Abstract
Three cohorts of farmed yellowtail kingfish (Seriola lalandi) from South Australia were examined for Chlamydia-like organisms associated with epitheliocystis. To characterize the bacteria, 38 gill samples were processed for histopathology, electron microscopy, and 16S rRNA amplification, sequencing, and phylogenetic analysis. Microscopically, the presence of membrane-enclosed cysts was observed within the gill lamellae. Also observed was hyperplasia of the epithelial cells with cytoplasmic vacuolization and fusion of the gill lamellae. Transmission electron microscopy revealed morphological features of the reticulate and intermediate bodies typical of members of the order Chlamydiales. A novel 1,393-bp 16S chlamydial rRNA sequence was amplified from gill DNA extracted from fish in all cohorts over a 3-year period that corresponded to the 16S rRNA sequence amplified directly from laser-dissected cysts. This sequence was only 87% similar to the reported “Candidatus Piscichlamydia salmonis” (AY462244) from Atlantic salmon and Arctic charr. Phylogenetic analysis of this sequence against 35 Chlamydia and Chlamydia-like bacteria revealed that this novel bacterium belongs to an undescribed family lineage in the order Chlamydiales. Based on these observations, we propose this bacterium of yellowtail kingfish be known as “Candidatus Parilichlamydia carangidicola” and that the new family be known as “Candidatus Parilichlamydiaceae.”
INTRODUCTION
The world's demand for seafood far exceeds the current supply (1), and significant increases in research and development of new species for aquaculture have been initiated in response to this demand. The yellowtail kingfish (YTK), Seriola lalandi, is an example of one such new aquaculture species, and for the last decade, efforts have focused on developing a commercial aquaculture industry for the species in Australia and New Zealand. Nine species are currently recognized in the genus Seriola, and three are cultured commercially around the world: S. dumerili, S. quinqueradiata, and S. lalandi (2–4). However, the establishment and commercial production of YTK aquaculture, although considered successful, has been beset by an increased incidence of disease within the cultured populations (5–8).
The order Chlamydiales is a constantly evolving taxonomic classification, and in the early 1990s, it was considered to be a closely related group of only four species of bacteria, all contained within one genus. However, with the increased use of molecular techniques and phylogenetic analysis, the Chlamydiales have undergone considerable revision. In 1999, classification rules for Chlamydiales bacteria were reviewed, which subsequently resulted in significant changes in the taxonomic classification of organisms within the order (9). These proposed rules were based upon the 16S rRNA sequence similarity, and it was subsequently accepted that a sequence similarity of <95% would constitute a new genus, a sequence similarity of <90% would constitute a new family, and a sequence similarity of <80% would constitute a different order (9).
Chlamydia-like organisms (CLO) are pathogens that are known to cause disease and mortality in a wide range of species, including humans, sheep, cattle, koalas, bats, birds, insects, and, most recently, fish (10), including the cutaneous and branchial infection referred to as epitheliocystis (11). Although epitheliocystis is often benign, it can cause epithelial hyperplasia and inflammation of infected tissues, resulting in significantly reduced growth (12, 13). In severe cases of hyperinfection, the cyst-like branchial lamellar lesions can result in increased lamellar fusion with consequent respiratory distress and death (14, 15). Epitheliocystis outbreaks have been reported in many aquaculture species, including barramundi, Lates calcarifer (16); white sturgeon, Acipenser transmontanus (17); silver perch, Bidyanus bidyanus (18); Atlantic salmon, Salmo salar (19); red sea bream, Pagrus major (20); carp, Cyprinus carpio (21); and yellowtail kingfish, S. lalandi (6). It has been suggested that increased water temperature increases the risk of infection and the incidence of mortality, although additional experimental work is required (11). Many of the reported losses in aquaculture attributed to epitheliocystis occur in the larval or juvenile stage (11, 19, 20, 22). Although this condition has been reported for over 80 years, the causative agent or agents of epitheliocystis have yet to be successfully cultured in vitro. Only recently have three of the etiological agents of epitheliocystis been more completely described as belonging to the order Chlamydiales, a group of obligate intracellular bacteria sharing unique and complex developmental cycles (15, 23–27), whereas four additional organisms have been characterized using molecular methods and phylogenetic analyses (15, 24, 27).
The objective of this study was to identify, characterize, and compare Chlamydia-like 16S rRNA genetic sequence data isolated and amplified from epitheliocystis-affected gills of YTK over three different cohorts from commercial farms in South Australia and from archival material held at the University of Tasmania.
MATERIALS AND METHODS
Ethics statement.
Sampling of animals for this study was conducted opportunistically and after commercial harvest. Animals were killed by commercial staff and subject to standard industry harvest practices.
Sample collection.
A total of 38 YTK, S. lalandi, from three cohorts were analyzed. They were sampled during commercial harvests from sea cages located throughout the YTK commercial production zone in South Australia. Samples were taken from the following years: 2008 (n = 8), 2009 (n = 10), and 2010 (n = 20) cohort YTK. The second gill arch on the sinistral side was sampled and fixed in 10% neutral buffered formalin for histology, with a small subsample fixed in RNAlater (Epicentre, Wisconsin) for molecular testing. Archival samples from 2002 were used for the transmission electron microscopy (TEM) and laser-dissected cyst analyses (87.5% of kingfish from 2002 were epitheliocystis positive; infection was previously reported on the basis of histology [6]).
Fish sampled from the 2009 YTK cohort were approximately 3 kg in size and exhibited clinical signs that included heavy infection with the monogenean Benedenia seriolae, slow swimming, and poor condition. Gills were swollen, with shortened filaments, and white streaking was observed along the filaments. Fish from the 2010 YTK cohort were commercially harvested at a size of 3.5 kg at the time of sampling, and all fish appeared to be clinically healthy. Fish sampled from the 2008 YTK cohort were clinically healthy.
Histopathology.
Formalin-fixed gills were trimmed and routinely processed for histology. The gills were sectioned at 5 μm and stained with hematoxylin and eosin. The sections were examined by light microscopy to identify epitheliocystis inclusions and associated lesions.
Transmission electron microscopy.
Selected areas of gill tissue from archival samples were procured from the paraffin blocks and deparaffinized in xylene overnight prior to rehydration through a graded ethanol series. Tissues were placed in Sorenson's phosphate buffer (0.1 M; pH 7.2) prior to fixation in Karnovsky's solution for 4 h at 4°C. Following a brief rinse in phosphate buffer, the tissues were postfixed in 1% aqueous osmium tetroxide, dehydrated through a graded acetone series, infiltrated, and embedded in epoxy resin. Thin sections cut at 60 to 90 nm were stained with 4% uranyl acetate and lead citrate prior to examination with a Philips EM 400 transmission electron microscope at 80 kV (Philips Electronic Instruments, Mahwah, NJ).
Laser dissection of epitheliocystis cysts.
Paraffin-embedded blocks were sectioned and mounted unstained on polyethylene naphthalate (PEN) membrane slides. The slides were then examined, and cysts were cut using a Leica LMD 6500 microscope (Leica, Wetzlar, Germany). The laser-cut cysts were dropped by gravity into a PCR tube cap and were used for further processing.
DNA extraction.
DNA was extracted from all cohorts tested and the laser-dissected cysts using a commercial DNA extraction kit (Epicentre MasterPure Complete DNA and RNA Purification Kit) according to the manufacturer's instructions. The DNA pellet was rinsed with 70% ethanol and resuspended in 100 μl of Tris-EDTA (TE) buffer.
Chlamydiales-specific 16S rRNA PCR amplification and sequencing.
The presence of chlamydial DNA was confirmed by Chlamydiales-specific 16S rRNA PCR using primers 16SIGF (5′-CGG CGT GGA TGA GGC AT-3′) and 16SIGR (5′-TCA GTC CCA GTG TTG GC-3′), resulting in a 298-bp signature sequence, as described previously (9). The amplification reaction was performed with initial denaturation (95°C; 10 min), followed by 35 cycles of denaturation (95°C; 30 s), annealing (54°C; 30 s), and extension (72°C; 1.5 min), and a final extension (72°C; 7 min). Distilled-H2O negative controls were performed in triplicate. For the expanded 800-bp 16S rRNA sequence from selected samples, the 16SIGF primer was matched with the 806R 16S rRNA primer (28). The amplification reaction was initiated by denaturation (95°C; 10 min), followed by 35 cycles of denaturation (95°C; 30 s), annealing (55°C; 45 s), and extension (72°C; 45 s) and a final extension (72°C; 7 min). For the nearly full-length 16S rRNA sequence, the 16SIGF primer was matched with the 16SBI primer (29). The amplification reaction was initiated by denaturation (95°C; 10 min), followed by 35 cycles of denaturation (95°C; 1 min), annealing (55°C; 1 min), and extension (72°C; 1.5 min) and a final extension (72°C; 7 min).
After each PCR, separation of PCR products by agarose gel electrophoresis was performed, followed by visualization of a band at the expected size by UV transillumination (254 nm). The amplified PCR products were purified using a PureLink PCR Purification Kit (Invitrogen) before being sent for sequencing at the Australian Genome Research Facility, Brisbane, Australia. Sequences were analyzed with Geneious Pro (30). The identities of amplified 16S rRNA sequences were determined by the BLAST-n algorithm (31) against sequences available in the GenBank database (http://www.ncbi.nlm.nih.gov).
Primer design, validation, and PCR of laser-dissected cysts.
After the samples for the three cohorts were screened using the Chlamydiales primer pair (16SIGF/16SIGR), YTK-specific primers were designed to validate the specificity of the results. A sequence alignment was performed using the ClustalW alignment algorithm with the YTK sequence reported here and from additional Chlamydiales species obtained from GenBank. The resulting primers, YTKfor (5′-GGG CCT TGC GGA TCG T-3′) and YTKrev (5′-CCG CTA CTC TCA AGT TC-3′), were designed to amplify a YTK epitheliocystis agent-specific 16S rRNA sequence with an expected PCR product size of 280 bp. The YTKfor/YTKrev primer pair was validated against known epitheliocystis-positive samples from other fish species (data not shown).
To confirm that the chlamydial DNA detected from the YTK gill sample was the same as that within the epitheliocystis cysts, 16S rRNA PCR of the laser-dissected cysts was performed using primers YTKfor and YTKrev. The amplification reaction performed was the same as the initial PCR screening reaction described above.
Molecular phylogenetic analysis.
The partial 16S rRNA region sequenced for the taxon reported here and data from additional Chlamydiales species and outgroup taxa obtained from GenBank (see Table S1 in the supplemental material) were initially aligned using MUSCLE version 3.7 (32) with ClustalW sequence weighting and unweighted-pair group method using average linkages (UPGMA) clustering for iterations 1 and 2. The resultant alignment was refined by eye using MESQUITE (33). After the alignment of the 16S data set was edited, the ends of each fragment were trimmed to match the shortest sequence in the alignment.
The software jModelTest version 0.1.1 (34, 35) was used to estimate the best nucleotide substitution models for this data set. Bayesian inference analysis of the 16S rRNA data set was performed using MrBayes version 3.1.2 (36) run on the CIPRES portal (37) to explore relationships among these taxa. Bayesian inference analysis was conducted on the 16S rRNA data set using the GTR + I + G model predicted as the best estimator by the Akaike information criterion (AIC) and Bayesian information criterion (BIC) in jModelTest. Bayesian inference analysis was run over 10,000,000 generations (ngen = 10,000,000) with two runs each containing four simultaneous Markov chain Monte Carlo (MCMC) chains (nchains = 4), and every 1,000th tree was saved (samplefreq = 1,000). Bayesian analysis used the following parameters: nst = 6, rates = invgamma, ngammacat = 4, and the priors parameters of the combined data set were set to a ratepr of variable. Samples of substitution model parameters and tree and branch lengths were summarized using the parameters “sump burnin = 3,000” and “sumt burnin = 3,000.” These “burnin” parameters were chosen because the log likelihood scores “stabilized” well before 3,000,000 replicates in the Bayesian inference analyses.
Maximum-likelihood analysis was performed on the 16S data set using the RAxML algorithm (38) on the CIPRES portal with the gamma rate model of heterogeneity and maximum-likelihood search estimating the proportion of invariable site parameters. Nodal support was inferred based on 100 bootstrap replicates.
Epidemiology.
The prevalence of CLO detected by PCR was calculated as a percentage of all samples tested for a particular cohort (Table 1). The prevalence and intensity of epitheliocystis infection in YTK was calculated after light microscopy examination of H&E-stained gill sections. Absolute counts of cysts and filaments for each fish were recorded. The mean cyst count and mean intensity (intensity = cysts per section/filaments per section) were calculated. Statistics were conducted with the Tinn-R 2.3.7.1 statistical package (2001; GUI for R language and environment; https://sourceforge.net/projects/tinn-r/). A residual plot and Bartlett test were used to test the assumption of homogeneity of variances, and the mean intensity variable was square root transformed to meet this assumption. A one-way analysis of variance (ANOVA) was performed, and a Tukey honestly significant difference (HSD) test was used to detect any differences between cohorts (Table 1).
Table 1.
Prevalence and intensity of infection of yellowtail kingfish, S. lalandi, with chlamydial DNA using both histology and PCR amplification
| Cohort | n | Histology |
PCR prevalence (%) | |
|---|---|---|---|---|
| Prevalence (%) | Mean intensity (±SE)a | |||
| 2008 | 8 | 100 | 0.32 (0.062) A | 100 |
| 2009 | 10 | 20 | 0.006 (0.004) B | 80 |
| 2010 | 20 | 30 | 0.013 (0.005) B | 80 |
SE, standard error. Different letters denote significant differences (P < 0.001).
Nucleotide sequence accession number.
The 16S rRNA gene sequence of the YTK epitheliocystis agent is available on GenBank under accession number JQ673516.
RESULTS
Histopathology.
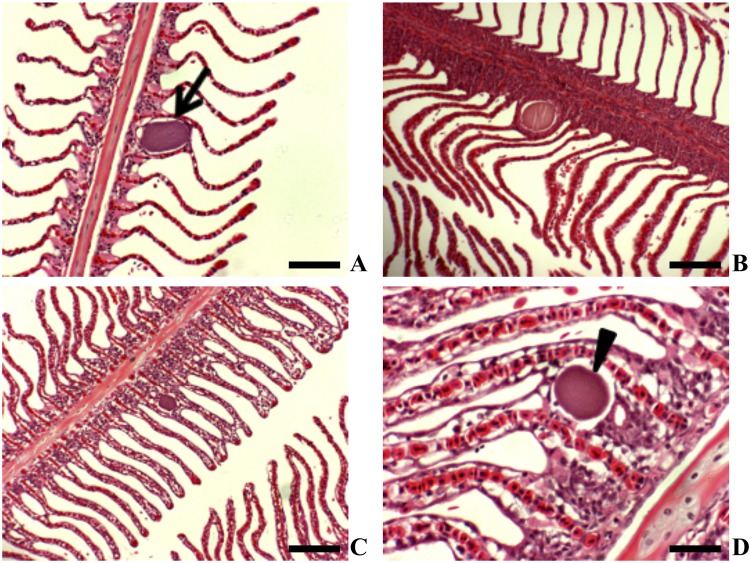
Epitheliocystis was present in fish from all three cohorts. Bacterial cysts were manifested as membrane-enclosed granulated basophilic inclusions. These basophilic inclusions were not always associated with cellular proliferation, and the majority were at the base of the gill lamellae (Fig. 1A and B). In gills with evidence of epitheliocystis, histopathological changes included cellular hyperplasia and vacuolation of cells with lamellar fusion (Fig. 1C and D).
Fig 1.
Yellowtail kingfish gills showing epitheliocystis (H&E staining). (A) Basophilic inclusion with no associated host response (scale bar = 100 μm) (2008 cohort). (B) Membrane-enclosed inclusion at base of lamellae (scale bar = 100 μm) (2010 cohort). (C) Basophilic inclusion with an associated lamellar fusion (scale bar = 200 μm) (2008 cohort). (D) Higher magnification of basophilic inclusion at base of lamella associated with proliferation and vacuolation of the basal lamellar epithelium (scale bar = 50 μm) (2009 cohort).
Transmission electron microscopy.
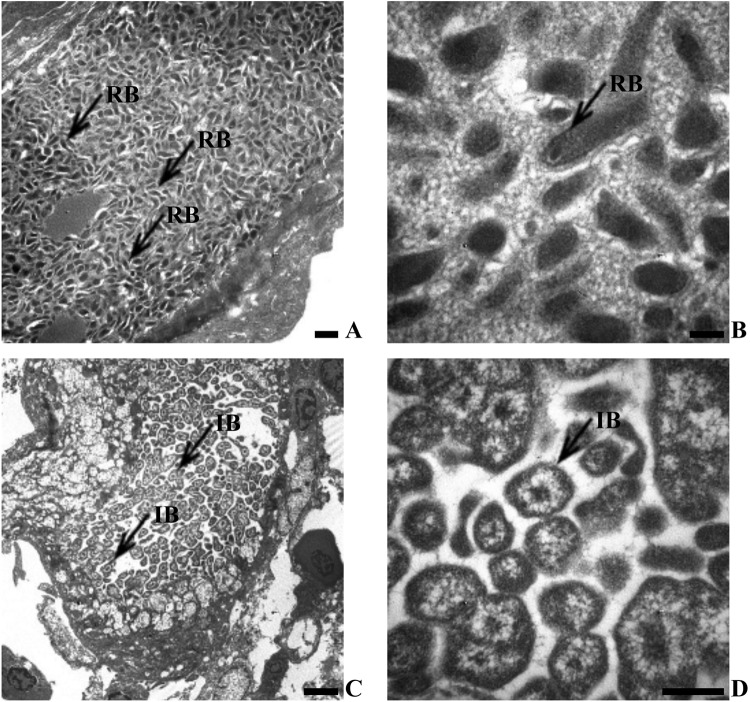
Transmission electron microscopy of the epitheliocystis inclusions revealed that the organisms were tightly packed within the membrane-bound vacuole. These vacuoles contained elongated reticulate bodies (RBs) (Fig. 2A and B) and spherical intermediate bodies (IBs) (Fig. 2C and D). Head and tail bodies were observed in association with the inclusions (Fig. 2C), although elementary bodies (EBs) were not observed. The RBs ranged in size from 83 by 211 nm to 100 by 361 nm, while the IBs ranged in size from 353 by 470 nm to 924 by 941 nm.
Fig 2.
Transmission electron microscopy of Chlamydia-like epitheliocystis agent inclusions in gill epithelium of yellowtail kingfish (2002 cohort), S. lalandi. (A) Membrane-bound epitheliocystis inclusion body containing reticulate bodies (scale bar = 500 nm). (B) Higher magnification of reticulate bodies (RB) (scale bar = 100 nm). (C) Membrane-bound epitheliocystis inclusion body containing intermediate bodies (IB) (scale bar = 2 μm). (D) Higher magnification of intermediate bodies (scale bar = 500 nm). Note the absence of elementary bodies.
Epidemiology of a novel YTK Chlamydia-like organism.
The prevalence of infection based on the results from two methods (PCR and histopathology) varied greatly. PCR proved much more sensitive for detection of epitheliocystis, with cohort positivity ranging from 80 to 100% for PCR compared to 20 to 100% for histopathology (Table 1). Significant differences in the mean intensities of epitheliocystis inclusions were observed between the 2008 and 2009–2010 cohorts only (F = 64.702; df = 2,116; P < 0.001) (Table 1).
Molecular identification and phylogenetic analysis of a novel Chlamydia-like organism.
Initial order Chlamydiales-specific 16S rRNA PCR assay screening revealed that 100% of the 2008 (n = 8) and 2009 (n = 10) cohorts sampled and 80% of the 2010 cohort (16/20) screened PCR positive for chlamydial DNA. Sequences from five randomly selected samples per cohort were aligned and analyzed and found to be identical within and between cohorts. Therefore, a single representative sample from each cohort was selected to identify the chlamydial species present by additional PCR amplification of extended and nearly complete 16S rRNA sequences. An expanded and nearly full-length sequence was amplified from a representative sample from each of the 2008 to 2010 cohorts (inclusive), and multiple-sequence alignment of the resulting 1,393-bp 16S rRNA sequence from all three samples indicated that they were 100% identical. The 16S rRNA PCR assay of the laser-dissected cysts was PCR positive for chlamydial DNA and was identical to the chlamydial 16S rRNA sequence obtained directly from the gill tissue.
BLAST-n analysis of the YTK epitheliocystis agent sequence against the NCBI database revealed the sequence to be novel, sharing only a distant 87% sequence similarity to the next closest 16S rRNA sequences from “Candidatus Piscichlamydia salmonis” (AY462243.1 and AY462244.1 [24]; EU326495.1 [39]).
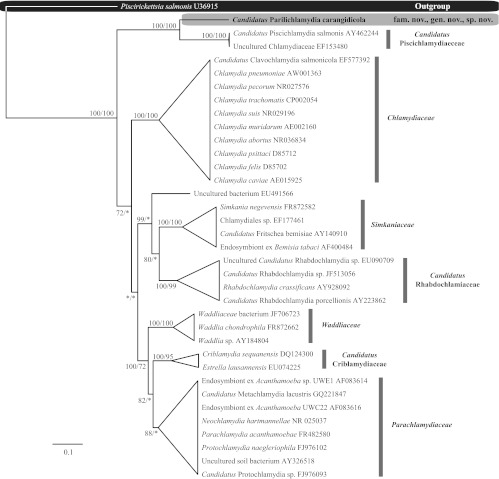
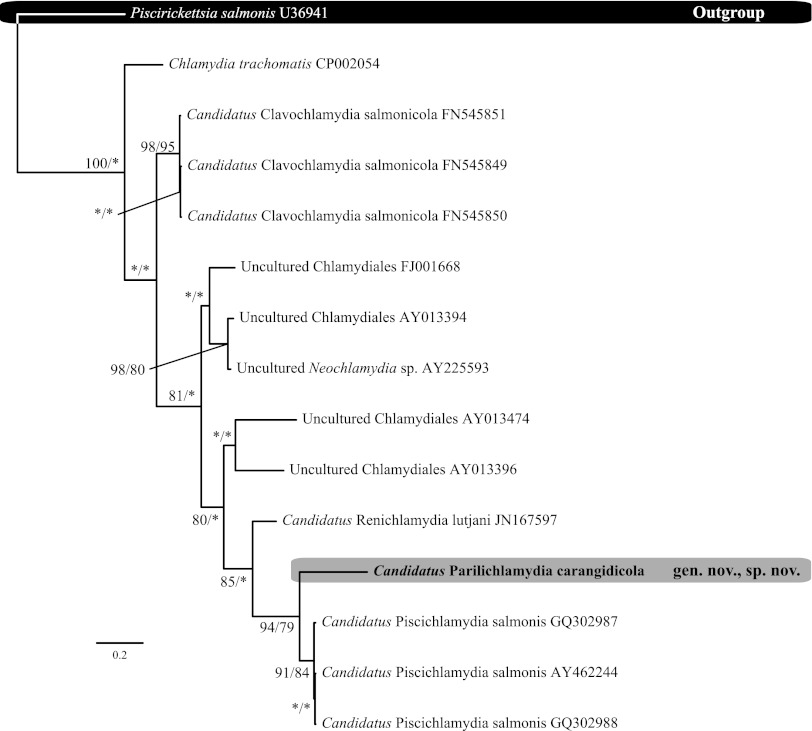
Alignment of the 16S rRNA data generated for the epitheliocystis agents isolated from YTK and the remainder of the Chlamydiales taxa and outgroups examined (see Table S1 in the supplemental material) yielded 1,136 characters of analysis. The percentages of pairwise identities observed between the family level taxa analyzed over the 16S data set are shown in Table 2. Bayesian inference and maximum-likelihood analyses resulted in phylograms with identical topologies, which displayed all of the currently recognized and candidate families within the Chlamydiales, forming relatively well-supported clades (Fig. 3). The sequence obtained from the novel epitheliocystis agent reported here was a sister taxon to the sequences available for “Ca. Piscichlamydia salmonis” and an uncultured Chlamydiaceae-like organism on GenBank (Fig. 3). Phylogenetic comparisons between known epitheliocystis 16S rRNA signature sequences from GenBank also confirm the novel lineage of the novel CLO reported here (Fig. 4). Furthermore, the chlamydial agent of epitheliocystis in YTK represents a novel family lineage in the order Chlamydiales.
Table 2.
Estimates of evolutionary divergence between sequences
| Family (no., name) | % base similarity with family no.a: |
|||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| 1, “Candidatus Piscichlamydiaceae” | ||||||||
| 2, “Candidatus Parilichlamydiaceae” fam. nov. | 86–86.1 | |||||||
| 3, Chlamydiaceae | 80.9–82.1 | 80.9–81.6 | ||||||
| 4, “Candidatus Rhabdochlamydiaceae” | 80.3–81.2 | 81.1–81.7 | 83–84.9 | |||||
| 5, Simkaniaceae | 80.8–82.3 | 81.6–82 | 83.6–85.2 | 85.4–88.4 | ||||
| 6, Waddliaceae | 81.5–82.5 | 80.8–81.7 | 86–87.3 | 84–86.2 | 84.4–86.8 | |||
| 7, “Candidatus Criblamydiaceae” | 82.1–82.6 | 81.5–82 | 86.1–87.9 | 84.6–85.2 | 84.3–85.8 | 87–89.1 | ||
| 8, Parachlamydiaceae | 80.8–82.2 | 81.2–82.7 | 85.2–87.5 | 85–88.4 | 85.5–88.6 | 88.4–90.6 | 87.2–91.3 | |
The percentages of base similarities between families are shown. Sequences were trimmed, and there were a total of 1,136 nucleotide positions in the final data set. Evolutionary analyses were conducted in MEGA5 (49).
Fig 3.
Relationships between the epitheliocystis agent detected in yellowtail kingfish gills and the remainder of the Chlamydiales taxa and outgroups examined based on Bayesian inference maximum-likelihood analyses of the 16S rRNA data set. Posterior probability/bootstrap support values are given at the nodes, with values of less than 70% indicated by asterisks. Family clades have been collapsed.
Fig 4.
Relationship between the 298-bp signature sequence of the epitheliocystis agent detected in yellowtail kingfish gills and epitheliocystis agents detected in other fish species, Chlamydia trachomatis and Piscirickettsia salmonis. Bayesian inference maximum-likelihood analyses of the 298-bp signature sequence 16S rRNA data set were performed. Posterior probability/bootstrap support values are given at the nodes, with values of less than 70% indicated by asterisks.
DISCUSSION
The novel YTK epitheliocystis bacterial agent from YTK has >80% sequence similarity to other members of the Chlamydiales, placing it within the order (16). At this stage, Koch's postulates have not been satisfied to definitively identify the sequenced agent as the cause of epitheliocystis, since this sequenced agent from YTK, and also other epitheliocystis CLO agents, has not been successfully cultured in vitro (24). However, the nearly full-length sequence of the CLO reported and confirmed here as identical in three different-year cohorts (sampled throughout the South Australian YTK farming zone) and the archival samples from 2002 from both the gill tissue and the cysts is strong evidence that it is a novel CLO bacterial agent causing epitheliocystis in YTK.
Epitheliocystis has been noted as a recurrent problem in the culture of different yellowtail species, including S. dumerili (40) and S. lalandi from Ecuador (formally Seriola mazatlana) (41). Epitheliocystis infection in both S. dumerili and Ecuadorian S. lalandi resulted in a proliferative response, with enlarged cells packed with basophilic granules. However, lamellar fusion occurred only in Ecuadorian S. lalandi (41). Severe infection of epitheliocystis has been reported in cultured S. lalandi from South Australia, although the infection resulted in the presence and absence of a proliferative host response (6, 11). This difference in the host response may be attributed to the age of the fish. Older, larger S. lalandi fish appear to have less of a host response to epitheliocystis infections than juveniles. These reports of epitheliocystis in species of Seriola occurred prior to the widespread application of molecular techniques to the study of epitheliocystis, and molecular analyses have not been performed on the material to further characterize the epitheliocystis agent(s) involved. It will be of interest to determine if the epitheliocystis agent in these Seriola species, especially the Ecuadorian S. lalandi, is the same as the agent reported in this study.
This study also highlighted the discrepancies that can be found when using histopathology (20 to 100%) versus PCR (80 to 100%) for detection of epitheliocystis in fish. A similar discrepancy was also observed in a recent study of epitheliocystis in Atlantic salmon, where reverse transcription (RT)-PCR positivity ranged from 75 to 100% compared to 20 to 100% when using histopathology (26). Since histological methods continue to be the primary tool for detection of gill diseases in fish, these observations emphasize the fact that epitheliocystis, as a gill disease, is likely underdiagnosed in aquaculture settings, particularly in fish without any clinical signs of infection. While infection is not synonymous with disease, PCR is of course more sensitive for the detection of nonclinical and subclinical infections and possible infections that result in only a mild clinical disease. This detection is important, as it may provide the opportunity to institute changes in husbandry management to prevent a more serious condition within the population in the future.
While the cyst location at the base of the lamellae and the general morphology of the cysts are consistent with previous reports (12, 13, 42–46), the proliferative response of the YTK in this study was different than the response in Atlantic salmon. In salmonids, epitheliocystis infection was associated only with mild lesions of the branchial epithelium, and any proliferative response was not associated with epitheliocystis (25, 33). The histological and electron microscopic features observed in YTK support the corresponding molecular result of a CLO being present within the epitheliocystis cysts. This agrees with previous reports for the molecular description of “Ca. Piscichlamydia salmonis” in Atlantic salmon (24, 47) and Neochlamydia-like bacteria in Arctic charr (14).
Although there have been two previous reports of nearly full-length 16S rRNA sequences, “Ca. Piscichlamydia salmonis” (24) and “Ca. Clavochlamydia salmonicola” (25), there is no single genus for Chlamydia-like bacteria associated with epitheliocystis (15). With the addition of this novel CLO, there are now three nearly full-length 16S rRNA sequences, all with <90% sequence similarity. According to accepted criteria, these bacteria belong to three separate family lineages within the Chlamydiales (9). Based on its novel 16S rRNA sequence, the percentage of sequence divergence from other Chlamydiales species, and the observed phylogenetic relationships of the bacteria to other taxa within the order, the name “Candidatus Parilichlamydia carangidicola” (gen. nov., sp. nov.) (Order Chlamydiales) is proposed to identify this Chlamydia-like epitheliocystis agent, whereas the name “Candidatus Parilichlamydiaceae” is proposed to identify the family lineage. Additional morphological and genetic data, including the sequencing of additional genes, will be required to formally characterize and classify this novel pathogen.
With this report, evidence continues to accrue concerning the taxonomic diversity of epitheliocystis agents from farmed and wild fish populations in marine and freshwater environments in both hemispheres. Further research will be required, not only to characterize the agents of the disease, but to evaluate the effects of risk factors, such as increased stocking densities (11), in response to increased demand for fish products, and environmental factors, such as water temperature (46, 48), that may promote and exacerbate epitheliocystis-related diseases.
Taxonomy.
“Candidatus Parilichlamydia carangidicola” gen. nov., sp. nov. (“Candidatus Parilichlamydiaceae” fam. nov.), recovered from yellowtail kingfish (Seriola lalandi).
Parilichlamydiaceae fam. nov.; Parili-, L. adj. parilis, equal or similar; Chlamydiaceae, N.L. fem. n., a bacterial family name. Parilichlamydia gen. nov.; Parili-, L. adj. parilis, equal or similar; Chlamydia, N.L. fem. n., a bacterial genus name. Parilichlamydia carangidicola sp. nov., carangidicola N.L. gen. sing. n., of Carangidae, the family to which the fish host belongs.
Obligate intracellular bacteria. Cells present tightly packed within a membrane-bound inclusion in vacuolated gill epithelial cells. These vacuoles contained elongated reticulate bodies and spherical intermediate bodies. Head and tail bodies were observed in the inclusions. There was a marked absence of elementary bodies. The reticulate bodies ranged in size from 83 by 211 nm to 100 by 361 nm, while the intermediate bodies ranged in size from 353 by 470 nm to 924 by 941 nm. The new family, genus, and species are distinguished from all other species of formally described and candidate Chlamydiales taxa based on a combination of the morphological and genetic differences (i.e., only 86 to 86.1% similarity to other Chlamydiales over the 16S rRNA data set examined) observed here. Fish are infected in the gills, with cellular hyperplasia and resulting vacuolation of cells and fusion of gill lamellae observed in histology.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following people for their assistance with this work: Nicole Kirchhoff and Ken Cain for assistance with sampling; Karine Cadoret for assistance and training in DNA extractions and histology; Brian Jones for laser dissection microscopy; and James Marsh, Eileen Roulis, and Martina Jelocnik for assisting with additional PCRs.
Footnotes
Published ahead of print 28 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02899-12.
REFERENCES
- 1. Delgado CL, Wada N, Rosegrant MW, Meijer S, Ahmed M. 2003. Outlook for fish to 2020: meeting global demand. International Food Policy Research Institute, Washington, DC [Google Scholar]
- 2. Egusa S. 1983. Disease problems in Japanese yellowtail, Seriola quinqueradiata, culture: a review. Rapp. P. V. Reun. 182:10–18 [Google Scholar]
- 3. Jover M, Garcia-Gomez A, Tomas A, De la Gandara F, Perez L. 1999. Growth of Mediterranean yellowtail (Seriola dumerilii) fed extruded diets containing different levels of protein and lipid. Aquaculture 179:25–33 [Google Scholar]
- 4. Poortenaar C, Hooker SH, Sharp NJ. 2001. Assessment of yellowtail kingfish (Seriola lalandi lalandi) reproductive physiology, as a basis for aquaculture development. Aquaculture 201:271–286 [Google Scholar]
- 5. Cobcroft JM, Pankhurst PM, Poortenaar C, Hickman B, Tait M. 2004. Jaw malformation in cultured yellowtail kingfish (Seriola lalandi) larvae. N. Z. J. Mar. Freshw. Res. 38:67–71 [Google Scholar]
- 6. Mansell B, Powell MD, Ernst I, Nowak BF. 2005. Effects of the gill monogenean Zeuxapta seriolae (Meserve, 1938) and treatment with hydrogen peroxide on pathophysiology of kingfish, Seriola lalandi Valenciennes, 1833. J. Fish Dis. 28:253–262 [DOI] [PubMed] [Google Scholar]
- 7. Sharp NJ, Poortenaar CW, Diggles BK, Willis TJ. 2003. Metazoan parasites of yellowtail kingfish, Seriola lalandi lalandi, in New Zealand: prevalence, intensity, and site preference. N. Z. J. Mar. Freshw. Res. 37:273–282 [Google Scholar]
- 8. Stephens FJ, Savage A. 2010. Two mortality events in sea-caged yellowtail kingfish Seriola lalandi Valenciennes, 1833 (Nannopercidae) from Western Australia. Aust. Vet. J. 88:414–416 [DOI] [PubMed] [Google Scholar]
- 9. Everett KDE, Bush RM, Anderson AA. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415–440 [DOI] [PubMed] [Google Scholar]
- 10. Corsaro D, Greub G. 2006. Pathogenic potential of novel Chlamydiae and diagnostic approaches to infections due to these obligate intracellular bacteria. Clin. Microbiol. Rev. 19:283–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nowak BF, LaPatra SE. 2006. Review: epitheliocystis in fish. J. Fish Dis. 29:573–588 [DOI] [PubMed] [Google Scholar]
- 12. Crespo S, Zarza C, Padros F, Marin de Mateo M. 1999. Epitheliocystis agents in sea bream Sparus aurata: morphological evidence for two distinct Chlamydia-like developmental cycles. Dis. Aquat. Organ. 37:61–72 [DOI] [PubMed] [Google Scholar]
- 13. Lewis EJ, McLaughlin SM, Bodammer JE, Sawyer TK. 1992. Epitheliocystis in ten new host species of marine fish. J. Fish Dis. 15:267–271 [Google Scholar]
- 14. Draghi A, II, Bebak J, Popov VL, Noble AC, Geary SJ, West A, Byrne P, Frasca S., Jr 2007. Characterization of a Neochlamydia-like bacterium associated with epitheliocystis in cultured Arctic charr Salvelinus alpinus. Dis. Aquat. Organ. 76:27–38 [DOI] [PubMed] [Google Scholar]
- 15. Meijer A, Roholl PJM, Ossewaarde JM, Jones B, Nowak BF. 2006. Molecular evidence for association of Chlamydiales bacteria with epitheliocystis in leafy seadragon (Phycodurus eques), silver perch (Bidyanus bidyanus), and barramundi (Lates calcarifer). Appl. Environ. Microbiol. 72:284–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anderson I, Prior HC. 1992. Subclinical epitheliocystis in barramundi, Lates calcarifer, reared in sea cages. Aust. Vet. J. 69:226–227 [DOI] [PubMed] [Google Scholar]
- 17. Groff JM, LaPatra SE, Munn RJ, Anderson ML, Osburn BI. 1996. Epitheliocystis infection in cultured white sturgeon, Acipenser transmontanus: antigenic and ultrastructural similarities of the causative agent to the chlamydiae. J. Vet. Diagn. Invest. 8:172–180 [DOI] [PubMed] [Google Scholar]
- 18. Frances J, Tennent R, Nowak BF. 1997. Epitheliocystis in silver perch, Bidyanus bidyanus (Mitchell). J. Fish Dis. 20:453–457 [Google Scholar]
- 19. Nylund A, Kvenseth AM, Isdal E. 1998. A morphological study of the epitheliocystis agent in farmed Atlantic salmon. J. Aquat. Anim. Health 10:43–55 [Google Scholar]
- 20. Syasina I, Park IS, Kim JM. 2004. Epitheliocystis disease in red sea bream Pagrus major and induced hybrid, red sea bream Pagrus major female black sea bream Acanthopagrus schlegeli male, cultured in Korea. Bull. Eur. Assoc. Fish Pathol. 24:260–267 [Google Scholar]
- 21. Kim DJ, Park JH, Seok SH, Cho SA, Baek MW, Lee HY. 2005. Epitheliocystis in carp (Cyprinus carpio) in South Korea. J. Vet. Med. Sci. 67:119–120 [DOI] [PubMed] [Google Scholar]
- 22. Katharios P, Papadaki M, Papandroulakis N, Divanach P. 2008. Severe mortality in mesocosm-reared sharpsnout sea bream Diplodus puntazzo larvae due to epitheliocystis infection. Dis. Aquat. Organ. 82:55–60 [DOI] [PubMed] [Google Scholar]
- 23. Draghi A., II 2006. Molecular characterization of chlamydia-like bacteria associated with epitheliocystis in farmed salmonids, Atlantic salmon (Salmo salar) and Arctic char (Salvelinus alpinus), and environmental chlamydiae from aquatic environments. Ph.D. thesis, University of Connecticut, Storrs, CT [Google Scholar]
- 24. Draghi A, Popov VL, Kahl MM, Stanton JB, Brown CC, Tsongalis GJ, West A, Frasca S. 2004. Characterization of “Candidatus Piscichlamydia salmonis” (order Chlamydiales), a Chlamydia-like bacterium associated with epitheliocystis in farmed Atlantic Salmon (Salmo salar). J. Clin. Microbiol. 42:5286–5297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Karlsen M, Nylund A, Watanabe K, Helvik JV, Nylund S, Plarre H. 2008. Characterisation of “Candidatus Clavochlamydia salmonicola”: an intracellular bacterium infecting salmonid fish. Environ. Microbiol. 10:208–218 [DOI] [PubMed] [Google Scholar]
- 26. Mitchell SO, Steinum T, Rodger H, Holland C, Falk K, Colquhoun DJ. 2010. Epitheliocystis in Atlantic salmon, Salmo salar L., farmed in fresh water in Ireland is associated with “Candidatus Clavochlamydia salmonicola” infection. J. Fish Dis. 33:665–673 [DOI] [PubMed] [Google Scholar]
- 27. Polkinghorne A, Schmidt-Posthaus H, Meijer A, Lehner A, Vaughan L. 2010. Novel Chlamydiales associated with epitheliocystis in a leopard shark Triakis semifasciata. Dis. Aquat. Organ. 91:75–81 [DOI] [PubMed] [Google Scholar]
- 28. Relman DA. 1993. Universal bacterial 16S rDNA amplification and sequencing, p 489–495 In Pershing DH, Smith TF, Tenover FC, White TJ. (ed), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, DC [Google Scholar]
- 29. Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T. 2006. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 4:1841–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Drummond AJ, Ashton B, Buxton S, Cheung M, Cooper A, Duran C, Field M, Heled J, Kearse M, Markowitz S, Moir R, Stones-Havas S, Sturrock S, Thierer T, Wilson A. 2011. Geneious v5.4. http://www.geneious.com/
- 31. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maddison WP, Maddison DR. 2009. Mesquite: a modular system for evolutionary analysis, version 2.72. http://mesquiteproject.org/
- 34. Guindon S, Gascuel O. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 35. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 36. Ronquist F, Hulelsenbeck JP. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 37. Miller MA, Holder MT, Voc R, Midford PE, Liebowitz T, Chan L, Hoover P, Warnow T. 2009. The CIPRES portals. CIPRES. 2009-08-04. http://www.phylo.org/sub_sections/portal
- 38. Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57:758–771 [DOI] [PubMed] [Google Scholar]
- 39. Nylund A, Watanabe K, Nylund S, Karlsen M, Saether PA, Arnesen CE, Karlsbakk E. 2008. Morphogenesis of salmonid gill poxvirus associated with proliferative gill disease in farmed Atlantic salmon (Salmo salar) in Norway. Arch. Virol. 153:1299–1309 [DOI] [PubMed] [Google Scholar]
- 40. Grau A, Crespo S, Pastor E, Gonzalez P, Carbonell E. 2003. High infection by Zeuxapta seriolae (Monogenea: Heteraxinidae) associated with mass mortalities of amberjack Seriola dumerili Risso reared in sea cages in the Balearic Islands (western Mediterranean). Bull. Eur. Assoc. Fish Pathol. 23:139–142 [Google Scholar]
- 41. Venizelos A, Benetti DD. 1996. Epitheliocystis disease in cultured yellowtail Seriola mazatlana in Ecuador. J. World Aquaculture Soc. 27:223–227 [Google Scholar]
- 42. Bradley TM, Newcomer CE, Maxwell KO. 1988. Epitheliocystis associated with massive mortalities of cultured lake trout Salvelinus namaycush. Dis. Aquat. Organ. 4:9–17 [Google Scholar]
- 43. Costa G, Lom J, Andrade C, Barradas R. 1998. First report of Ceratomyxa sparusaurati (Protozoa: Myxosporea) and the occurrence of epitheliocystis in cultured sea bream, Sparus aurata L. from Madeira. Bull. Eur. Assoc. Fish Pathol. 18:165–167 [Google Scholar]
- 44. Goodwin AE, Park E, Nowak BF. 2005. Short communication: successful treatment of largemouth bass, Micropterus salmoides (L.), with epitheliocystis hyperinfection. J. Fish Dis. 28:623–625 [DOI] [PubMed] [Google Scholar]
- 45. Nowak BF. 1996. Health of red morwong, Cheilodactylus fuscus, and rock cale, Crinodus lophodon, from Sydney cliff-face sewage outfalls. Mar. Pollut. Bull. 33:281–292 [Google Scholar]
- 46. Nowak BF, Clark A. 1999. Prevalence of epitheliocystis in Atlantic salmon, Salmo salar L., farmed in Tasmania, Australia. J. Fish Dis. 22:73–78 [Google Scholar]
- 47. Draghi A, Bebak J, Daniels S, Tulman ER, Geary SJ, West A, Popov VL, Frasca S., Jr 2010. Identification of “Candidatus Piscichlamydia salmonis” in Arctic charr Salvelinus alpinus during a survey of charr production facilities in North America. Dis. Aquat. Organ. 89:39–49 [DOI] [PubMed] [Google Scholar]
- 48. Schmidt-Posthaus H, Polkinghorne A, Nufer L, Schifferli A, Zimmermann DR, Segner H, Steiner P, Vaughan L. 2012. A natural freshwater origin for two chlamydial species, Candidatus Piscichlamydia salmonis and Candidatus Clavochlamydia salmonicola, causing mixed infections in wild brown trout (Salmo trutta). Environ. Microbiol. 14:2048–2057 [DOI] [PubMed] [Google Scholar]
- 49. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.