Abstract
A heterotrimeric flavoprotein-cytochrome c complex fructose dehydrogenase (FDH) of Gluconobacter japonicus NBRC3260 catalyzes the oxidation of d-fructose to produce 5-keto-d-fructose and is used for diagnosis and basic research purposes as a direct electron transfer-type bioelectrocatalysis. The fdhSCL genes encoding the FDH complex of G. japonicus NBRC3260 were isolated by a PCR-based gene amplification method with degenerate primers designed from the amino-terminal amino acid sequence of the large subunit and sequenced. Three open reading frames for fdhSCL encoding the small, cytochrome c, and large subunits, respectively, were found and were presumably in a polycistronic transcriptional unit. Heterologous overexpression of fdhSCL was conducted using a broad-host-range plasmid vector, pBBR1MCS-4, carrying a DNA fragment containing the putative promoter region of the membrane-bound alcohol dehydrogenase gene of Gluconobacter oxydans and a G. oxydans strain as the expression host. We also constructed derivatives modified in the translational initiation codon to ATG from TTG, designated TTGFDH and ATGFDH. Membranes of the cells producing recombinant TTGFDH and ATGFDH showed approximately 20 times and 100 times higher specific activity than those of G. japonicus NBRC3260, respectively. The cells producing only FdhS and FdhL had no fructose-oxidizing activity, but showed significantly high d-fructose:ferricyanide oxidoreductase activity in the soluble fraction of cell extracts, whereas the cells producing the FDH complex showed activity in the membrane fraction. It is reasonable to conclude that the cytochrome c subunit is responsible not only for membrane anchoring but also for ubiquinone reduction.
INTRODUCTION
Fructose dehydrogenase (FDH; EC 1.1.99.11) of Gluconobacter japonicus NBRC3260 (formerly Gluconobacter industrius IFO3260), which catalyzes the oxidation of d-fructose to produce 5-keto-d-fructose, is a heterotrimeric membrane-bound enzyme with a molecular mass of ca. 140 kDa, consisting of subunits I (67 kDa), II (51 kDa), and III (20 kDa). The enzyme, purified for the first time in 1981, is a flavoprotein-cytochrome c complex, since subunits I and II have covalently bound flavin adenine dinucleotide (FAD) and heme C as prosthetic groups, respectively (1).
FDH shows strict substrate specificity to d-fructose and thus is used in diagnosis and food analysis and is commercially available (2). This enzyme is also used in a number of basic research projects to examine the electrochemical properties of enzyme-catalyzed electrode reactions, which is called bioelectrocatalysis (3). The reaction is classified into two types. One is the direct electron transfer (DET)-type system, in which electrons are transferred directly between the enzyme and electrode. The other is the mediated electron transfer (MET)-type system, in which mediators transfer electrons between the enzyme and electrode. As far as we know, FDH has the highest ability of DET-type bioelectrocatalysis on the anode (4). The DET-type system is convenient for the construction of compact bioelectrochemical devices and is utilized to develop biosensors, biofuel cells, and bioreactors. However, DET-type bioelectrocatalysis occurs only at some limited kinds of electrodes suitable for individual redox enzymes, such as FDH (3), alcohol dehydrogenase (5), cellobiose dehydrogenase (6), bilirubin oxidase (7), and Cu efflux oxidase (8). Although DET-type bioelectrocatalysis is attractive for applications, mechanisms for the reaction have not been fully described yet. For the first step to explore the mechanisms of the DET-type bioelectrocatalytic reaction of FDH, we sequenced the genes encoding each subunit of the FDH complex from G. japonicus NBRC3260 and constructed an expression system to highly produce FDH in a Gluconobacter oxydans strain.
MATERIALS AND METHODS
Materials.
Fructose dehydrogenase of Gluconobacter japonicus NBRC3260 was both a gift from and purchased from Toyobo (Osaka, Japan). Restriction endonucleases and modification enzymes for genetic engineering were kind gifts from Toyobo (Osaka, Japan) and were also purchased from TaKaRa Shuzo (Kyoto, Japan) and Agilent Technologies (Santa Clara, CA). Yeast extract was a generous gift from Oriental Yeast (Osaka, Japan). All other materials were purchased from commercial sources and were of a guaranteed grade.
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Gluconobacter japonicus NBRC3260 and Gluconobacter oxydans ATCC 621H and NBRC12528 and its ΔadhA::Kmr derivative (9) were used in this study. The broad-host-range vector pBBR1MCS-4 was used for the heterologous expression of the fdhSCL genes in G. oxydans. Gluconobacter spp. were grown on ΔP medium, consisting of 5 g of glucose, 20 g of glycerol, 10 g of polypeptone, and 10 g of yeast extract per liter, at 30°C with vigorous shaking, unless otherwise stated. Kanamycin and ampicillin were used at final concentrations of 50 μg ml−1 and 250 μg ml−1, respectively.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or referencea |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F− endA1 hsdR17(rk− mk+) supE44 thi-1 λ− recAl gyrA96 relA1 deoR Δ(lacZYA-argF)U169 ϕ80dlacZΔM15 | 10 |
| HB101 | F− thi-1 hsdS20(rB mB) supE44 recA13 ara14 leuB6 proA2 lacY1 galK2 rpsL20 (Strr xyl-5 mtl-1 λ−) | 34 |
| Gluconobacter japonicus NBRC3260 | Wild type | NBRC |
| Gluconobacter oxydans | ||
| NBRC12528 | Wild type | NBRC |
| ΔadhA mutant | NBRC12528 ΔadhA::Kmr | 9 |
| ATCC 621H | Wild type | ATCC |
| Plasmids | ||
| pKR2013 | Plasmid mediates plasmid transfer; Kmr | 11 |
| pBBR1MCS-4 | Broad-host-range plasmid; mob Apr | 35 |
| pSHO8 | pBBR1MCS-4, a 0.7-kb fragment of a putative promoter region of the adhAB gene of G. oxydans 621H | This study |
| pSHO12 | pSHO8, a 3.8-kb fragment of the fdhSCL genes of G. japonicus NBRC3260 | This study |
| pSHO13 | pSHO8, a 3.7-kb fragment of the fdhATGSCL genes | This study |
| pSHO16 | pSHO8, a 2.4-kb fragment of the fdhATGSL genes (in-frame deletion of fdhC) | This study |
The URL addresses of NBRC and ATCC are http://www.nbrc.nite.go.jp/ and http://www.atcc.org/, respectively.
Escherichia coli DH5α was used for plasmid construction (10). E. coli HB101 harboring pRK2013 was used as a helper strain for conjugative plasmid transfer, using a triparental mating method (11). E. coli strains were grown on modified Luria-Bertani medium, consisting of 10 g of polypeptone, 5 g of yeast extract, and 5 g of NaCl, filled to 1 liter with distilled water and with the pH adjusted to 7.0 with NaOH. Ampicillin was used at a final concentration of 50 μg ml−1.
Determination of the N-terminal amino acid sequence of purified FDH.
Commercially available FDH was subjected to SDS-PAGE (10% acrylamide). The proteins in the gel were transferred electrophoretically onto a polyvinylidene difluoride membrane at 2 mA cm−2 for 40 min. Proteins were stained with Coomassie brilliant blue (CBB) stain one (Nacalai Tesque, Japan) and destained with 5% (vol/vol) methanol, followed by the excision and drying of bands. The N-terminal amino acid sequence was analyzed with the peptide sequencer Procise 491 (Life Technologies, Carlsbad, CA).
Sequencing of the fdhSCL genes.
Degenerate primers, forward primer A and reverse primer B, were designed for PCR-based gene amplification (see Table S1 in the supplemental material). The genomic DNA of G. japonicus was isolated from cells grown to the mid-exponential phase of growth by the method of Marmur (12), with some modifications: i.e., we used cetyltrimethylammonium bromide at a final concentration of 1% (wt/vol) to remove polysaccharides but omitted the perchlorate step in the original procedure. PCR was performed with the genomic DNA of G. japonicus as the parental DNA molecule using KOD Dash polymerase (Toyobo, Japan) and the MyCycler thermal cycler (Bio-Rad, CA). The amplified DNA fragment was sequenced using the same primers. The thermal asymmetric interlaced PCR (TAIL-PCR) method was repeatedly conducted to extend sequencing to the 5′ and 3′ directions using one of the three arbitrary degenerate primers AD1, AD2, or AD3 and KOD Dash polymerase, according to Liu et al. (13). The product of TAIL-PCR was sequenced to be homologous to the 3′ region of the gene encoding the cytochrome c subunit of sorbitol dehydrogenase. Thus, degenerate primers were designed from the conserved amino acid sequence in the heme C binding motifs in the cytochrome c subunits of other dehydrogenases to extend sequencing. We repeated the TAIL-PCR method to further obtain the complete structural genes for the FDH complex.
Construction of plasmids.
For plasmid construction, we used Herculase II fusion DNA polymerase (Agilent Technologies, Santa Clara, CA) to amplify the designed DNA fragments. A putative promoter region of the adhAB genes, which encode two major subunits of the pyrroloquinoline quinone-dependent alcohol dehydrogenase, was amplified with Herculase II fusion DNA polymerase using a genomic DNA preparation of G. oxydans 621H and two primers, 621H-adh-pro(+) and 621H-adh-pro(−) (see Table S1 in the supplemental material). The PCR product was inserted into pBBR1MCS-4 (9) treated with KpnI and XhoI to yield pSHO8. The fdhSCL genes were amplified with DNA polymerase using the genome DNA of G. japonicus NBRC3260 and two primer sets, fdhS-5-Eco(+) and fdhL-3-PstBam(−) and fdhS-370-ATG-Xho(+) and fdhL-3-PstBam(−) (see Table S1), respectively. The PCR products were inserted into pSHO8 treated with EcoRI and BamHI and with XhoI and BamHI to yield pSHO12 and pSHO13, respectively. To construct a plasmid to express only the fdhSL genes, an in-frame deletion in the fdhC gene was introduced in pSHO13 by fusion PCR as follows. The 5′ and 3′ fragments for a deletion derivative of pSHO13, in which most of subunit II (from His11 to Trp451, the amino acid number of the putative mature subunit II) is lost in frame, were amplified with the DNA polymerase with the primer sets fdhS-370-ATG-Xho(+) and delta-fdhC(−) and delta-fdhC(+) and fdhL-3-PstBam(−), respectively (see Table S1). The two PCR products were purified, and fusion PCR was conducted with the primers fdhS-370-ATG-Xho(+) and fdhL-3-PstBam(−). The amplified 2.4-kb DNA fragment was inserted into pSHO8 treated with XhoI and BamHI to yield pSHO16. All nucleotide sequences for PCR cloning were confirmed by cycle sequencing techniques using a 310 DNA sequencer (Applied Biosystems, CA).
Expression of recombinant FDH and preparation of the membrane fraction.
G. oxydans NBRC12528 ΔadhA::Kmr was transformed with the plasmids via a triparental mating method using the HB101 strain harboring pRK2013 (11). Acetic acid was added to the medium for selection at a final concentration of 0.1% (wt/vol) to eliminate E. coli growth. Acetic acid- and ampicillin-resistant conjugant colonies were screened twice on ΔP agar medium containing 0.1% (wt/vol) acetic acid and 250 μg ml−1 ampicillin. Finally, the transconjugants were screened in liquid ΔP medium containing 250 μg ml−1 ampicillin.
Gluconobacter cells were cultivated in ΔP medium with or without 250 μg ml−1 ampicillin to the late exponential growth phase. Cells were collected by centrifugation at 10,000 × g for 10 min and washed twice with 20-fold-diluted McIlvaine buffer (McB [pH 6.0]: a mixture of 0.1 M citric acid and 0.2 M disodium hydrogen phosphate). Preparation of the membrane fraction was carried out as described by Ameyama et al. (1), with some modifications, as follows. Cells were suspended in 20-fold-diluted McB (pH 6.0) and were disrupted by two passages through a French pressure cell press (Thermo Fisher Scientific, Waltham, MA). After cell debris was sedimented by low-speed centrifugation (10,000 × g, 10 min, 4°C), the supernatant was ultracentrifuged (100,000 × g, 1 h, 4°C). The supernatant was used as the soluble fraction, and precipitates were resuspended in 20-fold-diluted McB (pH 6.0) and used as the membrane fraction.
Purification of recombinant FDH.
The solubilization and purification of FDH were performed as described previously (1), with some modifications, as follows. Membranes were suspended in 20-fold-diluted McB (pH 6.0) at a concentration of 10 mg membrane protein ml−1 containing 1 mM 2-mercaptoethanol and 1.0% (wt/vol) Triton X-100 and gently stirred for 10 h at 4°C. FDH was obtained in the supernatant fraction of ultracentrifugation at 100,000 × g for 1.5 h. The supernatant fraction was applied to a DEAE-Sepharose column equilibrated with 20-fold-diluted McB (pH 6.0) containing 1 mM 2-mercaptoethanol and 0.1% (wt/vol) Triton X-100. The elution of FDH from a DEAE-Sepharose column was carried out by a concentration gradient of McB: i.e., from 20-fold-diluted McB (pH 6.0) to the original concentrations of McB (pH 6.0) containing 1 mM 2-mercaptoethanol and 0.1% (wt/vol) Triton X-100. The purities of recombinant FDH were judged by Coomassie brilliant blue R-250 staining of SDS-PAGE.
Oxygen consumption rates by intact cells.
Oxygen consumption of intact Gluconobacter cells was measured at 25°C with a Clark-type oxygen electrode (Opto Science, Tokyo, Japan). Cell suspensions were prepared at concentrations of an optical density at 600 nm (OD600) of 1.0 with 50 mM sodium phosphate buffer (pH 6.0). d-Glucose and d-fructose were added at 200 mM as the respiration substrate. Oxygen concentrations were recorded amperometrically as the reduction current of oxygen at −600 mV versus the Ag|AgCl reference electrode.
Other analytical methods.
Global identity between predicted amino acid sequences was calculated by the software GENETYX-MAC (version 14; Genetyx, Tokyo, Japan). Protein concentrations were determined with the DC protein assay kit (Bio-Rad, CA) using bovine serum albumin as a standard. FDH activity was measured spectrophotometrically with potassium ferricyanide and the ferric dupanol reagent as described previously (1). One FDH unit was defined as the amount of enzyme oxidizing 1 μmol of d-fructose per min. Covalently bound heme C on protein separated by SDS-PAGE was stained by heme-catalyzed peroxidase activity (14). Heme C content was determined spectrophotometrically as described previously (15).
Nucleotide sequence accession number.
The nucleotide sequence and predicted amino acid sequence of G. japonicus NBRC3260 FDH have been deposited into the DNA Data Bank of Japan (DDBJ) under accession number AB728565.
RESULTS
Sequencing of the fdhSCL genes.
We determined the N-terminal amino acid sequence of subunit I of the commercially available FDH complex purified from G. japonicus NBRC3260 (Gluconobacter sp. in the instructions provided by Toyobo) to be SNETLSADVVIIGAGICGSLLAH (in an amino-to-carboxyl direction), as shown in Fig. S1 in the supplemental material. Basic Local Alignment Search Tool (BLAST) analysis of the determined amino acid sequence revealed that subunit I of sorbitol dehydrogenase (SLDH) of Gluconobacter frateurii THD32 shows the highest identity to the N-terminal amino acid sequence of subunit I of FDH (16). We thus designed degenerate primers for PCR (see Table S1 in the supplemental material) based on the N-terminal sequence and conserved the amino acid sequence in SLDH subunit I, respectively. To obtain sequence information upstream and downstream of the PCR product, the TAIL-PCR method was conducted as described in Materials and Methods. We also designed degenerate primers from the heme C binding consensus sequence and further repeated the TAIL-PCR method. We determined the nucleotide sequence of the 4,208-base PCR product containing the complete structural genes for the FDH complex.
The nucleotide and predicted amino acid sequences of FDH and the flanking regions are shown in Fig. S1 in the supplemental material. Three open reading frames (ORFs) were found for fdhSCL, encoding the small, cytochrome c, and large subunits, or subunits III, II, and I, respectively. They may be in the same transcriptional unit. A sequence of SRRKLLA, similar to the consensus motif SRRXFLK (where X is any polar amino acid) for the twin-arginine translocation (Tat) system of E. coli that translocates secretory proteins across the cytoplasmic membrane, was found in the N terminus of FdhS (17). Since there was no ATG or GTG codon between the Tat signal and nonsense codon in the upstream region, a TTG codon at nucleotide (nt) 93 can be the start codon for fdhS. A possible Shine-Dalgarno (SD) sequence was found at 6 bp upstream of this start codon. We did not find a rho-independent terminator-like sequence around the termination codon for fdhL, rather there seems to have been another ORF from nt 3794 of which the product is homologous to the hypothetical protein GDI_0857 of Gluconacetobacter diazotrophicus PAI5 and the hypothetical protein GMO_23960 of Gluconobacter morbifer G707. Since we failed to obtain the sequence for upstream and downstream regions of the fdhSCL genes, the whole structure of the fdh operon is uncertain. A 35-amino-acid stretch in the predicted N terminus of subunit III can be recognized as a signal sequence by the SOSUIsignal program (18), whereas no signal sequence was found for the N terminus of subunit I, suggesting that subunit I may be translocated together with subunit III by the Tat system. The fdhS gene encoded 183 amino acids but there were 148 for the mature protein, of which the calculated molecular mass was approximately 16 kDa.
The ORF corresponding to subunit II, fdhC, started at the position nt 663. A possible SD sequence, AGGA, was found 15 nt upstream of the start codon. The 25-amino-acid sequence of the predicted N terminus of FdhC was suggested as a Sec-dependent signal sequence by the SOSUIsignal program (18). The molecular mass of the mature protein could be calculated as approximately 49 kDa, with the sequence composed of 461 amino acids, but it should be higher because the deduced amino acid sequence was revealed to have three CXXCH sequence motifs for heme C binding sites.
The coding region of subunit I was started at position 2145 with the ATG codon. There was a possible SD sequence, AGG, 9 nt upstream of the initiation codon. No signal sequence for translocation was found in the predicted sequence, consistent with the result of the N-terminal amino acid sequencing of purified FDH, which started at the second Ser residue. The fdhL gene encoded a polypeptide of 544 amino acid residues with a calculated molecular mass of approximately 60 kDa being assembled with and covalently bound to FAD. The deduced amino acid sequence was found to have the sequence GAGICG at a position between the 14th and 19th residues, corresponding to the binding motif of FAD (GXGXXG) (19).
Global identity between the predicted amino acid sequences of each subunit of FDH and SLDH from G. frateurii (16) was calculated as follows using the putative mature forms of protein: subunit I, 52% identity; subunit II, 44% identity; subunit III, 24% identity. Even though there are high percentages of identity, the SLDH of Gluconobacter thailandicus NBRC3254 (formally Gluconobacter suboxydans subsp. α IFO3254), closely related to that of G. frateurii (16), has been shown to be inert on sugars but active on d-mannitol at only 5% of the rate of d-sorbitol (20). The global identities of each subunit of FDH with those of GDH from Burkholderia cepacia (21) were 52%, 45%, and 32% for subunit I, subunit II, and subunit III, respectively. B. cepacia GDH shows relatively wide substrate specificity; i.e., this enzyme oxidizes maltose at half the rate of d-glucose (22). On the other hand, since thorough substrate specificity has not been reported so far, it is not clear yet whether B. cepacia GDH oxidizes other monosaccharides.
The putative mature form of the predicted amino acid sequence of fdhC showed considerable identity to those of the cytochrome c subunits of ADH of G. oxydans (36% [23]) and aldehyde dehydrogenase of Gluconacetobacter europaeus (31% [24]).
Construction of Gluconobacter strains for fdhSCL expression.
Since G. oxydans NBRC12528 highly produces c-type cytochromes and flavoproteins (25) but does not have FDH activity (1), we tried heterologous expression of the fdhSCL genes in this strain. Moreover, because ADH is one of major membrane proteins in NBRC12528 and may disturb protein purification, its derivative, which has gene replacement in the adhA gene encoding a large subunit of ADH (ΔadhA::Kmr strain), was used in this study. The broad-host-range plasmid vector pBBR1MCS-4 was stable in G. oxydans NBRC12528 and easy to manipulate; thus, we used this plasmid vector to express the fdhSCL genes. To ensure heterologous expression, a putative promoter region for the adhAB genes of G. oxydans 621H was inserted at the upstream region of the fdhSCL genes.
Judging from the multiple alignment analysis of subunit III of several flavoprotein-cytochrome c complex dehydrogenases (data not shown), the start codon of the FdhS subunit seemed to be TTG and not ATG. In addition to simple cloning of the native fdhSCL genes, in order to confirm the translational start site of subunit III and examine translation efficiency, we constructed modified fdhSCL genes, designated fdhATGSCL, where the TTG codon was replaced with ATG and a termination codon (TAA) was inserted just before the ATG codon. The ΔadhA strain was transformed with the constructed plasmids by conjugation-based gene transfer.
Comparison between wild-type and recombinant FDHs.
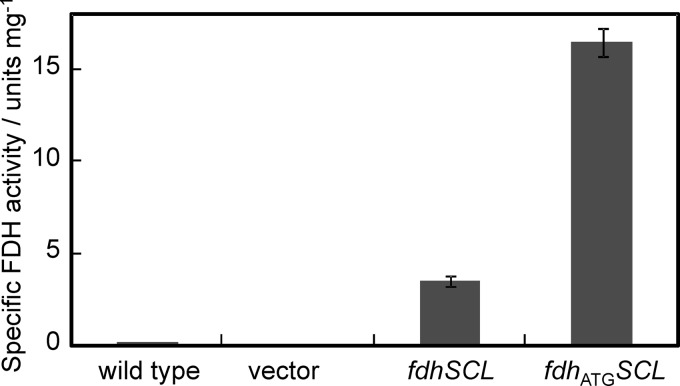
The G. japonicus NBRC3260 strain, which produces wild-type FDH, showed FDH activity of 0.15 U (mg membrane protein)−1 in the membranes. Although we did not examine this in detail, the low specific FDH activity in the membranes of G. japonicus NBRC3260 may be attributed to the difference in the media used in the present and former studies (i.e., ΔP medium was used in this study, while the former study used synthetic medium). Membranes of the ΔadhA cells harboring pSHO12 carrying the wild-type fdhSCL genes showed activity of 3.5 ± 0.3 U mg−1, activity approximately 20 times higher than that of G. japonicus NBRC3260 (Fig. 1). Furthermore, membranes of the ATGFDH strain showed activity of 16 ± 0.8 U mg−1, approximately 5 times higher than that of the TTGFDH strain. We could not detect FDH activity in the membranes of the Δadh strain harboring pSHO8 carrying the putative promoter region only.
Fig 1.
Comparison of specific FDH activity in the membranes of G. japonicus NBRC3260 (wild type) and the ΔadhA strains harboring pSHO8 (vector), pSHO12 (native fdhSCL), or pSHO13 (fdhATGSCL). Data are shown as mean values with 90% confidence intervals (error bars; n = 3).
Heme-catalyzed peroxidase staining of the SDS-PAGE gel revealed that both membranes with TTGFDH and ATGFDH showed approximately 51-kDa bands, while the Δadh strain harboring pSHO8 did not. The apparent intensity of staining of ATGFDH was the highest in the samples examined in this study, and that of TTGFDH was also higher than that of G. japonicus NBRC3260 (data not shown). These results clearly indicate that the initiation codon for subunit III is TTG at nt 93 and also suggest that levels of expression of not only subunit III, but also the whole FDH complex, are increased by changing the initiation codon to ATG.
Characterization of purified ATGFDH.
The specific activity of ATGFDH purified in this study was 260 U (mg protein)−1 at 25°C, which is approximately 1.5 times higher than that reported in the previous study (1). The purified ATGFDH had three main bands of 68, 51, and 18 kDa on SDS-PAGE (see Fig. S2 in the supplemental material), which are similar sizes to those reported previously (1) and correspond to the expected molecular masses from the fdhSCL genes determined in this study. At least two smaller bands could be seen in the CBB-stained SDS-PAGE of our FDH preparation (see Fig. S2). However, we did not find these bands when we used other detergents for preliminary FDH purification, such as n-dodecyl-β-d-maltoside and n-octyl-β-d-glucoside (S. Kawai, T. Yakushi, K. Matsushita, and K. Kano, unpublished data). Thus, we consider them likely to be contaminants.
The purified ATGFDH showed a reduced cytochrome c-like absorption spectrum (data not shown), which is derived from the heme C moieties in subunit II. Based on the FDH complex being a heterotrimeric structure, the number of heme C molecules was determined to be 2.1 per complex, which was calculated from spectrometric heme C contents and protein contents as described in Materials and Methods. We suggest that FDH has three heme C moieties, as predicted from the deduced amino acid sequence of subunit II, although the estimated value is more than 2 but much less than 3, because some protein impurities can be seen in the CBB-stained SDS-PAGE of our FDH preparation (see Fig. S2 in the supplemental material), and minor invisible contaminations are also possible.
The purified ATGFDH transferred electrons to the electrodes directly, as commercially available FDH does (Kawai, Yakushi, Matsushita, and Kano, unpublished).
Characterization of the subunit I/III subcomplex.
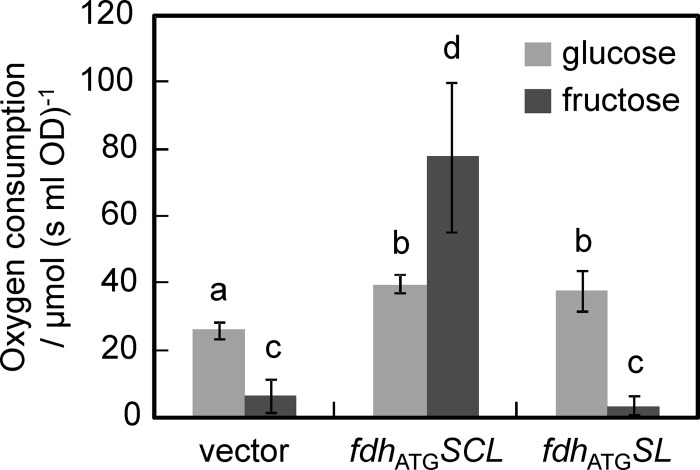
To examine the roles of subunit II in the electron transfer to ubiquinone, the physiological electron acceptor, and ferricyanide, an artificial electron acceptor, and in the subcellular localization of the FDH complex, we constructed a strain to produce only subunits I and III. Oxygen consumption with d-glucose and d-fructose by the ΔadhA cells harboring pSHO8 (vector), pSHO13 (fdhATGSCL), or pSHO16 (fdhATGSL) was measured (Fig. 2). d-Fructose-dependent oxygen consumption by the ΔadhA cells harboring the empty vector was much lower than oxygen consumption with glucose by the same cells (P < 0.01, Student's t test; n = 6), suggesting that the ΔadhA strain and even the parental strain G. oxydans NBRC12528 have the glucose-oxidizing respiratory chain as previously reported (26), but do not have the fructose-oxidizing respiratory chain. They presumably have the ability to metabolize d-fructose to produce NADH being reoxidized by the respiratory chain. d-Glucose-dependent oxygen consumption rates by the ΔadhA cells harboring pSHO13 (fdhATGSCL) and pSHO16 (fdhATGSL) were increased by approximately 1.5-fold that of the cells harboring pSHO8 (vector) by a mechanism that has yet to be elucidated (P < 0.01, Student's t test; n = 6). The cells harboring pSHO13 (fdhATGSCL) showed an ability to consume oxygen, depending on fructose, at approximately a 10-fold higher rate than that of the cells harboring the empty vector (P < 0.01, Student's t test; n = 6), which is much higher than the rate observed with glucose, suggesting that the fructose-oxidizing respiratory chain was heterologously reconstituted in the ΔadhA cells. On the other hand, the difference in rates of d-fructose-dependent oxygen consumption between the cells harboring pSHO16 (fdhATGSL) and those harboring the empty vector may be considered negligible (P > 0.1, Student's t test; n = 6).
Fig 2.
d-Fructose-dependent oxygen consumption (dark gray bars) of the whole-cell preparations of the ΔadhA strains harboring pSHO8 (vector), pSHO13 (fdhATGSCL), or pSHO16 (fdhATGSL). Control experiments were also conducted with d-glucose (light gray bars). The rates of oxygen consumption were normalized by optical density of the cell preparations. Data are shown as mean values with 90% confidence intervals (error bars; n = 3). Significance can be seen between columns labeled with a and b, a and c, and c and d (P < 0.01, Student's t test; n = 6). Columns with the same letters were not significantly different (P > 0.1, Student's t test; n = 6).
In order to know whether the functional subunit I/III subcomplex is expressed, we examined the in vitro fructose dehydrogenase activity of the cell extract of the ΔadhA cells harboring pSHO16 (fdhATGSL). The activity of the cells that express whole FDH complex could be found mostly in the membrane fraction at a specific activity of 20 ± 5 U mg−1 at pH 5.0 (Fig. 3A). However, the activity of the cells that express the subunit I/III subcomplex (I/III) was detected mostly in the soluble fraction at a specific activity of 3.8 ± 0.4 U mg−1, indicating that functional I/III is produced and subunit II is a membrane-anchoring subunit for the FDH complex. Because I/III had significant activity to oxidize fructose but failed to link the respiratory chain, it is reasonable to conclude that subunit II is responsible for ubiquinone reduction. By using the purified FDH complex and partially purified I/III, we determined bimolecular rate constants for the reduction of several artificial electron acceptors. I/III had no selectivity for electron acceptors, while the FDH complex reacted specifically with 2,3-dimethoxy-5-methyl-1,4-benzoquinone (Q-0) and 2,3-dimethoxy-5-farnesyl-1,4-benzoquinone (Q-1) (Kawai, Yakushi, Matsushita, and Kano, unpublished). We examined the pH dependency of fructose dehydrogenase activity for the FDH complex in the membrane fraction and I/III in the soluble fraction (Fig. 3B). They were different from each other: i.e., the optimum pH of the FDH complex in the membrane fraction was pH 4.0, whereas I/III showed the highest activity at pH 6.0.
Fig 3.
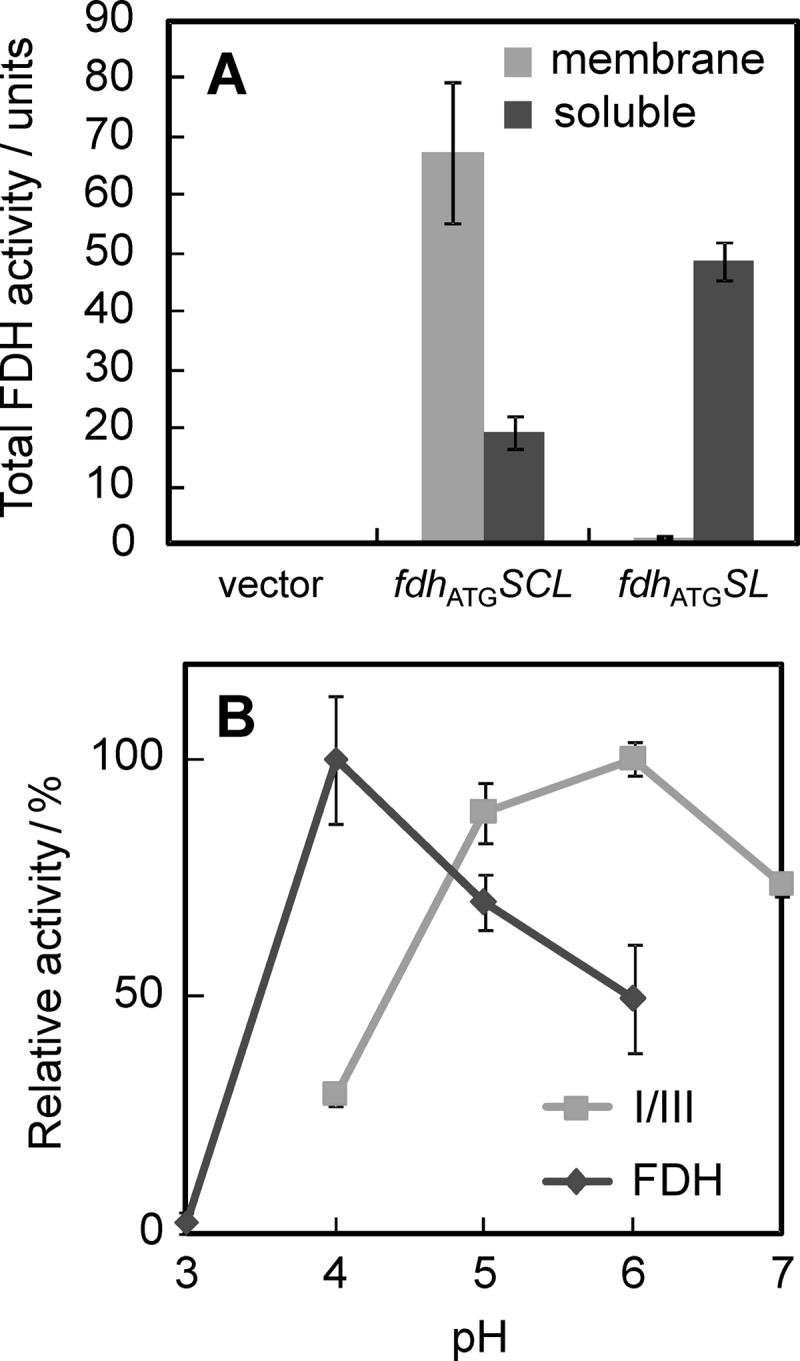
Comparison of the FDH complex (FDH) and I/III. (A) The membrane (light gray bars) and soluble (heavy gray bars) fractions of the ΔadhA strains harboring pSHO8 (vector), pSHO13 (fdhATGSCL), or pSHO16 (fdhATGSL) were prepared, and FDH activities in the membrane and soluble fractions were measured at pH 5.0 and pH 6.0, respectively. Total activity in each fraction was shown. (B) FDH activities of the membrane fraction for the FDH complex and the soluble fraction for I/III were measured under various pH conditions. Comparisons of relative activity to the highest activity are shown individually. Data are shown as mean values with 90% confidence intervals (error bars; n = 3). The specific FDH activities of the membrane fraction of the cells harboring pSHO13 (fdhATGSCL) and the soluble fraction of the cells harboring pSHO16 (fdhATGSL) were 20 ± 5 and 3.8 ± 0.4 U mg−1, respectively.
DISCUSSION
We purified the FDH complex from the membranes of G. japonicus NBRC3260 (formerly G. industrius IFO3260) in 1981 (1). This enzyme is useful for the determination of d-fructose, which can be applied to diagnosis (2) and, more recently, for basic research to understand the properties of enzyme electrodes that can transfer electrons directly (3). Here, we sequenced whole structural genes for the heterotrimeric complex for the first time. The genes for several kinds of flavoprotein-cytochrome c complexes have been sequenced so far, such as gluconate 2-dehydrogenase from Erwinia cypripedii (27), glucose dehydrogenase from Burkholderia cepacia (BcGDH) (21), and sorbitol dehydrogenase from G. frateurii (16). Each enzyme, including FDH, has strict substrate specificity; the genes determined in this study offer the 4th new member of the flavoprotein-cytochrome c complex family as a fructose-specific enzyme. The genes coding for 2-ketogluconate 5-dehydrogenase, a flavoprotein-cytochrome c complex, remain to be determined (28). The gene organization of the fdh genes is unique compared to those of the others, i.e., the order of the genes is from the small to large and cytochrome c subunits in the 5′-to-3′ direction for those reported so far, whereas that of the fdh genes is from the small to cytochrome c and large subunits. However, we anticipate that there is less physiological significance for the difference in gene organization because we qualitatively reconstituted an FDH complex from partially purified I/III and the cytochrome subunit independently expressed (Kawai, Yakushi, Matsushita, and Kano, unpublished). Another unique feature predicted from the primary sequence of the FDH complex is a hydrophobic patch in the C terminus of subunit II for integration into the membrane by a hydrophobic helical structure, which can be predicted by a hydropathy plot using the SOSUI program (29; data not shown). However, we ran the secondary structure prediction program Jpred 3 (30), and the hydrophobic patch would be part of a sheet structure rather than a helix with a relatively high probability (data not shown). Thus, it is an interesting issue whether the hydrophobic patch has a role in the membrane localization of subunit II or not.
We successfully overexpressed the FDH complex in the G. oxydans ΔadhA strain. This strain is beneficial for the expression of the heterologous protein in the host strain for purification because it fails to produce the ADH complex, one of the major membrane proteins in Gluconobacter. In addition, compared to E. coli, our expression system does not need to consider heme C assembly because Gluconobacter produces large amounts of c-type cytochromes naturally (25). Tsuya et al. reported on the heterologous expression of BcGDH in the E. coli strain harboring a plasmid to express the heme C assembly system (21). When pSHO13 (fdhATGSCL) was used, the membranes contained a specific FDH activity of 16 U mg−1, suggesting that approximately 5% of the membrane proteins were the FDH complex, taking into account purified FDH, which had a specific activity of 260 U mg−1. We suggest that G. oxydans can produce the FDH complex at such high productivity because it is a related species of G. japonicus.
The translation of subunit III was found to start at the TTG codon, by construction of the plasmid derivative pSHO13 (fdhATGSCL) containing a termination codon in frame just before the initiation codon, which was replaced with the ATG codon. The G. oxydans strain harboring pSHO13 not only produced the FDH complex, but also a much larger amount of FDH. Translation initiation by the TTG codon is enhanced by the T signal (ATTT) in the 5′ side of the initiation codon (31). However, we did not find a candidate for the T signal in the nucleotide sequence near the initiation codon. The results in this study suggest that the TTG codon is a less efficient codon even in Gluconobacter, and its replacement with the ATG codon improves translation efficiency.
Since we reconstituted the d-fructose-oxidizing respiratory chain in G. oxydans ΔadhA cells, we suggest that the FDH complex is a d-fructose:ubiquinone 5-oxidoreductase functioning as the primary dehydrogenase in the respiratory chain of G. japonicus. Moreover, the ΔadhA cells harboring pSHO16 (fdhATGSL) producing I/III only failed to support the d-fructose-oxidizing ability, even though these cells showed significantly high d-fructose:ferricyanide oxidoreductase activity in the soluble fraction. Thus, we suggest that subunit II is responsible for anchoring the FDH complex to the cytoplasmic membrane and transferring electrons to ubiquinone. Another Gluconobacter membrane-bound enzyme, ADH, consists of three subunits; one of which is a triheme cytochrome c subunit (AdhB) responsible for ubiquinone reduction and membrane anchoring (32). The cytochrome c subunit of heterotrimeric BcGDH has a functionally critical role in the ubiquinone reaction and membrane localization (21). Indeed, a significant homology was observed among FdhC, AdhB, and the β subunit of BcGDH (see the Results section). Our results suggest an analogy to the cytochrome c function of other Gluconobacter enzymes, such as molybdopterin-dependent aldehyde dehydrogenase (24, 33) and other heterotrimeric flavoprotein-cytochrome c complexes that can be found in many kinds of bacterial genomes.
As described earlier, the FDH complex was characterized by its ability to transfer electrons to electrodes directly. As far as we know, this ability is unique to this enzyme, and details of the mechanisms remain unknown. We can start creating FDH derivatives through genetic engineering procedures to characterize their electrochemical properties and discuss the mechanism underlining direct electron transfer. Indeed, we observed large differences in the pH dependencies of the FDH complex and I/III (Fig. 3B). These findings suggest that I/III has a different intramolecular electron transport pathway and different electrochemical properties from the FDH complex.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yuichi Yoshino for technical assistance. We are grateful to Osao Adachi, who continuously encourages us to proceed with our FDH study.
This work was supported in part by a Grant-in-Aid from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print 28 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03152-12.
REFERENCES
- 1. Ameyama M, Shinagawa E, Matsushita K, Adachi O. 1981. d-Fructose dehydrogenase of Gluconobacter industrius: purification, characterization, and application to enzymatic microdetermination of d-fructose. J. Bacteriol. 145:814–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nakashima K, Takei H, Adachi O, Shinagawa E, Ameyama M. 1985. Determination of seminal fructose using d-fructose dehydrogenase. Clin. Chim. Acta 151:307–310 [DOI] [PubMed] [Google Scholar]
- 3. Kamitaka Y, Tsujimura S, Kano K. 2007. High current density bioelectrolysis of d-fructose at fructose dehydrogenase-adsorbed and Ketjen black-modified electrodes without a mediator. Chem. Lett. 36:218–219 [Google Scholar]
- 4. Kamitaka Y, Tsujimura S, Setoyama N, Kajino T, Kano K. 2007. Fructose/dioxygen biofuel cell based on direct electron transfer-type bioelectrocatalysis. Phys. Chem. Chem. Phys. 9:1793–1801 [DOI] [PubMed] [Google Scholar]
- 5. Ikeda T, Kobayashi D, Matsushita F, Sagara T, Niki K. 1993. Bioelectrocatalysis at electrodes coated with alcohol dehydrogenase, a quinohemoprotein with heme c serving as a built-in mediator. J. Electroanal. Chem. 361:221–228 [Google Scholar]
- 6. Gorton L, Lindgren A, Larsson T, Munteanu FD, Ruzgas T, Gazaryan I. 1999. Direct electron transfer between heme-containing enzymes and electrodes as basis for third generation biosensors. Anal. Chim. Acta 400:91–108 [Google Scholar]
- 7. Tsujimura S, Nakagawa T, Kano K, Ikeda T. 2004. Kinetic study of direct bioelectrocatalysis of dioxygen reduction with bilirubin oxidase at carbon electrodes. Electrochemistry 72:437–439 [Google Scholar]
- 8. Miura Y, Tsujimura S, Kamitaka Y, Kurose S, Kataoka K, Sakurai T, Kano K. 2007. Bioelectrocatalytic reduction of O2 catalyzed by CueO from Escherichia coli adsorbed on a highly oriented pyrolytic graphite electrode. Chem. Lett. 36:132–133 [Google Scholar]
- 9. Habe H, Shimada Y, Yakushi T, Hattori H, Ano Y, Fukuoka T, Kitamoto D, Itagaki M, Watanabe K, Yanagishita H, Matsushita K, Sakaki K. 2009. Microbial production of glyceric acid, an organic acid that can be mass produced from glycerol. Appl. Environ. Microbiol. 75:7760–7766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grant SG, Jessee J, Bloom FR, Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. U. S. A. 87:4645–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Figurski DH, Helinski DR. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Marmur J. 1963. A procedure for the isolation of deoxyribonucleic acid from microorganisms. Methods Enzymol. 6:726–738 [Google Scholar]
- 13. Liu YG, Mitsukawa N, Oosumi T, Whittier RF. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8:457–463 [DOI] [PubMed] [Google Scholar]
- 14. Goodhew CF, Brown KR, Pettigrew GW. 1986. Haem staining in gels, a useful tool in the study of bacterial c-type cytochromes. Biochim. Biophys. Acta 852:288–294 [Google Scholar]
- 15. Berry EA, Trumpower BL. 1987. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal. Biochem. 161:1–15 [DOI] [PubMed] [Google Scholar]
- 16. Toyama H, Soemphol W, Moonmangmee D, Adachi O, Matsushita K. 2005. Molecular properties of membrane-bound FAD-containing D-sorbitol dehydrogenase from thermotolerant Gluconobacter frateurii isolated from Thailand. Biosci. Biotechnol. Biochem. 69:1120–1129 [DOI] [PubMed] [Google Scholar]
- 17. Berks BC, Palmer T, Sargent F. 2005. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr. Opin. Microbiol. 8:174–181 [DOI] [PubMed] [Google Scholar]
- 18. Gomi M, Sonoyama M, Mitaku S. 2004. High performance system for signal peptide prediction: SOSUIsignal. Chem. Bio Informatics J. 4:142–147 [Google Scholar]
- 19. Dym O, Eisenberg D. 2001. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 10:1712–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shinagawa E, Matsushita K, Adachi O, Ameyama M. 1982. Purification and characterization of d-sorbitol dehydrogenase from membrane of Gluconobacter suboxydans var. α. Agric. Biol. Chem. 46:135–141 [Google Scholar]
- 21. Tsuya T, Ferri S, Fujikawa M, Yamaoka H, Sode K. 2006. Cloning and functional expression of glucose dehydrogenase complex of Burkholderia cepacia in Escherichia coli. J. Biotechnol. 123:127–136 [DOI] [PubMed] [Google Scholar]
- 22. Yamaoka H, Yamashita Y, Ferri S, Sode K. 2008. Site directed mutagenesis studies of FAD-dependent glucose dehydrogenase catalytic subunit of Burkholderia cepacia. Biotechnol. Lett. 30:1967–1972 [DOI] [PubMed] [Google Scholar]
- 23. Kondo K, Horinouchi S. 1997. Characterization of the genes encoding the three-component membrane-bound alcohol dehydrogenase from Gluconobacter suboxydans and their expression in Acetobacter pasteurianus. Appl. Environ. Microbiol. 63:1131–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Thurner C, Vela C, Thony-Meyer L, Meile L, Teuber M. 1997. Biochemical and genetic characterization of the acetaldehyde dehydrogenase complex from Acetobacter europaeus. Arch. Microbiol. 168:81–91 [DOI] [PubMed] [Google Scholar]
- 25. Matsushita K, Toyama H, Adachi O. 1994. Respiratory chains and bioenergetics of acetic acid bacteria. Adv. Microb. Physiol. 36:247–301 [DOI] [PubMed] [Google Scholar]
- 26. Matsushita K, Shinagawa E, Adachi O, Ameyama M. 1989. Reactivity with ubiquinone of quinoprotein d-glucose dehydrogenase from Gluconobacter suboxydans. J. Biochem. 105:633–637 [DOI] [PubMed] [Google Scholar]
- 27. Yum DY, Lee YP, Pan JG. 1997. Cloning and expression of a gene cluster encoding three subunits of membrane-bound gluconate dehydrogenase from Erwinia cypripedii ATCC 29267 in Escherichia coli. J. Bacteriol. 179:6566–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shinagawa E, Matsushita K, Adachi O, Ameyama M. 1981. Purification and characterization of 2-keto-d-gluconate dehydrogenase from Gluconobacter melanogenus. Agric. Biol. Chem. 45:1079–1085 [Google Scholar]
- 29. Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378–379 [DOI] [PubMed] [Google Scholar]
- 30. Cole C, Barber JD, Barton GJ. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197–W201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ganoza MC, Marliere P, Kofoid EC, Louis BG. 1985. Initiator tRNA may recognize more than the initiation codon in mRNA: a model for translational initiation. Proc. Natl. Acad. Sci. U. S. A. 82:4587–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Matsushita K, Yakushi T, Toyama H, Shinagawa E, Adachi O. 1996. Function of multiple heme c moieties in intramolecular electron transport and ubiquinone reduction in the quinohemoprotein alcohol dehydrogenase-cytochrome c complex of Gluconobacter suboxydans. J. Biol. Chem. 271:4850–4857 [DOI] [PubMed] [Google Scholar]
- 33. Adachi O, Tayama K, Shinagawa E, Matsushita K, Ameyama M. 1980. Purification and characterization of membrane-bound aldehyde dehydrogenase from Gluconobacter suboxydans. Agric. Biol. Chem. 44:503–515 [Google Scholar]
- 34. Boyer HW, Roulland-Dussoix D. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459–472 [DOI] [PubMed] [Google Scholar]
- 35. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.