Abstract
SUMMARY
Restriction-modification (R-M) systems are ubiquitous and are often considered primitive immune systems in bacteria. Their diversity and prevalence across the prokaryotic kingdom are an indication of their success as a defense mechanism against invading genomes. However, their cellular defense function does not adequately explain the basis for their immaculate specificity in sequence recognition and nonuniform distribution, ranging from none to too many, in diverse species. The present review deals with new developments which provide insights into the roles of these enzymes in other aspects of cellular function. In this review, emphasis is placed on novel hypotheses and various findings that have not yet been dealt with in a critical review. Emerging studies indicate their role in various cellular processes other than host defense, virulence, and even controlling the rate of evolution of the organism. We also discuss how R-M systems could have successfully evolved and be involved in additional cellular portfolios, thereby increasing the relative fitness of their hosts in the population.
INTRODUCTION
One of the attributes for success in microbial evolution and diversity is the ability of bacteria to recognize and distinguish incoming foreign DNA from self DNA. The organisms have evolved strategies to limit constant exposure to extraneous foreign DNA elements. Mechanisms involving restriction-modification (R-M) systems directly target invading DNA elements. To begin with, this review covers the various aspects of R-M systems that target invading DNA elements and counterstrategies employed by the invading genomes to evade restriction. From analyses of these defense and counterdefense measures, it is apparent that the cellular defense function does not comprehensively provide an explanation for (i) the uneven distribution of R-M systems in the bacterial kingdom, (ii) the high level of specificity in sequence recognition, and (iii) the independent evolution of restriction endonucleases (REases) with respect to methyltransferases (MTases). The present review deals with new developments that provide insights into the roles of R-M systems in other aspects of cellular function. The review is not intended to cover the vast literature on structure-function studies, modes of recognition, catalytic motifs, or mechanisms of catalysis by these enzymes. Instead, the major emphasis is to understand the reasons for their diversity and to discuss additional biological roles.
Background
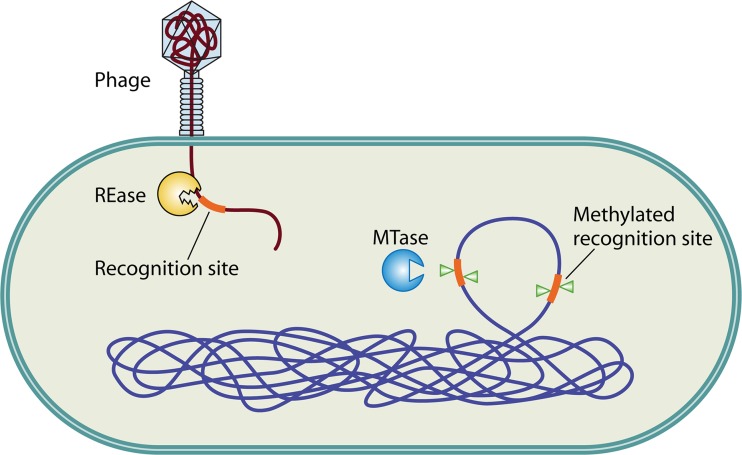
Restriction-modification (R-M) systems are important components of prokaryotic defense mechanisms against invading genomes. They occur in a wide variety of unicellular organisms, including eubacteria and archaea (1, 2), and comprise two contrasting enzymatic activities: a restriction endonuclease (REase) and a methyltransferase (MTase). The REase recognizes and cleaves foreign DNA sequences at specific sites, while MTase activity ensures discrimination between self and nonself DNA, by transferring methyl groups to the same specific DNA sequence within the host's genome (Fig. 1). Functionally, REases cleave endonucleolytically at phosphodiester bonds, generating 5′ or 3′ overhangs or blunt ends. MTases transfer the methyl group from S-adenosyl methionine to the C-5 carbon or the N4 amino group of cytosine or to the N6 amino group of adenine (3).
Fig 1.
Restriction-modification (R-M) systems as defense mechanisms. R-M systems recognize the methylation status of incoming foreign DNA, e.g., phage genomes. Methylated sequences are recognized as self, while recognition sequences on the incoming DNA lacking methylation are recognized as nonself and are cleaved by the restriction endonuclease (REase). The methylation status at the genomic recognition sites is maintained by the cognate methyltransferase (MTase) of the R-M system.
R-M systems are classified mainly into four different types based on their subunit composition, sequence recognition, cleavage position, cofactor requirements, and substrate specificity (4). Type I enzymes consist of a hetero-oligomeric protein complex encompassing both restriction and modification activities. These enzymes bind to a bipartite DNA sequence and cleave from ∼100 bp to tens of thousands of base pairs away from the target (5). Typical examples are EcoKI and EcoR124I (5, 6). In contrast, most type II systems contain separate REase and MTase enzymes. Generally, type II REases are homodimeric or homotetrameric and cleave DNA within or near their target site. These enzymes are highly diverse and are known to utilize at least five types of folds: PD-(D/E)XK, PLD, HNH, GIY-YIG, and halfpipe, e.g., R.EcoRI, R.BfiI, R.KpnI, R.Eco29kI, and R.PabI enzymes, respectively (2, 7 –10). Type II enzymes are the most widely studied and are also extensively utilized nucleases in genetic engineering. Type III enzymes are heterotrimers (M2R1) (11) or heterotetramers (M2R2) (12) containing restriction-, methylation-, and DNA-dependent NTPase activities. As a consequence, they compete within themselves for modification or restriction in the same catalytic cycle (13). These enzymes recognize short asymmetric sequences of 5 to 6 bp, translocate along DNA, and cleave the 3′ side of the target site at a distance of ∼25 bp (1, 5). Restriction is elicited only when two recognition sequences are in an inverse orientation with respect to each other. Typical examples are EcoP1I and EcoP15I (5, 14). In contrast to the above-described three groups, the type IV systems cleave only DNA substrates containing methylated, hydroxymethylated, or glucosyl-hydroxymethylated bases at specific sequences (4). For example, EcoKMcrBC, a well-studied type IV enzyme, targets A/GmC (methylated cytosine, either m4C or m5C) separated by ∼40 to 3,000 bases (15). The recently discovered type IV enzyme GmrSD specifically digests DNAs containing sugar-modified hydroxymethylated cytosine (16). However, the sequence specificity of the enzyme is not well studied. In addition to the above-described four groups, a number of genomes are also known to encode MTases that are not associated with REases and are thus termed “orphan/solitary MTases.” Examples of this group of enzymes are the N6-adenine MTases Dam and CcrM (cell cycle-regulated MTase) and the C-5–cytosine MTase Dcm (17–19). Interestingly, unlike the vast majority of REases, which are accompanied by MTases to protect the genomic DNA from self-digestion, some of the rare-cutting REases, viz., R.PacI and R.PmeI, seem to be solitary enzymes with no cognate MTase (http://rebase.neb.com/rebase/rebase.html) (20). It appears that genome protection in these organisms is dependent on the underrepresentation of the recognition sequences in the genome (20). However, the biological significance of “solitary REases” is not known.
Prevalence and Distribution
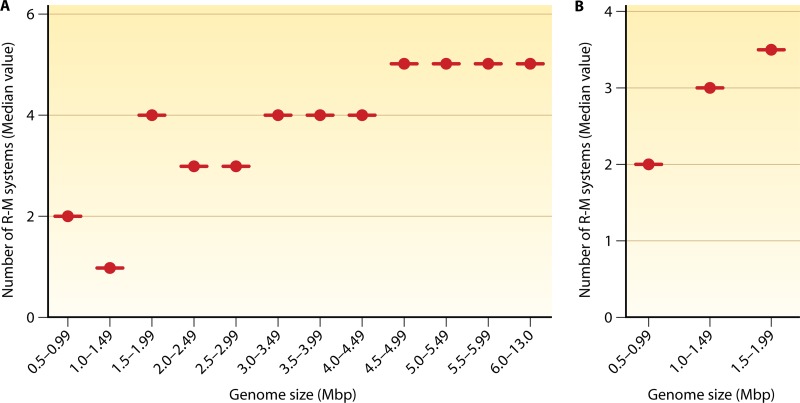
R-M systems are an extremely diverse group of enzymes and are ubiquitous among prokaryotes. To date, nearly 4,000 enzymes are known, with about 300 different specificities (21). The sequencing of more than 2,450 bacterial and archaeal genomes has only reaffirmed their vast diversity in the prokaryotic kingdom (21). In contrast to REases, MTases show highly conserved features, a surprising finding initially. The diversity and prevalence of R-M systems indicate their success in the bacterial world as a defense mechanism. To a large extent, the distribution of MTases among sequenced genomes seems to reflect the distribution of R-M systems. It has been observed that ∼90% of the genomes contain at least one R-M system and that ∼80% contain multiple R-M systems (http://rebase.neb.com/rebase/rebase.html). Interestingly, a positive correlation can be observed with respect to the number of R-M genes and the size of the genome (see the supplemental material). A general trend is an increase in the number of R-M systems with an increase in genome size (Fig. 2; see also Fig. S1 in the supplemental material). For example, organisms with a genome size of 2 to 3 Mbp have a median number of 3 R-M systems per genome, those with a genome size of 3 to 4 Mbp have 4 R-M systems per genome, and those with a genome size of 4 to 5 Mbp have 5 R-M systems per genome. However, an anomalous decrease in the 1- to 1.5-Mbp genome size class can be seen in the distribution of R-M systems because of many Brucella species harboring single R-M systems per chromosome (Fig. 2A). In contrast, the presence of multiple R-M systems among Helicobacter and Campylobacter species brings an anomalous increase in the 1.5- to 2-Mbp genome size class (Fig. 2A). A linear correlation can be observed when the above-mentioned bacterial species are omitted from the 1- to 1.49-Mbp and the 1.5- to 1.99-Mbp genome size classes (Fig. 2B). The significance of the presence of multiple R-M systems per organism observed for many bacterial species is discussed below in this review (see “Immigration Control, Maintenance of Species Identity, and Control of Speciation”). A further anomaly was observed for certain organisms wherein the correlation of the number of R-M systems to the genome size is not apparent. For instance, genomes of Buchnera, Borrelia, Chlamydia, Chlamydophila, Coxiella, Rickettsia, and Synechococcus vary in size (ranging from 1 to 2.5 Mb), and they do not appear to encode R-M systems. Notably, some of these organisms are obligate intracellular pathogens or endosymbiotic and therefore occupy the intracellular niche of infected cells. Hence, they may seldom encounter bacteriophages, obviating the need for R-M systems. Alternatively, a low frequency of horizontal gene transfer (HGT) in such species living in closed environments could account for the observed small number or total absence of the systems (see below).
Fig 2.
Distribution of R-M systems. (A) Genome-wide analysis for the presence of conserved MTase genes among bacteria with genome sizes ranging from 0.5 to 13 Mbp. The plot shows the median value of the distribution of the number of MTase genes with the specified class interval of genome size. A correlation of an increase in the number of modification systems with an increase in the genome size can be observed. The anomalous decrease (in the 1- to 1.5-Mbp genome size class) in the distribution of R-M systems is because of many Brucella species harboring a single R-M system per chromosome. The presence of multiple R-M systems among Helicobacter and Campylobacter species brings an anomalous increase in the 1.5- to 2-Mbp genome size class. (B) A linear correlation can be observed when the above-mentioned bacterial species are omitted from the 1- to 1.49-Mbp and the 1.5- to 1.99-Mbp classes.
Another peculiarity is seen with respect to the occurrence of REases recognizing long or short palindromic DNA sequences. Some of the sequenced genomes belonging to the genera Bacillus, Nocardia, Pseudomonas, and Streptomyces have a larger proportion of R-M systems that recognize longer palindromic DNA sequences. Many of these genomes have a relatively large genome of >5 Mbp. As a larger genome would have more 4-bp and 6-bp recognition sites than 8-bp sites, the utilization of an R-M system that recognizes the latter sites might prevent accidental double-stranded DNA (dsDNA) breaks inflicted by REases. For example, the probable occurrence of a particular 4-bp sequence in a 5-Mbp genome would be 19,531 times, while an 8-bp recognition sequence would be represented only 76 times, assuming equal base composition and an even 4-base distribution. Continuous selection against REases recognizing smaller target sequences could have resulted in the enrichment of enzymes recognizing longer sequences in the larger genomes. The preference for enzymes recognizing longer recognition sites in larger genomes appears to be an outcome of minimizing accidental double-strand breaks on the host DNA. However, the GC contents of the organism and the recognition site also play an important role. For example, an 8-base GC-rich recognition sequence (such as GGCCGGCC) would occur with a normal frequency in a highly GC-rich genome (e.g., Frankia species [∼73%]). The probable occurrence of a 4- or 6-base GC-rich sequence in the same genome would be greater than that of the 8-base sequence, and thus, in such a scenario, the organism may employ other mechanisms to protect the genome from accidental double-strand breaks.
BACTERIAL DEFENSE SYSTEMS
R-M Systems
Restriction was first observed in the 1950s, when phage λ (propagated in Escherichia coli B) was found to grow poorly on E. coli K-12 (22, 23). Restriction is achieved by the cleavage of the phage DNA (foreign), which is unmethylated, while the genome of the host (self) remains protected due to methylation by the cognate MTase (Fig. 1). Because of their ability to recognize self versus nonself, a property observed for the immune systems of higher organisms, R-M systems are considered to function as primitive immune systems (24, 25). Various studies have demonstrated a 10- to 108-fold protection of the host cell from phages by different R-M systems (reviewed in reference 26). Their role in curtailing the spread of phage is also evident from the fact that a number of phages have evolved to evade restriction, viz., modification (methylation, glucosylation, and other modified nucleotides) of the phage DNA (1). These modifications of the phage genome directly affect DNA cleavage by REases and thus ensure the evasion of restriction. In turn, bacteria are known to express modification-specific endonucleases to restrict these adapted phages, resulting in a “coevolutionary arms race” (1, 27).
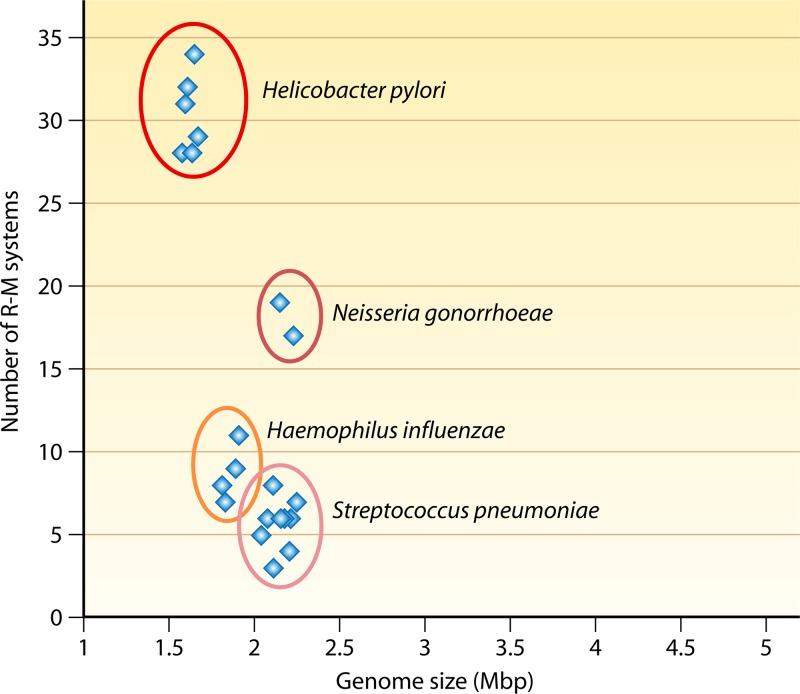
The “cellular defense” function of R-M systems does not comprehensively provide an explanation for the following. (i) It does not provide an explanation for the high specificity in sequence recognition (28). A highly sequence-specific REase or a “rare cutter” would be less efficient in targeting an incoming DNA. Hence, it is not clear whether selection pressure on bacteria due to phages would be sufficient to maintain the high sequence specificity of the R-M systems (28). (ii) It does not provide an explanation for the presence of multiple R-M systems per organism in many bacterial species. While the antirestriction strategies evolved by phages may lead to the generation of multiple specificities, it is unclear why only certain organisms (e.g., naturally competent bacteria) have an abundance of R-M systems. For example, Neisseria gonorrhoeae contains 16 different biochemically identified systems (29). Moreover, some organisms, such as Helicobacter pylori, N. gonorrhoeae, Haemophilus influenzae, and Streptococcus pneumoniae, have an abundance of R-M systems (Fig. 3). (iii) It does not provide an explanation for the poor sequence homology of REases. While MTases share considerable homology and could be identified by primary sequence analysis, REases have very low levels of sequence similarity among themselves. A faster evolution of REases, if occurring, could be one way to account the low level of similarity among themselves. Alternatively, the evolution of REases could have taken place multiple times from different catalytic/structural folds. Although there is no sufficient evidence for the independent origins of REases and cognate MTases, such a scenario, rather than a coevolutionary strategy, would explain their diversification.
Fig 3.
Abundance of R-M systems in naturally competent bacteria. Whole-genome sequence analyses of some of the naturally competent bacteria show that they are rich in R-M genes (5 to 34 genes) compared to other noncompetent bacteria (e.g., a single R-M system in many Bacillus anthracis strains).
Non-R-M Defense Systems
In addition to the R-M systems, other gene loci are also involved in limiting the entry of invasive DNA elements. Short stretches of direct repeats interrupted by unique sequences, termed “clustered regularly interspaced short palindromic repeats” (CRISPRs), are found in many eubacteria and archaea (30). The nonrepetitive sequences of the CRISPR loci exhibit homology to previously encountered phage genomes (31). These loci were proposed to serve as memory for the bacteria with respect to earlier phage encounters (32). Recent evidence suggests that CRISPRs along with their associated genes (cas genes) are involved in adaptive immunity against phages (33). It appears that R-M systems and CRISPRs are the strategies employed by bacteria to serve as innate and adaptive immune systems, respectively, to evade invading genomes. It would be interesting to study the functional cooperation, if any, between the CRISPR and the R-M systems.
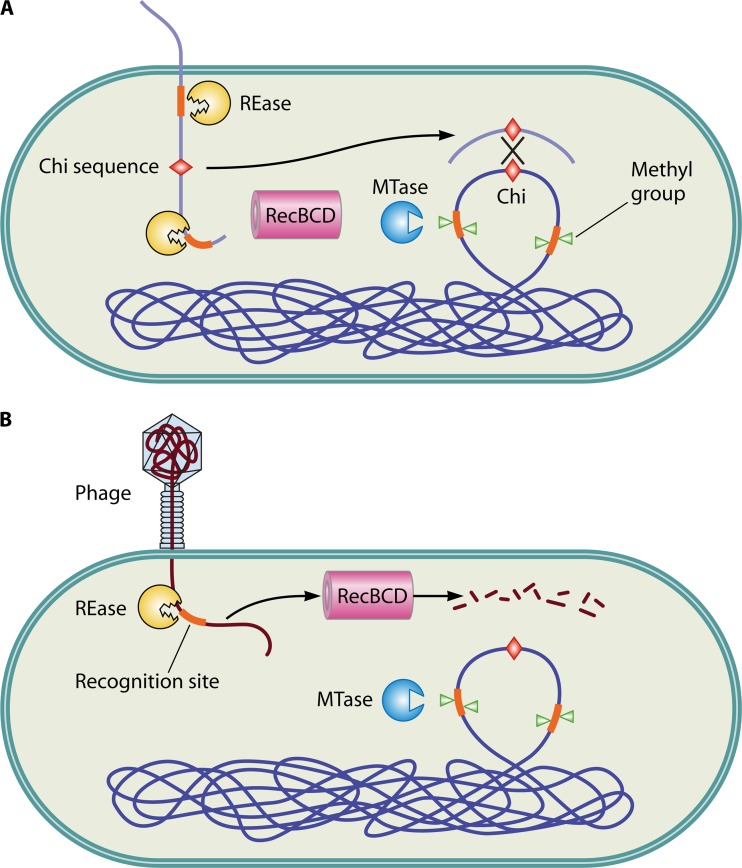
Another cellular machine which functions similarly to REases in limiting invasive genome elements is RecBCD. RecBCD functions both in restricting foreign genomes and in host DNA repair by recombination (34). The DNA repair function on the phage genome or the restriction function on host DNA could potentially be lethal to the host. RecBCD distinguishes the host genome from the phage DNA by means of a cis element, the Chi sequence, which is absent in phages but present at high frequencies in bacterial genomes (35). RecBCD is a bipolar helicase with nuclease activity that hydrolyzes DNA from a double-strand end (36). When RecBCD reaches a Chi sequence, the hydrolysis of DNA is arrested, and recombination is initiated (37) (Fig. 4). The Chi sequences differ among bacteria and serve as a bar code (38). The recognition of Chi sequences by the RecBCD enzyme is now understood at an atomic resolution (39). The RecBCD enzyme degrades phage DNA after restriction breakage but repairs chromosomal DNA after restriction (40). Alternative roles of RecBCD systems have been proposed by Kobayashi et al. (40; reviewed in reference 41). Similar to RecBCD, the RecFOR pathway has also been shown to repair lethal double-strand breaks on the chromosomes generated by REase and degrade restricted nonself DNA (40, 42).
Fig 4.
Role of R-M systems in recombination. R-M systems effectively restrict incoming DNA. (A) Restriction of incoming DNA from a closely related bacterium (harboring similar Chi sequences) generates DNA fragments which can be utilized as substrates for homologous recombination by the RecBCD pathway. (B) In contrast, the fragments generated by the restriction of phage DNA (lacking the Chi sequence) are recognized as nonself and subjected to further degradation by the RecBCD pathway.
In contrast to the defense systems discussed above, abortive infection of phage (termed “Abi”) systems or phage exclusion mechanisms enable bacteria to resist phage multiplication through programmed cell death of the infected cell (reviewed in reference 43). The Rex system of E. coli, which contains two components, viz., RexA and RexB, is one of the most well-studied abortive infection systems to date (43, 44). Upon phage infection, a replication complex intermediate activates RexA. Two activated molecules of RexA in turn activate RexB, an ion channel protein (43). The activated RexB reduces the membrane potential of the cell, resulting in the death of the host and the phage. Other Abi systems of E. coli include the Lit protein and the polypeptide encoded by prr (43).
In addition to the above-mentioned defense systems, bacteria also keep track of invasive elements by transcriptional silencing. The transcription termination factor Rho and nucleoid-associated proteins are two such factors that are implicated in the silencing of foreign genomic elements. Nucleoid-associated proteins selectively bind to xenogeneic DNA with AT contents higher than that of the genome and silence them (45, 46). The Rho protein of E. coli was recently implicated in causing the premature transcription termination of horizontally acquired genes and prophages in the genome (47). It appears that differences in genetic code utilization could facilitate the recognition of these genomic islands. These two strategies, however, do not limit the acquisition of foreign DNA, but rather, they prevent the expression of any lethal genes.
Several studies indicated that the toxin-antitoxin (T-A) systems could also protect against invading genomes (reviewed in reference 48). The protection conferred by T-A systems could be through either a direct or an indirect pathway. The direct mechanism is exemplified by the toxIN system (49). The system encodes the Abi protein, which abrogates the maturation of phage particles (49). An indirect mechanism was proposed in the case of the prevention of plasmid establishment (50). According to this model, a host cell harboring a chromosomally encoded antitoxin would neutralize the toxin of a plasmid-encoded T-A system and prevent plasmid addiction (50).
Strategies against R-M Systems
An R-M system would assist bacteria in populating a new habitat containing phages (51). However, the host barrier can be overcome by a phage which escapes restriction to become refractory to that particular REase of the host. The restriction barrier is overcome either by chance alone, with low probability, or by phages with specially evolved strategies (1, 26). A question that is often asked is whether restriction is indeed an efficient mechanism to limit foreign DNA (52). The basis for this question is that bacteriophages and other invasive genome elements employ a number of strategies to evade restriction by host REases (reviewed in references 24, 1, and 26). In this section, several mechanisms by which phages and other invading genomes evade restriction by REases are illustrated. Only the well-characterized antirestriction systems employed by invading genomes are described below.
(i) Many of the phages encode MTases (1, 26). These enzymes are known to have broad specificity and can thus protect their own genome from multiple REases (1, 26). For example, phages of Bacillus subtilis, SPR, φ3T, δl1, and SPβ, have evolved protection mechanisms against host restriction enzymes by self-methylating their DNA at various sequences (53). The methylation of phage genomes is caused by the acquisition of host-encoded MTases by the phages (54).
(ii) In addition, phages have evolved a plethora of modifications as effective antirestriction strategies to evade entire groups of host defense mechanisms. Some of these modifications involve the attachment of bulky groups, e.g., hydroxymethylation (53), glycosylation (55), and acetamidation (56, 57). The mom gene of bacteriophage Mu encodes a protein that catalytically transfers an acetamide group to the N-6 position of adenine in the sequence context 5′-G/C-A-G/C-N-C/T-3′ (58). Modification by Mom (“momification”) confers resistance against a wide range of REases (59).
(iii) A number of phages and plasmids are equipped with proteins that block restriction. A well-studied example is the OCR (overcome classical restriction) protein of T7 phages (60). The antirestriction protein OCR is an exquisite mimic of a 24-bp B-form DNA (61, 62). This structure of OCR prevents the type I R-M complexes from binding to DNA. Similar DNA mimics, termed Ard (alleviation of restriction of DNA) proteins, are expressed by many plasmids. To illustrate, ArdA, which is rich in Asp and Glu residues, mimics a 42-bp DNA and inhibits type I enzymes (63). In addition, some of the phages and conjugating plasmids are known to encode antirestriction proteins, which alleviate the restriction function (26). These antirestriction proteins are usually coinjected with the DNA in order to impede the restriction enzyme transiently (26). ArdC is another antirestriction protein specific for type I R-M systems and is encoded by the E. coli IncW conjugative plasmid pSa. ArdC protects the incoming T strand during conjugation (64). Similarly, P1 phage encodes the DarA and DarB (defense against restriction) proteins, which are coinjected along with the phage genome to avoid the restriction of the DNA by the host type I REases (65). Although the mechanism of inhibition is not completely understood, it was proposed that the Dar proteins coat phage DNA and inhibit the translocation of type I enzymes (26). Another example is a hydrolase from T3 phage that cleaves S-adenosyl methionine and acts as part of the defense of the phage against the host type I restriction systems, which are dependent on this molecule as a cofactor for DNA cleavage (66, 67).
(iv) Continuous selection against specific recognition sites leading to a reduction in the number of sites in the phage genomes would also avoid restriction by the REases. Such a decrease in the palindromic sequences recognized by REases is usually observed for a number of phages (68 –71). For example, the restriction sites CCGG, CGCG, and GGCC are underrepresented in B. subtilis phage PZA compared with other lytic dsDNA phages (68). An examination of the phage PZA genome indicated the presence of many 1-nucleotide variants of the recognition sequences (68). If one were to consider the prevalence of such single-nucleotide variants in different phages, the mechanism could be one of the oldest outcomes of selection observed in the invading genomes.
(v) The typical length of the recognition site of a REase varies from 4 to 8 bp. Because of their smaller size, phage genomes have a lower frequency of palindromic sequences of ≥8 bp (28). Thus, the frequency of restriction of a phage is lower for a REase recognizing 8 bp than for a REase recognizing 4 bp. Furthermore, R-M systems that require two copies of the recognition sites (type IIE, type IIF, and many type IIS sites) have a much lower probability of restricting incoming phage genomes (72). R.EcoRII, a type IIE REase, needs an interaction with two or three recognition sites for efficient DNA cleavage (73, 74). T7 phage evades restriction by R.EcoRII due to an underrepresentation of EcoRII sites (75).
(vi) A unique antirestriction mechanism is employed by T7 phage, by exploiting the cleavage mechanism of type III enzymes. EcoP1I, a well-characterized type III enzyme, requires two copies of its asymmetric recognition sites to be oriented in a head-to-head manner for successful cleavage (76). T7 phage exhibits strand bias; i.e., all the EcoP1I sites in T7 DNA are in the same orientation rather than in the head-to-head formation, which is required for cleavage (1, 77). As a result, EcoP1I does not efficiently cleave the phage DNA. Thus, this example serves to illustrate the evolution of a phage genome to avoid the recognition sites of the host enzyme.
Interestingly, the above-mentioned observation also led to the understanding of single-stranded asymmetric host methylation by type III MTases (76). The hemimethylation of asymmetric sequences would give rise to unmodified sites after replication. In type IIS R-M systems, this problem is usually overcome by utilizing two MTases, each recognizing either the top or the bottom strand of the DNA. For example, R.MboII recognizes the asymmetric sequence 5′-GAAGA-3′/3′-CTTCT-5′. M1.MboII modifies the last adenine of the top-strand recognition sequence, and M2.MboII transfers the methyl group to the internal cytosine in the bottom strand (4, 78). In contrast, type III R-M systems utilize only one MTase to discriminate self from nonself (e.g., hemimethylation of the internal adenine in the sequence 5′-AGACC-3′ by M.EcoP1I) (14). The cognate REase requires the recognition sites on both the strands of the genome to be oriented convergently; i.e., the orientation of the two enzyme molecules in a head-to-head manner is required for efficient cleavage, and hemimethylation would inhibit enzyme binding on the methylated strand. The postreplicatively generated unmodified sites in the daughter strands would be in one orientation. Since these substrates are poorly cleaved by the type III REases, a single MTase could be sufficient to protect the genome (see Fig. S2 in the supplemental material). Further genome analyses are required to corroborate these findings.
(vii) Barring a few exceptions, most REases require dsDNA for recognition and cleavage (http://rebase.neb.com/rebase/rebase.html). Phages that harbor single-stranded DNA (ssDNA) genomes are thus resistant to cleavage by REases (79). However, the double-stranded replicative forms of these ssDNA viruses were found to be sensitive to restriction in vitro (80). It is conceivable that mechanisms that exist to protect the genomic DNA may help alleviate restriction. Hence, the ssDNA genomes would offer greater protection against REases and facilitate their propagation in the bacterial host.
(viii) Phages appear to have evolved mechanisms to utilize the rod-shape morphology of Gram-negative bacteria. A recent study showed that a number of temperate and virulent phages, viz., T4, T7, λ, KVP40, P1, and φA1122, localize to bacterial poles (81). However, contrary to those studies, in a very old report, T6 phage was found to be present all over the cell surface (82). It is known that DNA uptake in bacteria that exhibit natural competence usually occurs at the poles (83). If the uptake of DNA at the poles is to escape from the host restriction machinery targeting the incoming DNA, then, by injecting the DNA at or near the poles, the phages may evade the host REases. However, the differential intracellular distribution of REases has not been explored, and hence, the strategies that are developed by bacteria against the phages that invade at poles are areas that need further investigation.
Fitness Cost Incurred by R-M Systems on Host Bacteria
It is evident that the above-mentioned diverse strategies employed by the invading genomes ensure the evasion of restriction and thus increase their survival. Moreover, the maintenance of an active defense system in bacteria could incur a fitness cost to the organism. The following points describe some of the possible ways by which an active REase escalates the cost/benefit ratio for the bacteria.
(i) It has been observed that bacteria containing an R-M system have a decreased restriction site frequency in the genome for that particular recognition sequence, a phenomenon called restriction site avoidance (84). This strategy is employed by phages to evade restriction (see “Strategies against R-M Systems”). This strategy also protects host bacteria from attack by its own REase. Interestingly, palindrome avoidance is observed to a greater extent in bacteria than in phages (84). Palindrome/restriction site avoidance by mutations, however, may affect other cellular functions. For example, missense mutations arising due to palindrome avoidance could affect an essential cellular function and lower the fitness of bacteria. However, bacterial fitness measurements with predefined genomic palindrome avoidance have not been explored and await further experimental studies.
(ii) Studies have shown the extensive hydrolysis of ATP by the type I enzyme EcoR124I prior to the restriction of the foreign DNA to facilitate translocation (85). The enzyme progresses in small steps of 1 bp along the DNA, with 1 ATP unit consumed per step (85). This is clearly an extravagant way of spending energy if it occurs in vivo. Since cleavage often occurs at a site distant from the initial recognition sequence, the defense mechanism is energetically inefficient. To circumvent this burden, the organism appears to control the intracellular enzyme concentration. As shown for E. coli, efficient phage restriction occurs by using about 60 molecules of EcoKI, which would consume ∼0.2% of the total ATP pool (86, 87).
ADDITIONAL FUNCTIONS OF R-M SYSTEMS
Selfish Genes
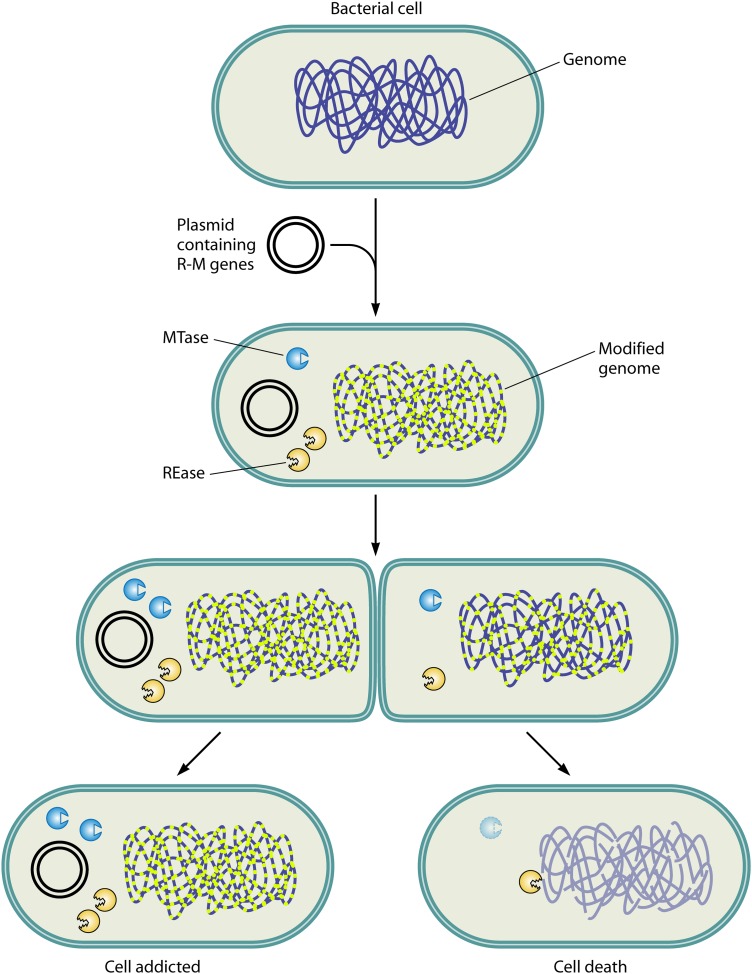
Generally, as described above, the gene for a given REase is linked to the gene of its cognate MTase, and together, they form an R-M complex. Despite their function in cellular defense, these gene complexes tend to propagate as selfish elements to promote their own survival and increase their relative frequency (28, 41, 88). For example, the failure to segregate R-M-encoding plasmids equally during cell division results in cell death for the progeny lacking these plasmids (28, 89). Intact copies of the gene complex survive in other cells of the clone. Therefore, the bacteria become dependent on the resident R-M system and are thus addicted (Fig. 5). The basis for this behavior is postsegregational killing, wherein a cell cannot afford to lose an established R-M system (28). When the resident R-M gene complex is cured from the genome, the intracellular MTase and REase concentrations would be diluted with every cell division. The dilution of similar numbers of REase and MTase molecules causes cell death because a single unmethylated site is enough to cause a DNA double-strand break by a REase, while all of the recognition sites should be methylated to protect the genome from cleavage. This would require many more molecules of MTase than REase. Indeed, in the EcoRI R-M system, the REase and MTase exhibit similar half-lives, but when genes encoding the R-M system are lost from the cell, REase-mediated cell death is observed (90).
Fig 5.
Postsegregational cell killing. Plasmid-harbored R-M gene complexes tend to propagate as selfish genetic elements to promote their own survival. The R-M system present in a cell expresses both REase and MTase: the REase restricts the foreign DNA, and the MTase protects the host genome against cleavage by the cognate REase. The postsegregational loss of the R-M gene complex results in the loss of methylation. The REase, owing to its higher level of stability, attacks the unmodified host genome, resulting in cell death (see “Selfish Genes”). The R-M gene complex thus propagates in the clonal population, resulting in the addiction of the host cell.
Kobayashi has reviewed in detail the behavior of R-M systems as selfish gene loci and the effect of their mobility on the genomes (41, 88). Evidence supporting the selfish gene model was observed for many type II enzymes, such as Bsp6I (91), EcoRI (92), EcoRII (93), EcoRV (94), PaeR7I (28), PvuII (95), and SsoII (93). The selfish gene model explains the phenomenon that type II R-M systems could not be lost due to random fluctuations in plasmid segregation. Host cell killing was also shown with the type IV enzyme McrBC by the introduction of a DNA methylation gene (96). Although such selfish behavior appears to be a widespread phenomenon with type II R-M systems, it is not common in type I or III enzymes (97). In the latter systems, the MTase and the REase are subunits of the same protein complex, and the intracellular ratios of MTase to REase do not change over time upon the loss of the R-M locus, a prerequisite for function as an addiction module. Studies of EcoKI, one of the earliest-characterized type I enzymes, revealed that the enzyme does not behave as a selfish element (97). Similarly, EcoR124I, another well-studied type IC enzyme encoded on a large plasmid, does not seem to exhibit postsegregational killing (98). The importance of the selfish gene behavior of R-M systems in maintaining the methylation status of the genome is discussed below (see “Enforcing Methylation on the Genome”).
There are many characteristics of type II R-M systems that are similar to T-A systems: (i) both systems encode a stable toxin and a labile antitoxin, (ii) both systems are associated with mobile elements, and (iii) the genetic loci exhibit selfish behavior (88, 99, 100). The mobility of these genetic elements appears to allow the R-M systems to invade new genomes, thus contributing to their propagation. Moreover, Kobayashi's group showed that postsegregational killing favors the spread of genetic elements in the presence of a spatial structure (101). In the case of T-A addiction modules, bacteria appear to counteract the addiction of plasmids by neutralizing the toxin (see “Non-R-M Defense Systems” above). Although mechanisms that counter the addiction of type II R-M systems have been proposed (41), further experimental studies are awaited.
Stabilization of Genomic Islands
R-M systems prevent the loss of the episome by postsegregational killing, because daughter cells that do not acquire the plasmid undergo cell death due to the induction of dsDNA breaks by the stable REase. Similarly, it was observed that an R-M gene complex residing on a bacterial chromosome cannot be replaced easily by a homologous sequence of DNA (102, 103), indicating that they are difficult to eliminate. Furthermore, it was observed that R-M systems are often linked to mobile elements or acquired through HGT (99). This raises a question regarding the fitness advantage conferred by these systems to the host bacteria. It is conceivable that in addition to their function in cellular defense, genomic R-M systems may also play a role in stabilizing the host chromosome, similar to their role in plasmid stabilization; i.e., genomic islands acquired by the bacterium through HGT are not lost. A similar role in the stabilization of genomic islands was proposed for T-A systems. It was observed that chromosomally encoded T-A systems stabilized neighboring regions of the genome (104). It is rather enigmatic that some genomes have an abundance of R-M components. Perhaps, the importance of having multiple R-M systems can be partly explained by their role in the stabilization of genomic islands.
Role in Nutrition
The chlorella viruses, which infect algae, harbor R-M systems (105). These viruses encode a number of DNA MTases and site-specific endonucleases. The biological function of the chlorella virus-encoded REases is unknown, but they are hypothesized to play a role in host chromosome degradation and/or the prevention of virus superinfection (106). It was observed that the in vivo degradation of host nuclear DNA coincides with the appearance of site-specific endonuclease activity (107). In contrast, MTase activity is manifested at 60 to 90 min postinfection, correlating with viral DNA replication (107). Hence, while endonucleases help in degrading host DNA and providing deoxyribonucleotides for incorporation into viral DNA, the methylation of newly replicated viral DNA by the cognate MTases would protect the genome from self-digestion. Recently, a giant “Marseille” virus that infects amoeba was isolated, and its genome was found to encode 10 gene loci belonging to HNH endonucleases and REase-like enzymes (108). It is possible that some of these nucleases may function in the reallocation of host resources to the virus.
Immigration Control, Maintenance of Species Identity, and Control of Speciation
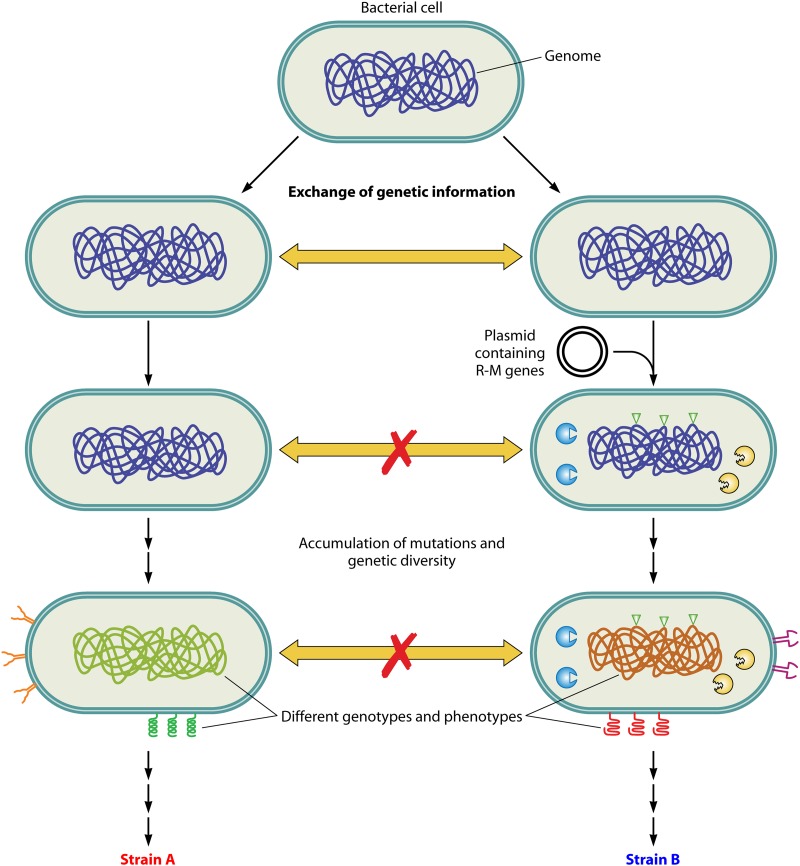
Species are defined as groups of organisms that can interbreed to generate offspring. In higher organisms, species are maintained by reproductive and/or geographical isolation. In the case of prokaryotes, however, the horizontal transfer as well as the vertical transfer of DNA are rampant. Thus, in bacteria, species are maintained by genetic isolation. REases, by the restriction of foreign DNA (that possesses nonnative methylation patterns), function in immigration control (52). Such a barrier would also serve the function of the maintenance of species in bacteria (52). In support of this, many E. coli and Salmonella enterica serovar Typhimurium strains harbor specific genomic loci rich in R-M systems, termed “immigration control regions” (109). Accordingly, R-M systems facilitate genetic isolation, which is required for the acquisition of new biological properties. Genetic isolation is provided by controlling the uptake of DNA from the environment. The methylation pattern provides a specific identity to that particular strain distinct from those of other closely related species and thus distinguishes self from nonself. According to this model, the presence of different recognition specificities in various strains of the same species further divides the species into different variant strains of bacteria, termed “biotypes.” These variant strains would not exchange genetic material among each other due to differences in methylation patterns. With a sufficient accumulation of genetic variation, biotypes might evolve into different species (Fig. 6).
Fig 6.
Role of R-M systems in the evolution of new strains. The horizontal transfer of DNA in bacteria increases the genetic diversity among them. A bacterial cell which acquires a new R-M gene complex (right) becomes genetically isolated from its clonal population (left). The MTase component of the newly acquired R-M system modifies the genome. Owing to this change in the methylation pattern, the REase prevents the genetic exchange of alleles between closely related strains. Furthermore, mutations acquired in these populations would facilitate genetic diversity, resulting in different genotypes. These populations would further evolve into different strains.
HGT of genetic material represents a substantial source of novel genetic information in prokaryotes (110, 111). Notably, the uptake of foreign genes along with their establishment and maintenance are often biased toward the acquisition of traits that contribute directly to the fitness of the bacteria, such as virulence or resistance to toxins (112, 113). The horizontal transfer of DNA occurs in prokaryotes via transduction, transformation, or conjugation. Staphylococcus aureus is a major pathogen that relies on HGT for the modulation of virulence. In this species, the specificities of the type I enzymes among the strains vary (due to differences in the HsdS subunit), and this impedes the transfer of mobile genetic elements among different strains (114). Thus, type I gene complexes appear to function in controlling the evolution of S. aureus strains. Recent studies of type III-like enzymes also revealed the role of these REases as a major barrier to HGT in clinical strains of methicillin-resistant S. aureus (115). Strains deficient in these enzymes were hypersusceptible to the horizontal acquisition of DNA from other species, such as E. coli, and could easily acquire a vancomycin resistance gene from enterococci (115). Subsequent studies, however, indicated that the type III-like enzyme is actually a type IV REase recognizing 5-methylcytosine/5-hydroxymethyl cytosine-modified DNA (116). Additionally, a whole-genome sequencing analysis of 20 Neisseria meningitidis strains revealed that R-M systems generate phylogenetically distinct clades, suggesting a regulation of HGT by REases (117). An emerging picture from all these studies is that a variety of R-M systems may indeed function as immigration controllers.
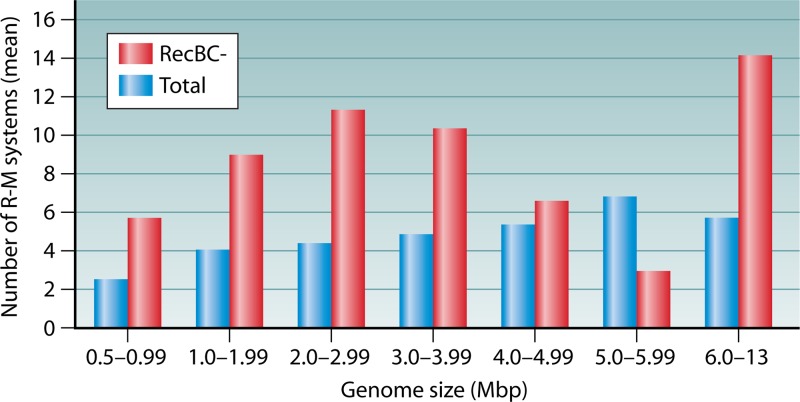
An analysis of genomes for the distribution of R-M systems in organisms lacking RecBC (see the supplemental material) revealed an interesting correlation. It was observed that the mean number of R-M systems in organisms lacking RecBC is higher than the mean number of total genomes (Fig. 7; see also Fig. S3 in the supplemental material). Since organisms lacking RecBC genes would have a higher frequency of HGT, it appears that in these organisms, the REases might serve to regulate genome flux in addition to their primary defense mechanism (see “Non-R-M Defense Systems” above).
Fig 7.
Distribution of R-M systems in RecBC− organisms. Shown are data from a genome-wide analysis of the presence of conserved methyltransferase genes among bacteria with genome sizes ranging from 0.5 to 13 Mb (see the supplemental material). The plot shows the mean values for the distributions of numbers of R-M systems with the specified class intervals of genome size. The list of organisms lacking RecBC was taken from data reported previously (229, 230). A correlation of an increase in the number of R-M systems in RecBC− organisms compared to the total distribution of R-M systems can be observed.
Recombination and Genome Rearrangements
REases cleave foreign DNA into small fragments, which inside the host could either be further degraded by exonucleases or act as substrates for the recombination machinery. Several studies have revealed that foreign DNA restriction by REases generates products that could stimulate homologous recombination with the host genome (118–124; reviewed in references 52 and125). The stimulation of homologous recombination was proposed to have two possible cellular roles: (i) rescuing accidental R-M-mediated lethality in the host and/or (ii) providing genetic variation by enhancing recombination between similar species (125). However, it was argued that the enhancement of recombination by REases might be a by-product rather than a primary function of these systems (126). This is illustrated by the fact that the role of R-M systems in recombination does not completely explain why they exist with so many different recognition specificities.
Restriction was also identified to play a role in nonhomologous recombination, in which a small stretch of homologous DNA sequences at one end is utilized to recombine and integrate a large foreign DNA (lacking homology) into recipient genomes (127). EcoKI, a well-studied type I enzyme, was shown to promote this homology-facilitated illegitimate recombination (127). In addition, there is evidence that R-M systems could bring in genome rearrangements (99, 102, 128–132). Thus, by promoting homologous recombination and functioning in nonhomologous genome rearrangements, R-M systems may play a role in generating genomic diversity.
Evolution of Genomes
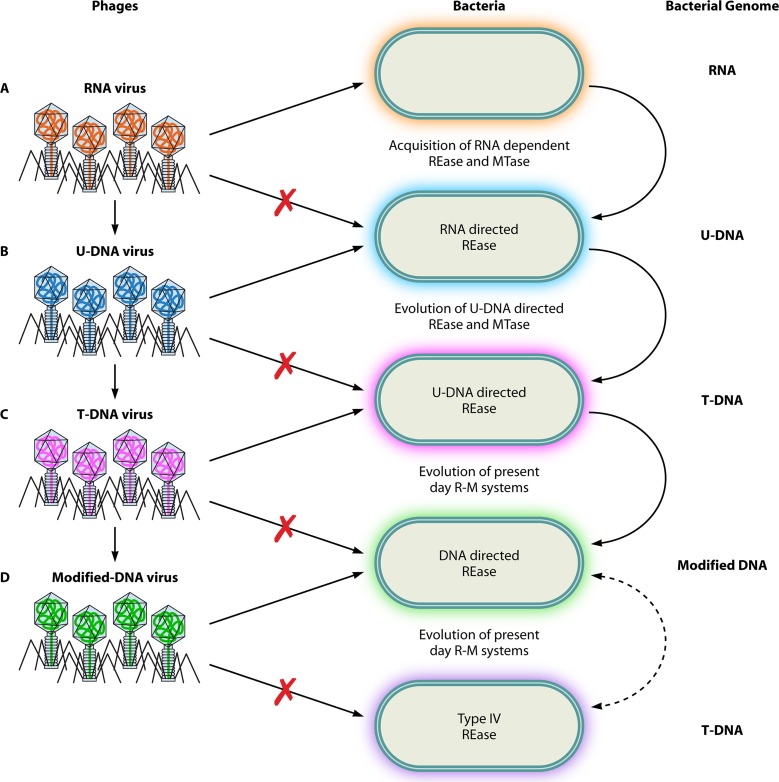
According to Arber, R-M systems modulate genetic variation and thus modulate the rate of evolution (118, 133). Thus, it was proposed that defense systems constitute precious tools for natural genetic engineering (134). Recent advances in viral and microbial genomics have greatly stimulated the interest in the origin and evolution of genomes. It has been proposed that the initial transition of RNA to DNA genomes might have occurred in phages conferring an advantage to evade primitive defense systems (135). These genes might have been further taken up by bacteria. Similar coevolutionary arms races could explain the transition from uracil-containing DNA to thymine-containing DNA genomes (Fig. 8) (135). Phages PBS1 and PBS2, which infect B. subtilis, contain uracil in their DNA genomes, which, in all likelihood, could be an intermediary form during the evolution into thymine-containing DNA genomes (136). Likewise, the modification of nucleotides in phage and bacterial genomes could be a consequence of the coevolution between them. Recent studies revealed variant type I systems that utilize DNA backbone modifications by phosphorothioation to distinguish self from nonself (137, 138). In this host-specific restriction system, instead of methylation, the host genome is protected due to a nonbridging sulfur atom attached to the backbone phosphorus at rare but specific sites (138). The unmodified foreign DNA is recognized as “nonself” and subjected to degradation. These observations imply a major role for R-M systems in the evolution of novel base and epigenetic modifications in host and phage genomes.
Fig 8.
Role of R-M systems in genome evolution. The probable role of defense systems in the evolution of genomes is depicted. (A) Initially, RNA viruses coexisted with bacteria containing RNA genomes. With the evolution of uridine-containing DNA (U-DNA) genomes in bacteria and the acquisition of RNA-dependent endonucleases, a primitive R-M system could have ensured the restriction of the RNA viruses. (B) Such a selection pressure would enforce the evolution of a U-DNA genome in viruses to evade this primitive R-M system. This in turn would result in the evolution of thymidine-containing DNA (T-DNA) genomes in bacteria to evade phage infection. (C) The phage adapts to the host defense strategy by evolving a T-DNA genome. (D) Continuous selection would result in an “arms race” between bacteria and viruses, resulting in the utilization of modified DNA bases in phage and bacterial genomes.
Promiscuity in Cofactor Utilization and Substrate Specificity
Analyses of diverse enzyme superfamilies have shown that many enzymes are capable of catalyzing reactions with noncanonical substrates (139, 140). In contrast to these enzymes, REases were initially considered to be very-high-fidelity enzymes with exquisite site specificity (at least for type II systems) from the time of their discovery. However, with the identification of “star” activity in these enzymes, it became apparent that some REases had the ability to recognize and cleave noncanonical sites under experimental conditions that are not totally optimum. Hence, to a large extent, the REases continued to enjoy the status of highly sequence-specific enzymes. However, it is now becoming increasingly apparent that REases may not exhibit exquisite sequence specificity after all (141). Instead, many of them may show promiscuous DNA cleavage to various degrees. The fidelity index value, which quantitatively measures the star activity of REases, identified a significant number of REases as being “star prone” (141). Additionally, the cleavage of heteroduplex substrates containing mismatches in the target site, when tested with 14 REases, indicated that many enzymes were indeed capable of cleaving mispaired recognition sites (142). It appears that relaxed specificity is a common phenomenon for these enzymes.
Studies of R.KpnI, a well-characterized type II REase, have provided interesting insights into the mechanism and biological role of catalytic versatility in the enzyme. The enzyme exhibits promiscuous DNA cleavage characteristics in the presence of the natural cofactor Mg2+ (143, 144). Notably, the promiscuous activity is triggered by the binding of an additional metal ion to the enzyme (144). The catalytic promiscuity exhibited by the enzyme under in vivo conditions suggests a functional role. Studies carried out with R.KpnI revealed that retaining broad specificity in DNA cleavage characteristics provided a selective advantage to the host by better targeting foreign invading genetic elements (145). It is possible that a relaxed specificity of REases might also serve a hitherto-unknown function(s) in the organism. The additional biological roles and the cellular conditions that trigger the promiscuous activity are yet to be determined.
A recent study showed that the REases R.AvaII, R.AvrII, R.BanI, R.HaeIII, R.HinfI, and R.TaqI can cleave RNA-DNA heteroduplex oligonucleotides in a site-specific manner (146). The ability to cleave RNA-DNA hybrids could be a strategy developed by bacteria to target phage genomes that utilize uracil in place of thymine. For example, as stated above, many phages that infect Bacillus species utilize uracil in their genomes (136). The promiscuous REases that cleave RNA-DNA hybrids could acquire additional functions to increase the fitness of the organism. Since the cleavage of both strands of the RNA-DNA hybrid by promiscuous REases releases the RNA and generates a single-strand break in the DNA, some of the molecular processes that require the cleavage of RNA-DNA duplexes may utilize this property as a backup strategy. The processes where RNA-DNA hybrid intermediates are found are (i) the release of mRNA from mRNA-DNA hybrids, (ii) the removal of R loops generated during transcription (147), and (iii) priming reactions during phage/plasmid replication (148).
A vast number of REases, with the exception of BfiI and PabI, show an absolute requirement for Mg2+ for DNA cleavage (8, 9). Other divalent ions with similar atomic radii are a poor replacement at the catalytic center. A closely related metal ion, Ca2+, belonging to the same group in the periodic table, indeed inhibits the catalytic activity of most enzymes when bound at the active site (95). However, a comparison of the cofactor preferences of PD-(D/E)XK and HNH REases indicated that HNH enzymes have a greater flexibility for metal ion coordination. For example, R.KpnI, the first REase member of the HNH superfamily to be identified, exhibits broad cofactor utilization for DNA cleavage (10, 149, 150). This utilization is also seen in nonspecific endonucleases belonging to the superfamily, e.g., colicin E7, colicin E9, and Serratia nuclease (151–153). The broad cofactor preference of the HNH enzymes might confer a fitness advantage to the genomes that harbor them by retaining their activity with different metal ions in vivo. Alternatively, it is possible that this unique feature is preserved during the evolution of the HNH REases to serve some as-yet-unknown function(s) in the cell.
Genetic Variation by Cytosine-to-Thymine Transitions
MTases modify DNA by transferring a methyl group to either cytosine or adenine. 5-Methyl-cytosine is highly susceptible to deamination, resulting in C-to-T mutational sites (154). For example, M.HpaII expression results in a 104-fold increase in the C-to-T mutation frequency (155). Thus, in genomes which utilize m5C MTases, evolution has tinkered with this pitfall in three ways, viz., palindrome avoidance, replacement with a different methylation mechanism, and acquisition of repair machinery (1). The C-to-T mutational load would progressively decrease the numbers of cytosine-containing palindromes in the genomes. In addition to palindrome avoidance, bacteria have also adapted to modulate the genetic variation caused by C-to-T transitions by utilizing different methylation positions, i.e., m4C rather than m5C. This change in the methylation pattern is more evident in thermophiles, where the frequency of deamination is severalfold higher. Thus, it was proposed that the transition from m5C to m4C was an adaptation to higher temperatures, which would also avoid to some extent the hypermutability associated with m5C (156). Furthermore, it was also observed that m5C MTases are often linked with the DNA repair locus vsp, which encodes the very short patch repair endonuclease (157, 158). Recent studies of N. gonorrhoeae revealed that the organism codes for two different Vsr endonucleases. The enzyme V.NgoAXIII recognizes all T-to-G mismatches, and V.NgoAXIV recognizes these mismatches only in the context of specific 4-bp sequences (GTGG, CTGG, GTGC, ATGC, and CTGC) (159). An analysis of the N. gonorrhoeae FA 1090 genome revealed the presence of eight m5C MTases. To add to this unusual burden, some of these MTases are known to methylate DNA at noncanonical sites, resulting in an increased number of potential mutational sites in the genome (160). Thus, it was proposed that the presence of V.NgoAXIII or V.NgoAXIV could prevent mutations arising from the increased frequency of deamination of m5C to thymine (due to the increased presence of m5C) in the genome (159). Interestingly, C-to-T transition mutations are modulated depending on the S-adenosyl methionine levels in E. coli (161). The small changes in alleles which arise in response to C-to-T mutations as well as the corrective measures explained above would enhance genetic variation in the population. Alleles that are advantageous essentially get fixed during evolution, allowing bacteria to adapt to new environments rapidly to increase the fitness of the host.
Functions of DNA Adenine Methyltransferases
Now, it is well established that the epigenetic modification of genomic DNA by MTases is important in defining the transcriptome. Although studies of eukaryotes provided the impetus, it is apparent that it is also a common theme in other kingdoms of life. DNA methylation, which discriminates self from nonself in prokaryotes, is brought about either by solitary MTases or by MTases associated with REases. In bacteria, of the three kinds of DNA base modifications observed, m5C, m4C, and m6A, adenine-specific methylation has been well studied and shown to have diverse cellular roles (17–19, 162). Functions carried out by this class of enzymes are self-versus-nonself discrimination during the restriction of phages, the downregulation or silencing of transposition events, the regulation of conjugation, the regulation of DNA replication initiation, cell cycle control, nucleoid reorganization, DNA mismatch repair, the transcriptional regulation of housekeeping and virulence genes, and posttranscriptional gene regulation (reviewed in references 162 and163). Thus, in those genomes that possess m6A MTases, the organisms seem to have taken full advantage of this exocyclic methylation. Indeed, these enzymes might function in generating a genomic bar code specific for the species hosting the R-M or “orphan MTase.” The CcrM MTase, encoded by a cell cycle-regulated gene of Caulobacter crescentus, fits into this category (164). The function of Dam MTase in oriC replication, transposition and virulence gene regulation, mismatch repair, and phage P1 packaging, etc., is another classic well-illustrated example (17, 164). Although most type II MTases are monomeric in nature, possibly due to their preference for hemimethylated substrates, a few studies suggested that some of them function as dimers (165–167). However, the significance of dimeric MTases is unknown. Dimerization is essential for MTase function, at least in some cases (165, 167), suggesting that the oligomeric status is optimized during evolution and hinting at an additional biological role(s) for these enzymes.
Enforcing Methylation on the Genome
Bacteria use DNA MTases as switches to systematically change their transcriptome (see “Functions of Phase-Variable R-M Systems” below). Alterations of methylation levels have been shown to cause changes in gene expression (168, 169). Since HGT between prokaryotic genomes is common, and the DNA MTase genes are known to undergo rampant transfer among organisms (41, 170), untimely methylation represents a possible threat to the methylation pattern of the host genomes. Recently, Ishikawa et al. dealt with this topic in detail and discussed various ways by which the epigenetic methylation of a genome is protected (171). The underlying mechanisms in the resolution of conflicts between different methylation systems in such a scenario have been reviewed (171). According to this model, the function of R-M systems is to impose a particular methylation pattern in the host. Cells that exhibit lower-level or altered methylation are removed from the population by cell death. The enforcement of the genome methylation status has been shown in the case of type I, II, and IV R-M systems. In the case of type I enzymes, the specific methylation status is imposed when methylation is disturbed by DNA damage and repair, wherein the REase may target an arrested replication fork (172). Postsegregational killing is triggered by REases belonging to type II R-M systems when methylation by the cognate MTase is decreased (see “Selfish Genes”). Type IV enzymes maintain the genome methylation status by initiating cell death when an additional foreign genome encoding an MTase gene enters the cell (96). Thus, cell death caused by type I, II, and IV systems in host bacteria exhibiting lower-level or altered methylation would ensure the maintenance of the epigenetic status in the remaining clonal population.
Functions of Phase-Variable R-M Systems
Phase variation is the heritable, interchangeable, and high-frequency on-or-off switching of transcription mediated by several mechanisms (173, 174). Host-adapted bacterial pathogens frequently use phase variation to generate diversity in antigenic surface structures such as pili, capsules, lipopolysaccharides, and flagella (174, 175). Pathogenic bacteria use the phase variability of gene expression to evade the host immune system. Based on sequence analysis and biochemical evidence, many type I and III enzymes of Bacteroides fragilis as well as Haemophilus, Helicobacter, Mycoplasma, and Neisseria species were found to be potentially phase variable (176–181). The biological significance of R-M systems that exhibit phase variability is not completely understood. However, the phenomenon appears to play a role in increasing the fitness of the bacteria under certain environmental conditions. Principal ways by which a phase-variable defense system could confer a fitness advantage to the host are as follows.
(i) REases protect the bacterial genome against phage infection and the incorporation of other selfish elements. However, in certain cases, a lysogenic phage infection could benefit the population if the phage encodes a factor that increases virulence (182–184). For example, infection of the Gram-negative bacterium Vibrio cholerae with the lysogenic CTX phage results in its toxinogenicity, as the lysogen codes for cholera toxin (184). Conversely, lytic infection could be beneficial when the lysed population benefits the survivors (clonal cells) by providing essential nutrients, an altruistic behavior comparable to kin selection. Phase variability in R-M systems can be utilized as a way to modulate these effects within a clonal population.
(ii) The phase variability of R-M systems could also function in the fine-tuning of the uptake of foreign DNA (185). To illustrate, in a restriction-off phase (r−), the uptake of nonself DNA could bring in variation. On the other hand, in a restriction-on phase (r+), REase would limit invasive xenogeneic DNA. It was proposed that the phase-variable expression of R-M systems might fine-tune these two phases in the population (179, 185). The fine-tuning of foreign DNA uptake might be an important process in naturally competent bacteria, viz., B. subtilis, H. influenzae, H. pylori, N. gonorrhoeae, and S. pneumoniae. Interestingly, whole-genome sequencing revealed that these bacteria harbor abundant R-M systems (Fig. 3). The phase-variable R-M systems in these organisms may function in the regulation of genome flux in addition to their role in defense.
(iii) Whole-genome sequencing of B. fragilis revealed that many R-M systems of bacteria undergo inversions (176). It has been proposed that phase variation in these bacteria increases the diversity of R-M systems, generating up to eight different recognition subunits (176). Similarly, Bayliss et al. proposed that diversity and phase variability in H. influenzae type III enzymes have evolved in order to confer a fitness advantage to the bacteria against diverse bacteriophage populations (186).
(iv) Fox et al. observed that H. influenzae, N. gonorrhoeae, and N. meningitidis strains contain phase-variable MTase genes often associated with an inactive REase component (180, 181). It was proposed that phase-variable modification systems play a role in regulating distinct set of genes, called “phase-varions” (180, 181), a function independent of the defense role. Phase variability would generate different phenotypes and might confer a fitness advantage to the organism by controlling the phase-varion.
EVOLUTION OF MOONLIGHTING ROLES IN R-M SYSTEMS
The widespread occurrence of R-M systems among eubacteria, archaea, and certain viruses of unicellular algae (46, 97, 187, 188) may hint at their other functions in addition to cellular defense. These additional roles could be better understood by looking into the evolutionary route by which a particular R-M system acquired a new biological function. As described above, it is well established that bacteriophages employ a plethora of strategies to escape restriction by resident REases (1, 55, 64). This raises the possibility that the restriction process alone could be an incomplete defense barrier against invading DNA. In evolution, R-M systems that exhibit additional cellular roles along with their defensive role could have been selected for and, hence, retained in the genomes. Thus, additional roles could be considered a result of distinct selective forces that could have driven the maintenance of these enzymes in bacterial genomes. This hypothesis would explain the presence of other defense systems against phages and the retention of REases by the host organism even after constant evolutionary pressure.
A new function in an R-M system could be achieved as a consequence of an evolutionary by-product. For example, a gene fusion event with one of the components of the R-M system would result in an additional function. The type II MTases EcoRII and SsoII belong to this category, as these enzymes double as transcription regulators in addition to their MTase activity (189, 190). These MTases harbor distinct helix-turn-helix motifs (not required for the MTase activity) and recognize operator sequences which are different from their respective methylation recognition sequences (189, 190).
Bacteria employ various strategies to impede phage infection. These strategies are in turn counteracted by the phages. As a result, a constant coevolutionary process is operational between bacteria and phages. In order to counter phage invasions, R-M systems seem to evolve faster (191, 192) and, hence, are frequently exchanged between species (95, 193). In such a scenario, genes which hinder phage infection would be selected and are fixed in the population. Novel substrate specificities are thus generated in response to phage evasion strategies. For example, the occurrence of methyl-directed REases (e.g., EcoKMcrBC [15] and GmrSD [16]) could be a strategic adaptation against phages with modified genomes.
R-M SYSTEMS OF HELICOBACTER PYLORI
H. pylori is a Gram-negative human pathogen. It is estimated that half of the world's population harbors this pathogen and that nearly 20% of the infected population develops disease (194). H. pylori strains exhibit natural competence and are known for their genome plasticity and diversity. The organism has attained a unique status in the bacterial kingdom because of its unusually large number of diverse R-M systems (21, 195). Owing to this peculiar feature of H. pylori and its unique niche in the human gut, this section aims at an understanding of the biological roles of R-M systems in this organism. Emphasis is placed on new developments addressing the role of R-M systems in the regulation of genetic exchange, adaptation to hostile environments, gene regulation, and the virulence of the organism.
Comparative analyses of the genomes of H. pylori strains predicted the presence of a large number of putative R-M systems but with a high degree of heterogeneity (196, 197). Further analyses revealed that only some of these systems retain enzyme activity (195, 198). In some of the inactive systems, REases are truncated while retaining the functional MTase gene. The presence of a functional MTase in the absence of REase suggests that these MTases might have other biological roles. For example, the hp0050 gene encodes a solitary MTase that is highly specific in H. pylori strains PG227 and 128 but exhibits relaxed specificity in H. pylori 26695 (199). Notably, an MTase gene (hp0050) deletion in clinical strain PG227 results in impaired growth (199).
Coliform bacteria like E. coli colonize the gut and adapt to the environment by acquiring adaptive alleles through HGT among different species. However, for species like H. pylori, which colonize the acidic regions of the stomach, diversification through interspecies gene transfer appears to be difficult. Gene diversification in these organisms is brought about by having a hypermutator phenotype and natural competence (187, 200, 201). These properties would impart a high level of genome plasticity and might facilitate the pathogen to adapt to new hostile environments (202). Unlike other naturally competent organisms that acquire larger DNAs by transformation, H. pylori strains were found to incorporate DNAs of only ∼1.3 kbp into their genomes (203). Since H. pylori lacks some of the key DNA recombination/repair function homologs, it was suggested that REases might play a role in limiting the recombination length (203). The limit for the size of the DNA to be integrated perhaps indicates that REases regulate genetic exchange in H. pylori (203). However, the unequivocal establishment of such a mechanism awaits further experimentation.
In addition to the possible role of R-M systems in the regulation of genetic exchange, recent whole-genome and transcriptome analyses of virulent strains of H. pylori have revealed new insights into the novel functions of these defense systems. For example, by genome comparisons of eight chronic atrophic gastritis strains, it was found that many R-M systems are associated with this condition (204, 205). Some of these chronic atrophic gastritis-associated enzymes were also found to be pH regulated (205). These genes might help the organism adapt to an acidic environment (205). Similarly, an analysis of the H. pylori transcriptome profile in infected mice resulted in the identification of several R-M genes that contribute to the colonization of the gut (206). These studies point toward a role for R-M systems in the adaptation of the bacteria to hostile environments.
The HpyC1I R-M system was first described by Lin et al. in a study designed to identify factors required for H. pylori cell adherence (207). Interestingly, the knockout of the hpyC1IR gene results in an elongated-cell morphology and decreased adherence to epithelial cells (207). Similarly, iceA1, the gene for a CATG-specific REase, is associated with H. pylori infection. While some of the H. pylori strains harbor a full-length open reading frame, the gene is truncated in other strains (208). Interestingly, the expression of the truncated iceA1 gene was shown to be upregulated upon contact with epithelial cells (209), suggesting a possible additional role other than the endonuclease function.
The regulation of genes involved in virulence is of crucial importance for every pathogen. While the above-described examples reveal the function of R-M systems in the adherence, colonization, and adaptation of H. pylori in the host environment, other studies have identified a role for R-M systems in the regulation of virulence gene expression (210–212). For example, MTases of H. pylori are known to selectively alter transcript levels of some genes, e.g., the genes of the dnaK operon and catalase (210, 212). Similarly, the target sites of an acid-adaptive MTase (HP0593) have been found to be in the promoter regions of physiologically important genes (213). Recent studies have also shown a regulatory role for the phase-variable modH gene of a type III R-M system in H. pylori (214). These studies suggest that the presence or absence of H. pylori MTases among different isolates might give rise to strain-specific differences in methylation patterns and alterations in gene expression. This difference in gene expression would in turn alter the extent of virulence of these bacterial strains. It is conceivable that in addition to their function in cellular defense, the regulation of genetic exchange, adaptation to host environments, and virulence, the chromosomally encoded R-M systems of H. pylori may also play a role in stabilizing genomic islands acquired by the bacterium through HGT (see “Stabilization of Genomic Islands” above).
FUTURE PERSPECTIVES
Evolutionary forces appear to act differentially on the two components of R-M systems. REases are toxic in the absence of their cognate MTases. Consequently, the endonucleases seldom deviate from their recognition specificity. In contrast, a specific MTase could evolve to possess broad specificity without altering the modification of the cognate recognition site. Indeed, it was observed that many MTase clones exhibit promiscuous activity, viz., M.HaeIII, M.EcoRI, M.EcoRV, and M.FokI (160, 215–217). However, contrary to those studies, recent data from single-molecule, real-time (SMRT) DNA sequencing of bacterial genomes showed that many of the MTases tested exhibited high fidelity (218). Interestingly, that study also revealed that the E. coli Dam MTase is highly promiscuous (218). Thus, those enzymes which have a broad specificity could have evolved to serve other roles in the cell, viz., the regulation of transcription and the modulation of DNA transaction processes. Moreover, low-level methylation at the promiscuous sites might provide protection to host genomes acquiring/evolving REases with novel substrate specificities. Studies of the in vivo methylation status at promiscuous sites could aid in an understanding of the generation of novel R-M specificities.
Several studies showed bacterial programmed cell death as a mechanism to counteract phage infection (reviewed in reference 43). However, a direct role for R-M systems in cell death mechanisms has not been reported. Recent work by Kobayashi's group provided evidence for a new function of the methyl-specific endodeoxyribonuclease McrBC (96). In that study, McrBC of E. coli was shown to limit the invasion of exogenous methyltransferase by host cell death (96). Interestingly, in the absence of exogenous methylation, the type IV REase Mrr enzymes of E. coli and S. Typhimurium were shown to restrict their own genomes under stress conditions (219, 220), indicating a cellular role distinct from the classical defense function of R-M systems. It is possible that Mrr enzymes have an additional role in the stress response, a view also supported by the fact that some of the Mrr REases are cryptic and expressed only due to spontaneous activation mutations (221). It is well established that bacteria live as communities and resemble multicellular organisms in many features. It was proposed that the programmed cell death of infected or damaged clonal cells within the community is beneficial to the population as a whole (222). Artificially designed systems that trigger suicide by R-M components upon phage entry have been shown to function as a defense strategy against infection (223–225). However, the natural occurrence of such R-M systems has not been explored. Phages are the most abundant microbes in the biosphere and outnumber bacteria in the environment (226). Hence, it would be interesting to investigate the role of R-M systems in cell death mechanisms that can be triggered to limit the spread of phages.
Arber referred to R-M components as “evolution genes” which modulate the rate of evolution (118). Studies carried out by Kobayashi's group indeed support this view, and these components have been shown to accelerate evolutionary changes in the genome (128). The application of such a function in genomic rearrangements for a beneficial phenotype awaits further investigation.
The uptake and maintenance of extracellular DNA by means of genetic transformation are well recognized as major forces in microbial evolution. In addition, it was also proposed that natural transformation could also provide DNA as a nutrient (188, 227). Studies of E. coli showed that mutants that are defective in such a process could be isolated (228). In addition, it has been established that REases could function in nutrition (see “Role in Nutrition”). It would be interesting to test a similar role of REases in providing DNA as a nutrient for bacteria, especially under conditions of nutrient starvation.
From a review of the vast literature, it is evident that the function of R-M components as an innate immune mechanism does not completely explain their diversity. While these enzymes are widespread, the rationale for the presence of multiple R-M systems in a single host is also not clear. An understanding of their additional biological roles might shed light on deciphering the basis for their diversity and redundancy. Although it is apparent that components of R-M systems function in multiple cellular roles, it is not clear whether the additional functions are manifested at different times. It is possible that while some of the functions occur simultaneously, other “moonlighting” roles could be induced under certain conditions.
CONCLUDING REMARKS
It is well established that the undisputed role of R-M systems is to serve as a defense strategy against the invasion of foreign DNA. From the vast literature, it is apparent that they have successfully evolved and gained additional roles. The nonrandom distribution of R-M systems could indeed be an indicator of their additional roles in the cell. Emerging data indicate their role in recombination, nutrition, and the generation of genetic diversity, etc. In addition to safeguarding genomic integrity, it appears that in many organisms, they may regulate genomic flux, stabilize genomic islands, or maintain methylation patterns. Thus, cells seem to utilize R-M systems in various biological processes to increase their relative fitness in the population. The study of noncanonical roles for this large group of diverse enzymes would open up avenues for a better understanding of bacterial genome dynamics and evolution.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. T. F. Dryden and members of the laboratory of V.N. for suggestions and critical reading of the manuscript.
K.V. is a recipient of a Shyama Prasad Mukherjee fellowship from the Council of Scientific and Industrial Research, Government of India. V.N. is a J. C. Bose fellow of the Department of Science and Technology, Government of India.
Biographies
Kommireddy Vasu is currently a research associate in the laboratory of Professor V. Nagaraja, Department of Microbiology and Cell Biology, Indian Institute of Science. He received his M.S. degree in Biological Sciences from the Indian Institute of Science in 2007. While there, he obtained a Ph.D. from the Department of Microbiology and Cell Biology, researching mechanistic details of protein-nucleic acid interactions and the evolution of recognition specificity in endonucleases. He was awarded the Shyama Prasad Mukherjee Fellowship from the Council of Scientific and Industrial Research for carrying out his predoctoral research. Dr. Vasu's interests are focused on protein-nucleic acid interactions and understanding how bacteria evolve strategies to gain the upper hand over viruses in a coevolutionary arms race.

Valakunja Nagaraja received a Ph.D. from the Indian Institute of Science for his work on mycobacteriophage I3 and the role of DNA gyrase in mycobacteria. He was a research associate at Biozentrum, University of Basel, Switzerland, and at the Department of Biology, University of Rochester, working on type I restriction enzymes and the regulation of the antirestriction system in phage Mu, respectively. He is presently a Professor and Chairman of Microbiology and Cell Biology, Indian Institute of Science. He has been involved in teaching microbiology and molecular biology and carrying out research in the areas of R-M systems, the regulation of gene expression, DNA topoisomerases, and DNA-protein interactions. His research efforts have culminated in several important findings, applications, and patents. He has authored 140 publications in reputed journals and mentored a number of graduate students.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MMBR.00044-12.
REFERENCES
- 1. Bickle TA, Kruger DH. 1993. Biology of DNA restriction. Microbiol. Rev. 57: 434–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pingoud A, Fuxreiter M, Pingoud V, Wende W. 2005. Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 62: 685–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dryden DTF. 1999. Bacterial DNA methyltransferases, p 283–340 In Cheng X, Blumenthal RM. (ed), S-Adenosylmethionine-dependent methyltransferases: structures and functions. World Scientific Publishing, Singapore [Google Scholar]
- 4. Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev SK, Dryden DTF, Dybvig K, Firman K, Gromova ES, Gumport RI, Halford SE, Hattman S, Heitman J, Hornby DP, Janulaitis A, Jeltsch A, Josephsen J, Kiss A, Klaenhammer TR, Kobayashi I, Kong H, Krüger DH, Lacks S, Marinus MG, Miyahara M, Morgan RD, Murray NE, Nagaraja V, Piekarowicz A, Pingoud A, Raleigh E, Rao DN, Reich N, Repin VE, Selker EU, Shaw PC, Stein DC, Stoddard BL, Szybalski W, Trautner TA, Van Etten JL, Vitor JM, Wilson GG, Xu SY. 2003. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 31: 1805–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dryden DTF, Murray NE, Rao DN. 2001. Nucleoside triphosphate-dependent restriction enzymes. Nucleic Acids Res. 29: 3728–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Murray NE. 2000. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle). Microbiol. Mol. Biol. Rev. 64: 412–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ibryashkina EM, Zakharova MV, Baskunov VB, Bogdanova ES, Nagornykh MO, Den'mukhamedov MM, Melnik BS, Kolinski A, Gront D, Feder M, Solonin AS, Bujnicki JM. 2007. Type II restriction endonuclease R.Eco29kI is a member of the GIY-YIG nuclease superfamily. BMC Struct. Biol. 7: 48 doi:10.1186/1472-6807-7-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miyazono K, Watanabe M, Kosinski J, Ishikawa K, Kamo M, Sawasaki T, Nagata K, Bujnicki JM, Endo Y, Tanokura M, Kobayashi I. 2007. Novel protein fold discovered in the PabI family of restriction enzymes. Nucleic Acids Res. 35: 1908–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sapranauskas R, Sasnauskas G, Lagunavicius A, Vilkaitis G, Lubys A, Siksnys V. 2000. Novel subtype of type IIs restriction enzymes. BfiI endonuclease exhibits similarities to the EDTA-resistant nuclease Nuc of Salmonella typhimurium. J. Biol. Chem. 275: 30878–30885 [DOI] [PubMed] [Google Scholar]
- 10. Saravanan M, Bujnicki JM, Cymerman IA, Rao DN, Nagaraja V. 2004. Type II restriction endonuclease R.KpnI is a member of the HNH nuclease superfamily. Nucleic Acids Res. 32: 6129–6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wyszomirski KH, Curth U, Alves J, Mackeldanz P, Moncke-Buchner E, Schutkowski M, Krüger DH, Reuter M. 2012. Type III restriction endonuclease EcoP15I is a heterotrimeric complex containing one Res subunit with several DNA-binding regions and ATPase activity. Nucleic Acids Res. 40: 3610–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gupta YK, Yang L, Chan SH, Samuelson JC, Xu SY, Aggarwal AK. 2 May 2012, posting date. Structural insights into the assembly and shape of type III restriction-modification (R-M) EcoP15I complex by small-angle X-ray scattering. J. Mol. Biol. doi:10.1016/j.jmb.2012.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McCleland SE, Sczcelkun MD. 2004. The type I and III restriction endonucleases: structural elements in molecular motors that process DNA, p 111–135 In Pingoud A. (ed), Restriction endonucleases. Springer, Berlin, Germany [Google Scholar]
- 14. Bourniquel AA, Bickle TA. 2002. Complex restriction enzymes: NTP-driven molecular motors. Biochimie 84: 1047–1059 [DOI] [PubMed] [Google Scholar]
- 15. Stewart FJ, Panne D, Bickle TA, Raleigh EA. 2000. Methyl-specific DNA binding by McrBC, a modification-dependent restriction enzyme. J. Mol. Biol. 298: 611–622 [DOI] [PubMed] [Google Scholar]
- 16. Bair CL, Black LW. 2007. A type IV modification dependent restriction nuclease that targets glucosylated hydroxymethyl cytosine modified DNAs. J. Mol. Biol. 366: 768–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lobner-Olesen A, Skovgaard O, Marinus MG. 2005. Dam methylation: coordinating cellular processes. Curr. Opin. Microbiol. 8: 154–160 [DOI] [PubMed] [Google Scholar]
- 18. Low DA, Weyand NJ, Mahan MJ. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect. Immun. 69: 7197–7204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reisenauer A, Kahng LS, McCollum S, Shapiro L. 1999. Bacterial DNA methylation: a cell cycle regulator? J. Bacteriol. 181: 5135–5139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen BW, Heiter DF, Chan SH, Wang H, Xu SY, Morgan RD, Wilson GG, Stoddard BL. 2010. Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI. Structure 18: 734–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Roberts RJ, Vincze T, Posfai J, Macelis D. 2010. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38: D234–D236 doi:10.1093/nar/gkp874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bertani G, Weigle JJ. 1953. Host controlled variation in bacterial viruses. J. Bacteriol. 65: 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luria SE, Human ML. 1952. A nonhereditary, host-induced variation of bacterial viruses. J. Bacteriol. 64: 557–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bickle TA. 2004. Restricting restriction. Mol. Microbiol. 51: 3–5 [DOI] [PubMed] [Google Scholar]
- 25. Loenen WAM. 2003. Tracking EcoKI and DNA fifty years on: a golden story full of surprises. Nucleic Acids Res. 31: 7059–7069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tock MR, Dryden DTF. 2005. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 8: 466–472 [DOI] [PubMed] [Google Scholar]
- 27. Nechaev S, Severinov K. 2008. The elusive object of desire—interactions of bacteriophages and their hosts. Curr. Opin. Microbiol. 11: 186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Naito T, Kusano K, Kobayashi I. 1995. Selfish behavior of restriction-modification systems. Science 267: 897–899 [DOI] [PubMed] [Google Scholar]
- 29. Stein DC, Gunn JS, Radlinska M, Piekarowicz A. 1995. Restriction and modification systems of Neisseria gonorrhoeae. Gene 157: 19–22 [DOI] [PubMed] [Google Scholar]
- 30. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315: 1709–1712 [DOI] [PubMed] [Google Scholar]
- 31. Horvath P, Romero DA, Coute-Monvoisin AC, Richards M, Deveau H, Moineau S, Boyaval P, Fremaux C, Barrangou R. 2008. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J. Bacteriol. 190: 1401–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. 2005. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 151: 2551–2561 [DOI] [PubMed] [Google Scholar]
- 33. Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321: 960–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dillingham MS, Kowalczykowski SC. 2008. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 72: 642–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stahl MM, Kobayashi I, Stahl FW, Huntington SK. 1983. Activation of Chi, a recombinator, by the action of an endonuclease at a distant site. Proc. Natl. Acad. Sci. U. S. A. 80: 2310–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dillingham MS, Spies M, Kowalczykowski SC. 2003. RecBCD enzyme is a bipolar DNA helicase. Nature 423: 893–897 [DOI] [PubMed] [Google Scholar]
- 37. Handa N, Bianco PR, Baskin RJ, Kowalczykowski SC. 2005. Direct visualization of RecBCD movement reveals cotranslocation of the RecD motor after Chi recognition. Mol. Cell 17: 745–750 [DOI] [PubMed] [Google Scholar]
- 38. Halpern D, Chiapello H, Schbath S, Robin S, Hennequet-Antier C, Gruss A, Karoui ME. 2007. Identification of DNA motifs implicated in maintenance of bacterial core genomes by predictive modeling. PLoS Genet. 3: 1614–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Handa N, Yang L, Dillingham MS, Kobayashi I, Wigley DB, Kowalczykowski SC. 2012. Molecular determinants responsible for recognition of the single-stranded DNA regulatory sequence, χ, by RecBCD enzyme. Proc. Natl. Acad. Sci. U. S. A. 109: 8901–8906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Handa N, Ichige A, Kusano K, Kobayashi I. 2000. Cellular responses to postsegregational killing by restriction-modification genes. J. Bacteriol. 182: 2218–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kobayashi I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29: 3742–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Handa N, Ichige A, Kobayashi I. 2009. Contribution of RecFOR machinery of homologous recombination to cell survival after loss of a restriction-modification gene complex. Microbiology 155: 2320–2332 [DOI] [PubMed] [Google Scholar]
- 43. Snyder L. 1995. Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents? Mol. Microbiol. 15: 415–420 [DOI] [PubMed] [Google Scholar]
- 44. Parma DH, Snyder M, Sobolevski S, Nawroz M, Brody E, Gold L. 1992. The Rex system of bacteriophage lambda: tolerance and altruistic cell death. Genes Dev. 6: 497–510 [DOI] [PubMed] [Google Scholar]
- 45. Gordon BR, Li Y, Wang L, Sintsova A, van Bakel H, Tian S, Navarre WW, Xia B, Liu J. 2010. Lsr2 is a nucleoid-associated protein that targets AT-rich sequences and virulence genes in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 107: 5154–5159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313: 236–238 [DOI] [PubMed] [Google Scholar]
- 47. Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. 2008. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science 320: 935–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van Melderen L. 2010. Toxin-antitoxin systems: why so many, what for? Curr. Opin. Microbiol. 13: 781–785 [DOI] [PubMed] [Google Scholar]
- 49. Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salmond GP. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc. Natl. Acad. Sci. U. S. A. 106: 894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. van Melderen L, Saavedra De Bast M. 2009. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 5: e1000437 doi:10.1371/journal.pgen.1000437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Korona R, Levin BR. 1993. Phage-mediated selection and the evolution and maintenance of restriction-modification. Evolution 47: 556–575 [DOI] [PubMed] [Google Scholar]
- 52. Murray NE. 2002. 2001 Fred Griffith review lecture. Immigration control of DNA in bacteria: self versus non-self. Microbiology 148: 3–20 [DOI] [PubMed] [Google Scholar]
- 53. Warren RA. 1980. Modified bases in bacteriophage DNAs. Annu. Rev. Microbiol. 34: 137–158 [DOI] [PubMed] [Google Scholar]
- 54. Hill C, Miller LA, Klaenhammer TR. 1991. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J. Bacteriol. 173: 4363–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kruger DH, Bickle TA. 1983. Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol. Rev. 47: 345–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Swinton D, Hattman S, Crain PF, Cheng CS, Smith DL, McCloskey JA. 1983. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2′-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc. Natl. Acad. Sci. U. S. A. 80: 7400–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wilson GG, Murray NE. 1991. Restriction and modification systems. Annu. Rev. Genet. 25: 585–627 [DOI] [PubMed] [Google Scholar]
- 58. Kahmann R. 1984. The mom gene of bacteriophage Mu. Curr. Top. Microbiol. Immunol. 108: 29–47 [DOI] [PubMed] [Google Scholar]
- 59. Hattman S. 1999. Unusual transcriptional and translational regulation of the bacteriophage Mu mom operon. Pharmacol. Ther. 84: 367–388 [DOI] [PubMed] [Google Scholar]
- 60. Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DTF. 2002. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell 9: 187–194 [DOI] [PubMed] [Google Scholar]
- 61. Atanasiu C, Byron O, McMiken H, Sturrock SS, Dryden DTF. 2001. Characterisation of the structure of ocr, the gene 0.3 protein of bacteriophage T7. Nucleic Acids Res. 29: 3059–3068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Blackstock JJ, Egelhaaf SU, Atanasiu C, Dryden DTF, Poon WC. 2001. Shape of Ocr, the gene 0.3 protein of bacteriophage T7: modeling based on light scattering experiments. Biochemistry 40: 9944–9949 [DOI] [PubMed] [Google Scholar]
- 63. McMahon SA, Roberts GA, Johnson KA, Cooper LP, Liu H, White JH, Carter LG, Sanghvi B, Oke M, Walkinshaw MD, Blakely GW, Naismith JH, Dryden DTF. 2009. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res. 37: 4887–4897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wilkins BM. 2002. Plasmid promiscuity: meeting the challenge of DNA immigration control. Environ. Microbiol. 4: 495–500 [DOI] [PubMed] [Google Scholar]
- 65. Iida S, Streiff MB, Bickle TA, Arber W. 1987. Two DNA antirestriction systems of bacteriophage P1, darA, and darB: characterization of darA− phages. Virology 157: 156–166 [DOI] [PubMed] [Google Scholar]
- 66. Bandyopadhyay PK, Studier FW, Hamilton DL, Yuan R. 1985. Inhibition of the type I restriction-modification enzymes EcoB and EcoK by the gene 0.3 protein of bacteriophage T7. J. Mol. Biol. 182: 567–578 [DOI] [PubMed] [Google Scholar]
- 67. Studier FW, Movva NR. 1976. SAMase gene of bacteriophage T3 is responsible for overcoming host restriction. J. Virol. 19: 136–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Blaisdell BE, Campbell AM, Karlin S. 1996. Similarities and dissimilarities of phage genomes. Proc. Natl. Acad. Sci. U. S. A. 93: 5854–5859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gelfand MS, Koonin EV. 1997. Avoidance of palindromic words in bacterial and archaeal genomes: a close connection with restriction enzymes. Nucleic Acids Res. 25: 2430–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Karlin S, Mrazek J, Campbell AM. 1997. Compositional biases of bacterial genomes and evolutionary implications. J. Bacteriol. 179: 3899–3913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sharp PM. 1986. Molecular evolution of bacteriophages: evidence of selection against the recognition sites of host restriction enzymes. Mol. Biol. Evol. 3: 75–83 [DOI] [PubMed] [Google Scholar]
- 72. Bilcock DT, Daniels LE, Bath AJ, Halford SE. 1999. Reactions of type II restriction endonucleases with 8-base pair recognition sites. J. Biol. Chem. 274: 36379–36386 [DOI] [PubMed] [Google Scholar]
- 73. Kruger DH, Barcak GJ, Reuter M, Smith HO. 1988. EcoRII can be activated to cleave refractory DNA recognition sites. Nucleic Acids Res. 16: 3997–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tamulaitis G, Sasnauskas G, Mucke M, Siksnys V. 2006. Simultaneous binding of three recognition sites is necessary for a concerted plasmid DNA cleavage by EcoRII restriction endonuclease. J. Mol. Biol. 358: 406–419 [DOI] [PubMed] [Google Scholar]
- 75. Kruger DH, Schroeder C, Reuter M, Bogdarina IG, Buryanov YI, Bickle TA. 1985. DNA methylation of bacterial viruses T3 and T7 by different DNA methylases in Escherichia coli K12 cells. Eur. J. Biochem. 150: 323–330 [DOI] [PubMed] [Google Scholar]
- 76. Meisel A, Bickle TA, Kruger DH, Schroeder C. 1992. Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature 355: 467–469 [DOI] [PubMed] [Google Scholar]
- 77. Schroeder C, Jurkschat H, Meisel A, Reich JG, Kruger D. 1986. Unusual occurrence of EcoP1 and EcoP15 recognition sites and counterselection of type II methylation and restriction sequences in bacteriophage T7 DNA. Gene 45: 77–86 [DOI] [PubMed] [Google Scholar]
- 78. McClelland M, Nelson M, Cantor CR. 1985. Purification of MboII methylase (GAAGmA) from Moraxella bovis: site-specific cleavage of DNA at nine and ten base pair sequences. Nucleic Acids Res. 13: 7171–7182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Levin BR. 1993. The accessory genetic elements of bacteria: existence conditions and (co)evolution. Curr. Opin. Genet. Dev. 3: 849–854 [DOI] [PubMed] [Google Scholar]
- 80. Horiuchi K, Zinder ND. 1975. Site-specific cleavage of single-stranded DNA by a Hemophilus restriction endonuclease. Proc. Natl. Acad. Sci. U. S. A. 72: 2555–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Edgar R, Rokney A, Feeney M, Semsey S, Kessel M, Goldberg MB, Adhya S, Oppenheim AB. 2008. Bacteriophage infection is targeted to cellular poles. Mol. Microbiol. 68: 1107–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Begg KJ, Donachie WD. 1977. Growth of the Escherichia coli cell surface. J. Bacteriol. 129: 1524–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen I, Christie PJ, Dubnau D. 2005. The ins and outs of DNA transfer in bacteria. Science 310: 1456–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rocha EP, Danchin A, Viari A. 2001. Evolutionary role of restriction/modification systems as revealed by comparative genome analysis. Genome Res. 11: 946–958 [DOI] [PubMed] [Google Scholar]
- 85. Seidel R, Bloom JG, Dekker C, Szczelkun MD. 2008. Motor step size and ATP coupling efficiency of the dsDNA translocase EcoR124I. EMBO J. 27: 1388–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kelleher JE, Raleigh EA. 1994. Response to UV damage by four Escherichia coli K-12 restriction systems. J. Bacteriol. 176: 5888–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Roberts GA, Cooper LP, White JH, Su TJ, Zipprich JT, Geary P, Kennedy C, Dryden DTF. 2011. An investigation of the structural requirements for ATP hydrolysis and DNA cleavage by the EcoKI type I DNA restriction and modification enzyme. Nucleic Acids Res. 39: 7667–7676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kobayashi I. 2004. Restriction-modification systems as minimal forms of life, p 19–62 In Pingoud A. (ed), Restriction endonucleases, vol 14. Springer, Berlin, Germany [Google Scholar]
- 89. Handa N, Kobayashi I. 1999. Post-segregational killing by restriction modification gene complexes: observations of individual cell deaths. Biochimie 81: 931–938 [DOI] [PubMed] [Google Scholar]
- 90. Ichige A, Kobayashi I. 2005. Stability of EcoRI restriction-modification enzymes in vivo differentiates the EcoRI restriction-modification system from other postsegregational cell killing systems. J. Bacteriol. 187: 6612–6621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kulakauskas S, Lubys A, Ehrlich SD. 1995. DNA restriction-modification systems mediate plasmid maintenance. J. Bacteriol. 177: 3451–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kusano K, Naito T, Handa N, Kobayashi I. 1995. Restriction-modification systems as genomic parasites in competition for specific sequences. Proc. Natl. Acad. Sci. U. S. A. 92: 11095–11099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chinen A, Naito Y, Handa N, Kobayashi I. 2000. Evolution of sequence recognition by restriction-modification enzymes: selective pressure for specificity decrease. Mol. Biol. Evol. 17: 1610–1619 [DOI] [PubMed] [Google Scholar]
- 94. Nakayama Y, Kobayashi I. 1998. Restriction-modification gene complexes as selfish gene entities: roles of a regulatory system in their establishment, maintenance, and apoptotic mutual exclusion. Proc. Natl. Acad. Sci. U. S. A. 95: 6442–6447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Kobayashi I, Nobusato A, Kobayashi-Takahashi N, Uchiyama I. 1999. Shaping the genome—restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev. 9: 649–656 [DOI] [PubMed] [Google Scholar]
- 96. Fukuda E, Kaminska KH, Bujnicki JM, Kobayashi I. 2008. Cell death upon epigenetic genome methylation: a novel function of methyl-specific deoxyribonucleases. Genome Biol. 9: R163 doi:10.1186/gb-2008-9-11-r163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. O'Neill M, Chen A, Murray NE. 1997. The restriction-modification genes of Escherichia coli K-12 may not be selfish: they do not resist loss and are readily replaced by alleles conferring different specificities. Proc. Natl. Acad. Sci. U. S. A. 94: 14596–14601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kulik EM, Bickle TA. 1996. Regulation of the activity of the type IC EcoR124I restriction enzyme. J. Mol. Biol. 264: 891–906 [DOI] [PubMed] [Google Scholar]
- 99. Furuta Y, Abe K, Kobayashi I. 2010. Genome comparison and context analysis reveals putative mobile forms of restriction-modification systems and related rearrangements. Nucleic Acids Res. 38: 2428–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kobayashi I. 2004. Genetic addiction—a principle in symbiosis of genes in a genome, p 105–144 In Phillips G, Funnell B. (ed), Plasmid biology. ASM Press, Washington, DC [Google Scholar]
- 101. Mochizuki A, Yahara K, Kobayashi I, Iwasa Y. 2006. Genetic addiction: selfish gene's strategy for symbiosis in the genome. Genetics 172: 1309–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Handa N, Nakayama Y, Sadykov M, Kobayashi I. 2001. Experimental genome evolution: large-scale genome rearrangements associated with resistance to replacement of a chromosomal restriction-modification gene complex. Mol. Microbiol. 40: 932–940 [DOI] [PubMed] [Google Scholar]
- 103. Sadykov M, Asami Y, Niki H, Handa N, Itaya M, Tanokura M, Kobayashi I. 2003. Multiplication of a restriction-modification gene complex. Mol. Microbiol. 48: 417–427 [DOI] [PubMed] [Google Scholar]
- 104. Szekeres S, Dauti M, Wilde C, Mazel D, Rowe-Magnus DA. 2007. Chromosomal toxin-antitoxin loci can diminish large-scale genome reductions in the absence of selection. Mol. Microbiol. 63: 1588–1605 [DOI] [PubMed] [Google Scholar]
- 105. Xia Y, Burbank DE, van Etten JL. 1986. Restriction endonuclease activity induced by NC-1A virus infection of a Chlorella-like green alga. Nucleic Acids Res. 14: 6017–6030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. van Etten JL, Xia YN, Burbank DE, Narva KE. 1988. Chlorella viruses code for restriction and modification enzymes. Gene 74: 113–115 [DOI] [PubMed] [Google Scholar]
- 107. van Etten JL. 2003. Unusual life style of giant chlorella viruses. Annu. Rev. Genet. 37: 153–195 [DOI] [PubMed] [Google Scholar]
- 108. Boyer M, Yutin N, Pagnier I, Barrassi L, Fournous G, Espinosa L, Robert C, Azza S, Sun S, Rossmann MG, Suzan-Monti M, La Scola B, Koonin EV, Raoult D. 2009. Giant Marseillevirus highlights the role of amoebae as a melting pot in emergence of chimeric microorganisms. Proc. Natl. Acad. Sci. U. S. A. 106: 21848–21853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sibley MH, Raleigh EA. 2004. Cassette-like variation of restriction enzyme genes in Escherichia coli C and relatives. Nucleic Acids Res. 32: 522–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Koonin EV, Makarova KS, Aravind L. 2001. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 55: 709–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Lerat E, Daubin V, Ochman H, Moran NA. 2005. Evolutionary origins of genomic repertoires in bacteria. PLoS Biol. 3: e130 doi:10.1371/journal.pbio.0030130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Nakamura Y, Itoh T, Matsuda H, Gojobori T. 2004. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat. Genet. 36: 760–766 [DOI] [PubMed] [Google Scholar]
- 113. Sorek R, Zhu Y, Creevey CJ, Francino MP, Bork P, Rubin EM. 2007. Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318: 1449–1452 [DOI] [PubMed] [Google Scholar]
- 114. Lindsay JA. 2010. Genomic variation and evolution of Staphylococcus aureus. Int. J. Med. Microbiol. 300: 98–103 [DOI] [PubMed] [Google Scholar]
- 115. Corvaglia AR, Francois P, Hernandez D, Perron K, Linder P, Schrenzel J. 2010. A type III-like restriction endonuclease functions as a major barrier to horizontal gene transfer in clinical Staphylococcus aureus strains. Proc. Natl. Acad. Sci. U. S. A. 107: 11954–11958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Xu SY, Corvaglia AR, Chan SH, Zheng Y, Linder P. 2011. A type IV modification-dependent restriction enzyme SauUSI from Staphylococcus aureus subsp. aureus USA300. Nucleic Acids Res. 39: 5597–5610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Budroni S, Siena E, Dunning Hotopp JC, Seib KL, Serruto D, Nofroni C, Comanducci M, Riley DR, Daugherty SC, Angiuoli SV, Covacci A, Pizza M, Rappuoli R, Moxon ER, Tettelin H, Medini D. 2011. Neisseria meningitidis is structured in clades associated with restriction modification systems that modulate homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 108: 4494–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Arber W. 2000. Genetic variation: molecular mechanisms and impact on microbial evolution. FEMS Microbiol. Rev. 24: 1–7 [DOI] [PubMed] [Google Scholar]
- 119. Chang S, Cohen SN. 1977. In vivo site-specific genetic recombination promoted by the EcoRI restriction endonuclease. Proc. Natl. Acad. Sci. U. S. A. 74: 4811–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. King G, Murray NE. 1994. Restriction enzymes in cells, not Eppendorfs. Trends Microbiol. 2: 465–469 [DOI] [PubMed] [Google Scholar]
- 121. Kobayashi I. 1998. Selfishness and death: raison d'etre of restriction, recombination and mitochondria. Trends Genet. 14: 368–374 [DOI] [PubMed] [Google Scholar]
- 122. McKane M, Milkman R. 1995. Transduction, restriction and recombination patterns in Escherichia coli. Genetics 139: 35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Milkman R, Raleigh EA, McKane M, Cryderman D, Bilodeau P, McWeeny K. 1999. Molecular evolution of the Escherichia coli chromosome. V. Recombination patterns among strains of diverse origin. Genetics 153: 539–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Radding CM. 1973. Molecular mechanisms in genetic recombination. Annu. Rev. Genet. 7: 87–111 [DOI] [PubMed] [Google Scholar]
- 125. Price C, Bickle TA. 1986. A possible role for DNA restriction in bacterial evolution. Microbiol. Sci. 3: 296–299 [PubMed] [Google Scholar]
- 126. Jeltsch A. 2003. Maintenance of species identity and controlling speciation of bacteria: a new function for restriction/modification systems? Gene 317: 13–16 [DOI] [PubMed] [Google Scholar]
- 127. Kusano K, Sakagami K, Yokochi T, Naito T, Tokinaga Y, Ueda E, Kobayashi I. 1997. A new type of illegitimate recombination is dependent on restriction and homologous interaction. J. Bacteriol. 179: 5380–5390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Asakura Y, Kojima H, Kobayashi I. 2011. Evolutionary genome engineering using a restriction-modification system. Nucleic Acids Res. 39: 9034–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Chinen A, Uchiyama I, Kobayashi I. 2000. Comparison between Pyrococcus horikoshii and Pyrococcus abyssi genome sequences reveals linkage of restriction-modification genes with large genome polymorphisms. Gene 259: 109–121 [DOI] [PubMed] [Google Scholar]
- 130. Furuta Y, Kawai M, Yahara K, Takahashi N, Handa N, Tsuru T, Oshima K, Yoshida M, Azuma T, Hattori M, Uchiyama I, Kobayashi I. 2011. Birth and death of genes linked to chromosomal inversion. Proc. Natl. Acad. Sci. U. S. A. 108: 1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Furuta Y, Kobayashi I. 2011. Restriction-modification systems as mobile epigenetic elements. In Roberts AP, Mullany P. (ed), Madame Curie bioscience database. Landes Bioscience, Austin, TX: http://www.ncbi.nlm.nih.gov/books/NBK63963 [Google Scholar]
- 132. Nobusato A, Uchiyama I, Ohashi S, Kobayashi I. 2000. Insertion with long target duplication: a mechanism for gene mobility suggested from comparison of two related bacterial genomes. Gene 259: 99–108 [DOI] [PubMed] [Google Scholar]
- 133. Arber W. 2009. Genetically encoded generators of genetic variants. J. Proteomics 72: 836–837 [DOI] [PubMed] [Google Scholar]
- 134. Arber W. 1991. Elements in microbial evolution. J. Mol. Evol. 33: 4–12 [DOI] [PubMed] [Google Scholar]
- 135. Forterre P. 2002. The origin of DNA genomes and DNA replication proteins. Curr. Opin. Microbiol. 5: 525–532 [DOI] [PubMed] [Google Scholar]
- 136. Hitzeman RA, Price AR. 1978. Relationship of Bacillus subtilis DNA polymerase III to bacteriophage PBS2-induced DNA polymerase and to the replication of uracil-containing DNA. J. Virol. 28: 697–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. He X, Ou HY, Yu Q, Zhou X, Wu J, Liang J, Zhang W, Rajakumar K, Deng Z. 2007. Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Mol. Microbiol. 65: 1034–1048 [DOI] [PubMed] [Google Scholar]
- 138. Xu T, Yao F, Zhou X, Deng Z, You D. 2010. A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res. 38: 7133–7141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Khersonsky O, Roodveldt C, Tawfik DS. 2006. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 10: 498–508 [DOI] [PubMed] [Google Scholar]
- 140. Khersonsky O, Tawfik DS. 2010. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79: 471–505 [DOI] [PubMed] [Google Scholar]
- 141. Wei H, Therrien C, Blanchard A, Guan S, Zhu Z. 2008. The fidelity index provides a systematic quantitation of star activity of DNA restriction endonucleases. Nucleic Acids Res. 36: e50 doi:10.1093/nar/gkn182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Langhans MT, Palladino MJ. 2009. Cleavage of mispaired heteroduplex DNA substrates by numerous restriction enzymes. Curr. Issues Mol. Biol. 11: 1–12 [PMC free article] [PubMed] [Google Scholar]
- 143. Saravanan M, Vasu K, Kanakaraj R, Rao DN, Nagaraja V. 2007. R.KpnI, an HNH superfamily REase, exhibits differential discrimination at non-canonical sequences in the presence of Ca2+ and Mg2+. Nucleic Acids Res. 35: 2777–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Saravanan M, Vasu K, Nagaraja V. 2008. Evolution of sequence specificity in a restriction endonuclease by a point mutation. Proc. Natl. Acad. Sci. U. S. A. 105: 10344–10347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Vasu K, Nagamalleswari E, Nagaraja V. 2012. Promiscuous restriction is a cellular defense strategy that confers fitness advantage to bacteria. Proc. Natl. Acad. Sci. U. S. A. 109: E1287–E1293 doi:10.1073/pnas.1119226109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Murray IA, Stickel SK, Roberts RJ. 2010. Sequence-specific cleavage of RNA by type II restriction enzymes. Nucleic Acids Res. 38: 8257–8268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. von Hippel PH. 1998. An integrated model of the transcription complex in elongation, termination, and editing. Science 281: 660–665 [DOI] [PubMed] [Google Scholar]
- 148. Masukata H, Tomizawa J. 1990. A mechanism of formation of a persistent hybrid between elongating RNA and template DNA. Cell 62: 331–338 [DOI] [PubMed] [Google Scholar]
- 149. Chandrashekaran S, Saravanan M, Radha DR, Nagaraja V. 2004. Ca(2+)-mediated site-specific DNA cleavage and suppression of promiscuous activity of KpnI restriction endonuclease. J. Biol. Chem. 279: 49736–49740 [DOI] [PubMed] [Google Scholar]
- 150. Saravanan M, Vasu K, Ghosh S, Nagaraja V. 2007. Dual role for Zn2+ in maintaining structural integrity and inducing DNA sequence specificity in a promiscuous endonuclease. J. Biol. Chem. 282: 32320–32326 [DOI] [PubMed] [Google Scholar]
- 151. Friedhoff P, Kolmes B, Gimadutdinow O, Wende W, Krause KL, Pingoud A. 1996. Analysis of the mechanism of the Serratia nuclease using site-directed mutagenesis. Nucleic Acids Res. 24: 2632–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Keeble AH, Hemmings AM, James R, Moore GR, Kleanthous C. 2002. Multistep binding of transition metals to the H-N-H endonuclease toxin colicin E9. Biochemistry 41: 10234–10244 [DOI] [PubMed] [Google Scholar]
- 153. Pommer AJ, Wallis R, Moore GR, James R, Kleanthous C. 1998. Enzymological characterization of the nuclease domain from the bacterial toxin colicin E9 from Escherichia coli. Biochem. J. 334: 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Lindahl T. 1993. Instability and decay of the primary structure of DNA. Nature 362: 709–715 [DOI] [PubMed] [Google Scholar]
- 155. Shen JC, Rideout WM, Jones PA. 1992. High frequency mutagenesis by a DNA methyltransferase. Cell 71: 1073–1080 [DOI] [PubMed] [Google Scholar]
- 156. Ehrlich M, Gama-Sosa MA, Carreira LH, Ljungdahl LG, Kuo KC, Gehrke CW. 1985. DNA methylation in thermophilic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res. 13: 1399–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Bhagwat AS, McClelland M. 1992. DNA mismatch correction by very short patch repair may have altered the abundance of oligonucleotides in the E. coli genome. Nucleic Acids Res. 20: 1663–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Glasner W, Merkl R, Schellenberger V, Fritz HJ. 1995. Substrate preferences of Vsr DNA mismatch endonuclease and their consequences for the evolution of the Escherichia coli K-12 genome. J. Mol. Biol. 245: 1–7 [DOI] [PubMed] [Google Scholar]
- 159. Kwiatek A, Luczkiewicz M, Bandyra K, Stein DC, Piekarowicz A. 2010. Neisseria gonorrhoeae FA1090 carries genes encoding two classes of Vsr endonucleases. J. Bacteriol. 192: 3951–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Cohen HM, Tawfik DS, Griffiths AD. 2002. Promiscuous methylation of non-canonical DNA sites by HaeIII methyltransferase. Nucleic Acids Res. 30: 3880–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Macintyre G, Atwood CV, Cupples CG. 2001. Lowering S-adenosylmethionine levels in Escherichia coli modulates C-to-T transition mutations. J. Bacteriol. 183: 921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Wion D, Casadesus J. 2006. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 4: 183–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Low DA, Casadesús J. 2008. Clocks and switches: bacterial gene regulation by DNA adenine methylation. Curr. Opin. Microbiol. 11: 106–112 [DOI] [PubMed] [Google Scholar]
- 164. Collier J. 2009. Epigenetic regulation of the bacterial cell cycle. Curr. Opin. Microbiol. 12: 722–729 [DOI] [PubMed] [Google Scholar]
- 165. Bheemanaik S, Bujnicki JM, Nagaraja V, Rao DN. 2006. Functional analysis of amino acid residues at the dimerisation interface of KpnI DNA methyltransferase. Biol. Chem. 387: 515–523 [DOI] [PubMed] [Google Scholar]
- 166. Bheemanaik S, Chandrashekaran S, Nagaraja V, Rao DN. 2003. Kinetic and catalytic properties of dimeric KpnI DNA methyltransferase. J. Biol. Chem. 278: 7863–7874 [DOI] [PubMed] [Google Scholar]
- 167. Malygin EG, Evdokimov AA, Hattman S. 2009. Dimeric/oligomeric DNA methyltransferases: an unfinished story. Biol. Chem. 390: 835–844 [DOI] [PubMed] [Google Scholar]
- 168. Li E, Bestor TH, Jaenisch R. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- 169. Srikhanta YN, Maguire TL, Stacey KJ, Grimmond SM, Jennings MP. 2005. The phasevarion: a genetic system controlling coordinated, random switching of expression of multiple genes. Proc. Natl. Acad. Sci. U. S. A. 102: 5547–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Bujnicki JM, Radlinska M. 1999. Molecular evolution of DNA-(cytosine-N4) methyltransferases: evidence for their polyphyletic origin. Nucleic Acids Res. 27: 4501–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Ishikawa K, Fukuda E, Kobayashi I. 2010. Conflicts targeting epigenetic systems and their resolution by cell death: novel concepts for methyl-specific and other restriction systems. DNA Res. 17: 325–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Ishikawa K, Handa N, Kobayashi I. 2009. Cleavage of a model DNA replication fork by a type I restriction endonuclease. Nucleic Acids Res. 37: 3531–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Hallet B. 2001. Playing Dr Jekyll and Mr Hyde: combined mechanisms of phase variation in bacteria. Curr. Opin. Microbiol. 4: 570–581 [DOI] [PubMed] [Google Scholar]
- 174. van der Woude MW, Baumler AJ. 2004. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 17: 581–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Weiser JN, Williams A, Moxon ER. 1990. Phase-variable lipopolysaccharide structures enhance the invasive capacity of Haemophilus influenzae. Infect. Immun. 58: 3455–3457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Cerdeño-Tárraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, Barron A, Clark L, Corton C, Doggett J, Holden MT, Larke N, Line A, Lord A, Norbertczak H, Ormond D, Price C, Rabbinowitsch E, Woodward J, Barrell B, Parkhill J. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307: 1463–1465 [DOI] [PubMed] [Google Scholar]
- 177. De Bolle X, Bayliss CD, Field D, van de Ven T, Saunders NJ, Hood DW, Moxon ER. 2000. The length of a tetranucleotide repeat tract in Haemophilus influenzae determines the phase variation rate of a gene with homology to type III DNA methyltransferases. Mol. Microbiol. 35: 211–222 [DOI] [PubMed] [Google Scholar]
- 178. de Vries N, Duinsbergen D, Kuipers EJ, Pot RG, Wiesenekker P, Penn CW, van Vliet AH, Vandenbroucke-Grauls CM, Kusters JG. 2002. Transcriptional phase variation of a type III restriction-modification system in Helicobacter pylori. J. Bacteriol. 184: 6615–6623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Dybvig K, Sitaraman R, French CT. 1998. A family of phase-variable restriction enzymes with differing specificities generated by high-frequency gene rearrangements. Proc. Natl. Acad. Sci. U. S. A. 95: 13923–13928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Fox KL, Dowideit SJ, Erwin AL, Srikhanta YN, Smith AL, Jennings MP. 2007. Haemophilus influenzae phasevarions have evolved from type III DNA restriction systems into epigenetic regulators of gene expression. Nucleic Acids Res. 35: 5242–5252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Fox KL, Srikhanta YN, Jennings MP. 2007. Phase variable type III restriction-modification systems of host-adapted bacterial pathogens. Mol. Microbiol. 65: 1375–1379 [DOI] [PubMed] [Google Scholar]
- 182. Brussow H, Canchaya C, Hardt WD. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68: 560–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Waldor MK. 1998. Bacteriophage biology and bacterial virulence. Trends Microbiol. 6: 295–297 [DOI] [PubMed] [Google Scholar]
- 184. Waldor MK, Mekalanos JJ. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272: 1910–1914 [DOI] [PubMed] [Google Scholar]
- 185. van der Woude MW. 2006. Re-examining the role and random nature of phase variation. FEMS Microbiol. Lett. 254: 190–197 [DOI] [PubMed] [Google Scholar]
- 186. Bayliss CD, Callaghan MJ, Moxon ER. 2006. High allelic diversity in the methyltransferase gene of a phase variable type III restriction-modification system has implications for the fitness of Haemophilus influenzae. Nucleic Acids Res. 34: 4046–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Kersulyte D, Chalkauskas H, Berg DE. 1999. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol. Microbiol. 31: 31–43 [DOI] [PubMed] [Google Scholar]
- 188. Stewart GJ, Carlson CA. 1986. The biology of natural transformation. Annu. Rev. Microbiol. 40: 211–235 [DOI] [PubMed] [Google Scholar]
- 189. Karyagina A, Shilov I, Tashlitskii V, Khodoun M, Vasil'ev S, Lau PC, Nikolskaya I. 1997. Specific binding of Sso II DNA methyltransferase to its promoter region provides the regulation of sso II restriction-modification gene expression. Nucleic Acids Res. 25: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Som S, Friedman S. 1994. Regulation of EcoRII methyltransferase: effect of mutations on gene expression and in vitro binding to the promoter region. Nucleic Acids Res. 22: 5347–5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Jeltsch A, Pingoud A. 1996. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J. Mol. Evol. 42: 91–96 [DOI] [PubMed] [Google Scholar]
- 192. Lauster R. 1989. Evolution of type II DNA methyltransferases. A gene duplication model. J. Mol. Biol. 206: 313–321 [DOI] [PubMed] [Google Scholar]
- 193. Kita K, Tsuda J, Kato T, Okamoto K, Yanase H, Tanaka M. 1999. Evidence of horizontal transfer of the EcoO109I restriction-modification gene to Escherichia coli chromosomal DNA. J. Bacteriol. 181: 6822–6827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Kusters JG, van Vliet AH, Kuipers EJ. 2006. Pathogenesis of Helicobacter pylori infection. Clin. Microbiol. Rev. 19: 449–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Kong H, Lin LF, Porter N, Stickel S, Byrd D, Posfai J, Roberts RJ. 2000. Functional analysis of putative restriction-modification system genes in the Helicobacter pylori J99 genome. Nucleic Acids Res. 28: 3216–3223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Nobusato A, Uchiyama I, Kobayashi I. 2000. Diversity of restriction-modification gene homologues in Helicobacter pylori. Gene 259: 89–98 [DOI] [PubMed] [Google Scholar]
- 197. Xu Q, Morgan RD, Roberts RJ, Blaser MJ. 2000. Identification of type II restriction and modification systems in Helicobacter pylori reveals their substantial diversity among strains. Proc. Natl. Acad. Sci. U. S. A. 97: 9671–9676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Lin LF, Posfai J, Roberts RJ, Kong H. 2001. Comparative genomics of the restriction-modification systems in Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 98: 2740–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Kumar R, Mukhopadhyay AK, Rao DN. 2010. Characterization of an N6 adenine methyltransferase from Helicobacter pylori strain 26695 which methylates adjacent adenines on the same strand. FEBS J. 277: 1666–1683 [DOI] [PubMed] [Google Scholar]
- 200. Ando T, Xu Q, Torres M, Kusugami K, Israel DA, Blaser MJ. 2000. Restriction-modification system differences in Helicobacter pylori are a barrier to interstrain plasmid transfer. Mol. Microbiol. 37: 1052–1065 [DOI] [PubMed] [Google Scholar]
- 201. Tsuda M, Karita M, Nakazawa T. 1993. Genetic transformation in Helicobacter pylori. Microbiol. Immunol. 37: 85–89 [DOI] [PubMed] [Google Scholar]
- 202. Israel DA, Lou AS, Blaser MJ. 2000. Characteristics of Helicobacter pylori natural transformation. FEMS Microbiol. Lett. 186: 275–280 [DOI] [PubMed] [Google Scholar]
- 203. Lin EA, Zhang XS, Levine SM, Gill SR, Falush D, Blaser MJ. 2009. Natural transformation of Helicobacter pylori involves the integration of short DNA fragments interrupted by gaps of variable size. PLoS Pathog. 5: e1000337 doi:10.1371/journal.ppat.1000337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Gressmann H, Linz B, Ghai R, Pleissner KP, Schlapbach R, Yamaoka Y, Kraft C, Suerbaum S, Meyer TF, Achtman M. 2005. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 1: e43 doi:10.1371/journal.pgen.0010043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Oh JD, Kling-Backhed H, Giannakis M, Xu J, Fulton RS, Fulton LA, Cordum HS, Wang C, Elliott G, Edwards J, Mardis ER, Engstrand LG, Gordon JI. 2006. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc. Natl. Acad. Sci. U. S. A. 103: 9999–10004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Baldwin DN, Shepherd B, Kraemer P, Hall MK, Sycuro LK, Pinto-Santini DM, Salama NR. 2007. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect. Immun. 75: 1005–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Lin TL, Shun CT, Chang KC, Wang JT. 2004. Isolation and characterization of a HpyC1I restriction-modification system in Helicobacter pylori. J. Biol. Chem. 279: 11156–11162 [DOI] [PubMed] [Google Scholar]
- 208. Xu Q, Morgan RD, Roberts RJ, Xu SY, van Doorn LJ, Donahue JP, Miller GG, Blaser MJ. 2002. Functional analysis of iceA1, a CATG-recognizing restriction endonuclease gene in Helicobacter pylori. Nucleic Acids Res. 30: 3839–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209. Peek RM, Thompson SA, Donahue JP, Tham KT, Atherton JC, Blaser MJ, Miller GG. 1998. Adherence to gastric epithelial cells induces expression of a Helicobacter pylori gene, iceA, that is associated with clinical outcome. Proc. Assoc. Am. Physicians 110: 531–544 [PubMed] [Google Scholar]
- 210. Donahue JP, Israel DA, Torres VJ, Necheva AS, Miller GG. 2002. Inactivation of a Helicobacter pylori DNA methyltransferase alters dnaK operon expression following host-cell adherence. FEMS Microbiol. Lett. 208: 295–301 [DOI] [PubMed] [Google Scholar]
- 211. Humbert O, Salama NR. 2008. The Helicobacter pylori HpyAXII restriction-modification system limits exogenous DNA uptake by targeting GTAC sites but shows asymmetric conservation of the DNA methyltransferase and restriction endonuclease components. Nucleic Acids Res. 36: 6893–6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Skoglund A, Bjorkholm B, Nilsson C, Andersson AF, Jernberg C, Schirwitz K, Enroth C, Krabbe M, Engstrand L. 2007. Functional analysis of the M.HpyAIV DNA methyltransferase of Helicobacter pylori. J. Bacteriol. 189: 8914–8921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Banerjee A, Rao DN. 2011. Functional analysis of an acid adaptive DNA adenine methyltransferase from Helicobacter pylori 26695. PLoS One 6: e16810 doi:10.1371/journal.pone.0016810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Srikhanta YN, Gorrell RJ, Steen JA, Gawthorne JA, Kwok T, Grimmond SM, Robins-Browne RM, Jennings MP. 2011. Phasevarion mediated epigenetic gene regulation in Helicobacter pylori. PLoS One 6: e27569 doi:10.1371/journal.pone.0027569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Friedrich T, Fatemi M, Gowhar H, Leismann O, Jeltsch A. 2000. Specificity of DNA binding and methylation by the M.FokI DNA methyltransferase. Biochim. Biophys. Acta 1480: 145–159 [DOI] [PubMed] [Google Scholar]
- 216. Jeltsch A, Christ F, Fatemi M, Roth M. 1999. On the substrate specificity of DNA methyltransferases. Adenine-N6 DNA methyltransferases also modify cytosine residues at position N4. J. Biol. Chem. 274: 19538–19544 [DOI] [PubMed] [Google Scholar]
- 217. Woodbury CP, Downey RL, von Hippel PH. 1980. DNA site recognition and overmethylation by the EcoRI methylase. J. Biol. Chem. 255: 11526–11533 [PubMed] [Google Scholar]
- 218. Clark TA, Murray IA, Morgan RD, Kislyuk AO, Spittle KE, Boitano M, Fomenkov A, Roberts RJ, Korlach J. 2012. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res. 40: e29 doi:10.1093/nar/gkr1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Aertsen A, Michiels CW. 2005. Mrr instigates the SOS response after high pressure stress in Escherichia coli. Mol. Microbiol. 58: 1381–1391 [DOI] [PubMed] [Google Scholar]
- 220. Aertsen A, Van Houdt R, Vanoirbeek K, Michiels CW. 2004. An SOS response induced by high pressure in Escherichia coli. J. Bacteriol. 186: 6133–6141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Aertsen A, Mebrhatu MT, Michiels CW. 2008. Activation of the Salmonella typhimurium Mrr protein. Biochem. Biophys. Res. Commun. 367: 435–439 [DOI] [PubMed] [Google Scholar]
- 222. Lewis K. 2000. Programmed death in bacteria. Microbiol. Mol. Biol. Rev. 64: 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Djordjevic GM, Klaenhammer TR. 1997. Bacteriophage-triggered defense systems: phage adaptation and design improvements. Appl. Environ. Microbiol. 63: 4370–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Djordjevic GM, O'Sullivan DJ, Walker SA, Conkling MA, Klaenhammer TR. 1997. A triggered-suicide system designed as a defense against bacteriophages. J. Bacteriol. 179: 6741–6748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Fukuyo M, Sasaki A, Kobayashi I. 2012. Success of a suicidal defense strategy against infection in a structured habitat. Sci. Rep. 2: 238 doi:10.1038/srep00238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. 2010. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 18: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Solomon JM, Grossman AD. 1996. Who's competent and when: regulation of natural genetic competence in bacteria. Trends Genet. 12: 150–155 [DOI] [PubMed] [Google Scholar]
- 228. Finkel SE, Kolter R. 2001. DNA as a nutrient: novel role for bacterial competence gene homologs. J. Bacteriol. 183: 6288–6293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Montague M, Barnes C, Smith HO, Chuang RY, Vashee S. 2009. The evolution of RecD outside of the RecBCD complex. J. Mol. Evol. 69: 360–371 [DOI] [PubMed] [Google Scholar]
- 230. Rocha EP, Cornet E, Michel B. 2005. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 1: e15 doi:10.1371/journal.pgen.0010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.