Abstract
The population dynamics of Aspergillus flavus, shaped in part by intraspecific competition, influence the likelihood and severity of crop aflatoxin contamination. Competition for nutrients may be one factor modulating intraspecific interactions, but the influences of specific types and concentrations of nutrients on competition between genotypes of A. flavus have not been investigated. Competition between paired A. flavus isolates on agar media was affected by varying concentrations of carbon (sucrose or asparagine) and nitrogen (nitrate or asparagine). Cocultivated isolate percentages from conidia and agar-embedded mycelia were quantified by measurements of isolate-specific single-nucleotide polymorphisms with quantitative pyrosequencing. Compositions and concentrations of nutrients influenced conidiation resulting from cocultivation, but the percentages of total conidia from each competing isolate were not predicted by sporulation of isolates grown individually. Success during sporulation did not reflect the outcomes of competition during mycelial growth, and the extents to which isolate percentages from conidia and mycelia differed varied among both isolate pairs and media. Whether varying concentrations of sucrose, nitrate, or asparagine increased, decreased, or had no influence on competitive ability was isolate dependent. Different responses of A. flavus isolates to nutrient variability suggest genotypes are adapted to different nutrient environments that have the potential to influence A. flavus population structure and the epidemiology of aflatoxin contamination.
INTRODUCTION
Aspergillus flavus is a ubiquitous filamentous fungus that is both a saprophyte and an opportunistic pathogen of plants and animals (1). Though A. flavus has the ability to infect humans (2), its most serious threat to human health is through production of aflatoxins that contaminate food crops, including maize, peanuts, and tree nuts (3, 4). Consumption of food contaminated with these potent mycotoxins can lead to acute aflatoxicosis and death; chronic dietary exposure to sublethal aflatoxin concentrations can result in liver cancer, growth impairment, and immune suppression (3, 5–7). Most developed nations impose strict regulations on maximum allowable concentrations of aflatoxins in food and feed (8), but regulations are either nonexistent or unenforced in many developing regions (3, 4). Regulations impose an economic burden on growers of crops susceptible to contamination (9, 10), whereas the costs of aflatoxin contamination in human life are exemplified by recent outbreaks of fatal aflatoxicosis in Kenya (5).
Incidences and severities of crop contamination are influenced by the genetic structure and average aflatoxin-producing potential of A. flavus populations (11, 12). The species A. flavus is subdivided into genetically distinct vegetative compatibility groups (VCGs) by a heterokaryon incompatibility system (13, 14). Populations of A. flavus are composed of many interacting sympatric VCGs (15–18), and phenotypic characteristics, including the ability to produce aflatoxins, are usually conserved within a VCG (17, 19–21). Individual agroecosystems harbor highly diverse assemblages of A. flavus VCGs, with genotypes ranging from highly aflatoxigenic to being unable to produce aflatoxins (22, 23). Crop aflatoxin levels are positively correlated with proportions of crop-associated A. flavus populations comprised of highly aflatoxigenic genotypes (5, 12). In contrast, modification of the aflatoxin-producing potential of fungal populations through application of nonaflatoxigenic biocontrol strains of A. flavus is currently the most effective management strategy for limiting crop aflatoxin contamination (10, 24, 25).
In a previous study, a competitive advantage was conferred on the first A. flavus isolate to contact the crop host, suggesting nutrient sequestration plays a role in competitive exclusion by A. flavus (26). This reinforces suggestions that one mechanism by which nonaflatoxigenic strains exclude aflatoxigenic genotypes and reduce the production of aflatoxins is through competition for nutrients (26–28). The success of various A. flavus genotypes in diverse nutrient environments, including host tissues and soils, may also influence the structure of A. flavus populations across agroecosystems.
The ability of A. flavus to produce a broad spectrum of hydrolytic enzymes (1, 29–31) allows the fungus to utilize both complex and simple nutrients within diverse and frequently heterogeneous nutrient environments. The extent to which variation among nutrient environments influences the structures of A. flavus populations is unknown. However, such effects could impact which fungi colonize and infect plant and animal hosts. Though influences of nutrients on A. flavus competition have gone unstudied, differential rates of nutrient sequestration, efficiency of nutrient utilization, and nutrient preferences have been studied in other fungi (32–34). Variation in production of specific hydrolases among A. flavus isolates confers differential abilities to invade hosts and utilize substrates, and this suggests certain isolates may be adapted to different niches (e.g., plant versus soil) and/or nutrient environments (30).
Responses to and competition for nutrients during A. flavus growth and reproduction may dictate the success of individual genotypes in the environment. Nutrients influence germination (35, 36), as well as production of mycelia, conidia, and aflatoxins by A. flavus (37–42), but the influence of nutrients on intraspecific interactions among A. flavus genotypes has not been studied. Outcomes of competition among morphologically indistinct A. flavus individuals are difficult to quantify with culture-based techniques, but quantitative pyrosequencing, a DNA-based method for measuring proportions of genotype-specific single-nucleotide polymorphisms (SNPs) within pools of DNA, has provided insights into factors dictating intraspecific interactions (26, 43, 44). Fungal responses to nutrients may influence the success of specific A. flavus genotypes in agriculturally important niches and, in so doing, affect both the efficacy of aflatoxin biocontrol and the epidemiology of contamination. The current work utilized pyrosequencing to quantify nutrient influences on competitive interactions among A. flavus VCGs during sporulation and mycelial growth.
MATERIALS AND METHODS
Fungal isolates.
Eight genetically distinct isolates, each representing a different VCG and known to differ in competitive abilities and aflatoxin production on maize (26, 43), were examined in the current study (Table 1). CG136, YV04, and MR17 were isolated from agricultural soils in Arizona, and WM01, EB01, DV901, RB04, and MN902 were isolated from cottonseed in Texas (43). All eight VCGs are sympatric and compete on crops and in soils in populations that extend across both states (18). VCG membership was confirmed by complementation of nitrate-nonutilizing auxotrophs derived from wild-type isolates, with tester pairs corresponding to previously identified VCGs (45). Single-spore purified wild-type isolates were cultivated on 5/2 agar (5% V8 juice, 2% agar, pH 5.2), and plugs of agar with abundant sporulation were suspended in 4-ml vials containing sterile distilled water. Conidial suspensions from water vials were used to centrally seed 5/2 agar. After 7 days (31°C), the conidia were dislodged from the plates with sterile cotton swabs and suspended in sterile distilled water. The numbers of conidia were estimated by measuring the turbidity of the suspensions with a turbidity meter (Turbidimeter; Orbeco Analytical Systems, Farmingdale, NY) and using a nephalometric turbidity unit (NTU)-versus-CFU standard curve, where y is equal to 49,937x (x is NTU, and y is conidia/ml) (12). The conidia were adjusted to the concentrations described below in sterile distilled water.
Table 1.
Characteristics of isolates and SNPs used to distinguish between A. flavus isolates in different VCGs
| VCGa | Isolate | SNP positionb |
Aflatoxin (ppm)c | Competitive ability on maize (rank out of 38)d |
||
|---|---|---|---|---|---|---|
| 1 | 2 | Seed infection | Sporulation | |||
| CG136 | 136 | T | A | 141 | NAe | NA |
| YV04 | Red-4-E | A | C | NDf | 4 | 5 |
| MR17 | MR2-17 | A | A | 33 | 38 | 36 |
| WM01 | Woolam-G | T | A | 59 | 3 | 16 |
| EB01 | E. Bernard B-c | A | A | 35 | 1 | 9 |
| DV901 | Danevang B-f | T | A | 138 | 37 | 33 |
| RB04 | Robstown B-d | A | A | 64 | 15 | 38 |
| MN902 | Moreman D-a | A | C | 24 | 35 | 13 |
In the text, isolates are referred to by the VCG to which they belong.
SNPs are located in a portion of the aflatoxin biosynthesis gene cluster.
Aflatoxin production during maize kernel infection was evaluated previously (43).
Previously, isolates from each of 38 VCGs were coinoculated with CG136 and ranked for competitive ability during both seed infection and sporulation on maize (43).
NA, not applicable. CG136 was the isolate against which the competitive abilities of the other isolates were assessed.
ND, none detected.
Growth media.
Each liter of full-strength (1×) Czapek's agar contained 0.5 g each of K2HPO4, KH2PO4, MgSO4 · 7H2O, and KCl; 30 g sucrose (88 mM); 3 g NaNO3 (35 mM); 1 ml micronutrient solution containing 0.11 g MnSO4, 0.5 g (NH4)6Mo7O24 · 4H2O, 10.0 g Fe2(SO4)3 · 6H2O, 17.6 g ZnSO4 · 7H2O, 0.7 g Na2B4O7 · 10H2O, and 0.3 g CuSO4 · 5H2O per liter; and 20 g Bacto agar. All reagents except sucrose and NaNO3 were added prior to autoclaving (121°C; 20 min); sucrose and NaNO3 solutions were filter sterilized (0.22-μm filter) and added just prior to pouring. To test the influences of sucrose and nitrate on A. flavus competition, either one or both of the nutrients were added at 1/10 (0.1×) the concentration present in 1× Czapek's agar (0.1× Czapek's agar contains 8.8 mM sucrose, 3.5 mM NaNO3; 0.1× sucrose contains 8.8 mM sucrose, 35 mM NaNO3; 0.1× nitrate contains 88 mM sucrose, 3.5 mM NaNO3). The concentrations of the other ingredients remained constant. Czapek's agar was also prepared with 2, 1, 0.5, 0.1, and 0.05 times the concentration of sucrose and NaNO3 in 1× Czapek's agar.
Asparagine agar contained asparagine as the sole carbon and nitrogen source and all of the Czapek's agar ingredients except sucrose and nitrate. The medium was autoclaved (121°C; 20 min), and filter-sterilized asparagine solution was added just prior to pouring. Full-strength (1×) and 0.1× asparagine agar contained 20 g (150 mM) and 2 g (15 mM) of asparagine, respectively.
For host competition treatments, intact, uniformly sized kernels of Pioneer hybrid 33B54 were surface sterilized by submersion in hot water (80°C; 45 s) as previously described (43). A subsample of kernels were plated and incubated on agar to confirm their ability to germinate and the lack of contaminating fungi.
Competition experiments.
Competition on a host was compared to competition on media with different nutrient concentrations. Interactions between A. flavus isolates YV04 and CG136 were examined on 1× and 0.1× Czapek's agar and on maize kernels. In order to evaluate the influences of contact sequence on competition, the seeding order was included as a treatment. Agar media were prepared as described above and seeded with 105 conidia of each isolate. Plates were spread evenly with either 105 conidia of CG136 in 50 μl followed 1 h later by 105 conidia of YV04 in 50 μl, YV04 followed 1 h later by CG136, or a mixture of the two isolates in 100 μl. Surface-sterilized kernels (5 g) were placed in 250-ml Erlenmeyer flasks sealed with gas-permeable BugStopper plugs (Whatman, Piscataway, NJ). The moisture content of the seed was determined with an HB43 Halogen Moisture Analyzer (Mettler Toledo, Columbus, OH), and 5 × 104 conidia of each isolate were inoculated sequentially or simultaneously as described above in the appropriate volume of water to bring seed moisture to 25%.
To test the influence of a range of nutrient concentrations on competition between YV04 and CG136, the isolates were cocultivated on plates of Czapek's agar prepared with 2, 1, 0.5, 0.1, and 0.05 times the concentration of sucrose and NaNO3 in 1× Czapek's agar, as described above. One hundred microliters of a suspension containing 105 conidia of both isolates (2 × 105 total) was spread on each plate.
Additional competition experiments were conducted with six different pairs of A. flavus isolates (Table 2). All isolate pairings were examined previously on maize and were selected for the current study to represent a range of competitive abilities during host infection and reproduction (Table 1) (26, 43, 44). As described above, plates were spread evenly with a 100-μl suspension containing 105 conidia of each isolate in the pair. To compare the influences of nutrient concentrations on isolates with and without the presence of a competing genotype, six isolate pairs were grown on 0.1× and 1× Czapek's agar, and in a parallel experiment, each of the isolates from the pairs were grown individually on the two media. To test the influence of the nutrient type (e.g., sucrose, nitrate, or asparagine) and concentration on A. flavus competition, the six pairs of isolates (Table 2) were cocultivated on 1× and 0.1× Czapek's agar, 0.1× sucrose agar, 0.1× nitrate agar, and 1× and 0.1× asparagine agar.
Table 2.
Predicted and measured percentages of conidia from paired A. flavus isolates cocultivated on 0.1× and 1× Czapek's agara
| Isolate pair | Isolates | % Conidia on Czapek's agar |
|||
|---|---|---|---|---|---|
| 0.1×b |
1×c |
||||
| Predictedd | Measurede | Predicted | Measured | ||
| 1 | CG136 | 47b | 47b | 58a | 65a |
| YV04 | 53x | 53x | 42y | 35y | |
| SE | 1 | 3 | 1 | 4 | |
| P valuef | 0.06 | 0.36 | <0.001 | 0.01 | |
| 2 | CG136 | 52b | 44c | 70a | 48c |
| MR17 | 48y | 56x | 30z | 52x | |
| SE | 1 | 2 | 3 | 1 | |
| P valuef | 0.22 | 0.05 | 0.002 | 0.009 | |
| 3 | YV04 | 55b | 54ab | 62a | 60a |
| MR17 | 45x | 46xy | 38y | 40y | |
| SE | 1 | 6 | 4 | 1 | |
| P valuef | 0.007 | 0.47 | 0.03 | <0.001 | |
| 4 | WM01 | 52a | 56a | 30b | 54a |
| MR17 | 48y | 44y | 70x | 46y | |
| SE | 4 | 3 | 2 | 3 | |
| P valuef | 0.69 | 0.08 | <0.001 | 0.08 | |
| 5 | EB01 | 54b | 47bc | 80a | 42c |
| DV901 | 46y | 53xy | 20z | 58x | |
| SE | 2 | 3 | 3 | 3 | |
| P valuef | 0.1 | 0.41 | <0.001 | 0.05 | |
| 6 | RB04 | 53b | 39c | 84a | 86a |
| MN902 | 47y | 61x | 16z | 14z | |
| SE | 1 | 1 | 3 | 1 | |
| P valuef | 0.23 | <0.001 | <0.001 | <0.001 | |
For each of the examined isolates, percentages followed by the same letter are not significantly different by Tukey's Studentized range test (n = 6).
Czapek's agar with 1/10 concentration (0.1×) of sucrose (8.8 mM) and nitrate (3.5 mM).
Full-strength (1×) Czapek's agar (88 mM sucrose, 35 mM nitrate).
Predicted percentages were calculated based on the quantities of conidia produced by each isolate grown individually on each medium (e.g., predicted % isolate A = 100% × isolate A conidia/[isolate A conidia + isolate B conidia]). Predicted percentages were calculated for each replicate (n = 6).
Measured percentages of total conidia from each isolate were measured by quantifying isolate-specific SNPs within conidial DNA using pyrosequencing (n = 6).
The percentages of the two isolates were compared with a paired t test (n = 6).
Quantification of conidia and isolation of DNA.
Following incubation (31°C; 7 days), the plates were flooded with 10 ml 0.1% Tween 80, and the conidia were dislodged with a sterile triangle spreader. Each plate was washed three times, and the washings were combined. Maize kernels were washed with 20 ml 0.1% Tween 80 followed by 20 ml water. The conidial suspensions were filtered through Miracloth, and the total numbers of conidia were measured using turbidity as described above. The agar plates were dried (60°C; 72 h) and then pulverized to a powder with an analytical mill (IKA Works, Wilmington, NC). Fungal DNA was isolated, using the FastDNA SPIN Kit and the FastPrep Instrument (MP Biomedicals, Santa Ana, CA), both from the total quantity of conidia and from a 200-mg subsample of the powdered agar. Fungal DNA from agar was assumed to be of mycelial origin.
PCR and pyrosequencing.
A quantitative pyrosequencing assay that distinguishes isolates using two SNPs was used to quantify the percentages of cocultivated isolates from conidia and mycelia (Table 1) (26, 43). The PCR conditions for primer pair CG136-Afl-F/CG136-Afl-R were described previously. The forward primer was biotinylated and high-performance liquid chromatography (HPLC) purified.
Amplicons were prepared for pyrosequencing as described previously (26, 43, 46). The biotinylated amplicons (40 μl) were immobilized on streptavidin-coated beads (GE Healthcare, Bio-Sciences AB, Uppsala, Sweden) and captured on the filter probes of the Vacuum Prep Tool (Qiagen, Valencia, CA) before being washed with 70% ethanol, denatured with 0.2 M NaOH, and washed with buffer (10 mM Tris-acetate, pH 7.6). Single-stranded amplicons were released into the wells of a pyrosequencing plate containing 0.5 μM sequencing primer CG136-Afl-S2 in 40 μl annealing buffer (Qiagen), and then the plate was heated (90°C; 10 min) and cooled to room temperature to anneal the primer.
Pyrosequencing was performed with a PSQ 96MA pyrosequencer using a PyroMark Gold Q96 reagent kit according to the manufacturer's instructions (Qiagen). The pyrosequencing reaction produces light proportional to the quantity of nucleotide incorporated at each position on the single-stranded template. Nucleotide percentages for isolate-specific SNPs were calculated using the allele quantification option of the PSQMA 2.1 software (Qiagen).
Experimental design and data analysis.
Each experiment was performed twice, with either three or four replicates, and the treatments utilized a randomized complete block design. The numbers of conidia were log transformed, and percentage data were arcsine transformed prior to analyses. Nutrient influences on the quantities of conidia produced and isolate percentages from either conidia or agar-embedded mycelia were each tested with an analysis of variance (ANOVA). For Czapek's agar treatments, the influences of sucrose, nitrate, and the interaction between the two nutrients on isolate competition were tested with a factorial ANOVA. Mean separations were performed following significant ANOVAs with Tukey's Studentized range test. Unless otherwise noted, trial-treatment interactions were not significant (P > 0.05), and data from the two trials were pooled. Correlations between dependent variables were performed on treatment means. Statistical analyses were performed using JMP 8.0.1 software (SAS Institute, Cary, NC).
RESULTS
Nutrients influence the success of competing A. flavus isolates.
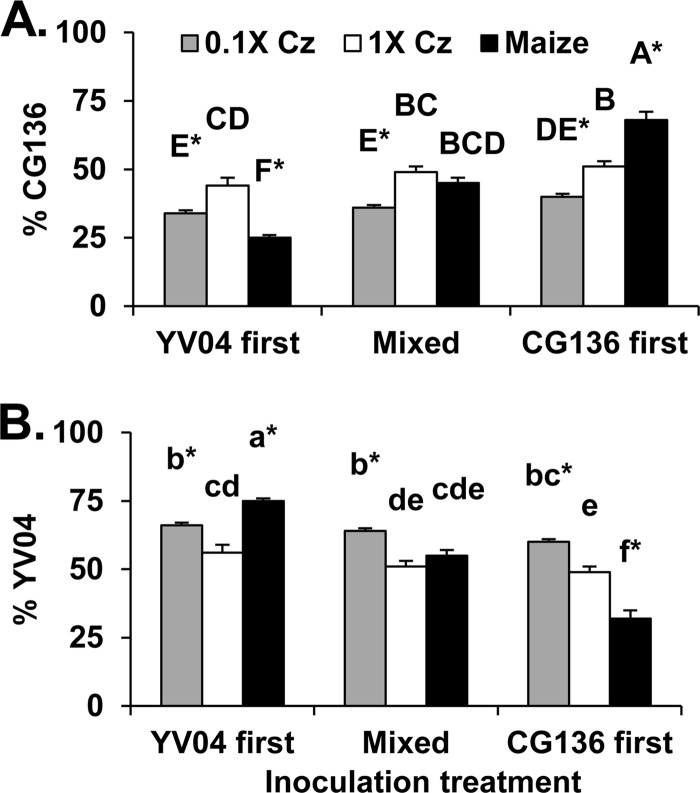
Percentages of cocultivated isolates CG136 and YV04 from conidia produced on either maize kernels, 1× Czapek's agar, or 0.1× Czapek's agar were influenced by both the substrate (P < 0.001) and the sequence of isolate contact with the substrate (P < 0.001), as well as an interaction between the two factors (P < 0.001) (Fig. 1). CG136 and YV04 were equally competitive on 1× Czapek's agar, but YV04 produced a greater percentage of the conidia than CG136 on 0.1× Czapek's agar, regardless of the order in which isolates were introduced to the substrate (Fig. 1). Contact with the host 1 h prior to the competing isolate conferred an advantage on both YV04 and CG136 during sporulation on maize kernels, but a similar advantage was not observed on either 0.1× or 1× Czapek's agar. When isolates were mixed prior to contact with either the host or agar media, the two isolates competed similarly during sporulation on maize and 1× Czapek's agar.
Fig 1.
Influences of nutrients and the sequence of isolate contact with the substrate on outcomes of competition between A. flavus isolates CG136 and YV04. The isolates were cocultivated on either full-strength Czapek's agar (1× Cz) (88 mM sucrose, 35 mM nitrate) or Czapek's agar with 1/10 strength sucrose (8.8 mM) and nitrate (3.5 mM) (0.1× Cz). The isolates were concurrently inoculated on surface-sterilized maize kernels. Equal quantities of the two isolates were applied either simultaneously (mixed) or sequentially with 1 h between isolates (YV04 first or CG136 first). The percentages of total A. flavus conidia produced by CG136 (A) and YV04 (B) are shown. Means labeled with the same letter are not significantly different by Tukey's Studentized range test (n = 8). The asterisks indicate percentages significantly different from 50%. The error bars indicate 1 standard error (SE).
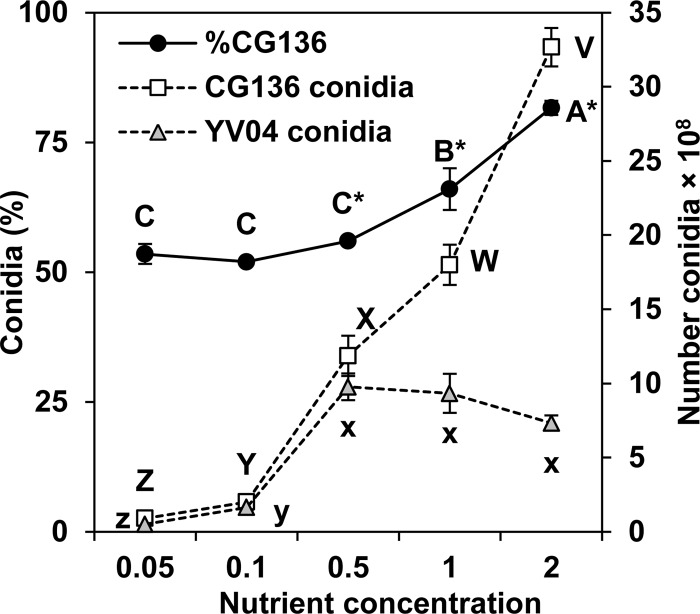
The quantities and percentages of conidia from cocultivated A. flavus isolates CG136 and YV04 varied with increasing sucrose and nitrate concentrations (Fig. 2). In these tests, sucrose and nitrate were varied simultaneously and maintained at the same ratio found in 1× Czapek's agar. Isolate percentages differed between the two trials (P = 0.04), but the influences of nutrient concentrations on competition were similar (trial-concentration interaction; P = 0.051). The results of a single trial are presented in Fig. 2. The percentages of total conidia from CG136 were correlated with the nutrient concentrations (r2 = 0.88; P < 0.001; n = 20), and CG136 comprised a greater percentage of the total conidia than YV04 when nitrate and sucrose concentrations were equal to or greater than half of those used in 1× Czapek's agar (17.5 and 44 mM, respectively). Yields of conidia for both CG136 and YV04 were directly proportional to an increase in nutrients, but sporulation by YV04 leveled out over the three highest concentrations (Fig. 2).
Fig 2.
Outcomes of competition between A. flavus isolates CG136 and YV04 during sporulation on Czapek's agar with variable concentrations of carbon (sucrose) and nitrogen (nitrate). Nutrient concentrations increase from left to right along the x axis and are multiples of the sucrose (88 mM) and nitrate (35 mM) in 1× Czapek's agar. The experiment was performed twice with similar results, and the results of a single trial are presented. Conidia (%) (primary y axis), the percentage of total conidia from CG136; number conidia (secondary y axis), the quantity of conidia produced by either CG136 or YV04 during cocultivation on each of the agar media. Means labeled with the same letter are not significantly different by Tukey's Studentized range test (n = 4). The asterisks indicate percentages significantly different from 50%. The error bars indicate 1 SE.
Responses of individual isolates to nutrients do not always predict the outcomes of competition.
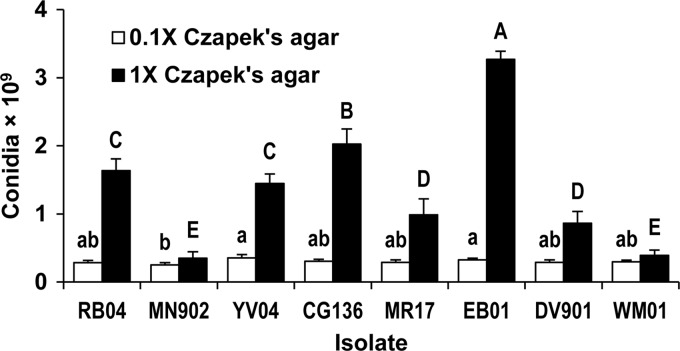
Isolates grown individually varied in production of conidia on 0.1× (P = 0.003) and 1× (P < 0.001) Czapek's agar (Fig. 3). Predicted isolate percentages during cocultivation, based on sporulation by isolates cultivated individually, are compared to measured percentages in Table 2. The quantities of conidia produced by isolates grown individually did not always reflect the ability of the isolates to compete during cocultivation. For example, when grown individually, RB04 and MN902 produced similar quantities of conidia on 0.1× Czapek's agar (Fig. 3), but when cocultivated on this medium, MN902 outcompeted RB04 (Table 2). On 1× Czapek's agar, EB01 produced almost four times as many conidia as DV901 (Fig. 3), but DV901 comprised a greater percentage of conidia when isolates were cocultivated (Table 2). In contrast, sporulation by CG136 and YV04 grown individually on 0.1× and 1× Czapek's agar (Fig. 3) predicted outcomes of competition between these two isolates on each type of medium (Table 2).
Fig 3.
Sporulation by A. flavus isolates grown individually on 0.1× (8.8 mM sucrose, 3.5 mM nitrate) and 1× (88 mM sucrose, 35 mM nitrate) Czapek's agar. Means labeled with the same letter (lowercase, 0.1× Czapek's agar; uppercase, 1× Czapek's agar) are not significantly different by Tukey's Studentized range test (n = 6). The error bars indicate 1 SE.
Nutrients influence competition in an isolate-specific manner.
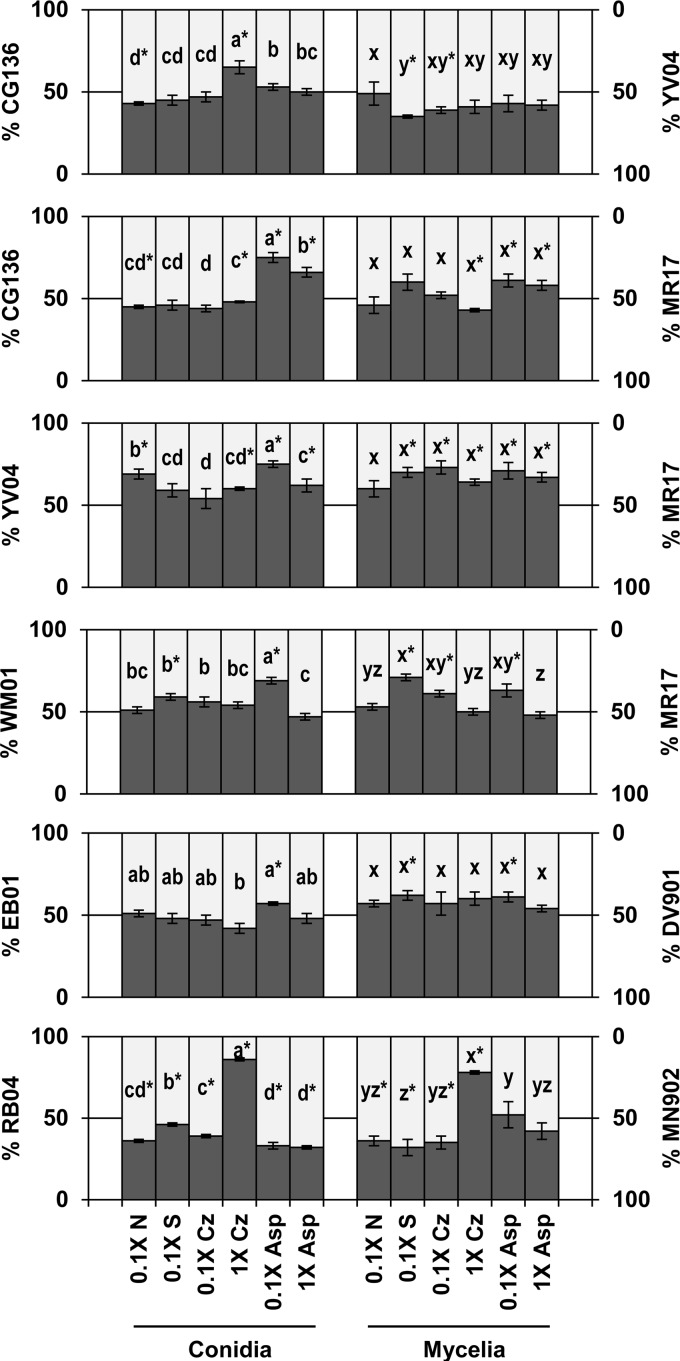
The six isolate pairs differed in their responses to sucrose, nitrate, and asparagine (Table 3 and Fig. 4). For example, isolate percentages from conidia during cocultivation of EB01 and DV901 were not influenced by changes in sucrose or nitrate, but they were influenced by a 10-fold increase in asparagine (Table 3). When cocultivated with MR17, the percentages of conidia from CG136 increased when sucrose and nitrate concentrations were increased (0.1× versus 1× Czapek's agar) but decreased with a 10-fold increase in asparagine (Table 3 and Fig. 4). MR17 responded similarly to 10-fold increases in nutrients whether it was cocultivated with CG136 or YV04 (Fig. 4) (cocultivated isolate-nutrient interaction: sucrose, P = 0.26; nitrate, P = 0.50; asparagine, P = 0.26). In contrast, CG136 responded differently to changes in nutrient concentrations when cocultivated with YV04 than when cocultivated with MR17 (cocultivated isolate-nutrient interaction: sucrose, P < 0.001; nitrate, P < 0.001; asparagine, P = 0.02), and YV04 responded differently to nutrients when cocultivated with CG136 than when cocultivated with MR17 (cocultivated isolate-nutrient concentration interaction: sucrose, P < 0.001; nitrate, P < 0.001; asparagine, P < 0.001) (Fig. 4).
Table 3.
Statistical significance of influences of nutrients on proportions of conidia and mycelia produced by A. flavus isolates during cocultivation on agar media with varying sources and concentrations of carbon and nitrogen
| Isolate pair |
P valuea |
|||||||
|---|---|---|---|---|---|---|---|---|
| Sucroseb |
Nitratec |
Sucrose-nitrated |
Asparaginee |
|||||
| Conidia | Mycelia | Conidia | Mycelia | Conidia | Mycelia | Conidia | Mycelia | |
| CG136/YV04 | <0.001 | 0.028 | <0.001 | 0.07 | <0.001 | 0.63 | 0.14 | 0.77 |
| CG136/MR17 | 0.009 | 0.009 | 0.01 | 0.69 | 0.86 | 0.1 | <0.001 | 0.56 |
| YV04/MR17 | <0.001 | 0.03 | 0.046 | 0.99 | <0.001 | 0.43 | 0.001 | 0.4 |
| WM01/MR17 | 0.05 | <0.001 | 0.36 | 0.16 | 0.81 | 0.007 | <0.001 | 0.02 |
| EB01/DV901 | 0.91 | 0.66 | 0.18 | 0.69 | 0.08 | 0.99 | 0.01 | 0.04 |
| RB04/MN902 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.78 | 0.19 |
Probability that the percentage of mycelia or conidia for each isolate changed in response to a 10-fold increase in the indicated nutrient(s) (ANOVA; n = 6).
Sucrose was the carbon source in Czapek's agar. The sucrose was adjusted to either 8.8 or 88 mM.
Nitrate was the nitrogen source in Czapek's agar. The nitrate was adjusted to either 3.5 or 35 mM.
The interaction between sucrose (8.8 or 88 mM) and nitrate (3.5 or 35 mM) concentrations in Czapek's agar was tested with a factorial analysis of variance.
Asparagine, the sole carbon and nitrogen source in asparagine agar, was adjusted to either 15 or 150 mM.
Fig 4.
Outcomes of competition between paired A. flavus isolates during cocultivation on agar media with various sources and concentrations of carbon and nitrogen. For Czapek's (Cz) treatments, sucrose (S) and nitrate (N) were the carbon and nitrogen sources, respectively (1× Cz, 88 mM sucrose, 35 mM nitrate; 0.1× Cz, 8.8 mM sucrose, 3.5 mM nitrate; 0.1× N, 88 mM sucrose, 3.5 mM nitrate; 0.1× S, 8.8 mM sucrose, 35 mM nitrate). Asparagine (Asp) treatments contained asparagine as both the sole carbon and nitrogen sources (1× Asp, 150 mM asparagine; 0.1× Asp, 15 mM asparagine). The percentages of isolates from either conidia or mycelia are indicated along the y axes. For each pair, the isolate indicated on the primary y axis is represented by dark-gray bars and the isolate indicated on the secondary y axis is represented by light-gray bars. Percentages labeled with the same letter are not significantly different by Tukey's Studentized range test (n = 6). The agar was seeded with equal quantities of each isolate (50%), and the asterisks indicate isolate percentages significantly different from 50% following cocultivation. The error bars indicate 1 SE.
Nutrient influences on competition during sporulation do not predict competition during mycelial growth.
There was a lack of correlation between the outcomes of competition during sporulation and mycelial growth for all pairings and nutrient media, except for Czapek's agar with 0.1× nitrate (Table 4). The extents to which 10-fold increases in nutrients influenced competition between cocultivated isolates differed between production of conidia and mycelia (Table 3 and Fig. 4). For example, when cocultivated with MR17, the percentages of total conidia from WM01 did not vary on Czapek's agar in response to changes in sucrose concentrations, but the percentages of total mycelia from WM01 were inversely proportional to increasing sucrose concentrations (Fig. 4). In certain other cases, only isolate percentages from conidia responded to changes in nutrient concentrations (e.g., YV04 with MR17).
Table 4.
Coefficients of determination (r2) and probabilities (P) for relationships of proportions of conidia to proportions of mycelia produced by A. flavus isolates during cocultivation for all nutrient treatments to which an isolate pair was subjected and for all isolate pairs cultivated on each nutrient treatment
| Category | r2 | P value |
|---|---|---|
| Isolate paira | ||
| CG136/YV04 | 0.007 | 0.87 |
| CG136/MR17 | 0.37 | 0.20 |
| YV04/MR17 | 0.08 | 0.59 |
| WM01/MR17 | 0.49 | 0.12 |
| EB01/DV901 | 0.02 | 0.79 |
| RB04/MN902 | 0.65 | 0.05 |
| Nutrient treatmentb | ||
| 0.1× nitrate | 0.77 | 0.02 |
| 0.1× sucrose | 0.58 | 0.08 |
| 0.1× Czapek's | 0.65 | 0.05 |
| 1× Czapek's | 0.32 | 0.24 |
| 0.1× asparagine | 0.47 | 0.13 |
| 1× asparagine | 0.61 | 0.07 |
For each pair of cocultivated isolates, the percentages of conidia and mycelia produced by each isolate were quantified on six agar media, each representing a different nutrient treatment; the coefficients of determination reflect the relationships of mycelial values to conidial values (n = 6) for each isolate pair.
The coefficients of determination reflect the relationships of mycelial values to conidial values (n = 6) in each nutrient treatment. The asparagine treatments contained asparagine as the sole source of both carbon and nitrogen (0.1×, 15 mM asparagine; 1×, 150 mM asparagine). For Czapek's treatments, sucrose and nitrate were the carbon and nitrogen source, respectively (0.1× nitrate, 88 mM sucrose and 3.5 mM nitrate; 0.1× sucrose, 8.8 mM sucrose and 35 mM nitrate; 0.1× Czapek's agar, 8.8 mM sucrose and 3.5 mM nitrate; 1× Czapek's agar, 88 mM sucrose and 35 mM nitrate).
DISCUSSION
Genetically diverse A. flavus individuals coexist in heterogeneous nutrient environments, including soils (15, 47), plant debris (16, 48, 49), and plant or animal hosts (15, 50). Competition for limited resources, including nutrients, influences the composition and size of fungal populations (51). In the current study, competition was influenced by the type and concentration of nutrients within chemically defined agar media. A. flavus genotypes varied in response to specific nutrients, suggesting differentiation among genotypes in the ability to compete within diverse nutrient environments. Divergent adaptation to nutrient environments provides a mechanism for niche partitioning of competing A. flavus genotypes within the environment (52, 53). Furthermore, differential allocation of resources to production of mycelia or conidia in response to both nutrient conditions and competition within various nutrient environments suggests diverse ecological strategies among A. flavus genotypes.
Innate differences among individual A. flavus isolates in response to nutrients in the absence of competition explained some, but not all, of the competitive differences among isolates (Fig. 3 and Table 2), suggesting several mechanisms are involved in nutrient modulation of competition. Certain genotypes of A. flavus, such as YV04, may have an innate ability to sporulate under nutrient-limiting conditions, a characteristic that confers an advantage on that genotype whether or not competitors are present. Modulation of indirect (e.g., nutrient sequestration) (51) and direct (e.g., antagonism or signaling) interactions (54) between genotypes by the nutrient environment may explain additional portions of the observed nutrient influences on competition. For example, MN902, which outcompeted RB04 during sporulation on 0.1× Czapek's agar though the two isolates produced similar quantities of conidia when grown individually (Fig. 3 and Table 2), may have superior ability to capture and efficiently utilize resources in environments where nutrients are scarce. More efficient capture and utilization of limited nutrients may allow survival and superior competitive saprophytic ability in low-nutrient environments, such as within soil matrices (33). Direct interactions between genotypes that either inhibit or stimulate growth and reproduction during competition were also observed. For example, when grown individually on 1× Czapek's agar, EB01 produced more conidia while DV901 produced fewer (Fig. 3); however, during cocultivation of these strains on 1× Czapek's agar, DV901 produced the greater percentage of the total conidia (Table 2). Since the isolates were equally competitive on the low-nutrient medium, the advantage conferred on DV901 on 1× Czapek's agar may indicate an antagonistic interaction rather than superior capture of limited nutrients. One possible mechanism is production of secondary metabolites that inhibit sporulation by competitors during competition for resources on high-nutrient media (55). Further work is needed to determine potential roles of fungal metabolites in the modulation of intraspecific competition by nutrient environments.
A. flavus isolates varied in competitive ability in response to both qualitative and quantitative changes in nutrients, indicating differential adaptation to nutrient environments. For example, competition between some isolate pairs (e.g., RB04/MN902) was influenced by nitrate and sucrose medium enrichment, but not by the addition of carbon and nitrogen in the form of asparagine. Conversely, competition between EB01 and DV901 was influenced by asparagine enrichment, but not by the addition of nitrate or sucrose. Thus, specific nutrient environments appear to support the success of specific A. flavus genotypes, and this specificity is not solely a function of nutrient richness. Previously, isolates of A. flavus and related fungi were found to differ in their abilities to utilize nutrients (30, 52, 53), but the influences of nutrient preference on competitive interactions were not examined.
Genotype-specific responses to nutrient environments may influence competition during host infection. Competitive abilities vary among A. flavus genotypes in a host-dependent manner (44), and since hosts vary in nutrient composition (56–58), these differences may influence competitive interactions among A. flavus genotypes during invasion of host tissues and sporulation. The sequence of contact with the substrate had little influence on competition between cocultivated isolates on Czapek's agar. However, during maize kernel infection, an advantage is conferred on the first isolate to contact the host (Fig. 1) (26). The dynamics of competition for nutrients may be different on homogeneous substrates (e.g., agar medium) than on the heterogeneous nutrient environments present in host tissues. The first isolate to contact the host surface may quickly sequester available nutrients, leaving fewer nutrients for competitors.
In the current study, competition during sporulation was quantified independently from competition resulting from mycelial interaction, and competitive advantage during sporulation was not correlated with success during mycelial growth (Table 4). Differential allocation of resources between vegetative growth (mycelia) and dispersal propagules (conidia) in response to diverse nutrient environments may allow divergent adaptive advantages to A. flavus genotypes. The extent to which A. flavus produces vegetative mycelia or conidia is influenced by both abiotic (e.g., pH, nutrients, and temperature) (23, 37, 42, 59) and biotic (e.g., host and microbial community) factors (44). A. flavus isolates differ in innate ability to produce conidia (23), but the results of the current study suggest that the extent to which resources are allocated to mycelia or conidia is influenced by both nutrient availability and the presence of competing genotypes. Mycelia ramify host tissues/substrates and sequester nutrients, whereas conidia allow dispersal (60, 61). In the environment, allocation of resources to sporulation when nutrients are limited may be an adaptive trait that allows rapid spread to new substrates or hosts. In contrast, vegetative growth provides an advantage in capture and defense of nutrients. However, not all A. flavus isolates respond similarly to either nutrients or competition, indicating differential adaptation and a potential mechanism for niche partitioning that allows the coexistence of multiple genetic types within the environment (15, 16, 62).
Improved understanding of both mechanisms through which A. flavus genotypes adapt to nutrient environments and the dominant nutrients supporting shifts in population structure may allow the development of improved strategies and technologies for aflatoxin mitigation. Nutrient environments associated with different crops, soils, and cropping practices may in part explain the effects of crop rotation and other agronomic practices on populations of aflatoxin-producing fungi (63–65). Nutrient influences on sporulation demonstrate a mechanism through which nutrient environments may shape populations of aflatoxin-producing fungi. Characterizing nutrient influences on specific genotypes of A. flavus may provide criteria for selecting optimal nonaflatoxigenic biocontrol strains that are strong competitors on target hosts and in soil matrices. Though additional work is needed to verify the role of nutrients in A. flavus soil population dynamics, the current study suggests some genotypes may be adapted to low-nutrient environments, a characteristic that would confer competitive advantage in soils (32–34) and, potentially, result in long-term persistence in the environment.
ACKNOWLEDGMENTS
We thank the members of the Cotty laboratory, especially Lauren Price, Stephen Ngo, and Amanda Lee, for technical assistance with this work.
This work was supported by the Agricultural Research Service, U.S. Department of Agriculture, CRIS project 5347-42000-020-00D.
The use of trade, firm, or corporation names in this paper does not constitute official endorsement or approval by the U.S. Department of Agriculture or the Agricultural Research Service.
Footnotes
Published ahead of print 21 December 2012
REFERENCES
- 1. St Leger RJ, Joshi L, Roberts DW. 1997. Adaptation of proteases and carbohydrates of saprophytic, phytopathogenic and entomopathogenic fungi to the requirements of their ecological niches. Microbiology 143:1983–1992 [DOI] [PubMed] [Google Scholar]
- 2. Hedayati MT, Pasqualotto AC, Warn PA, Bowyer P, Denning DW. 2007. Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology 153:1677–1692 [DOI] [PubMed] [Google Scholar]
- 3. Shephard GS. 2008. Risk assessment of aflatoxins in food in Africa. Food Addit. Contam. 25:1246–1256 [DOI] [PubMed] [Google Scholar]
- 4. Wu F, Khlangwiset P. 2010. Evaluating the technical feasibility of aflatoxin risk reduction strategies in Africa. Food Addit. Contam. 27:658–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Probst C, Njapau H, Cotty PJ. 2007. Outbreak of an acute aflatoxicosis in Kenya in 2004: identification of the causal agent. Appl. Environ. Microbiol. 73:2762–2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Khlangwiset P, Shephard GS, Wu F. 2011. Aflatoxins and growth impairment: a review. Crit. Rev. Toxicol. 41:740–755 [DOI] [PubMed] [Google Scholar]
- 7. Wu F, Khlangwiset P. 2010. Health economic impacts and cost-effectiveness of aflatoxin-reduction strategies in Africa: case studies in biocontrol and post-harvest interventions. Food Addit. Contam. 27:496–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rustom IYS. 1997. Aflatoxin in food and feed: occurrence, legislation and inactivation by physical methods. Food Chem. 59:57–67 [Google Scholar]
- 9. Robens J, Cardwell FK. 2003. The costs of mycotoxin management to the U. S. A.: management of aflatoxins in the United States. J. Toxicol. 22:139–152 [Google Scholar]
- 10. Wu F, Liu Y, Bhatnagar D. 2008. Cost-effectiveness of aflatoxin control method: economic incentives. Toxin Rev. 27:203–225 [Google Scholar]
- 11. Cotty PJ, Probst C, Jaime-Garcia R. 2008. Etiology and management of aflatoxin contamination, p 287–299 In Leslie JF, Bandyopadhyay R, Visconti A. (ed), Mycotoxins: detection methods, management, public health and agricultural trade. CAB International, Oxfordshire, United Kingdom [Google Scholar]
- 12. Probst C, Schulthess F, Cotty PJ. 2010. Impact of Aspergillus section Flavi community structure on the development of lethal levels of aflatoxins in Kenyan maize (Zea mays). J. Appl. Microbiol. 108:600–610 [DOI] [PubMed] [Google Scholar]
- 13. Leslie JF. 1993. Fungal vegetative compatibility. Annu. Rev. Phytopathol. 31:127–150 [DOI] [PubMed] [Google Scholar]
- 14. Papa KE. 1986. Heterokaryon incompatibility in Aspergillus flavus. Mycologia 78:98–101 [Google Scholar]
- 15. Bayman P, Cotty PJ. 1991. Vegetative compatibility and genetic diversity in the Aspergillus flavus population of a single field. Can. J. Bot. 69:1707–1711 [Google Scholar]
- 16. Horn BW, Greene RL. 1995. Vegetative compatibility within populations of Aspergillus flavus, A. parasiticus, and A. tamarii from a peanut field. Mycologia 87:324–332 [Google Scholar]
- 17. Pildain MB, Vaamonde G, Cabral D. 2004. Analysis of population structure of Aspergillus flavus from peanut based on vegetative compatibility, geographic origin, mycotoxin and sclerotia production. Int. J. Food Microbiol. 93:31–40 [DOI] [PubMed] [Google Scholar]
- 18. Grubisha L, Cotty PJ. 2010. Genetic isolation among sympatric vegetative compatibility groups of the aflatoxin-producing fungus Aspergillus flavus. Mol. Ecol. 19:269–280 [DOI] [PubMed] [Google Scholar]
- 19. Bayman P, Cotty PJ. 1993. Genetic diversity in Aspergillus flavus: association with aflatoxin production and morphology. Can. J. Bot. 71:23–31 [Google Scholar]
- 20. Horn BW, Greene RL, Sobolev VS, Dorner JW, Powell JH, Layton RC. 1996. Association of morphology and mycotoxin production with vegetative compatibility groups in Aspergillus flavus, A. parasiticus, and A. tamarii. Mycologia 88:574–587 [Google Scholar]
- 21. Novas MV, Cabral D. 2002. Association of mycotoxin and sclerotia production with compatibility groups in Aspergillus flavus from peanut in Argentina. Plant Dis. 86:215–219 [DOI] [PubMed] [Google Scholar]
- 22. Schroeder HW, Boller RA. 1973. Aflatoxin production by species and strains of the Aspergillus flavus group isolated from field crops. Appl. Microbiol. 25:885–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cotty PJ. 1989. Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology 79:808–814 [Google Scholar]
- 24. Cotty PJ, Antilla L, Wakelyn PJ. 2007. Competitive exclusion of aflatoxin producers: farmer driven research and development, p 241–253 In Vincent C, Goettel N, Lazarovits G. (ed), Biological control: a global perspective. CAB International, Oxfordshire, United Kingdom [Google Scholar]
- 25. Dorner JW, Cole RJ, Connick WJ, Daigle DJ, McGuire MR, Shasha BS. 2003. Evaluation of biological control formulations to reduce aflatoxin contamination in peanuts. Biol. Control 26:318–324 [Google Scholar]
- 26. Mehl HL, Cotty PJ. 2011. Influence of the host contact sequence on the outcome of competition among Aspergillus flavus isolates during host tissue invasion. Appl. Environ. Microbiol. 77:1691–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cotty PJ, Bayman P. 1993. Competitive exclusion of a toxigenic strain of Aspergillus flavus by an atoxigenic strain. Phytopathology 83:1283–1287 [Google Scholar]
- 28. Horn BW, Greene RL, Dorner JW. 2000. Inhibition of aflatoxin B1 production by Aspergillus parasiticus using nonaflatoxigenic strains: role of vegetative compatibility. Biol. Control 17:147–154 [Google Scholar]
- 29. St Leger RJ, Screen SE, Shams-Pirzadeh B. 2000. Lack of host specialization in Aspergillus flavus. Appl. Environ. Microbiol. 66:320–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mellon JE, Cotty PJ, Dowd MK. 2007. Aspergillus flavus hydrolases: their roles in pathogenesis and substrate utilization. Appl. Microbiol. Biotechnol. 77:497–504 [DOI] [PubMed] [Google Scholar]
- 31. Kothary MH, Chase T, Jr, Macmillan JD. 1984. Correlation of elastase production by some strains of Aspergillus fumigatus with ability to cause pulmonary invasive aspergillosis in mice. Infect. Immun. 43:320–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Celar F. 2003. Competition for ammonium and nitrate forms of nitrogen between some phytopathogenic and antagonistic soil fungi. Biol. Control 28:19–24 [Google Scholar]
- 33. Couteaudier Y, Alabouvette C. 1990. Quantitative comparison of Fusarium oxysporum competitiveness in relation to carbon utilization. FEMS Microbiol. Lett. 74:261–267 [Google Scholar]
- 34. Baker R. 1968. Mechanisms of biological control of soil-borne pathogens. Annu. Rev. Phytopathol. 6:263–294 [Google Scholar]
- 35. Pass T, Griffin GJ. 1972. Exogenous carbon and nitrogen requirements for conidial germination by Aspergillus flavus. Can. J. Microbiol. 18:1453–1461 [DOI] [PubMed] [Google Scholar]
- 36. Osherov N, May GS. 2001. The molecular mechanisms of conidial germination. FEMS Microbiol. Lett. 199:153–160 [DOI] [PubMed] [Google Scholar]
- 37. Hesseltine CW, Sorenson WG, Smith M. 1970. Taxonomic studies of the aflatoxin-producing strains in the Aspergillus flavus group. Mycologia 62:123–132 [PubMed] [Google Scholar]
- 38. Payne GA, Hagler WM., Jr 1983. Effect of specific amino acids on growth and aflatoxin production by Aspergillus parasiticus and Aspergillus flavus in defined media. Appl. Environ. Microbiol. 46:805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thapar GS. 1988. Metabolic behaviour of aflatoxin producing strain and non-toxigenic strain of Aspergillus flavus to different sources of nitrogen and glucose concentration. Mycopathologia 102:9–12 [DOI] [PubMed] [Google Scholar]
- 40. Klich MA. 2007. Aspergillus flavus: the major producer of aflatoxin. Mol. Plant Pathol. 8:713–722 [DOI] [PubMed] [Google Scholar]
- 41. Fanelli C, Fabbri AA, Passi S. 1980. Growth requirements and lipid metabolism of Aspergillus flavus. Br. Mycol. Soc. 75:371–375 [Google Scholar]
- 42. Olutiola PO. 1976. Some environmental and nutritional factors affecting growth and sporulation of Aspergillus flavus. Br. Mycol. Soc. 66:131–136 [Google Scholar]
- 43. Mehl HL, Cotty PJ. 2010. Variation in competitive ability among isolates of Aspergillus flavus from different vegetative compatibility groups during maize infection. Phytopathology 100:150–159 [DOI] [PubMed] [Google Scholar]
- 44. Mehl HL, Cotty PJ. 2010. Host and life strategy adaptations mediate competition among isolates of Aspergillus flavus. Phytopathology 100:S81. [DOI] [PubMed] [Google Scholar]
- 45. Cotty PJ. 1994. Influence of field application of an atoxigenic strain of Aspergillus flavus on the populations of A. flavus infecting cotton bolls and on the aflatoxin content of cottonseed. Phytopathology 84:1270–1277 [Google Scholar]
- 46. Das MK, Ehrlich KC, Cotty PJ. 2008. Use of pyrosequencing to quantify incidence of a specific Aspergillus flavus strain within complex fungal communities associated with commercial cotton crops. Phytopathology 98:282–288 [DOI] [PubMed] [Google Scholar]
- 47. Horn BW, Dorner JW. 1999. Regional differences in production of aflatoxin B1 and cyclopiazonic acid by soil isolates of Aspergillus flavus along a transect within the United States. Appl. Environ. Microbiol. 65:1444–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jaime-Garcia R, Cotty PJ. 2004. Aspergillus flavus in soils and corncobs in South Texas: implications for management of aflatoxins in corn-cotton rotations. Plant Dis. 88:1366–1371 [DOI] [PubMed] [Google Scholar]
- 49. Nesci A, Etcheverry M. 2002. Aspergillus section Flavi populations from field maize in Argentina. Lett. Appl. Microbiol. 34:343–348 [DOI] [PubMed] [Google Scholar]
- 50. Rudramurthy SM, de Valk HA, Chakrabarti A, Meis JFGM, Klaassen CHW. 2011. High resolution genotyping of clinical Aspergillus flavus isolates from India using microsatellites. PLoS One 6:e16086 doi:10.1371/journal.pone.0016086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lockwood JL. 1992. Exploitation competition, p 243–263 In Carroll GC, Wicklow DT. (ed), The fungal community: its organization and role in the ecosystem, 2nd ed Marcel Dekker, New York, NY [Google Scholar]
- 52. Lee HB, Magan N. 1999. Environmental factors and nutritional utilization patterns affect niche overlap indices between Aspergillus ochraceus and other spoilage fungi. Lett. Appl. Microbiol. 28:300–304 [DOI] [PubMed] [Google Scholar]
- 53. Giorni P, Magan N, Battilani P. 2009. Environmental factors modify carbon nutritional patterns and niche overlap between Aspergillus flavus and Fusarium verticillioides strains from maize. Int. J. Food Microbiol. 130:213–218 [DOI] [PubMed] [Google Scholar]
- 54. Wicklow DT. 1992. Interference competition, p 265–274 In Carroll GC, Wicklow DT. (ed), The fungal community: its organization and role in the ecosystem, 2nd ed Marcel Dekker, New York, NY [Google Scholar]
- 55. Losada L, Ajayi O, Frisvad JC, Yu J, Nierman WC. 2009. Effect of competition on the production and activity of secondary metabolites in Aspergillus species. Med. Mycol. 47(Suppl. 1):S88–S96 [DOI] [PubMed] [Google Scholar]
- 56. Mellon JE, Cotty PJ, Dowd MK. 2000. Influence of lipids with and without other cottonseed reserve materials on aflatoxin B1 production by Aspergillus flavus. J. Agric. Food Chem. 48:3611–3615 [DOI] [PubMed] [Google Scholar]
- 57. Mellon JE, Dowd MK, Cotty PJ. 2005. Substrate utilization by Aspergillus flavus in inoculated whole corn kernels and isolated tissues. J. Agric. Food Chem. 53:2351–2357 [DOI] [PubMed] [Google Scholar]
- 58. Solomon PS, Tan KC, Oliver RP. 2003. The nutrient supply of pathogenic fungi; a fertile field for study. Mol. Plant Pathol. 4:203–210 [DOI] [PubMed] [Google Scholar]
- 59. Cotty P. 1988. Aflatoxin and sclerotial production by Aspergillus flavus: influence of pH. Phytopathology 78:1250–1253 [Google Scholar]
- 60. Amaike S, Keller NP. 2011. Aspergillus flavus. Annu. Rev. Phytopathol. 49:107–133 [DOI] [PubMed] [Google Scholar]
- 61. Cotty PJ, Bayman P, Egel DS, Elias KS. 1994. Agriculture, aflatoxins and Aspergillus, p 1–27 In Powell KA, Renwick A, Peverdy JF. (ed), The genus Aspergillus: from taxonomy and genetics to industrial application. Plenum Press, New York, NY [Google Scholar]
- 62. Fitt BD, Huang YJ, van den Bosch F, West JS. 2006. Coexistence of related pathogen species on arable crops in space and time. Annu. Rev. Phytopathol. 44:163–182 [DOI] [PubMed] [Google Scholar]
- 63. Jaime-Garcia R, Cotty PJ. 2006. Spatial relationships of soil texture and crop rotation to Aspergillus flavus community structure in South Texas. Phytopathology 96:599–607 [DOI] [PubMed] [Google Scholar]
- 64. Jaime-Garcia R, Cotty PJ. 2010. Crop rotation and soil temperature influence the community structure of Aspergillus flavus in soil. Soil Biol. Biochem. 42:1842–1847 [Google Scholar]
- 65. Griffin GJ, Garren KH, Taylor JD. 1981. Influence of crop rotation and minimum tillage on the population of Aspergillus flavus group in peanut field soil. Plant Dis. 65:898–900 [Google Scholar]