Abstract
Escherichia coli-mycobacterium shuttle vectors are important tools for gene expression and gene replacement in mycobacteria. However, most of the currently available vectors are limited in their use because of the lack of extended multiple cloning sites (MCSs) and convenience of appending an epitope tag(s) to the cloned open reading frames (ORFs). Here we report a new series of vectors that allow for the constitutive and regulatable expression of proteins, appended with peptide tag sequences at their N and C termini, respectively. The applicability of these vectors is demonstrated by the constitutive and induced expression of the Mycobacterium tuberculosis pknK gene, coding for protein kinase K, a serine-threonine protein kinase. Furthermore, a suicide plasmid with expanded MCS for creating gene replacements, a plasmid for chromosomal integrations at the commonly used L5 attB site, and a hypoxia-responsive vector, for expression of a gene(s) under hypoxic conditions that mimic latency, have also been created. Additionally, we have created a vector for the coexpression of two proteins controlled by two independent promoters, with each protein being in fusion with a different tag. The shuttle vectors developed in the present study are excellent tools for the analysis of gene function in mycobacteria and are a valuable addition to the existing repertoire of vectors for mycobacterial research.
INTRODUCTION
Mycobacterium tuberculosis is the single largest cause of morbidity and mortality by a bacterial pathogen. Its complex physiology and ability to survive in hostile environments, coupled with the serious trend of a rise in multidrug-resistant (MDR) and extensively drug-resistant (XDR) tuberculosis cases, has necessitated the renewal of efforts to understand the molecular basis of its pathogenesis (1). The analysis of gene expression patterns and the availability of molecular genetics tools are central to the study of the physiology and pathogenesis of any pathogen.
The molecular genetics of mycobacteria was first investigated in 1979, with the identification of plasmids from Mycobacterium avium (2, 3). Several plasmids in various other species of mycobacteria, like Mycobacterium scrofulaceum (4), Mycobacterium chelonae (5), and Mycobacterium fortuitum (6), and linear plasmids from Mycobacterium xenopi, Mycobacterium branderi, and Mycobacterium celatum (7) have since been identified. The origin of replication identified from M. fortuitum plasmid pAL5000 was widely explored and quickly became a foundation for the development of Escherichia coli-mycobacterium shuttle vectors (6, 8–10).
Routinely used shuttle vectors, such as pMV261, express genes constitutively under the control of the hsp60 gene promoter (Phsp60) (11–13), while the pMF series of vectors carry M. tuberculosis furA promoters to allow different levels of gene expression (14). However, when the expressed proteins are toxic, or when conditional gene replacement mutants are sought, it is necessary to tightly regulate the expression of the genes of interest. A number of systems are currently available for regulatable expression of genes in mycobacteria. Among these, the acetamidase-inducible systems controlled by two positive regulators, AmiC and AmiD, and a negative regulator, AmiA, were the first inducible promoter systems to be developed (15–17). Other inducible systems developed over the years include pGB-T7 RNA polymerase (RNAP), which utilizes the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible T7 promoter system, pNIT-1, which carries the isovaleronitrile-inducible gene expression system (18), pMY696, which has a pristinamycin-inducible system (19), and tetracycline (Tet)-inducible systems (20–22).
The Tet-inducible expression systems rely on induction by tetracycline or its analogs (23). Based on the source of these tetracycline regulatory systems, they are further classified as E. coli Tn10-derived pMC1s, pTACT21 and pSE100 plasmids (21, 22, 24), the Corynebacterium glutamicum TetZ locus-derived pMind vector (20), and the Streptomyces coelicolor tcp830 promoter and operator-derived pMHA series of vectors (23). The promoter is modulated by the interaction of the Tet repressor (TetR) with its operator sequence within the promoter, resulting in inhibition of transcription. Subsequent to the interaction of tetracycline or its analogs with TetR, the repressor-operator interaction is lost, resulting in the activation of transcription. The Tet system has proven successful in regulating inducible expression in the pathogen even within the host macrophages (20, 23). The efficacy of the TetR-operator interaction has been increased ∼50-fold by mutation of the gene in accordance with codon usage in M. tuberculosis in order to improve TetR expression (25). In addition, the tetR gene has been engineered to produce reverse TetR, wherein the addition of tetracycline or its analogs induces interaction of the repressor with the operator sequence, thus resulting in gene repression (25). Together, the two systems provide powerful means to regulate the expression of genes and aid in generating conditional gene replacement mutants for investigation of the events of molecular physiology in mycobacteria.
Unlike the commercially available E. coli expression vectors, M. tuberculosis shuttle vectors lack extended multiple cloning sites (MCSs), convenient epitope tags, or smaller size. These vectors also do not always allow the expression of genes under different physiological conditions. This report presents the results of our efforts to create a series of vectors possessing expanded MCSs that allow the constitutive or inducible expression of genes in fusion with hexahistidine (6×His) and FLAG tags at the N and C termini, respectively. In addition, the vectors have been further modified for expression of the cloned gene under hypoxic conditions. Thus, these vectors are an important addition to the assortment of vectors presently available for mycobacterial research.
MATERIALS AND METHODS
Reagents, bacterial strains, and growth conditions.
Restriction/modification enzymes were obtained from NEB and MBI-Fermentas. DNA oligomers (see Table S2 in the supplemental material) and analytical-grade chemicals were purchased from Sigma. Mycobacterium smegmatis mc2155 was grown on Difco Middlebrook 7H10 agar (Becton, Dickinson) supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC) and 0.2% glycerol at 37°C. For suspension culture, M. smegmatis was grown in 7H9 broth (Becton, Dickinson) supplemented with 10% ADC, 0.2% glycerol, and 0.05% Tween 80. E. coli DH5α was grown in Difco Luria-Bertani broth (Becton Dickinson) at 37°C. LBCT (Luria-Bertani broth, 0.2% dextrose, 0.2% glycerol, and 0.05% Tween 80) and LBC agar (Luria-Bertani broth, 0.2% dextrose, 0.2% glycerol, 1.5% agar) media were used to grow M. smegmatis cells wherever stated. The following antibiotic supplements were used: kanamycin (50 μg/ml for E. coli and 25 μg/ml for mycobacteria), hygromycin (150 μg/ml for E. coli and 100 μg/ml for mycobacteria), apramycin (30 μg/ml), and ampicillin (100 μg/ml). The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material.
Mycobacterial transformation and expression of recombinant proteins.
M. smegmatis mc2155 and M. tuberculosis H37Rv competent cells were prepared as described previously (26, 27). Freshly made competent cells (200 μl) were electroporated with 200 to 400 ng of plasmid DNA. For the analysis of constitutive expression of proteins, M. smegmatis cultures were incubated for 24 h. For analysis of inducible expression, cultures were grown to an optical density at 600 nm (OD600) of ∼0.6, induced with different concentrations of anhydrotetracycline (ATc; 0 to 1,000 ng/ml), and further incubated for 14 h and 72 h for M. smegmatis and M. tuberculosis, respectively. Cells were harvested, resuspended in lysis buffer (phosphate-buffered saline [PBS] containing 5% glycerol [PBSG]), and lysed using 0.1-mm zirconia beads, in a Mini-BeadBeater (BioSpec Products). The lysates were clarified by centrifugation at 16,100 × g for 30 min at 4°C, and protein concentrations were estimated. Clarified lysates containing 10 to 50 μg total protein were resolved on 8-to-12% SDS-PAGE gels and transferred to polyvinylidene difluoride (PVDF)/nitrocellulose membranes. Western blotting was performed as described earlier (28).
Generation of constitutive and inducible shuttle vectors.
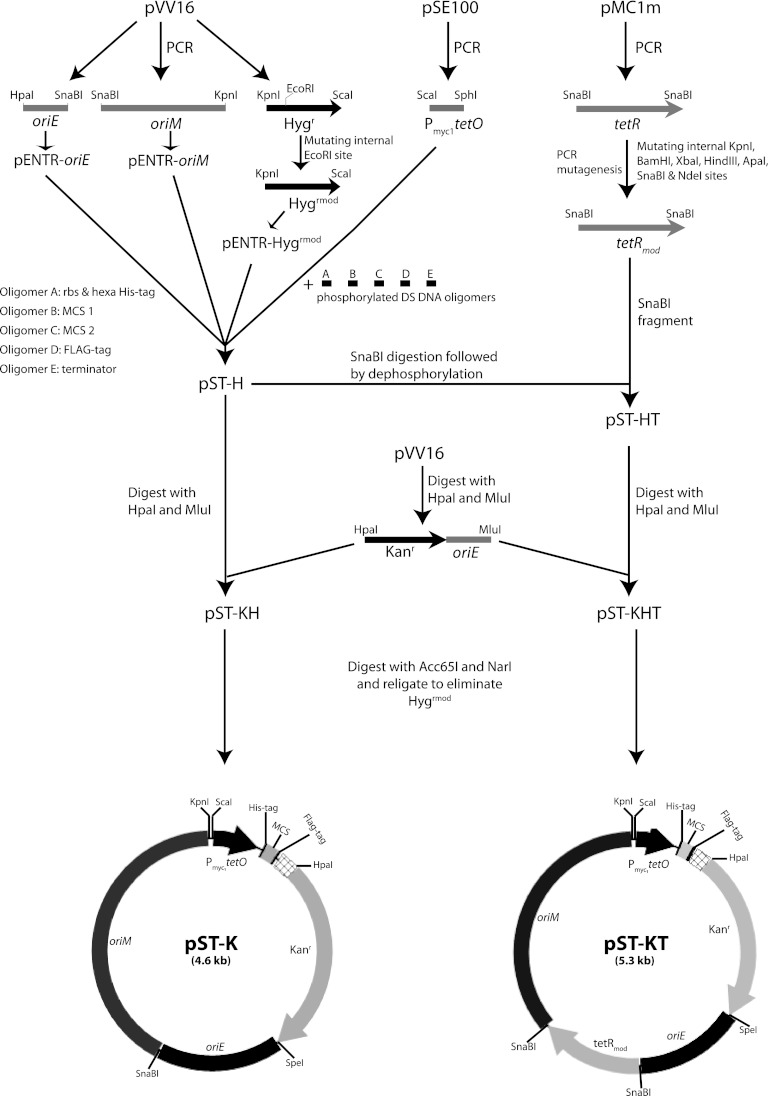
The E. coli origin of replication (oriE), M. tuberculosis origin of replication (oriM), and hygromycin resistance (Hygr) gene (hph) cassettes were amplified from plasmid pVV16 (TBVRM, Colorado State University), using gene-specific primers and Phusion DNA polymerase. The vector pSE100 (24) was used as the template for amplification of sequence of Pmyc1tetO (a promoter with a tet operator). The amplicons obtained were then cloned into the pENTR/D-TOPO vector (Invitrogen). The internal EcoRI site in hph (Hygr) was destroyed with the help of overlapping PCR mutagenesis to produce pENTR-Hygrmod. The oriM and Hygrmod cassettes were released by SnaBI-KpnI and KpnI-EcoRV digestions, respectively, and subcloned into SnaBI-EcoRV sites on pENTR-oriE, generating pENTR-EMH (containing oriE, oriM, and Hygrmod). The Pmyc1tetO cassette was released by ScaI-EcoRV digestion and cloned into corresponding sites on pENTR-EMH to create plasmid pENTR-EMHPmyc1tetO[r]. The nucleic acid sequence specific to pENTR was removed by HpaI digestion followed by recircularization, to generate pEMHPmyc1tetO[r]. Adaptor primer duplexes containing the ribosomal binding site (RBS), hexahistidine tag, MCS, FLAG tag, and transcription terminator sequence were cloned into SphI-HpaI-digested pEMHPmyc1tetO[r] to generate shuttle vector pST-H (Fig. 1).
Fig 1.
Schematic depiction of creation of constitutive and inducible shuttle vectors. rbs, ribosome binding site.
A cassette encoding tetracycline repressor (tetR) under the control of an intermediate-strength promoter, PimyctetR, was amplified from pMC1m (kind gift from S. Ehrt) and cloned into pENTR/D-TOPO vector. Several sites were mutated either in the promoter or in the wobble base of the codons (by overlapping PCR mutagenesis) to obliterate internal BamHI, KpnI, XbaI, HindIII, ApaI, SnaBI, and NdeI restriction sites without altering the protein's coding sequence. The modified cassette was released by SnaBI and cloned into the pST-H vector to generate the pST-HT construct. All vectors generated were verified by restriction digestion and DNA sequencing analysis.
In order to generate shuttle vectors with the kanamycin resistance gene (aphA), the MluI-HpaI fragment containing oriE and the aphA (Kanr) gene from pVV16 was cloned into the corresponding sites in pST-H and pST-HT to generate vectors pST-KH and pST-KHT, containing both Hygrmod and aphA (Kanr) genes. Subsequently, the Hygrmod gene was removed by Acc65I-NarI digestion followed by filling in with Klenow polymerase and religation, to generate pST-K and pST-KT. (The Acc65I/KpnI site is regenerated.) The XbaI-HindIII fragment from pENTR-pknK containing a 3.3-kb protein kinase K gene (pknK) (29) was subcloned into corresponding sites in pST-K and pST-KT to create the pST-K-pknK and pST-KT-pknK plasmids, respectively.
Generation of constitutive and inducible integrating vectors.
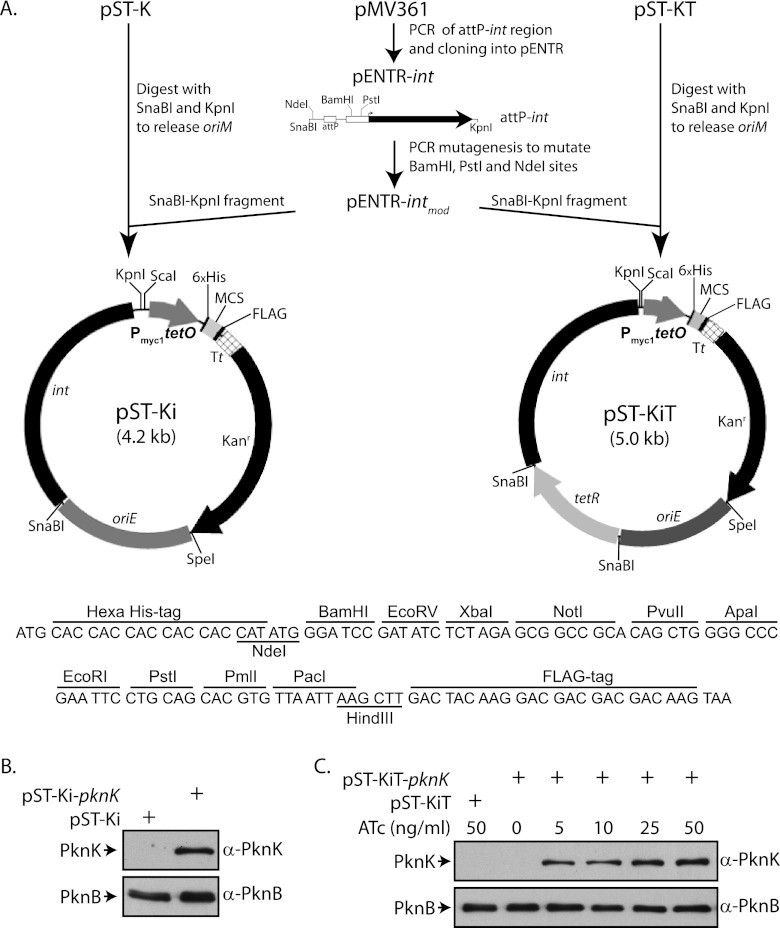
The attP site and the integrase gene (int) of mycobacteriophage L5 were amplified from pMV361 (12) using specific primers, and the amplicon obtained was cloned into pENTR/D-TOPO vector to generate pENTR-int. Overlapping PCR mutagenesis was performed to eliminate the internal restriction sites BamHI, PstI, and NdeI in the attP-int fragment. The oriM gene was released from the pST-K and pST-KT vectors by SnaBI-KpnI digestion and replaced with the SnaBI-KpnI attP-intmod fragment from pENTR-intmod to generate pST-Ki and pST-KiT, respectively. The M. tuberculosis H37Rv pknK gene was subcloned from pENTR-pknK to pST-Ki and pST-KiT, as described above, to generate pST-Ki-pknK and pST-KiT-pknK, respectively.
Generation of a suicide delivery vector for gene replacement studies.
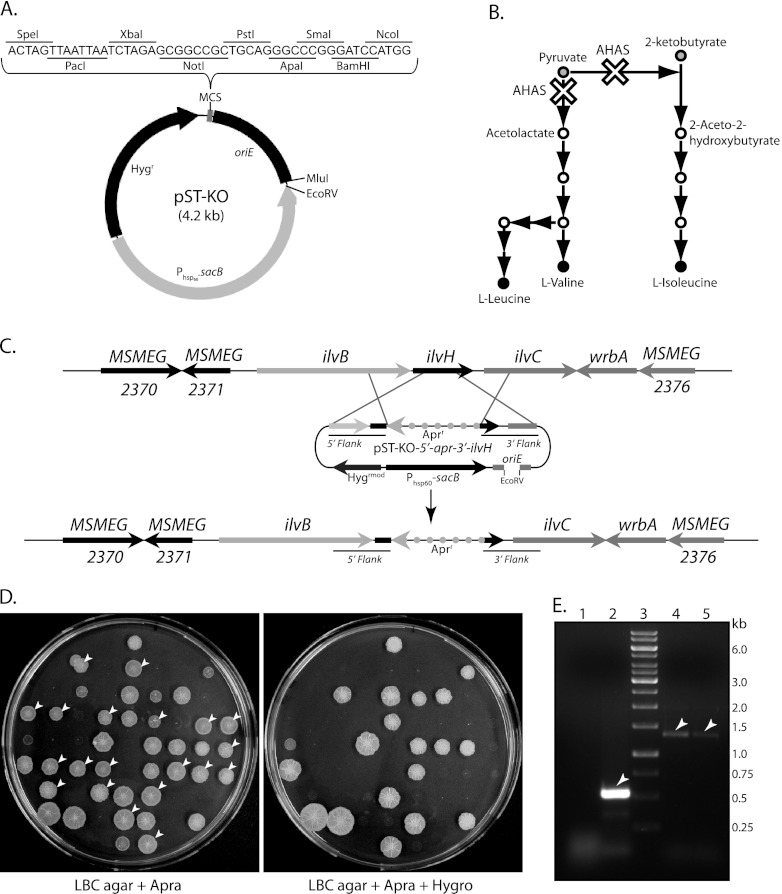
The Acc65I-SnaBI oriM fragment in pENTR-EMH was replaced by Phsp60-sacB, amplified from pGOAL17 (30), resulting in pENTR-ESH. Subsequently, pENTR sequence was deleted and T4-PNK phosphorylated DNA oligomer duplex containing MCS sequence was cloned into the SpeI site to generate the suicide delivery vector pST-KO. To validate the utility of pST-KO, we produced an M. smegmatis mc2155 ilvH gene replacement mutant (mc2ΔilvH). Approximately ∼1.1 kb of 5′ and 3′ flank sequences of the ilvH gene (including ∼100 bp of 5′- and 3′-end sequences of the gene) was amplified and cloned in the pENTR-D-TOPO vector. The aacC41 gene (1.1 kb) exhibiting the apramycin resistance (Aprr) phenotype was amplified from pMV261apra and cloned into pENTR-D-TOPO vector. The 5′ flank, aacC41 gene sequence, and 3′ flank sequence were released by PstI-ScaI, ScaI-ScaI, and ScaI-NotI, respectively, and cloned into pQEII (Qiagen) vector digested with PstI-NotI. The entire cassette was digested with SmaI-NotI enzymes and cloned into corresponding sites on pST-KO. The EcoRV-linearized pST-KO-ΔilvH construct was electroporated into M. smegmatis and selected for Hygr transformants.
Two of the Hygr transformants were grown in LBCT broth in the absence of hygromycin for 24 h, serially diluted, and plated on LBC agar plates containing 10% sucrose, followed by incubation at 37°C for 36 h to select for homologous recombinants (double-crossover strains [DCOs]). Sixty sucrose-resistant colonies were inoculated in apramycin-containing LBCT broth, incubated at 37°C for 24 h, and subsequently spotted on LBC agar plates containing only apramycin or both apramycin and hygromycin. To confirm the deletion of the ilvH gene, we tested for auxotrophy for branched-chain amino acids (BCAAs; Leu, Ile, and Val). This was done by replica spotting apramycin-resistant and hygromycin-sensitive (Aprr Hygs) colonies onto 7H10 agar plates containing only apramycin, apramycin and BCAA, or hygromycin and BCAA. Colonies that appeared only in apramycin with BCAA were expected to be the desired knockouts (see Fig. S1 in the supplemental material). The genomic deletion was verified by PCR amplification across the deletion junctions using appropriate primers and analyzing the PCR products for expected fragment length polymorphisms in comparison with the wild type.
Generation of hypoxia-inducible shuttle vector.
The hypoxia-inducible narK2 promoter (PnarK2) was amplified from pTrc-narK2 vector (31) and cloned into pENTR-D-TOPO vector. pENTR-narK2 vector was digested with ScaI-SphI, and the 304-bp PnarK2 promoter thus released was cloned into corresponding sites on pST-K, replacing Pmyc1tetO with PnarK2 to generate the pST-KnarK2 vector. In order to demonstrate hypoxia-attributed responsiveness of the narK2 promoter in pST-KnarK2, the ∼1-kb EcoRI-HindIII fragment from pMV-gfpaav (31, 32), which codes for adeno-associated virus green fluorescent protein (GFPaav) along with a short tag that confers a half-life of ∼ 40 min, was cloned into the corresponding sites in pST-KnarK2 and transformed into M. smegmatis. Transformants were inoculated into Dubos medium with 10% ADC and grown to an OD600 of ∼0.6, and the cultures were diluted (1:100) in 20 ml of fresh medium and 10 ml of air space (headspace ratio of 0.5 [33]) in flat-bottom screw-cap tubes with magnetic beads (34). Methylene blue indicator dye was added to a final concentration of 1.5 μg/ml. The tubes were sealed and incubated on a magnetic stirrer at 37°C for 10 days (35).
For the ascorbic acid treatment, methylene blue indicator dye was added to the early-log-phase culture grown to an OD600 of ∼0.3, followed by the addition of ascorbic acid solution to a final concentration of 10 mM. The tubes were sealed with Parafilm and incubated at 37°C for 12 h. Cells were harvested after 12 h and processed as described before. Briefly, cells were collected by centrifugation at 10,000 × g at 4°C. The bacteria were fixed in PBS (pH 7.2) containing 1.5% paraformaldehyde, and the fixed samples were stored at 4°C until further use. Fixed samples were subjected to flow cytometry analysis and microscopy to check the expression of adeno-associated virus green fluorescent protein (GFPaav).
Generation of dual-expression vector.
Double-stranded DNA oligomers carrying Shine-Dalgarno sequence 1 (SD1), 6×His tag, multiple cloning site 1 (MCS1), SD2, MCS2, FLAG tag, and transcriptional terminator sequence 2 (Term2) were systematically cloned into SphI-HpaI-treated pEMHPmyc1tetO[r]. The antibiotic selection marker was switched from Hygr to Kanr, as described earlier. The vector pMC1s (22) was used as the template for amplification of Psmyc (a promoter without tet operator sequences for constitutive expression), and the amplicon was cloned into an entry vector. The Psmyc promoter was released from the entry vector and subcloned in the NotI-ApaI sites upstream of SD2. Additional transcriptional terminator sequence Term1 was introduced after the NotI site in the first MCS by DNA oligomer duplex cloning, to generate the pST-2K vector. The pknB-KD (990-bp) and garA (498-bp) gene fragments were amplified from M. tuberculosis H37Rv bacterial artificial chromosome (BAC) clones (a kind gift from S. T. Cole [36]) using gene-specific primers. The NdeI-NotI-digested pknB-KD amplicon was cloned into the corresponding sites in MCS1, and the HindIII-digested garA amplicon was cloned into the HindIII site in MCS2. The amplification of pknK and virS was carried out as described previously (29). The pknK gene was cloned into the XbaI site in MCS1, and the virS gene was cloned into the HindIII site in MCS2.
Nucleotide sequence accession numbers.
The sequences of vectors reported in this article have been deposited in GenBank under accession no. KC153033 (pST-K), KC153038 (pST-KT), KC153034 (pST-Ki), KC153035 (pST-KiT), KC153037 (pST-KO), KC153036 (pST-narK2), and KC153032 (pST-2K).
RESULTS AND DISCUSSION
Vectors pST-K and pST-KT permit constitutive and inducible expression of proteins in M. tuberculosis.
Several mycobacterial vectors permitting constitutive or inducible expression of proteins are currently available from various sources. However, one of the major limitations of most of these vectors is the unavailability of expanded restriction endonuclease sites for cloning of the gene(s) of interest, in addition to the lack of epitope tags at one or both ends of the expressed proteins. The present study was undertaken to expand and improve upon the existing array of mycobacterial expression vectors. The essential features of the shuttle vectors, the origins of replication from E. coli (oriE) and M. tuberculosis (oriM), antibiotic resistance markers (Hygr or Kanr), promoters, ribosome binding sites, epitope tags, multiple cloning sites (MCS), and transcription terminator sequences were put together by amplifying the different components from various plasmids and cloning them stepwise or by cloning DNA oligomers carrying the desired sequences. The approach used is outlined in Fig. 1, and the details are described in Materials and Methods. Briefly, the oriE, oriM, and Hygr DNA fragments were amplified from shuttle vector pVV16 (TBVRM, Colorado State University). The tetracycline-inducible promoter Pmyc1tetO was amplified from pSE100 (24), and the tetracycline repressor (tetR) along with its promoter was amplified from pMC1m, and all fragments were cloned into the entry vector. In order to abolish the restriction sites within the Hygr and tetR genes, we have introduced silent point mutations by overlapping PCR mutagenesis. To introduce the ribosome binding site (RBS) sequence, His tag, MCS, FLAG tag, and transcriptional terminator sequence, we cloned in appropriately designed DNA oligomers (see Table S2 in the supplemental material). The constitutive shuttle vector (pST-H) thus generated (Fig. 1 and see Materials and Methods) had a significantly expanded MCS, with 12 restriction enzyme sites and defined peptide sequences (6×His and FLAG) fused at both ends (Fig. 2A). The modified tetR gene (tetRmod) was then introduced in pST-H to generate the tet-inducible pST-HT vector. As hygromycin resistance often serves as a marker when a creating gene replacement mutant(s) (37–40), pST-K and pST-KT were created from pST-H and pST-HT by introducing the kanamycin resistance gene in place of the hygromycin resistance gene (Fig. 1).
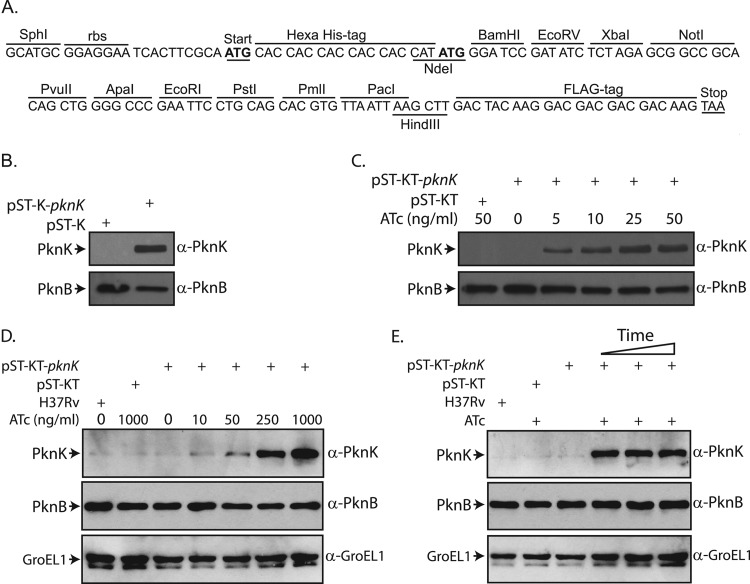
Fig 2.
Expression of M. tuberculosis PknK using replication-proficient vectors in mycobacteria. (A) Sequence of multiple cloning site (MCS) of pST-K and pST-KT vectors. The ribosomal binding site (rbs), peptide tags, and restriction enzyme sites are as indicated. (B) Constitutive expression of PknK in M. smegmatis. Whole-cell extracts were prepared from M. smegmatis mc2155 electroporated with pST-K or pST-K-pknK. Ten micrograms and 25 μg of extracts were resolved on 8% SDS-PAGE gels, transferred onto PVDF membranes, and probed with rabbit polyclonal anti-PknB (α-PknB) and anti-PknK (α-PknK) antibodies, respectively. (C) Regulated expression of PknK in M. smegmatis. Cultures of M. smegmatis transformed with pST-KT or pST-KT-pknK were grown to an OD600 of ∼0.6 and induced with various concentrations of ATc (as indicated). Cell lysates were processed as described above. (D) Regulated expression of PknK in M. tuberculosis H37Rv. Cultures of M. tuberculosis H37Rv transformed with pST-KT or pST-KT-pknK were grown to an OD600 of ∼0.6 and induced with various concentrations of ATc as indicated for 72 h. Ten micrograms (for PknB) and 50 μg (for PknK and GroEL1) of extracts were resolved on 10% SDS-PAGE gels, transferred to nitrocellulose membranes, and probed with rabbit anti-PknB, anti-PknK, and anti-GroEL1 (α-GroEL1) antibodies. (E) Cultures of M. tuberculosis H37Rv transformed with pST-KT or pST-KT-pknK were grown to an OD600 of ∼0.6 and induced with 1 μg/ml ATc for 24, 48, and 72 h (lanes 4, 5, and 6, respectively). All of the control cultures grown to an OD600 of ∼0.6 were allowed to grow for 72 h with or without inducer. Cell lysates were processed as described above.
M. tuberculosis codes for 11 eukaryotic-like serine/threonine protein kinases (eSTPKs), which have been shown to modulate various cellular functions (41). Protein kinase K (PknK) is the largest kinase in mycobacteria and has been reported to be nonessential to the cell's survival (29, 42). To determine if the vectors pST-K and pST-KT could be successfully used for protein expression, we cloned the 3.3-kb pknK gene into the plasmids. While the promoter Pmyc1tetO contains the tet operator sequence (tetO), in the absence of the repressor, it remains constitutively active; thus, the protein must be constitutively expressed from pST-K and expressed only upon induction from pST-KT. As evident from Western blot analysis of lysates made from M. smegmatis transformed with pST-K-pknK, the protein is robustly expressed (Fig. 2B) from this plasmid. While the endogenous PknK could not be detected under the experimental conditions, it can be visualized when larger amounts of lysates are loaded (data not shown). As expected, in pST-KT vector (containing tetRmod) we could not detect expression of PknK in the absence of the inducer anhydrotetracycline (ATc) (Fig. 2C). PknK expression could be detected in M. smegmatis extracts made from cells induced by concentrations of ATc as low as 5 ng/ml (Fig. 2C). The expression levels increased with increasing concentrations of the inducer, with maximum expression seen at 25 ng/ml ATc, which is in keeping with an earlier report (22). In order to check the efficacy of the inducible vector in M. tuberculosis H37Rv, we transformed the strain with pST-KT and pST-KT-pknK. We found that the ATc concentration required for robust expression was about 10-fold higher in M. tuberculosis than that in M. smegmatis (Fig. 2D). Furthermore, we observed that expression of PknK could be detected as early as 24 h after induction, and expression was sustained at similar levels until 72 h after induction (Fig. 2E, lanes 4, 5, and 6). In a previous study using β-galactosidase as the reporter gene, investigators demonstrated effective expression from a Tet-inducible promoter in M. tuberculosis at a 25-ng/ml ATc concentration (22). This apparent difference in suitable inducer concentrations may be due to the sensitivity of the β-galactosidase assay compared to the immunoblot assay for PknK. However, it is clear that saturating levels of expression are being attained only at higher ATc concentrations of about 250 ng/ml. This may also be related to the different levels of expression of TetR from the different plasmids.
The integrating vectors pST-Ki and pST-KiT control constitutive and inducible expression of proteins.
The copy numbers of episomal vectors having the pAL5000 origin of replication (oriM) have been reported to be 3 by Southern hybridization (8) and 8 by single-cell antibiotic resistance assay (43). However, using a more reliable quantitative PCR (qPCR) method, the copy number in M. smegmatis was shown to be ∼23 (44). The expression levels of proteins vary, depending on the strain, suggesting that the copy number of the vector is also dependent on the host strain (45). When expressing genes that are detrimental to cell growth if overexpressed, it is disadvantageous to express them from multicopy episomal plasmids. In these cases, it is beneficial to express the protein from single-copy chromosomal integrants. Integrase- or recombinase-directed site-specific chromosomal integrations have been used in many organisms. The mycobacteriophage L5 (ϕL5) integrase-based system has been extensively used to generate integration proficient vectors (12, 46). To derive integrating vectors from pST-K and pST-KT, we swapped the region containing the oriM with a fragment containing the modified integrase gene (intmod) and the attP sites of the L5 phage to create vectors pST-Ki and pST-KiT (Fig. 3A), respectively. The efficacy of these vectors was checked by cloning the pknK gene into the MCS of either pST-Ki or pST-KiT and transforming the resulting constructs into M. smegmatis. In keeping with results obtained with the expression vector pST-K-pknK, we observed constitutive expression of PknK when pST-Ki-pknK was electroporated into M. smegmatis (Fig. 3B). Upon transformation of M. smegmatis with pST-KiT-pknK, we detected expression of PknK at ATc concentrations as low as 5 ng/ml (Fig. 3C). Apart from ϕL5-based integrase, various integrating systems have been adopted from ϕMs6, ϕTweety, and Streptomyces ϕC31. Vectors derived from ϕMs6 and ϕTweety integrate at the tRNAAla and tRNALys loci in the genome, respectively, different from the ϕL5-based vectors that integrate at tRNAGly (47–49). The integrating pST-KiT vector for inducible expression is a useful addition to the existing range of integrating vectors and may prove to be an excellent means for investigating the functional role of essential genes and for the creation of conditional gene replacement mutants. The creation of additional vectors using integration systems derived from other phages would enable the insertion of multiple genes at different loci, and expression of proteins in fusion with specific epitopes would allow analyses of protein-protein interactions without raising antibodies to the proteins.
Fig 3.
Stable expression of PknK in mycobacteria using integrative plasmids. (A) Schematic outline of the creation of constitutive and inducible integrative shuttle vectors and sequence of the multiple cloning site (MCS) showing the peptide tags and restriction enzyme sites. Tt, transcription terminator. (B) Western blots showing constitutive expression of PknK from pST-Ki-pknK in M. smegmatis. Extracts (10- and 20-μg quantities) were resolved on 8% SDS-PAGE gels, transferred to PVDF membranes, and probed for endogenous PknB and PknK expressed using the integrated vector, respectively. (C) Western blots showing inducible expression of PknK from integrative pST-KiT-pknK vector. Cultures of M. smegmatis were grown to an OD600 of ∼0.6 and induced with the indicated ATc concentrations for 14 h. Extracts were resolved and probed as described for panel B.
The delivery vector pST-KO can be used for gene knockout studies.
Understanding of the complexities of infection and disease progression relies largely on the identification of important pathogenic determinants. The creation of gene knockout mutants and the characterization of these mutants play an important role in these studies. Gene knockout/replacement can be performed by multistep recombination (30), by employing nonreplicating (suicide) vectors (50), by using incompatible plasmids (51), by using long linear DNA fragments (52), or by mycobacteriophage-mediated single-step delivery (38). The approach of using homologous recombination for replacement of a gene(s) has always been associated with substantial in vitro engineering of suicide delivery vectors. This is mostly due to the availability of only a limited number of restriction sites, coupled to the large size of the vehicle vector, which results in cumbersome cloning options and lower transformation efficiencies, respectively. To circumvent these shortcomings, a suicide delivery vector, pST-KO (4.2 kb), which can be used with minimal cloning steps, was designed (Fig. 4A). While this vector is autonomously replicated in E. coli, it cannot replicate in mycobacteria. It carries an extended MCS along with functional Hygr and sacB genes.
Fig 4.
Gene replacement studies in mycobacteria. (A) Pictorial representation of pST-KO vector and its MCS. (B) Illustration of the biosynthetic pathways of branched-chain amino acids (BCAAs). The white crosses show the impact of ilvH deletion on the BCAA pathway. (C) An overview of the genomic organization of the ilvBHC locus in M. smegmatis before (upper panel) and after (lower panel) ilvH deletion. Homologous recombination of pST-KO-5′-apr-3′-ilvH with the corresponding chromosomal region is depicted. (D) Replica plating for the selection of DCOs. Sixty sucrose-resistant colonies were inoculated in LBCT broth containing apramycin (+ Apra) and incubated at 37°C for 24 h. Four microliters of cultures was replica spotted onto LBC agar plates containing only apramycin or apramycin with hygromycin (Apra + Hygro). Colonies that grew only on apramycin (left panel) but failed to do so in plates containing apramycin plus hygromycin (right panel) are indicated by white arrowheads. (E) Confirmation of mc2ΔilvH knockout strains using PCR. Deletion of the targeted region was confirmed by PCR across the deletion junctions using ilvH gene-specific primers. The templates used for the PCR were as follows: lane 1, E. coli DH10B genomic DNA; lane 2, M. smegmatis mc2155 genomic DNA; and lanes 4 and 5, genomic DNA from probable M. smegmatis mc2ΔilvH mutants 2 and 8, respectively.
To establish the successful use of this vector, we took the approach of utilizing auxotrophic complementation of branched-chain amino acids (BCAA) as a selectable marker in M. smegmatis. Acetohydroxyacid synthase (AHAS; also known as acetolactate synthase) catalyzes the first universal step of BCAA biosynthesis. The reaction involves the irreversible decarboxylation of pyruvate and condensation of the acetaldehyde moiety with either a second pyruvate molecule or a 2-ketobutyrate molecule to form 2-acetolactate or 2-aceto-2-hydroxybutyrate, respectively (53). Eventually, isoleucine (Ile), valine (Val), and leucine (Leu) are formed in parallel reactions (Fig. 4B). AHAS is composed of the IlvB (large) and IlvH (small) subunits encoded by the ilvB and ilvH genes, respectively (54, 55). The genomic organization of ilvH and its flanking regions is illustrated in Fig. 4C. As the ilvH gene codes for the regulatory subunit of acetohydroxyacid synthase, ilvH deletion is expected to completely stall BCAA synthesis (Fig. 4B). The delivery construct pST-KO-ΔilvH, which contains the 5′ and 3′ flanks of the ilvH gene (along with the first and last 100 bp of the gene, respectively) with the apramycin selection marker between the flanks, was made. Figure 4C depicts how the recombination-mediated targeted gene replacement was expected to occur. The hygromycin-resistant colonies obtained after transformation of M. smegmatis with the linearized pST-KO-ΔilvH construct were further screened for sucrose resistance. Among the 60 sucrose-resistant colonies that were selected for analysis, only 20 were probable double crossovers (DCOs), as only these colonies were hygromycin sensitive (Fig. 4D; indicated by white arrowheads). The 20 probable DCOs were spotted in replicates on 7H10 agar plates containing (i) only apramycin, (ii) apramycin with BCAA, and (iii) apramycin with BCAA and hygromycin. None of the probable M. smegmatis ΔilvH mutants (DCOs) grew in the absence of BCAA or in the presence of hygromycin (see Fig. S1 in the supplemental material). The authenticity of two of the probable mutants was verified by PCR using ilvH gene-specific primers. While we could not detect the ilvH amplicon (∼500 bp) when we used E. coli DH10B genomic DNA as the template, the amplicon was observed when M. smegmatis mc2155 genomic DNA was used as the template (Fig. 4E, lanes 1 and 2). When genomic DNA from probable mutants was used as the template, an amplicon of size ∼1.3 kb was detected, in keeping with the increase in size due to the insertion of the apramycin gene marker (Fig. 4E, lanes 4 and 5). This confirmed that the selected clones were mc2ΔilvH mutants. The inactivation of the large subunit of acetohydroxyacid synthase, ilvB1 has been shown to be auxotrophic (54). Our results suggest that the small subunit of acetohydroxyacid synthase ilvH is also essential in the absence of BCAA complementation. Thus, our data clearly demonstrate the successful use of pST-KO as a one-step tool for the creation of gene replacement mutants.
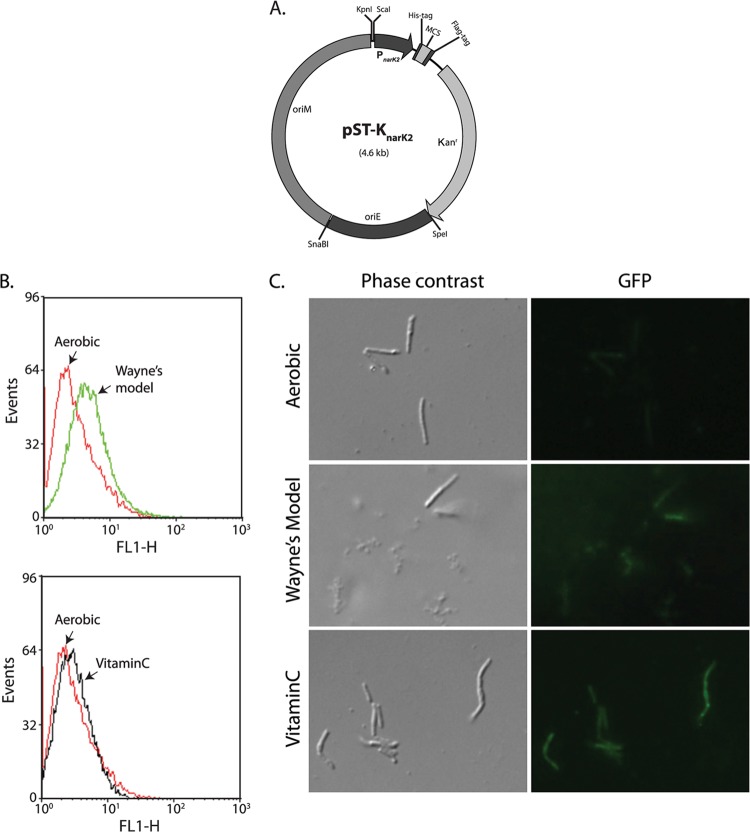
Protein expression can be induced under hypoxic conditions using vector pST-KnarK2.
Mycobacteria reside in phagosomes of host macrophages, wherein they face hypoxic conditions along with other stresses (56). Numerous virulence-associated genes, which are essential for the survival of mycobacteria in this hostile environment, are activated under these conditions. Due to the complications involved in analyses under in vivo infection conditions, in vitro conditions mimicking the in vivo environment are often used to demarcate specific roles of genes in bacterial pathogenesis. Studies involving the expression of genes in mycobacteria under hypoxic conditions so as to simulate the in vivo infection environment involve the use of vectors carrying promoters that are active under hypoxic conditions. We modified the pST-K shuttle vector by replacing the Pmyc1tetO promoter with the hypoxia-inducible PnarK2 promoter (31, 57), thus creating the plasmid pST-KnarK2 (Fig. 5A). To ensure that expression from the PnarK2 promoter in the vector is indeed hypoxia inducible, we cloned gfpaav (coding for a GFP variant having a shorter half-life [32], which would allow us to monitor gene expression changes) into the EcoRI-HindIII sites of the vector. M. smegmatis transformed with pST-KnarK2-gfpaav plasmid was cultured aerobically without ascorbic acid (vitamin C), subjected to hypoxic growth (56), or grown in the presence of vitamin C (58). Expression of GFP was analyzed by flow cytometry and immunofluorescence microscopy using Wayne's model of hypoxia (59) or in the presence of vitamin C (an inducer of hypoxia) (Fig. 5B and C). The results of flow cytometry analysis and microscopy indicate that GFP expression could not be detected under aerobic conditions of growth (Fig. 5B and C). However, when we mimicked Wayne's model of hypoxic growth or used vitamin C to induce hypoxia, the expression of GFPaav could be detected (Fig. 5B and C). These results confirm that protein expression using this vector is indeed hypoxia inducible.
Fig 5.
Generation of hypoxia-inducible shuttle vector. (A) Map of hypoxia-inducible pST-Knark2 shuttle vector. (B) Flow cytometry analysis to check the expression of GFPaav. The samples were analyzed using the BD FACSCalibur flow cytometer (BD Biosciences) using the excitation wavelength (488 nm), and the FL1 readout corresponding to GFP fluorescence intensity was plotted on a log scale using WinMDI (version 2.9) software. (C) Fixed bacterial cells were observed microscopically using a Carl Zeiss image analyzer system equipped with the Axioplan MPM-400 microscope (Carl Zeiss, Inc., Germany) and KS 300 software, under a magnification of ×1,282. Fluorescence was observed with Wayne's model of hypoxia induction as well as upon providing vitamin C as an inducer of hypoxia.
The dual-expression vector pST-2K allows robust coexpression of two proteins.
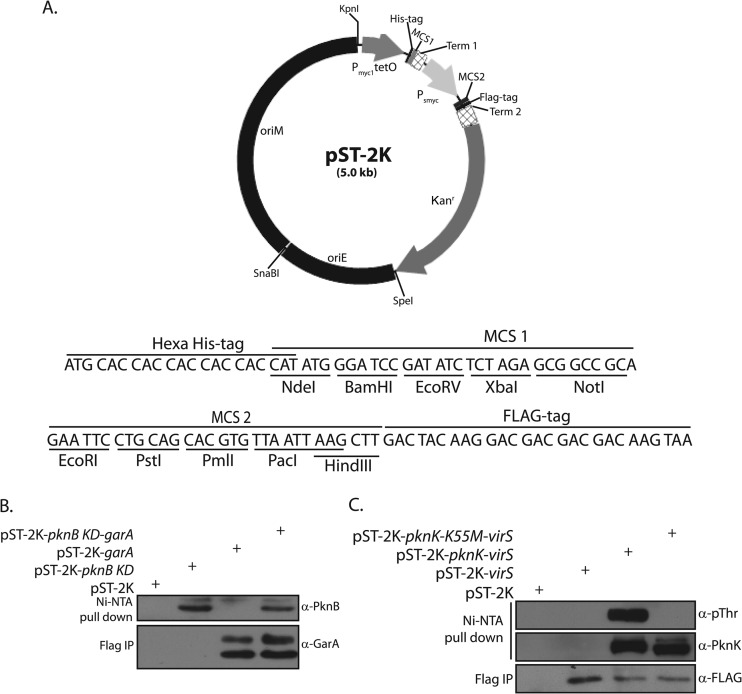
Various methods such as yeast two-hybrid assays, bacterial two-hybrid assays, coimmunoprecipitation, and pulldown studies, are routinely used to investigate protein-protein interactions. Dual-expression vectors, such as pET-Duet1 (Novagen), are available for tandem expression of proteins with different epitope tags in E. coli. We have previously used the pDuet expression system to determine the phosphorylation of enoyl-acyl carrier protein reductase (InhA) by multiple kinases (60). However, options for investigating interactions between mycobacterial proteins in their native environment are limited. A shuttle vector for the constitutive expression of two proteins controlled by independent promoters has been made as part of the present study. This was achieved by the successive cloning of various components of the vector, as described in Materials and Methods, creating a vector, pST-2K, that expresses proteins using the Pmyc1tetO and Psmyc promoters, in which each MCS carries five unique restriction sites (Fig. 6A). Ribosome binding sites and transcription terminator sequences have been coupled to both promoters or MCSs. Proteins expressed from Pmyc1tetO carry an N-terminal His tag, and those expressed from Psmyc carry a C-terminal FLAG tag (Fig. 6A).
Fig 6.
Coexpression of proteins in mycobacteria. (A) Map of the shuttle vector pST-2K. Sequences of multiple cloning sites 1 and 2 (MCS1 and -2) of pST-2K vector are also shown. Peptide tags and restriction enzyme sites are as indicated. (B) Western blot showing the coexpression of PknB-KD (330 amino acids) and GarA. Cultures of M. smegmatis transformed with various constructs grown for 24 h were used for preparation of whole-cell extracts. Extracts were mixed with equilibrated anti-FLAG (Sigma) and Ni Sepharose (GE Healthcare) affinity beads for 2 and 4 h at 4°C, respectively, using a Nutator mixer. The beads were washed with lysis buffer (composition) and resuspended in SDS sample buffer, resolved, transferred to PVDF membrane, and probed with rabbit anti-PknB (α-PknB) and anti-GarA (α-GarA) antibodies. (C) Western blots showing coexpression of PknK or PknK-K55M with VirS. Whole-cell extracts prepared from cultures of M. smegmatis transformed with various constructs were processed as described above. IP, immunoprecipitate.
The essential pknA and pknB genes, encoding protein kinases A and B, respectively, are part of the same operon (36, 61). PknA and PknB have been shown to be involved in modulating cell shape, and possibly cell division, of M. tuberculosis (61). GarA has been identified as a substrate of PknA and PknB by resolving in vitro kinase reactions performed with mycobacterial lysates and purified kinase on two-dimensional (2D) gel electrophoresis followed by mass spectrometry (62). In an effort to analyze the usefulness of pST-2K, we cloned the pknB gene to express it under the control of the Pmyc1tetO promoter and cloned its cognate substrate garA gene to express it using the Psmyc promoter. Plasmids carrying one or both genes were electroporated into M. smegmatis, and Ni-nitrilotriacetic acid (NTA) affinity pulldowns and FLAG immunoprecipitation were performed on whole-cell lysates. Western blot analysis of these pulldowns and immunoprecipitates revealed that PknB and GarA are expressed independently as well as concurrently from pST-2K, indicating that both promoters are active (Fig. 6B). We also coexpressed protein kinase K (PknK) and its substrate, VirS, using pST-2K. VirS was coexpressed with both wild-type and kinase-dead (inactive) PknK. Robust coexpression of PknK and VirS was observed in cells transformed with a plasmid carrying both genes (Fig. 6C). It was also apparent from Western blot analysis using antiphosphothreonine antibodies that while we could not detect autophosphorylation of PknK-K55M (inactive kinase) (29), the wild-type PknK was autophosphorylated efficiently (Fig. 6C, upper panel). Our results thus conclusively demonstrate the constitutive and concomitant expression of two proteins from pST-2K. As the first MCS is under the Pmyc1tetO promoter, the cloning of tetRmod into the unique SnaBI site in pST-2K will generate a vector where one protein may be expressed constitutively and the other only upon induction with anhydrotetracycline inducer.
This study was undertaken with the aim of expanding the collection of E. coli-mycobacterium shuttle vector systems presently available. Vectors with expanded multiple cloning sites have been constructed. These vectors allow proteins to be expressed in fusion with short epitopes. The shuttle vectors reported in this study use kanamycin as the antibiotic selection marker. However, vectors with Hygr as the antibiotic selection marker have also been made in the laboratory, so that one can use either plasmid based on the need of the experiment. As plasmid size largely influences transformation efficiency, we have attempted to keep the size of the vectors to a minimum by using only the essential features of shuttle vectors.
To generate gene replacement mutants of essential genes or to overexpress a gene whose product is toxic, it is necessary to express the target gene under the regulation of inducible promoters. Though the ectopic and inducible vectors created in the study are inducible by ATc, they can easily be converted from a tet-on to tet-off system using the recently engineered reverse tetR repressor (tetRrev) (25). A simple replacement of tetRmod with reverse tetR (tetRrev) will convert Tet-responsive inducible pST-KT and pST-KiT plasmids into Tet-responsive silencing plasmids. In addition to ATc-regulated expression systems, four other systems (acetamide inducible, IPTG inducible, nitrile inducible, and pristinamycin inducible) have been reported for the controlled expression of proteins in mycobacteria (16, 18, 19, 63). The pST series of vectors reported here have been constructed by stitching together essential features. Features of other such inducible systems can be integrated into pST-KT and/or pST-HT using molecular manipulations to generate a gamut of vectors with expanded MCS and flexible epitope tags. The range of the vectors generated in the present study can be extended by replacing the N-terminal 6×His tag with other epitope tags, such as glutathione S-transferase (GST), maltose binding protein (MBP), or GFP. The plasmids made as part of this study have thus appreciably increased the options to investigators and are an important addition to the expression vectors presently available.
Supplementary Material
ACKNOWLEDGMENTS
We thank William R. Jacobs, Jr. (Department of Microbiology and Immunology, Albert Einstein College of Medicine, New York, NY), Sabine Ehrt (Department of Microbiology and Immunology, Weill Medical College of Cornell University, New York, NY), Tanya Parish (Infectious Disease Research Institute, Seattle, WA), and TBVRM, Colorado State University, Fort Collins, CO, for providing various vectors used as starting materials in this study. We also thank Stewart T. Cole (Global Health Institute, Lausanne) for providing the M. tuberculosis bacterial artificial chromosome (BAC) library.
This work was supported by generous funding provided by the National Institute of Immunology, a National Bioscience Award (NBA/34/01/2010), and the Department of Biotechnology (DBT), India, to V.K.N. A.P. was a DBT Postdoctoral Fellow, and D.K. was a Senior Research Fellow. Y.C. is a Senior Research Fellow of the Council of Scientific and Industrial Research. K.K. and S.K. were Senior Research Fellows of the Council of Scientific and Industrial Research.
Footnotes
Published ahead of print 11 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03695-12.
REFERENCES
- 1. Lawn SD, Zumla AI. 2011. Tuberculosis. Lancet 378:57–72 [DOI] [PubMed] [Google Scholar]
- 2. Crawford JT, Bates JH. 1979. Isolation of plasmids from mycobacteria. Infect. Immun. 24:979–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Crawford JT, Cave MD, Bates JH. 1981. Characterization of plasmids from strains of Mycobacterium avium-intracellulare. Rev. Infect. Dis. 3:949–952 [DOI] [PubMed] [Google Scholar]
- 4. Meissner PS, Falkinham JO., III 1984. Plasmid-encoded mercuric reductase in Mycobacterium scrofulaceum. J. Bacteriol. 157:669–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Labidi A, Guibourdenche PM, Riou JY. 1984. Epidemiological surveillance of beta-lactamase-producing gonococci. II. Characterization of plasmids of 66 strains isolated in France (May 1979–March 1983). Pathol. Biol. (Paris) 32:1013–1018 (In French.) [PubMed] [Google Scholar]
- 6. Labidi A, David HL, Roulland-Dussoix D. 1985. Cloning and expression of mycobacterial plasmid DNA in Escherichia coli. FEMS Microbiol. Lett. 30:221–225 [Google Scholar]
- 7. Picardeau M, Vincent V. 1997. Characterization of large linear plasmids in mycobacteria. J. Bacteriol. 179:2753–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ranes MG, Rauzier J, Lagranderie M, Gheorghiu M, Gicquel B. 1990. Functional analysis of pAL5000, a plasmid from Mycobacterium fortuitum: construction of a “mini” mycobacterium-Escherichia coli shuttle vector. J. Bacteriol. 172:2793–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Matsumoto S, Tamaki M, Yukitake H, Matsuo T, Naito M, Teraoka H, Yamada T. 1996. A stable Escherichia coli-mycobacteria shuttle vector 'pSO246′ in Mycobacterium bovis BCG. FEMS Microbiol. Lett. 135:237–243 [DOI] [PubMed] [Google Scholar]
- 10. Stolt P, Stoker NG. 1996. Functional definition of regions necessary for replication and incompatibility in the Mycobacterium fortuitum plasmid pAL5000. Microbiology 142:2795–2802 [DOI] [PubMed] [Google Scholar]
- 11. Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF, Snapper SB, Barletta RG, Jacobs WR, Jr, Bloom BR. 1991. New use of BCG for recombinant vaccines. Nature 351:456–460 [DOI] [PubMed] [Google Scholar]
- 12. Parker AE, Bermudez LE. 1997. Expression of the green fluorescent protein (GFP) in Mycobacterium avium as a tool to study the interaction between mycobacteria and host cells. Microb. Pathog. 22:193–198 [DOI] [PubMed] [Google Scholar]
- 13. Hickey MJ, Arain TM, Shawar RM, Humble DJ, Langhorne MH, Morgenroth JN, Stover CK. 1996. Luciferase in vivo expression technology: use of recombinant mycobacterial reporter strains to evaluate antimycobacterial activity in mice. Antimicrob. Agents Chemother. 40:400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fan XY, Ma H, Guo J, Li ZM, Cheng ZH, Guo SQ, Zhao GP. 2009. A novel differential expression system for gene modulation in mycobacteria. Plasmid 61:39–46 [DOI] [PubMed] [Google Scholar]
- 15. Gordon S, Parish T, Roberts IS, Andrew PW. 1994. The application of luciferase as a reporter of environmental regulation of gene expression in mycobacteria. Lett. Appl. Microbiol. 19:336–340 [DOI] [PubMed] [Google Scholar]
- 16. Triccas JA, Parish T, Britton WJ, Gicquel B. 1998. An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol. Lett. 167:151–156 [DOI] [PubMed] [Google Scholar]
- 17. Roberts G, Muttucumaru DG, Parish T. 2003. Control of the acetamidase gene of Mycobacterium smegmatis by multiple regulators. FEMS Microbiol. Lett. 221:131–136 [DOI] [PubMed] [Google Scholar]
- 18. Pandey AK, Raman S, Proff R, Joshi S, Kang CM, Rubin EJ, Husson RN, Sassetti CM. 2009. Nitrile-inducible gene expression in mycobacteria. Tuberculosis (Edinb.) 89:12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Forti F, Crosta A, Ghisotti D. 2009. Pristinamycin-inducible gene regulation in mycobacteria. J. Biotechnol. 140:270–277 [DOI] [PubMed] [Google Scholar]
- 20. Blokpoel MC, Murphy HN, O'Toole R, Wiles S, Runn ES, Stewart GR, Young DB, Robertson BD. 2005. Tetracycline-inducible gene regulation in mycobacteria. Nucleic Acids Res. 33:e22 doi:10.1093/nar/gni023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carroll P, Muttucumaru DG, Parish T. 2005. Use of a tetracycline-inducible system for conditional expression in Mycobacterium tuberculosis and Mycobacterium smegmatis. Appl. Environ. Microbiol. 71:3077–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ehrt S, Guo XV, Hickey CM, Ryou M, Monteleone M, Riley LW, Schnappinger D. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21 doi:10.1093/nar/gni013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hernandez-Abanto SM, Woolwine SC, Jain SK, Bishai WR. 2006. Tetracycline-inducible gene expression in mycobacteria within an animal host using modified Streptomyces tcp830 regulatory elements. Arch. Microbiol. 186:459–464 [DOI] [PubMed] [Google Scholar]
- 24. Guo XV, Monteleone M, Klotzsche M, Kamionka A, Hillen W, Braunstein M, Ehrt S, Schnappinger D. 2007. Silencing Mycobacterium smegmatis by using tetracycline repressors. J. Bacteriol. 189:4614–4623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klotzsche M, Ehrt S, Schnappinger D. 2009. Improved tetracycline repressors for gene silencing in mycobacteria. Nucleic Acids Res. 37:1778–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Goude R, Parish T. 2008. Electroporation of mycobacteria. J. Vis. Exp. 15:e761 doi:10.3791/761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Venkatesh J, Kumar P, Krishna PS, Manjunath R, Varshney U. 2003. Importance of uracil DNA glycosylase in Pseudomonas aeruginosa and Mycobacterium smegmatis, G+C-rich bacteria, in mutation prevention, tolerance to acidified nitrite, and endurance in mouse macrophages. J. Biol. Chem. 278:24350–24358 [DOI] [PubMed] [Google Scholar]
- 28. Parikh A, Verma SK, Khan S, Prakash B, Nandicoori VK. 2009. PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity. J. Mol. Biol. 386:451–464 [DOI] [PubMed] [Google Scholar]
- 29. Kumar P, Kumar D, Parikh A, Rananaware D, Gupta M, Singh Y, Nandicoori VK. 2009. The Mycobacterium tuberculosis protein kinase K modulates activation of transcription from the promoter of mycobacterial monooxygenase operon through phosphorylation of the transcriptional regulator VirS. J. Biol. Chem. 284:11090–11099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parish T, Stoker NG. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969–1975 [DOI] [PubMed] [Google Scholar]
- 31. Kurthkoti K, Varshney U. 2010. Detrimental effects of hypoxia-specific expression of uracil DNA glycosylase (Ung) in Mycobacterium smegmatis. J. Bacteriol. 192:6439–6446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dick T, Lee BH, Murugasu-Oei B. 1998. Oxygen depletion induced dormancy in Mycobacterium smegmatis. FEMS Microbiol. Lett. 163:159–164 [DOI] [PubMed] [Google Scholar]
- 34. Kurthkoti K, Kumar P, Jain R, Varshney U. 2008. Important role of the nucleotide excision repair pathway in Mycobacterium smegmatis in conferring protection against commonly encountered DNA-damaging agents. Microbiology 154:2776–2785 [DOI] [PubMed] [Google Scholar]
- 35. Sohaskey CD. 2008. Nitrate enhances the survival of Mycobacterium tuberculosis during inhibition of respiration. J. Bacteriol. 190:2981–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, III, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, Rajandream MA, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544 [DOI] [PubMed] [Google Scholar]
- 37. Howard NS, Gomez JE, Ko C, Bishai WR. 1995. Color selection with a hygromycin-resistance-based Escherichia coli-mycobacterial shuttle vector. Gene 166:181–182 [DOI] [PubMed] [Google Scholar]
- 38. Bardarov S, Bardarov S, Jr, Pavelka MS, Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR., Jr 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007–3017 [DOI] [PubMed] [Google Scholar]
- 39. Borsuk S, Mendum TA, Fagundes MQ, Michelon M, Cunha CW, McFadden J, Dellagostin OA. 2007. Auxotrophic complementation as a selectable marker for stable expression of foreign antigens in Mycobacterium bovis BCG. Tuberculosis (Edinb.) 87:474–480 [DOI] [PubMed] [Google Scholar]
- 40. Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. 2007. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat. Med. 13:1515–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chakraborti PK, Matange N, Nandicoori VK, Singh Y, Tyagi JS, Visweswariah SS. 2011. Signalling mechanisms in mycobacteria. Tuberculosis (Edinb.) 91:432–440 [DOI] [PubMed] [Google Scholar]
- 42. Malhotra V, Arteaga-Cortes LT, Clay G, Clark-Curtiss JE. 2010. Mycobacterium tuberculosis protein kinase K confers survival advantage during early infection in mice and regulates growth in culture and during persistent infection: implications in immune modulation. Microbiology 156:2829–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gavigan JA, Ainsa JA, Perez E, Otal I, Martin C. 1997. Isolation by genetic labeling of a new mycobacterial plasmid, pJAZ38, from Mycobacterium fortuitum. J. Bacteriol. 179:4115–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Huff J, Czyz A, Landick R, Niederweis M. 2010. Taking phage integration to the next level as a genetic tool for mycobacteria. Gene 468:8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Griffin S, Williamson AL, Chapman R. 2009. Optimisation of a mycobacterial replicon increases foreign antigen expression in mycobacteria. Tuberculosis (Edinb.) 89:225–232 [DOI] [PubMed] [Google Scholar]
- 46. Lee MH, Pascopella L, Jacobs WR, Jr, Hatfull GF. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc. Natl. Acad. Sci. U. S. A. 88:3111–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pham TT, Jacobs-Sera D, Pedulla ML, Hendrix RW, Hatfull GF. 2007. Comparative genomic analysis of mycobacteriophage Tweety: evolutionary insights and construction of compatible site-specific integration vectors for mycobacteria. Microbiology 153:2711–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Murry J, Sassetti CM, Moreira J, Lane J, Rubin EJ. 2005. A new site-specific integration system for mycobacteria. Tuberculosis (Edinb.) 85:317–323 [DOI] [PubMed] [Google Scholar]
- 49. Vultos TD, Mederle I, Abadie V, Pimentel M, Moniz-Pereira J, Gicquel B, Reyrat JM, Winter N. 2006. Modification of the mycobacteriophage Ms6 attP core allows the integration of multiple vectors into different tRNAala T-loops in slow- and fast-growing mycobacteria. BMC Mol. Biol. 7:47 doi:10.1186/1471-2199-7-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Husson RN, James BE, Young RA. 1990. Gene replacement and expression of foreign DNA in mycobacteria. J. Bacteriol. 172:519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pashley CA, Parish T, McAdam RA, Duncan K, Stoker NG. 2003. Gene replacement in mycobacteria by using incompatible plasmids. Appl. Environ. Microbiol. 69:517–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. El Karoui M, Amundsen SK, Dabert P, Gruss A. 1999. Gene replacement with linear DNA in electroporated wild-type Escherichia coli. Nucleic Acids Res. 27:1296–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Duggleby RG, Pang SS. 2000. Acetohydroxyacid synthase. J. Biochem. Mol. Biol. 33:1–36 [Google Scholar]
- 54. Awasthy D, Gaonkar S, Shandil RK, Yadav R, Bharath S, Marcel N, Subbulakshmi V, Sharma U. 2009. Inactivation of the ilvB1 gene in Mycobacterium tuberculosis leads to branched-chain amino acid auxotrophy and attenuation of virulence in mice. Microbiology 155:2978–2987 [DOI] [PubMed] [Google Scholar]
- 55. Choi KJ, Yu YG, Hahn HG, Choi JD, Yoon MY. 2005. Characterization of acetohydroxyacid synthase from Mycobacterium tuberculosis and the identification of its new inhibitor from the screening of a chemical library. FEBS Lett. 579:4903–4910 [DOI] [PubMed] [Google Scholar]
- 56. Rustad TR, Harrell MI, Liao R, Sherman DR. 2008. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One 3:e1502 doi:10.1371/journal.pone.0001502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rao SP, Camacho L, Huat Tan B, Boon C, Russel DG, Dick T, Pethe K. 2008. Recombinase-based reporter system and antisense technology to study gene expression and essentiality in hypoxic nonreplicating mycobacteria. FEMS Microbiol. Lett. 284:68–75 [DOI] [PubMed] [Google Scholar]
- 58. Taneja NK, Dhingra S, Mittal A, Naresh M, Tyagi JS. 2010. Mycobacterium tuberculosis transcriptional adaptation, growth arrest and dormancy phenotype development is triggered by vitamin C. PLoS One 5:e10860 doi:10.1371/journal.pone.0010860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wayne LG, Hayes LG. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Khan S, Nagarajan SN, Parikh A, Samantaray S, Singh A, Kumar D, Roy RP, Bhatt A, Nandicoori VK. 2010. Phosphorylation of enoyl-acyl carrier protein reductase InhA impacts mycobacterial growth and survival. J. Biol. Chem. 285:37860–37871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kang CM, Abbott DW, Park ST, Dascher CC, Cantley LC, Husson RN. 2005. The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19:1692–1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Villarino A, Duran R, Wehenkel A, Fernandez P, England P, Brodin P, Cole ST, Zimny-Arndt U, Jungblut PR, Cervenansky C, Alzari PM. 2005. Proteomic identification of M. tuberculosis protein kinase substrates: PknB recruits GarA, a FHA domain-containing protein, through activation loop-mediated interactions. J. Mol. Biol. 350:953–963 [DOI] [PubMed] [Google Scholar]
- 63. Lee BY, Clemens DL, Horwitz MA. 2008. The metabolic activity of Mycobacterium tuberculosis, assessed by use of a novel inducible GFP expression system, correlates with its capacity to inhibit phagosomal maturation and acidification in human macrophages. Mol. Microbiol. 68:1047–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.