Abstract
Q fever is a zoonotic disease caused by inhalation of the bacterium Coxiella burnetii. Ruminant livestock are common reservoirs for C. burnetii, and bacteria present in aerosols derived from the waste of infected animals can infect humans. The significance of infection from material deposited in the environment versus transmission directly from infected animals is not known. In 2011, an outbreak of Q fever cases on farms in Washington and Montana was associated with infected goats. A study was undertaken to investigate the quantity and spatial distribution of C. burnetii in the environment of these goat farms. Soil, vacuum, and sponge samples collected on seven farms epidemiologically linked to the outbreak were tested for the presence of C. burnetii DNA by quantitative PCR. Overall, 70.1% of the samples were positive for C. burnetii. All farms had positive samples, but the quantity of C. burnetii varied widely between samples and between farms. High quantities of C. burnetii DNA were in goat housing/birthing areas, and only small quantities were found in samples collected more than 50 m from these areas. Follow-up sampling at one of the farms 1 year after the outbreak found small quantities of C. burnetii DNA in air samples and large quantities of C. burnetii persisting in soil and vacuum samples. The results suggest that the highest concentrations of environmental C. burnetii are found in goat birthing areas and that contamination of other areas is mostly associated with human movement.
INTRODUCTION
Coxiella burnetii is an intracellular bacterium that causes the disease Q fever, which presents in both acute and chronic forms in humans (1). The acute stage of the disease generally has flu-like symptoms but can also cause pneumonia or hepatitis and sometimes requires hospitalization. The chronic stage of the disease occurs in <2% of infected persons, can occur months to years after the primary infection, and most commonly presents as blood culture-negative infectious endocarditis (2). Treatment of the acute disease relies on a 2-week course of doxycycline, whereas chronic Q fever is treated with 18 to 24 months of combination antibiotic therapy and often requires heart valve replacement surgery. Chronic Q fever endocarditis is fatal if left untreated. Q fever is a notifiable disease in the United States, and although fewer than 200 cases per year have been reported to the CDC in the last decade, large outbreaks are possible. For example, the Netherlands experienced over 4,000 cases from 2007 to 2010 (3). A recent seroprevalence study in the United States found that 3.1% of the adult population in the United States has antibodies against C. burnetii (4). The discrepancy between low numbers of reported cases and relatively high seroprevalence is probably due to a combination of asymptomatic cases, nonspecific symptoms, and underutilized diagnostics.
Q fever is transmitted by inhalation of contaminated aerosols, and infectious aerosols are typically contaminated with C. burnetii from the waste products of infected animals. Ruminant livestock commonly serve as reservoirs of C. burnetii, and goats, sheep, and cattle are the animals most often associated with Q fever outbreaks (5). C. burnetii can be shed from urine, feces, and milk, but the highest numbers of C. burnetii isolates come from infected birth products. C. burnetii replicates to very high levels in the placental tissue of infected animals, and aerosols with high concentrations of C. burnetii can be generated during parturition (6). Most animals do not have symptoms when they are infected with C. burnetii; however, abortion and stillbirth can occur, especially in goats (5).
C. burnetii is classified as a category B agent of bioterrorism by the CDC. This is due to its transmission by inhalation, low infectious dose, high stability, and prior weaponization. C. burnetii is resistant to a variety of disinfectants, and when the bacteria are not replicating, they can form a spore-like small cell variant (SCV) that remains viable in the face of temperature changes and desiccation (7, 8). Release of large numbers of C. burnetii cells during the birthing of infected livestock has the potential to place a large burden of infectious agent into the environment that may remain infectious for long periods. Environmental C. burnetii also has the potential to spread for great distances. In a 1989 outbreak in the United Kingdom, Q fever cases were reported up to 11 miles (∼18 km) away from the farm that was the source of the outbreak (9). A study that examined a large number of environmental samples across the United States detected C. burnetii DNA by PCR in 23.4% of samples tested (10). Although the bacterial DNA was more likely to be detected in locations where livestock were housed or treated, a low level of DNA was found in many samples that were distant from animal housing locations. These places included post offices, banks, schools, and retail stores. Possible mechanisms for the bacteria to be transported to places with no livestock and relatively high human activity include wind, birds, small mammals, vehicular traffic, and humans.
Although anecdotal evidence suggests that it is possible for C. burnetii to travel long distances from the animals in which it replicates, it is not clear how common such spreading is. Although cases of wind-borne C. burnetii spreading widely have been reported (9, 11), most Q fever outbreaks occur when people are in fairly close proximity to the birthing of infected animals. In a 2008 outbreak in the Netherlands, it was reported that persons living within 2 km of an infected goat farm were at higher risk for Q fever than persons living 5 to 10 km from the farm (12). C. burnetii is also expected to remain in the environment once it is released. The stability of the SCV can theoretically result in a viable organism remaining in the environment for years after it is shed from infected animals. However, there are very few quantitative data available describing the contamination of the environment with C. burnetii by infected animals (13).
In 2011, C. burnetii was detected on a Washington farm that had recently experienced goat abortions. Soon thereafter, Q fever cases were detected in humans that had lived on or visited this farm. As this farm was involved in breeding and sale of goats to other farms, it was discovered that 20 farms in Washington, Montana, and Oregon had recently received goats from this farm, and a total of 21 people were identified as Q fever cases that had an epidemiologic link to this farm (14). During the outbreak investigation, environmental samples were obtained from several of the epidemiologically linked farms. The presence of C. burnetii in these environmental samples was evaluated, with detailed examination on two of the farms.
MATERIALS AND METHODS
Environmental sampling and mapping.
During the outbreak investigation, it was possible to collect environmental samples from 13 of the involved farms, but at 6 of those 13 farms, it was possible to collect only small numbers of samples. At the other 7 farms, between 14 and 138 samples were collected, and these 7 farms have been included in this study. The majority of samples were taken at farms 5, 6, and 7 (Table 1) because of farmer cooperation and a suspicion of higher levels of goat involvement due to goat abortion history and known clinical cases. Follow-up sampling at farm 5 was possible 11 months after the initial sampling. At that time, it was also possible to collect air samples from farm 5. A stratified convenience sampling approach was used, with samples collected in farmhouses, yards, animal birthing pens, animal enclosures, and equipment storage areas. Samples were bulk, sponge, HEPA vacuum, or air samples. Bulk samples were scraped from the surface of the ground using 50-ml conical tubes and were composed largely of soil. Vacuum samples were taken from within the farm houses and on and around a truck. Sponge samples were wiped from solid surfaces in animal areas. Air samples were collected using an SAS Super 180 bioaerosol sampler (Bioscience International, Rockville, MD). Five hundred liters of air was collected in 3 min for each sample onto a 0.45-μm filter membrane made of cellulose acetate and cellulose nitrate (Fisher Scientific, Pittsburgh, PA), cut to a diameter of 60 mm, and held in place by a 65-mm polystyrene contact plate. The filters were prewet with an aqueous 1% peptone solution. After collection, filters were placed in sterile, sealed plastic bags for transport to the laboratory. The location for each environmental sample was collected using a handheld global positioning satellite (GPS) receiver. GPS locations were projected onto satellite imagery of the farms using ArcGIS Explorer Online (ESRI, Redlands, CA). Relevant geographic features were manually digitized from the satellite imagery using Adobe Illustrator CS5 (Adobe Systems, Inc., San Jose, CA) to produce the final maps.
Table 1.
Detection and quantitation of C. burnetii DNA on goat farmsa
| Farm no. | No. of samples | No. of PCR-positive samples | % positive | Mean no. of GE ± SD | Median no. of GE | No. of associated Q fever cases |
|---|---|---|---|---|---|---|
| 1 | 14 | 2 | 14.3 | 0.79 ± 2.1 | 0 | 0 |
| 2 | 14 | 6 | 42.9 | 1.71 ± 3.2 | 0 | 0 |
| 3 | 20 | 19 | 95 | 117 ± 295 | 8 | 0 |
| 4 | 30 | 29 | 96.7 | 21.6 ± 23.9 | 11 | 5 |
| 5 | 64 | 54 | 84.4 | 34,273 ± 194,514 | 7 | 7 |
| 6 | 111 | 56 | 50.5 | 7.32 ± 24.5 | 1 | 2 |
| 7 | 138 | 108 | 78.3 | 107,491 ± 589,519 | 27 | 4 |
GE, genome equivalents; SD, standard deviation.
Sample processing.
The bulk, sponge, and vacuum environmental samples were processed in a manner similar to a published method (15). Material collected on sponges was extracted using 10 ml phosphate-buffered saline (PBS) for 1 h at room temperature on a Stomacher 80 laboratory blender (Seward, Port Saint Lucie, FL). The PBS was then removed and centrifuged at 20,000 × g for 15 min to pellet microorganisms. The pellet was resuspended in 200 μl PBS, and then DNA was extracted using the QIAamp DNA minikit tissue protocol (Qiagen, Valencia, CA). For the bulk and vacuum samples, five grams of each sample was mixed with 10 to 30 ml of PBS. The mixtures were incubated at room temperature with rocking for 1 h and then centrifuged at 125 × g for 5 min. The supernatant was saved and then centrifuged for 15 min at 20,000 × g to concentrate the microorganisms. Pellets were resuspended in 0.7 to 1 ml PBS, and DNA was purified from 700 μl of this microbial suspension using the QIAamp DNA stool kit (Qiagen). The resulting DNA was then tested for inhibition of PCR as described previously (15). None of the samples inhibited control PCRs. For air samples, the filters were mixed with 10 ml PBS and incubated at room temperature with rocking for 1 h, followed by centrifugation of the PBS at 20,000 × g for 15 min to pellet microorganisms. The pellet was resuspended in 200 μl PBS, and then DNA was extracted using the QIAamp DNA minikit tissue protocol (Qiagen).
PCR.
Quantitative PCR to detect the IS1111a insertion sequence was performed as described previously (16). One microliter of DNA extract was used in each 25-μl PCR. Samples were run in duplicate, and samples for which both reactions crossed the cycle threshold (CT) in less than 40 cycles were considered positive. Samples that had one positive and one negative reaction were run a second time in duplicate, and only samples for which both reactions then had a CT of less than 40 were considered positive. Genomic equivalents (GE) of C. burnetii were calculated by comparing experimental CTs to a standard curve of C. burnetii Nine Mile phase I genomic DNA. It was determined that the outbreak strain had approximately 60 copies of IS1111a, whereas Nine Mile phase I has only 20 copies of the PCR target. The numbers of GE were then adjusted accordingly, with the assumption that all C. burnetii strains detected in the environmental samples had 60 copies of IS1111a.
Isolation.
C. burnetii strains were isolated from environmental and animal samples by passage through mice and culture on a rabbit kidney cell line (RK13). Five grams of environmental material was mixed with 10 ml PBS, and samples were incubated at room temperature with rocking for 1 h. After centrifugation for 5 min at 200 × g to remove the larger particles, the supernatants were saved and then centrifuged for 15 min at 20,000 × g to concentrate the microorganisms. The supernatants were then discarded, and pellets were resuspended in 1 ml PBS. Five hundred microliters of this suspension was then injected intraperitoneally into male BALB/c mice. After 12 days, the mice were euthanized; spleens were removed, and single-cell suspensions were made in 6 ml PBS. One milliliter of the suspension was then placed on RK13 cells in Dulbecco's modified Eagle medium (DMEM) with 5% fetal bovine serum. The culture was passed weekly, and a 200-μl sample was taken at each passage and tested by PCR for the quantity of C. burnetii DNA. Cultures that appeared infected visually and had 2 weeks of increasing amounts of C. burnetii DNA were considered to be infected by a new C. burnetii isolate.
Genotyping.
Newly derived C. burnetii isolates were grown on confluent monolayers of RK13 cells until the host cells were heavily infected. Supernatants taken from these cultures contained C. burnetii with limited contamination from the RK13 host cells. DNA was purified from the isolates using the QIAamp DNA minikit tissue protocol (Qiagen). Base pair assignments at 14 single nucleotide polymorphisms (SNPs) were made using SNP genotyping assays as described previously (17).
RESULTS
As part of the 2011 Washington/Montana Q fever outbreak investigation, environmental samples were collected at many of the 21 total farms involved in the investigation. At 7 of the farms, 14 or more bulk/vacuum/sponge samples were collected (range, 14 to 138), and results from those samples are described in Table 1. For all of the samples combined, 70.1% were PCR positive for C. burnetii DNA, with the percentage positive at the 7 farms ranging from 14.3% to 96.7%. On 4 of the farms, more than 75% of the samples were positive (farms 3, 4, 5, and 7).
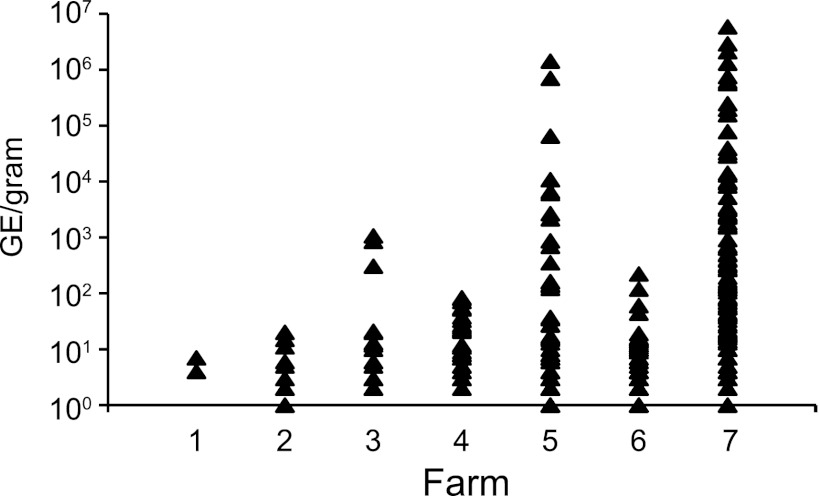
Because the PCR used to detect C. burnetii was quantitative, it was possible to estimate the number of genome equivalents (GE) in each sample. The mean number of GE detected on the farms was calculated (Table 1). Two of the farms had a very high mean number of GE in the environmental samples. Farms 5 and 7 had mean numbers of GE greater than 30,000 GE per gram of material, whereas the other farms had less than 120 GE per gram. The very high mean levels of C. burnetii DNA at farms 5 and 7 were largely a reflection of a few samples having extremely large amounts of C. burnetii DNA. Four samples at farm 7 had greater than 1 million GE per gram, and one sample at farm 5 had over 1 million GE per gram. For some of the farms, the percentage of positive samples did not reflect the true burden of C. burnetii on those farms. Although farms 1, 2, and 6 had between 14.3 and 50.5% of their samples positive, there were fewer than 10 GE of C. burnetii per gram of sample on these farms (Table 1). The most concentrated sample on these three farms contained only 220 GE per gram. The distribution of GE for all of the positive samples is displayed in Fig. 1. The number of Q fever cases associated with each of these seven farms is shown in Table 1.
Fig 1.
Environmental bulk and vacuum samples were collected on 7 farms in Washington and Montana. Genomic DNA was extracted from the samples and tested for the presence of C. burnetii DNA by quantitative PCR. The graph shows the number of genome equivalents per gram for each positive sample collected at each farm.
Although the majority of samples taken at farms were bulk samples, there were also vacuum and sponge samples taken at three of the farms. Table 2 shows the number of samples taken of each type, along with the number of positive samples and the percentage positive. A total of 15 vacuum samples were taken, and 86.7% of these were positive. Of 19 sponge samples taken, 84.2% were positive. The majority of samples taken were bulk samples, and these were 68.6% positive overall.
Table 2.
Detection of C. burnetii DNA on farms using different sampling methods
| Farm no. | Detection of C. burnetii DNA in each sample type |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Bulk |
Vacuum |
Sponge |
|||||||
| No. of samples | No. of PCR-positive samples | % positive | No. of samples | No. of PCR-positive samples | % positive | No. of samples | No. of PCR-positive samples | % positive | |
| 1 | 14 | 2 | 14.3 | 0 | 0 | NAa | 0 | 0 | NA |
| 2 | 14 | 6 | 42.9 | 0 | 0 | NA | 0 | 0 | NA |
| 3 | 20 | 19 | 95 | 0 | 0 | NA | 0 | 0 | NA |
| 4 | 30 | 29 | 96.7 | 0 | 0 | NA | 0 | 0 | NA |
| 5 | 56 | 46 | 82.1 | 3 | 3 | 100 | 5 | 5 | 100 |
| 6 | 92 | 42 | 45.7 | 5 | 3 | 60 | 14 | 11 | 78.6 |
| 7 | 131 | 101 | 77.1 | 7 | 7 | 100 | 0 | 0 | NA |
NA, not applicable.
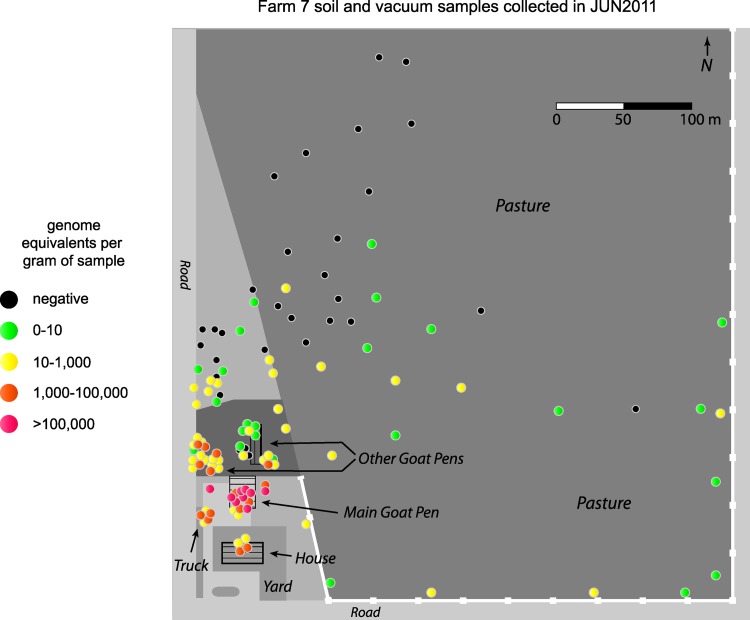
For farm 7, 138 samples were collected in June 2011, and GPS coordinates for the locations of the samples were recorded. This information was used to construct a map with color-coded markers indicating the position and number of GE for each environmental sample on the farm (Fig. 2). Most of the negative samples taken on the farm were located in the pasture to the northeast of the goat pens, with positive samples having increasing numbers of GE as the samples were closer to the main goat pen just above the house. The main goat pen had most of the samples on the farm that contained more than 100,000 GE per gram (range, 3,000 to 5,785,000), and most of the goat birthing took place in this area. Samples with more than 1,000 GE per gram were also found in the farmhouse (range, 28,000 to 39,000) and some of the other goat housing areas. The small group of positive samples to the west of the house was taken from a truck used on the farm.
Fig 2.
GPS coordinates were recorded for environmental samples collected on farm 7. The positions of the samples were mapped onto a diagram of the farm and color coded to indicate the number of genome equivalents of C. burnetii detected in the samples by quantitative PCR. Samples from the house and the truck were vacuum samples, and all others were bulk samples.
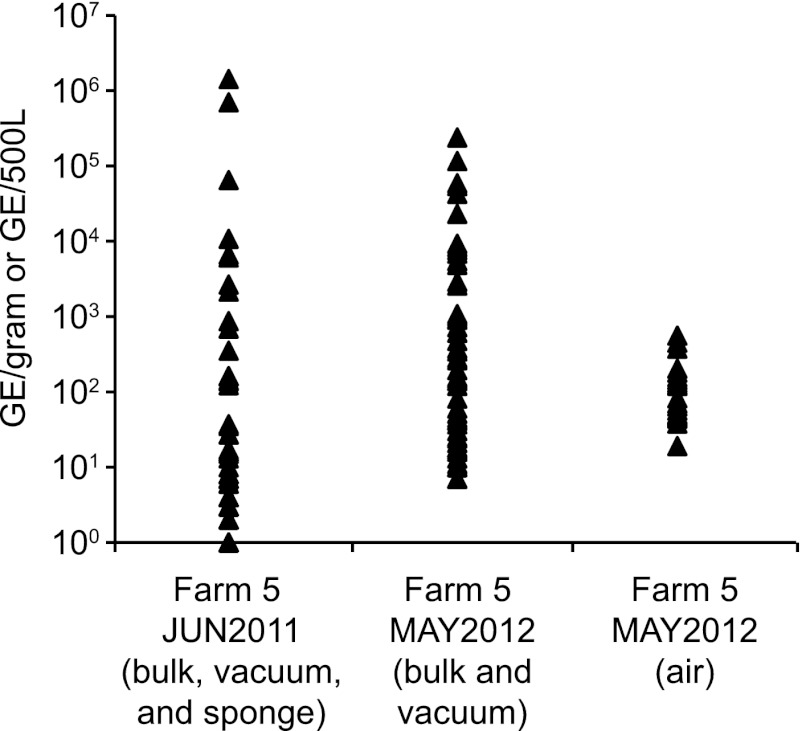
At farm 5, 64 samples were collected in June 2011, and in May 2012, a second round of 78 environmental samples was collected. This allowed a comparison of C. burnetii in the environment during the outbreak to C. burnetii in the environment 1 year postoutbreak (Table 3). By some measures, the environmental burden decreased from 2011 to 2012. For example, the percentage of positive samples decreased from 84.4% to 69.2% and the mean number of GE/gram for all samples went from 34,273 to 7,508 (Table 3). However, the decrease in the mean was largely due to the lack of a few samples with very high numbers of GE/gram. The median number of GE increased from 7 to 28 between 2011 and 2012. Figure 3 displays the number of GE/gram for all of the positive samples taken at farm 5 in 2011 and 2012. It was also possible to collect air samples in May 2012 (Table 3). A high percentage of the air samples were positive on the farm (66.7%); however, they contained only small amounts of C. burnetii. The mean number of GE per 500 liters of air was less than 100 GE, and this is near the limit of detection for the assay.
Table 3.
Detection and quantitation of C. burnetii DNA on farm 5a
| Date of sampling | Sample type(s) | No. of samples | No. of PCR-positive samples | % positive | Mean no. of GE ± SD | Median no. of GE |
|---|---|---|---|---|---|---|
| June 2011 | Bulk, sponge, vacuum | 64 | 54 | 84.4 | 34,273 ± 194,514 | 7 |
| May 2012 | Bulk, sponge, vacuum | 78 | 54 | 69.2 | 7,508 ± 30,969 | 28 |
| May 2012 | Air | 30 | 20 | 66.7 | 98.4 ± 138.1 | 49 |
GE, genome equivalents; SD, standard deviation.
Fig 3.
Environmental bulk, sponge, and vacuum samples were collected on farm 5 in June 2011 and May 2012. Air samples were collected in May 2012. Genomic DNA was extracted from the samples and tested for the presence of C. burnetii DNA by quantitative PCR. The graph shows the number of genome equivalents per gram for each positive bulk and vacuum sample, the number of genome equivalents per sponge for each sponge sample, and the number of genome equivalents per 500 liters for the air samples.
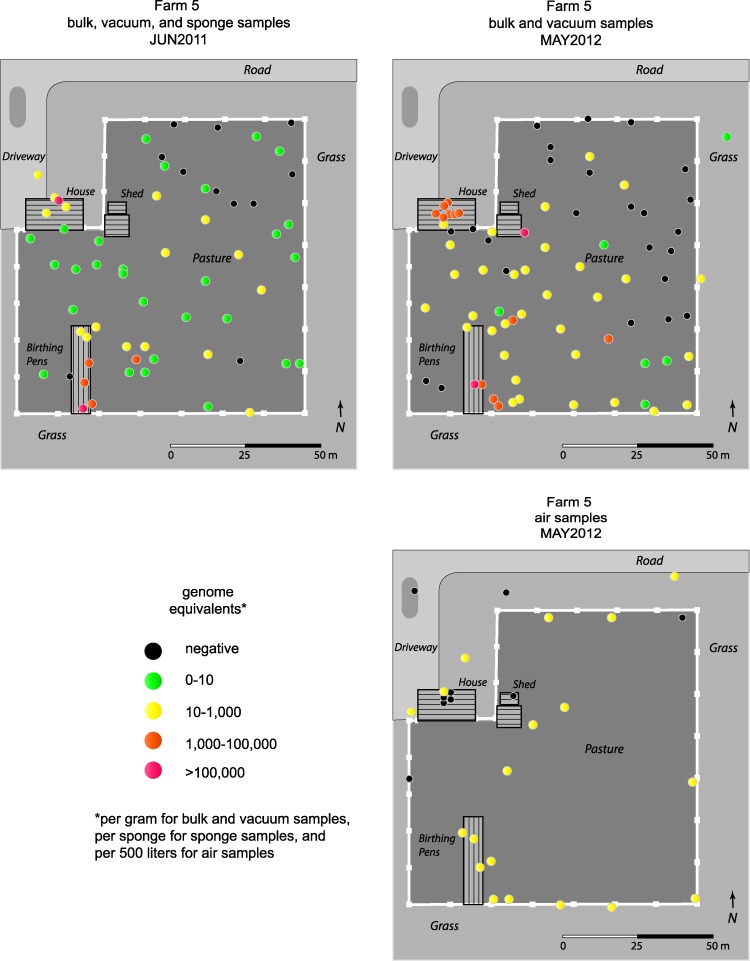
For samples taken at farm 5, maps indicating the locations of the samples were constructed (Fig. 4). In June 2011, the highest numbers of GE were observed at the goat birthing area (range, 6,000 to 1,400,000 GE/gram), with one additional highly concentrated sample taken from the farm house (692,000 GE/gram). The samples from the northeast corner of the pasture tended to be negative and were collected the furthest from the goat birthing area. This farm covered a much smaller area than farm 7 (17,000 m2 for farm 5 versus 165,000 m2 for farm 7), and most of the samples taken at the perimeter of the pasture were less than 100 m from the goat birthing area. Despite this short distance, high concentrations of C. burnetii were not found at the edges of the pasture. In May 2012, the most highly positive samples were either in the birthing area (range, 900 to 116,000 GE/gram) or in the house. Seven out of eight samples taken from the farm house had more than 1,000 GE/gram (range, 3,000 to 58,000 GE/gram). These samples were vacuum samples taken from the floors and furniture of the house. In 2012, samples taken from the pasture within 30 m of the birthing pens were mostly positive and were more likely to have more than 10 GE/gram than similar samples taken in 2011. Samples taken from the northeast corner of the pasture continued to be mostly negative in 2012. Mapping of the air samples shows that almost all of the samples taken from the goat birthing area and the pasture were positive. Only 1 out of 5 air samples taken in the house were positive, and 2 out of 5 samples taken outside the pasture were negative.
Fig 4.
GPS coordinates were recorded for the bulk, sponge, and vacuum samples collected on farm 5 in June 2011 and the bulk, vacuum, and air samples collected in May 2012. The positions of the samples were mapped onto diagrams of the farm and color coded to indicate the number of genome equivalents of C. burnetii detected in the samples by quantitative PCR. For the bulk and vacuum samples, the numbers of GE/gram are reported, for the sponge samples, the numbers of GE/sponge are reported, and for air samples, the numbers of GE/500 liters are reported. Three vacuum samples were taken from the house, four sponge samples were taken from goat pens, and all other samples were bulk.
Attempts were made to isolate viable C. burnetii from both animal and environmental samples. A total of six C. burnetii isolates were obtained during this investigation. One isolate was obtained from a bulk environmental sample taken from a goat pen, and two isolates came from vacuum samples taken from farm houses (Table 4). SNP-based genotyping (17) identified all three isolates as sequence type 8 (ST8), a genogroup that has also been designated by multilocus sequence typing (MLST) (18). Three animal isolates were also obtained in association with this outbreak: one from a goat placenta collected in April 2011, another from a goat vaginal swab taken in June 2011, and the third from a goat placenta collected in September 2011 (Table 4). All three of the animal isolates were also typed as ST8.
Table 4.
Viable isolates obtained from animal and environmental sourcesa
| Isolate | Source | Collection date | No. of GE in sample |
|---|---|---|---|
| GP-WA1 | Goat placenta | April 2011 | 1.5 × 108/gram |
| GP-MT2 | Goat placenta | September 2011 | 2.5 × 108/gram |
| GS-MT1 | Goat vaginal swab | June 2011 | 673,000/swab |
| ES-WA1 | Bulk sample from goat pen | June 2011 | 2,480/gram |
| ES-WA2 | Vacuum sample from house | June 2011 | 38,800/gram |
| ES-MT1 | Vacuum sample from house | June 2011 | 692,000/gram |
All six isolates contained the plasmid QpRS and were sequence type 8. GE, genome equivalents.
DISCUSSION
The data presented here show that C. burnetii can easily spread from an infected goat herd into the farm environment and persist there for at least 1 year. Evidence of environmental contamination was found on all 7 farms described in this study. Most of the organisms were found in areas of the farms where the goats give birth. Based on the replication of C. burnetii to very high levels in the placenta of infected goats (6) and the detection of very large amounts of C. burnetii in goat placental tissue collected at these farms (Table 3), it is not surprising that the largest amounts of organism were found in places where birth products would be deposited. C. burnetii was also detected in other areas of the farms where goats have access, but the numbers of organisms were much lower than they were in the birthing areas.
Previous studies have demonstrated that infectious C. burnetii can travel long distances on the wind, perhaps as far as 18 km (9, 19). However, Q fever cases occurring at such long distances from the animal source appears to be uncommon, and other studies have seen cases dropping to near zero at distances greater than 400 m from the infected animals (19). In this study, samples were taken in the farm environment at various distances from the animal birthing areas. This allowed a spatial analysis of C. burnetii on these farms at a fine scale. The numbers of organisms detected dropped off fairly quickly with the distance from the goat birthing areas. On the farms analyzed in the greatest detail (farms 5 and 7), only small quantities of C. burnetii were found at distances greater than 50 m from goat housing/birthing areas. High concentrations of C. burnetii DNA were found only in the goat pens and in the farm houses. Many factors will influence transfer of C. burnetii from animals to the environment, but it is expected that hot and dry conditions will favor wind-borne spread of C. burnetii aerosols. The relatively short distances traveled by C. burnetii on these farms suggest that conditions at these two farms did not favor wind-borne spread.
Although wind did not appear to be able to transport large amounts of C. burnetii from the goat pens to the perimeter of the farms, C. burnetii DNA was found in 67% of air samples taken at farm 5 in May 2012. This high percentage was surprising given that many of the samples were taken at the perimeter of the farm away from birthing areas and that shedding from the goats had declined between 2011 and 2012 (our unpublished data). Despite the high percentage of positives, the numbers of genomes detected in most of the samples were near the limits of detection, and most of the PCRs were detecting close to a single organism. Because such a large volume of air (500 liters) can be concentrated on a small filter, the air sampling technique is very sensitive for detection of C. burnetii in the farm environment. Air sampling for this study was performed only at this farm, which had a history of goat abortion and Q fever cases. It is not clear whether air sampling would detect such low levels of C. burnetii DNA even on farms with no recent history of C. burnetii contamination. A recent study from the Netherlands did compare aerosol samples from farms with known Q fever abortion waves to samples from farms without a Q fever history and found a higher percentage of positive samples on the farms with Q fever history (13). However, 66.7% of samples from farms with no Q fever history were positive, and the differences between infected and noninfected farms were not statistically significant (13). The data suggest that low levels of aerosols containing detectable amounts of C. burnetii DNA may be common on any farm environment.
With such low levels of C. burnetii detected by PCR, contamination is a concern. However, several points argue against contamination. First, samples taken outside the pasture area were mostly negative, whereas all of the samples taken in the goat birthing area were positive. Second, of the 5 samples taken inside the house, only one was positive, as would be expected from the relatively static air found inside the home. Finally, the last sample taken was at the perimeter of the pasture and it was negative. Therefore, the results show that a low level of airborne C. burnetii exists on the farm, but these organisms did not seem to travel long distances and there is no evidence regarding the viability of the airborne organisms on these farms.
If wind did not play a major role in spreading the bacteria, then it might be expected that the goats themselves would be an effective means to spread C. burnetii through the environment. Vaginal swabs taken from goats on these farms indicated that high levels of C. burnetii were being shed. It is possible that shedding by these infected goats can contaminate the environment over a large area. However, C. burnetii was primarily concentrated in and around animal pens and birthing areas. The trafficking patterns of the goats on these farms are not known, so it is possible that the high burden of C. burnetii in goat pens compared to that in pastures reflects the fact that goats may have spent most of their time in the pens. In addition to the goat pens, the other areas on the farms that had a high burden of C. burnetii were places that had human activity. The relatively strong PCR signals in samples from houses on farms 5 and 7 and the truck on farm 7 suggest that human activity played an important role in environmental contamination. On farm 5, the organisms in the house persisted into 2012 despite efforts by the homeowners to decontaminate the area. Although the effectiveness of the decontamination efforts was not evaluated, the high levels of organism in the goat pens and birthing areas and the persistence of high levels of C. burnetii in the home on farm 5 suggest that there was repeated introduction of C. burnetii into the home, most likely by people tracking C. burnetii back into the house after farm work. In a large environmental study published in 2010, many C. burnetii-positive samples were found at locations that were not near animals (10). A likely explanation for the introduction of C. burnetii into these locations is foot traffic from people that had recently visited areas directly contaminated by infected livestock. Such spreading by human foot traffic emphasizes the need for careful decontamination of shoes and clothing after working in or visiting an area with C. burnetii-infected livestock.
Six new isolates of C. burnetii were established from samples associated with this outbreak. All six of the isolates were estimated to have approximately 60 copies of IS1111a, contain the QpRS plasmid, and fall into the ST8 genotype. The method used for the genotyping was an SNP-based rapid typing method (17) that can define sequence types that are compatible with previously reported MLST sequence types (18). The ST8 genotype has been found previously in numerous human heart valve samples and in other goats from the western United States (18). Although the samples from this outbreak used for genotyping were all from animals and environmental samples, the close association with human acute Q fever cases suggests that the ST8 genotype also caused the acute Q fever cases associated with this outbreak with a range of symptoms and severity of disease.
This analysis demonstrates that C. burnetii can spread to the environment from infected goats. C. burnetii found in the environment can remain viable and infectious. In the farms examined, the organisms did not appear to spread for great distances and were confined to the vicinity of goat birthing areas. Because most of the organisms were found in goat birthing areas, it is likely that release of C. burnetii during parturition is the primary means for contamination of the environment, and ongoing shedding from infected goats plays a minor role. Although C. burnetii was able to be detected in the air on the farm, the very low numbers of C. burnetii isolates found away from the animal housing areas suggest that wind-borne spread under these conditions was not significant. The pattern of positive samples suggests that humans on the farms serve as an important vehicle for transportation of viable C. burnetii from the goat pens to the farm house, other parts of the farm, and perhaps beyond.
ACKNOWLEDGMENTS
We thank the farm owners for their cooperation in this study.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the CDC or the Department of Health and Human Services.
Footnotes
Published ahead of print 11 January 2013
REFERENCES
- 1. Maurin M, Raoult D. 1999. Q fever. Clin. Microbiol. Rev. 12:518–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hartzell JD, Wood-Morris RN, Martinez LJ, Trotta RF. 2008. Q fever: epidemiology, diagnosis, and treatment. Mayo Clin. Proc. 83:574–579 [DOI] [PubMed] [Google Scholar]
- 3. van der Hoek W, Dijkstra F, Schimmer B, Schneeberger PM, Vellema P, Wijkmans C, ter Schegget R, Hackert V, van Duynhoven Y. 2010. Q fever in the Netherlands: an update on the epidemiology and control measures. Euro Surveill. 15(12):pii=19520. http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19520 [PubMed] [Google Scholar]
- 4. Anderson AD, Kruszon-Moran D, Loftis AD, McQuillan G, Nicholson WL, Priestley RA, Candee AJ, Patterson NE, Massung RF. 2009. Seroprevalence of Q fever in the United States, 2003-2004. Am. J. Trop. Med. Hyg. 81:691–694 [DOI] [PubMed] [Google Scholar]
- 5. McQuiston JH, Childs JE. 2002. Q fever in humans and animals in the United States. Vector Borne Zoonotic Dis. 2:179–191 [DOI] [PubMed] [Google Scholar]
- 6. Welsh HH, Lennette EH, Abinanti FR, Winn JF. 1958. Air-borne transmission of Q fever: the role of parturition in the generation of infective aerosols. Ann. N. Y. Acad. Sci. 70:528–540 [DOI] [PubMed] [Google Scholar]
- 7. Scott GH, Williams JC. 1990. Susceptibility of Coxiella burnetii to chemical disinfectants. Ann. N. Y. Acad. Sci. 590:291–296 [DOI] [PubMed] [Google Scholar]
- 8. McCaul TF, Williams JC. 1981. Developmental cycle of Coxiella burnetii: structure and morphogenesis of vegetative and sporogenic differentiations. J. Bacteriol. 147:1063–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hawker JI, Ayres JG, Blair I, Evans MR, Smith DL, Smith EG, Burge PS, Carpenter MJ, Caul EO, Coupland B, Desselberger U, Farrell ID, Saunders PJ, Wood MJ. 1998. A large outbreak of Q fever in the West Midlands: windborne spread into a metropolitan area? Commun. Dis. Public Health 1:180–187 [PubMed] [Google Scholar]
- 10. Kersh GJ, Wolfe TM, Fitzpatrick KA, Candee AJ, Oliver LD, Patterson NE, Self JS, Priestley RA, Loftis AD, Massung RF. 2010. Presence of Coxiella burnetii DNA in the environment of the United States, 2006 to 2008. Appl. Environ. Microbiol. 76:4469–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tissot-Dupont H, Amadei MA, Nezri M, Raoult D. 2004. Wind in November, Q fever in December. Emerg. Infect. Dis. 10:1264–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schimmer B, Ter Schegget R, Wegdam M, Zuchner L, de Bruin A, Schneeberger PM, Veenstra T, Vellema P, van der Hoek W. 2010. The use of a geographic information system to identify a dairy goat farm as the most likely source of an urban Q-fever outbreak. BMC Infect. Dis. 10:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Bruin A, van der Plaats RQ, de Heer L, Paauwe R, Schimmer B, Vellema P, van Rotterdam BJ, van Duynhoven YT. 2012. Detection of Coxiella burnetii DNA on small-ruminant farms during a Q fever outbreak in the Netherlands. Appl. Environ. Microbiol. 78:1652–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Centers for Disease Control and Prevention (CDC) 2011. Notes from the field: Q fever outbreak associated with goat farms—Washington and Montana, 2011. MMWR Morb. Mortal. Wkly. Rep. 60:1393. [PubMed] [Google Scholar]
- 15. Fitzpatrick KA, Kersh GJ, Massung RF. 2010. Practical method for extraction of PCR-quality DNA from environmental soil samples. Appl. Environ. Microbiol. 76:4571–4573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kersh GJ, Lambourn DM, Self JS, Akmajian AM, Stanton JB, Baszler TV, Raverty SA, Massung RF. 2010. Coxiella burnetii infection of a Steller sea lion (Eumetopias jubatus) found in Washington State. J. Clin. Microbiol. 48:3428–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hornstra HM, Priestley RA, Georgia SM, Kachur S, Birdsell DN, Hilsabeck R, Gates LT, Samuel JE, Heinzen RA, Kersh GJ, Keim P, Massung RF, Pearson T. 2011. Rapid typing of Coxiella burnetii. PLoS One 6:e26201 doi:10.1371/journal.pone.0026201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Glazunova O, Roux V, Freylikman O, Sekeyova Z, Fournous G, Tyczka J, Tokarevich N, Kovacava E, Marrie TJ, Raoult D. 2005. Coxiella burnetii genotyping. Emerg. Infect. Dis. 11:1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gilsdorf A, Kroh C, Grimm S, Jensen E, Wagner-Wiening C, Alpers K. 2008. Large Q fever outbreak due to sheep farming near residential areas, Germany, 2005. Epidemiol. Infect. 136:1084–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]