated and untreated pigs. These observations are in agreement with the results of previous micro- and mesocosm studies (13, 23, 25) and indicate that besides a selection of resistant bacteria by SDZ, the initial increase in sul abundance relative to rrn was caused by the input of sul genes and resistant bacterial populations with manure. Intriguingly, neither an accumulation of sul genes nor an accumulation of easily extractable SDZ (6) was observed in the field soil after repeated application of manure containing SDZ, as observed in a previous microcosm experiment. In this microcosm experiment it was shown that the repeated application of manure, which is common agricultural practice (36), might lead to a gradual accumulation of resistance genes (24). In the field study presented here, the relative abundance of the sul genes decreased over time, concomitantly with decreasing SDZ concentrations, and fell below initial levels in the bulk soils during the 6-month experimental period (Fig. 1). In contrast to the microcosm study in which SDZ was spiked to manure in high concentrations, manures from SDZ-treated or untreated pigs were used in order to obtain a realistic composition regarding concentrations of SDZ, its metabolites, resistance gene abundance, and microbial community. This in turn had the disadvantage that the microbial community structure and the abundance of sul genes but also the range of physicochemical parameters likely differed between the control and SDZ manures. Bacterial populations from manure carrying sul1 and sul2 genes might not be well adapted to the soil environment and might decrease in abundance relative to soil bacteria (3, 20). Further, the application of reduced amounts of manure, which were applied on days 49 and 133 to approach German regulations for manure application (the maximum allowable amount of N applied per year was exceeded by approximately 80 kg/ha), may have contributed to the decrease in sul abundance over time through the application of reduced amounts of nutrients and sul genes to the soil. Selection by bioavailable SDZ from SDZ manure counteracted the decrease of sul-carrying populations, but the easily extractable concentration of SDZ, which accounts for the desorbable and hence potentially bioaccessible fraction of SDZ (37), decreased rapidly after each application of SDZ manure, with a dissipation half-life ranging from 2 to 12 days (6). Compared to results in a laboratory experiment, the dissipation was faster under field conditions for the first and second applications of manure but comparatively slow after the third application (6). The faster dissipation of easily extractable SDZ under field conditions compared to that in the laboratory experiment (24) might have resulted in a lower selective pressure contributing to a decrease in the relative sul abundance over time. Indications were found that temperature and moisture were important drivers of dissipation, with changes in soil moisture affecting the sorption and favoring sequestration and formation of nonextractable residues (6).
Effect of SDZ manure in the rhizosphere compared to bulk soil.
In the rhizosphere, significantly lower levels of relative sul abundance were reached over the experimental period than in bulk soil (Fig. 1 and Table 1), which can be partly explained by a higher abundance of 16S rRNA genes observed for this compartment but might additionally be associated with a lower selective pressure due to a faster dissipation of easily extractable SDZ (6), as described previously (13). However, the regression analysis showed that the difference between SDZ treatment and control samples increased over time after manuring in the rhizosphere soil, while the difference in bulk soil stayed constant (Fig. 2). This finding may be partially due to a higher abundance of sul genes repeatedly applied to the soil by the manure from SDZ-treated pigs but also may indicate that in the rhizosphere a selective pressure is prevailing that favors bacteria carrying SDZ resistance, even when the SDZ concentration of the easily extractable fraction is far below the published effective dose of 48 mg/kg for the inhibition of microbial activity by SDZ (38). Recently, it was shown that selection of resistant bacteria may occur at concentrations down to 150 μg/kg (3) and even down to several hundred-fold below the MIC (39). This is supported by a study using soils of the same origin as in our study, in which the SDZ concentration decreased over time while the SDZ effect on microbial community structure increased (20). Hence, despite the faster dissipation of easily extractable SDZ in the rhizosphere, the combination of a constant but low release of sequestered SDZ (40, 41), together with a potentially higher microbial activity in the rhizosphere due to root exudation, may have caused a low but continuing selective pressure of the bacteriostatic SDZ. In bulk soil, microbial activity may have been lower due to the lack of additional carbon provided by root exudates (42), resulting in lower effects of SDZ on the microbial community because the mode of action of SDZ is directed only at growing cells (21, 22). The constant difference between results for the SDZ-treated and control cells observed for bulk soil (Fig. 2), despite smaller amounts of manure applied on days 49 and 133, may have been an effect of the antibiotic resistance genes which were introduced to the soil via manure and of short-term selection of resistant bacteria by providing nutrients and SDZ to the soil, leading to a slight initial increase in relative sul abundance, which could best be observed between day 1 and day 7 of the experiment (Fig. 1).
Rhizosphere effect on transferability of SDZ resistance.
The sul gene transferability was assessed on days 48 and 132 from the frequencies of plasmid capture from manure-treated soil into E. coli. Day 48 was selected for the exogenous plasmid isolation as the longest incubation period before the second application of manure on day 49 to allow a sufficient development of the maize rhizosphere. On day 132, 83 days after the second application of manure, the maize plants were fully developed and the effect of the rhizosphere on the transfer frequency at the end of the growth season was assessed. SDZ-resistant transconjugants were obtained from all bulk soil and rhizosphere samples. This was expected since other studies showed that manuring itself, regardless of the presence of antibiotics in manure, enhanced the horizontal gene transfer in soil or even provided mobilizing genetic elements to the soil (13, 19, 23, 43). From almost all of the plasmids, a PCR product was obtained using primers specific for the LowGC-type plasmid group, indicating its high abundance and potential involvement in SDZ resistance. Although only plasmids able to transfer to and replicate in E. coli were captured, these results support previous findings suggesting an important role of plasmids belonging to the LowGC-type group in spreading antibiotic resistance in manure and manure-fertilized soils (13, 25).
In this study, an increase in transferability of sul genes following the application of manure containing SDZ might be expected due to the potential increase in selective pressure and availability of nutrients. Additionally, microbial communities differing in the proportion of plasmid-carrying populations might have been applied to the soil by the different manures, which could have contributed to an increased transferability in the SDZ manure-treated soil. Consequently, in the rhizosphere of grass and maize a clear increase in TF compared to that in the control was observed on day 48 but intriguingly on day 132 for grass only. This indicated that the developmental stage of the plant might have influenced the stimulation of microbial activity and transferability of resistance genes, as the rhizosphere of maize developed during the course of the experiment while the grass was already seeded about 2 months before the start of the experiment. Further, plant species-specific factors, such as soil moisture affected by water uptake as well as root growth and exudate production, may have differently affected the microbial community and activity as well as the fate of the antibiotic in the respective soil compartment. This observation is in line with previous studies demonstrating a plant species-specific rhizosphere effect on the conjugal transfer process for pea, barley, and wheat (44) whereas a plant species-dependent shift of the relative abundance of bacterial populations in the rhizosphere was revealed by denaturing gradient gel electrophoresis (45). These differences in plant species (maize, grass) and the developmental stage could explain the higher TF of the grass compared to the maize plots on day 48. The active incorporation of manure and SDZ to a depth of 12 cm on the maize plots at the first manure application may have additionally increased the nutrition and selection pressure in the bulk soil, leading to an increase in TF in the SDZ treatment compared to that in the control for maize. A decreased TF in the bulk soil of maize plots compared to the grass plots on day 48 might be due to the densely grown grass roots extending the rhizosphere effect to the bulk soil as well.
The increase in transfer and abundance of resistance genes in bulk soil and rhizosphere by the field application of manure from SDZ-treated pigs, as shown for the first time in this study, may contribute to the serious problem of antimicrobial resistance, since sul1 genes are frequently associated with class 1 integrons which can acquire, exchange, and express genes embedded within gene cassettes and are broadly distributed among Gram-negative bacteria of clinical interest (46, 47) and sul2 is often found to be clustered with strA-strB genes (48–50). In summary, the effects of SDZ manure on antibiotic resistance differed considerably between bulk soil and rhizosphere, a finding which needs to be considered when assessing risks to human health associated with the spreading of antibiotic resistance genes in the environment.
ACKNOWLEDGMENTS
Christoph Kopmann, Sven Jechalke, Ingrid Rosendahl, and Holger Heuer were funded by the Deutsche Forschungsgemeinschaft (DFG FOR566, SM59/5-3, and AM134/6-3).
We thank Ilse-Marie Jungkurth for proofreading the manuscript.
Footnotes
Published ahead of print 11 January 2013
REFERENCES
- 1. Burkhardt M, Stamm C, Waul C, Singer H, Müller S. 2005. Surface runoff and transport of sulfonamide antibiotics and tracers on manured grassland. J. Environ. Qual. 34:1363–1371 [DOI] [PubMed] [Google Scholar]
- 2. Sarmah AK, Meyer MT, Boxall ABA. 2006. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 65:725–759 [DOI] [PubMed] [Google Scholar]
- 3. Heuer H, Focks A, Lamshöft M, Smalla K, Matthies M, Spiteller M. 2008. Fate of sulfadiazine administered to pigs and its quantitative effect on the dynamics of bacterial resistance genes in manure and manured soil. Soil Biol. Biochem. 40:1892–1900 [Google Scholar]
- 4. Lamshöft M, Sukul P, Zühlke S, Spiteller M. 2007. Metabolism of 14C-labelled and non-labelled sulfadiazine after administration to pigs. Anal. Bioanal. Chem. 388:1733–1745 [DOI] [PubMed] [Google Scholar]
- 5. Lamshöft M, Sukul P, Zühlke S, Spiteller M. 2010. Behaviour of 14C-sulfadiazine and 14C-difloxacin during manure storage. Sci. Total Environ. 408:1563–1568 [DOI] [PubMed] [Google Scholar]
- 6. Rosendahl I, Siemens J, Groeneweg J, Linzbach E, Laabs V, Herrmann C, Vereecken H, Amelung W. 2011. Dissipation and sequestration of the veterinary antibiotic sulfadiazine and its metabolites under field conditions. Environ. Sci. Technol. 45:5216–5222 [DOI] [PubMed] [Google Scholar]
- 7. Förster M, Laabs V, Lamshöft M, Groeneweg J, Zühlke S, Spiteller M, Krauss M, Kaupenjohann M, Amelung W. 2009. Sequestration of manure-applied sulfadiazine residues in soils. Environ. Sci. Technol. 43:1824–1830 [DOI] [PubMed] [Google Scholar]
- 8. Heuer H, Schmitt H, Smalla K. 2011. Antibiotic resistance gene spread due to manure application on agricultural fields. Curr. Opin. Microbiol. 14:236–243 [DOI] [PubMed] [Google Scholar]
- 9. Sköld O. 2000. Sulfonamide resistance: mechanisms and trends. Drug Resist. Updat. 3:155–160 [DOI] [PubMed] [Google Scholar]
- 10. Swedberg G, Sköld O. 1980. Characterization of different plasmid-borne dihydropteroate synthases mediating bacterial resistance to sulfonamides. J. Bacteriol. 142:1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sköld O. 2001. Resistance to trimethoprim and sulfonamides. Vet. Res. 32:261–273 [DOI] [PubMed] [Google Scholar]
- 12. Perreten V, Boerlin P. 2003. A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 47:1169–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kopmann C, Jechalke S, Rosendahl I, Groeneweg J, Krögerrecklenfort E, Zimmerling U, Weichelt V, Siemens J, Amelung W, Heuer H, Smalla K. 2013. Abundance and transferability of antibiotic resistance as related to the fate of sulfadiazine in maize rhizosphere and bulk soil. FEMS Microbiol. Ecol. 83:125–134 [DOI] [PubMed] [Google Scholar]
- 14. Rahube TO, Yost CK. 2012. Characterization of a mobile and multiple resistance plasmid isolated from swine manure and its detection in soil after manure application. J. Appl. Microbiol. 112:1123–1133 [DOI] [PubMed] [Google Scholar]
- 15. Binh CTT, Heuer H, Kaupenjohann M, Smalla K. 2008. Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol. Ecol. 66:25–37 [DOI] [PubMed] [Google Scholar]
- 16. Binh CTT, Heuer H, Kaupenjohann M, Smalla K. 2009. Diverse aadA gene cassettes on class 1 integrons introduced into soil via spread manure. Res. Microbiol. 160:427–433 [DOI] [PubMed] [Google Scholar]
- 17. Heuer H, Krögerrecklenfort E, Wellington EMH, Egan S, van Elsas JD, van Overbeek L, Collard JM, Guillaume G, Karagouni AD, Nikolakopoulou TL, Smalla K. 2002. Gentamicin resistance genes in environmental bacteria: prevalence and transfer. FEMS Microbiol. Ecol. 42:289–302 [DOI] [PubMed] [Google Scholar]
- 18. van Overbeek LS, Wellington EMH, Egan S, Smalla K, Heuer H, Collard JM, Guillaume G, Karagouni AD, Nikolakopoulou TL, van Elsas JD. 2002. Prevalence of streptomycin-resistance genes in bacterial populations in European habitats. FEMS Microbiol. Ecol. 42:277–288 [DOI] [PubMed] [Google Scholar]
- 19. Götz A, Smalla K. 1997. Manure enhances plasmid mobilization and survival of Pseudomonas putida introduced into field soil. Appl. Environ. Microbiol. 63:1980–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hammesfahr U, Heuer H, Manzke B, Smalla K, Thiele-Bruhn S. 2008. Impact of the antibiotic sulfadiazine and pig manure on the microbial community structure in agricultural soils. Soil Biol. Biochem. 40:1583–1591 [Google Scholar]
- 21. Achari A, Somers DO, Champness JN, Bryant PK, Rosemond J, Stammers DK. 1997. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat. Struct. Biol. 4:490–497 [DOI] [PubMed] [Google Scholar]
- 22. Bock L, Miller GH, Schaper KJ, Seydel JK. 1974. Sulfonamide structure-activity-relationships in a cell-free system. 2. Proof for formation of a sulfonamide-containing folate analog. J. Med. Chem. 17:23–28 [DOI] [PubMed] [Google Scholar]
- 23. Heuer H, Smalla K. 2007. Manure and sulfadiazine synergistically increased bacterial antibiotic resistance in soil over at least two months. Environ. Microbiol. 9:657–666 [DOI] [PubMed] [Google Scholar]
- 24. Heuer H, Solehati Q, Zimmerling U, Kleineidam K, Schloter M, Müller T, Focks A, Thiele-Bruhn S, Smalla K. 2011. Accumulation of sulfonamide resistance genes in arable soils due to repeated application of manure containing sulfadiazine. Appl. Environ. Microbiol. 77:2527–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heuer H, Kopmann C, Binh CTT, Top EM, Smalla K. 2009. Spreading antibiotic resistance through spread manure: characteristics of a novel plasmid type with low %G+C content. Environ. Microbiol. 11:937–949 [DOI] [PubMed] [Google Scholar]
- 26. van Elsas JD, Turner S, Bailey MJ. 2003. Horizontal gene transfer in the phytosphere. New Phytol. 157:525–537 [DOI] [PubMed] [Google Scholar]
- 27. Heuer H, Smalla K. 2012. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 36:1083–1104 doi:10.1111/j.1574-6976.2012.00337.x [DOI] [PubMed] [Google Scholar]
- 28. Mølbak L, Molin S, Kroer N. 2007. Root growth and exudate production define the frequency of horizontal plasmid transfer in the rhizosphere. FEMS Microbiol. Ecol. 59:167–176 [DOI] [PubMed] [Google Scholar]
- 29. Kuzyakov Y, Domanski G. 2000. Carbon input by plants into the soil. J. Plant Nutr. Soil Sci. 163:421–431 [Google Scholar]
- 30. Doornbos RF, van Loon LC, Bakker P. 2012. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 32:227–243 [Google Scholar]
- 31. Brandt KK, Sjøholm OR, Krogh KA, Halling-Sørensen B, Nybroe O. 2009. Increased pollution-induced bacterial community tolerance to sulfadiazine in soil hotspots amended with artificial root exudates. Environ. Sci. Technol. 43:2963–2968 [DOI] [PubMed] [Google Scholar]
- 32. Kuchta SL, Cessna AJ. 2009. Fate of lincomycin in snowmelt runoff from manure-amended pasture. Chemosphere 76:439–446 [DOI] [PubMed] [Google Scholar]
- 33. Finck A. 1991. Düngung. Ertragssteigernd, qualitätsverbessernd, umweltgerecht. Ulmer, Stuttgart, Germany [Google Scholar]
- 34. Anonymous 2007. Verordnung über die Anwendung von Düngemitteln, Bodenhilfsstoffen, Kultursubstraten und Pflanzenhilfsmitteln nach den Grundsätzen der guten fachlichen Praxis beim Düngen (Düngeverordnung - DüV). http://www.gesetze-im-internet.de/bundesrecht/d_v/gesamt.pdf
- 35. Götz A, Pukall R, Smit E, Tietze E, Prager R, Tschäpe H, Van Elsas JD, Smalla K. 1996. Detection and characterization of broad-host-range plasmids in environmental bacteria by PCR. Appl. Environ. Microbiol. 62:2621–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Montforts M, Kalf DF, van Vlaardingen PLA, Linders J. 1999. The exposure assessment for veterinary medicinal products. Sci. Total Environ. 225:119–133 [DOI] [PubMed] [Google Scholar]
- 37. Thiele-Bruhn S, Aust MO. 2004. Effects of pig slurry on the sorption of sulfonamide antibiotics in soil. Arch. Environ. Contam. Toxicol. 47:31–39 [DOI] [PubMed] [Google Scholar]
- 38. Thiele-Bruhn S. 2005. Microbial inhibition by pharmaceutical antibiotics in different soils—dose-response relations determined with the iron(III) reduction test. Environ. Toxicol. Chem. 24:869–876 [DOI] [PubMed] [Google Scholar]
- 39. Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, Andersson DI. 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7:e1002158 doi:10.1371/journal.ppat.1002158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wehrhan A, Streck T, Groeneweg J, Vereecken H, Kasteel R. 2010. Long-term sorption and desorption of sulfadiazine in soil: experiments and modeling. J. Environ. Qual. 39:654–666 [DOI] [PubMed] [Google Scholar]
- 41. Zarfl C, Klasmeier J, Matthies M. 2009. A conceptual model describing the fate of sulfadiazine and its metabolites observed in manure-amended soils. Chemosphere 77:720–726 [DOI] [PubMed] [Google Scholar]
- 42. Butler JL, Bottomley PJ, Griffith SM, Myrold DD. 2004. Distribution and turnover of recently fixed photosynthate in ryegrass rhizospheres. Soil Biol. Biochem. 36:371–382 [Google Scholar]
- 43. Smalla K, Heuer H, Götz A, Niemeyer D, Krögerrecklenfort E, Tietze E. 2000. Exogenous isolation of antibiotic resistance plasmids from piggery manure slurries reveals a high prevalence and diversity of IncQ-like plasmids. Appl. Environ. Microbiol. 66:4854–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schwaner NE, Kroer N. 2001. Effect of plant species on the kinetics of conjugal transfer in the rhizosphere and relation to bacterial metabolic activity. Microb. Ecol. 42:458–465 [DOI] [PubMed] [Google Scholar]
- 45. Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, Roskot N, Heuer H, Berg G. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stalder T, Barraud O, Casellas M, Dagot C, Ploy M-C. 2012. Integron involvement in environmental spread of antibiotic resistance. Front. Microbiol. 3:119 doi:10.3389/fmicb.2012.00119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G. 2012. The shared antibiotic resistome of soil bacteria and human pathogens. Science 337:1107–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vinue L, Saenz Y, Rojo-Bezares B, Olarte I, Undabeitia E, Somalo S, Zarazaga M, Torres C. 2010. Genetic environment of sul genes and characterisation of integrons in Escherichia coli isolates of blood origin in a Spanish hospital. Int. J. Antimicrob. Agents 35:492–496 [DOI] [PubMed] [Google Scholar]
- 49. Zhao J, Dang H. 2011. Identification of a globally distributed clinical streptomycin-resistance plasmid and other resistance determinants in a coastal bay of China. Lett. Appl. Microbiol. 52:1–8 [DOI] [PubMed] [Google Scholar]
- 50. Yau S, Liu XL, Djordjevic SP, Hall RM. 2010. RSF1010-like plasmids in Australian Salmonella enterica serovar Typhimurium and origin of their sul2-strA-strB antibiotic resistance gene cluster. Microb. Drug Resist. 16:249–252 [DOI] [PubMed] [Google Scholar]
- 51. Kuratorium für Technik und Bauwesen in der Landwirtschafts e.V. (KTBL) 2009. Faustzahlen für die Landwirtschaft. KTBL, Darmstadt, Germany [Google Scholar]