Abstract
Marine aggregates are naturally forming conglomerations of larvacean houses, phytoplankton, microbes, and inorganics adhered together by exocellular polymers. In this study, we show in vitro that the bacterial pathogen Vibrio vulnificus can be concentrated into laboratory-generated aggregates from surrounding water. We further show that environmental (E-genotype) strains exhibit significantly more integration into these aggregates than clinical (C-genotype) strains. Experiments where marine aggregates with attached V. vulnificus cells were fed to oysters (Crassostrea virginica) resulted in greater uptake of both C and E types than nonaggregated controls. When C- and E-genotype strains were cocultured in competitive experiments, the aggregated E-genotype strains exhibited significantly greater uptake by oyster than the C-genotype strains.
INTRODUCTION
Vibrio vulnificus is a Gram-negative, halophilic bacterium capable of causing gastroenteritis, wound infections, and fatal septicemia in humans (1–3). It is routinely found in waters of estuarine environments as part of the normal microflora, as well as in oysters and other shellfish inhabiting those estuaries (3). V. vulnificus infection is the leading cause of seafood-borne deaths in the United States, usually resulting from the consumption of raw or undercooked oysters (3). Infections caused by ingestion commonly result in primary septicemia, almost always require hospitalization, and have a fatality rate of greater than 50% (3, 4). Wound infections usually result from exposure of open wounds to seawater containing the bacterium and can progress to fatal necrotizing fasciitis (5, 6).
V. vulnificus exhibits high genotypic and phenotypic variation (3) and is divided into two genotypes, a difference originally discovered by randomly amplified polymorphic DNA PCR analysis of strains from both clinical and environmental sources (7). In this classification system, a gene designated vcg (virulence-correlated gene) was found to have two variations (8). One allele (vcgC) correlates highly with strains obtained from clinical sources and is designated the C genotype, while the other (vcgE) is correlated with environmentally isolated strains and is designated the E genotype (7, 8).
Over 95% of infections resulting in septicemia caused by V. vulnificus involve the consumption of raw oysters, with the remainder arising from ingestion of steamed oysters and clams (3). While millions of people in the United States eat raw oysters (9), if a consumer is afflicted with a predisposing condition, such as liver impairment or immune system dysfunction (10), the risk for infection increases 80-fold (11). Even considering these two facts, it is surprising that there are only ca. 40 primary septicemia cases reported per year (10). Usually, oysters predominately contain the E-genotype strains of V. vulnificus, which is likely a factor in the low number of infections (12–18).
A study comparing the population dynamics of the V. vulnificus genotypes revealed an interesting phenomenon. While Warner and Oliver (13) found a nearly even ratio of C-type strains to E-type strains in North Carolina and Florida seawater samples, strains isolated from oysters in those same waters were predominately (>84%) E type (13, 19). Presently, the reason for this incongruity has not been determined. It is especially odd considering that oysters are filter feeders, pumping water through their gills and straining food particles from the flow (20). Incredibly, Crassostrea virginica is able to pump water at a rate of 10 liters h−1 g−1 dry tissue weight (20). With such a high rate of water clearance, one would naturally expect the oyster's internal composition of V. vulnificus to mimic that of the surrounding water. However, in experiments where marked V. vulnificus strains of the C or E genotype were added to oysters and their entry into and exit out of the shellfish were followed, no difference between the uptake or depuration rates of the two types was observed (14, 21).
An oyster is able to select the particles of food that it eats on the basis of size. The gills act as a sieve, catching particles of optimum size and moving them toward the mouth, while particles that are too large are stopped and passed from the oyster as pseudofeces. Particles that are smaller than the optimum size pass through the gills without capture. These too are excreted from the oyster undigested. For C. virginica, the optimum particle size is 5 to 7 μm in diameter, with particles of this size being retained with 90% efficiency (22). Particle retention rates drop to 50% when the diameter is only 1.8 μm, and when particles are the size of a single V. vulnificus bacterium (ca. 1 μm), oysters retain only ca. 16% of what is passed through the gills (22). This size selection would likely limit the effectiveness of bacterial uptake experiments where bacterial cells are simply added to oyster tanks.
Marine aggregates, also known as marine snow, are a natural part of marine waters. These particles consist of fecal pellets, larvacean houses, phytoplankton, microbes, and inorganics brought together by shear forces and Brownian movement. These particles are conglomerated by exocellular polymers and physical/chemical forces (23) and, once achieving a critical size, sink to the ocean and estuary floor. Visible aggregates are termed “marine snow,” after the “long snowfall” of sedimentary material described by Rachel Carson (23–25).
The purpose of this study was to compare the integration of C- and E-genotype V. vulnificus cells into marine aggregates. These particles, with added V. vulnificus, were then fed to oysters to measure the uptake and depuration rates of this pathogen. We hypothesized that differences in the ability of V. vulnificus strains to incorporate into marine aggregates could play a role in the population disparity that we have observed within oysters.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
V. vulnificus CVD713 is a C-genotype strain possessing a stable (for at least 10 days) TnphoA transposon that confers kanamycin resistance and alkaline phosphatase activity (26–28). This strain forms blue colonies when grown on Tn agar, consisting of Luria agar with the addition of 0.2 g liter−1 kanamycin, 2 g liter−1 glucose, and 0.04 g liter−1 5-bromo-4-chloro-3-indolyl phosphate (BCIP). Tn agar selects for this TnphoA-possessing strain via its kanamycin resistance and is differential by means of the BCIP breakdown (28). V. vulnificus strain JDO-2 is a C-type strain derived from the parent strain C7184. This strain is resistant to chloramphenicol and grows on heart infusion agar in the presence of 2 mg/ml of the antibiotic. V. vulnificus strains pGTR-JY1305 and pGTR-Env1 are E-type strains that contain the stable pGTR plasmid which confers kanamycin resistance when the strain is grown on Luria agar containing 10 g liter−1 l-arabinose and 0.3 g liter−1 kanamycin (29). These genetically marked strains were used in aggregation and oyster uptake experiments to allow differentiation of the added strains from naturally occurring Vibrio spp. and other bacteria found in oysters and seawater.
In competition experiments, wherein both kanamycin-resistant C- and E-genotype V. vulnificus strains were used simultaneously, a selective mannitol detection medium consisting of 16 g of BBL phenol red broth base (BD, NJ) and 1.0% d-mannitol per liter of deionized water that was autoclaved for 5 min at 121°C and then supplemented with 0.3 g liter−1 kanamycin and 10 g liter−1 l-arabinose was employed. Distinct yellow colonies are formed by pGTR-JY1305 and pGTR-Env1 (E genotypes), and red colonies are formed by CVD713 and JDO-2 (C genotypes) (12).
Oyster maintenance.
Oysters (Crassostrea virginica) from the North Carolina coast were collected by hand in the intertidal zone, rinsed, and placed into holding aquaria to acclimate to laboratory conditions. The tanks contained a 1:1 mixture of artificial seawater (ASW; Instant Ocean; Aquarium Systems, Mentor, OH) and natural seawater (NSW; collected from the North Carolina coast) which had been passed through a 0.45-μm-pore-size filter (Millipore, Bedford, MA) and finally adjusted to 20‰ salinity with deionized water. Tank water was kept at 23°C, and oysters were fed an algal mixture of Skeletonema and Isochrysis species daily. The algal cultures were grown at room temperature in vented, 1-liter flasks containing F/2 medium and were provided with constant fluorescent light for 1 to 2 weeks until a dense population was achieved (30, 31). Serial transfer of algal cultures was used to generate new cultures.
Marine aggregates.
Laboratory-created aggregates (marine snow) were generated using the method described by Shanks and Edmondson (32) with the modifications suggested by Ward and Kach (33). Briefly, cells were added to seawater diluted to 15‰ salinity with deionized water in 250-ml roller bottles, 10 μg liter−1 hyaluronic acid was added, and the bottles were placed on a roller table at 15 rpm for 24 h. Static bottles were placed next to the roller table to serve as nonaggregated controls.
Bacterial incorporation into aggregates was measured by allowing the aggregates to settle for 20 min and removing a 750-μl sample including the aggregates (or nonaggregated particulate matter in the static controls), disrupting the aggregates via vortexing to disaggregate the bacterial cells for more accurate enumeration, and subsequent plating onto medium specific for the added V. vulnificus strain. Control bottles not rolled were inverted three times and allowed to settle for 20 min prior to sampling. Each experiment involved four static control bottles and four rolled bottles per bacterial strain, and each experiment was performed in triplicate. For uptake experiments, roller bottles containing aggregates and control bottles without aggregates were inverted three times before their contents were gently poured into the oyster aquaria.
Oyster uptake and depuration.
For each experiment, oysters were fed 24 h prior to being removed from maintenance tanks and placed in experimental tanks with 20‰ salinity ASW at 23°C. Twenty-five oysters were placed into each tank, and five oysters were sampled at each time point. Five oysters were removed from the tanks and sampled to establish a background population count of V. vulnificus (sampling methods are described below). V. vulnificus cells grown to a concentration of 108 to 109 CFU per ml were added to the experimental tanks to a final concentration of 7.5 × 104 CFU/ml of tank water. Oysters were incubated in the V. vulnificus-supplemented water for 24 h. After the initial 24-h exposure and every 48 h thereafter, the oysters were removed from the tanks and the aquaria were cleaned, sanitized, and refilled with fresh ASW (20‰ salinity). The oysters were then placed back into the clean tanks before selecting five oysters that were removed for sampling, allowing the determination of bacterial uptake and depuration rates. All studies were conducted in triplicate.
C- and E-genotype competition experiments.
To allow competition between the C- and E-genotype V. vulnificus cells, strain pGTR-JY1305 and strain CVD713 or strain pGTR-Env1 and strain JDO-2 were grown to a concentration of 108 to 109 CFU per ml and then combined in a 1:1 ratio. The mixture of cells was washed twice with phosphate-buffered saline (PBS), added to the roller bottles as described above. Oysters in competition uptake experiments were placed into individual 1-liter aquaria with the oyster resting on a raised metal platform. These individual oyster aquaria contained a stir bar to maintain water flow and prevent the settling of aggregates.
Oyster dissection and homogenization.
Oysters, once removed from experimental tanks, were rinsed with ethanol and patted dry with paper towels. The oysters were shucked with a flame-sterilized oyster knife, and the meat was washed with sterile ASW of 20‰ salinity and placed into a sterile test tube.
The oyster meat was homogenized in 20‰ ASW at a 1:1 (wt/vol) ratio (minimum, 5 ml ASW) using sterile blender cups (Waring, Torrington, CT) and a blending pattern of 3 bursts of 15 s each with a 5-s pause between the bursts.
Sampling methods for marked strains.
After homogenization, samples were serially diluted in sterile PBS and plated onto the appropriate medium for the specific detection of the inoculated strains of V. vulnificus, as described above. The total numbers of CFU per gram of oyster tissue were calculated.
Statistics.
Data were compared using a two-way analysis of variance (ANOVA) followed by post hoc tests with Bonferroni corrections for multiple comparisons (34). Data were analyzed using SigmaStat statistical software (version 2.0; Access Softek Inc.).
RESULTS AND DISCUSSION
Using the method modified from Ward and Kach (33), we formed aggregates of particles suspended in natural seawater by mixing the water using roller bottles. By adding C- or E-genotype strains of V. vulnificus to these aggregates as they formed, we were able to measure the incorporation of the bacteria into these particulate conglomerations. Macroscopic aggregates were always observed in the bottles placed onto the roller table, whereas the static control bottles never formed such aggregates. Aggregates ranged in size from centimeter scale to micrometer scale (Fig. 1). The concentration of V. vulnificus cells was significantly greater in the samples with aggregated marine snow than in the samples where aggregates did not form (P < 0.001), regardless of genotype (Fig. 2). In the environment, V. vulnificus and other vibrios have also been reported to be present in higher concentrations in natural marine aggregates than in the surrounding water (35, 36). Marine aggregates are believed to increase bacterial survival when moving between hosts (35). Furthermore, direct access to organic substrates and protection from chemical or physical stress can be gained by association with aggregates (36, 37). Thus, it is beneficial for the cells to concentrate in marine snow in a short time.
Fig 1.
Micrograph of disrupted aggregates. Magnification, ×1,000.
Fig 2.
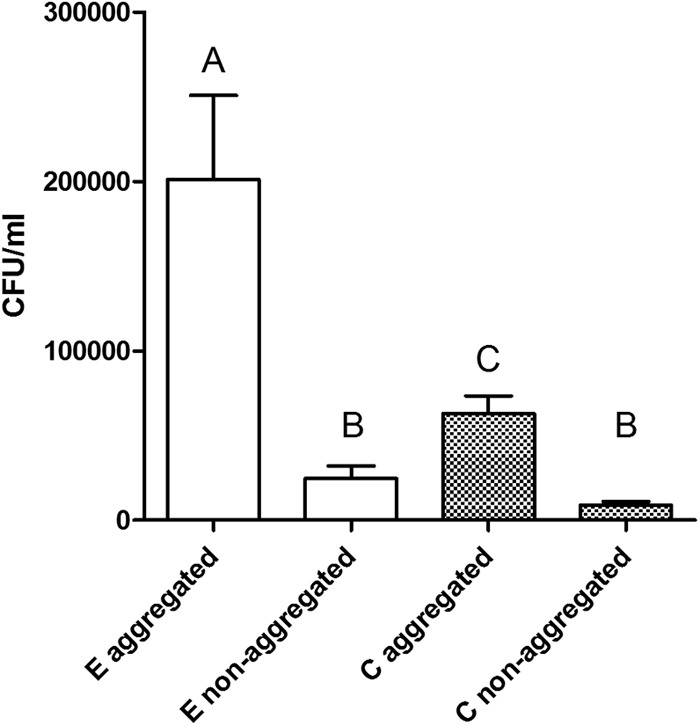
Concentrations of V. vulnificus cells by genotype recovered from microcosms allowed to form aggregates by rolling or from static control samples. Bars with different letters are significantly different (P < 0.05) from each other, as determined by two-way ANOVA with Bonferroni posttest comparisons.
E-type cells showed significantly more integration into marine aggregates than C-type cells (P < 0.001), while the nonaggregated V. vulnificus concentrations between the two genotypes were not statistically significantly different (P > 0.05; Fig. 2). The cause of the increased affinity for the E-genotype strain for marine snow is not known, but because aggregate affinity and assimilation involve cell-cell interactions, motility, chemokinetics, and exoenzyme production, we assume that one of these or other factors are different between the two genotypes (35, 37–39). Recently, sequencing data revealed genomic differences between C- and E-genotype strains, showing that E-type strains have genes linked to attachment proteins (40). A PKD domain present in the E types and absent from the C types could allow more effective binding and hydrolysis of chitin, a component of marine aggregates (40–42).
For decades, experiments looking at the uptake of V. vulnificus by oysters have been conducted by simply adding planktonic bacteria to tanks containing the oysters or, occasionally, by adding the bacteria to algae and feeding the algae to oysters. Because oysters sort particles on the basis of size, the efficiency of planktonic bacterial uptake could be so low that results do not mimic what would be expected to occur in situ. Thus, the planktonic model, while useful, is likely to be highly inefficient. Aggregates with integrated V. vulnificus cells were added to oyster tanks, and the uptake and depuration of the cells were recorded (Fig. 3). Oysters sampled after 1 day of incubation with the aggregated V. vulnificus treatments were found to have significantly more E-genotype cells (Fig. 3A) or more C-genotype cells (Fig. 3B) than oysters sampled after incubation with the nonaggregated control treatments (P = 0.025 and P = 0.002, respectively). The aggregated C-type cell numbers were also significantly higher than those for the control at 3 days after incubation (P = 0.03), but by day 6, aggregated C-type cells were undetectable (Fig. 3B). E-type cell numbers were not significantly different in aggregated samples and controls (P > 0.05; Fig. 3A) and were very low in both.
Fig 3.
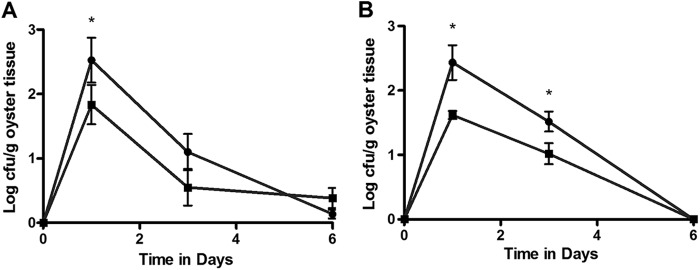
(A) Uptake and depuration of aggregated (circles) or nonaggregated (squares) E-genotype cells in oysters. *, time point at which the number for the experimental group was significantly different (P < 0.05) from that for the control. Cells were added immediately after recording the zero time point. (B) Uptake and depuration of aggregated (circles) or nonaggregated (squares) C-genotype cells in oysters. *, time points at which the number for the experimental group was significantly different (P < 0.05) from that for the control. Cells were added immediately after recording the zero time point.
When C- and E-genotype cells were added to aggregate bottles in competition, we observed significantly greater (P < 0.001) incorporation of the E-type strain than the C-type strain into the newly formed marine snow (Fig. 4). Furthermore, when comparing the uptake data presented in Fig. 2 with those presented in Fig. 4, it appears that the presence of the E-type strains caused the C-type strain to incorporate into the snow less than if the two strains types were cultured individually. Aggregates generated with C- and E-type strains in competitive coculture were subsequently fed to oysters (Fig. 5); we found that oysters exhibited significantly greater (P = 0.029) uptake of the E-type cells than the C-type cells that were added at the same time. Nonaggregated controls showed no significant (P > 0.05) differences in uptake between the two genotypes in these competitive studies. Thus, it appears that in a mixed-genotype environment, such as that which would be found in an estuarine system, E-genotype cells outcompete the C-type cells for integration into marine aggregates. Moreover, while both genotypes of V. vulnificus individually exhibit greater uptake by oysters when they are associated with these aggregates, the E types benefit from their advantage and can display increased uptake into oysters compared to the C-genotype counterparts. This finding may explain why oysters are consistently found to contain disproportionate ratios of E-genotype cells even when they are not dominant in the surrounding water column.
Fig 4.
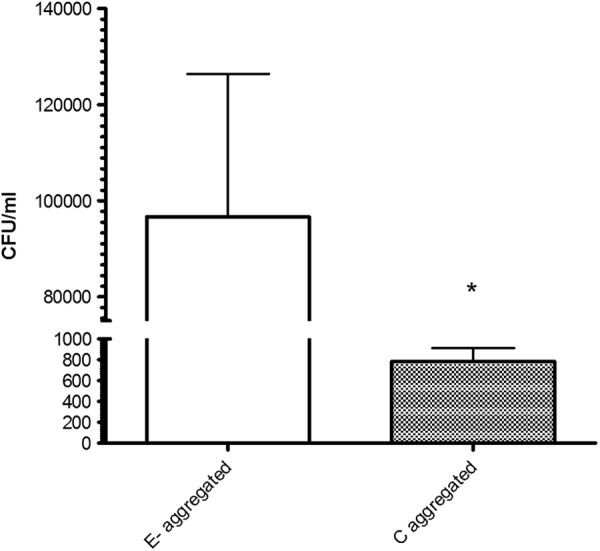
Incorporation of E- and C-type V. vulnificus into marine aggregates when incubated in competitive coculture. *, significantly different (P < 0.05) values.
Fig 5.
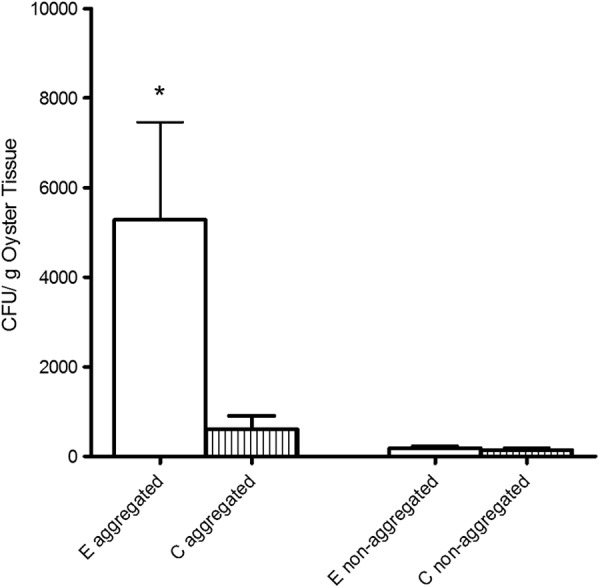
Uptake of C- and E-genotype V. vulnificus strains fed to oysters after competitive coculture in either rolled (aggregated) or static (nonaggregated) bottles. *, data that are significantly different (P < 0.05) from the others.
ACKNOWLEDGMENTS
We thank Melissa Jones for the creation of V. vulnificus strains pGTR-JY1305 and pGTR-Env1. We also thank Tiffany Williams for assistance with aquarium maintenance and oyster collection.
This material is based upon work supported by the Cooperative State Research, Education, and Extension Service, U.S. Department of Agriculture, under award no. 2007-35201-18381.
Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the U.S. Department of Agriculture.
Footnotes
Published ahead of print 21 December 2012
REFERENCES
- 1. Johnston JM, Becker SF, McFarland LM. 1986. Gastroenteritis in patients with stool isolates of Vibrio vulnificus. Am. J. Med. 80:336–338 [DOI] [PubMed] [Google Scholar]
- 2. Oliver JD. 1989. Vibrio vulnificus, p 569–599 In Doyle MP. (ed), Foodborne bacterial pathogens. Marcel Dekker, Inc., New York, NY [Google Scholar]
- 3. Oliver JD. 2006. Vibrio vulnificus, p 349–366 In Thompson FL, Austin B, Swings J. (ed), The biology of vibrios. ASM Press, Washington, DC [Google Scholar]
- 4. Mead PS, Slusker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RBV. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jones MK, Oliver JD. 2009. Vibrio vulnificus: disease and pathogenesis. Infect. Immun. 77:1723–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oliver JD. 2005. Wound infections caused by Vibrio vulnificus and other marine bacteria. Epidemiol. Infect. 133:383–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Warner JM, Oliver JD. 1999. Randomly amplified polymorphic DNA analysis of clinical and environmental isolates of Vibrio vulnificus and other Vibrio species. Appl. Environ. Microbiol. 65:526–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rosche T, Yano Y, Oliver JD. 2005. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical and environmental isolation. Microbiol. Immunol. 49:381–389 [DOI] [PubMed] [Google Scholar]
- 9. US Food and Drug Administration 2009. Vibrio vulnificus heath education kit. US Department of Health and Human Services, Silver Spring, MD [Google Scholar]
- 10. Oliver JD. 2005. Vibrio vulnificus, p 253–276 In Belkin S, Colwell RR. (ed), Oceans and health: pathogens in the marine environment. Springer, New York, NY [Google Scholar]
- 11. National Center for Emerging and Zoonotic Infectious Diseases 2009. Vibrio vulnificus. Centers for Disease Control and Prevention, Atlanta, GA [Google Scholar]
- 12. Froelich B, Oliver J. 2011. Orientation of mannitol related genes can further differentiate strains of Vibrio vulnificus possessing the vcgC allele. Adv. Stud. Biol. 3:151–160 [Google Scholar]
- 13. Warner EB, Oliver JD. 2008. Population structures of two genotypes of Vibrio vulnificus in oysters (Crassostrea virginica) and seawater. Appl. Environ. Microbiol. 74:80–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Froelich B, Oliver JD. 3 January 2013, posting date The interactions of Vibrio vulnificus and the oyster Crassostrea virginica. Microb. Ecol. [Epub ahead of print.] doi:10.1007/s00248-012-0162-3 [DOI] [PubMed] [Google Scholar]
- 15. Han F, Pu S, Hou A, Ge B. 2009. Characterization of clinical and environmental types of Vibrio vulnificus isolates from Louisiana oysters. Foodborne Pathog. Dis. 6:1251–1258 [DOI] [PubMed] [Google Scholar]
- 16. Staley C, Jones MK, Wright AC, Harwood VJ. 2011. Genetic and quantitative assessment of Vibrio vulnificus populations in oyster (Crassostrea virginica) tissues. Environ. Microbiol. Rep. 3:543–549 [DOI] [PubMed] [Google Scholar]
- 17. Nilsson WB, Paranjype RN, DePaola A, Strom MS. 2003. Sequence polymorphism of the 16S rRNA gene of Vibrio vulnificus is a possible indicator of strain virulence. J. Clin. Microbiol. 41:442–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vickery M, Nilsson W, Strom M, Nordstrom J, DePaola A. 2007. A real-time PCR assay for the rapid determination of 16 S rRNA genotype in Vibrio vulnificus. J. Microbiol. Methods 68:376–384 [DOI] [PubMed] [Google Scholar]
- 19. Krantz JA, Nordstrom JL, Bowers JC, Depaola A. 2008. An evaluation of CPC+, a new medium for isolation and enumeration of Vibrio vulnificus from U.S. market oysters, abstr P-121. Abstr. 108th Gen. Meet. Am. Soc. Micriobiol., Boston, MA. American Society for Microbiology, Washington, DC [Google Scholar]
- 20. Newell RIE, Langdon CJ. 1996. Mechanisms and physiology of larval and adult feeding, p 185–229 In Kennedy VS, Newell RIE, Eble AF. (ed), The eastern oyster: Crassostrea virginica. Maryland Sea Grant College, College Park, MD [Google Scholar]
- 21. Froelich B, Ringwood A, Sokolova I, Oliver J. 2010. Uptake and depuration of the C- and E-genotypes of Vibrio vulnificus by the Eastern Oyster (Crassostrea virginica). Environ. Microbiol. Rep. 2:112–115 [DOI] [PubMed] [Google Scholar]
- 22. Ward JE, Shumway SE. 2004. Separating the grain from the chaff: particle selection in suspension- and deposit-feeding bivalves. J. Exp. Mar. Biol. Ecol. 300:83–130 [Google Scholar]
- 23. Alldredge AL, Silver MW. 1988. Characteristics, dynamics and significance of marine snow. Prog. Oceanogr. 20:41–82 [Google Scholar]
- 24. Suzuki N, Kato K. 1953. Studies on suspended materials. Marine snow in the sea. I. Sources of marine snow. In Bulletin of the Faculty of Fisheries of Hokkaido University. Hokkaido University, Hokkaido, Japan [Google Scholar]
- 25. Carson R. 1951. The sea around us. Oxford University Press, New York, NY [Google Scholar]
- 26. Morris JG, Wright AC, Roberts DM, Wood PK, Simpson LM, Oliver JD. 1987. Identification of environmental Vibrio vulnificus isolates with a DNA probe for the cytotoxin-hemolysin gene. Appl. Environ. Microbiol. 53:193–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murphy SK, Oliver JD. 1992. Effects of temperature abuse on survival of Vibrio vulnificus in oysters. Appl. Environ. Microbiol. 58:2771–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wright AC, Simpson LM, Oliver JD, Morris JG. 1990. Phenotypic evaluation of acapsular transposon mutants of Vibrio vulnificus. Infect. Immun. 58:1769–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ochi K, Zhang D, Kawamoto S, Hesketh A. 1997. Molecular and functional analysis of the ribosomal L11 and S12 protein genes (rplK and rpsL) of Streptomyces coelicolor A3(2). Mol. Gen. Genet. 256:488–498 [DOI] [PubMed] [Google Scholar]
- 30. Miescier JJ, Hunt DA, Redman J, Salinger A, Lucas JP. 1992. Molluscan shellfish: oysters, mussels, and clams, p 901–907 In Splittstoesser DF. (ed), Compendium of methods for microbiological examination of food, 3rd ed American Public Health Association, Washington, DC [Google Scholar]
- 31. James DE. 1978. Culturing algae. Carolina Biological Supply Company, Burlington, NC [Google Scholar]
- 32. Shanks AL, Edmondson EW. 1989. Laboratory-made artificial marine snow: a biological model of the real thing. Mar. Biol. 101:463–470 [Google Scholar]
- 33. Ward JE, Kach DJ. 2009. Marine aggregates facilitate ingestion of nanoparticles by suspension-feeding bivalves. Mar. Environ. Res. 68:137–142 [DOI] [PubMed] [Google Scholar]
- 34. Sokal RR, Rohlf FJ. 1995. Biometry: the principles and practice of statistics in biological research. WH Freeman & Co., San Francisco, CA [Google Scholar]
- 35. Lyons MM, Lau Y-T, Carden WE, Ward JE, Roberts SB, Smolowitz R, Vallino J, Allam B. 2007. Characteristics of marine aggregates in shallow-water ecosystems: implications for disease ecology. EcoHealth 4:406–420 [Google Scholar]
- 36. Lyons MM, Ward JE, Gaff H, Hicks RE, Drake JM, Dobbs FC. 2010. Theory of island biogeography on a microscopic scale: organic aggregates as islands for aquatic pathogens. Aquat. Microb. Ecol. 60:1–13 [Google Scholar]
- 37. Tang KW, Dziallas C, Grossart HP. 2011. Zooplankton and aggregates as refuge for aquatic bacteria: protection from UV, heat and ozone stresses used for water treatment. Environ. Microbiol. 13:378–390 [DOI] [PubMed] [Google Scholar]
- 38. Kiorboe T, Jackson GA. 2001. Marine snow, organic solute plumes, and optimal chemosensory behavior of bacteria. Limnol. Oceanogr. 46:1309–1318 [Google Scholar]
- 39. DeLong EF, Franks DG, Alldredge AL. 1993. Phylogenetic diversity of aggregate-attached vs. free-living marine bacterial assemblages. Limnol. Oceanogr. 38:924–934 [Google Scholar]
- 40. Morrison SS, Williams T, Cain A, Froelich B, Taylor C, Baker-Austin C, Verner-Jeffreys D, Hartnell R, Oliver JD, Gibas CJ. 2012. Pyrosequencing-based comparative genome analysis of Vibrio vulnificus environmental isolates. PLoS One 7:e37553 doi:10.1371/journal.pone.0037553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith DC, Simon M, Alldredge AL, Azam F. 1992. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature 359:139–142 [Google Scholar]
- 42. Yatsunami R. 2004. Enzymatic syntheses of novel oligosaccharides using haloarchaeal glycosidases. NISR Research Grant. Department of Bioengineering, Tokyo Institute of Technology, Tokyo, Japan [Google Scholar]


