Abstract
Various forms of stress can cause an attenuation of bulk translation activity and the accumulation of nontranslating mRNAs into cytoplasmic messenger RNP (mRNP) granules termed processing bodies (P-bodies) and stress granules (SGs) in eukaryotic cells. Furfural and 5-hydroxymethylfurfural (HMF), derived from lignocellulosic biomass, inhibit yeast growth and fermentation as stressors. Since there is no report regarding their effects on the formation of cytoplasmic mRNP granules, here we investigated whether furfural and HMF cause the assembly of yeast P-bodies and SGs accompanied by translational repression. We found that furfural and HMF cause the attenuation of bulk translation activity and the assembly of cytoplasmic mRNP granules in Saccharomyces cerevisiae. Notably, a combination of furfural and HMF induced the remarkable repression of translation initiation and SG formation. These findings provide new information about the physiological effects of furfural and HMF on yeast cells, and also suggest the potential usefulness of cytoplasmic mRNP granules as a warning sign or index of the deterioration of cellular physiological status in the fermentation of lignocellulosic hydrolysates.
INTRODUCTION
Lignocellulosic biomass, including agricultural and wood residues, is a promising source for bioethanol production, because it is regenerable, abundant, and uncompetitive with food resources. Lignocellulosic biomass is often pretreated with acid or hot-compressed water for saccharification (1, 2). However, these pretreatments generate fermentation inhibitors such as furfural and 5-hydroxymethylfurfural (HMF) (1, 3). Furfural and HMF are furaldehydes and degradation products of pentoses and hexoses, respectively (4, 5). They inhibit the growth of yeast cells and subsequent fermentation in a dose-dependent manner (6). Although furfural and HMF can act synergistically, yeast cells are more sensitive to growth inhibition by furfural than by HMF at the same concentration (7–9). It is known that furfural affects glycolytic activity and the tricarboxylic acid cycle (10, 11). It has also been reported that furfural causes oxidative stress and reduces the activities of various dehydrogenases in yeast cells (12, 13). Although several genome-wide analyses regarding the response to furfural and HMF have been carried out (14, 15), the cellular stress response caused by furfural and HMF is not fully understood in yeast cells.
Various forms of stress can reduce bulk translation activity and consequently elevate levels of nontranslating mRNAs. In eukaryotic cells, nontranslating mRNAs can be recruited from polysomes and segregated into cytoplasmic messenger RNP (mRNP) granules via associations with various RNA-associated proteins under stressed conditions (16, 17). Saccharomyces cerevisiae as well as mammalian cells has processing bodies (P-bodies) and stress granules (SGs) as cytoplasmic mRNP granules. P-bodies generally contain the mRNA decay machinery, including decapping enzymes such as Dcp1 and Dcp2 (18), a decapping activator (Dhh1) (18), an exonuclease (Xrn1) (18), deadenylases (Pop2 and Ccr4) (19), and nontranslating mRNAs (16, 20, 21). On the other hand, SGs typically consist of a subset of RNA binding protein (Ngr1) (17, 22), a 40S ribosomal subunit (Rps30A) (17, 23), a Pab1 binding protein (Pbp1) (22, 23), translation initiation-related factors (Eap1, Prt1, Rpg1, and Nip1) (22, 23), and nontranslating mRNAs (17). Therefore, these proteins are used as representative markers of P-bodies and SGs (16–18, 20, 22–25).
In S. cerevisiae, several kinds of stress such as glucose depletion, severe heat shock, severe ethanol stress, and exposure to sodium azide (NaN3) induce the formation of SGs with a pronounced repression of translation (22–25). On the other hand, it has been reported that high Ca2+ levels, hyperosmotic shock, and acidic stress can induce P-body formation without pronounced repression of translation and SG formation (22, 26, 27). It has also been reported that yeast SGs are assembled following P-body formation during severe ethanol stress, glucose deprivation, and NaN3 treatment (22, 24, 25). These findings suggest that SG formation requires severer stress than P-body formation. It has been indicated that P-bodies and SGs serve as sites of nontranslating mRNA storage under stressed conditions and are responsible for translational repression (16, 17, 28–30). Additionally, both granules might play important roles in the rapid recovery from stress, since the segregated mRNAs in these granules can rapidly re-enter the translation initiation process when stress is eliminated (16, 17, 22, 24).
In order to understand posttranscriptional events in the response to furfural and HMF in yeast cells, here we investigated whether these furaldehydes induce the formation of cytoplasmic mRNP granules and repression of bulk translation activity. We found that furfural and HMF cause the attenuation of bulk translation activity and the assembly of cytoplasmic mRNP granules in S. cerevisiae. Notably, a combination of furfural and HMF induced the remarkable repression of translation initiation and SG formation. This is the first report about the formation of cytoplasmic mRNP granules by furaldehydes in S. cerevisiae, providing new information about the physiological effects of furfural and HMF on yeast cells.
MATERIALS AND METHODS
Strain, plasmids, and media.
The S. cerevisiae strain used in this study was W303-1A (MATa his3-11,15 leu2-3,112 trp1-1 ade2-1 ura3-1 can1-100) as a laboratory strain. Cells were cultured in 50 ml of SD medium (2% glucose and 0.67% yeast nitrogen base without amino acids) with appropriate supplements of amino acids and bases at 30°C with reciprocal shaking (120 rpm) in Erlenmeyer flasks (300 ml). Plasmids used for the observation of P-bodies and SGs were previously described (24, 27, 31). Briefly, the DCP1 gene amplified with primers (5′-AAAAAGCAGCTTGGGAATTTCTAGAGGAGA-3′ and 5′-CCTCTCTCGAGAAGCAAAAGAATCTTTTGGCTCATT-3′) was digested with XbaI and XhoI and cloned into the XbaI-XhoI sites of pAUR-ZRC1-GFP (pAUR-ZRC1-green fluorescent protein) (32) to construct pAUR-DCP1-GFP. The DCP2 gene was amplified using primers (5′-GAGCAGATTGATCCCTCTAGAGAGTTGCTG-3′ and 5′-CTTATTCTTGCTAGCCCTATGCAAAATGCT-3′), digested with XbaI and NheI, and cloned into the XbaI-NheI sites of pJK67 (33) to construct YIp-DCP2-GFP. The NGR1 gene amplified with primers (5′-AACCCTCTAGAACGTTATGGATGGGAGATT-3′ and 5′-CTTTTCTCGAGAATGTAGAAGAGAAGGATG-3′) was digested with XbaI and XhoI and cloned into the XbaI-XhoI sites of pJK67 to construct YIp-NGR1-GFP. The RPS30A gene was amplified (5′-GCACATCTAGACAGACGGCCGGCAGGAGGAA-3′ and 5′-TTAATCTCGAGATTGGACGGATGGACCTGGG-3′), digested with XbaI and XhoI, and cloned into the XbaI-XhoI sites of pAUR-ZRC1-GFP to construct pAUR-RPS30A-GFP. pAUR-DCP1-GFP and YIp-NGR1-GFP were linearized by StuI and introduced into the yeast cells. YIp-DCP2-GFP and pAUR-RPS30A-GFP were digested with Eco47III and BsiWI, respectively.
Chemicals and analysis methods.
Furfural and HMF were obtained from Wako (Osaka, Japan) and Tokyo Chemical Industry (Tokyo, Japan), respectively. The purity of both furaldehydes was >95.0%. To prevent oxidation, they were immediately dissolved in dimethyl sulfoxide (DMSO) as 2 M stock solutions after new bottles were opened (34). The stock solutions were stored at −30°C in small quantities. Exponentially growing cells (ca. 1 × 107 cells/ml) were further incubated with various concentrations of furaldehydes at 30°C. Since the common concentrations of furfural and HMF in bioethanol fermentors for lignocellulosic biomass and in previous reports are 20 to 50 mM (3, 6, 8–15), we examined the effects of 10 to 70 mM furfural and HMF. We verified that DMSO has no effect on the formation of mRNP granules and translation activity. The preparation of the yeast extract and sucrose gradient separation for the polysome analysis were carried out by the methods of Inada and Aiba (35) using a gradient master and fractionator (BioComp Instruments Inc.; catalog no. 107-201 M and 152-002). A Leica AF6500 (Leica Microsystems Vertrieb GmbH, Germany) fluorescence microscopic system was used for the microscopic analysis. Cells treated with furfural or HMF were immediately observed without fixation. All experiments were repeated more than 4 times.
RESULTS
Furfural and HMF cause the formation of P-bodies.
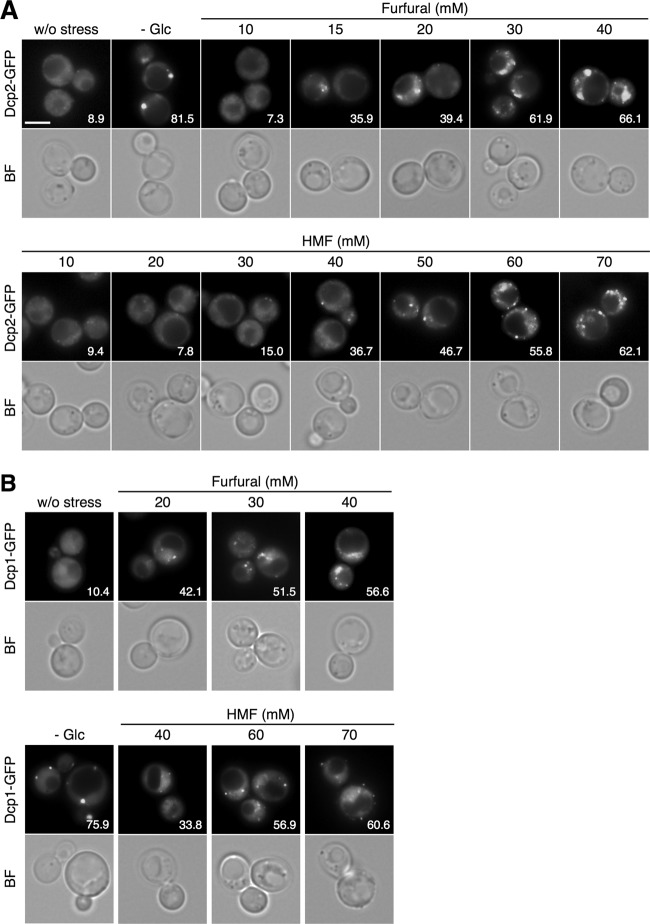
First we examined whether furfural and HMF can induce the assembly of P-bodies. Dcp1 and Dcp2 are decapping enzymes and the core components of yeast P-bodies (18). Since they have been used in numerous previous studies (18, 20, 22, 26–29, 31, 36), here we used Dcp1-GFP and Dcp2-GFP as P-body markers. As shown in Fig. 1, these P-body markers formed granules in the cytoplasm when cells were treated with furfural or HMF for 30 min at 30°C. Treatment with furaldehydes caused a dose-dependent increase in the number of cells containing P-bodies. Over half of the cells formed cytoplasmic foci of Dcp1-GFP and Dcp2-GFP after the treatment with 30 mM furfural or 60 mM HMF for 30 min. We observed similar phenomena using other P-body markers such as Dhh1-GFP (18, 19, 27, 31) and Xrn1-GFP (22, 25) (data not shown). These results indicate that these furaldehydes can induce the formation of P-bodies in yeast cells. No clear correlation was observed between the concentration of furaldehydes and the number or size of P-bodies.
Fig 1.
P-body formation is induced by furfural and HMF in S. cerevisiae. Cells in an exponential phase of growth in SD medium were treated with furfural (10 to 40 mM) or HMF (10 to 70 mM) at 30°C for 30 min or subjected to glucose deprivation stress (-Glc) for 15 min. The assembly of P-bodies was examined with Dcp2-GFP (A) and Dcp1-GFP (B). The white bar indicates 5 μm. BF, bright field. At least 150 cells under each condition were examined. Numbers in the panels indicate percentages of cells with GFP foci.
Furfural but not HMF causes the formation of stress granules.
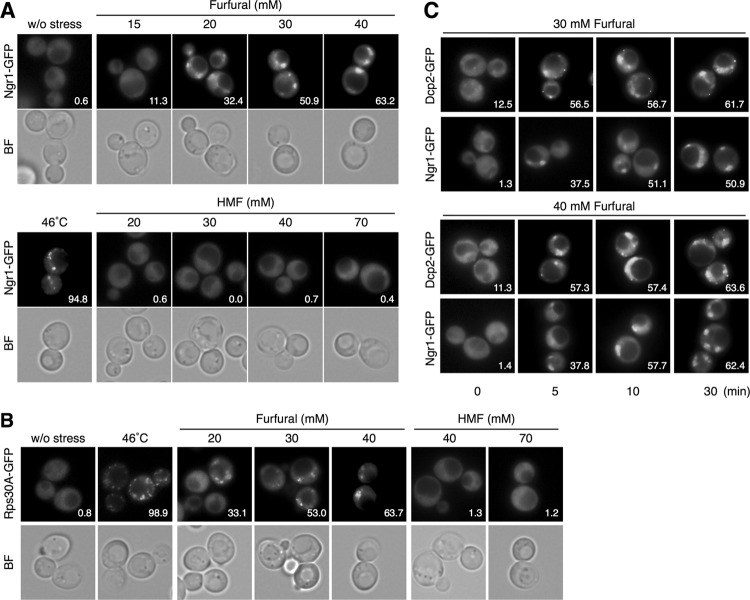
We next investigated the effects of furfural and HMF on the formation of SGs. Ngr1-GFP and Rps30A-GFP were used as SG markers. Ngr1 (negative growth regulatory protein) and Rps30A (ribosomal protein of the 40S small subunit) are core components of SG and representative SG markers (22–24). As shown in Fig. 2A and B, treatment with furfural caused the formation of Ngr1-GFP foci or Rps30A-GFP foci in the cytoplasm. Furfural caused a dose-dependent increase in the number of cells containing SGs, and over half of the cells formed cytoplasmic granules of Ngr1-GFP and Rps30A-GFP following treatment with more than 30 mM furfural for 30 min (Fig. 2A and B). The number and size of the granules did not show a clear correlation with the concentration of furfural. We verified the SG formation caused by furfural using other components such as Pbp1-GFP and Eap1-GFP (22, 23) (data not shown). These results clearly indicate that furfural can induce the formation of SGs.
Fig 2.
SG formation is induced by furfural but not HMF in S. cerevisiae. Cells in an exponential phase of growth in SD medium were treated with furfural (15 to 40 mM) or HMF (20 to 70 mM) at 30°C for 30 min or with robust heat shock at 46°C for 10 min. The assembly of SGs was examined with Ngr1-GFP (A) and Rps30A-GFP (B). (C) Time course of SG formation caused by furfural. Cells were treated with 30 or 40 mM furfural for 0 to 30 min. The assembly of P-bodies and SGs was examined with Dcp2-GFP and Ngr1-GFP, respectively. At least 150 cells under each condition were examined. Numbers in the panels indicate percentages of cells with GFP foci.
On the other hand, HMF treatment (20 to 70 mM for 30 min) did not cause the formation of SGs (Fig. 2A and B). We have reported that SG formation upon ethanol stress requires a more severe stress and a longer period of time than P-body formation (24). Additionally, Buchan et al. have reported that SGs are assembled following P-body formation during glucose deprivation or NaN3 treatment (22, 25). Therefore, we investigated the effects of prolonged treatment with HMF at higher concentrations (40 to 200 mM for 120 min). Even prolonged treatment with 200 mM HMF for 120 min did not have any significant effect on the formation of Ngr1-GFP foci (data not shown), indicating that HMF induces almost no SG formation.
When cells were treated with 30 or 40 mM furfural, it took around 10 min to form SGs in more than 50% of cells, while it took only 5 min to form P-bodies in over half of the cells (Fig. 2C). This result indicates that P-body formation was induced more quickly than SG formation in the presence of furfural.
Cycloheximide prevents the formation of cytoplasmic mRNP granules.
It is well established that cycloheximide prevents the assembly of P-bodies and SGs by trapping mRNAs in polysomes (22, 29). We also examined the effects of cycloheximide on the formation of P-bodies and SGs caused by furaldehydes. Cells were treated with cycloheximide (100 μg/ml) for 1 min prior to the treatment with furfural or HMF, and then the assembly of P-bodies and SGs was investigated using Dcp2-GFP and Ngr1-GFP, respectively. Consistent with previous studies (22, 29), pretreatment with cycloheximide prevented the formation of Dcp2-GFP foci upon glucose depletion and Ngr1-GFP foci upon robust heat shock (Fig. 3). Likewise, the formation of Dcp2-GFP and Ngr1-GFP foci caused by 40 mM furfural or 70 mM HMF was also prevented by the pretreatment with cycloheximide (Fig. 3). These results indicate that the furaldehydes cause the formation of cytoplasmic mRNP granules via the release of nontranslating mRNAs from polysomes.
Fig 3.
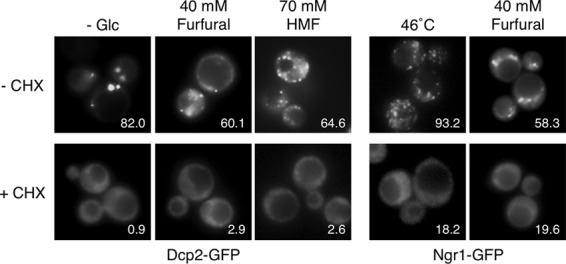
Cycloheximide prevents the formation of mRNP granules caused by the furaldehydes. Cells were treated with cycloheximide (CHX) (100 μg/ml) for 1 min prior to treatment with 40 mM furfural or 70 mM HMF for 30 min, glucose deprivation (- Glc) for 15 min, or robust heat shock at 46°C for 10 min. The assembly of P-bodies and SGs was examined with Dcp2-GFP and Ngr1-GFP, respectively. At least 150 cells under each condition were examined. Numbers in the panels indicate percentages of cells with GFP foci.
Furfural and HMF reduce bulk translation activity.
The formation of cytoplasmic mRNP granules is often accompanied by translational repression (20, 23, 25, 27). Since we found that furfural and HMF can induce the formation of cytoplasmic mRNP granules, we examined how these furaldehydes affect bulk translation activity using polysome profiles. Consistent with a previous study (37), 10% ethanol stress caused a pronounced reduction in the polysome fraction and a concomitant increase in the monosome fraction (80S), indicating the repression of translation initiation by ethanol stress (Fig. 4). Treatment with furfural (20 mM and 40 mM) for 30 min led to a clear increase in the monosome fraction (80S), indicating that furfural caused an attenuation of bulk translation initiation (Fig. 4). Additionally, HMF also caused a gradual reduction in the polysome fraction and a concomitant increase in the 80S monosome fraction. These results clearly indicate that furfural and HMF have negative effects on translation and subsequently increase the levels of nontranslating mRNA, although the repressive effects of these furaldehydes were apparently milder than that of 10% ethanol stress. Elevation of the nontranslating mRNA levels upon the treatment with furfural or HMF presumably contributes to the formation of mRNP granules.
Fig 4.
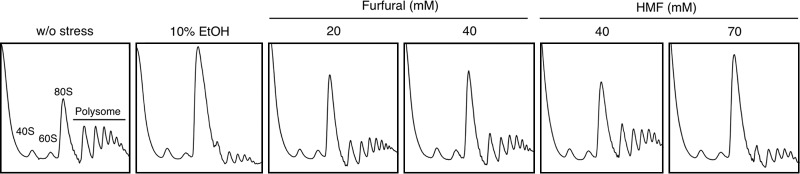
Polysome profiles of cells treated with furfural, HMF, or ethanol were determined. Cells in an exponential phase of growth were treated with furfural (20 mM or 40 mM for 30 min), HMF (40 mM or 70 mM for 30 min), or ethanol (10% for 30 min). The polysome, 40S (small ribosomal subunit), 60S (large ribosomal subunit), and 80S (monosome) peaks are labeled.
The combination of furfural and HMF causes severe repression of translation.
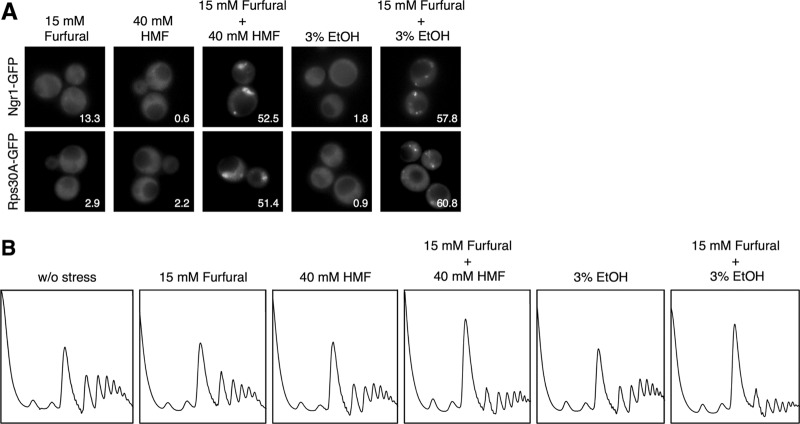
Furfural and HMF usually coexist during bioethanol production, and reportedly can act synergistically to suppress yeast cell growth even at low concentrations (8, 9). Therefore, we next examined the effects of combinations of these furaldehydes on the formation of cytoplasmic mRNP granules and bulk translation activity. Since the most common concentrations of furfural and HMF in bioethanol fermentors for lignocellulosic biomass are 20 to 50 mM (3, 6), we examined the effects of 15 mM furfural and 40 mM HMF. Although 15 mM furfural or 40 mM HMF alone did not cause SG formation, the assembly of SGs was induced when cells were treated with these furaldehydes in combination (Fig. 5A). Cytoplasmic foci of Ngr1-GFP and Rps30A-GFP were formed in over half of cells within 30 min upon the treatment with 15 mM furfural plus 40 mM HMF. Additionally, synergistic effects of furfural and HMF were observed on bulk translation activity. Although each furaldehyde alone (15 mM furfural or 40 mM HMF) had little effect on the polysome profile, in combination they caused a significant decrease in the polysome fraction and a concomitant increase in the 80S monosome fraction, indicating a definite repression of translation activity (Fig. 5B). Additionally, SG formation and severe repression of translation activity were induced within 30 min upon the treatment with 15 mM furfural plus 3% ethanol stress, although 3% ethanol alone did not cause SG formation or translational repression (Fig. 5) (24). These findings also suggest that translational repression in yeast cells might be caused in the fermentor for bioethanol production from lignocellulosic biomass.
Fig 5.
Furfural and HMF in combination induce SG formation. Cells were treated with the combination of furfural (15 mM) and HMF (40 mM) for 30 min. Cells were also treated with the combination of furfural (15 mM) and ethanol (3% [vol/vol]) for 30 min. (A) The assembly of SGs was examined with Ngr1-GFP and Rps30A-GFP. At least 150 cells under each condition were examined. Numbers in the panels indicate percentages of cells with SGs. (B) Polysome profiles of cells treated with furfural, HMF, and ethanol were determined.
DISCUSSION
Here we found that furfural and HMF can induce the formation of P-bodies and SGs in S. cerevisiae. The minimum concentration required for P-body formation in over half of the cells within 30 min was higher for HMF (60 mM) than furfural (30 mM) (Fig. 1). Additionally, SG formation was caused by furfural but not by HMF (Fig. 2). These results are not surprising because it has been reported that yeast cells are more sensitive to growth inhibition by furfural than by HMF at the same concentration (7–9). It is quite likely that yeast cells are also more sensitive to furfural than HMF regarding the formation of cytoplasmic mRNP granules.
We have reported that the formation of SGs upon ethanol stress requires a longer period of time and higher concentration than that of P-bodies (24). Furfural (30 or 40 mM) also required a longer period for SG formation (around 10 min) than P-body formation (within 5 min) (Fig. 2C). Parker and his colleagues have reported that, first, nontranslating mRNAs caused by stress form P-bodies with various proteins and then a portion of the P-bodies load translation initiation factors and form SGs in S. cerevisiae (16, 17, 22, 38). Since P-body formation was faster than SG formation upon furfural stress (Fig. 2C), our findings also seem to support this model.
Here we also demonstrated that furfural and HMF cause the repression of bulk translation initiation. The monosome fraction was clearly increased by the treatment with furfural or HMF (Fig. 4). Additionally, synergistic effects of furfural and HMF-ethanol on translational repression were observed (Fig. 5B). It is feasible that an attenuation of translation is one of the main reasons for the reduction in the growth rate by furfural and HMF.
Synergistic effects of furfural and HMF on SG formation were also observed (Fig. 5A). Yeast cells are usually exposed simultaneously to furfural and HMF during the industrial production of bioethanol from lignocellulosic biomass. Additionally, other stressors such as weak acids, phenolics, and ethanol also exist in the bioethanol fermentor of lignocellulosic hydrolysates and might have synergistic inhibitory effects with furfural and HMF on yeast cells (6). Therefore, it is conceivable that pronounced translational repression and SG formation occur frequently during industrial bioethanol production, having adverse effects on yeast cells.
Since the formation of the cytoplasmic mRNP granules reflects the deterioration of cellular physiological status, P-bodies and SGs might be useful as warning signs for the impairment of cell growth and fermentation during the process of bioethanol production from lignocellulosic biomass (Fig. 6). Additionally, tolerance toward furaldehydes is clearly strain dependent and obtained by the reduction of these compounds to less-toxic alcohols (6, 9, 39, 40). It is conceivable that yeast strains with greater tolerance of furaldehydes would require higher concentrations of furfural and HMF to cause the formation of P-bodies and/or SGs. Therefore, P-bodies and SGs might also be useful as indexes of furaldehyde tolerance in the isolation or development of yeast strains. GFP-tagged markers of P-bodies and SGs might be utilized as a powerful and easily handled tool for the control of fermentation conditions and breeding of suitable yeast strains.
Fig 6.
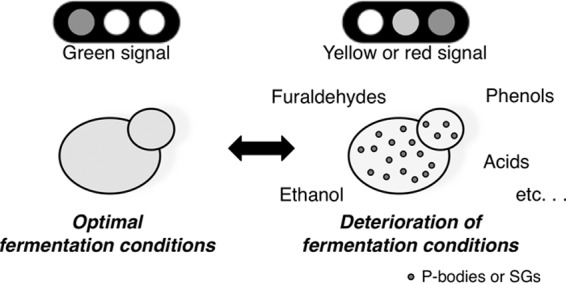
For the proper and efficient control of fermentation conditions for lignocellulosic hydrolysates, GFP-tagged markers of mRNP granules might be useful as warning signs of the deterioration of cellular physiological status.
ACKNOWLEDGMENT
This study was supported by a grant from the Japanese Ministry of Education, Culture, Sports, Science and Technology (no. 23580113).
Footnotes
Published ahead of print 28 December 2012
REFERENCES
- 1. Lu X, Yamauchi K, Phaiiboonsilpa N, Saka S. 2009. Two-step hydrolysis of Japanese beech as treated by semi-flow hot-compressed water. J. Wood Sci. 55:367–375 [Google Scholar]
- 2. Margeot A, Hahn-Hagerdal B, Edlund M, Slade R, Monot F. 2009. New improvements for lignocellulosic ethanol. Curr. Opin. Biotechnol. 20:372–380 [DOI] [PubMed] [Google Scholar]
- 3. Klinke HB, Thomsen AB, Ahring BK. 2004. Inhibition of ethanol producing yeast and bacteria by degradation product produced during pre-treatment of biomass. Appl. Microbiol. Biotechnol. 66:10–26 [DOI] [PubMed] [Google Scholar]
- 4. Antal MJ, Mok WS, Richards GN. 1990. Mechanism of formation of 5-(hydroxymethyl)-2-furaldehyde from d-fructose and sucrose. Carbohydr. Res. 199:91–109 [DOI] [PubMed] [Google Scholar]
- 5. Antal MJ, Leesomboon T, Mok WS, Richards GN. 1991. Mechanism of formation of 2-furaldehyde from d-xylose. Carbohydr. Res. 217:71–85 [DOI] [PubMed] [Google Scholar]
- 6. Almeida JRM, Modig T, Petersson A, Hähn-Hägerdal B, Lidén G, Gorwa-Grauslund MF. 2007. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J. Chem. Technol. Biotechnol. 82:340–349 [Google Scholar]
- 7. Sanchez B, Bautista J. 1988. Effects of furfural and 5-hydroxymethylfurfrual on the fermentation of Saccharomyces cerevisiae and biomass production from Candida guilliermondii. Enzyme Microb. Technol. 10:315–318 [Google Scholar]
- 8. Taherzadeh MJ, Gustafsson L, Niklasson C, Lidén G. 2000. Physiological effects of 5-hydroxymethylfurfural on Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 53:701–708 [DOI] [PubMed] [Google Scholar]
- 9. Liu ZL, Slininger PJ, Dien BS, Berhow MA, Kurtzman CP, Gorsich SW. 2004. Adaptive response of yeasts to furfural and 5-hydroxymethylfurfural and new chemical evidence for HMF conversion to 2,5-bis-hydroxymethylfuran. J. Ind. Microbiol. Biotechnol. 31:345–352 [DOI] [PubMed] [Google Scholar]
- 10. Sárvári Horváth I, Franzén CJ, Taherzadeh MJ, Niklasson C, Lidén G. 2003. Effects of furfural on the respiratory metabolism of Saccharomyces cerevisiae in glucose-limited chemostats. Appl. Environ. Microbiol. 69:4076–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gorsich SW, Dien BS, Nichols NN, Slininger PJ, Liu ZL, Skory CD. 2006. Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 71:339–349 [DOI] [PubMed] [Google Scholar]
- 12. Modig T, Liden G, Taherzadeh MJ. 2002. Inhibition effects of furfural on alcohol dehydrogenase, aldehyde dehydrogenase and pyruvate dehydrogenase. Biochem. J. 363:769–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Allen SA, Clark W, McCaffery JM, Cai Z, Lanctot A, Slininger PJ, Liu ZL, Gorsich SW. 2010. Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol. Biofuels 3:2 doi:10.1186/1754-6834-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin FM, Qiao B, Yuan YJ. 2009. Comparative proteomic analysis of tolerance and adaptation of ethanologenic Saccharomyces cerevisiae to furfural, a lignocellulosic inhibitory compound. Appl. Environ. Microbiol. 75:3765–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma M, Liu ZL. 2010. Comparative transcriptome profiling analyses during the lag phase uncover YAP1, PDR1, PDR3, RPN4, and HSF1 as key regulatory genes in genomic adaptation to the lignocellulose derived inhibitor HMF for Saccharomyces cerevisiae. BMC Genomics 11:660 doi:10.1186/1471-2164-11-660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Balagopal V, Parker R. 2009. Polysomes, P bodies and stress granules: states and fates of eukaryotic mRNAs. Curr. Opin. Cell Biol. 21:403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buchan JR, Parker R. 2009. Eukaryotic stress granules:the ins and outs of translation. Mol. Cell 36:932–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sheth U, Parker R. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reijns MAM, Alexander RD, Spiller MP, Beggs JD. 2008. A role for Q/N-rich aggregation-prone regions in P-body localization. J. Cell Sci. 121:2463–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Parker R, Sheth U. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25:635–646 [DOI] [PubMed] [Google Scholar]
- 21. Franks TM, Lykke-Andersen J. 2008. The control of mRNA decapping and P-body formation. Mol. Cell 32:605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Buchan JR, Muhlrad D, Parker R. 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183:441–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grousl T, Ivanov P, Frydlova I, Vasicová P, Janda F, Vojtová J, Malínská K, Malcová I, Nováková L, Janosková D, Valásek L, Hasek J. 2009. Robust heat shock induces eIF2α-phosphorylation-independent assembly of stress granules containing eIF3 and 40S ribosomal subunits in budding yeast, Saccharomyces cerevisiae. J. Cell Sci. 122:2078–2088 [DOI] [PubMed] [Google Scholar]
- 24. Kato K, Yamamoto Y, Izawa S. 2011. Severe ethanol stress induces assembly of stress granules in Saccharomyces cerevisiae. Yeast 28:339–347 [DOI] [PubMed] [Google Scholar]
- 25. Buchan JR, Yoon JH, Parker R. 2011. Stress-specific composition, assembly and kinetics of stress granules in Saccharomyces cerevisiae. J. Cell Sci. 124:228–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kilchert C, Weidner J, Prescianotto-Baschong C, Spang A. 2010. Defects in the secretory pathway and high Ca2+ induce multiple P-bodies. Mol. Biol. Cell 21:2624–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Iwaki A, Izawa S. 2012. Acidic stress induces the formation of P-bodies, but not stress granules, with mild attenuation of bulk translation in Saccharomyces cerevisiae. Biochem. J. 446:225–233 [DOI] [PubMed] [Google Scholar]
- 28. Coller J, Parker R. 2005. General translational repression by activators of mRNA decapping. Cell 122:875–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kedersha N, Anderson P. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Transact. 30(Pt 6):963–969 [DOI] [PubMed] [Google Scholar]
- 31. Izawa S, Kita T, Ikeda K, Miki T, Inoue Y. 2007. Formation of the cytoplasmic P-bodies in sake yeast during Japanese sake brewing and wine making. Biosci. Biotechnol. Biochem. 71:2800–2807 [DOI] [PubMed] [Google Scholar]
- 32. Izawa S, Ikeda K, Miki T, Wakai Y, Inoue Y. 2010. Vacuolar morphology of Saccharomyces cerevisiae during the process of wine making and Japanese sake brewing. Appl. Microbiol. Biotechnol. 88:277–282 [DOI] [PubMed] [Google Scholar]
- 33. Kahana JA, Schlenstedt G, Evanchuk DM, Geiser JR, Hoyt MA, Silver PA. 1998. The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol. Biol. Cell 9:1741–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miyakawa Y, Nishi Y, Kato K, Sato H, Takahashi M, Hayashi Y. 1991. Initiating activity of eight pyrolysates of carbohydrates in a two-stage mouse skin tumorigenesis model. Carcinogenesis 12:1169–1173 [DOI] [PubMed] [Google Scholar]
- 35. Inada T, Aiba H. 2005. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 24:1584–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shah KH, Zhang B, Ramachandran V, Herman PK. 2013. Processing body and stress granule assembly occur by independent and differentially regulated pathways in S. cerevisiae. Genetics 193:109–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Izawa S. 2011. Yeast mRNA flux and organelle morphology in the response to ethanol stress, p 41–49 In Kitamoto K. (ed), The new technology and functionality of fermentation foods II. CMC Press, Tokyo, Japan [Google Scholar]
- 38. Brengues M, Sheth U, Parker R. 2005. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310:486–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Petersson A, Almeida JRM, Modig T, Karhumaa K, Hähn-Hägerdal B, Gorwa-Grauslund MF, Lidén G. 2006. A 5-hydroxymethy furfural reducing enzyme encoded by the Saccharomyces cerevisiae ADH6 gene conveys HMF tolerance. Yeast 23:455–464 [DOI] [PubMed] [Google Scholar]
- 40. Modig T, Almeida JRM, Gorwa-Grauslund MF, Liden G. 2008. Variability of the response of Saccharomyces cerevisiae strains to lignocellulose hydrolysate. Biotechnol. Bioeng. 100:423–429 [DOI] [PubMed] [Google Scholar]