Abstract
Conditional gene expression is key for functional studies in any given microorganism. To allow tight regulation in the pathogenic mold Aspergillus fumigatus, improved versions of the doxycycline-dependent Tet-On system were generated by replacing functional elements of the precursor module, thereby circumventing the former problem of leakiness due to intramolecular recombination.
TEXT
Deletion or hyperexpression of a gene product is a central strategy for characterization of its function in any given organism. Conditional gene expression has evolved as a valuable tool of molecular biology when addressing the intracellular role of uncategorized proteins, as it allows the discovery of relevant phenotypes that might correlate with the expression level. In general, two major approaches have been followed to adjust or condition protein expression: expression of conditional alleles that result in functionally altered gene products dependent on external cues or manipulation of the expression level of the encoding gene. Especially the latter approach is advantageous, as it may allow various levels of expression, resulting in specific phenotypes based on gene dosage effects. For instance, the complete shutdown of expression can result in a pronounced null phenotype or demonstrate essentiality of the gene of interest (1), whereas forced overexpression could give additional hints on the function of the encoded protein and the involved cellular processes.
Conditional gene expression is generally based on the replacement of the endogenous gene promoter by an adjustable one that drives transcription of the gene in question. The controllable promoter responds to changing environmental conditions, such as alternative nutritional sources or the presence/absence of specific molecules. For the latter, tetracycline-dependent systems have been designed and validated in a widespread and highly successful manner, with relevant cellular systems ranging from viral ones over unicellular microbes to mammalian cell lines and transgenic animals (2–5).
For filamentous fungi of the genus Aspergillus, such a conditional expression system was established by expressing a tetracycline-binding transcriptional activator protein constitutively and fusing the coding sequence of interest behind a minimal promoter sequence preceded by several operator (tetO) sites that the transactivator would bind to (6). Two variants of this tetracycline-responsive transcriptional activator protein exist: the tetracycline transactivator (tTA) is inactivated through binding of tetracycline, whereas the reverse tetracycline transactivator (rtTA) will be activated through binding this molecule. These systems are generally referred to as Tet-Off and Tet-On systems, respectively. Due to its superior stability, doxycycline is commonly used instead of tetracycline.
While the initial system for A. fumigatus was assembled by two separate constructs that need to be integrated in the recipient's genome, a recently streamlined Tet-On version comprises all regulatory and coding elements in one cassette (7). Functionality of this module was proven in A. niger to allow tightly regulated, linear “gene expression on demand” and later again in the human-pathogenic mold A. fumigatus when demonstrating the essentiality of the Rho GTPase Rho1 (8).
In the course of testing essential genes, we became aware of an inherent drawback of this recent construct that resulted in an apparent leakiness of the expression module: when plating conidia from a strain carrying an essential gene under the control of the Tet-On system on selective media, a certain fraction (app. 0.01%) was able to germinate and form colonies that would grow in an unrestricted fashion (our unpublished results). Inspection of such clones by diagnostic PCR on genomic DNA revealed that a recombination event had taken place, based on a direct repeat sequence from the gpdA promoter region (pgpdA). This A. nidulans promoter drives constitutive transcription of the transactivator-encoding sequence, and a short segment (174 bp) of it also serves as the minimal promoter element downstream of the tetO sites and upstream of the gene of interest. As a consequence, homologous recombination of these direct repeats leads to excision and placement of the corresponding gene behind the reconstituted gpdA promoter to result in constitutive transcription at high levels. Depending on the frequency, such recombinants may interfere with reliable phenotype analysis of the transgenic strain as they dilute its clonality.
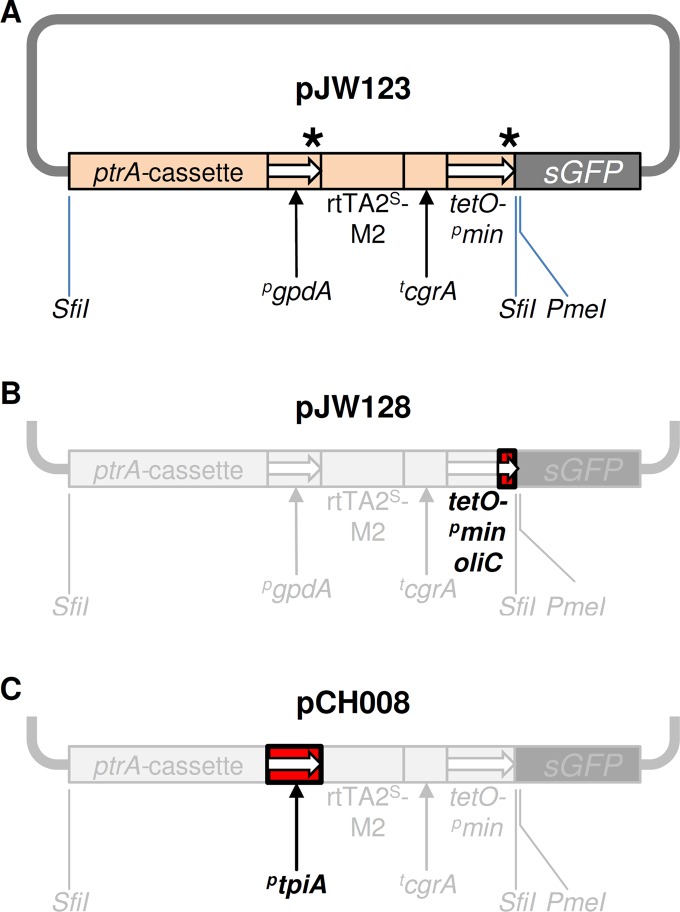
In order to circumvent this issue of leakiness due to recombination in the Tet-On module, we followed two distinct approaches. One aimed at the replacement of the constitutive gpdA promoter, the other, alternatively, of the minimal promoter segment derived from the same regulatory element. Accordingly, two derivative constructs were generated by conventional cloning techniques using plasmid pJW123 (8) as the starting template (Fig. 1A): in pJW128, the 174-bp pgpdA fragment downstream of the tetO sites was replaced by a 160-bp fragment from the A. nidulans oliC promoter that had been mapped as the minimal region to drive transcription of downstream sequences (9, 10). In parallel, the gpdA promoter region of pJW123 was replaced by the 506-bp upstream sequence of the A. nidulans tpiA gene, which encodes a triosephosphate isomerase and was shown to be constitutively expressed (11), yielding pCH008. After validation of both constructs, pJW128 (Fig. 1B) and pCH008 (Fig. 1C), by restriction mapping and sequence analysis, each one was incorporated into a replacement cassette for the promoter of the essential A. fumigatus rho1 gene. The cassettes were transformed and integrated at the homologous locus in the genome of recipient strain AfS35 (12). Several transgenic clonal isolates were screened for correct promoter replacement by diagnostic PCR (not shown), and two validated and independent strains for each construct were subjected to further analyses.
Fig 1.
Tet-On expression systems for conditional gene expression in Aspergillus. (A) The original Tet-On cassette in pJW123 is enclosed in two incompatible SfiI digestion sites and harbors the pyrithiamine resistance gene (ptrA cassette), the gpdA promoter of A. nidulans (pgpdA), the reverse tetracycline transactivator rtTA2S-M2, the terminating region of cgrA from A. fumigatus (tcgrA), and the chimeric pgpdA-based tetO-pmin promoter. Asterisks mark the sites of homologous recombination. (B) In pJW128, the chimeric pgpdA-based tetO-pmin promoter was replaced by a minimal promoter based in the oliC promoter of A. nidulans (tetO-pmin-oliC). (C) In pCH008, the pgpdA was exchanged for the tpiA promoter of A. nidulans (ptpiA).
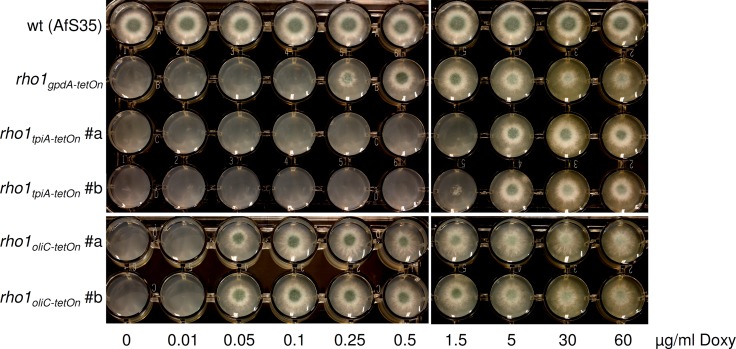
First, growth capacities in the presence or absence of the inducing agent doxycycline at various concentrations were monitored (Fig. 2). Whereas the wild-type progenitor displayed accurate radial extension of its mycelium accompanied by abundant conidiation, conidia from the rho1tetOn strains were unable to grow when doxycycline was omitted from the culture plates. Only in the presence of doxycycline were these strains able to form a mycelium from which mature conidia developed. A fundamental difference became apparent with respect to the threshold concentration that allowed germination and growth: the ptpiA derivatives required higher doses (5 μg · ml−1) than the pgpdA progenitor strain (0.25 to 0.5 μg · ml−1), while the poliCmin strains displayed growth at lower concentrations (0.05 μg · ml−1).
Fig 2.
Doxycycline-dependent growth of conditional rho1tetOn strains carrying different versions of the gene silencing module. From suspensions of 5 × 105 conidia · ml−1 of the indicated strains, aliquots of 3 μl (⩠1,500 conidia) were inoculated on AMM agar supplemented with the indicated amount of doxycycline (Doxy) and incubated at 37°C for 36 h.
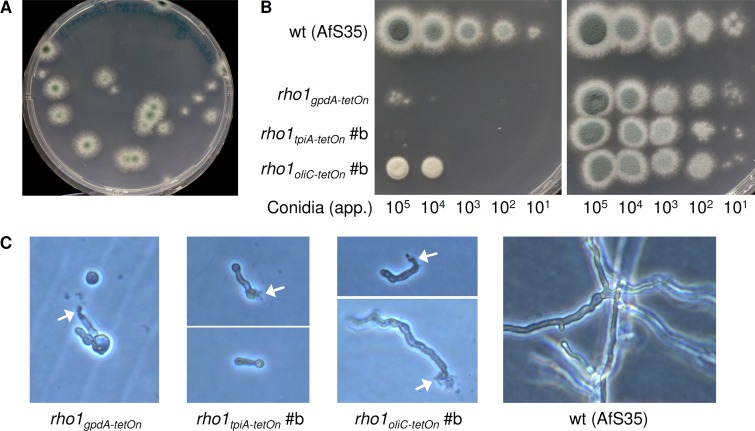
Furthermore, we were interested in validating the presumed absence of recombination in the new Tet-On derivatives due to the lack of direct repeats in the module. For this purpose, defined amounts of freshly harvested conidia from each rho1tetOn strain were plated on minimal culture medium containing 5 μg · ml−1 doxycycline or being devoid of it and incubated at 37°C. All plates were tracked for 96 h, and numbers of single colonies were determined to assess the frequency of recombination and degree of leakiness (Fig. 3A and data not shown). The initial isolate displayed a pronounced and reproducible proportion of spores that were able to grow like the wild type under restrictive conditions (approximately 1 out of 10,000 conidia), while conidia harvested from the derivative strain carrying the ptpiA fragment were unable to do so. Interestingly, we observed occasional growth of the strain carrying the oliC minimal promoter fragment. These colonies always had a prominent growth phenotype, i.e., slow growth and drastically reduced conidiation. Since conidia isolated from these colonies phenotypically did not differ from the parental strain (i.e., doxycycline dependency), we assume this growth is attributed to a higher basal expression driven by the minimal poliC region. Notably, this phenotype specifically depended on the inoculum density (Fig. 3B). As expected, microscopic inspection of freshly germinated spores of all rho1tetOn strains revealed frequent cytoplasmic leakage and death in the absence of doxycycline (8) (Fig. 3C).
Fig 3.
Evaluation of recombination rates and growth phenotypes. (A) Exemplary image. A total of 250,000 conidia of the rho1gpdA-tetOn strain were spread onAMM agar and incubated at 37°C for at least 48 h. (B) In a series of 10-fold dilutions derived from a starting suspension of 3 × 107 conidia · ml−1 of the indicated strains, 3-μl aliquots were inoculated on AMM agar supplemented with no (left) or 5 μg · ml−1 doxycycline (right) and incubated at 37°C for 36 h. (C) Exemplary microscopy images of the indicated strains. Conidia were inoculated in AMM (5 × 103 conidia · ml−1) and incubated at 37°C in the absence of doxycycline for 16 h. White arrows indicate cytoplasmic leakage.
In conclusion, we were able to generate improved versions of the recently introduced tetracycline-dependent system for adjustable gene expression in Aspergillus. Both constructs represent a further development of the validated system by V. Meyer and coworkers (7), as they do not contain repetitive sequences that entail the risk of intramolecular recombination. Accordingly, both Tet-On modules allow gene silencing, which is essential for studying the cellular function of any gene of interest by conditional promoter replacement. This screening approach has proven extremely useful for the identification of virulence determinants and drug targets in fungal pathogens (1, 13) and represents a superior alternative to the commonly used gene deletion strategy or the recent RNA interference (RNAi)-mediated gene silencing approach (14, 15). Of special interest is the fact that both Tet-On constructs differ in their response toward the endogenous inducer doxycycline, with the poliC version resulting in apparently sufficient expression of the rho1 gene product at lower concentrations at the costs of an apparently higher basal expression. In contrast, the precursor construct and the newly designed ptpiA version are more tightly regulated but also require higher concentrations of doxycycline. The exact reason for these differences is unclear and may be due to differing expression levels of the transactivator protein or binding affinities to the chimeric tetO-pmin promoters. However, both Tet-On constructs trigger transcription of a downstream sequence above a certain threshold level of doxycycline, although each at a different concentration range. As the ptpiA-derived construct requires higher doses of doxycycline, the resulting transcription rates of certain genes might not be sufficient for full complementation, as could be demonstrated for the glucan synthase-encoding fks1 gene (our unpublished observation). Accordingly, each module might be more or less appropriate for conditional gene silencing or expression purposes, but both expand the toolbox of Aspergillus molecular biology (16, 17).
ACKNOWLEDGMENTS
We thank Vera Meyer for providing the pVG2.2 construct, Michaela Dümig for excellent technical assistance, and Christian Bogdan for critical reading of the manuscript.
Support from the Institute of Molecular Infection Biology, University of Würzburg, is highly acknowledged, as is funding by the Research Center for Infectious Diseases, the University of Würzburg, the Free State of Bavaria, and the Microbiology Institute of the University Hospital of Erlangen. This work was further supported by the Förderprogramm für Forschung und Lehre of the Ludwig-Maximilians-Universität, the Münchner Medizinische Wochenschrift, the Deutsche Forschungsgemeinschaft (KR2294/1-3 and WA3016/1-1), and the European Science Foundation by its FUMINOMICS Research Networking Programme (06-RNP-132).
Footnotes
Published ahead of print 28 December 2012
REFERENCES
- 1. Hu W, Sillaots S, Lemieux S, Davison J, Kauffman S, Breton A, Linteau A, Xin C, Bowman J, Becker J, Jiang B, Roemer T. 2007. Essential gene identification and drug target prioritization in Aspergillus fumigatus. PLoS Pathog. 3:e24 doi:10.1371/journal.ppat.0030024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Corbel SY, Rossi FMV. 2002. Latest developments and in vivo use of the Tet system: ex vivo and in vivo delivery of tetracycline-regulated genes. Curr. Opin. Biotechnol. 13:448–452 [DOI] [PubMed] [Google Scholar]
- 3. Sprengel R, Hasan MT. 2007. Tetracycline-controlled genetic switches. Handb. Exp. Pharmacol. 2007:49–72 [DOI] [PubMed] [Google Scholar]
- 4. Bertram R, Hillen W. 2008. The application of Tet repressor in prokaryotic gene regulation and expression. Microb. Biotechnol. 1:2–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Urlinger S, Baron U, Thellmann M, Hasan MT, Bujard H, Hillen W. 2000. Exploring the sequence space for tetracycline-dependent transcriptional activators: novel mutations yield expanded range and sensitivity. Proc. Natl. Acad. Sci. U. S. A. 97:7963–7968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vogt K, Bhabhra R, Rhodes JC, Askew DS. 2005. Doxycycline-regulated gene expression in the opportunistic fungal pathogen Aspergillus fumigatus. BMC Microbiol. 5:1 doi:10.1186/1471-2180-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meyer V, Wanka F, van Gent J, Arentshorst M, van den Hondel CAMJJ, Ram AFJ. 2011. Fungal gene expression on demand: an inducible, tunable, and metabolism-independent expression system for Aspergillus niger. Appl. Environ. Microbiol. 77:2975–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dichtl K, Helmschrott C, Dirr F, Wagener J. 2012. Deciphering cell wall integrity signalling in Aspergillus fumigatus: identification and functional characterization of cell wall stress sensors and relevant Rho GTPases. Mol. Microbiol. 83:506–519 [DOI] [PubMed] [Google Scholar]
- 9. De Lucas JR, Gregory S, Turner G. 1994. Analysis of the regulation of the Aspergillus nidulans acuD gene, encoding isocitrate lyase, by construction of a hybrid promoter. Mol. Gen. Genet. 243:654–659 [DOI] [PubMed] [Google Scholar]
- 10. Turner G, Brown J, Kerry-Williams S, Bailey AM, Ward M, Punt PJ, van den Hondel CAMJJ. 1989. Analysis of the oliC promoter of Aspegillus nidulans. Found. Biotechnol. Ind. Ferment. Res. 6:101–109 [Google Scholar]
- 11. Upshall A, Kumar AA, Bailey MC, Parker MD, Favreau MA, Lewison KP, Joseph ML, Maraganore JM, McKnight GL. 1987. Secretion of active human tissue plasminogen activator from the filamentous fungus Aspergillus nidulans. Biotechnology 5:1301–1304 [Google Scholar]
- 12. Krappmann S, Sasse C, Braus GH. 2006. Gene targeting in Aspergillus fumigatus by homologous recombination is facilitated in a nonhomologous end-joining-deficient genetic background. Eukaryot. Cell 5:212–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roemer T, Jiang B, Davison J, Ketela T, Veillette K, Breton A, Tandia F, Linteau A, Sillaots S, Marta C, Martel N, Veronneau S, Lemieux S, Kauffman S, Becker J, Storms R, Boone C, Bussey H. 2003. Large-scale essential gene identification in Candida albicans and applications to antifungal drug discovery. Mol. Microbiol. 50:167–181 [DOI] [PubMed] [Google Scholar]
- 14. Henry C, Mouyna I, Latgé J-P. 2007. Testing the efficacy of RNA interference constructs in Aspergillus fumigatus. Curr. Genet. 51:277–284 [DOI] [PubMed] [Google Scholar]
- 15. Yamada O, Ikeda R, Ohkita Y, Hayashi R, Sakamoto K, Akita O. 2007. Gene silencing by RNA interference in the koji mold Aspergillus oryzae. Biosci. Biotechnol. Biochem. 71:138–144 [DOI] [PubMed] [Google Scholar]
- 16. Brakhage AA, Langfelder K. 2002. Menacing mold: the molecular biology of Aspergillus fumigatus. Annu. Rev. Microbiol. 56:433–455 [DOI] [PubMed] [Google Scholar]
- 17. Krappmann S. 2006. Tools to study molecular mechanisms of Aspergillus pathogenicity. Trends Microbiol. 14:356–364 [DOI] [PubMed] [Google Scholar]