Abstract
SUMMARY
Twenty-five years have passed since the discovery of cyclic dimeric (3′→5′) GMP (cyclic di-GMP or c-di-GMP). From the relative obscurity of an allosteric activator of a bacterial cellulose synthase, c-di-GMP has emerged as one of the most common and important bacterial second messengers. Cyclic di-GMP has been shown to regulate biofilm formation, motility, virulence, the cell cycle, differentiation, and other processes. Most c-di-GMP-dependent signaling pathways control the ability of bacteria to interact with abiotic surfaces or with other bacterial and eukaryotic cells. Cyclic di-GMP plays key roles in lifestyle changes of many bacteria, including transition from the motile to the sessile state, which aids in the establishment of multicellular biofilm communities, and from the virulent state in acute infections to the less virulent but more resilient state characteristic of chronic infectious diseases. From a practical standpoint, modulating c-di-GMP signaling pathways in bacteria could represent a new way of controlling formation and dispersal of biofilms in medical and industrial settings. Cyclic di-GMP participates in interkingdom signaling. It is recognized by mammalian immune systems as a uniquely bacterial molecule and therefore is considered a promising vaccine adjuvant. The purpose of this review is not to overview the whole body of data in the burgeoning field of c-di-GMP-dependent signaling. Instead, we provide a historic perspective on the development of the field, emphasize common trends, and illustrate them with the best available examples. We also identify unresolved questions and highlight new directions in c-di-GMP research that will give us a deeper understanding of this truly universal bacterial second messenger.
INTRODUCTION
This review discusses the current status of research on cyclic dimeric (3′→5′) GMP (cyclic di-GMP or c-di-GMP) (Fig. 1), a small molecule that was first described in 1987 as an allosteric activator of a bacterial cellulose synthase (1). During the past 25 years, c-di-GMP has been implicated in a growing number of cellular functions, including regulation of the cell cycle, differentiation, biofilm formation and dispersion, motility, virulence, and other processes (2–7). With enzymes of c-di-GMP synthesis and degradation identified in all major bacterial phyla, it is now recognized as a universal bacterial second messenger (Table 1).
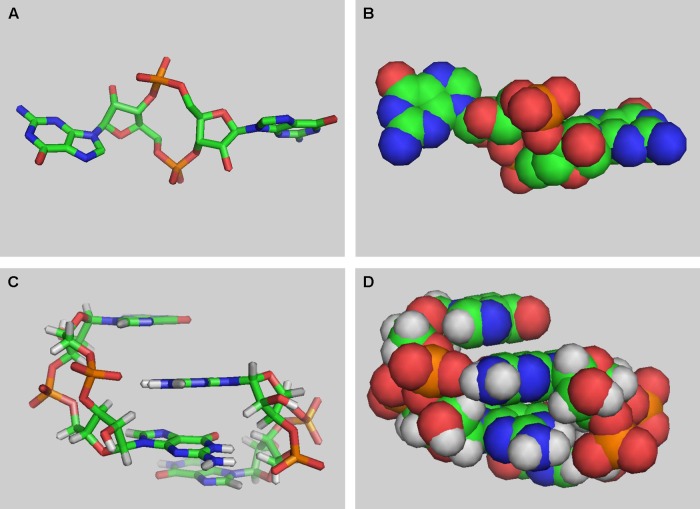
Fig 1.
Three-dimensional structures of cyclic di-GMP. Carbon atoms are shown in green, nitrogen in blue, oxygen in red, and phosphorus in orange. (A and B) Cyclic di-GMP monomer (from Protein Data Bank [PDB] entry 3N3T). This form is usually seen bound to the EAL domain, e.g., in PDB entries 3GG1, 3N3T, 2W27, and 3HV8 (63–65, 85). Note the characteristic 12-member ribose-phosphate ring in the center of the molecule. (C and D) Cyclic di-GMP dimer (from PDB entry 2L74). This form has been seen bound to the allosteric site of PleD (PDB entry 1W25), PilZ domains (PDB entries 2L74 and 3KYF), the transcriptional regulator VpsT (PDB entry 3KLO), and a riboswitch (PDB entry 3MUT) (36, 75, 82–84).
Table 1.
Phylogenetic distribution of GGDEF, EAL, and HD-GYP domains
| Bacterial phyluma | No. of proteins |
% of total | ||||
|---|---|---|---|---|---|---|
| Totalb | GGDEFc | EALc | GGDEF-EAL | HD-GYP | ||
| Well-sampled phyla | ||||||
| Acidobacteria (7) | 27,342 | 67 | 18 | 17 | 20 | 0.45 |
| Actinobacteria (177) | 564,041 | 430 | 105 | 377 | 51 | 0.17 |
| Aquificae (10) | 15,127 | 59 | 26 | 47 | 9 | 0.93 |
| Bacteroidetes (69) | 190,793 | 31 | 6 | 1 | 0 | 0.02 |
| Chlamydiae (38) | 23,262 | 1 | 0 | 0 | 0 | 0.00 |
| Chlorobi (11) | 23,163 | 19 | 0 | 0 | 7 | 0.11 |
| Chloroflexi (15) | 43,101 | 100 | 4 | 26 | 55 | 0.43 |
| Cyanobacteria (42) | 129,836 | 193 | 30 | 173 | 33 | 0.33 |
| Deferribacteres (5) | 9,699 | 71 | 8 | 17 | 19 | 1.19 |
| Deinococcus-Thermus (16) | 35,779 | 155 | 4 | 62 | 69 | 0.81 |
| Firmicutes (437) | 838,221 | 1,213 | 290 | 560 | 734 | 0.33 |
| Fusobacteria (5) | 12,723 | 17 | 4 | 8 | 8 | 0.29 |
| Planctomycetes (5) | 24,772 | 35 | 5 | 2 | 22 | 0.26 |
| Proteobacteria (794) | 2,283,662 | 7,029 | 2,461 | 4,867 | 1,453 | 0.69 |
| Spirochaetes (40) | 76,276 | 164 | 50 | 41 | 112 | 0.48 |
| Tenericutes (37) | 26,877 | 13 | 2 | 3 | 0 | 0.06 |
| Thermotogae (12) | 21,587 | 127 | 1 | 4 | 99 | 1.07 |
| Poorly sampled phyla | ||||||
| Chrysiogenetes (1) | 2,571 | 14 | 5 | 5 | 12 | 1.40 |
| Dictyoglomi (2) | 3,514 | 18 | 0 | 0 | 17 | 1.00 |
| Elusimicrobia (2) | 2,280 | 2 | 0 | 0 | 2 | 0.18 |
| Fibrobacteres (1) | 3,059 | 23 | 1 | 4 | 9 | 1.21 |
| Gemmatimonadetes (1) | 3,891 | 8 | 2 | 5 | 7 | 0.57 |
| Nitrospirae (2) | 6,330 | 11 | 3 | 1 | 14 | 0.46 |
| Synergistetes (3) | 5,489 | 25 | 0 | 0 | 22 | 0.86 |
| Thermodesulfobacteria (2) | 3,791 | 14 | 0 | 5 | 4 | 0.61 |
| Verrucomicrobia (4) | 12,206 | 2 | 0 | 0 | 1 | 0.02 |
The numbers in parentheses show the numbers of completely sequenced genomes from the respective phyla as of 1 January 2012. An updated version of this table with protein counts for representative genomes of 1,116 bacterial and archaeal species is available at http://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html.
According to the NCBI Reference Sequences (RefSeq) database (8).
Excluding proteins that contain both GGDEF and EAL domains.
Several researchers, including us, a few years ago proclaimed the dawning of the new signal transduction system (2, 3, 5). We can now confidently say that the dawning stage has ended and that c-di-GMP-related research is now in full swing. In the past several years, studies of c-di-GMP functions and mechanisms of action have been progressing at an ever-increasing pace, culminating in a number of thoughtful reviews (4, 7, 9–16) and a recently published comprehensive book that covered the entire field (17). What, then, is the purpose of yet another review?
We feel that there remains a need for a source of information on c-di-GMP that is comprehensive yet concise, not limited to a particular aspect of the c-di-GMP signaling field or only to recent advances in the field. In this review, we provide a historic perspective that will likely prove useful for numerous newcomers to this burgeoning field, discuss common trends, identify unique features of the c-di-GMP-mediated signaling systems in various organisms, and highlight the most exciting recent developments. We also emphasize the remaining questions and attempt to identify emerging directions in c-di-GMP research. The field of c-di-GMP signaling has grown so large and is developing so fast that an overview encompassing the whole body of data on c-di-GMP is no longer feasible. Our goal is therefore to organize the best available examples of experimental data into a set of common themes and concepts.
HISTORICAL PERSPECTIVE
As is true for most important scientific discoveries, the discovery of c-di-GMP was serendipitous, and the importance of its discovery was underappreciated for quite some time. Cyclic-di-GMP was originally identified by Moshe Benziman and colleagues at The Hebrew University of Jerusalem (1) as an allosteric factor required for activation of cellulose biosynthesis in the alphaproteobacterium Gluconacetobacter xylinus (at that time referred to as Acetobacter xylinum). The history of this discovery was described in a 1991 review by Benziman and his students (18), in a book chapter by Deborah Delmer (19), and, more recently, by Dorit Amikam and colleagues (20). Briefly, cellulose biosynthesis by acetic acid bacteria, including G. xylinus, was thought of as a useful model for understanding cellulose biosynthesis in plants and had been studied by Benziman's teachers and colleagues since the 1940s (Table 2).
Table 2.
The history of c-di-GMP: a timeline
| Time | Event | Reference(s) |
|---|---|---|
| ∼220 BC, Qin dynasty in China | Reportedly the first use of the Kombucha “tea mushroom,” a symbiotic culture of yeast and acetobacteria which produces a thick cellulose pellicle | |
| 1946 | First studies of bacterial cellulose synthesis at The Hebrew University | 21, 22 |
| 1987 | Discovery of c-di-GMP, its chemical synthesis, proof that c-di-GMP is the true activator of cellulose synthase | 1 |
| 1995 | Discovery that c-di-GMP suppresses replication of cancer cells | 23 |
| 1995 | Characterization of GGDEF domain in the C. crescentus response regulator PleD | 24 |
| 1998 | Characterization of DGC and c-di-GMP PDE genes (published in Journal of Bacteriology) | 25 |
| 1998 | Characterization of the EAL domain protein BvgR in Bordetella pertussis, alignment of the EAL domains | 26 |
| 1999 | Description of the HD-GYP domain, proposal of a c-di-GMP-related novel signal transduction system | 27 |
| 1999 | Characterization of the GGDEF-containing response regulators PleD and CelR | 28, 29 |
| 2000 | Involvement of AdrA, a transmembrane protein with a C-terminal GGDEF domain, in intercellular adhesion | 30 |
| 2000 | Involvement of the HD-GYP domain protein RpfG in regulation of pathogenicity in X. campestris | 31 |
| 2000 | The COG database identifies GGDEF, EAL, and HD-GYP domain genes in most bacteria but not in archaea | 32 |
| 2001 | Genetic proof that the GGDEF domain has DGC activity | 33 |
| 2001 | Detailed description of the GGDEF, EAL, and HD-GYP domains as components of bacterial signal transduction | 34 |
| 2001 | Binding of oxygen to its PAS domain regulates activity of the c-di-GMP PDE from G. xylinus | 35 |
| 2004 | Crystal structure of the GGDEF domain, experimental proof of its DGC activity, identification of the allosteric I site for feedback inhibition | 36, 37 |
| 2004 | Proposal that c-di-GMP is a universal second messenger | 3 |
| 2004 | c-di-GMP involvement in pathogenesis of Yersinia pestis and Vibrio cholerae | 38–40 |
| 2004 | c-di-GMP and transition from sessility to motility | 41 |
| 2005 | GGDEF-catalyzed c-di-GMP biosynthesis in various bacterial phyla | 42 |
| 2005 | Experimental proof of the PDE activity of the EAL domain | 43–46 |
| 2005 | Biofilm dispersal by c-di-GMP | 47 |
| 2006 | Description of the c-di-GMP-binding PilZ domain | 48 |
| 2006 | Description of global c-di-GMP network regulation by the stress sigma factor RpoS in E. coli | 49 |
| 2006–2007 | Experimental proof that the PilZ domain binds c-di-GMP | 50–52 |
| 2006–2007 | Characterization of GGDEF-EAL domain proteins in which both domains are enzymatically active | 53, 54 |
| 2007 | Description of immunostimulating activity of c-di-GMP | 55–58 |
| 2008 | Discovery of a c-di-GMP-sensing riboswitch | 59 |
| 2008–2010 | Description of global c-di-GMP network regulation by the RNA-binding protein CsrA and the quorum sensing system | 60–62 |
| 2009 | Crystal structure of the EAL domain | 63–65 |
| 2010 | Discovery of the second c-di-GMP-sensing riboswitch | 66 |
| 2011 | Molecular mechanism of regulation of LapG proteolytic activity through the c-di-GMP receptor LapD | 67, 68 |
| 2011–2012 | Identification and structural characterization of the first eukaryotic c-di-GMP receptor | 69–77 |
| 2012 | Discovery of a c-di-GMP signaling system in the eukaryote Dictyostelium, a social amoeba | 183a |
However, purified cellulose synthase consistently showed far lower activity than whole cells of G. xylinus or partially purified membrane fractions (19). A long search for the cofactor that may have been lost during purification resulted in its identification, first as a GTP derivative, then as guanyl nucleotide composed of guanine, ribose, and phosphate at a 1:1:1 ratio (78, 79), and finally as bis(3′→5′)-cyclic dimeric guanylic acid, or c-di-GMP (1) (Fig. 1). Cyclic di-GMP proved to be a very efficient regulator of cellulose synthase, activating it with submicromolar dissociation constant (Kd) values (1). The following year, cellulose synthase from another alphaproteobacterium, Agrobacterium tumefaciens, was demonstrated to be c-di-GMP dependent (80), thus indicating that c-di-GMP is not a G. xylinus-specific molecule but has a wider phylogenetic distribution.
Structural analysis of chemically synthesized c-di-GMP (81) showed that in addition to the monomeric form, it also forms a stable dimer with stacked self-intercalated guanine units (Fig. 1C and D). Both forms were subsequently found in crystal structures of c-di-GMP-binding and -metabolizing proteins (36, 63–65, 75, 82–86). Cyclic di-GMP can also form higher oligomers, tetramers, and even octamers (87); their physiological roles, if any, remain unknown.
Shortly after discovering c-di-GMP, Benziman's group identified and sequenced the genes encoding enzymes responsible for its synthesis and breakdown, i.e., the diguanylate cyclase (DGC) and c-di-GMP-specific phosphodiesterase (PDE), respectively. This work resulted in a patent application originally filed in 1991 but approved only much later, in 1998 (88), which delayed publication of the sequence data (25). Sequence analysis of six G. xylinus DGCs and PDEs, characterized in that work, revealed that they all had similar multidomain architectures, containing at least three common domains, PAS-GGDEF-EAL, which turned out to be the most common domain architecture of the c-di-GMP-metabolizing proteins (Table 3).
Table 3.
Most common domain architectures involving GGDEF, EAL, and HD-GYP domains
| Protein category and domain organization | Phylogenetic distribution | Total no. of proteinsa | Characterized example (reference)b |
|---|---|---|---|
| Cytoplasmic sensor proteins | |||
| PAS-GGDEF | Actinobacteria, Proteobacteria, Spirochaetes | 3,346 | NA |
| PAS-GGDEF-EAL | Actinobacteria, Cyanobacteria, Firmicutes, Proteobacteria | 5,855 | 35, 89, 90 |
| GAF-GGDEF | Acidobacteria, Actinobacteria, Chloroflexi, Cyanobacteria, Proteobacteria, Spirochaetes | 1,351 | NA |
| GAF-GGDEF-EAL | Actinobacteria, Cyanobacteria, Deinococcus-Thermus, Firmicutes, Proteobacteria | 504 | NA |
| Globin-GGDEF | Proteobacteria | 108 | E. coli DosP (89), Bordetella pertussis GReg (91) |
| Response regulatorsc | |||
| REC-GGDEF (WspR family) | Acidobacteria, Actinobacteria, Chloroflexi, Cyanobacteria, Nitrospirae, Proteobacteria, Spirochaetes, Thermotogae | 2,022 | P. aeruginosa WspR (92–95), B. burgdorferi Rrp1 (42) |
| REC-REC-GGDEF (PleD family) | Alphaproteobacteria, Deferribacteres, Thermotogae | 614 | C. crescentus PleD (24, 28, 36, 37, 86) |
| REC-EAL (PvrR family) | Acidobacteria, Actinobacteria, Cyanobacteria, Proteobacteria, Spirochaetes | 433 | P. aeruginosa PvrR (96, 97) |
| REC-HD-GYP (RpfG family) | Chloroflexi, Cyanobacteria, Nitrospirae, Proteobacteria, Spirochaetes, Thermotogae | 514 | X. campestris RpfG (98, 99) |
| REC-GGDEF-EAL | Chloroflexi, Cyanobacteria, Proteobacteria, Spirochaetes | 401 | |
| REC-PAS-GGDEF-EAL (FimX family) | Nitrospirae, Proteobacteria | 201 | P. aeruginosa FimX (85, 100, 101) |
NA, not available.
The central GGDEF domain in all DGCs and PDEs proved to be similar to protein domains previously seen in several other bacteria. This domain was originally described in 1995 by Hecht and Newton for the response regulator PleD from Caulobacter crescentus (genome locus tag CC_2462). These authors designated it the GGDEF domain, based on its highly conserved Gly-Gly-Asp-Glu-Phe sequence motif, but they did not follow up with biochemical characterization (24). The N-terminal domains of DGCs and PDEs, which are PAS domains (106), showed significant similarity to oxygen- and redox-sensing domains found in a variety of bacterial signaling proteins (25). The C-terminal domains of G. xylinus DGCs and PDEs comprised a new protein domain, which has been designated the EAL domain, again based on the highly conserved sequence motif (Glu-Ala-Leu) near the start of this domain. Tal and colleagues concluded their 1998 Journal of Bacteriology paper as follows: “…if these regions are specifically associated with c-di-GMP metabolism, the possibility arises that c-di-GMP has wider significance as a regulatory molecule for processes other than cellulose synthesis” (25).
We know now that this prediction proved to be visionary. In a subsequent paper, the last one authored by Benziman, Ausmees and colleagues showed that cellulose biosynthesis in the plant symbiont Rhizobium leguminosarum solely required the GGDEF domain, but not necessarily the GGDEF-EAL tandem, suggesting the potential involvement of GGDEF in c-di-GMP production (33). Only a short time later, GGDEF and EAL domains were specifically coupled to c-di-GMP synthesis and breakdown, respectively, and c-di-GMP signaling was directly associated with the regulation of phenotypes other than cellulose biosynthesis in different bacteria (37, 39, 41). This work, combined with the analysis of sequenced bacterial genomes that contained numerous GGDEF, EAL, and also HD-GYP domains (27, 34, 107), identified c-di-GMP as part of a potential new second messenger in bacteria and paved the way to studies of c-di-GMP-dependent signaling pathways in the 21st century.
BIOCHEMISTRY OF CYCLIC di-GMP SYNTHESIS, DEGRADATION, AND BINDING
Cyclic di-GMP Synthesis: the GGDEF Domain
The observation that DGCs and PDEs from G. xylinus contained a tandem arrangement of the GGDEF and EAL domains presented an enzymatic conundrum. Are both of these domains required for c-di-GMP synthesis and hydrolysis? If so, how is the prevailing enzymatic activity determined? Alternatively, if only one domain is sufficient for enzymatic activity, why are both domains present in the G. xylinus enzymes?
The genetic evidence presented by Ausmees and colleagues and by others suggested that the GGDEF domain may be sufficient for DGC activity (33, 40, 41, 108). A bioinformatic analysis of the GGDEF domain sequence and structure published in 2001 by Pei and Grishin (109) was also useful in connecting this domain to the cyclase activity. These authors discovered that the GGDEF domain is distantly related to the catalytic domain of adenylate/guanylate nucleotide cyclases (110, 111). While primary sequence similarity between these domains is low, the predicted secondary and tertiary structures of the GGDEF domain are remarkably similar to those of the type III adenylate cyclase. Pei and Grishin proposed that the GGDEF domain is a DGC and predicted the loop involving the most conserved signature motif, GG(D/E)EF, to be part of the substrate (GTP) binding site.
The first biochemical evidence solidifying this connection came from a study by Paul et al. (37), who showed that the phosphorylated form of PleD converts GTP into c-di-GMP in vitro. This was also observed by Hickman et al. (93) and Ryjenkov et al. (42). The latter study analyzed in vitro activities of six different GGDEF domain enzymes originating from representatives of diverse branches of the bacterial phylogenetic tree, including Alpha- and Gammaproteobacteria as well as Thermotogae, Deinococcus-Thermus, Cyanobacteria, and Spirochaetes. All of these GGDEF domain proteins possessed DGC activity and were incapable of utilizing nucleotide substrates other than GTP. Therefore, the ubiquity and evolutionary conservation of c-di-GMP envisioned earlier (25) were established experimentally (Fig. 2).
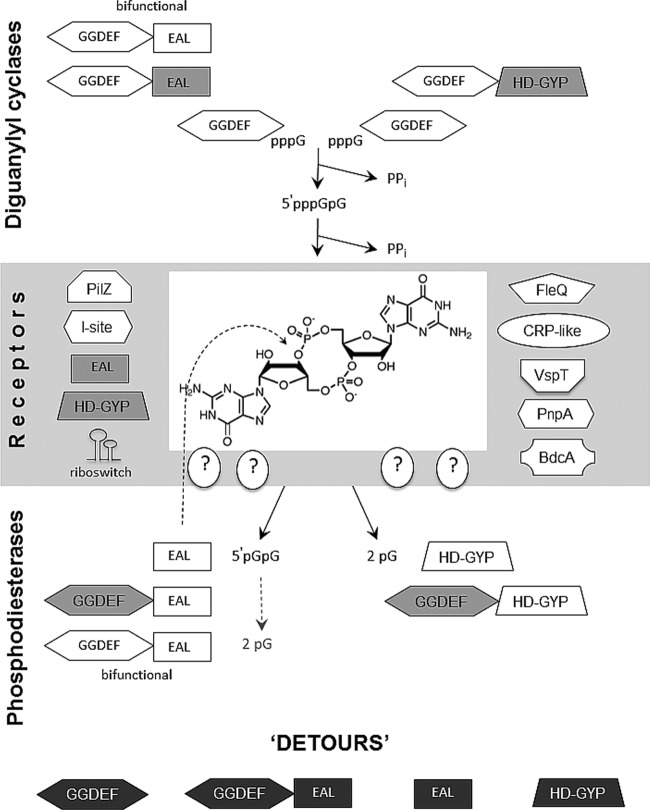
Fig 2.
Basic biochemistry of c-di-GMP synthesis, degradation, and c-di-GMP receptors. The diagrams show the protein domains involved in c-di-GMP metabolism and signaling. Enzymatically active GGDEF, EAL, and HD-GYP domains are shown on a white background. Enzymatically inactive domains involved in substrate binding are shown in light gray, and domains that are no longer associated with c-di-GMP are shown in dark gray. (Adapted from reference 456.)
How do GGDEF domain proteins catalyze c-di-GMP formation? The early insights into this question were obtained by Benziman and colleagues (1), who revealed that c-di-GMP formation from 2 molecules of GTP is a two-step reaction proceeding via 5′-pppGpG as a reaction intermediate (Fig. 2). Two molecules of pyrophosphate are reaction by-products. A further mechanistic understanding of c-di-GMP synthesis came from the biochemical and structural characterization of DGCs.
The apparent similarity of DGCs to type III nucleotide cyclases, as well as the dinucleotide nature of c-di-GMP, implied that GGDEF domains function as homodimers, where two monomers come together to form an active site at the dimer interface (112). Each GGDEF monomer contributes a GTP substrate to the formation of an intermolecular phosphoester bond to another molecule of GTP. It was observed that purified GGDEF domains by themselves form homodimers and, at high concentrations, show low-level DGC activity. This activity is significantly, usually 1 to 2 orders of magnitude, lower than the DGC activity of the full-length proteins. The prevailing activity of stand-alone GGDEF domains is a GTPase activity (42). In practice, even low DGC activity of purified GGDEF domains can serve as an indicator of whether or not the full-length proteins possess DGC activity. This is particularly useful when the full-length proteins either are recalcitrant to purification or display no activity because their activating signals are missing in vitro.
The pioneering collaborative work of Jenal's and Schirmer's groups produced crystal structures of the C. crescentus PleD protein, which provided valuable insights into the active and inactive conformations of DGCs and potential modes of enzyme activation, substrate binding, catalytic mechanism, and product inhibition (36, 86). PleD is composed of two response regulator receiver domains, REC, linked to a GGDEF domain, i.e., REC-REC-GGDEF (Table 3). The two GGDEF domains form an antiparallel homodimer (for an in-depth review of the structures of c-di-GMP-metabolizing enzymes and receptors, see reference 14).
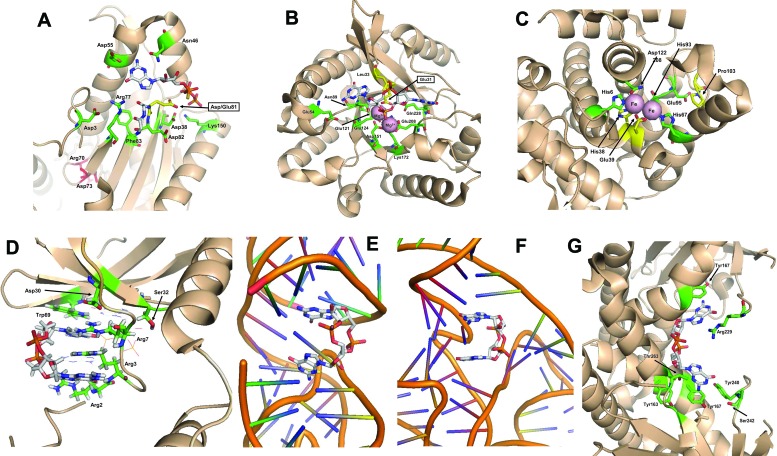
The active site, or A site, of the GGDEF domain is involved in GTP binding. Probing this site with the nonhydrolyzable GTP analog GTPαS revealed residues that bind to the β- and γ-phosphates and to the guanine base and helped to explain the specificity of the GGDEF domains for GTP (as opposed to ATP). Two Mg2+ or Mn2+ cations are required for phosphoester bond formation. The GG(D/E)EF signature motif (Fig. 3A, 4A, and 5A) forms a β-hairpin, consistent with the prediction from structural modeling (109). The first two (Gly) residues of this motif are involved in GTP binding, while the fourth residue (Glu) is involved in metal ion coordination. The third amino acid of the signature motif (Asp/Glu) is indispensable for catalysis and also plays a role in metal coordination (36, 86).
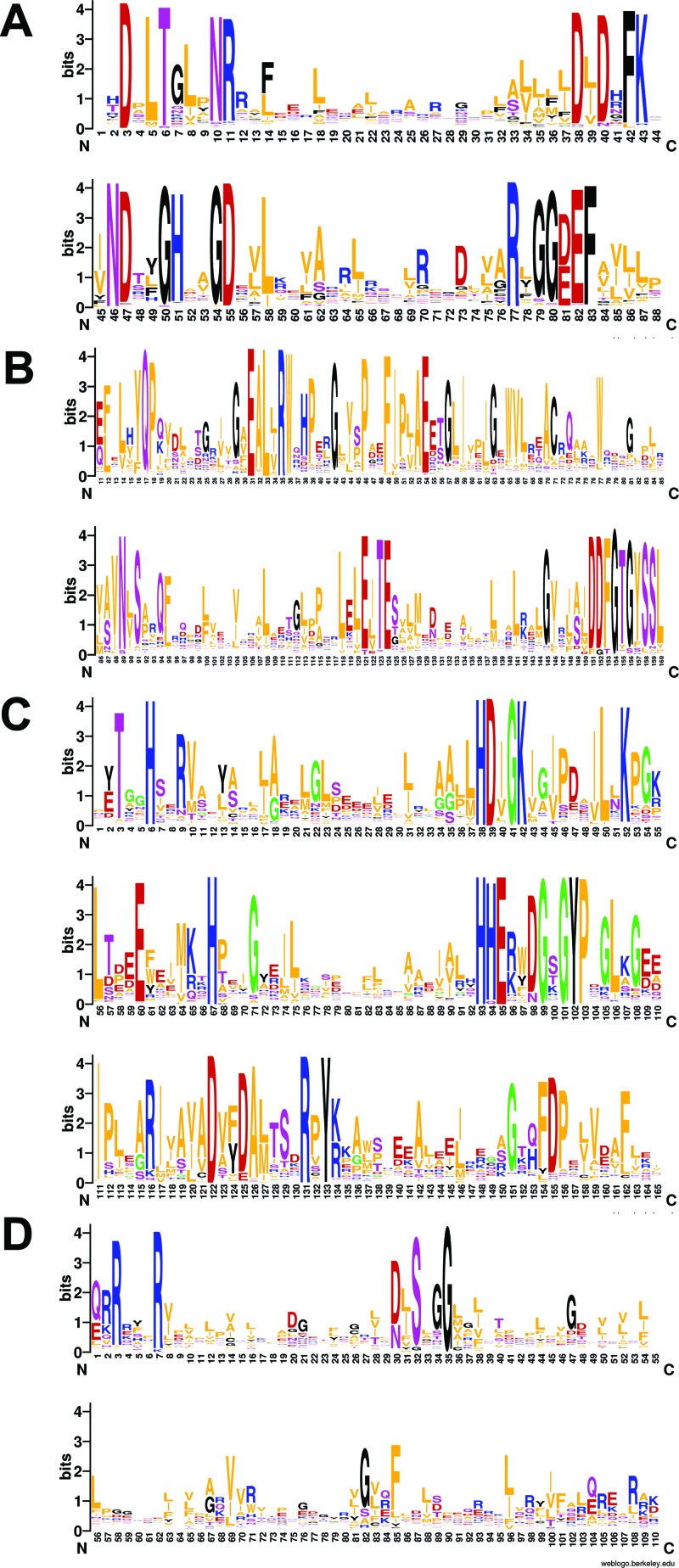
Fig 3.
Sequence conservation in cyclic di-GMP-related domains. Sequence logos of the GGDEF (A), EAL (B), HD-GYP (C), and PilZ (D) domains were generated with the WebLogo tool (457) from sequence alignments of Pfam (116) entries PF00990, PF00563, PF01966, and PF07238, respectively. Residue numbering is from Conserved Domain Database (140) entries cd01949, cd01948, cd00077, and cl01260, respectively. The height of each letter reflects the relative frequency of the corresponding amino acid at that position; the overall height of the column reflects the degree of sequence conservation at that position (measured in bits). The eponymous sequence motifs correspond to residues 79 to 83 in panel A, residues 31 to 33 in panel B, and residues 38, 39, and 101 to 103 in panel C.
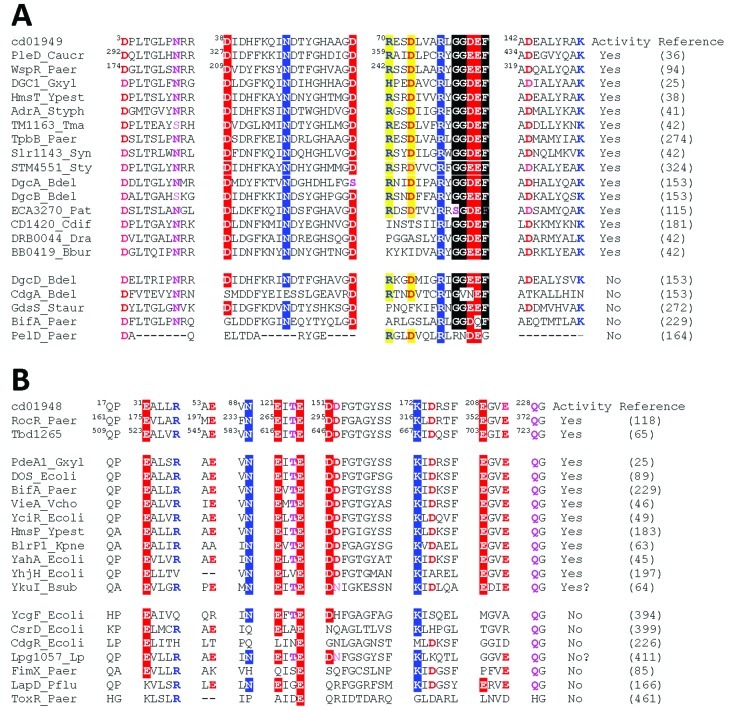
Fig 4.
Conservation of active site residues in various GGDEF and EAL domains. The residues that form the enzyme active sites and are required for the diguanylate cyclase activity of the GGDEF domain (A) or the c-di-GMP phosphodiesterase activity of the EAL domain (B) are shown in white on a red or blue background; other conserved residues in the vicinity of the active sites are shown in bold. Yellow shading in panel A indicates the residues forming the allosteric I site. The residue numbering shows positions of the respective amino acids in Conserved Domain Database (140) entries cd01949 (GGDEF) and cd01948 (EAL) and in Caulobacter crescentus PleD (UniProt entry Q9HX69), Pseudomonas aeruginosa WspR (UniProt entry Q3SJE6) and RocR (UniProt entry Q9HX69), and Thiobacillus denitrificans TBD1265 (UniProt entry Q3SJE6) (36, 65, 94, 118). (Modified from references 183 and 458 and based on previous data [38, 65, 94, 118, 125, 267].)
Fig 5.
Structural organization of the active sites of cyclic di-GMP-related molecules. The upper row shows enzymes of c-di-GMP metabolism, and the lower row shows c-di-GMP-binding proteins and riboswitches. The residues highlighted in Fig. 3 and 4 are shown with the same numbers. Residue coloring is as in Fig. 1, except that carbon atoms of GTPαS and c-di-GMP are in silver, and Mg and Fe atoms are shown as pink spheres. (A) Active site of the GGDEF domain of PleD with the bound substrate analog GTPαS (PDB entry 2V0N) (86). The catalytic Asp/Glu81 residue is shown in gold, Gly79 and Gly80 of the GGDEF motif are in silver, and Arg70 and Asp73 of the RxxD motif in the allosteric inhibitory I site (36) are shown in red. (B) Active site of the EAL domain of Tbd1265 with bound c-di-GMP (PDB entry 3N3T) (65). Glu31 and Leu33 residues of the EAL motif are shown in gold. (C) Active site of the HD-GYP domain of Bd1817 with bound c-di-GMP (PDB entry 3TM8) (129). His38, Asp39, Gly101, and Pro103 of the HD and GYP motifs are shown in gold (Tyr102 is missing in Bd1817). (D) c-di-GMP binding site of the PilZ domain of PA4608 (PDB entry 2L74) (82). For simplicity, one of the c-di-GMP molecules is shown only as lines. (E) c-di-GMP bound to riboswitch I (PDB entry 3IRW) (75). (F) c-di-GMP bound to riboswitch II (PDB entry 3Q3Z) (76). (G) c-di-GMP bound to the stimulator of interferon genes STING (PDB entry 4EMT) (74; see references 70 to 77 for further details). The figure was generated with PyMOL (Schrödinger, LLC).
Since it has proved difficult to capture an active cyclase homodimer in action, the catalytic mechanism of c-di-GMP formation remains murky. One important conclusion stemming from these structures is that most likely no large conformational changes in the GTP-binding half-sites of the GGDEF domains take place during catalysis (14), and therefore, the reason that GGDEF domains display DGC activity is that they come together and form a catalytically competent homodimer. This suggests that regulatory interactions that keep the GGDEF domains physically separated from each other would prevent their DGC activity.
Two mechanisms appear to affect formation of the catalytically competent GGDEF homodimer. One involves conformational rearrangements in response to changes in the sensory domains linked to the GGDEF domains. While biochemical evidence for activation of DGCs by various primary signals is growing, no structural information is currently available on how GGDEF domains are activated by environmental signals. However, DGC activation by secondary mechanisms derived from primary signals, e.g., protein phosphorylation, has been revealed using biochemical and structural biology approaches. Complex domain and protein subunit rearrangements that bring the GGDEF domains in close proximity have been observed by comparing X-ray structures of the (pseudo)phosphorylated and nonphosphorylated states of PleD and Pseudomonas aeruginosa WspR (PA3702; REC-GGDEF domain architecture) (86, 92). Phosphorylation is a common (Table 3) and powerful mechanism for GGDEF domain activation. For example, the sole DGC (REC-GGDEF) of the pathogenic spirochete Borrelia burgdorferi, Rrp1 (BB_0419), is completely inactive in vitro until its REC domain is phosphorylated (42).
The second mechanism affecting activation/inactivation of DGCs involves feedback inhibition. The PleD protein crystallized in the presence of c-di-GMP revealed a product-inhibited conformation where a base-intercalated dimer of c-di-GMP molecules (Fig. 1C and D) is bound to the inhibitory (I) site (36, 113). A four-residue motif constituting the I site, RxxD (where “x” is any residue), is positioned five amino acids upstream of the GG(D/E)EF motif. Despite primary sequence proximity between the I and A sites, they are located antipodal to each other (36, 86) (Fig. 5A). Additional residues coordinating binding of the c-di-GMP dimer to the I site come either from the regulatory domain, as in PleD (86), or from the GGDEF domain of another protein monomer, as in WspR or PleD (92). This allows the intercalated c-di-GMP dimer to block the GGDEF domain movement required for formation of the catalytically competent homodimer. The inhibition constant for DGCs containing the I site is in the low micromolar range. Therefore, the likely purpose of product inhibition is to limit the time of the (desired) c-di-GMP target activation and/or to prevent c-di-GMP spill to undesired downstream targets.
The I site is found in approximately half of the GGDEF domain DGCs (114) (Table 4). Are enzymes lacking I sites subject to product inhibition? Apparently some are. A recently solved structure of the GGDEF domain from the XCC4471 protein (locus tag XCC3486) of the plant pathogen Xanthomonas campestris may have an answer to the question of how DGCs lacking I sites can still be product inhibited. XCC4471 has been captured with a semi-intercalated c-di-GMP dimer in the A site (117). Therefore, whereas many DGCs contain I sites and are inhibited noncompetitively, some DGCs that do not contain I sites may be inhibited competitively by c-di-GMP bound to their A sites. How widespread the competitive inhibition of DGCs is remains unknown at present.
Table 4.
Conservation of active site residues in GGDEF domains
| A-site motifa | Activity | Count (%)b | No. (%) of proteins with RxxD in I sitec |
|---|---|---|---|
| RxGGDEF | Yes | 11,327 (40.8) | 5,815 (51.3) |
| RxGGEEF | Yes | 9,063 (32.6) | 5,066 (55.9) |
| RxSGDEF | Yes | 462 (1.7) | 146 (31.1) |
| RxAGDEF | Yes | 428 (1.5) | 194 (45.3) |
| HxGGDEF | ? | 320 (1.2) | 14 (4.4) |
| QxSGYDF | No | 228 (0.8) | None |
| RxHRSDF | No | 218 (0.8) | None |
| RxGSDEF | No? | 165 (0.6) | 47 (28.5) |
| RxGGEEL | No | 157 (0.6) | 112 (71.3) |
| RxEGEVF | No | 133 (0.5) | 122 (91.7) |
Activity data are as described previously (38, 94, 115). The RxGGDEF motif appears to tolerate a large variety of residues in the second (x) position, whereas the work reported in reference 94 suggests that the GGEEF motif is active only in the RYGGEEF variant, which is found in ∼1/3 of RxGGEEF contexts. A mutant variant of the Yersinia pestis HmsT protein with the RYAGEEF active site motif was inactive (38).
Number of occurrences of the motif among 27,782 full-length sequences of the GGDEF (PF00990) domain listed in the 26th release (November 2011) of the Pfam database (116).
Twenty-six percent of HxGGDDF motif proteins have either I or V in the second position and DxxD in the I site; QxSGYDF motif proteins have either I or V in the second position and either SxxM (64.5%), AxxM (32.9%), or PxxM (2.6%) in the I site; and RxHRSDF motif proteins always have Y in the second position and either MxxA (66.5%) or MxxS (32.6%) in the I site.
Cyclic di-GMP Hydrolysis: the EAL Domain
Since GGDEF domains function in c-di-GMP synthesis, it followed that EAL domains must be responsible for c-di-GMP hydrolysis. However, it was unclear whether or not EAL domains are sufficient for the c-di-GMP-specific PDE activity or whether both GGDEF and EAL domains are necessary. Like the case with DGCs, Benziman and coworkers laid the groundwork for PDE research. They purified PDEs from G. xylinus and showed that these proteins hydrolyze c-di-GMP into linear di-GMP, i.e., 5′-pGpG. The c-di-GMP-specific PDE activity required either Mn2+ or Mg2+ and was strongly inhibited by Ca2+. The product of c-di-GMP hydrolysis, 5′-pGpG, was subsequently degraded to monomeric pG, apparently by different enzymes that had Ca2+-independent activity (79).
Simm et al. (41) and Tischler and Camilli (39) provided strong pieces of genetic evidence that the EAL domains are sufficient for c-di-GMP-specific hydrolysis by showing that overexpressed EAL domain proteins inhibit biofilm phenotypes. Biochemical evidence that PDE activity is associated with the EAL domains was obtained shortly thereafter. Bobrov et al. (43) used a nonspecific PDE substrate, bis(p-nitrophenyl) phosphate, to show that the purified EAL domain protein HmsP from Yersinia pestis can break it down. Schmidt et al. (45) used the Escherichia coli EAL domain protein YahA as well as individual EAL domains from YahA and Dos (recently renamed DosP [89]) to show that EAL domains hydrolyze c-di-GMP and that this activity is c-di-GMP specific. Several phosphoester- and phosphodiester-containing compounds tested, including cyclic AMP (cAMP), were unaffected. The EAL domain was found to be capable of hydrolyzing 5′-pGpG, however, at a rate that was much lower than the rate of c-di-GMP hydrolysis. Therefore, in vivo 5′-pGpG is likely hydrolyzed not by the EAL domain PDEs but by alternative enzymes (Fig. 2) (also see “Open Questions in c-di-GMP Signaling”). The biochemical parameters of c-di-GMP hydrolysis, i.e., dependence on Mn2+ or Mg2+ and strong sensitivity to inhibitory Ca2+ cations (45), were consistent with the observations made earlier in the Benziman lab for preparations of G. xylinus c-di-GMP PDEs (79). Therefore, these features are common to hydrolysis by the EAL domain PDEs. Simultaneously with Schmidt et al. (45), the in vitro activities of the EAL domain proteins were reported by other groups (44, 46), thus solidifying the connection between EAL and the c-di-GMP-specific PDE activity.
Unlike GGDEF domains, which must function as homodimers, EAL domains appear to retain some PDE activity as monomers (45). However, the vast majority of EAL domain PDEs characterized thus far form dimers or higher-order oligomers in vitro (54, 63, 65, 118). The dimeric state appears to be critical for activation of PDEs by environmental stimuli (119, 120). Therefore, a dimer is the most probable functional unit of the EAL domain engaged in c-di-GMP hydrolysis in vivo.
Structures of several c-di-GMP PDEs have now been solved (63–65, 67, 85, 121). The structural work of Barends et al. (63) provided rich information about the c-di-GMP binding site, catalytic mechanism, pH dependence, choice of catalytic cations, inhibition by Ca2+, and mechanisms of activation by environmental stimuli. These authors crystallized the BLUF-EAL protein BlrP1 (KPN_01598) from Klebsiella pneumoniae, whose PDE activity is upregulated by blue light sensed via the flavin-containing BLUF domain (122, 123). The two antiparallel EAL domains of BlrP1 interact through three α-helices: one from each EAL domain and one “compound” helix made of two shorter helices originating from each of the EAL domains. c-di-GMP in the EAL domains is present in an extended (open) conformation (Fig. 1A), which differs from the bent, U-shaped (closed) conformation of c-di-GMP observed in the I sites of DGCs and c-di-GMP receptors (Fig. 1C). The extended conformation likely facilitates hydrolysis of one of the phosphoester bonds in c-di-GMP.
PDEs operating on cyclic mononucleotides typically use a two-metal catalytic mechanism (124). Consistent with this expectation, BlrP1 was found to bind c-di-GMP through two metal cations. While the issue of whether c-di-GMP hydrolysis involves a two- or one-metal mechanism has been somewhat controversial (64, 125), this controversy has now been resolved. Two-metal catalysis (63) appears to be the only catalytic mechanism of c-di-GMP hydrolysis by the EAL domain PDEs (65). Those EAL domain proteins that were crystallized with a single cation turned out to be enzymatically inactive.
The activity of the EAL domain proteins depends on the structure of a two-metal cation cluster in which the metals coordinate two water molecules, one of which is involved in a hydrolytic attack on a phosphoester bond of c-di-GMP. A higher pH and Mn2+ promote optimal bond lengths in the metal-water cluster, whereas a lower pH and Mg2+ distort the cluster away from the optimum required for catalysis. In BlrP1, blue light-induced conformational changes in the BLUF domain of one monomer affect the EAL-EAL dimer interface such that this optimizes the metal-water cluster configuration in the EAL domain of a partner monomer, thus stimulating its PDE activity. Ca2+ distorts the distances within the cluster, which explains its strong inhibitory effect. The BlrP1 structure (63) and mutagenesis work (65, 118, 125) helped to explain the nature of the conserved amino acid motifs (Fig. 3B) identified earlier (34) and used to distinguish enzymatically active from inactive EAL domains (45). Most of these conserved motifs proved to be involved in c-di-GMP binding or in two-metal catalysis (Fig. 4B and 5B). It is noteworthy that the Glu residue of the EAL motif is directly involved in coordination of one of the metals (63, 65), which explains its 100% conservation in the active enzymes.
Cyclic di-GMP Hydrolysis: the HD-GYP Domain
The HD-GYP domain is a subset of the larger HD family, whose members possess hydrolytic activities toward diverse substrates (27, 126). HD-GYP was predicted to have c-di-GMP-specific PDE activity primarily because of the frequent linkage between the GGDEF and HD-GYP domains, reminiscent of the GGDEF-EAL tandems (27, 34). Ryan et al. (99) used the HD-GYP domain protein RpfG from X. campestris (XC_2335) to test the hypothesis that the HD-GYP domain is involved in c-di-GMP degradation. When expressed in a heterologous host, RpfG functionally replaced an EAL domain phosphodiesterase. When it was purified, it had c-di-GMP-specific PDE activity. Interestingly, the main product of c-di-GMP hydrolysis by RpfG was GMP, not 5′-pGpG, the product of the EAL domain PDEs (Fig. 2). It is therefore possible that the HD-GYP domain PDEs either do not release the 5′-pGpG intermediate or readily rebind the released product for its full hydrolysis to GMP. It is also possible that 5′-pGpG was not detected in the original experiment because of the long reaction time and/or RpfG functioning as a dimer (99), so earlier time points in c-di-GMP hydrolysis by the HD-GYP domain may need to be analyzed to clarify the significance of the apparent difference between the products of EAL and HD-GYP PDEs.
The genetic evidence supporting engagement of HD-GYP proteins in c-di-GMP hydrolysis, in addition to Xanthomonas PDEs, includes representatives from Pseudomonas and Borrelia (127, 128). However, biochemical data on HD-GYP proteins remain scarce. Thus far, the HD-GYP domain proteins have resisted crystallization, and no structure of the active HD-GYP domain has been determined. Mechanistic insights into c-di-GMP hydrolysis by HD-GYP PDEs began to emerge only recently, when the first structure of an HD-GYP domain protein, Bd1817, from the bacterial predator Bdellovibrio bacteriovorus, was solved by Lovering et al. (129). The HD-GYP domain of Bd1817 has no enzymatic activity, possibly because it lacks a conserved tyrosine in the GYP motif, and does not appear to bind c-di-GMP in vitro (129). However, the structure of Bd1817 (Fig. 5C) still proved instructive. It showed several conserved residues of the HD-GYP family grouping around the binuclear metal center, where the catalytic metals are likely to be either Fe2+ or Mn2+. Furthermore, Lovering et al. modeled the protein with c-di-GMP and proposed a catalytic mechanism involving a water-derived hydroxide ion attack on the c-di-GMP phosphoester bond. While this model yielded important insights, more mechanistic studies are clearly needed to understand c-di-GMP hydrolysis by the HD-GYP domain PDEs.
Proteins with GGDEF and EAL or HD-GYP Domains Arranged in Tandem
The “enzymatic conundrum.”
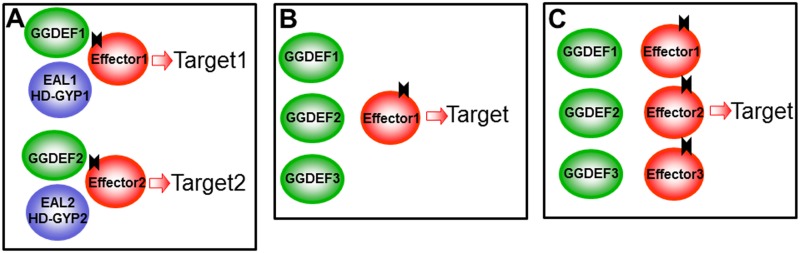
Genomic analyses show that GGDEF and EAL domains are often found on the same polypeptide chain as parts of multidomain proteins. As discussed above, the very first identified DGCs and PDEs of G. xylinus contained GGDEF-EAL domains arranged in tandem, but they had either DGC or PDE activity (25, 35, 130), implying that one of the two domains in each enzyme was catalytically inactive. It is noteworthy that the sheer number of GGDEF-EAL tandems is huge, e.g., as many as ∼1/3 of all GGDEF domains and ∼2/3 of all EAL domains are found on the same polypeptide chains (114; http://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html). Since the GGDEF domain is fully capable of DGC activity and either an EAL or HD-GYP domain is capable of c-di-GMP hydrolysis, why do so many proteins contain GGDEF-EAL and GGDEF–HD-GYP tandems (Table 1 and Fig. 2)?
Theoretically, two possibilities exist that may explain the “enzymatic conundrum” of proteins containing two domains with opposite enzymatic activities. One scenario is that while both domains are enzymatically active, they are differentially regulated by environmental and/or intracellular signals so that at any given point one activity is prevalent. The precedents of bifunctional signaling enzymes are well known and include protein His kinases/phosphatases of two-component regulatory systems (131) and the SpoT proteins, catalyzing synthesis and degradation of the bacterial alarmone (p)ppGpp (132). While almost half of all GGDEF-EAL proteins reportedly have intact active sites (114), only a few examples of truly bifunctional DGCs/PDEs have been described so far (54); some of these are discussed below.
By far more common is the situation where one of the two domains is enzymatically inactive or catalytically incompetent (44, 45). These “retired from active duty” domains have evolved to carry out new functions. One of these functions may involve binding (but not processing) of the substrate, e.g., GTP binding in the A sites of inactive GGDEF domains (44) or c-di-GMP binding in the substrate binding sites of enzymatically inactive EAL domains (85, 101, 133). Another set of functions of GGDEF, EAL, and HD-GYP domains that have “retired” from catalysis includes their participation in protein-protein or protein-RNA interactions. According to genomic analysis, mutations predicted to impair DGC activity are present in ∼40% of the GGDEF domains in proteins containing GGDEF-EAL modules (114). Some of the GGDEF, EAL, and HD-GYP proteins have completely lost their ties to c-di-GMP and represent “detours” from the mainstream c-di-GMP signaling pathways (Fig. 2). Several examples of these “retired” domains and “detours” are discussed in detail throughout this review.
Bifunctional enzymes with tandemly arranged GGDEF and EAL domains.
One of the few bifunctional proteins that contain enzymatically active GGDEF and EAL domains arranged in tandem is Rhodobacter sphaeroides BphG1 (RSP_4191), a bacteriophytochrome with a PAS-GAF-PHY photosensory module linked to a GGDEF-EAL output (54). The photosensory module binds a bilin chromophore and responds to red/near-infrared light in a reversible manner. However, despite light sensitivity of the photoreceptor module, the output PDE activity of BphG1 proved to be irresponsive to irradiation (54). It was observed that BphG1 overexpressed in E. coli underwent site-specific proteolysis that released the C-terminal EAL domain. Interestingly, the truncated PAS-GAF-PHY-GGDEF protein fragment lacking the EAL domain gained DGC activity, which was strongly activated by light. In this rather eccentric, apparently irreversible regulation, a constitutive PDE activity turns into the opposing, DGC, activity, which is responsive to light. It is unclear as yet whether proteolysis occurs in the native host, R. sphaeroides, and what controls the extent of proteolysis.
It cannot be excluded that instead of proteolysis, the switch between two opposite activities of BphG1 in R. sphaeroides is controlled by proteins interacting with BphG1, as is the case with another bifunctional GGDEF-EAL protein, ScrC (VPA1511) from Vibrio parahaemolyticus (134). ScrC has an N-terminal periplasmic sensor domain linked to a GGDEF-EAL module. The scrC gene belongs to the scrABC operon, which regulates the switch between motile swarmer cells and sessile biofilm cells producing capsular polysaccharide (135). When expressed by itself, ScrC shows DGC activity. However, this is switched to PDE activity in the presence of ScrC's protein partners, ScrA (VPA1513) and ScrB (VPA1512) (134). At high cell densities, the periplasmic domain of ScrB binds a novel autoinducer, which stimulates its interaction with ScrC and facilitates the DGC-to-PDE switch in ScrC (136).
The Mycobacterium smegmatis cytoplasmic protein MSDGC-1 (MSMEG_2196), which has a GAF-GGDEF-EAL domain architecture, has been shown to both synthesize and hydrolyze c-di-GMP in vitro (137). MSDGC-1 is widespread in the genus Mycobacterium and is the only functional DGC in M. smegmatis, Mycobacterium tuberculosis (locus tag Rv1354c), and Mycobacterium bovis (Mb1389c). Given the requirement of c-di-GMP for long-term mycobacterial survival under conditions of nutrient starvation (138), it will be important to understand the mechanism that regulates its DGC and PDE activities.
In the Lpl0329 protein from Legionella pneumophila, a phosphorylation-based switch appears to control the relative contributions of the DGC and PDE activities. Lpl0329 contains a receiver domain, REC, of the two-component regulatory systems linked to a GGDEF-EAL tandem (139). The atypical histidine kinase Lpl0330 phosphorylates Lpl0329, which lowers the DGC activity of the protein but leaves the PDE activity unaffected. The physiological significance of this phosphorylation-based switch in L. pneumophila, as well as the mechanisms and functions of bifunctional DGC/PDE enzymes from other bacteria, has yet to be investigated.
Active versus degenerate domains.
The availability of high-resolution crystal structures of GGDEF, EAL, and HD-GYP domains combined with site-directed mutagenesis studies allowed the formulation of general rules for distinguishing domains that are likely to be enzymatically active versus degenerate, inactive domains (Fig. 3 and 4). In the GGDEF domain, the active site includes the catalytic Asp/Glu residue surrounded on each side by two strongly conserved residues, which together form the eponymous 79GG(D/E)EF83 sequence motif in the A site (113) (residue numbering is from the GGDEF domain model in the NCBI's Conserved Domain Database [140]) (Fig. 3A, 4A, and 5A). In addition, the active site includes the Asp38 residue, which binds Mg2+, and Asn46 and Asp55, which bind the guanine base (36). Early studies suggested an absolute requirement of all five residues of the GG(D/E)EF motif for DGC activity (38). A detailed study of the P. aeruginosa response regulator WspR revealed an additional requirement for the Arg77 and Tyr78 residues immediately preceding this motif (94) (Fig. 4A and 5A; Table 4). However, subsequently, more relaxed residue conservation requirements were observed (115). It is possible that the RYGGEEF active site motif found in the PleD, WspR, and HmsT proteins does indeed require strict conservation of all residues surrounding the catalytic Glu81 residue (38, 94). For example, a mutant variant of Y. pestis HmsT with an RYAGEEF active site motif is inactive (38). In contrast, an RxGGDEF motif with a catalytic Asp81 residue may accommodate several different hydrophobic residues in the “x” position. In addition, it apparently retains some DGC activity even when the first Gly is replaced with Ala or Ser (115).
In the well-studied C. crescentus protein CC3396 (PAS-GGDEF-EAL domain composition), a GGDEF domain with a degenerate GEDEF motif in the A site had no DGC activity but was still able to bind GTP with a high affinity (Kd = 4 μM) and to regulate the PDE activity of the downstream EAL domain (44). GTP binding by this domain dramatically increased the affinity of the EAL domain for its substrate, bringing the Km for c-di-GMP from the physiologically irrelevant level of ∼100 μM to the physiologically relevant level of 0.42 μM. Therefore, this degenerate GGDEF domain may serve a structural role, and possibly even a regulatory role, under extreme starvation conditions when the GTP concentration drops to very low, micromolar levels.
The requirements for the c-di-GMP-specific PDE activity of the EAL domains have been studied in much detail through sequence comparisons, X-ray crystallography, and mutagenesis of the key residues (45, 63–65, 118, 125). The availability of high-resolution X-ray structures and the understanding of the mechanism of c-di-GMP hydrolysis discussed earlier in this review (63, 65) resulted in identification of the sets of residues involved in c-di-GMP binding as well as in coordination of catalytic Mg2+ or Mn2+ cations (Fig. 4B and 5B; Table 5). Analysis of the EAL domains from various bacterial genomes suggested that ∼85% of them are enzymatically active (114) (Table 5).
Table 5.
Conservation of active site residues in EAL domains
| Active site residuea |
Activity | Count (%)b | ||||||
|---|---|---|---|---|---|---|---|---|
| 31 | 89 | 121 | 124 | 151 | 172 | 208 | ||
| E | N | E | E | D | K | E | Yes | 13,821 (85.2) |
| E | Q | E | E | N | K | T | No | 132 (0.8) |
| E | N | L | E | G | M | G | No | 126 (0.8) |
| E | N | E | E | D | K | M | No | 102 (0.7) |
| E | N | E | E | D | K | K | No | 86 (0.5) |
Residue numbering is from the EAL domain entry (cd01948) in the NCBI Conserved Domain Database (CDD) (140) and corresponds to the E175, N233, E265, E268, D295, K316, and E352 positions in the P. aeruginosa protein PA3947 (RocR) (118) and the E523, N584, E616, E620, D646, K667, and E703 residues in the Thiobacillus denitrificans protein TBD1265 (65). Activity data are from references 65 and 118.
Number of occurrences of the residue combination among 16,211 full-length sequences of the EAL (PF00563) domain listed in the 26th release (November 2011) of the Pfam database (116).
Thus far, little biochemical work has been done on DGCs containing GGDEF-EAL tandems, and therefore the functions of the enzymatically inactive EAL domains present in such proteins remain largely unknown. Our unpublished data on the G. xylinus DgcA1 (GLX_04270) protein (25) revealed that deletion of the enzymatically inactive EAL domain destroys the DGC activity of the protein, which suggests that degenerate EAL domains in GGDEF-EAL proteins may have structural or important regulatory functions.
GGDEF–HD-GYP proteins.
Although less numerous than GGDEF-EAL domain fusions, fusions of the GGDEF and HD-GYP domains are also widespread in bacteria, particularly among the Aquificae, Deinococci, Firmicutes, and Planctomycetes (http://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html). As noted above, analysis of such a fusion in Aquifex aeolicus provided the first clue to the involvement of the HD-GYP domain in c-di-GMP metabolism (27, 34). While no such proteins have been characterized experimentally so far, they may follow the same logic of the interplay between enzymatically active and inactive domains as the GGDEF-EAL tandem proteins.
Types of c-di-GMP Receptors
Many bacterial species contain several dozen enzymes involved in c-di-GMP synthesis and breakdown (34, 141). While these enzymes are readily identifiable due to the characteristic GGDEF, EAL, and HD-GYP domains, identifying proteins that function as c-di-GMP receptors/effectors based on sequence information proved to be more challenging. We now know that c-di-GMP binds to diverse classes of proteins, many of which have no sequence or structural similarity to each other. Not surprisingly, at present, we know much less about c-di-GMP receptors and targets regulated by c-di-GMP than about c-di-GMP-metabolizing enzymes. Fortunately, this situation is rapidly changing, as the number of c-di-GMP-binding proteins has been growing rapidly (as summarized in recent reviews [15, 142, 143]). In addition to acting through protein receptors, c-di-GMP has been shown to bind to two types of riboswitches (Fig. 3 and 5E and F).
Several classes of c-di-GMP receptors have been predicted based on primary sequences. These include PilZ domain receptors, I-site receptors, inactive EAL domain receptors, and likely HD-GYP domain receptors. There are also less predictable or unpredictable c-di-GMP receptors, which include transcriptional regulators of various kinds and proteins of diverse and unrelated functions that are just beginning to be discovered.
PilZ domain c-di-GMP receptors.
The first c-di-GMP protein receptor type, designated the PilZ domain, was predicted by Amikam and Galperin (48) to be part of the glycosyltransferase protein of the G. xylinus cellulose synthase complex. While c-di-GMP was indeed extracted from the membrane preparations of cellulose synthase, and c-di-GMP added exogenously to the washed membrane preparations could stimulate cellulose synthase activity (1, 78, 79), the identity of the c-di-GMP-binding protein in G. xylinus has been somewhat controversial (144, 145). Amikam and Galperin noticed that an approximately 100-amino-acid C terminus of the glycosyltransferase BcsA subunit forms a separate protein domain that is also present downstream of some EAL or GGDEF-EAL proteins, thus making it a good candidate for a c-di-GMP binding domain. The domain name originated from the P. aeruginosa PilZ (PA2960) protein, which consists exclusively of this domain and is involved in pilus formation (146).
Soon after that, c-di-GMP binding to the PilZ domain was verified experimentally. The C terminus of G. xylinus BcsA, the PilZ domain protein YcgR from E. coli (52), the DgrA protein from C. crescentus (51), and the Vibrio cholerae PilZ domain proteins PlzC and PlzD (147) were shown to bind c-di-GMP in vitro with high specificities, demonstrating that the PilZ domain was indeed the long-sought-after c-di-GMP receptor. The ability of PilZ domains to bind c-di-GMP in vitro has now been demonstrated for PilZ domain proteins of numerous bacterial species (147–149).
YcgR and DgrA showed submicromolar affinities for c-di-GMP, which is consistent with intracellular c-di-GMP concentrations, which have been estimated to be in the sub- to low-micromolar range (37, 41, 145). It is noteworthy that PilZ domain c-di-GMP receptors appear to have the highest affinities for c-di-GMP compared to the majority of subsequently discovered c-di-GMP protein receptors, whose Kd values usually fall into the low- to medium-micromolar range.
Several X-ray and nuclear magnetic resonance (NMR) structures of the PilZ domain receptors have now been solved (50, 82, 83, 150, 151). These studies have confirmed the bioinformatic (48) and biochemical (52) prediction that two short stretches of residues comprising the PilZ domain consensus, RxxxRx20–30(D/N)x(S/A)xxG, are involved in c-di-GMP binding (Fig. 3D and 5D). In addition, the structures have revealed that the RxxxR motif is a primary binding loop that wraps around c-di-GMP and likely brings the remainder of the consensus residues, located on distant structural elements, in closer proximity. Cyclic di-GMP functions as an interdomain (or intermolecular) glue that brings together two protein moieties, and this initiates downstream signaling events. Unexpectedly, c-di-GMP can bind to the PilZ domain proteins in more than one way, i.e., either as an intercalated dimer (52, 83, 150) or as a monomer in the closed conformation (50). Furthermore, PilZ domain proteins were found to adopt different oligomeric states (Fig. 1), from monomeric to tetrameric (82, 83, 87, 152), indicating potentially different modes of downstream signal transduction.
While PilZ domain proteins are widespread c-di-GMP receptors, their numbers vary dramatically among species and do not directly correlate with the number of c-di-GMP-metabolizing proteins (http://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html). For example, E. coli has 29 GGDEF/EAL domain proteins and only 2 PilZ domain proteins, BcsA and YcgR (52), while B. bacteriovorus has 12 GGDEF/EAL/HD-GYP domain proteins but 15 PilZ domain proteins (153), which is close to the record among bacteria (http://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html). Approximately half of all PilZ domain proteins comprise stand-alone PilZ domains with short N-terminal extensions. The other half contain PilZ domains bound to other protein domains, including type 2 glycosyltransferases (as in BcsA), PilZN (YcgR_N) domains, response regulator (REC) domains, methyl-accepting protein (MCP) domains, DNA binding domains, adenylate/guanylate cyclases, and other domains. Some of the better-characterized PilZ domain proteins, involved in motility regulation (51, 52, 148, 154–156), polysaccharide synthesis and translocation (18, 149, 157), and DNA binding (120, 158), are described in this review. It is clear that the PilZ domain functions as a versatile module that can regulate diverse activities in a c-di-GMP-dependent manner.
Similarly to the enzymatic domains associated with c-di-GMP, PilZ domains come in “active” and “inactive” varieties, where inactivity means a lack of c-di-GMP binding. The inactive PilZ domain proteins are quite common (159). Ironically, the eponymous PilZ protein from P. aeruginosa also belongs to this category (160).
I sites and enzymatically inactive EAL and HD-GYP domains as c-di-GMP receptors.
Given that some bacterial species containing c-di-GMP-metabolizing enzymes do not carry any PilZ domain proteins (http://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html), it has been obvious that non-PilZ domain c-di-GMP receptors must exist (48). Genetic evidence confirmed this prediction. For example, deletion of all PilZ domain-encoding genes in V. cholerae did not abolish the effect of c-di-GMP on colony morphology (161). What do non-PilZ c-di-GMP receptors look like?
The most obvious candidate for such a receptor was the GGDEF domain, with its allosteric c-di-GMP-binding I site (36, 113), so even catalytically inactive GGDEF domains could still serve as c-di-GMP receptors (4, 162). A general scenario applicable to many systems has been that when enzymes “retire from active duty” (i.e., lose their catalytic functions), they often retain the ability to bind their substrates and/or products and can function as substrate/product-binding proteins.
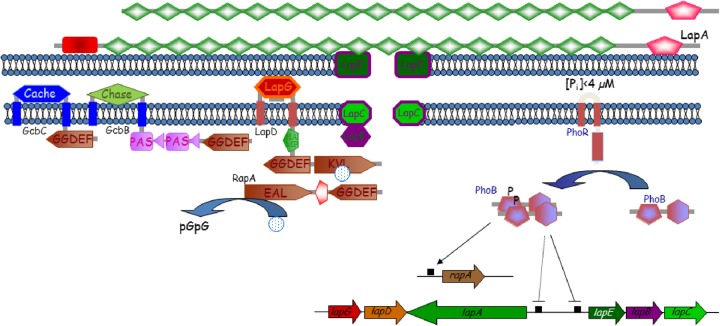
This concept fully applies to catalytically incompetent GGDEF domain proteins. Having lost the enzymatic activity and the characteristic GGDEF motif, these proteins often retain their product-inhibiting I site, the first identified c-di-GMP binding sequence (113) (Fig. 2). Thus, I sites not only prevent overproduction of c-di-GMP by DGCs, but they allow proteins that no longer possess the DGC activity to function as c-di-GMP receptors. One of the first examples of such a catalytically “retired” GGDEF domain protein which functions as a c-di-GMP receptor was the response regulator PopA (CC_1842), which promotes cell cycle progression in C. crescentus (162). Another such receptor is the hybrid histidine kinase SgmT (MXAN_4640) of Myxococcus xanthus (163). c-di-GMP binding to SgmT mediates spatial localization of this cytoplasmic histidine kinase, without any obvious change in functionality. Yet another example involves the c-di-GMP receptor CdgA (Bd3125) required for rapid entry of the bacterial predator B. bacteriovorus into prey cells (153). In some instances, the GGDEF domain sequences containing I sites have diverged so much that they are barely recognizable, which makes their identification nontrivial. One such example is P. aeruginosa PelD (PA3061), the protein that posttranslationally controls Pel polysaccharide synthesis in a c-di-GMP-dependent manner (164, 165).
The same scenario proved true for EAL domains that lost PDE activity but retained the ability to bind c-di-GMP. An interesting example of such a hybrid protein is P. aeruginosa FimX, involved in type IV pilus-based motility. The degenerate and enzymatically inactive C-terminal EAL domain of FimX serves as a high-affinity c-di-GMP receptor (85, 101, 133). Another example involves the GGDEF-EAL c-di-GMP receptor LapD from Pseudomonas fluorescens (166, 167). While neither the GGDEF nor EAL domain of this receptor is enzymatically active, c-di-GMP binds to the degenerate EAL domain of LapD with a high affinity (68, 166). The Bacillus subtilis EAL-YkuI_C protein YkuI (BSU14090) is another candidate receptor belonging to this class. YkuI has been shown to bind to but not hydrolyze c-di-GMP (64). Catalytically incapable HD-GYP domain proteins that function as c-di-GMP receptors have not yet been described. However, it is probably just a matter of time before such receptors are uncovered. Mother Nature rarely misses apparent biological solutions.
Cyclic di-GMP receptors not predicted by bioinformatics.
In the last few years, a plethora of new c-di-GMP-binding proteins belonging to diverse types have been discovered. None of these could have been predicted readily from sequence analysis. This exciting development helps to resolve the long-standing puzzle that bacteria seemed to have many more enzymes involved in c-di-GMP synthesis and breakdown than proteins responding to the actions of these enzymes, a situation akin to a dysfunctional army that has many more officers giving orders than soldiers executing these orders. In retrospect, the existence of diverse c-di-GMP receptor types could have been expected. In this regard, it is worth recalling that there are numerous ways through which proteins bind cAMP. Why would there not be as many or even more ways to bind c-di-GMP, a second messenger that is more widespread than cAMP?
The first protein that did not fall into the predictable c-di-GMP receptor categories was the enhancer-binding protein FleQ from P. aeruginosa (168) (Fig. 2). FleQ has an N-terminal receiver domain, an AAA+/ATPase σ54-interaction domain, and a C-terminal DNA binding domain and belongs to the NtrC/DctD family of transcriptional regulators. At present, it is not yet known how FleQ binds to and responds to c-di-GMP, but its N-terminal domain is not involved in binding. FleQ is not the sole member of the NtrC family that binds c-di-GMP. Recently, VpsR of V. cholerae was also identified as a c-di-GMP receptor (169). Subsequently to FleQ, several different transcription factors were discovered that also bind c-di-GMP. Among these are the c-di-GMP-binding CRP/FNR-type transcriptional activators from Xanthomonas and Burkholderia (170–173) and the CsgD/LuxR-type transcription factors from the Vibrio species (84, 174, 175), described in detail later in this review (Fig. 2). In addition, the ability to bind c-di-GMP, albeit with low affinity, has been reported for the E. coli protein BdcA (YjgJ), a member of the short-chain oxidoreductase family (176).
New and exciting high-throughput methods of identification of c-di-GMP receptors were recently described. One of these relies on capturing c-di-GMP-binding proteins from cell extracts by using a c-di-GMP-affinity resin (177) or a pulldown procedure (178). Another is based on identification of c-di-GMP-binding proteins by using an E. coli overexpression system without the need for protein purification (179). If these methods live up to expectations, we should see a burst in newly discovered c-di-GMP receptors that may balance the soldier/officer ratios in the c-di-GMP armies of bacteria.
Cyclic di-GMP-specific riboswitches.
While the discovery of every new type of c-di-GMP protein receptor has been exciting, the discovery of a c-di-GMP-specific riboswitch (59) was an unanticipated bonus. Riboswitches are RNA aptamers, noncoding segments of mRNA, that adopt specific secondary structures and bind small molecular ligands. Upon ligand binding, the mRNA secondary structures change, which results in changes in transcription, mRNA stability, or translation of the downstream genes (180). Breaker and colleagues (59) found that c-di-GMP binds specifically to a particular class of riboswitches, GEMM, that had been identified earlier but lacked known ligands. A second type of c-di-GMP-binding riboswitch was also identified by the Breaker group, and some representatives of this class are involved in c-di-GMP-induced RNA splicing (66). It is amazing that riboswitches bind c-di-GMP in vitro with extremely high affinities, i.e., with Kd values in the nanomolar range (Fig. 5E and F). Since intracellular c-di-GMP concentrations are usually higher than this, it is not yet clear what these high affinities mean in vivo. The abundance of c-di-GMP-specific riboswitches upstream of a large number of diverse genes (59) suggests that c-di-GMP controls many as yet unappreciated functions in a variety of bacteria (see “Cyclic di-GMP and RNA”).
CYCLIC di-GMP IN GENOMIC CONTEXT
Cyclic di-GMP Signaling Enzymes in Microbial Genomes
Following early genomic analyses predicting that GGDEF, EAL, and HD-GYP domains are involved in c-di-GMP metabolism in a variety of bacteria (27, 34, 107), experimental evidence for c-di-GMP signaling pathways was obtained for the major phylogenetic branches, including the Proteobacteria, Spirochetes, Cyanobacteria, Deinococcus-Thermus, Thermotogae, Actinobacteria, and Firmicutes (59, 181, 182). In the past several years, genome sequencing has revealed domains associated with c-di-GMP signaling even in the phyla previously thought to be devoid of c-di-GMP. For example, the first ∼20 genomes of the Mollicutes, sequenced between 1995 and 2007, did not encode GGDEF, EAL, or HD-GYP domains. However, the slightly larger genome of Acholeplasma laidlawii, sequenced at the end of 2007, was found to encode as many as 15 GGDEF/EAL proteins. The recently completed genome of Simkania negevensis is the first genome of a Chlamydia organism to encode a GGDEF domain, albeit a degenerate one that is likely devoid of DGC activity. With that exception, GGDEF, EAL, or HD-GYP domain-containing proteins have been found in representatives of all major bacterial phyla that have at least one completely sequenced genome. It is curious that the number of bacterial species containing GGDEF domains is far greater than the number of species containing the structurally similar adenylate and guanylate cyclase catalytic domain (PF00211 in the Pfam database [116]) (110, 111) and exceeds the number of bacterial species carrying any type of adenylate cyclase (141). Therefore, c-di-GMP appears to be a much more common second messenger in bacteria than its once more famous cousin, cAMP.
The distribution of c-di-GMP signaling among members of each particular phylum is usually skewed in such a way that free-living bacteria with complex environmental lifestyles carry far more c-di-GMP-metabolizing enzymes than obligate parasites do (141). The reductive evolution of c-di-GMP signaling pathways was recently nicely demonstrated by Bobrov et al. (183), who showed degeneration of the genes involved in c-di-GMP synthesis and hydrolysis in the plague-causing obligate pathogen Y. pestis but preservation of these genes in its close relative, Yersinia pseudotuberculosis, which can live not only in its hosts but also in the environment.
For reasons that we still do not completely understand, c-di-GMP-metabolizing enzymes do not seem to be encoded by archaea. The only GGDEF (and HD-GYP)-containing protein in archaea is encoded in the genome of the uncultured methanogenic archaeon Methanocella arvoryzae MRE50 (http://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html), so there is a possibility of bacterial contamination. It may well be that archaea using c-di-GMP signaling pathways have unusual niches and have yet to be discovered.
The distribution of c-di-GMP among eukaryotes is more complex. The current databases list dozens of GGDEF and EAL domain proteins encoded in plants (poplar and castor bean) and lower eukaryotes, such as hydra, sea anemone, Dictyostelium, and Trichoplax. One such protein, a PleD-like DGC from the social amoeba Dictyostelium discoideum, has been recently characterized, providing the first evidence for the role of c-di-GMP as a developmental regulator in lower eukaryotes (183a). Dictyostelium uses c-di-GMP as an extracellular signal to regulate the development of the stalk subsequently progressing into a multicellular spore-forming fruiting body. Surprisingly, while four different dictyostelia each carry a single DGC gene, one species, Dictyostelium fasciculatum, has 13 paralogous DGC genes; a c-di-GMP-specific PDE has not yet been identified. The functions of plant GGDEF and EAL domain proteins, if any, remain unknown; at least some of them appear to represent bacterial contamination. In any case, no GGDEF/EAL/HD-GYP domain-encoding genes seem to be present in mammals. Moreover, mammalian cells appear to monitor cytoplasmic c-di-GMP, perceived as a sign of bacterial infection, and to launch an innate immune response to counter the infection (see Practical Aspects of c-di-GMP).
The presence of c-di-GMP-metabolizing enzymes in numerous representatives of such early-diverging branches of bacteria as the Thermotogae, Deinococcus-Thermus, Cyanobacteria, Aquificae, and Chloroflexi (Table 1) strongly suggests that c-di-GMP was adapted as a secondary messenger at the early stages of bacterial evolution (42). Cyclic di-GMP signaling has been nearly lost in some lineages, often as a result of genome compaction during adaptation to obligate parasitism (e.g., in Chlamydia and Mycoplasma), but has been preserved in other lineages (e.g., Firmicutes and Chlorobi) and dramatically expanded in such phyla as the Proteobacteria, Acidobacteria, and Spirochetes. The emergence of two distinct classes of c-di-GMP hydrolases, based on EAL and HD-GYP domains (prevalent in the Proteobacteria and Thermotogae, respectively) (Table 1), may have contributed to diversification of the c-di-GMP networks.
Regulation by Sensory Domains
As noted in Historical Perspective, the first indications that c-di-GMP was part of the cellular signal transduction machinery came from the association of the GGDEF and EAL domains with the oxygen-sensing PAS domain in DGCs and PDE of G. xylinus (25, 35) and with the receiver (REC) domain of two-component signal transduction systems in PleD and CelR2 (24, 28, 33). It turns out that cytoplasmic proteins combining GGDEF, EAL, and HD-GYP domains with REC, PAS, and/or GAF domains show by far the most common domain architectures of c-di-GMP-metabolizing enzymes (Table 3). In terms of signal transduction, this association implies modulation of the enzymatic activity of the GGDEF, EAL, and HD-GYP domains by the upstream domains. For PAS- and GAF-linked domains, such activity modulation would occur because of the bound ligands, e.g., heme, flavin mononucleotide, flavin adenine dinucleotide, and various chromophores. These domains enable proteins to sense O2, NO, CO, the redox state of the electron transport chain components, light quorum-sensing molecules, and a variety of other signals (106, 184, 184a). In addition to PAS and GAF domains, other, more signal-specific sensory protein domains, e.g., globin domains involved in O2 sensing, are often bound to GGDEF and EAL domains (Table 3). Proteins containing such sensory domains monitor cytoplasmic levels of their respective ligands and respond by altering the synthesis or hydrolysis of c-di-GMP. Several DGCs and PDEs that sense O2 (35, 89, 91), NO (185), the redox state (130), and light (54, 63, 186, 187) have been characterized (Table 3). Note that stand-alone sensors, often encoded by neighboring genes, may also interact with DGCs and PDEs (188, 189).
The signaling proteins combining GGDEF and/or EAL or HD-GYP domains with the REC domain are response regulators of two-component signal transduction systems, modulating c-di-GMP levels in response to extracellular or intracellular signals received by their cognate sensor His kinases. According to the Response Regulator Census (http://www.ncbi.nlm.nih.gov/Complete_Genomes/RRcensus.html), proteins with an REC-GGDEF or REC-REC-GGDEF domain architecture (assigned to the WspR and PleD families, respectively) account for ∼2.3% of all response regulators encoded in bacterial genomes (104). Response regulators of the PvrR family (REC-EAL domain architecture) comprise 0.4% of all response regulators; proteins containing both GGDEF and EAL output domains (FimX family) add another 1.1%, whereas proteins with the REC–HD-GYP domain architecture (RpfG family) comprise ∼1.6% of all response regulators. Thus, c-di-GMP-metabolizing domains are found in at least 5.4% of all bacterial response regulators, making them a major constituent of the two-component signal transduction machinery.
Another important group of signaling proteins consists of membrane-bound sensors that combine cytoplasmic GGDEF, EAL, or HD-GYP domains with periplasmic (or extracellular) sensory domains, connecting them via one or more membrane-spanning fragments. In most cases, the ligand specificity of the periplasmic domains remains unknown, and they are recognized as sensors based on their presence in other signaling proteins, such as histidine kinases, adenylate cyclases, or MCP proteins (141, 190, 191).
In some cases, c-di-GMP-metabolizing enzymes are regulated by dynamic protein-protein interactions with other c-di-GMP-metabolizing enzymes or sensory domains. For example, in X. campestris and Xanthomonas axonopodis, not only do some GGDEF domain DGCs specifically interact with the HD-GYP domain PDE, RpfG, but these interactions proved important for regulating a subset of downstream processes (98, 192). The periplasmic protein YfiR (PA1121) from P. aeruginosa inhibits the activity of the DGC TpbB (YfiN; PA1120), which prevents the formation of small-colony variants of this species (193). The H-NOX domain, a selective NO sensor, interacts as a free-standing protein with DGCs to affect c-di-GMP metabolism (188, 189). The occurrence of H-NOX domains adjacent to c-di-GMP-metabolizing proteins in other bacteria suggests that modulation of biofilm formation by NO through c-di-GMP signaling is common.
PHYSIOLOGY AND MECHANISMS OF CYCLIC di-GMP SIGNALING
Scope of c-di-GMP Signaling
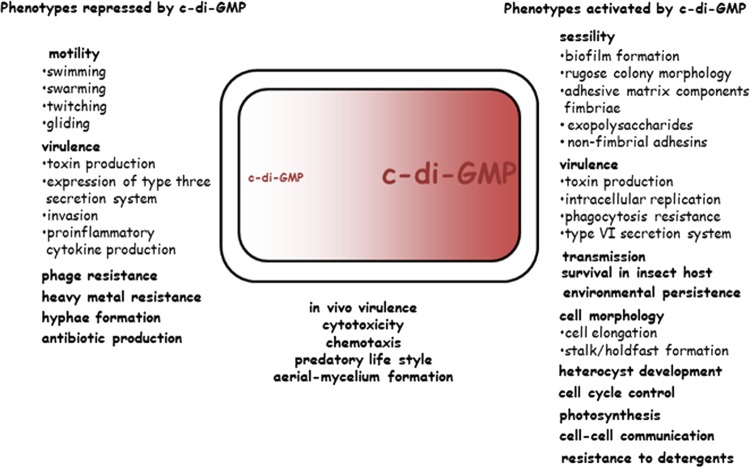
When c-di-GMP appeared above the horizon as a potential novel second messenger, the first phenotypes associated with c-di-GMP signaling were cell differentiation in C. crescentus and biofilm formation in G. xylinus, Salmonella enterica, V. cholerae, and P. aeruginosa (2, 3, 5, 37, 39, 41). A role for c-di-GMP in V. cholerae virulence was identified soon thereafter (40). Since then, not only has the list of bacteria relying on c-di-GMP signaling grown immensely, but so has the range of phenotypes affected by c-di-GMP. This range now includes such diverse phenomena as survival and transmission of obligate intracellular pathogens from the Proteobacteria and Spirochetes in insect and mammalian hosts (183, 194), predatory behavior of a bacterial killer (153), heterocyst formation in cyanobacteria (195), multicellular development and antibiotic production in streptomycetes (196), and long-term nutritional stress survival and lipid metabolism and transport in mycobacteria (137, 196a) (Fig. 6).
Fig 6.
Phenotypes that are regulated by cyclic di-GMP signaling. On the left are target outputs activated by low c-di-GMP or repressed by high c-di-GMP (require the absence of c-di-GMP binding to the cognate receptor for expression), and on the right are target outputs activated by high c-di-GMP (require c-di-GMP binding to the cognate receptor for expression). Some processes can be repressed and activated by c-di-GMP, depending on the bacterial species and conditions. (Adapted from reference 12 with permission of the publisher. Copyright © 2009 Karger Publishers, Basel, Switzerland.)
A key role of c-di-GMP in the transition between motile and sessile lifestyles of the Gram-negative bacteria G. xylinus, S. enterica, E. coli, and P. aeruginosa was appreciated early on (41), and the molecular mechanisms of this regulation are beginning to emerge (9, 155, 156, 197, 198). Importantly, the c-di-GMP-dependent motility-to-sessility transition occurs not only when a swimming cell approaches a surface for settling down but also upon building of three-dimensional (3D) biofilms and during biofilm dispersal. In addition to swimming, c-di-GMP regulates swarming, twitching, and gliding motility on colonized surfaces (174, 199–201), not only in diverse Proteobacteria but also in the Spirochetes, Firmicutes, and Cyanobacteria (128, 187, 202). Therefore, the motility-to-sessility transition is a very common, possibly universal phenomenon controlled by c-di-GMP.
Some pathogens do not seem to depend on c-di-GMP signaling pathways for acute infections (7, 183, 203), whereas other human, animal, and plant pathogens do (204, 205). It is becoming apparent that virulence may be both promoted and inhibited by c-di-GMP, depending on the pathogen, stage of infection, infection route, and other factors. How and when c-di-GMP signaling pathways affect acute infections is still poorly understood, in part because we do not have the means to observe or modulate bacteria inside their hosts during infection with sufficient spatiotemporal resolution. In contrast to acute infections, the vast majority of chronic infections involve biofilms, which are often associated with elevated c-di-GMP levels in bacterial pathogens (97, 206).
It has been noticed previously (5) that the number of phenotypes regulated by c-di-GMP appears low compared to the multitude of enzymes involved in c-di-GMP synthesis and breakdown. This apparent paradox has been partially demystified by the realization that individual biofilm components often involve specific, not necessarily overlapping, c-di-GMP signaling circuits. Furthermore, studies of c-di-GMP receptors that act as global transcriptional regulators (171, 207) helped to uncover a plethora of new processes directly or indirectly controlled by c-di-GMP. In addition, the c-di-GMP-dependent riboswitches present in various bacteria may regulate the expression of many unexpected target genes (59, 66) (Fig. 6).
In spite of the recent burst in information, our knowledge about c-di-GMP signaling pathways remains fragmentary. Only a few complete c-di-GMP signaling circuits have been described. More often than not, we know only of isolated components of c-di-GMP signaling networks, such as enzymes or receptors, but molecular mechanisms of regulation, signals affecting specific c-di-GMP-dependent circuits, or even targets of c-di-GMP action are often missing. Consequently, there is still a lot to discover in the field of c-di-GMP signaling (see Concluding Remarks and Perspectives).
Motility-to-Sessility Transition
The vast majority of bacteria spend a significant amount of time growing attached to surfaces of abiotic or biotic origin. The transition of single motile bacterial cells to a surface-attached, sessile state represents a drastic lifestyle change, which is commonly associated with subsequent multicellular growth as a colony and/or biofilm. The motility-to-sessility transition involves several stages. First, a cell needs to reach the surface to make a temporary contact and then permanently attach using its surface-adhesive components. Ultimately, if it is motile, the cell needs to inhibit the motility that helped it to reach the surface (208). Several mechanisms are involved in accomplishing this transition, some of which involve c-di-GMP.
We first consider how flagellated bacteria undergo the motility-to-sessility transition. One of the challenges of a motility-to-sessility transition for bacteria that swim using flagella is that they need rotating flagella not only to come into proximity with the surface but also to overcome surface repulsion at the liquid-surface interface. Therefore, flagella are expected to keep rotating until the surface contact is made. Furthermore, flagella themselves often serve as organelles involved in initial, temporary attachment (209, 210). However, flagella are responsive to chemotactic signals. For a cell committed to surface attachment, this may be problematic because it increases the chances of the cell swimming away from the surface following the initial contact. Therefore, bacteria might benefit from a mechanism that temporarily disengages flagella from chemotactic inputs during surface attachment. Such a mechanism would be expected to regulate flagella quickly, on the scale of seconds; thus, it should operate at the posttranslational level. One such mechanism that operates in enteric bacteria has been described at the mechanistic level. The molecular underpinnings in other bacteria are only beginning to emerge. Once cells are permanently attached, a second, slower mechanism may be involved in turning off rotating flagella and/or turning off flagellum synthesis. The slower mechanisms of c-di-GMP-dependent regulation of flagellar motility are also beginning to emerge.
YcgR, the c-di-GMP receptor of enteric bacteria.
E. coli and S. enterica have peritrichous flagella that rotate in either a counterclockwise (CCW) or clockwise (CW) direction. The CCW-rotating flagella form a bundle that propels the cell forward. Chemotactic signals result in changes in rotation direction of one or several flagella, from the default CCW to the CW direction, which results in flagellar bundle disassembly and cell tumbling. Ultimately, the CCW rotation resumes and propels the cell in a new, random direction. The protein delivering chemotaxis signals to the flagellum is the phosphorylated response regulator CheY (CheY∼P), which binds to the flagellum rotor subunit FliM to induce CW rotation (reviewed in references 211 and 212). The flagellar rotor, also known as a switch complex, is an approximately 4-MDa complex located at the cytoplasmic side of the flagellum. It is made of three subunits, FliG, FliM, and FliN, with the FliG subunit being critical for converting the transmembrane proton flow through MotBA to the torque that drives rotation of the flagellum body. FliG is also critical for setting the rotation direction, which is defined by its interactions with the FliM subunits.
Girgis et al. first noticed that elevated levels of intracellular c-di-GMP, brought about by deletion of the major c-di-GMP PDE of E. coli, YhjH, induced a strong CCW bias in flagellar rotation (213). The CCW bias promoted smooth swimming and inhibited bacterial spreading in semisolid agar, apparently because cells unable to switch swimming direction became trapped in the blind alleys of the semisolid medium (214). The c-di-GMP receptor relaying intracellular c-di-GMP levels to flagella was identified earlier as YcgR. The inactivation of ycgR in the yhjH mutant largely restored motility in semisolid agar (52, 213, 215). Römling and Amikam (6) and Wolfe and Visick (16) proposed that YcgR may deliver the c-di-GMP signal by direct interactions with the flagellum motor.
Two models have subsequently emerged to explain the YcgR–c-di-GMP-mediated effect. According to one model derived from studies of S. enterica and E. coli (155, 156), YcgR controls the direction of flagellar rotation by binding to the FliG and FliM subunits of the flagellum switch complex FliGMN. This primary binding is relatively weak and does not require c-di-GMP, but it ensures concentration of YcgR at the flagellum sites, which may facilitate a fast response to increased c-di-GMP levels. Upon such an increase, the interaction between YcgR–c-di-GMP and the flagellar switch complex is strengthened, which stabilizes the CCW conformation of the complex (155, 156). Cyclic di-GMP binding is known to result in drastic conformational changes in YcgR, i.e., the N-terminal domain is brought in close proximity to the C-terminal PilZ domain (52). In addition to the CCW bias, an ∼30% slowdown in flagellar rotation at high c-di-GMP levels was observed in S. enterica when YcgR was overexpressed (156). The block imposed by YcgR–c-di-GMP on the reversal of flagellar rotation to the CW direction has been named a “backstop brake” (156).
The second model (154) is focused on the c-di-GMP-induced deceleration of flagellar rotation and does not involve the CCW rotation bias. According to this model, instead of interacting with the rotor, YcgR–c-di-GMP interacts with the MotA subunit of the stator. By interfering with the stator-rotor energy transfer, YcgR–c-di-GMP slows down the rotating flagellum, i.e., it acts as a brake. A correlation between flagellum rotation velocity and intracellular c-di-GMP levels was recorded. This model is supported by genetic and fluorescence resonance energy transfer (FRET) data, whereas the former model (155, 156) also includes protein-protein interaction analyses. Additional experimentation to probe for the MotA-YcgR interaction directly would be helpful to verify YcgR interactions with the stator. However, the relatively modest reduction in rotation velocity (∼40% decrease) (154) in itself may not be sufficient to account for the drastic inhibition of swimming in semisolid agar of the ΔyhjH strains. Therefore, the c-di-GMP-induced CCW bias that can be set only by YcgR interactions with the rotor likely plays an essential role.
It has been proposed (155) that smooth swimming may decrease the distraction of chemotactic signals and set bacteria on a “crash course” with a surface, which promotes the transition to the sessile lifestyle. In semisolid media, which are typical for enteric bacteria (e.g., animal digestive tracts and fecal material), this may be a particularly effective strategy for settling down (214). Some deceleration in swimming velocity could be meaningful for surface attachment, although a drastic slowdown may impair the cell's ability to reach the surface, overcome surface tension, and initiate temporary attachment. Cells containing nonrotating flagella poorly colonize abiotic surfaces (216).
YcgR is a motility-specific c-di-GMP receptor. Expression of the ycgR gene, as well as the gene encoding the c-di-GMP-specific PDE, YhjH, is under the control of the FlhD4C2 flagellum master regulator (215). Several DGCs have been shown to contribute c-di-GMP to YcgR, suggesting that a variety of signals affect the motility-to-sessility transition (154, 197). Furthermore, YcgR–c-di-GMP has been proposed to function outside the motility-to-sessility transition, e.g., in adjusting swimming velocity to the energy status of liquid-grown cells during the exponential-to-stationary-phase transition (154).
Despite its key role in c-di-GMP-dependent motility regulation in enteric bacteria, YcgR is only part of the c-di-GMP-mediated regulation of motility, as judged by the fact that the ycgR deletion in yhjH mutants does not fully restore the size of the swim zone to wild-type levels (155). Motility, however, is fully restored by deletion of ycgR and the cellulose synthase gene bcsA (216a) whose product binds c-di-GMP via the C-terminal PilZ domain (52). Thereby, extracellular cellulose directly inhibits flagellar rotation. These findings provide clues as to how flagella of enteric bacteria stop rotating following cell attachment. Interestingly, a glycosyltransferase that inhibits motility and promotes exopolysaccharide biosynthesis in a c-di-GMP-independent way was recently discovered in B. subtilis (217). In this case, however, polysaccharide biosynthesis and motility inhibition could be genetically separated.
The YcgR–c-di-GMP-dependent regulation of flagellar function is not limited to E. coli and S. enterica but apparently is characteristic of other members of the Gamma- and Betaproteobacteria that express YcgR homologs (155). A much more distant homolog of YcgR from B. subtilis, YpfA, was also found to inhibit motility through its interaction with the flagellar motor protein MotA that interferes with flagellar rotation (217a). Further, PilZ domain c-di-GMP receptors unrelated to YcgR are also involved in regulating flagellar motility, albeit via different mechanisms. For example, C. crescentus DgrA and, to a lesser extent, DgrB have been shown to inhibit flagellar function (51). dgrA overexpression was reported to decrease the level of FliL, a protein required for flagellar rotation in C. crescentus. The mechanistic details of the DgrA-FliL connection have not yet been uncovered.
Cyclic di-GMP regulation of chemotaxis: an emerging theme.
One can envision that variations of the mechanism involving c-di-GMP-dependent flagellar rotation regulation may exist. For example, induction of the CCW bias can be achieved by modulating the chemotaxis machinery itself, e.g., by depleting the pool of CheY∼P by either decreasing CheY phosphorylation or increasing CheY∼P dephosphorylation. Both types of c-di-GMP-dependent motility regulation have now been documented.
The PilZ domain c-di-GMP receptor PlzA from the spirochete B. burgdorferi, which is propelled by a single periplasmic flagellum, has been proposed to bind to the CheY∼P phosphatase, CheX (128). Activation of CheX by PlzA–c-di-GMP would be expected to lower CheY∼P levels and promote smooth swimming. The details of this mechanism, which is thus far supported only by genetic data, have yet to emerge.
In the alphaproteobacterium Azospirillum brasilense, the MCP Tlp1 plays a key role in energy taxis and colonization of plant roots (218). Tlp1 contains a C-terminal PilZ domain (48) that binds c-di-GMP with low-micromolar affinity (218a). c-di-GMP binding to Tlp1 promotes smooth swimming as well as swimming velocity. Changing c-di-GMP levels serve as an intracellular cue that allows A. brasilense cells to optimize their search for microaerobic environmental niches optimal for energy generation. According to bioinformatic analysis, a number of proteobacteria contain MCP-PilZ fusions, and therefore c-di-GMP-dependent control of chemotaxis may not be limited to A. brasilense (218a). Given that c-di-GMP receptors may affect the chemotaxis machinery without being fused to chemotaxis proteins (128), the c-di-GMP–chemotaxis theme is likely poised for expansion.
It is noteworthy that in the cases described above, increased c-di-GMP levels promote (smooth) swimming instead of inhibiting swimming, which is expected from the current paradigm that associates high c-di-GMP levels with sessility. Two considerations may help in resolving this apparent contradiction. One is that c-di-GMP-dependent motility regulation does not always involve the motility-to-sessility transition (154, 218a). Second, a distinction needs to be made between transient increases and decreases in c-di-GMP levels and more permanent changes in intracellular c-di-GMP concentrations. While the example of A. brasilense nicely demonstrates that transient c-di-GMP changes do not promote immediate sessility, more permanently elevated c-di-GMP levels (e.g., as observed in PDE mutants) do promote sessile lifestyles, both in A. brasilense that forms cell aggregates (218a) and in enteric bacteria (52, 155, 213). There is a need to go beyond the static view of intracellular c-di-GMP levels as the sole deterministic factor and to consider the highly dynamic nature of c-di-GMP signaling, which we are just beginning to appreciate (219).
Cyclic di-GMP-dependent transcriptional regulation of flagellar genes.
In addition to the fast regulation of flagellar behavior by c-di-GMP, the cells already attached to a surface need to inhibit flagellar motility and/or block synthesis of new flagella, and these processes depend on c-di-GMP, at least in some bacteria. Several different c-di-GMP-dependent transcriptional regulators of motility genes have evolved. Most of these appear to be engaged in inverse regulation of flagellar genes and genes involved in biosynthesis of polysaccharides, adhesive pili, and/or protein adhesins, i.e., genes characteristic of the surface-attached biofilm lifestyle.
The first factor regulating flagellar gene expression in a c-di-GMP-dependent manner, FleQ, was identified in P. aeruginosa by Hickman and Harwood (168). FleQ controls expression of the polysaccharide pel biosynthesis genes involved in biofilm formation, as well as the flagellar regulon (220). The molecular understanding of FleQ's action is beginning to emerge. The c-di-GMP-dependent binding of FleQ to the pel operon promoter has been demonstrated; however, regulation of the flagellar genes by c-di-GMP does not seem to be mediated by FleQ, the master regulator of flagellar gene expression (221).
In addition to P. aeruginosa FleQ, two unrelated transcription factors from proteobacteria were recently shown to inhibit flagellar gene expression in a c-di-GMP-dependent manner. In Xanthomonas, the CRP-type transcriptional regulators (Clp proteins) have evolved to bind c-di-GMP. DNA binding by Clp is abolished in the presence of c-di-GMP (170, 173, 222). A clp mutation has been reported to result in lower expression of the flagellin fliC gene, likely via an indirect mechanism (223). Consistent with the inverse regulation of biofilm and motility genes, V. cholerae VspT, yet another c-di-GMP-dependent transcription factor, has been shown to repress flagellar genes. A genomewide transcriptional analysis revealed that a number of flagellar genes are upregulated in a vspT mutant and that the mutant migrates better than the wild type in semisolid agar (84).
Interestingly, the above-described control via various c-di-GMP-dependent transcription factors is modest, i.e., only 1.5- to 3-fold downregulation of flagellar gene expression (84, 220, 223). Such modest downregulation may not be sufficient to account for the nonrotating or absent flagella in biofilm-grown cells, for which experimental evidence is plentiful (16, 60, 224, 225). Therefore, additional molecular mechanisms of motility control involved in the motility-to-sessility transition remain to be uncovered.
Thus far, no c-di-GMP-responsive transcription factor has been found in enteric bacteria. However, the “retired,” enzymatically inactive, and non-c-di-GMP-binding EAL domain protein YdiV has been shown to act as an anti-transcriptional activator of the master flagellar regulator FlhD4C2 (226, 227).
Cyclic di-GMP-dependent control of motility-to-sessility transition on surfaces.
Some bacteria utilize the same flagellar apparatuses for swimming in liquid or semisolid media and swarming on wet surfaces, while others use specialized flagella for swarming or engage flagellum-independent motility modes. For those bugs that use the same flagella for both swimming and swarming, the association of high c-di-GMP levels with sessility and low c-di-GMP levels with surface swarming appears to hold (52).
The O'Toole group identified several critical pieces of the c-di-GMP signaling module involved in swarming motility regulation in P. aeruginosa PA14. The DGC SadC (PA4332), the PDE BifA (PA4367), and the protein of unknown function SadB (PA5346) act upstream of the chemotaxis-like system that affects the frequency of directional reversals of swarming cells (201, 228–230). sadC and sadB mutations promote surface swarming by increasing the frequency with which cells change their swarming direction, whereas a bifA mutation apparently results in decreased directional reversal. Note that while the reversal frequency is changed, no change in swarming speed has been detected. In P. aeruginosa PA14, a chemotaxis-like pathway has been identified as a mediator of swarming behavior. This situation is reminiscent of the regulation of directional reversals in the enteric bacteria and A. brasilense. It is tempting to speculate that a component of this chemotaxis pathway is regulated by c-di-GMP synthesized by SadC and degraded by BifA.
McCarter and colleagues have begun to unravel molecular details of the c-di-GMP-based control of surface swarming in Vibrio parahaemolyticus (174). This gammaproteobacterium lives in the sea and occasionally acts as an opportunistic seafood-borne human pathogen (231). A decrease in c-di-GMP levels activates and an increase in c-di-GMP levels lowers expression of laf genes that encode lateral flagella involved in swarming. The transcription factor controlling laf gene expression has not yet been identified. However, it is known that at the top of the regulatory hierarchy stands the ScrABC quorum sensing system. ScrA, a predicted pyridoxal-dependent aminotransferase, is involved in synthesis of a novel autoinducer. ScrB, a predicted periplasmic protein, is believed to bind the autoinducer. ScrC is a bifunctional enzyme possessing both DGC and PDE activities. The ScrB-autoinducer complex, expected to be present at high cell densities, interacts with the large periplasmic domain of ScrC and switches ScrC from the DGC mode to the PDE mode (136). The ScrC-mediated decrease in c-di-GMP levels is likely sensed by one or more of the V. parahaemolyticus transcription factors, i.e., CpsC, CpsR, or VP2710, predicted to bind c-di-GMP and involved in laf expression regulation and swarming (232). One can envision that on the surface of a shellfish densely populated by V. parahaemolyticus, bacteria would sense overcrowding via high autoinducer levels, which would decrease intracellular c-di-GMP and promote lateral flagellation, thus allowing bacteria to swarm away from the colony and expand the colonized surface. Spreading over the surface may be a more sensible strategy than swimming away into the dangerous surroundings of an open sea.
Cyclic di-GMP-dependent control of nonflagellar motility.
Beyond flagellum-mediated swarming, type IV pilus-mediated twitching motility is also regulated by c-di-GMP. This mechanism is discussed later in this review. Here, we briefly mention the recently discovered regulation of these two surface-stimulated motility modes by c-di-GMP in P. aeruginosa (232a). Three proteins involved in pilus biogenesis elevate the levels of c-di-GMP provided by the DGC SadC and degraded by the PDE BifA and thereby inhibit swarming motility. These findings are consistent with the notion that twitching motility requires c-di-GMP for pilus polymerization, while swarming motility is only inhibited by c-di-GMP.
Less is known about c-di-GMP-dependent regulation of gliding motility, but in those species that rely on gliding, this type of motility also appears to be controlled by c-di-GMP. In B. bacteriovorus, cell gliding is required for the escape of progeny of the bacterial predator from the exhausted prey and for preying on surfaces. Hobley et al. recently showed that a mutation in one of three active DGCs of B. bacteriovorus, DgcA (Bd0367), impairs gliding motility (153). In another social predator of bacteria, Myxococcus xanthus, increased c-di-GMP levels have been observed to inhibit surface gliding (Wall and Gomelsky, unpublished data). At present, mechanistic insights into the regulation of these processes is lacking.
Regulation of Biofilms
The bottom-line message of numerous studies on the role of c-di-GMP signaling pathways in biofilm formation is that c-di-GMP promotes biofilms (38, 41, 49, 201, 204, 234–239). This view holds true for various biofilm models, e.g., pellicles at the air-surface interface; rugose, wrinkled, or rdar (red, dry, and rough) colony morphotypes on agar plates (240–242); and bacterial adhesion to abiotic surfaces under steady-state or continuous-flow conditions. In P. aeruginosa, mucoid colonies and small-colony variants commonly isolated from the airways of cystic fibrosis patients are associated with elevated biofilm formation and c-di-GMP levels (97, 206, 243–245).
All extracellular matrix components known to contribute to biofilm formation, including diverse exopolysaccharides, adhesive pili, and nonfimbrial adhesins, as well as extracellular DNA, can be regulated by c-di-GMP (13). Biofilm-related targets can be controlled by c-di-GMP on the transcriptional, posttranscriptional, and posttranslational levels. Below, we describe the current status of knowledge of c-di-GMP's involvement in biofilm formation.
Cellulose biosynthesis as a c-di-GMP target.
The capacity for cellulose biosynthesis is present in many bacteria from diverse branches of the phylogenetic tree, such as Thermotogae, Proteobacteria, and Cyanobacteria (246). Cellulose is a common component of environmental bacterial biofilms (247–250) and a component of interkingdom biofilms, i.e., bacteria attached to plants, fungi, and human intestinal cells (251–254). One may recall that cellulose biosynthesis in the fruit-rotting bacterium G. xylinus was the first process shown to be regulated by c-di-GMP (1).
Bacterial cellulose synthases contain a c-di-GMP-binding PilZ domain at the C terminus, suggesting a common allosteric regulatory mechanism. Indeed, c-di-GMP binds to the PilZ domain of the G. xylinus cellulose synthase (52), and in vitro cellulose biosyntheses using membrane fractions of G. xylinus and E. coli require only the substrate, UDP-glucose, c-di-GMP, and no other cytoplasmic components (1, 253). Since the cellulose biosynthesis operon is expressed constitutively in G. xylinus, E. coli, and S. enterica (250, 255, 256), this posttranslational regulation is possibly the major mechanism of cellulose biosynthesis activation in these organisms.
In G. xylinus, three highly similar, albeit nonidentical, DGCs and PDEs affect cellulose expression simultaneously (25). The DGCs monitor oxygen levels via the heme-containing PAS domains (35), while PDEs monitor the cellular redox state via the FAD-containing PAS domains (130). In S. enterica and E. coli, a single DGC feeds in c-di-GMP for activation of cellulose biosynthesis under standard laboratory conditions (30, 257, 258) (Fig. 7). However, under different growth conditions or upon binding of an IgA antibody, which protects mice against Salmonella infection, additional DGCs may contribute (30, 259, 259a, 461).
Fig 7.
Regulation of exopolysaccharides by c-di-GMP signaling. (A) Bacterial cellulose biosynthesis is positively regulated by c-di-GMP signaling on the posttranslational level, as the cellulose synthase BcsA contains a PilZ domain at its C-terminal end which binds c-di-GMP (1, 52). For example, in S. enterica, the DGC AdrA provides the c-di-GMP to activate cellulose biosynthesis (30). The DGC WspR regulates transcription of cellulose biosynthesis operons upon constitutive activation in P. fluorescens (261). (B) Alginate polymerization by the alginate synthase Alg8 requires activation by the c-di-GMP receptor protein Alg44 (149). (C) The PelD protein is the c-di-GMP receptor that activates biosynthesis of the exopolysaccharide PelD on the posttranslational level, with regulation of the c-di-GMP pool through the DGCs RoeA (PA1107) and SadC (PA4332) and the PDE BifA (PA4367) (229, 230). Repression of pel transcription by the transcriptional regulator FleQ is relieved upon c-di-GMP binding with the DGCs WspR (PA3702) and YfiN (PA1120), providing the c-di-GMP (93, 168). See the text for further details.
In species where cellulose biosynthesis is not expressed under laboratory conditions, transcription of the cellulose biosynthesis operon usually requires c-di-GMP. In the plant growth-promoting strain P. fluorescens SBW25, the wss operon, encoding the biosynthesis genes for acetyl-substituted cellulose (95), is expressed only on plant surfaces (260). However, constitutive activation of the DGC WspR can stimulate wss operon transcription with subsequent cellulose production under laboratory conditions (261). Similarly, in Rhizobium spp. and K. pneumoniae, where cellulose biosynthesis is silent under laboratory conditions, c-di-GMP-dependent up-mutations resulting in cellulose synthesis can be isolated (29, 262). Importantly, for most species, neither complete c-di-GMP circuits for regulation of cellulose biosynthesis have been identified nor signals activating DGCs and PDEs are known. Furthermore, we do not understand what exactly happens when c-di-GMP binds to the PilZ domain of the glycosyltransferase subunit of cellulose synthase. Does it affect the reaction rate or secretion of the nascent polysaccharide chain across the cytoplasmic membrane? The recently solved cellulose synthase structure (PDB 4HG6) suggests that c-di-GMP binding to the PilZ domain in the vicinity of the active site induces a conformational change which allows access by the substrate UDP-glucose (262a).
PAG as a c-di-GMP target.
The poly-β-1,6-N-acetylglucosamine (PAG; also called PNAG or polysaccharide intercellular adhesin [PIA]) is another exopolysaccharide produced by a wide variety of Gram-positive and Gram-negative bacteria as a biofilm matrix component (263–265). In Y. pestis, E. coli, and Pectinobacterium atrosepticum, PAG biosynthesis is activated by c-di-GMP (38, 266, 267). In Y. pestis, the hmsHFRS locus encodes the structural proteins required for PAG biosynthesis. The membrane complex containing the putative glycosyltransferase HmsR interacts with the c-di-GMP PDE HmsP and the DGC HmsT (38, 268). Thus, c-di-GMP-metabolizing proteins appear to colocalize with the target. Temperature-dependent proteolysis of the DGC HmsT and other Hms proteins is known to be responsible for PAG production at 26°C but not at 37°C (269).
In E. coli K-12, deletion of csrA highly upregulates PAG-dependent biofilm formation (270). Two DGCs, DosC (YddV) and YdeH, are required for PAG-dependent biofilm formation. While DosC affects transcription of the PAG biosynthesis operon, pgaABCD, the activity of YdeH stabilizes the PAG biosynthesis protein PgaD posttranscriptionally (266, 271). The molecular mechanism of PAG activation by c-di-GMP has recently been elucidated (271a). Cyclic di-GMP binding to the inner membrane components PgaC and PgaD promotes their interaction and stimulates glycosyltransferase activity. This mechanism is an intriguing example of c-di-GMP-promoted protein-protein interactions.
Interestingly, Staphylococcus epidermidis and Staphylococcus aureus also produce PAG (232), yet PAG biosynthesis in staphylococci is not regulated by c-di-GMP. Indeed, c-di-GMP signaling does not even exist in this genus, as the only potentially functional GGDEF domain protein, GdpS, was experimentally shown to be enzymatically inactive (272).
Alginate, Pel, and Psl polysaccharides as c-di-GMP targets.
Although the genetic capacity to produce alginate and the Pel and Psl polysaccharides appears to be present in diverse bacterial species, activation of biosynthesis of these exopolysaccharides by c-di-GMP has been studied mostly in P. aeruginosa.
The Pel and Psl polysaccharides are major extracellular matrix components of nonmucoid P. aeruginosa biofilms. While the genetic capacity to produce Pel polysaccharide seems to be ubiquitous among P. aeruginosa strains, not all strains (including the well-characterized strain P. aeruginosa PA14) harbor the psl operon. Pel and Psl exopolysaccharides are positively regulated by c-di-GMP on the transcriptional level, which is readily observed upon constitutive activation of the REC-GGDEF response regulator-DGC WspR (93). Phosphorylation of WspR is carried out by the hybrid His kinase WspE, which receives a surface-derived signal from the membrane-bound MCP domain protein WspA (273).
The transcriptional regulator FleQ is a c-di-GMP receptor that, upon binding c-di-GMP, promotes pel and psl transcription (168). In the absence of c-di-GMP, FleQ forms a complex with the accessory ATP-binding FleN protein and binds to two sites up- and downstream of the pel promoter (221). This complex bends the DNA and inhibits transcription. In the presence of c-di-GMP, bending is relieved, which activates pel transcription. It is intriguing to speculate that cytoplasmic WspR clusters may be formed in proximity to the c-di-GMP receptor FleQ for efficient and localized c-di-GMP signaling (Fig. 7).
Biosynthesis of the Pel exopolysaccharide is also regulated on the posttranslational level by c-di-GMP (Fig. 7). The pel operon encodes an I-site c-di-GMP receptor, PelD, which is part of a macromolecular biosynthetic complex (164). In strain PA14, the DGC RoeA, the PDE BifA, and partially the DGC SadC are involved in the posttranslational regulation of Pel synthesis (229, 230).
Besides being stimulated by c-di-GMP, Psl exopolysaccharide elevates c-di-GMP levels in a positive feedback loop through the DGCs SiaD and SadC (273a). This mechanism also promotes the expression of other biofilm components and stimulates biofilm formation in general.
Overexpression of the Pel and Psl polysaccharides is a characteristic of some rugose small-colony variants that arise upon prolonged persistence in the lungs of cystic fibrosis patients (206, 245). In some isolates, this phenotype depends on enhanced c-di-GMP output by the DGC TpbB (YfiN), which elevates transcription of the pel operon (97, 245, 274). The activity of TpbB is repressed posttranslationally by dephosphorylation through the tyrosine phosphatase TpbA (PA3885) (274, 275) and by the negative regulator YfiR, through an unknown mechanism (245). Because YfiR and TpbA are both located in the periplasm, there exists the possibility that YfiR is involved in the regulation of the phosphorylation status of TpbB. While phosphorylation/dephosphorylation of a tyrosine residue is a well-known mechanism of controlling activities of bacterial proteins, the functionality of such a mechanism in the periplasm remains controversial.
Alginate has evoked interest mainly as the exopolysaccharide overproduced by mucoid P. aeruginosa strains adapted to long-term colonization of the cystic fibrosis lung. Alg44 (PA3542) is a PilZ domain c-di-GMP receptor required for alginate polymerization or transport (48, 149) (Fig. 7). Alg44 is localized to the inner membrane, most likely as part of the large alginate synthase complex (149, 157). The N-terminal cytoplasmic PilZ domain of Alg44 is separated from the long periplasmic C terminus by a single transmembrane domain. The molecular mechanism of activation of alginate synthesis upon c-di-GMP binding to Alg44 has not been addressed. However, the membrane fusion protein domain present in the periplasmic fragment of Alg44 suggests that it is involved in protein-protein interactions within the alginate translocation complex. Deletion of a short C-terminal sequence abolished alginate polymerization (157), suggesting that c-di-GMP binding to the cytoplasmic PilZ domain of Alg44 can be transferred via an inside-out signaling mechanism (67, 68) to the periplasmic domain to activate alginate polymerization.
The source of the c-di-GMP required for alginate production remains poorly characterized. In the alginate-overproducing P. aeruginosa strain PDO300, the putative DGC MucR (PA1727) affects alginate biosynthesis (243). This activation, however, seems to be strain specific, as MucR overexpression in a different strain, PAO1, leads to wrinkled colonies indicative of Pel or Psl polysaccharide biosynthesis but not to alginate overproduction. This observation emphasizes the need to understand specificity determinants allowing DGCs to communicate with specific c-di-GMP targets and highlights the fact that these determinants may be different even in closely related species and even strains.
In general, we poorly understand how c-di-GMP signaling pathways activate specific target exopolysaccharides. A study by Bassis and Visick (276) shed light on how this is achieved in Vibrio fischeri, a bacterium that produces two different types of biofilms, one based on cellulose and another based on the symbiosis polysaccharide Syp. The transcription factor SypG activates expression of the Syp polysaccharide genes along with the binA gene, encoding a c-di-GMP PDE (276). BinA controls the degradation of c-di-GMP that is required for cellulose biosynthesis, but it has no effect on the Syp polysaccharide. By coordinating expression of the syp and binA genes, SypG regulates which type of polysaccharide will be produced.
Pili as c-di-GMP targets.
Pili or fimbriae are the nonflagellar long filamentous appendages built of protein subunits on the outer surfaces of bacteria. The major assembly classes of fimbriae include those assembled by the chaperone/usher-dependent pathway or by the extracellular nucleation/precipitation pathway (curli fimbriae), type IV fimbriae, and conjugative fertility fimbriae (F pili). Representatives of all fimbrial classes have been associated with biofilm formation on abiotic and biotic surfaces (277–281), suggesting a potential regulation by c-di-GMP signaling. Expression of fimbriae is tightly controlled, with most fimbriae being cryptic under laboratory conditions. If they are expressed, a highly regulated expression pattern is usually observed. Consistently, negative regulation of fimbrial expression by a c-di-GMP PDE has been found for several classes of fimbriae (158, 282, 283).
Klebsiella pneumoniae is an opportunistic pathogen that frequently causes hospital-acquired urinary and respiratory tract infections associated with indwelling devices. In K. pneumoniae, type 3 fimbriae facilitate biofilm formation on abiotic surfaces and on human extracellular matrix-coated surfaces (158, 284, 285). The type 3 fimbrial cluster is activated by c-di-GMP on the transcriptional level (120, 158, 286) (Fig. 8). The DGC YfiN stimulates expression of type 3 fimbriae, while the PDE MrkJ downregulates their expression. Transcriptional activation by c-di-GMP is mediated by MrkH, the first characterized transcriptional factor containing a PilZ domain linked to an LytTR-type DNA binding domain. Upon binding of c-di-GMP, MrkH interacts with the promoter of the type 3 fimbrial cluster and turns on its expression.
Fig 8.
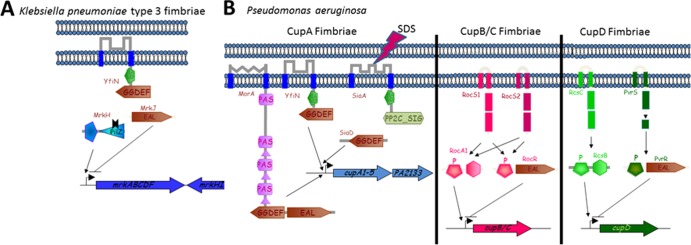
Regulation of type 3 fimbriae and Cup fimbriae by c-di-GMP signaling. (A) Type 3 fimbriae of K. pneumoniae and Cup fimbriae of P. aeruginosa are positively regulated on the transcriptional level by c-di-GMP signaling. An entire regulatory circuit of c-di-GMP signaling has been identified for transcriptional regulation of type 3 fimbriae. The transcriptional regulator MrkH binds c-di-GMP produced by the DGC YfiN and subsequently activates transcription of the type 3 fimbria mrkABCDF operon (120, 158), while the PDE MrkJ represses mrkABCDF transcription by degrading c-di-GMP (158, 286). (B) Transcription of CupA fimbriae is activated in response to detergent (sodium dodecyl sulfate [SDS]) exposure by the DGC SiaA (PA0172) (290) or in small-colony variants of P. aeruginosa by the DGCs MorA (PA2474) and YfiN (PA1120) (97). Transcription of CupB/C fimbriae is repressed by the response regulator PDE RocR (PA3947) (282), while repression of CupD fimbriae requires the response regulator PDE PvrR (283). See the text for further explanation.
The genes encoding the PDE MrkJ and the transcriptional factor MrkH are located immediately downstream of the Mrk type 3 fimbrial gene cluster and are transcribed in opposite directions. Type 3 fimbrial gene clusters are present not only in K. pneumoniae but also in various species of Enterobacteriaceae, and the EAL proteins are found downstream of several but not all of these operons (287), suggesting a similar regulation of type 3 fimbriae by c-di-GMP in some, but perhaps not all, strains.
It is noteworthy that potential EAL domain PDE genes have been found immediately downstream of the sfa and pap fimbrial operons in those E. coli strains that cause newborn meningitis and urinary tract infections, respectively (288). This finding suggests that PDE-mediated downregulation of fimbrial genes may be a common regulatory mechanism of fimbrial expression. In addition, it has recently been shown in a uropathogenic E. coli strain that the expression of type 1 fimbriae, which are commonly produced under laboratory conditions, is modulated by PDEs and DGCs (268).
Cup fimbriae as c-di-GMP targets.
In P. aeruginosa, five fimbrial gene clusters of the chaperon/usher pathway (Cup) have been identified to date and are designated cupA to -E (289). Although specific functions of these fimbriae are unknown, all of them have been shown to either alter adhesive properties of P. aeruginosa cells or contribute to biofilm formation. All Cup fimbriae, except for CupE, are regulated by c-di-GMP on the transcriptional level (Fig. 8).
Certain P. aeruginosa strains with a small-colony variant phenotype, isolated from cystic fibrosis patients, show c-di-GMP-dependent enhanced expression of CupA fimbriae through the DGCs YfiN and MorA (97). Detergent stress also leads to a c-di-GMP-dependent protective upregulation of CupA fimbriae through SiaD (PA0169) (97, 290). The cupB and cupC gene clusters are coregulated by the three-component system Roc1, which consists of one sensor kinase and two response regulators (282). The sensor kinase RocS1 balances the activities of the two response regulators, which have opposite effects on cupB and cupC gene expression. The RocR response regulator has an EAL domain and possesses PDE activity (118); it reduces cupC gene expression by an unknown mechanism. RocR can also be phosphorylated by the sensor kinase RocS2, which also contributes to cupB and cupC gene expression.
CupD fimbriae were horizontally acquired by P. aeruginosa strains through acquisition of the PAPI-1 pathogenicity island. Two distinct two-component systems, including the response regulator PvrR with an EAL PDE output, are encoded among the four genes adjacent to cupD. The products of these genes regulate the cupD fimbrial gene cluster in a similar way to that for the cupB and cupC genes (283).
Type IV pili as c-di-GMP targets.
No other pili are as diverse and ubiquitous as type IV pili (291). Type IV pili have the unique ability to polymerize and retract, thereby conferring twitching motility (292, 293). The presence of type IV pili and twitching motility has been shown to be required for biofilm formation and maturation resulting in a three-dimensional biofilm architecture (199, 210). Type IV pilus biogenesis and twitching motility in P. aeruginosa are controlled by c-di-GMP signaling. Type IV pilus biogenesis, but not gene expression, requires the GGDEF-EAL domain FimX response regulator localized at one cell pole (100). FimX senses c-di-GMP via its degenerate EAL domain (85, 133). Although the molecular mechanisms by which FimX affects type IV pilus biogenesis and twitching motility are unknown for P. aeruginosa, studies of the FimX homolog in X. axonopodis suggest that FimX interacts with a PilZ protein (Fig. 9). Similarly to FimX, PilZ is required for surface localization and assembly of pilin (146), but it does not bind c-di-GMP (149, 159). PilZ subsequently interacts with PilB, an ATPase required for type IV pilus polymerization. This cascade of protein-protein interactions likely conveys the presence of c-di-GMP to PilB. Interestingly, suppressor mutations of a P. aeruginosa fimX mutant which restored type IV pilus biogenesis and partially restored twitching motility were located in genes associated with elevated c-di-GMP levels (294). Those pili were not assembled at the cell pole, however, but produced peritrichously by the cell.
Fig 9.
Regulation of virulence, type IV pilus motility, and biofilm formation in X. campestris. The transcriptional regulator Clp controls a virulence regulon of more than 300 genes, among them engXCA, encoding endoglucanase. Cyclic di-GMP binding to Clp prevents transcription and abolishes virulence. The c-di-GMP PDEs RavR and RpfG additively create an environment low in c-di-GMP, which is required for the activity of Clp. Hypoxia and the diffusible signaling factor DSF activate the respective sensor kinases RavS and RpfC, which phosphorylate RavR and RpfG. RpfF is required for the synthesis of DSF. Besides activation of virulence, RpfG but not RavR is required for type IV pilus-mediated motility and repression of biofilm formation. PilZ domain-containing proteins XC_2249 and XC_3221 appear to function as an adaptor in the interaction of two different protein complexes with two different ATPases. RpfG in a complex with the diguanylate cyclases XC_0249 and XC_0420 recruits the XC_2249 adaptor and interacts with the PilT/PilU ATPases required for pilus retraction (295a). This interaction stimulates motility, as does XC_3221-mediated interaction of the c-di-GMP binding protein FimX with the pilus polymerization ATPase PilB. Mechanisms of RpfG suppression of biofilm formation involve inhibition of transcription of the putative exopolysaccharide operon xagABC and activation of dispersion (222).
Since a positive role for c-di-GMP in type IV pilus polymerization has been established, might pilus polymerization/retraction be affected oppositely by c-di-GMP? Such a scenario would require c-di-GMP concentrations to oscillate, with a wave period in the second scale. While the sources of c-di-GMP required for pilus biogenesis have not been identified unambiguously, mutations in the P. aeruginosa HD-GYP PDEs PA4108 and PA4781 and in Xanthomonas RpfG have been found to prevent twitching motility (127, 295). In Xanthomonas, the ATPases required for pilus retraction, PilU and PilT, interact with another PilZ protein (XC_2249) recruited by a complex of RpfG with two GGDEF domain proteins (295a). This complex, but not either GGDEF protein alone, affects type IV pilus motility (296). Paradoxically, the PilZ protein, which interacts with PilU and PilT, binds c-di-GMP, although ligand binding is not required for twitching motility (295a). Similarly, in P. aeruginosa, an additional PilZ domain protein is involved in positive regulation of type IV pilus-based twitching motility (149). Cumulatively, these findings imply molecular mechanisms of pilus protrusion and retraction regulated by fast local fluctuations of c-di-GMP levels.
Phototactic cyanobacteria show type IV pilus-dependent phototaxis toward white light, and this is inhibited by blue light. Differentiated regulation of phototaxis of Synechocystis sp. PCC 6803 to light of different wavelengths is mediated by Cph2, a complex hybrid photoreceptor with c-di-GMP-metabolizing output (187). Thereby, perception of blue light by the C-terminal light sensor stimulates the DGC activity of the adjacent GGDEF domain and inhibits twitching motility. In this case, as well as in Pseudomonas and Xanthomonas, elevated c-di-GMP levels inhibit twitching motility.
Curli fimbriae as c-di-GMP targets.
Amyloid fibers termed curli fimbriae have been studied intensively in E. coli and S. enterica, where they mediate biofilm formation at low temperatures and are also involved in colonization of host tissues (277). Curli fimbriae are directly and indirectly regulated by c-di-GMP signaling pathways. First, a panel of c-di-GMP-synthesizing and -metabolizing proteins regulate expression of the orphan response regulator CsgD on the transcriptional and posttranscriptional levels. CsgD is a key transcription factor that controls expression of the csgBAC operon, encoding the structural subunits of curli fimbriae (197, 234, 297, 298). Second, the csgBAC operon is subject to specific transcriptional regulation by a DGC and PDE pair, DosC and DosP, that is independent of csgD expression (298, 299). The DosC-DosP pair activates transcription of the csgB promoter under aerobic conditions, consistent with the enzyme activities being oppositely regulated by oxygen, but not under anaerobic conditions (299). In addition to the transcriptional component, c-di-GMP may affect curli biosynthesis posttranscriptionally (234), through mechanisms that have yet to be uncovered.
Adhesins as c-di-GMP targets.
Besides pili, nonfimbrial adhesins contribute to biofilm formation. Large multirepeat adhesins often participate in biofilm formation through cell surface adherence and as components stabilizing the extracellular matrix. In Pseudomonas putida and P. fluorescens, the large adhesive protein LapA is required for biofilm formation (300, 301). Specifically, LapA mediates the irreversible surface attachment and cell-to-cell interconnection in the biofilm, which stabilize the biofilm and prevent its dispersal. P. fluorescens Pfl0-1 LapA is transported out of the cell by the type 1 secretion system (Fig. 10). LapA can exist as a cell surface-associated form, which promotes bacterial adhesion, and an extracellular form, i.e., released into the medium. Proteolytic processing of LapA at its N terminus transforms one form into the other. Elegant studies by the O'Toole group have uncovered the molecular mechanisms regulating LapA proteolysis in a c-di-GMP-dependent manner. Specifically, at low c-di-GMP levels, LapA is proteolytically processed by the periplasmic protease LapG (68). The proteolytic activity of LapG is regulated by the transmembrane protein LapD, which binds c-di-GMP via its degenerate EAL domain (166). Upon c-di-GMP binding, LapD transmits the signal by an inside-out mechanism through its HAMP domain to the periplasmic domain (67, 68). The conformational change of the periplasmic domain, initiated by c-di-GMP binding to the EAL domain, allows LapD to sequester LapG and likely to inhibit its proteolytic activity, thus preventing proteolytic processing of LapA.
Fig 10.
Cyclic di-GMP signaling circuit controlling surface location of the P. fluorescens adhesin LapA. LapA can exist in a cell surface-associated or proteolytically processed supernatant released form. Cyclic di-GMP binding to the c-di-GMP receptor LapD (Pfl01_0131) sequesters the periplasmic protease LapG and prevents LapA cleavage (67, 68). Cyclic di-GMP dedicated to binding to LapD is produced by the DGCs GcbB (Pfl01_1789) and GcbC (Pfl01_4666), while RapA (Pfl01_1678) degrades the respective c-di-GMP (167, 302). LapEBC proteins constitute components of the type I secretion system. The PhoR-PhoB two-component system responds to alterations in environmental phosphate levels to regulate LapA's surface location (302). Low phosphate activates the response regulator PhoB, which represses the expression of the LapA type I secretion system and activates expression of the c-di-GMP PDE RapA. RapA activity favors the supernatant released form of LapA and subsequently leads to biofilm dispersal.
The LapA surface transition is regulated by a specific c-di-GMP signaling network responding to low levels of inorganic phosphate through the PhoR/PhoB two-component system, which upregulates the PDE RapA (302). This system represents a rare example where a complete signal transduction cascade has been elucidated at the molecular level, from primary signal (phosphate) sensing to surface attachment via LapA. The LapD/LapG proteolysis regulatory system is conserved in a wide variety of bacteria, some of which are pathogenic (67, 303). Interestingly, in the phytopathogen Pectobacterium atrosepticum, the expression of the LapA adhesin is regulated by a DGC and a PDE encoded downstream of the operon (115). These enzymes are not conserved in P. fluorescens, which suggests that they respond to different environmental signals. Besides LapA, RTX-like bacterial toxins have been identified as putative substrates based on predicted LapG cleavage site sequences (67, 303).
In P. aeruginosa, the CdrAB two-partner secretion system, which includes a large nonfimbrial β-helical adhesin, is positively regulated on the transcriptional level by c-di-GMP signaling (304). Upon posttranslational derepression of its DGC activity, YfiN activates transcription of cdrAB (274).
Complex regulation of biofilm formation by c-di-GMP via CsgD-like transcriptional regulators.
Regulation of biofilm formation by c-di-GMP may occur at various levels. Therefore, multiple c-di-GMP-metabolizing enzymes are often identified as being involved in biofilm formation in genetic screens (167, 197, 204, 234, 242, 297, 298). However, it is worth remembering that individual targets contributing to biofilm formation are often regulated by distinct DGC-PDE signaling circuits, different DGC-PDE signaling circuits may contribute under different conditions, and a single important target may be regulated on multiple levels (Fig. 11).
Fig 11.
Specificity and redundancy of c-di-GMP-mediated regulation of target output. The apparent redundancy of c-di-GMP signaling proteins in biofilm formation and other phenotypes might be caused by distinct molecular mechanisms. (A) More than one target is required for phenotype output, and individual targets are affected by distinct c-di-GMP signaling pathways consisting of a pair of c-di-GMP-metabolizing proteins and an effector protein. (B) One target causes the phenotype output. Cyclic di-GMP from several cyclic di-GMP-metabolizing proteins simultaneously contributes to target output through one effector. (C) Alternatively, target output is affected by c-di-GMP signaling on several levels through different effector proteins. Specific c-di-GMP-metabolizing proteins provide the c-di-GMP for each effector.
In V. cholerae, rugose colony formation is regulated by at least seven DGCs and four PDEs (161, 305–307) (Fig. 12). Expression of the Vps (Vibrio polysaccharide) is the major characteristic of the rugose colony morphology. The positive transcriptional regulators VpsR and VpsT activate vps gene expression, whereby the major biofilm regulator VpsR is required for vpsT transcription. Most of the c-di-GMP turnover enzymes affect rugose colony morphology via its positive regulators, VpsT and VpsR, both of which work as c-di-GMP receptors and are activated upon c-di-GMP binding. Note that not all V. cholerae DGCs and PDEs affect Vps expression, which highlights the issue of target specificity of c-di-GMP-metabolizing enzymes. However, as many as five DGCs have been shown to contribute to activity and localization of VpsT (307).
Fig 12.
Regulation of biofilm formation in V. cholerae El Tor. The transcriptional regulators VpsR and VpsT activate expression of the Vps exopolysaccharide required for rugose colony morphology and biofilm formation upon c-di-GMP binding (242). The c-di-GMP signaling network that regulates biofilm formation consists of various DGCs (CdgA, CdgG, CdgH, and VpvC) and PDEs (RocS, MbaA, and CdgC). V. cholerae quorum sensing represses biofilm formation through the quorum sensing regulator HapR, which regulates c-di-GMP-metabolizing proteins, among them CdgA and CdgG, and the transcription of vpsT (62). On the other hand, HapR is repressed by c-di-GMP through VpsT and VpsR via the virulence regulator AphA (459).
In addition to Vps polysaccharide, matrix proteins may contribute to the development of rugose colonies in a c-di-GMP-dependent manner (308). Also, at least one of five PilZ domain proteins is required for c-di-GMP-mediated biofilm formation in V. cholerae, by an as yet unknown mechanism (147).
Another example of multifactorial c-di-GMP control is the rdar morphotype of S. enterica, which is regulated by at least five DGCs and four PDEs (30, 226, 234, 297; I. Ahmad and U. Römling, unpublished data). In this case, the main target of c-di-GMP is the master regulator of biofilm formation CsgD, which is controlled on the transcriptional and posttranscriptional levels by largely unknown mechanisms. In the closely related organism E. coli, a distinct set of DGCs and PDEs regulates csgD transcription (49, 197, 298). CsgD expression is tightly coupled to the activities of the PDE YhjH and several DGCs involved in motility control (197, 297). CsgD in turn directly activates the csgBAC operon, encoding the subunits of curli fimbriae, as well as adrA, encoding the DGC required for activation of cellulose biosynthesis (309). Although CsgD does not appear to affect flagellar motility in S. enterica in a major way, it does affect the expression of distinct genes of the flagellar regulon (309, 310) and is involved in flagellum-mediated stimulation of the innate immune response (311).
VpsT of V. cholerae and CsgD are members of the CsgD family of LuxR-type helix-turn-helix DNA-binding proteins. CsgD family members are found mainly in the Gammaproteobacteria, in representatives of the families Vibrionaceae, Enterobacteriaceae, and Aeromonadaceae. However, the c-di-GMP binding motif [W(F/L/M)(T/S)R] of VpsT is not present in the enterobacterial proteins, suggesting that not all CsgD family members bind c-di-GMP (84, 309). While the genomes of most enterobacterial species harbor one CsgD homolog gene, up to five CsgD paralog genes are found in Vibrio species. In V. parahaemolyticus, which harbors four CsgD paralogs, at least two, CpsQ and CpsS, affect expression of the capsular polysaccharide operon, which mediates biofilm formation. CpsS represses transcription of the cps operon, while CpsQ binds c-di-GMP and activates the cps operon in response to elevated c-di-GMP levels (174, 312).
Additional aspects of biofilm regulation by c-di-GMP signaling.
Although activation of biofilm components by elevated levels of c-di-GMP is the main theme in biofilm formation, 3D biofilm structures and maintenance may be affected by lower c-di-GMP levels. For example, cell lysis-dependent production of extracellular DNA (eDNA), which is necessary for mature biofilm structures in many species, is stimulated by low c-di-GMP levels (313). Furthermore, the 3D architecture of mature biofilms is affected by motile cells on the biofilm surface, and lower c-di-GMP levels contribute to the sessility-to-motility transition of biofilm cells. Last but not least, biofilm dispersion, an integral part of long-term biofilm maintenance, requires the presence of specific PDEs (200, 314, 315). A recent study provided valuable clues to the molecular mechanism of dispersion in response to starvation signals (315a). In P. aeruginosa, the dispersion-specific PDE DipA was activated by interaction with the active form of the chemotaxis transducer protein BdlA. Surprisingly, BdlA activation required c-di-GMP-dependent phosphorylation and subsequent nonprocessive proteolysis (315a).
Regulation of Cell Cycle and Differentiation
Cell cycle and swimming- to stalked-cell differentiation in C. crescentus.
The freshwater bacterium C. crescentus undergoes a morphological development from a free-swimming motile swarmer to a sessile stalked cell (316). While the swarmer cell possesses polar flagella and adhesive pili at one cell pole, an adhesive holdfast and a stalk are subsequently developed on the same pole to promote surface adhesion. This cell differentiation into a stalked cell is tightly coupled with the initiation of the cell cycle. The stalked cell becomes competent for DNA replication and subsequently initiates an asymmetric cell division to produce a swarmer cell. After cell division, the stalked cell starts a new round of replication, while the swarmer cell is replication resistant unless it attaches to a surface and develops into a stalked cell. The stem-cell-like sessile reproductive lifestyle of C. crescentus makes the requirement for progeny with different behavior obvious. While the mother cell stays anchored, the daughter cell always swims away to explore new ecological niches. In C. crescentus, phosphotransfer signaling intimately integrates with c-di-GMP signaling into the regulation of cell cycle progression and cell differentiation (Fig. 13).
Fig 13.
Components of the c-di-GMP signaling network regulate cell cycle progression and cell differentiation. Only components proficient in metabolizing c-di-GMP and c-di-GMP-dependent processes are shown. Elevated c-di-GMP levels in cells upon G1- to S-phase transition are indicated by a gray background. PleD, which is dispersed in the cytoplasm of swarmer cells due to dephosphorylation by PleC, becomes DGC proficient upon phosphorylation and localizes to the differentiating pole (37). DgcB displays DGC activity, presumably throughout the cell cycle, whereby the PDE PdeA antagonizes DgcB activity in swarmer cells (318). Recruited to the cell pole by a c-di-GMP-independent mechanism, PopA remains localized at the differentiating pole upon swarmer- to stalked-cell transition, where it mediates S-phase entry and CtrA degradation upon c-di-GMP binding to its I site (162). DgrA and DgrB are PilZ domain c-di-GMP receptor proteins (51) which stall motility upon elevated c-di-GMP levels. DgrA and DgrB potentially act in the predivisional cell, stalling motility before the completion of cytokinesis and/or upon surface sensing, when the flagellar rotation slows down (460).
In accordance with its positive role in promoting sessility (5), c-di-GMP peaks upon transition from a swarmer cell to a stalked cell (317). Collectively, two DGCs, PleD and DgcB, drive the differentiation of the swarmer cell into a stalked cell through holdfast biogenesis and stalk initiation and elongation (24, 108, 318). In parallel, these DGCs share the task of counteracting motility, whereby DgcB controls motility inhibition and PleD controls the ejection of the flagellum (24, 108, 318). The PDE PdeA modulates the timing of the swarmer- to stalked-cell transition by preventing premature initiation of the holdfast (318). While many of the molecular mechanisms of the downstream events remain elusive, remarkable progress has been made in understanding the regulation of c-di-GMP signaling during the swarmer- to stalked-cell transition in C. crescentus.
The required peak in c-di-GMP concentration is achieved by at least two events. First, the DGC PleD (REC-REC-GGDEF) becomes activated in the stalked cell, and second, the PDE PdeA, which counteracts the activity of the constitutively expressed DgcB protein, is proteolytically degraded upon entry into S phase. Both events are coordinated by complex phosphorylation cascades (317, 318). PleD requires phosphorylation of the conserved aspartate in the REC domain for stimulation of its DGC activity (37). Two histidine kinases, DivJ and PleC, which are temporarily localized at the stalked pole during the swarmer- to stalked-cell transition, contribute to PleD phosphorylation. The single-domain response regulator DivK, which is sequestered by DivJ to the cell pole, stimulates the autokinase activity of both DivJ and PleC. This interaction allows efficient phosphotransfer to PleD (317, 319). Subsequently, activated PleD accumulates at the stalked cell pole and promotes the swarmer- to stalked-cell transition. Because DivK is also essential for cell cycle progression through regulation of CtrA (320), DivK couples cell differentiation with cell cycle progression.
Degradation of the PDE PdeA by the ClpXP protease upon swarmer- to stalked-cell transition occurs upon recruitment of both proteins to the cell pole (318). The single-domain response regulator CpdR, in its unphosphorylated form, recruits PdeA and ClpXP to the cell pole and acts as an adaptor protein for PdeA degradation. The CpdR-ClpXP complex couples cell differentiation with cell cycle progression, as polarly recruited ClpXP also degrades the essential cell cycle regulator CtrA. CtrA controls more than 100 cell cycle genes and directly binds to the chromosomal origin of replication to repress replication initiation. Proteolysis of CtrA requires c-di-GMP. The response regulator PopA, a PleD paralog, is enzymatically inactive but strongly binds c-di-GMP through its intact I site (162). PopA specifically localizes to the stalked pole and recruits CtrA as well as ClpXP. Several DGCs redundantly contribute c-di-GMP to PopA. One of them is PleD, as upon constitutive expression of the PDE PdeA, deletion of PleD is sufficient to prevent CtrA degradation (318).
Low c-di-GMP levels seem to be required for correct development of swarmer cells (225, 318). In swarmer cells, PleD is inactivated as it becomes dephosphorylated by PleC, which is switched from a kinase to a phosphatase mode. The predicted EAL domain PDE TipF is required for flagellum assembly (225). A tipF mutant lacks flagella, whereby the translation and secretion of the basal body hook subunit FlgE and the flagellin FljK are severely impaired. TipF is localized at the undifferentiated pole during swarmer and stalked cell development, where upon cell division it aids flagellum biogenesis in the swarmer cell. Subsequently, TipF is recruited to the division plane where the newborn undifferentiated pole is formed. TipN is required for recruitment and proper localization of TipF, with subsequent flagellum assembly. In addition, the PDE PdeA is expressed primarily in the swarmer cells, where it counteracts the DGC activity of DgcB, presumably via protein-protein interaction. Although a flagellum is still formed upon deletion of PdeA, motility is partially impaired. The PilZ domain c-di-GMP receptor proteins DgrA and DgrB potentially mediate stalled motility upon elevated c-di-GMP levels in swarmer cells (51). DgrA, but not DgrB, is involved in downregulating FliL, which is required for flagellum rotation.
The above-described genetic and biochemical studies unraveled a distinct distribution of c-di-GMP-metabolizing proteins and c-di-GMP receptors in the swarmer cell and stalked cell upon cell division. Altogether, a body of evidence suggests that c-di-GMP is asymmetrically distributed between the dividing swarmer cell and stalked cell. However, can these spatial and temporal differences in c-di-GMP concentrations be measured directly? Christen et al. developed just such a tool. They made a c-di-GMP sensor based on the PilZ domain c-di-GMP receptor YcgR from S. enterica (219). As YcgR undergoes a dramatic conformational change upon c-di-GMP binding (50), the distance between two different fluorescent proteins, coupled to the N-terminal PilZN (YcgR1) domain and the C-terminal PilZ domain of YcgR, is altered, which leads to changes in FRET. Using this c-di-GMP sensor, a transient 5-fold drop in c-di-GMP concentration was observed in the swarmer cell compared to the stalked cell immediately after cell division. Low c-di-GMP levels in both cell types were observed for the pleD mutant, while the pleC mutant displayed high concentrations of c-di-GMP in the swarmer cell, consistent with PleC negatively regulating PleD activity in those cells (108). Similar asymmetric distributions of c-di-GMP concentrations upon cell division were observed in P. aeruginosa, S. enterica, and K. pneumoniae, suggesting that cell division is sufficient to create phenotypic diversity.
Axenic- to predatory-lifestyle transition in B. bacteriovorus.
Bdellovibrio bacteriovorus represents yet another example of cell differentiation gone wrong as a result of impaired c-di-GMP signaling. This deltaproteobacterium has two lifestyles, axenic and predatory. It predates the proteobacteria, including important human, animal, and plant pathogens, and is considered a potential “live antibiotic.” After entering the periplasm of a prey bacterium, B. bacteriovorus undergoes a transition to a bdelloplast stage, where it replicates inside the periplasm, feeding on the prey nutrients. Upon prey exhaustion, several motile progeny of the attack-phase cells emerge (321). Hobley et al. (153) demonstrated that two specific c-di-GMP signaling cascades play unique, nonoverlapping roles in the axenic- to predatory-lifestyle transition. One of the three DGCs of B. bacteriovorus, DgcB (Bd0742), controls invasion of prey bacteria. It probably signals through the I-site c-di-GMP receptor CdgA, located at the prey-interacting “nose” of Bdellovibrio. A dgcB mutant is predation impaired and therefore locked in the axenic lifestyle. In contrast, a mutant in the second DGC, DgcC (Bd1434), is an obligate predator, incapable of living outside prey, even in rich media. This obligately predatory mutant is a good candidate for a live antibiotic. Notably, deletion of the third DGC, DgcA, impairs the gliding motility of B. bacteriovorus required for newly “hatched” bacteria to leave their exhausted prey. Therefore, DgcA also contributes, in a very distinct way, to the axenic- to predatory-lifestyle transition of B. bacteriovorus. Aside from the unique c-di-GMP-dependent lifestyle transition, the story of a miniature bacterial parasite provides irrefutable evidence of the extreme specificity of c-di-GMP signaling pathways in this bacterium. Each of the three functional DGCs in B. bacteriovorus has a unique role that does not overlap the roles of other DGCs (153).
Cyclic di-GMP in cell differentiation in multicellular bacteria.
The first observation linking c-di-GMP to cell differentiation in multicellular bacteria was described by Neunuebel and Golden (195). Upon nitrogen starvation, the filamentous cyanobacterium Anabaena sp. PCC 7120 forms specialized cells, called heterocysts. The main function of heterocysts is to perform oxygen-sensitive nitrogen fixation as opposed to oxygen-evolving photosynthesis, and thus to supply the multicellular filament with fixed nitrogen. In wild-type Anabaena, the interval between two heterocysts is approximately 25 vegetative, photosynthesizing cells. However, inactivation of one of the GGDEF domain DGCs, All2874, resulted in a large increase in the intervals between the heterocysts, to approximately 200 vegetative cells. Interestingly, this phenotype was most pronounced at a high light intensity. The authors of this study showed that All2874 acts upstream of HetR, the master transcriptional regulator in differentiating cells, but how c-di-GMP affects the differentiation gene program remains to be investigated.
Another piece of evidence documenting the involvement of c-di-GMP in multicellular differentiation comes from Streptomyces coelicolor, a nonmotile filamentous soil actinobacterium known for its complex morphological differentiation. Streptomycetes have significant pharmacological importance because they produce over two-thirds of currently used antibiotics. The life cycle of S. coelicolor begins with germination of a free spore that produces a long, branching vegetative filament. Vegetative filaments grow into and on the substrate surface but rarely divide, yielding a network of multinucleated hyphae. These hyphae undergo a sporulation-like cell division process, resulting in an aerial mycelium comprised of prespores, each of which contains only one set of genomic DNA, that subsequently metamorphose into mature spores (322). den Hengst et al. first discovered that overexpression of the DGC designated CdgA (SCO2817) blocks formation of aerial hyphae and results in a bald colony appearance (182). Overexpression of yet another DGC, CdgB (SCO4281), resulted in the impaired formation of aerial mycelium. It is peculiar that the cdgB deletion also showed impaired hypha formation (323). Two EAL domain PDEs, RmdA (SCO0928) and RmdB (SCO5495), were also identified as responsible for hypha formation. Their individual knockouts delayed hypha formation, while a double knockout resulted in a hypha-less, bald phenotype (196). It is worth noting that not only was mycelium formation affected by c-di-GMP in S. coelicolor, but pigmentation and antibiotic actinorhodin production, both of which are known markers of cell development in this species, were also affected. The studies in multicellular bacteria extend the importance of c-di-GMP-dependent signal transduction in bacterial differentiation.
Cyclic di-GMP and Virulence
Specific c-di-GMP signaling pathways may affect various aspects of virulence.
Genetic screens demonstrated that c-di-GMP signaling pathways affect virulence in numerous animal and plant pathogens (7, 183, 203–205, 311).
The first observations of a role of c-di-GMP signaling in virulence were in V. cholerae, as elevated expression of the cholera toxin, a major virulence factor, was promoted by low c-di-GMP levels in vitro, and elevated c-di-GMP levels attenuated virulence in the infant mouse model of cholera (40). These early observations helped to shape the view that an acute infection requires low c-di-GMP levels or, as an extreme, does not require c-di-GMP at all (7). Several recent studies of pathogens with few c-di-GMP-metabolizing proteins support this view. Deletion of the functional DGCs demonstrated that c-di-GMP signaling is not required for virulence of Yersinia pestis in the plague mouse model (183). The Lyme disease spirochete B. burgdorferi caused infection in mice when its only DGC was deleted (194). The deletion of all GGDEF domain genes did decrease the virulence of Salmonella enterica serovar Enteritidis in a mouse model of systemic infection; however, virulence was restored upon expression of the enzymatically inactive mutant of the GGDEF domain gene, STM4551, suggesting that c-di-GMP plays no role in virulence in this model (324). Deletion of all PDEs reduced virulence, while deletion of all DGCs increased virulence, in a mouse model of Brucella melitensis infection (203). Furthermore, high c-di-GMP levels inhibited acute infections of Y. pestis and B. burgdorferi because of the expression of extracellular biofilm matrix (128, 183). Also in Francisella novicida, c-di-GMP inhibits virulence in mice and intracellular replication in macrophages (324a). F. novicida, which is essentially avirulent, contains a gene cluster with two GGDEF-EAL domain proteins, in contrast to its close relative Francisella tularensis, which is highly pathogenic for humans. These cumulative data suggest that constitutively elevated c-di-GMP levels are detrimental for acute infections.
However, subsequent genetic screens in pathogens containing sophisticated c-di-GMP signaling networks have uncovered a more complex picture in which deletion of specific DGCs and PDEs affects virulence in an unpredictable fashion (204, 205, 311). In the murine model of burn wound infection, c-di-GMP was found generally inhibiting the virulence of P. aeruginosa; however, virulence was reduced in some deletion mutants of PDEs as well as in a mutant of the DGC SadC (204). A systematic screen for virulence phenotypes in the plant pathogen X. campestris did not find a correlation between the predicted changes in c-di-GMP concentrations and virulence (205). Colonization of the mouse gastrointestinal tract by S. enterica revealed that c-di-GMP-metabolizing enzymes with opposite functions contribute to virulence in the same direction (311). These data cumulatively suggest that highly regulated c-di-GMP signaling is required for acute virulence.
What could be the reason for opposite functional c-di-GMP-metabolizing proteins working in the same direction? Possibly, certain biofilm-related phenotypes positively stimulated by c-di-GMP signaling are required at some stage of acute infection. As an example, an investigation into the V. cholerae infection process revealed a temporal and spatial requirement for opposite c-di-GMP-metabolizing processes at different disease stages. In contrast to early infection, at the late stage, a group of GGDEF/EAL genes were induced, suggesting an increased c-di-GMP concentration, but mutants in individual genes did not have growth disadvantages in the small intestines of mice (325, 326). A mutant in three of these GGDEF genes was, however, defective in survival in watery stool from the large bowel, suggesting that bacteria prepare for their further transition through the host without a disease phenotype (325). A temporal and spatial requirement of opposite c-di-GMP-metabolizing processes was also indicated for S. enterica, as deletion of a putative PDE abolished colonization of the mouse gastrointestinal tract after the first day, while DGC mutants were defective in colonization after day 10 (311). These findings suggest that processes inhibited as well as activated by c-di-GMP may play unique roles in the course of acute infections and that virulence may be considered a “phenotype” only at first approximation. In reality, every acute infection consists of complex interactions between various host systems and a pathogen constantly adapting to changing environments inside the host.
The increased virulence of pathogen mutants expected to have elevated c-di-GMP levels in vivo suggests that certain biofilm-related functions may be beneficial during the course of acute infections. This has been demonstrated best for the related obligate intracellular pathogens Anaplasma phagocytophilum and Ehrlichia chaffeensis. These pathogens infect immune cells (327), where they replicate as intracellular aggregates called morulae, which resemble biofilms. Expression of a gene encoding a single GGDEF domain protein in A. phagocytophilum or E. chaffeensis coincides with formation of morulae in host cells (328, 329). Treatment with a hydrophobic c-di-GMP analog suggested that c-di-GMP signaling is required for formation of morulae, intracellular proliferation, and enhanced bacterial intra-inclusion movement (328, 329). The expression of the GGDEF domain protein during human infection by A. phagocytophilum (329) provides convincing evidence that c-di-GMP signaling is required for acute infection by this bacterium.
Mechanisms of c-di-GMP signaling affecting virulence.
The following phenotypes have been associated with c-di-GMP signaling: adherence to host cells, host cell invasion, cytotoxicity, intracellular infection, secretion of virulence factors, resistance to oxidative stress, and modulation of immune responses (40, 164, 204, 311, 329–335) (Table 6).
Table 6.
Virulence phenotypes affected by c-di-GMP
| Phenotype | Species | Host or cell line | c-di-GMP level | Cyclic di-GMP-metabolizing enzymes involved | Reference(s) |
|---|---|---|---|---|---|
| In vivo virulence | X. campestris | Chinese radish | Variable | PDE RpfG and 12 additional PDEs/DGCs | 205 |
| P. aeruginosa | Thermal injury mouse model | Variable | 5 DGCs/PDEs | 204 | |
| S. Typhimurium | Enterocolitis in streptomycin-treated mouse | Variable | PDE STM3615, DGC STM2672, DGC STM4551 | 311 | |
| A. phagocytophilum | Human granulocytic anaplasmosis | High | DGC PleDAp | 329 | |
| B. melitensis | Mouse spleen infection | Low | PDEs BpdA and BpdB, DGC CgsB | 203 | |
| F. novicida | Mouse | Low | Overexpression of DGC | 324a | |
| Adherence to host cells | Adherent-invasive E. coli | Intestinal epithelial cell line intestine-407 | Low | PDE YhjH, DGC AdrA | 336 |
| Uropathogenic E. coli CFT073 | Bladder epithelial cell line UM-UC-3 | High | DGC YdeH, PDE C1610, and 10 additional DGCs/PDEs | 288a | |
| Host cell invasion | B. pseudomallei | Lung epithelial cells (A549) | Low | PDE CdpA | 337 |
| Adherent-invasive E. coli | Intestine-407 cells | Low | PDE YhjH | 336 | |
| E. chaffeensis | Macrophage-like cell line THP-1 | Elevated | PDE PleDEc | 331 | |
| S. Typhimurium | Intestinal cell line HT-29 | Variable | DGCs STM1987 and STM4551, PDEs STM3611 and STM4264 | 311, 333 | |
| Cytotoxicity to host cells | B. pseudomallei | A549 cells | Low | PDE CdpA | 337 |
| L. pneumophila | THP-1 cells | Low | Overexpression of DGCs | 338 | |
| P. aeruginosa | Chinese hamster ovary cell line CHO | Variable | DGCs, PDEs | 204 | |
| Intracellular infection | A. phagocytophilum | Human myelocytic leukemia HL-60 cells | Elevated | DGC PleDAp | 329 |
| E. chaffeensis | THP-1 cells | Elevated | DGC PleDEc | 328 | |
| L. pneumophila | THP-1 cells | Low | DGCs | 338 | |
| F. novicida | Macrophage-like cell line J774 | Low | Overexpression of DGCs | 324a | |
| Modulation of immune response | S. Typhimurium | HT-29 cells | Low | DGC STM1283, PDEs STM4264 and STM2503 | 311 |
All types of secretion systems have been shown to be subject to c-di-GMP-mediated regulation (Table 7). Secretion of such type II secretion system substrates as the cholera toxin in V. cholerae, endomannanases and endoglucanases in X. campestris, and pectate lyase production components in Dickeya dadantii (formerly Erwinia chrysanthemi) is inhibited by c-di-GMP (40, 205, 339). Direct and indirect transcriptional repression by c-di-GMP of genes encoding secreted proteins has been observed. In V. cholerae, the PDE VieA is required for optimal transcriptional expression of toxT, encoding a transcriptional activator of the cholera toxin (40). In X. campestris, Clp is a transcriptional activator of genes coding for many extracellular enzymes and exopolysaccharides (341), including engA, encoding an endoglucanase that functions as a major virulence factor of X. campestris (Fig. 9). Binding of c-di-GMP to Clp abolished binding to and activation of the engXCA promoter (170, 222).
Table 7.
Secretion systems
| Secretion system type and phenotype | Species | Molecular mechanism of c-di-GMP | Cyclic di-GMP-metabolizing enzymes involved | Reference |
|---|---|---|---|---|
| Type 1 | ||||
| Surface association vs secretion of LapA adhesin | P. fluorescens | Inhibition of proteolysis of LapA by LapG through c-di-GMP receptor LapD sequestration | DGCs GcbB and GcbC, PDE RapA | 68 |
| Secretion of LapA-like surface adhesin | P. atrosepticum | Transcriptional activation | DGC ECA3270, PDE ECA3271 | 115 |
| Type 2 | ||||
| Expression of CtxAB cholera toxin | V. cholerae | Inhibition of transcription of transcriptional activator ToxT | PDE VieA | 40 |
| Secretion of endoglucanase and endomannanase | X. campestris | c-di-GMP receptor Clp-mediated relief of transcription | PDE RpfG | 222 |
| Secretion of pectate lyase | D. dadantii | Unknown | PDEs EcpB and EcpC | 339 |
| Type 3 | ||||
| Transcription of TTSS | D. dadantii | Inhibition of expression of the sigma factor RpoN | PDEs EcpB and EcpC | 339 |
| Expression of TTSS proteins | P. aeruginosa | Unknown | DGC WspR | 340 |
| Type 4 | ||||
| Expression of T4SS protein VirB6-2 | E. chaffeensis | Proteolysis | DGC PleDEc | 331 |
| Type 5a | ||||
| Transcription of cdrAB | P. aeruginosa | c-di-GMP receptor FleQ-mediated transcription | DGC YfiN | 304 |
| Type 5b | ||||
| No c-di-GMP-affected function known | ||||
| Type 6 | ||||
| Expression of T6SS proteins | P. aeruginosa | Unknown | DGC WspR | 340 |
| Flagella | ||||
| Expression of flagella | B. melitensis | Transcription | PDE BpdA | 203 |
| Secretion of flagellin | S. Typhimurium | Unknown | DGC STM1283, PDEs STM4264 and STM2503 | 333 |
Type III secretion systems are also negatively affected by c-di-GMP (127, 311, 339, 340). Two PDEs are required for expression of the type III secretion system genes of D. dadantii. This regulation occurs through c-di-GMP-dependent transcriptional regulation of the σN factor (RpoN). RpoN activates transcription of the alternative sigma factor HrpL, which is required for the activation of transcription of type III secretion system genes (339). In the same vein, expression of the type III secretion system is inhibited by overexpression of DGCs in P. aeruginosa (340), and secretion of the effector protein ExoS is inhibited by deletion of HD-GYP PDEs (127). In S. enterica, GGDEF/EAL proteins regulate the secretion of type III secretion system effector proteins, but their c-di-GMP-metabolizing activity is not required (311).
Cyclic di-GMP signaling has a stimulating effect on type IV secretion. In E. chaffeensis, expression of VirB6-2, a component of the type IV secretion system, requires c-di-GMP (331). The expression of this and other surface-exposed proteins, not related to type IV secretion, is regulated by c-di-GMP-dependent proteolysis. TRP120, a protein exposed on the bacterial surface and a component of the matrix of morulae, is degraded by the surface-exposed protease HtrA, in a c-di-GMP-dependent manner (331).
Flagella are virulence factors required for pathogenicity, immunostimulation, and invasion into epithelial cells (342, 343). In B. melitensis, deletion of the PDE-encoding gene, bpdA, causes avirulence in a mouse model of infection (203). The bpdA deletion is associated with decreased transcription of flagellar genes in vitro. In line with this observation, the virulence of a flagellar gene deletion mutant was also partially impaired. A nonflagellated CdpA PDE mutant of Burkholderia pseudomallei was less cytotoxic to THP-1 human macrophage cells and showed reduced invasion of A549 human lung epithelial cells (337). Since flagellar expression is required for invasion and cytotoxicity of B. pseudomallei (344), the lack of flagellum expression in the cdpA mutant most likely contributed to the downregulation of virulence. Flagellin, the monomeric subunit of flagella, is a pathogen-associated molecular pattern (PAMP) recognized by Toll-like receptor 5 (TLR5) (345). The binding of flagellin to TLR5 triggers a subsequent immune response in the host cells. High levels of c-di-GMP inhibit inflammation caused by S. enterica, as judged by the decreased induction of the proinflammatory cytokine interleukin-8 (IL-8) in the intestinal cell line HT-29 (311, 333). Repression of flagellin secretion is most likely responsible for the downregulation of the immune response (333).
Adherence to and invasion of intestinal cells by the Crohn's disease-associated adherent-invasive E. coli strain LF82 depend on flagella and type 1 pili (346). Expression of type 1 pili partially requires the PDE YhjH and is inhibited by the DGC AdrA (336). Type 1 pilus expression was stimulated by low c-di-GMP levels. Although the role of c-di-GMP signaling in the biogenesis of type 1 fimbriae has not been elucidated, the observed phenotype is unconventional because expression of adhesive pili in other strains is stimulated by elevated c-di-GMP levels (158).
A genetic screen for P. aeruginosa mutants altered in cytotoxicity to CHO cells revealed that mutants in c-di-GMP-metabolizing enzymes with opposite activities operate in the same direction (204). This may be possible because cytotoxicity depends on both type III secretion (negatively regulated by c-di-GMP) and type IV pili (positively regulated by c-di-GMP) (347). Although type III secretion system-mediated invasion of epithelial cells by S. enterica is inhibited by high c-di-GMP levels (333), some DGCs were found to stimulate invasion (311). This may be another case where c-di-GMP-inhibited type III secretion is countered by c-di-GMP-stimulated adhesion via type IV pili, which are required for invasion (348–350).
Another level of complexity comes from the fact that in vitro and in vivo virulence phenotypes do not necessarily correlate. For P. aeruginosa, a lack of correlation between CHO cell cytotoxicity and virulence was reported for a mouse model of burn wounds. Although mutants in fimX and wspR showed cytotoxicity phenotypes, they had no virulence phenotypes in vivo (204). Different activities of c-di-GMP signaling pathways in vitro and in vivo may be responsible for this phenomenon. In support of this view, endoglucanase and endomannanase in Xanthomonas were found to be regulated by c-di-GMP signaling in one growth medium but not in another (205). Different levels of importance of c-di-GMP signaling pathways in vivo and in vitro have also been observed in Y. pestis, where stimulation of PAG-based biofilm formation in the arthropod host, the flea, required a DGC that played only a minor role in biofilm formation in vitro (351). Thus, contributions of c-di-GMP signaling pathways to virulence depend on the environmental conditions, so direct extrapolation of in vitro findings to the in vivo situation is not warranted.
Contributions of c-di-GMP signaling pathways to chronic infections.
The majority of chronic infections involve a biofilm stage; therefore, c-di-GMP signaling pathways play important roles in chronic infections (7). After a year-long persistence in cystic fibrosis lungs, P. aeruginosa develops small-colony variants (352, 353) with elevated intracellular c-di-GMP levels (97, 206, 354). These variants are characterized by enhanced biofilm formation, high fimbrial expression, repression of flagellar genes, resistance to phagocytosis, and enhanced antibiotic resistance (96, 97, 206, 245). Small-colony variants generated in vitro as well as obtained from clinical isolates contained mutations that upregulate the activity of the DGC TpbB (YfiN), suggesting a key role of this enzyme (193, 245). The importance of c-di-GMP for enhanced persistence of P. aeruginosa has also been demonstrated in a chinchilla model of middle ear infection (355).
In P. aeruginosa, the RetS/LadS/GacS signaling cascade regulates the transition from acute to chronic infection. In a ladS mutant, which promotes chronic infection and exopolysaccharide production, the DGCs WspR and PA0290 are specifically associated with suppression of type III secretion and enhanced expression of type VI secretion, which was shown to be important in chronic infections (340).
Role of c-di-GMP signaling in pathogen transmission.
Besides their direct contribution to virulence, c-di-GMP signaling pathways play important roles in pathogen transmission. Y. pestis and B. burgdorferi are transmitted by arthropod vectors. Although c-di-GMP signaling has no role in the virulence of Y. pestis and B. burgdorferi in mice, it plays a determinative role in growth in the arthropod vectors and in transmission between the arthropod and human hosts. For Y. pestis, transmission from the flea to the human host involves the c-di-GMP-activated PAG-based biofilm (351, 356). B. burgdorferi requires its sole DGC, Rrp1, for activation of the glycerol transport/catabolism operon in the tick (194). Glycerol produced by ticks may serve B. burgdorferi as an antifreeze as well as a carbon source.
While low c-di-GMP levels are necessary for cholera toxin synthesis and acute infection by V. cholerae, it was found that expression of several DGCs is induced in the late stages of disease, suggesting a role of increased c-di-GMP synthesis. While mutants in the DGC genes had no growth disadvantages in the small intestines of mice (325, 326), a mutant lacking three GGDEF genes was defective in survival in watery stool and pond water. Therefore, c-di-GMP is involved in the preparation of V. cholerae to exit the host, which is important for the spread of the pathogen to new hosts (325).
The natural hosts of the human pathogen L. pneumophila are amoebae, e.g., Acanthamoeba castellanii. During its intracellular life cycle, L. pneumophila alternates between an intracellular replicative form and an infectious nonreplicative form, which promotes transmission to a new host. Transition from the replicative to the transmissive phase requires coordination of functions critical for motility, contact-dependent cytotoxicity, host cell entry, survival, and other virulence traits. Strikingly, 18 of 24 potential c-di-GMP-metabolizing enzymes are upregulated in the transmissive phase, suggesting that a complex interplay of the c-di-GMP-regulated processes is involved in L. pneumophila transmission (338, 357).
Cyclic di-GMP and RNA
Cyclic di-GMP-dependent RNA degradation.
Cyclic di-GMP affects RNA turnover via several different mechanisms. One of the most intriguing and least understood mechanisms was discovered by Tuckerman et al., who found that the DosC-DosP DGC-PDE complex from E. coli (40) was copurified with a large ribonucleoprotein complex (RNC) that contained components of the RNA degradosome (90). Tailing with homopolymeric 3′ poly(A) tails to destabilize mRNAs, a catalytic activity performed by the RNA degradosome component polynucleotide phosphorylase (PNPase), was found to be oxygen dependent, in agreement with the oxygen-dependent activities of DosC and DosP. Subsequent analysis revealed that both tailing and phosphoryl exchange, another reaction catalyzed by PNPase, are positively regulated by c-di-GMP, and that PNPase binds c-di-GMP with a low-micromolar affinity. Cyclic di-GMP is believed to accumulate locally under microaerobic/anaerobic conditions, where the DosC DGC activity is high and the DosP PDE activity is low (89). This results in PNPase activation, while highly aerobic conditions inhibit PNPase (90). Cyclic di-GMP joins a growing group of small molecules that allosterically regulate PNPase activity and RNA turnover (358, 359).
While homopolymeric 3′ poly(A) tails are known to destabilize E. coli mRNAs, heteropolymeric 3′ poly(AG) tails are thought to render transcripts more stable (360). Furthermore, small RNAs are stabilized in the presence of PNPase, via an unknown mechanism (361). Therefore, it appears that c-di-GMP may differentially regulate RNA turnover. What are the RNA targets of c-di-GMP-regulated PNPase activity? Although no direct evidence exists, phenotypes of dosC/dosP mutants and overexpression strains suggest that mRNAs whose products are involved in biofilm formation, e.g., curli subunits and the PAG biosynthesis enzymes, as well as mRNAs related to cell division, may be involved (235, 271, 299). Revealing the intricacies of c-di-GMP-dependent RNA turnover will be critical to our understanding of c-di-GMP-dependent regulation in general.
Cyclic di-GMP-dependent riboswitches.
Two classes of distinct short RNA molecules, c-di-GMP-I and c-di-GMP-II, have been shown to represent c-di-GMP-binding aptamers (riboswitches), which help to regulate gene expression (59, 66). Depending on the genetic context, positive regulation (“on” switch) or negative regulation (“off” switch) upon c-di-GMP binding can occur.
c-di-GMP riboswitches are widespread in bacteria. At least 322 c-di-GMP type I candidate riboswitches have been identified in various species, including many representatives of the Proteobacteria and Firmicutes. The current record holder, the deltaproteobacterium Geobacter uraniireducens Rf4, encodes as many as 30 type I c-di-GMP riboswitches (66). Type II c-di-GMP riboswitches have a more restricted distribution, mostly within the Chloroflexi and Clostridia, with Clostridium difficile 630 encoding 12 c-di-GMP-II riboswitches. The genes affiliated with those riboswitches appear to be involved in biofilm-related functions, such as c-di-GMP synthesis and degradation, motility, pili, and transcription regulation, but also in nonribosomal peptide synthesis. Many genes have unknown functions (59, 362). Some riboswitches in G. uraniireducens suggest a c-di-GMP involvement in regulation of metal reduction. Indeed, many novel c-di-GMP-controlled processes will likely be discovered upon exploration of the c-di-GMP riboswitches.
Several c-di-GMP riboswitches have been tested experimentally. The C. difficile Cd1 riboswitch preceding the flagellum operon works as an “off” switch which, in the presence of c-di-GMP, causes transcriptional termination in vitro and decreases gene expression in a heterologous host, B. subtilis. On the other hand, the Vc2 riboswitch from V. cholerae is an “on” switch. Vc2 precedes the VC1722 gene, encoding a homolog of the transcription factor TfoX, which is required for the natural competence of V. cholerae associated with biofilms on chitin surfaces. VC1722 mRNA levels are upregulated at elevated c-di-GMP levels. Interestingly, a dual-layer control of gene expression is provided by the tandem arrangement of a c-di-GMP-II riboswitch with a self-splicing ribozyme upstream of CD3246, encoding a putative surface protein in C. difficile (66, 363). The riboswitch allosterically controls alternative self-splicing of the ribozyme whereby, in the presence of c-di-GMP, a product with an accessible ribosome binding site is favored. Upon c-di-GMP dissociation from the aptamer, however, the ribosome binding site is occluded, thus preventing translation. The riboswitch-ribozyme tandem may constitute a two-signal RNA-based input control system where the second signal, GTP, must be present in sufficient amounts to promote ribozyme self-splicing.
CYCLIC di-GMP AS PART OF A GENERAL SIGNALING MACHINERY
Coping with a “Regulatory Nightmare”: Specificity of c-di-GMP Signaling Pathways
The presence of numerous DGCs and PDEs as well as c-di-GMP receptors in many bacteria raises questions about the mechanisms that establish specificity of the c-di-GMP signaling pathways, i.e., the selective regulation of c-di-GMP target outputs by individual c-di-GMP-metabolizing proteins. Maintenance of the intracellular pool of c-di-GMP, to which dozens of DGCs and PDEs are potentially contributing, must be a “regulatory nightmare.” How do cells cope with numerous c-di-GMP signaling systems? Is there indeed specificity in c-di-GMP signaling? The more we learn about c-di-GMP-dependent signaling pathways, the better we realize that there is a hierarchical logic governing c-di-GMP regulatory systems, that individual components are controlled by specific environmental and intracellular stimuli, and that they often serve specific targets. However, we also see that interfering with just a single piece of this integrative network may be sufficient to create a bacterial cell with a radically altered physiology.
Regulation of expression of c-di-GMP-related genes.
An important mechanism that helps to shape the c-di-GMP signaling network for a particular lifestyle or growth phase is differential transcriptional regulation of c-di-GMP-metabolizing proteins. The powerful control of c-di-GMP signaling by the general stress sigma factor RpoS in E. coli is a good example. Quantitative analysis of the expression patterns of all 28 GGDEF/EAL domain genes in E. coli W3110 revealed that the majority of them (21 genes) are expressed under laboratory conditions (298). Nevertheless, distinct expression patterns of GGDEF/EAL domain genes were observed in response to growth phase, solid versus liquid medium, and temperature. In particular, the stress sigma factor RpoS positively or negatively regulated 15 of the 21 genes upon entry into the stationary phase. Regulation of the GGDEF/EAL domain-encoding genes by RpoS has also been observed in other bacteria (325, 364) and seems to represent a common trait. While six of the RpoS-controlled genes regulate CsgD and curli gene expression, downstream targets of most of the other GGDEF/EAL domain-encoding genes remain to be determined.
The regulation of c-di-GMP-metabolizing proteins through the global regulator RpoS also implies that factors affecting RpoS may also alter c-di-GMP signaling. For example, MqsA, the antitoxin component of the MqsR-MqsA toxin-antitoxin system, downregulates RpoS by directly binding to the RpoS promoter. Therefore, MqsA affects c-di-GMP levels, biofilm formation, stress resistance, and motility (365). MqsA probably affects biofilm formation through a negative-feed-forward loop, as an MqsA binding site is also found in the csgD promoter region. Toxin-antitoxin systems such as MqsA-MqsR are involved in formation of persister cells, which survive antibiotic challenges much better than regular cells do (366). Thus, biofilm formation and antibiotic resistance may be closely regulated through toxin-antitoxin systems and c-di-GMP (367).
While lifestyle issues such as the transition to stationary phase discussed above are important in shaping c-di-GMP network architectures, environmental and intracellular signals affecting individual c-di-GMP-metabolizing enzymes are also very important. As discussed above (see “Regulation by Sensory Domains”), many enzymes involved in c-di-GMP signaling contain sensory domains and respond to specific signals. Some enzymes respond to environmental signals by interacting with sensor proteins (136, 368). At present, a relatively limited number of signals affecting activities of the c-di-GMP-metabolizing proteins have been investigated experimentally.
Colocalization of DGCs and their targets.
Specific responses of c-di-GMP targets to individual DGCs and/or PDEs are another important aspect contributing to the specificity of c-di-GMP signaling (Fig. 11). Numerous examples of signaling specificity have been reported for diverse bacteria. One of the most striking recent examples involves a miniature bacterial parasite in which knockouts in each individual DGC resulted in distinct, nonoverlapping phenotypes (153) (see “Axenic- to predatory-lifestyle transition in B. bacteriovorus”). Another striking example comes from P. aeruginosa, where overexpression of the DGC PA2870 led to a 100% increase in c-di-GMP levels yet biofilm formation was not enhanced (204). In contrast, overexpression of SadC (PA4332) resulted in biofilm formation, although no change in the total c-di-GMP concentration was observed (204). Similarly, while overexpression of the DGC YcdT in E. coli led to higher c-di-GMP levels than those with YdeH overexpression, flagellum and pilus biosynthesis was abolished only upon YdeH overexpression. In contrast, YcdT overexpression led to slightly more pilus expression, while the number of flagella remained unchanged (60, 266). These observations were recently rationalized as no correlation was found between intracellular c-di-GMP concentrations created by seven different DGC and the biofilm and gene expression phenotype of V. cholerae, whereas such a correlation was seen for individual DGCs expressed at different levels (368a). Colocalization of an oxygen-sensing DGC and PDE, DosC and DosP, and their target, PNPase from E. coli (90), was discussed earlier in this review (see “c-di-GMP-dependent RNA degradation”).
On the other hand, global pools of c-di-GMP may be sensed by cytoplasmic c-di-GMP receptors such as transcription factors. For example, in V. cholerae, five DGCs cumulatively contribute to the activity of the c-di-GMP-dependent transcription factor VpsT (307).
Strains in which all DGCs are deleted represent useful examples in considering the specificity of c-di-GMP signaling. In an S. enterica strain where all 12 GGDEF domain-encoding genes were deleted, only three of eight potential DGCs, among them STM1987, were found to stimulate cellulose biosynthesis (259). In contrast, in the wild type, deletion of only one DGC, STM1987, affected cellulose production in the same minimal medium (259). It is still unknown whether STM1987 somehow prevents other DGCs from stimulating cellulose production, or, perhaps more likely, altered c-di-GMP degradation patterns in the all-GGDEF-protein deletion strain may have stimulated cellulose production by DGCs other than STM1987 (259).
Another interesting yet controversial finding emerged from the study that used the 12-GGDEF-protein deletion strain, in which virulence, CsgD-mediated rdar morphotype expression, and long-term survival were partially restored by the GGDEF domain protein STM4551 lacking DGC activity (324). Since STM4551 is not present in the nonpathogenic Salmonella relative E. coli K-12, it is possible that this Salmonella-specific gene has a specific, c-di-GMP-independent role(s) in regulating certain aspects of Salmonella virulence.
Impact of phosphodiesterases.
In considering the specificity of c-di-GMP signaling cascades, it is important to consider the role of PDEs (Fig. 14). These enzymes are expected to guard their targets from spillovers of undesired c-di-GMP. Therefore, a mutation in a PDE may drastically change the c-di-GMP landscape of a cell by exposing c-di-GMP receptors/targets to regulation by both specific and nonspecific DGCs. In S. enterica, deletion of the PDE STM1703 led to a 10-fold higher expression level of the major biofilm regulator CsgD (297) and to constitutive CsgD expression in all cells of the biofilm, in contrast to biphasic CsgD expression in less than half of the wild-type cells (369). Also, a deletion of the PDE PvrR (PA14_59790) totally abolished the virulence of P. aeruginosa strain PA14 in a murine model of burn wound infection (204).
Fig 14.
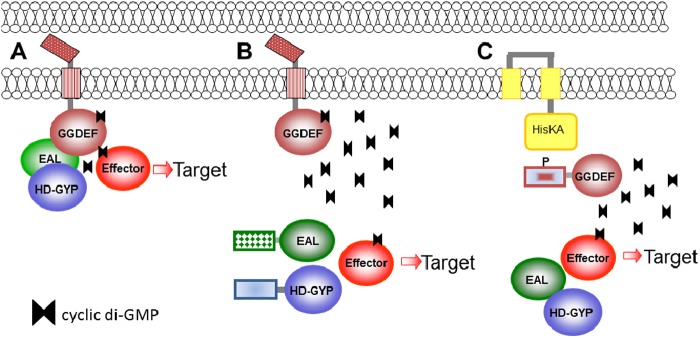
Cyclic di-GMP signaling models. (A) Spatial proximity of c-di-GMP-metabolizing proteins, the effector, and/or target output is required for effective signaling. In this case, c-di-GMP synthesis and degradation are probably low. (B) The DGC and/or PDE is distant from the effector/target. Cyclic di-GMP synthesis of the DGC is probably high. (C) Processes inhibited by c-di-GMP signaling are more likely to be affected by signaling from a distance.
Binding affinity of c-di-GMP receptors.
The dissociation constant of receptors for c-di-GMP can vary over 1,000-fold. Specificity of c-di-GMP signaling might therefore also be regulated simply through the binding affinity of the c-di-GMP receptors. That such a mechanism is indeed acting in cells has been recently demonstrated in S. enterica serovar Typhimurium (369a). Deletion of the PDE YhjH (STM3611) led to a c-di-GMP concentration sufficient to inhibit motility through the c-di-GMP receptor YcgR but not to stimulate the cellulose synthase BcsA, which harbors a PilZ domain with a 40-fold-lower affinity for c-di-GMP.
Connection to Other Signaling Systems
The regulation of many DGCs and PDEs by two-component signal transduction (Table 3) and the RpoS-dependent control of expression of numerous c-di-GMP signaling enzymes in E. coli are just two examples showing interconnectivity between regulatory systems. Details revealing relationships between various other regulatory systems are emerging. Some of them are discussed below.
Regulation by CsrA.
The carbon storage regulator protein CsrA of E. coli is the central component of a global regulatory circuit that poises metabolism for rapid growth and inhibits processes associated with the stationary phase. CsrA controls carbon metabolism and inversely regulates cell motility and biofilm formation as a major regulator of the motility-to-sessility transition. CsrA is a small homodimeric RNA-binding protein that exerts its effect by either repressing or activating expression of target mRNAs posttranscriptionally. In E. coli and S. enterica, CsrA plays an important role in the regulation of c-di-GMP metabolism by regulating mRNA transcripts of c-di-GMP-metabolizing proteins (60, 61). In E. coli, the mRNA transcripts of two DGCs, YcdT and YdeH, are strongly repressed by CsrA. YdeH is required for PAG-mediated biofilm formation, while both YcdT and YdeH repress motility (60, 266). Tight regulation of the motility-to-sessility transition is ensured, as CsrA exerts multilayer effects on motility and biofilm formation. CsrA directly stabilizes the transcript of the master regulator FlhD4C2 (370) and destabilizes the pgaA transcript (371). CsrA also exerts repressive effects on additional biofilm components, such as SdiA, the quorum-sensing receptor for N-acyl-l-homoserine lactone (372).
In S. enterica, CsrA regulates the expression of at least 8 of 20 c-di-GMP-metabolizing proteins (61). Multilayer control of motility is exerted, as CsrA not only activates FlhD4C2 expression but also represses the degenerate EAL domain protein YdiV (STM1344), which functions as an antiactivator of FlhD4C2, and hence as a motility inhibitor (226, 227, 373). In addition, CsrA directly regulates the flagellar class III gene yhjH, encoding a PDE that reciprocally controls motility and CsgD-mediated biofilm formation. In addition, CsrA inversely regulates c-di-GMP-metabolizing proteins required for infection processes. Several CsrA-suppressed c-di-GMP-metabolizing proteins inhibit invasion, while the CsrA-activated PDE YhjH stimulates invasion of host epithelial cells. This finding is in line with the suggestion that CsrA controls the switch between different physiological states in the infection process (374, 375). CsrA regulation of c-di-GMP metabolism seems to be conserved in unrelated species, as a CsrA homolog, RsmA (regulator of secondary metabolism), in X. campestris also inhibits biofilm formation by directly binding to GGDEF protein transcripts (375a).
In P. aeruginosa, RsmA seems to have a similar regulatory role to that in S. enterica, as it inversely regulates components involved in chronic versus acute infection processes (376–378). Whether c-di-GMP metabolism is also a target for RsmA regulation remains to be tested, but the inverse regulation of expression of the RsmA targets (type III and type VI secretion systems) by c-di-GMP signaling (340) points in this direction.
Cyclic di-GMP and quorum sensing.
Quorum sensing, the cell-cell communication between bacteria, is a process that allows bacteria to coordinate population-wide activities such as biofilm formation, swarming, and virulence, by secreting into the medium and detecting small diffusible molecules (autoinducers). Many languages of bacterial communication exist, as signals of diverse structural classes are produced by various bacteria. Extra- and intracellular signaling pathways coordinate quorum sensing and c-di-GMP signaling pathways to control bacterial behavior (379, 380).
Vibrio cholerae biotype El Tor, the cause of the current cholera pandemic, is an example where quorum sensing and c-di-GMP signaling regulate biofilm formation in opposite ways. At a high cell density, high levels of autoinducer collectively trigger a signaling cascade that results in the expression of HapR (VC395_0600), the major regulator of quorum sensing. HapR regulates, directly and indirectly, the expression of at least 14 of 52 GGDEF/EAL domain proteins and 4 HD-GYP domain proteins, which leads to an overall reduction of the intracellular c-di-GMP concentration (62, 381) and to inhibition of biofilm formation. As in the case of CsrA, HapR shows multilayer control of physiological processes, as it also directly downregulates transcription of the response regulator VpsT, the c-di-GMP-dependent transcription activator of the vps exopolysaccharide synthesis genes (84). In contrast, c-di-GMP positively affects transcription of vpsT through the c-di-GMP-binding transcription regulator VpsR (169). The VpsR binding sites overlap HapR binding sites on the vpsT promoter, suggesting that information delivered by quorum sensing and c-di-GMP signaling is combined to determine the biofilm output response. Furthermore, transcription of the virulence activator AphA is oppositely regulated by HapR and VpsR, whose binding sites overlap, although quorum sensing and c-di-GMP signaling both downregulate virulence. Because AphA regulates functions other than virulence, e.g., acetoin production, which is coregulated by AphA and c-di-GMP, it is likely that c-di-GMP inhibition of virulence occurs downstream of AphA or via a parallel pathway. A quorum system regulating c-di-GMP signaling and surface phenotypes in V. parahaemolyticus was described above (see “c-di-GMP-dependent control of motility-to-sessility transition on surfaces”).
The diffusible signaling factor (DSF), a cis-unsaturated fatty acid, is required for virulence in the plant pathogen X. campestris, the causal agent of black rot of crucifers. DSF synthesis is completely dependent on RpfF. The response to DSF requires the signal to be sensed by the histidine kinase RpfC and the response regulator RpfG. Upon perception by RpfC, the REC domain of RpfG is believed to be phosphorylated, which increases the activity of an HD-GYP output domain that functions as a PDE. RpfG has broad activity, as it not only affects the secretion of virulence factors and the production of exopolysaccharide required for virulence but also stimulates type IV pilus-mediated motility and represses biofilm formation (Fig. 9). This suggests that RpfG is a highly active PDE and/or that it affects processes with broad effects on cell physiology or affects localization of several DGCs (98), as shown for RpoG from X. axonopodis. Reductions of c-di-GMP concentrations by RpfG are thought to regulate virulence through the activation of the transcriptional regulator Clp, which is repressed upon c-di-GMP binding. Clp subsequently regulates a plethora of genes associated with virulence function (341).
In parallel with the RpfC-RpfG system, the RavS-RavR two-component system cumulatively affects virulence through Clp (382). RavR (Xcc1958), a response regulator with a GGDEF-EAL-REC domain architecture that has PDE activity, is believed to respond to low oxygen concentrations. RavR affects Clp expression, and presumably its activity, through removal of c-di-GMP from the Clp binding site. Thus, concerted degradation of c-di-GMP by two different PDEs may lead to full activation of Clp as a transcription activator of virulence.
Recently, the first example of a cytosolic c-di-GMP-metabolizing protein that is directly activated by a quorum-sensing signal was uncovered. In Burkholderia cenocepacia, the PAS domain of the RpfR hybrid GGDEF-EAL domain protein senses the fatty acid signaling molecule BDSF (184a). BDSF is a quorum-sensing signaling molecule required for virulence, swarming motility, and biofim formation structurally similar to the diffusible signaling factor DSF signal which activates twitching motility and virulence in X. campestris (see above). BDSF binding to the PAS domain activates the phosphodiesterase activity of the EAL domain of RpfR.
Note that the quorum (i.e., autoinducer concentration) is sensed and interpreted by bacterial c-di-GMP signaling systems as one of many primary inputs, so the interactions between quorum sensing and c-di-GMP signaling are mainly unidirectional, i.e., the autoinducer concentration (primary signal) regulates synthesis and/or hydrolysis of the c-di-GMP (second messenger). It is interesting to look at how various second messengers “talk” to each other.
Cyclic di-GMP and other second messengers.
Surprisingly little is known about whether and how the common nucleotide second messengers cAMP and (p)ppGpp interact with c-di-GMP signaling (383). Cyclic AMP and (p)ppGpp control various processes in bacteria, from responses to nutrient starvation to regulation of biofilm formation to virulence (132, 384), and have been studied for decades (185, 385). Cyclic AMP signaling in proteobacteria occurs via the cAMP-binding transcription activator CRP. In V. cholerae, cAMP-CRP has been found to regulate the expression of several genes involved in c-di-GMP turnover (386). Cyclic AMP-mediated repression of biofilm formation is mediated mainly via cAMP-CRP inhibition of transcription of the DGC gene cdpA. In E. coli, c-di-GMP and ppGpp appear to regulate PAG-dependent biofilms in opposite directions, where c-di-GMP activates and ppGpp inhibits biofilm formation (266). Both messengers act on the posttranscriptional level by affecting the expression of PAG biosynthesis proteins.
Dinucleoside polyphosphates, which are side products of amino acyl tRNA synthesis, also function as ubiquitous signaling molecules with pleiotropic effects on cellular physiology. Impaired degradation of di-adenosine tetraphosphate (Ap4A) in P. fluorescens leads to elevated c-di-GMP levels and increased biofilm formation by two distinct pathways (387). It will be interesting to see whether dinucleoside polyphosphates intersect with c-di-GMP signaling pathways in other bacteria.
Open Questions in c-di-GMP Signaling
Is there yet another signaling molecule? Hydrolysis of c-di-GMP proceeds through pGpG, which may potentially function as a signaling molecule in its own right (5). It was established early on that in G. xylinus, pGpG is hydrolyzed into two GMP molecules by a separate PDE, termed PDE-B, not by the c-di-GMP-specific PDE (1, 25). pGpG belongs to the class of “nanoRNA” molecules, which are 2 to 5 nucleotides long. NanoRNAs control gene expression, as they can serve as templates in the promoter-specific initiation of RNA synthesis by DNA-dependent RNA polymerases in vitro and in vivo (388). Thus, the possibility exists that pGpG may have a role in gene expression, presumably under conditions of high c-di-GMP concentrations. NanoRNAs are converted to mononucleotides by the highly conserved oligoribonuclease Orn (389), which may represent the enigmatic PDE-B protein. Interestingly, Orn was least active against a GpG dinucleotide compared to a panel of other tested dinucleotides (390). This suggests that the half-life of pGpG may be prolonged selectively in vivo to allow priming of transcription. In this context, it is worth noting that cyclic dinucleotides have been shown to inhibit the activity of RNA polymerase in vitro under certain conditions (391).
As noted above, the HD-GYP domain PDE may hydrolyze pGpG more readily than the EAL domain PDE, which suggests another potential regulatory mechanism. While relative numbers of encoded EAL and HD-GYP domain proteins vary, many organisms contain PDEs of both types (http://www.ncbi.nlm.nih.gov/Complete_Genomes/c-di-GMP.html). As DGCs and PDEs interact with each other and with target proteins (89, 98, 192), these two nonhomologous c-di-GMP hydrolases and their catalytically inactive derivatives could provide distinct protein interaction platforms to regulate a variety of c-di-GMP-dependent processes.
Variable impacts of various c-di-GMP concentrations.
Another puzzling observation concerns the lack of correlation of c-di-GMP concentrations with the phenotypic output. Although there is no doubt that high c-di-GMP concentrations promote sessility, while low c-di-GMP concentrations promote motility, there is no absolute correlation between c-di-GMP concentrations and the degree of phenotype output (230, 233, 234, 297). A local versus global effect of c-di-GMP signaling is a possible explanation, where in one instance the DGC is located in close vicinity to the effector and in another the spatial distance requires a more globally acting DGC and higher c-di-GMP levels to obtain the desired effect (Fig. 14). Another aspect of this problem is that the change in c-di-GMP concentration does not necessarily correlate with the breadth of the physiological response. Recent studies provided convincing evidence that local and global c-di-GMP signaling exists. A very tight DGC-effector constellation coupled to a potentially physiologically very broad target output response was found with the DGC DosC in complex with the RNA degradosome (90). In such an arrangement, a broad physiological response can be achieved with a minimal absolute change in the c-di-GMP concentration. On the other hand, c-di-GMP is cumulatively fed into the c-di-GMP receptor VpsT, a transcriptional activator of biofilm formation in V. cholerae (see above), by five DGCs (307). At least two of these DGCs, the membrane-bound CdgA (VCA0074) and CdgH (VC1067) proteins, do not interact with VpsT, which is dispersed in the cytoplasm, but promote c-di-GMP-dependent localization and target promoter activation.
Deletions of individual c-di-GMP-metabolizing proteins can have a clear-cut, “all-or-nothing” phenotype indicating the dependence of a phenotype on one specific c-di-GMP source or sink (Fig. 11) (30). However, more frequently, the relative regulation of a phenotype is observed with additive effects from different c-di-GMP sources or sinks. In addition, a phenotype for a given c-di-GMP protein also depends on the appropriate c-di-GMP level. If c-di-GMP is effectively removed by a nearby PDE, the c-di-GMP-inhibited phenotype cannot reveal the corresponding DGC or the c-di-GMP-binding proteins unless the c-di-GMP sink is deleted.
Atypical behavior of c-di-GMP-metabolizing proteins.
While the basic principles of c-di-GMP signaling are known, several experimental observations do not fit the general scheme. These and other observations which are inconsistent with the established paradigm will remain puzzling for as long as we do not understand the underlying molecular mechanisms.
One example of the unorthodox behavior of c-di-GMP was recorded for P. aeruginosa, where the PDE Arr (PA2818) appeared to induce biofilm formation instead of inhibiting it (233). Biofilm formation was reportedly induced by the aminoglycoside antibiotics tobramycin, amikacin, streptomycin, and gentamicin, and the effect of tobramycin was abolished by mutation of the glutamate residue in the EAL motif of Arr (233).
Other examples of “odd” behavior have been uncovered in X. campestris, where c-di-GMP-metabolizing enzymes were found to work in the opposite direction from the expectation. It has been established on the molecular level that c-di-GMP inhibits endoglucanase secretion, because the transcription factor Clp in complex with c-di-GMP cannot activate the engXCA operon (222) (Fig. 13). However, secretion of endoglucanase was found to be downregulated by the deletion of PDEs as well as, strangely, potential DGCs (205). Curiously, endoglucanase secretion was also found to be downregulated by deletion of the PilZ-containing YcgR homolog XC_2317 (294). Another counterintuitive observation is that deletion of two GGDEF domain proteins that interact with the RpfG HD-GYP PDE, as well as RpfG, abolished instead of activated type IV pilus motility in X. campestris (192).
CYCLIC di-GMP-INDEPENDENT LIFE OF GGDEF, EAL, AND PILZ DOMAIN PROTEINS
A significant fraction of GGDEF, EAL, and HD-GYP domains lack not only catalytic activity but also the ability to bind their respective substrate, GTP or c-di-GMP (see “I sites and enzymatically inactive EAL and HD-GYP domains as c-di-GMP receptors”). Interestingly, many of these degenerate domain proteins have functions normally associated with c-di-GMP signaling. Some of these examples are presented below.
GdpS of S. aureus and S. epidermidis is the only GGDEF domain protein of staphylococci with a conserved GGEEF motif, but biochemical analysis showed that GdpS does not synthesize c-di-GMP (272). Thus, staphylococci lack the c-di-GMP signaling pathway. Nevertheless, GdpS has a significant effect on biofilm formation in S. epidermidis. In a gdpS mutant, the transcript levels of the icaADBC operon, required for synthesis of the PAG polysaccharide, virulence factors, and the protein A regulator SarS, were downregulated (272). Similarly, the enzymatically inactive GdpS protein of S. aureus proved necessary for secretion of virulence factors such as proteases, fibronectin-binding proteins, and protein A (272, 392). Restoration of mRNA downregulation in both biofilm formation and virulence factor production required the N-terminal 5TMR-LYT (PF07694) membrane domain of GdpS, but not its GGDEF domain. Interestingly, GdpP (YybT; SE0013), the second identifiable GGDEF domain protein, with highly degenerate characteristic signature motifs, is involved in the degradation of the secondary messenger c-di-AMP (393) (see below).
The EAL domain of the E. coli BLUF-EAL protein YcgF is degenerate and does not metabolize or bind c-di-GMP (63, 394). YcgF interacts with the Mer-like transcriptional regulator YcgE and relieves DNA binding by YcgE upon blue light exposure (394). This relief allows the expression of a group of genes encoding mainly small proteins of less than 100 amino acids, predominantly at the low temperature of 16°C. Upon overexpression, at least two of these proteins promote production of colanic acid and repress curli fimbria biosynthesis, suggesting that YcgF contributes to the switch in biofilm matrix expression in E. coli upon light exposure (123).
Another example of an enzyme “retired from active duty” is the stand-alone EAL domain protein YdiV (STM1344) of S. enterica and E. coli. Biochemical studies showed that YdiV neither hydrolyzes nor binds c-di-GMP (226). The structure of YdiV revealed replacements with alanines for several amino acid residues required for coordination of the divalent cations (394a). A ydiV deletion promotes motility and inhibits biofilm formation, in contrast to a deletion in yhjH, encoding an enzymatically competent PDE that is the closest homolog of YdiV in S. enterica (226). This puzzling observation was explained by studies that showed that expression of the YhjH PDE is inversely coupled to YdiV activity through the flagellar regulon. In vitro studies determined that YdiV binds to the FlhD component of the master regulator FlhD4C2 (394a). Structural studies revealed that four YdiV molecules subsequently bind to each of the FlhD subunits. Binding of more than two YdiV molecules opens up the ring-like structure of the FlhD4C2 complex formed on DNA and thereby prevents activation of class II flagellar genes. FlhD4C2 released from the promoter is targeted by YdiV for degradation to the ClpXP protease (227, 373).
The positive effect of YdiV on biofilm formation is indirect, as inhibition of the flagellar regulon blocks expression of the PDE YhjH, which is needed for downregulating the major biofilm regulator CsgD (226). CsgD expression is further activated by YdiV through downregulation of the second PDE, STM1703, through a still unknown pathway. YdiV strongly inhibits swarming, thus promoting the surface motility-to-sessility transition. It prevents flagellar expression upon nutrient limitation and contributes to bistable expression of flagellin in cell populations (226, 227, 395). In line with that, expression of YdiV is repressed by glucose and the carbon storage regulator protein CsrA (61, 396). YdiV downregulates flagellum expression upon S. enterica host infection to avoid the inflammatory host response and to establish successful colonization in the mouse model (395). Enhanced expression of immunostimulatory flagellin in the YdiV mutant triggered an enhanced inflammatory cytokine response and cytotoxicity toward macrophages (330, 395) and led to a disadvantage in deeper tissue colonization compared with an fliC deletion mutant.
While YcgF and YdiV seem to act exclusively through protein-protein interactions, other EAL domain proteins have evolved to bind RNA. The small RNAs CsrB and CsrC counteract the activity of the RNA-binding protein CsrA in E. coli (397). To balance the concentration of CsrA with its antagonists, active CsrA indirectly activates CsrB and CsrC synthesis. An additional component of the Csr circuit is the GGDEF-EAL domain protein CsrD (398, 399), in which both the GGDEF and EAL domains are degenerate. Consequently, CsrD does not display activities related to c-di-GMP metabolism as determined by biochemical and genetic studies (399). However, CsrD affects the degradation of CsrB and CsrC, which also requires the essential endonuclease RNase E and the exonuclease PNPase (399). CsrD binds CsrB with high affinity; however, it does not contribute directly to its degradation. Thus, CsrD might function as a specificity factor or adaptor that selects CsrB and CsrC and targets them for decay.
Ironically, another example of a protein that no longer involves c-di-GMP is PilZ (PA2960), a type IV pilus assembly factor in P. aeruginosa (146) and the eponymous protein for a superfamily of c-di-GMP receptor domains (48). P. aeruginosa PilZ does not bind c-di-GMP because it lacks conserved residues essential for c-di-GMP binding (149). However, it interacts with the c-di-GMP-binding protein FimX and with PilB, an ATPase required for type IV pilus polymerization (159). Therefore, PilZ serves as an adaptor to transmit c-di-GMP-induced changes in FimX to polymerization of pilin subunits, i.e., type IV pilus biogenesis. In general, a loss of c-di-GMP binding capacity appears to be fairly common among PilZ domain proteins (159).
PRACTICAL ASPECTS OF CYCLIC di-GMP
The widespread distribution of c-di-GMP in pathogenic bacteria and its involvement in a variety of processes that contribute to virulence make c-di-GMP signaling pathways potential targets for antibacterial interventions, particularly because c-di-GMP is absent in mammals. A number of recent studies investigated the effects of c-di-GMP on bacterial and eukaryotic cells. Although c-di-GMP proved not to be bactericidal and could not be used to kill bacterial pathogens, adding c-di-GMP to some bacterial cell cultures stimulated dispersal of bacterial biofilms (47). Importantly, c-di-GMP has been shown to have an immunomodulating effect in mice and has been used as an immune enhancer, vaccine adjuvant, and anticancer agent (56–58, 400, 401). Most remarkably, it turned out that human and mouse cells are already familiar with c-di-GMP and use it in their immunosurveillance mechanisms. The presence of c-di-GMP in the cytoplasm of mammalian cells is recognized as a sign of bacterial invasion, which triggers an innate immune response (56, 57, 69, 334, 402).
Use of c-di-GMP for Biofilm Dispersal
Biofilm formation plays a key role in disease processes by making bacteria refractory to traditional antibiotics, which is why, by some estimates, up to 80% of human infections involve biofilms. This makes preventing biofilm formation and/or accelerating dispersal of existing biofilms a potential component of antimicrobial therapies. Can we use the current knowledge of c-di-GMP signaling to devise ways of fighting bacterial biofilms (403–405, 405a)?
Biofilm formation is also a problem in industrial settings, for example, in the form of biofouling. On the other hand, the engineering of strains with a controlled biofilm formation capacity has potential applications from disease treatment to controlled production of biofuel (405).
The requirement of c-di-GMP for biofilm formation means that decreasing intracellular levels of c-di-GMP can cause (or accelerate) biofilm dispersal. Indeed, inducible expression of the E. coli PDE YhjH in Shewanella oneidensis resulted in detachment of the cells from the biofilm matrix upon induction, apparently by lowering the c-di-GMP levels (239). For P. putida, the mechanism of detachment initiated by the decrease in the c-di-GMP level is now understood. It occurs through the LapD-LapG-LapA mechanism (see “Adhesins as c-di-GMP targets”), whereby the LapA adhesin is cleaved from the cell surface (68, 406).
Overexpression of the E. coli YjgI protein, which lacks PDE activity and instead belongs to the short-chain dehydrogenase family related to 3-ketoacyl-ACP reductase FabG, increases biofilm dispersal in E. coli. YjgI, renamed BdcA (biofilm dispersal via c-di-GMP), binds c-di-GMP, albeit with a high KD, 11 μM (176). An engineered E50Q mutant of this protein binds c-di-GMP with a much lower KD, 3 μM (176). BdcA overexpression in P. aeruginosa and Rhizobium meliloti leads to biofilm dispersal in these organisms (407). Most likely, BdcA acts as c-di-GMP sink, whose overexpression effectively decreases the intracellular c-di-GMP pool available to other receptors. These data show that decreasing cellular c-di-GMP levels could be an effective method of combatting bacterial biofilms.
Controlling biofilm formation in native, nonengineered cultures may be achieved by manipulating external signals that inhibit DGCs and/or activate PDEs. An attractive signal for biofilm dispersal is nitric oxide (NO), which is naturally produced by many prokaryotes and eukaryotes. While it is bactericidal at high concentrations, NO at nanomolar levels functions as a signaling molecule, triggering biofilm dispersal in a wide range of microorganisms (408). In P. aeruginosa, the NO effect has been linked to the stimulation of a c-di-GMP-specific PDE activity, both direct, through increased expression of the PDEs DipA (PA5017) and RbdA (PA0861) (315, 409), and indirect, mediated through the unusual chemotaxis receptor protein PA1423 (termed BdlA for biofilm dispersion locus A), which consists of two tandem PAS domains and a C-terminal MCP domain (410). Reduction of biofilm formation by NO modulation of c-di-GMP levels has been observed in other bacteria as well. In L. pneumophila and Shewanella woodyi, NO is sensed by an H-NOX domain protein which inhibits the DGC activity (411). Interestingly, NO controls hydrolysis of yet another cyclic dinucleotide, c-di-AMP, i.e., the c-di-AMP-specific PDE YybT of B. subtilis is activated by NO (412). Thus, the freely diffusible molecule NO is a promising antibiofilm compound that works through modulation of the c-di-GMP level and, possibly, c-di-AMP levels. Surprisingly, the upregulation of biofilm formation at intermediate NO levels is also mediated via c-di-GMP signaling (189), indicating that NO levels need to be adjusted tightly to achieve a biofilm-inhibiting effect.
As an alternative to targeting c-di-GMP network components, which are structurally diverse, it has been suggested to target the signaling molecule itself (413). Intercalators have been identified which trigger c-di-GMP to form higher-order aggregates. Those complexes are predicted to be biologically inactive, as only monomers and dimers of c-di-GMP have been found in complexes with c-di-GMP receptors.
Finally, it is important that extracellular c-di-GMP could also be used for biofilm dispersal. In 2005, a study by Karaolis and colleagues showed that treatment of S. aureus cells with 200 μM c-di-GMP had no effect on the growth rate but inhibited cell adhesion in liquid medium, inhibited adherence of S. aureus to human epithelial cells, and reduced biofilm formation >2-fold (47). These effects have been confirmed for a bovine mastitis isolate of S. aureus and a human methicillin-resistant S. aureus (MRSA) isolate, indicating their relevance to clinical settings. However, the high levels of c-di-GMP (at least 20 to 50 μM) required for these experiments make them impractical, at least until highly efficient methods of producing c-di-GMP have been developed (see below). Very recently, a high-throughput screen for DGC inhibitors has uncovered several small molecules that inhibit DGCs at submicromolar concentrations. These compounds, which proved active against diverse DGCs, hold the highest promise yet, as a means of preventing biofilm formation by bacterial pathogens (413a).
Cyclic di-GMP as an Immunomodulator
The first experiments on the potential effects of c-di-GMP on animal cells used human cancer cells and showed that addition of 50 μM c-di-GMP inhibited proliferation of several tumor cell lines: the human acute lymphoblastic leukemia cell line Molt 4 and the human T-cell lymphoblastoid CD4+ Jurkat cell line (23, 414). These observations were attributed to the binding of c-di-GMP to p21ras, a member of the Ras superfamily of GTPases (23). Several years later, c-di-GMP was shown to inhibit proliferation of human colon cancer cells and was suggested as a potential anticancer agent (401).
Subsequent studies in live mice confirmed that c-di-GMP had an immunostimulatory effect, affecting infection by S. aureus (including MRSA) and bacterial pneumonia caused by K. pneumoniae or Streptococcus pneumoniae (56–58, 400, 415). Administration of c-di-GMP to mice induced secretion of a variety of cytokines and chemokines, including interferons and interleukins (Table 8), and altered the expression of several chemokine receptors (56, 58). c-di-GMP also stimulated other innate immune responses, such as the recruitment of innate immune cells such as neutrophils, monocytes, and granulocytes (56, 57). As a result, c-di-GMP improved the antibacterial resistance of treated mice and accelerated the clearance of bacteria from the spleen and liver (400). In addition, c-di-GMP has been shown to work as an adjuvant, increasing the immune response to such commonly used antigens as S. aureus clumping factor A (ClfA), mutant staphylococcal enterotoxin, pneumolysin toxoid, and pneumococcal surface protein A (PspA) (56, 58). These data prompted proposals to use c-di-GMP clinically in humans and animals as an immunomodulator, immune enhancer, immunotherapeutic, immunoprophylactic, or vaccine adjuvant (55, 56, 58). Such a proposal was included in several patent applications by David Karaolis, two of which were granted in 2009, as U.S. patents 7,569,555 and 7,592,326 (418). The ability of c-di-GMP to work as a mucosal adjuvant was recently used for intranasal and sublingual administration of live attenuated vaccines of influenza H5N1 virus (419, 420).
Table 8.
Immunostimulating properties of c-di-GMP
| Cell type | Response to 200 μM c-di-GMP |
Reference | |
|---|---|---|---|
| Cytokines and chemokines | Other proteins | ||
| Human or mouse immature dendritic cells | Expression of gamma interferon (IFN-γ), IL-12, IL-1β, tumor necrosis factor (TNF), IFN-α, IL-8/CXCL8, monokine induced by IFN-γ (MIG)/CXCL9, IFN-γ-inducible protein 10/CXCL10, IFN-inducible T cell α chemoattractant/CXCL11, MCP-1/CCL2, MIP-1a/CCL3, MIP-1b/CCL4, and RANTES/CCL5 | Expression of costimulatory molecules CD80/CD86, maturation marker CD83, major histocompatibility complex (MHC) class II, chemokine receptors CCR1, CCR7, and CXCR4, p38 mitogen-activated protein kinase (MAPK); downregulation of CCR1 and CCR5 | 56 |
| Mouse bronchoalveolar lavage cells | Expression of KC, MCP-1, MIP-1α, MIP-2, RANTES | Expression of IgA, IgG | 416, 417 |
Since stimulation of the immune response of dendritic cells by c-di-GMP has been shown to be independent of Toll-like receptor- and nucleotide-binding oligomerization domain (Nod)-dependent pathways (56), the detailed mechanism of immunostimulation by c-di-GMP long remained unclear (334). Finally, a recent study identified the cytoplasmic domain of the transmembrane protein STING as a direct sensor of intracellular c-di-GMP (69, 335). STING binds c-di-GMP with a KD of 5 μM and stimulates production of type I interferons by signaling through the TANK binding kinase 1 (TBK1) and the interferon regulatory transcription factor 3 (IRF3) (69). At least five groups have solved the crystal structure of STING (70, 72, 73, 177, 421) in complex with c-di-GMP (Fig. 5G). Subsequent studies uncovered yet another, slightly more sensitive, mammalian c-di-GMP receptor, the DEAD-box helicase DDX41 (421a). Although it can be debated whether c-di-GMP ever reaches micromolar concentration levels inside mammalian cells, additional candidate eukaryotic c-di-GMP receptors exist (421). It remains to be seen how c-di-GMP makes its way into the cytosol of macrophages, but overexpression of a DGC in the intracellular pathogen L. pneumophila was correlated with increased type I interferon expression (421). These results highlight an important role for sensing c-di-GMP in the immunosurveillance by mammalian cells.
Thus, it appears that c-di-GMP (and other cyclic dinucleotides [see below]) belongs to a select group of compounds that are being kept under constant immunosurveillance. This group includes microbe-specific essential macromolecules such as the lipid A component of lipopolysaccharide, cell wall components, and bacterial DNA, as well as flagellin, which is nonessential but encoded by many pathogenic bacteria. These compounds represent conserved molecular signatures of bacteria that are used by the innate immune system to recognize “nonself” (i.e., bacterial infection) and elicit immune responses on the cellular and humoral levels. It could be argued that our cells recognized the importance of c-di-GMP long before we did.
Synthesis of c-di-GMP
Studies of the molecular mechanisms of c-di-GMP signaling and its potential use as an adjuvant or an anticancer agent require substantial amounts of this compound. For the first several years, c-di-GMP research relied largely on relatively small amounts of the substance synthesized enzymatically in the Benziman lab and kindly shared by Benziman's colleague Haim Weinhouse. The growing interest in c-di-GMP studies worldwide encouraged researchers to look for new methods of obtaining this compound.
The first chemical synthesis of c-di-GMP and its analogs was performed by Benziman's colleagues to prove that the artificially produced molecule was identical to the natural activator of cellulose synthase (422). This synthesis, as well as most of the subsequent approaches to chemically synthesize cyclic dinucleotides, used a derivatized nucleoside as a starting material (423–425). An alternative approach first synthesized the cyclic backbone, with subsequent introduction of the guanine base (426). The general drawbacks of these early approaches to chemically synthesize c-di-GMP were the laborious synthesis and purification procedure and the low yield, typically resulting in only a few milligrams of the product.
Recent advances in c-di-GMP synthesis followed two separate tracks: one included improvements in organic synthesis, whereas the other relied on streamlining the enzymatic production of c-di-GMP. The improved chemical synthesis procedure takes advantage of solid-phase synthesis and employs a one-flask synthesis protocol that gets rid of intermediate washing and purification steps (427, 428). This protocol allows for economical production of gram-scale amounts of c-di-GMP and its derivatives starting from commercially available and relatively low-cost guanosine phosphoramidite.
Enzymatic synthesis of c-di-GMP was hampered by product inhibition via the I sites of DGCs, which led to relatively low yields. To solve this problem, the Jenal lab mutated the I site in the constitutively active PleD mutant (113). Later, the Liang lab engineered TM1788, a single-domain GGDEF domain protein from the thermophilic organism Thermotoga maritima, for solubility and high product yields by mutating the I site (429). The optimized reaction conditions enable synthesis of hundreds of milligrams of c-di-GMP in vitro. The drawback of using DGCs to produce c-di-GMP from GTP is that GTP is relatively expensive. A solution to this problem has been found by Spehr and colleagues, who added two enzymatic steps to produce GTP from cheaper substrates (430). In their protocol, GTP is produced from GMP and ATP by the subsequent action of GMP kinase (which converts GMP into GDP) and nucleoside diphosphate kinase (which produces GTP from GDP). GTP is subsequently converted into c-di-GMP by an engineered DGC with a mutated I site. In a large-scale batch reaction, the authors were able to obtain gram amounts of c-di-GMP, with an overall yield of up to 44% of added GMP (430).
Quantification of c-di-GMP
Quantification of cellular c-di-GMP concentrations allows assessment of the activity of c-di-GMP-metabolizing proteins in vivo. Separation of cell extracts by high-pressure liquid chromatography combined with quantification by mass spectrometry has been the gold standard for measuring c-di-GMP concentrations (17, 431, 432). Alternative detection and quantification methods for in vitro and in vivo applications are being developed, although the required sensitivity and/or applicability for high-throughput analysis has not yet been reached. One line of development uses the unique property of c-di-GMP to form stable complexes with some planar intercalators (433, 434). In the presence of proflavine and hemin, c-di-GMP forms a supramolecular complex with peroxidase activity. Since the number of nucleotide-enzyme complexes correlates with substrate turnover, c-di-GMP is conveniently quantified with a substrate which changes the absorbance spectrum upon oxidation (435). Furthermore, significant changes in the absorption and fluorescence spectra upon interaction with c-di-GMP can be used for detection and quantification (436). Thiazole orange, a known fluorescent intercalator of nucleic acids, can specifically detect c-di-GMP in a concentration-dependent manner. Other quantification approaches use the high affinity of RNA aptamers for c-di-GMP to develop c-di-GMP-sensitive ribozymes (437). c-di-GMP concentrations down to 10 nM can be detected. On the other hand, the conformational change upon c-di-GMP binding to its receptor molecules has been used to design fluorescent biosensors for in vitro and in vivo use (219, 438). Also, transcription-based methods of monitoring c-di-GMP levels in vivo have been developed (413a, 438a). These latter methods may facilitate evaluation of c-di-GMP levels in bacteria under physiological conditions (438b). Although the sensitivity of the system remains a major drawback of current detection methods, these approaches have the potential to be developed into highly sensitive and versatile methods for detection of c-di-GMP. Another potential approach would be to develop a specific antibody to detect c-di-GMP. In this way, c-di-GMP could be detected conveniently by enzyme-linked immunosorbent assay (ELISA), as is the case for the cyclic mononucleotide second messengers cAMP and cGMP.
THE NOVEL CYCLIC DINUCLEOTIDE SECOND MESSENGERS CYCLIC di-AMP and CYCLIC AMP-GMP
Cyclic di-AMP.
In an unexpected development, the crystal structure of the DNA integrity scanning protein DisA, which responds to DNA double-strand breaks in Bacillus subtilis, was found to be associated with cyclic di-adenosine nucleotide (c-di-AMP) (439). Since c-di-AMP had never been observed in living cells previously and had only been synthesized chemically (440), this finding suggested secondary messenger signaling in bacteria by yet another cyclic dinucleotide (441). A diadenylate cyclase activity of the N-terminal DAC domain of DisA was verified by biochemical studies (439). Subsequently, a bioinformatic search for proteins that co-occur with DAC domain proteins identified members of the DHH domain family, which have documented phosphatase or phosphoesterase activity (442), as candidates for c-di-AMP-specific PDE activity. Physiologically relevant c-di-AMP hydrolysis was demonstrated for B. subtilis YybT (alternatively called GdpP), a multidomain protein that contains DHH, DHHA1, and a degenerate GGDEF domain (393).
In the past 2 years, c-di-AMP has been fully established as a second messenger in several Gram-positive bacteria, including S. aureus, Streptococcus pyogenes, Listeria monocytogenes, and M. tuberculosis (119, 443–447). In B. subtilis, elevated intracellular c-di-AMP levels serve as a signal for entry into sporulation. The checkpoint protein DisA scans the chromosome for lesions prior to sporulation (448) and synthesizes c-di-AMP, which apparently signals that DNA is undamaged and allows the progression to sporulation (439, 446). Upon DNA damage, however, DisA stalls and its diadenylate cyclase activity is inhibited. The drop in c-di-AMP level is probably further steepened as the c-di-AMP PDE YybT is concomitantly upregulated (446).
The diadenylate cyclase domain (DAC, or DisA_N; entry PF02457 in the Pfam database [116]) is found in most bacterial phyla (albeit not in Alpha-, Beta-, or Gammaproteobacteria) and in the Euryarchaeota (but not in Cren- or Thaumarchaeota). This suggests that like c-di-GMP, c-di-AMP is part of an ancient signaling system that has additional functions beyond monitoring DNA integrity.
In S. aureus, c-di-AMP has a role in resistance to extreme cell wall stress (443). A mutant in gdpP (yybT), encoding the c-di-AMP PDE, suppressed the lethal effect caused by the absence of lipoteichoic acid in the cell wall, enhanced the cross-linking of the peptidoglycan, and altered the expression of autolysin. Furthermore, elevated c-di-AMP levels were associated with increased resistance to some cell wall-active antimicrobials and with reduced cell size. Decreased resistance to β-lactam antibiotics upon decreased c-di-AMP levels was also observed in B. subtilis (445), where deletion of YbbP, one of three diadenylate cyclases, had the most pronounced effect. Screening studies of various Gram-positive bacteria provided further evidence for a more general role of c-di-AMP in resistance to various stresses, such as acid and oxidative stress (393, 449, 450). The next challenge is to identify c-di-AMP receptors, and there has also already been some progress. DarR, a TetR family transcriptional regulator in M. smegmatis, has been identified as the first bacterial c-di-AMP receptor (450a). Three identified DarR-regulated genes indicate that c-di-AMP has a role in cold shock and fatty acid synthesis and transport.
Like c-di-GMP, c-di-AMP is detected by mammalian immune systems. Cyclic di-AMP secreted into the cytosol of host cells by multiefflux pumps of the intracellular pathogen L. monocytogenes activates a cytosolic surveillance pathway in host immune cells such as macrophages (447). This potential role of c-di-AMP in virulence and stress resistance, in combination with the fact that genes coding for diadenylate cyclases are essential in intracellular pathogens (451–454), makes c-di-AMP signaling pathways an attractive target for antimicrobial therapy.
Cyclic AMP-GMP.
Yet another cyclic dinucleotide, the c-AMP-GMP hybrid, was recently discovered in V. cholerae (455). Cyclic AMP-GMP is synthesized by the novel dinucleotide cyclase DncV (VC0179), a member of the nucleotidyltransferase superfamily that is distantly related to 2′,5′-oligoadenylate synthetases (OAS1) and poly(A) polymerase (455) but distinct from either c-di-GMP-synthesizing GGDEF or c-di-AMP-synthesizing DAC domains. The key active site residues [G(G/S)x9-13Dx(D/E)] of the nucleotidyltransferase superfamily are conserved in DncV and required for its catalytic activity.
DncV homologs are present in a subset of proteobacteria, including such enterobacterial species as diarrheagenic E. coli strain DEC8D. In V. cholerae El Tor, the dncV gene is located on the Vibrio 7th pandemic island 1 (Vsp-1), which is thought to contribute to the success of this biotype in causing pandemics. Indeed, deletion of dncV affects intestinal colonization of V. cholerae in an infant mouse model. As intestinal colonization requires downregulation of chemotaxis and DncV represses chemotaxis, the colonization defect of the dncV mutant may be caused by deregulated chemotaxis.
Cyclic AMP-GMP and c-di-GMP signaling pathways are distinct and most likely are inversely coupled during infection, as low c-di-GMP enhances transcription of the master virulence regulator ToxT, which indirectly activates dncV expression (455). Remarkably, c-AMP-GMP has recently been found to be a signaling molecule in higher eukaryotes (455a).
CONCLUDING REMARKS AND PERSPECTIVES
In the first 10 years of its 25-year history, c-di-GMP remained on the sidelines of microbiological research. It was mostly perceived as an idiosyncratic regulator of a rare process in an unusual bacterium and had been studied only by the Benziman group and their collaborators (1, 18, 25, 33, 35, 80, 145, 414; see also references 19 and 20 and Table 2). Compared to the much-better-known cyclic nucleotide second messenger cAMP, c-di-GMP was about as popular as Kombucha tea compared to black or green tea. This all changed at the turn of this century, with the identification of the c-di-GMP-metabolizing GGDEF and EAL domains (25) and the discovery of multiple copies of these domains in the genomes of E. coli and a variety of other bacteria (27, 34, 107). It became clear that c-di-GMP was part of a widespread signal transduction system, even though the scale and importance of that system proved far greater than anybody could have imagined at that time.
The conversion of c-di-GMP from an ugly duckling into a white swan was a consequence of several other important developments, some of which are highlighted in Table 2. As we have noted previously (5, 20), these extraordinary developments bring several lessons that might be of general significance for (micro)biologists. (i) Although the goal of Benziman's work was applied, i.e., creating an effective system to produce bacterial cellulose by using a purified enzyme, his studies led to a fundamental discovery that dramatically improved our understanding of bacterial signal transduction. (ii) Cellulose biosynthesis, a supposedly arcane process carried out by an unusual bacterium, turned out to be widespread in the bacterial world, occurring even in the most-studied bacteria, such as E. coli and S. enterica. This “arcane” process has helped us to understand bacterial multicellular behavior. (iii) A study of a benign and harmless bacterium (which can even be consumed in the form of Kombucha tea and nata de coco) has resulted in major advances relevant to clinical microbiology and immunology, leading to a better understanding of the virulence and transmission of major pathogens, including V. cholerae, Y. pestis, P. aeruginosa, and S. enterica. (iv) Exploration of the c-di-GMP-dependent mechanisms of biofilm formation opened entirely novel avenues of research in antibacterial drug design.
These lessons highlight yet another example of the serendipitous nature of most discoveries and emphasize the futility of efforts to accelerate progress by narrowing down scientific inquiry to a limited number of applied/translational directions.
Where do we go from here? We believe that the most important gaps in our understanding of c-di-GMP-dependent signaling and, accordingly, the key directions of c-di-GMP research are as follows.
Inputs into c-di-GMP-dependent signal transduction.
So far, few environmental signals that regulate c-di-GMP-mediated signaling pathways have been identified. Even less is known about the intracellular signals sensed by the enzymes involved in c-di-GMP metabolism. Identifying the inputs into c-di-GMP signaling pathways will be necessary to understand the roles of these pathways in bacterial physiology and host-pathogen interactions.
Outputs of c-di-GMP signaling.
Although c-di-GMP has been shown to act on the transcriptional, posttranscriptional, and posttranslational levels, the number of identified c-di-GMP targets and mechanisms of c-di-GMP-mediated regulation remains relatively low. For example, despite many studies, the exact mechanisms through which c-di-GMP activates polysaccharide synthesis, one of the most common c-di-GMP targets and a key component of biofilms, remain unknown, nor have we fully resolved the issue of the apparent imbalance in the numbers of c-di-GMP-metabolizing enzymes and c-di-GMP receptors (the “c-di-GMP army” problem).
Understanding c-di-GMP signaling with spatial precision.
Several c-di-GMP signaling systems have distinct locations in the cell, yet the reasons and mechanisms responsible for their localization remain poorly understood. Furthermore, many c-di-GMP signaling modules are spatially restricted and secluded from other modules. How is the spatial separation of c-di-GMP signaling pathways achieved?
Understanding c-di-GMP signaling at sufficient temporal resolution.
Several recent studies highlighted the importance of transient c-di-GMP gradients. We need to move beyond the static view of the average intracellular c-di-GMP concentration as the sole deterministic factor of cell physiology and begin exploring c-di-GMP signaling on a much shorter time scale (e.g., seconds or faster).
Role of c-di-GMP in host-pathogen interactions.
The dual roles of c-di-GMP as a factor promoting long-term survival of pathogenic bacteria in chronic diseases and an alarmone that alerts the immune system to bacterial infection should account for a very interesting dynamics of c-di-GMP signaling in pathogens during infections.
In this review, we described the progress made during the first 25 years of exploration of the c-di-GMP world, the period when c-di-GMP emerged from obscurity to the limelight of bacterial regulation. We expect the next 25 years to be no less exciting for the c-di-GMP field and the broader field of cyclic dinucleotide second messengers.
ACKNOWLEDGMENTS
We apologize to those authors whose studies may have been inadvertently overlooked or discussed only briefly because of space limitations.
The work of our groups has been supported by the European Commission, the Swedish Research Council of Natural Sciences, the Petrus and Augusta Hedlund Foundation, the Carl Trygger Foundation, the Swedish Research Council of Medicine and Health, and the Karolinska Institutet (U.R.); by the Intramural Research Program of the National Institutes of Health at the National Library of Medicine (M.Y.G.); and by grants from the NSF (MCB 1052575), the USDA (AFRI 2010-65201-20599), and the University of Wyoming Agricultural Experiment Station (M.G.).
REFERENCES
- 1. Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281 [DOI] [PubMed] [Google Scholar]
- 2. D'Argenio DA, Miller SI. 2004. Cyclic di-GMP as a bacterial second messenger. Microbiology 150:2497–2502 [DOI] [PubMed] [Google Scholar]
- 3. Jenal U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185–191 [DOI] [PubMed] [Google Scholar]
- 4. Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385–407 [DOI] [PubMed] [Google Scholar]
- 5. Römling U, Gomelsky M, Galperin MY. 2005. c-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629–639 [DOI] [PubMed] [Google Scholar]
- 6. Römling U, Amikam D. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9:218–228 [DOI] [PubMed] [Google Scholar]
- 7. Tamayo R, Pratt JT, Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61:131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pruitt KD, Tatusova T, Brown GR, Maglott DR. 2012. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 40:D130–D135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 10. Liberman JA, Wedekind JE. 2011. Base ionization and ligand binding: how small ribozymes and riboswitches gain a foothold in a protein world. Curr. Opin. Struct. Biol. 21:327–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mills E, Pultz IS, Kulasekara HD, Miller SI. 2011. The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell. Microbiol. 13:1122–1129 [DOI] [PubMed] [Google Scholar]
- 12. Römling U, Simm R. 2009. Prevailing concepts of c-di-GMP signaling. Contrib. Microbiol. 16:161–181 [DOI] [PubMed] [Google Scholar]
- 13. Römling U. 2012. Cyclic di-GMP, an established secondary messenger still speeding up. Environ. Microbiol. 14:1817–1829 [DOI] [PubMed] [Google Scholar]
- 14. Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 7:724–735 [DOI] [PubMed] [Google Scholar]
- 15. Sondermann H, Shikuma NJ, Yildiz FH. 2012. You've come a long way: c-di-GMP signaling. Curr. Opin. Microbiol. 15:140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wolfe AJ, Visick KL. 2008. Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J. Bacteriol. 190:463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wolfe AJ, Visick KL. (ed). 2010. The second messenger cyclic di-GMP. ASM Press, Washington, DC [Google Scholar]
- 18. Ross P, Mayer R, Benziman M. 1991. Cellulose biosynthesis and function in bacteria. Microbiol. Rev. 55:35–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Delmer DP. 2000. Structure and biosynthesis of cellulose. Part II: biosynthesis, p 199–216 In Kung S-D, Yang S-F. (ed), Discoveries in plant biology, vol 3 World Scientific Publishing Co, Singapore, Singapore [Google Scholar]
- 20. Amikam D, Weinhouse H, Galperin MY. 2010. Moshe Benziman and the discovery of c-di-GMP, p 11–23 In Wolfe AJ, Visick KL. (ed), The second messenger cyclic di-GMP. ASM Press, Washington, DC [Google Scholar]
- 21. Aschner M, Hestrin S. 1946. Fibrillar structure of cellulose of bacterial and animal origin. Nature 157:659. [DOI] [PubMed] [Google Scholar]
- 22. Hestrin S, Aschner M, Mager J. 1947. Synthesis of cellulose by resting cells of Acetobacter xylinum. Nature 159:64–65 [DOI] [PubMed] [Google Scholar]
- 23. Amikam D, Steinberger O, Shkolnik T, Ben-Ishai Z. 1995. The novel cyclic dinucleotide 3′-5′ cyclic diguanylic acid binds to p21ras and enhances DNA synthesis but not cell replication in the Molt 4 cell line. Biochem. J. 311:921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hecht GB, Newton A. 1995. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 177:6223–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tal R, Wong HC, Calhoon R, Gelfand DH, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Merkel TJ, Barros C, Stibitz S. 1998. Characterization of the bvgR locus of Bordetella pertussis. J. Bacteriol. 180:1682–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galperin MY, Natale DA, Aravind L, Koonin EV. 1999. A specialized version of the HD hydrolase domain implicated in signal transduction. J. Mol. Microbiol. Biotechnol. 1:303–305 [PMC free article] [PubMed] [Google Scholar]
- 28. Aldridge P, Jenal U. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 32:379–391 [DOI] [PubMed] [Google Scholar]
- 29. Ausmees N, Jonsson H, Höglund S, Ljunggren H, Lindberg M. 1999. Structural and putative regulatory genes involved in cellulose synthesis in Rhizobium leguminosarum bv. trifolii. Microbiology 145:1253–1262 [DOI] [PubMed] [Google Scholar]
- 30. Römling U, Rohde M, Olsen A, Normark S, Reinköster J. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10–23 [DOI] [PubMed] [Google Scholar]
- 31. Slater H, Alvarez-Morales A, Barber CE, Daniels MJ, Dow JM. 2000. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol. Microbiol. 38:986–1003 [DOI] [PubMed] [Google Scholar]
- 32. Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ausmees N, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Lindberg M. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163–167 [DOI] [PubMed] [Google Scholar]
- 34. Galperin MY, Nikolskaya AN, Koonin EV. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11–21 [DOI] [PubMed] [Google Scholar]
- 35. Chang AL, Tuckerman JR, Gonzalez G, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Gilles-Gonzalez MA. 2001. Phosphodiesterase A1, a regulator of cellulose synthesis in Acetobacter xylinum, is a heme-based sensor. Biochemistry 40:3420–3426 [DOI] [PubMed] [Google Scholar]
- 36. Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. U. S. A. 101:17084–17089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kirillina O, Fetherston JD, Bobrov AG, Abney J, Perry RD. 2004. HmsP, a putative phosphodiesterase, and HmsT, a putative diguanylate cyclase, control Hms-dependent biofilm formation in Yersinia pestis. Mol. Microbiol. 54:75–88 [DOI] [PubMed] [Google Scholar]
- 39. Tischler AD, Camilli A. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tischler AD, Camilli A. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73:5873–5882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123–1134 [DOI] [PubMed] [Google Scholar]
- 42. Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bobrov AG, Kirillina O, Perry RD. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247:123–130 [DOI] [PubMed] [Google Scholar]
- 44. Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280:30829–30837 [DOI] [PubMed] [Google Scholar]
- 45. Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tamayo R, Tischler AD, Camilli A. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280:33324–33330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Karaolis DK, Rashid MH, Chythanya R, Luo W, Hyodo M, Hayakawa Y. 2005. c-di-GMP (3′-5′-cyclic diguanylic acid) inhibits Staphylococcus aureus cell-cell interactions and biofilm formation. Antimicrob. Agents Chemother. 49:1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6 [DOI] [PubMed] [Google Scholar]
- 49. Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. 2006. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol. Microbiol. 62:1014–1034 [DOI] [PubMed] [Google Scholar]
- 50. Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. 2007. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 26:5153–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Christen M, Christen B, Allan MG, Folcher M, Jeno P, Grzesiek S, Jenal U. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 104:4112–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ryjenkov DA, Simm R, Römling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP. The PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281:30310–30314 [DOI] [PubMed] [Google Scholar]
- 53. Kim YK, McCarter LL. 2007. ScrG, a GGDEF-EAL protein, participates in regulating swarming and sticking in Vibrio parahaemolyticus. J. Bacteriol. 189:4094–4107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tarutina M, Ryjenkov DA, Gomelsky M. 2006. An unorthodox bacteriophytochrome from Rhodobacter sphaeroides involved in turnover of the second messenger c-di-GMP. J. Biol. Chem. 281:34751–34758 [DOI] [PubMed] [Google Scholar]
- 55. Ebensen T, Schulze K, Riese P, Morr M, Guzman CA. 2007. The bacterial second messenger cdiGMP exhibits promising activity as a mucosal adjuvant. Clin. Vaccine Immunol. 14:952–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Karaolis DK, Means TK, Yang D, Takahashi M, Yoshimura T, Muraille E, Philpott D, Schroeder JT, Hyodo M, Hayakawa Y, Talbot BG, Brouillette E, Malouin F. 2007. Bacterial c-di-GMP is an immunostimulatory molecule. J. Immunol. 178:2171–2181 [DOI] [PubMed] [Google Scholar]
- 57. Karaolis DK, Newstead MW, Zeng X, Hyodo M, Hayakawa Y, Bhan U, Liang H, Standiford TJ. 2007. Cyclic di-GMP stimulates protective innate immunity in bacterial pneumonia. Infect. Immun. 75:4942–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ogunniyi AD, Paton JC, Kirby AC, McCullers JA, Cook J, Hyodo M, Hayakawa Y, Karaolis DK. 2008. c-di-GMP is an effective immunomodulator and vaccine adjuvant against pneumococcal infection. Vaccine 26:4676–4685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321:411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Jonas K, Edwards AN, Simm R, Romeo T, Römling U, Melefors Ö. 2008. The RNA binding protein CsrA controls cyclic di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol. Microbiol. 70:236–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jonas K, Edwards AN, Ahmad I, Romeo T, Römling U, Melefors O. 2010. Complex regulatory network encompassing the Csr, c-di-GMP and motility systems of Salmonella Typhimurium. Environ. Microbiol. 12:524–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Waters CM, Lu W, Rabinowitz JD, Bassler BL. 2008. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 190:2527–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Barends TR, Hartmann E, Griese JJ, Beitlich T, Kirienko NV, Ryjenkov DA, Reinstein J, Shoeman RI, Gomelsky M, Schlichting I. 2009. Structure and mechanism of a bacterial light-regulated cyclic nucleotide phosphodiesterase. Nature 459:1015–1018 [DOI] [PubMed] [Google Scholar]
- 64. Minasov G, Padavattan S, Shuvalova L, Brunzelle JS, Miller DJ, Basle A, Massa C, Collart FR, Schirmer T, Anderson WF. 2009. Crystal structures of YkuI and its complex with second messenger cyclic di-GMP suggest catalytic mechanism of phosphodiester bond cleavage by EAL domains. J. Biol. Chem. 284:13174–13184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tchigvintsev A, Xu X, Singer A, Chang C, Brown G, Proudfoot M, Cui H, Flick R, Anderson WF, Joachimiak A, Galperin MY, Savchenko A, Yakunin AF. 2010. Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases. J. Mol. Biol. 402:524–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. 2010. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 329:845–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Navarro MV, Newell PD, Krasteva PV, Chatterjee D, Madden DR, O'Toole GA, Sondermann H. 2011. Structural basis for c-di-GMP-mediated inside-out signaling controlling periplasmic proteolysis. PLoS Biol. 9:e1000588 doi:10.1371/journal.pbio.1000588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Newell PD, Boyd CD, Sondermann H, O'Toole GA. 2011. A c-di-GMP effector system controls cell adhesion by inside-out signaling and surface protein cleavage. PLoS Biol. 9:e1000587 doi:10.1371/journal.pbio.1000587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478:515–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. 2012. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat. Struct. Mol. Biol. 19:728–730 [DOI] [PubMed] [Google Scholar]
- 71. Kulshina N, Baird NJ, Ferre-D'Amare AR. 2009. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat. Struct. Mol. Biol. 16:1212–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, Li Y, Zhang R, Cheng G, Liu ZJ. 2012. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity 36:1073–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, Gu L. 2012. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat. Struct. Mol. Biol. 19:725–727 [DOI] [PubMed] [Google Scholar]
- 74. Shu C, Yi G, Watts T, Kao CC, Li P. 2012. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat. Struct. Mol. Biol. 19:722–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. 2009. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat. Struct. Mol. Biol. 16:1218–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Smith KD, Shanahan CA, Moore EL, Simon AC, Strobel SA. 2011. Structural basis of differential ligand recognition by two classes of bis-(3′-5′)-cyclic dimeric guanosine monophosphate-binding riboswitches. Proc. Natl. Acad. Sci. U. S. A. 108:7757–7762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. 2012. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol. Cell 46:735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ross P, Aloni Y, Weinhouse C, Michaeli D, Weinberger-Ohana P, Mayer R, Benziman M. 1985. An unusual guanyl oligonucleotide regulates cellulose synthesis in Acetobacter xylinum. FEBS Lett. 186:191–196 [DOI] [PubMed] [Google Scholar]
- 79. Ross P, Aloni Y, Weinhouse C, Michaeli D, Weinberger-Ohana P, Mayer R, Benziman M. 1986. Control of cellulose synthesis in Acetobacter xylinum. A unique guanyl oligonucleotide is the immediate activator of the cellulose synthase. Carbohydr. Res. 149:101–117 [Google Scholar]
- 80. Amikam D, Benziman M. 1989. Cyclic diguanylic acid and cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 171:6649–6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Egli M, Gessner RV, Williams LD, Quigley GJ, van der Marel GA, van Boom JH, Rich A, Frederick CA. 1990. Atomic-resolution structure of the cellulose synthase regulator cyclic diguanylic acid. Proc. Natl. Acad. Sci. U. S. A. 87:3235–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Habazettl J, Allan MG, Jenal U, Grzesiek S. 2011. Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout. J. Biol. Chem. 286:14304–14314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Ko J, Ryu KS, Kim H, Shin JS, Lee JO, Cheong C, Choi BS. 2010. Structure of PP4397 reveals the molecular basis for different c-di-GMP binding modes by Pilz domain proteins. J. Mol. Biol. 398:97–110 [DOI] [PubMed] [Google Scholar]
- 84. Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Navarro MV, De N, Bae N, Wang Q, Sondermann H. 2009. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure 17:1104–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wassmann P, Chan C, Paul R, Beck A, Heerklotz H, Jenal U, Schirmer T. 2007. Structure of BeF3−-modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure 15:915–927 [DOI] [PubMed] [Google Scholar]
- 87. Gentner M, Allan MG, Zaehringer F, Schirmer T, Grzesiek S. 2012. Oligomer formation of the bacterial second messenger c-di-GMP: reaction rates and equilibrium constants indicate a monomeric state at physiological concentrations. J. Am. Chem. Soc. 134:1019–1029 [DOI] [PubMed] [Google Scholar]
- 88. Tal R, Gelfand DH, Calhoon RD, Ben-Bassat A, Benziman M, Wong HC. 2 June 1998. Cyclic di-guanylate metabolic enzymes. US patent 5,759,828 [Google Scholar]
- 89. Tuckerman JR, Gonzalez G, Sousa EH, Wan X, Saito JA, Alam M, Gilles-Gonzalez MA. 2009. An oxygen-sensing diguanylate cyclase and phosphodiesterase couple for c-di-GMP control. Biochemistry 48:9764–9774 [DOI] [PubMed] [Google Scholar]
- 90. Tuckerman JR, Gonzalez G, Gilles-Gonzalez MA. 2011. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J. Mol. Biol. 407:633–639 [DOI] [PubMed] [Google Scholar]
- 91. Wan X, Tuckerman JR, Saito JA, Freitas TA, Newhouse JS, Denery JR, Galperin MY, Gonzalez G, Gilles-Gonzalez MA, Alam M. 2009. Globins synthesize the second messenger bis-(3′-5′)-cyclic diguanosine monophosphate in Bacteria. J. Mol. Biol. 388:262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. De N, Navarro MV, Raghavan RV, Sondermann H. 2009. Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J. Mol. Biol. 393:619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. U. S. A. 102:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Malone JG, Williams R, Christen M, Jenal U, Spiers AJ, Rainey PB. 2007. The structure-function relationship of WspR, a Pseudomonas fluorescens response regulator with a GGDEF output domain. Microbiology 153:980–994 [DOI] [PubMed] [Google Scholar]
- 95. Spiers AJ, Bohannon J, Gehrig SM, Rainey PB. 2003. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 50:15–27 [DOI] [PubMed] [Google Scholar]
- 96. Drenkard E, Ausubel FM. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740–743 [DOI] [PubMed] [Google Scholar]
- 97. Meissner A, Wild V, Simm R, Rohde M, Erck C, Bredenbruch F, Morr M, Römling U, Häussler S. 2007. Pseudomonas aeruginosa cupA-encoded fimbriae expression is regulated by a GGDEF and EAL domain-dependent modulation of the intracellular level of cyclic diguanylate. Environ. Microbiol. 9:2475–2485 [DOI] [PubMed] [Google Scholar]
- 98. Andrade MO, Alegria MC, Guzzo CR, Docena C, Rosa MC, Ramos CH, Farah CS. 2006. The HD-GYP domain of RpfG mediates a direct linkage between the Rpf quorum-sensing pathway and a subset of diguanylate cyclase proteins in the phytopathogen Xanthomonas axonopodis pv citri. Mol. Microbiol. 62:537–551 [DOI] [PubMed] [Google Scholar]
- 99. Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He YW, Zhang LH, Heeb S, Camara M, Williams P, Dow JM. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U. S. A. 103:6712–6717 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 100. Huang B, Whitchurch CB, Mattick JS. 2003. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 185:7068–7076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Kazmierczak BI, Lebron MB, Murray TS. 2006. Analysis of FimX, a phosphodiesterase that governs twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 60:1026–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Geer LY, Domrachev M, Lipman DJ, Bryant SH. 2002. CDART: protein homology by domain architecture. Genome Res. 12:1619–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Galperin MY. 2006. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J. Bacteriol. 188:4169–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Galperin MY. 2010. Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 13:150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. D'Argenio DA, Calfee MW, Rainey PB, Pesci EC. 2002. Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184:6481–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Henry JT, Crosson S. 2011. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu. Rev. Microbiol. 65:261–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Galperin MY. 2001. Conserved ‘hypothetical’ proteins: new hints and new puzzles. Comp. Funct. Genomics 2:14–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Aldridge P, Paul R, Goymer P, Rainey P, Jenal U. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47:1695–1708 [DOI] [PubMed] [Google Scholar]
- 109. Pei J, Grishin NV. 2001. GGDEF domain is homologous to adenylyl cyclase. Proteins 42:210–216 [DOI] [PubMed] [Google Scholar]
- 110. Linder JU. 2006. Class III adenylyl cyclases: molecular mechanisms of catalysis and regulation. Cell. Mol. Life Sci. 63:1736–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sinha SC, Sprang SR. 2006. Structures, mechanism, regulation and evolution of class III nucleotidyl cyclases. Rev. Physiol. Biochem. Pharmacol. 157:105–140 [DOI] [PubMed] [Google Scholar]
- 112. Paul R, Abel S, Wassmann P, Beck A, Heerklotz H, Jenal U. 2007. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J. Biol. Chem. 282:29170–29177 [DOI] [PubMed] [Google Scholar]
- 113. Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. 2006. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 281:32015–32024 [DOI] [PubMed] [Google Scholar]
- 114. Seshasayee AS, Fraser GM, Luscombe NM. 2010. Comparative genomics of cyclic-di-GMP signalling in bacteria: post-translational regulation and catalytic activity. Nucleic Acids Res. 38:5970–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Perez-Mendoza D, Coulthurst SJ, Humphris S, Campbell E, Welch M, Toth IK, Salmond GP. 2011. A multi-repeat adhesin of the phytopathogen, Pectobacterium atrosepticum, is secreted by a type I pathway and is subject to complex regulation involving a non-canonical diguanylate cyclase. Mol. Microbiol. 82:719–733 [DOI] [PubMed] [Google Scholar]
- 116. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. 2012. The Pfam protein families database. Nucleic Acids Res. 40:D290–D301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Yang CY, Chin KH, Chuah ML, Liang ZX, Wang AH, Chou SH. 2011. The structure and inhibition of a GGDEF diguanylate cyclase complexed with (c-di-GMP)2 at the active site. Acta Crystallogr. D Biol. Crystallogr. 67:997–1008 [DOI] [PubMed] [Google Scholar]
- 118. Rao F, Yang Y, Qi Y, Liang ZX. 2008. Catalytic mechanism of cyclic di-GMP-specific phosphodiesterase: a study of the EAL domain-containing RocR from Pseudomonas aeruginosa. J. Bacteriol. 190:3622–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Bai Y, Yang J, Zhou X, Ding X, Eisele LE, Bai G. 2012. Mycobacterium tuberculosis Rv3586 (DacA) is a diadenylate cyclase that converts ATP or ADP into c-di-AMP. PLoS One 7:e35206 doi:10.1371/journal.pone.0035206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Johnson JG, Murphy CN, Sippy J, Johnson TJ, Clegg S. 2011. Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in Klebsiella pneumoniae. J. Bacteriol. 193:3453–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Chen MW, Kotaka M, Vonrhein C, Bricogne G, Rao F, Chuah ML, Svergun D, Schneider G, Liang ZX, Lescar J. 2012. Structural insights into the regulatory mechanism of the response regulator RocR from Pseudomonas aeruginosa in cyclic di-GMP signaling. J. Bacteriol. 194:4837–4846 doi:10.1128/JB.00560-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gomelsky M, Klug G. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27:497–500 [DOI] [PubMed] [Google Scholar]
- 123. Gomelsky M, Hoff WD. 2011. Light helps bacteria make important lifestyle decisions. Trends Microbiol. 19:441–448 [DOI] [PubMed] [Google Scholar]
- 124. Salter EA, Wierzbicki A. 2007. The mechanism of cyclic nucleotide hydrolysis in the phosphodiesterase catalytic site. J. Phys. Chem. B 111:4547–4552 [DOI] [PubMed] [Google Scholar]
- 125. Rao F, Qi Y, Chong HS, Kotaka M, Li B, Li J, Lescar J, Tang K, Liang ZX. 2009. The functional role of a conserved loop in EAL domain-based c-di-GMP specific phosphodiesterase. J. Bacteriol. 191:4722–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Galperin MY, Koonin EV. 2012. Divergence and convergence in enzyme evolution. J. Biol. Chem. 287:21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ryan RP, Lucey J, O'Donovan K, McCarthy Y, Yang L, Tolker-Nielsen T, Dow JM. 2009. HD-GYP domain proteins regulate biofilm formation and virulence in Pseudomonas aeruginosa. Environ. Microbiol. 11:1126–1136 [DOI] [PubMed] [Google Scholar]
- 128. Sultan SZ, Pitzer JE, Boquoi T, Hobbs G, Miller MR, Motaleb MA. 2011. Analysis of the HD-GYP domain cyclic dimeric GMP phosphodiesterase reveals a role in motility and the enzootic life cycle of Borrelia burgdorferi. Infect. Immun. 79:3273–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Lovering AL, Capeness MJ, Lambert C, Hobley L, Sockett RE. 2011. The structure of an unconventional HD-GYP protein from Bdellovibrio reveals the roles of conserved residues in this class of cyclic-di-GMP phosphodiesterases. mBio 2:e00163–11 doi:10.1128/mBio.00163-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Qi Y, Rao F, Luo Z, Liang ZX. 2009. A flavin cofactor-binding PAS domain regulates c-di-GMP synthesis in AxDGC2 from Acetobacter xylinum. Biochemistry 48:10275–10285 [DOI] [PubMed] [Google Scholar]
- 131. Perego M, Hoch JA. 1996. Protein aspartate phosphatases control the output of two-component signal transduction systems. Trends Genet. 12:97–101 [DOI] [PubMed] [Google Scholar]
- 132. Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu. Rev. Microbiol. 62:35–51 [DOI] [PubMed] [Google Scholar]
- 133. Qi Y, Chuah ML, Dong X, Xie K, Luo Z, Tang K, Liang ZX. 2011. Binding of cyclic diguanylate in the non-catalytic EAL domain of FimX induces a long-range conformational change. J. Biol. Chem. 286:2910–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ferreira RB, Antunes LC, Greenberg EP, McCarter LL. 2008. Vibrio parahaemolyticus ScrC modulates cyclic dimeric GMP regulation of gene expression relevant to growth on surfaces. J. Bacteriol. 190:851–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Boles BR, McCarter LL. 2002. Vibrio parahaemolyticus scrABC, a novel operon affecting swarming and capsular polysaccharide regulation. J. Bacteriol. 184:5946–5954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Trimble MJ, McCarter LL. 2011. Bis-(3′-5′)-cyclic dimeric GMP-linked quorum sensing controls swarming in Vibrio parahaemolyticus. Proc. Natl. Acad. Sci. U. S. A. 108:18079–18084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Bharati BK, Sharma IM, Kasetty S, Kumar M, Mukherjee R, Chatterji D. 2012. A full length bifunctional protein involved in c-di-GMP turnover is required for long term survival under nutrient starvation in Mycobacterium smegmatis. Microbiology 158:1415–1427 [DOI] [PubMed] [Google Scholar]
- 138. Gupta K, Kumar P, Chatterji D. 2010. Identification, activity and disulfide connectivity of c-di-GMP regulating proteins in Mycobacterium tuberculosis. PLoS One 5:e15072 doi:10.1371/journal.pone.0015072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Levet-Paulo M, Lazzaroni JC, Gilbert C, Atlan D, Doublet P, Vianney A. 2011. The atypical two-component sensor kinase Lpl0330 from Legionella pneumophila controls the bifunctional diguanylate cyclase-phosphodiesterase Lpl0329 to modulate bis-(3′-5′)-cyclic dimeric GMP synthesis. J. Biol. Chem. 286:31136–31144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39:D225–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Galperin MY. 2005. A census of membrane-bound and intracellular signal transduction proteins in bacteria: bacterial IQ, extroverts and introverts. BMC Microbiol. 5:35 doi:10.1186/1471-2180-5-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Krasteva PV, Giglio KM, Sondermann H. 2012. Sensing the messenger: the diverse ways that bacteria signal through c-di-GMP. Protein Sci. 21:929–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Ryan RP, Tolker-Nielsen T, Dow JM. 2012. When the PilZ don't work: effectors for cyclic di-GMP action in bacteria. Trends Microbiol. 20:235–242 [DOI] [PubMed] [Google Scholar]
- 144. Mayer R, Ross P, Weinhouse H, Amikam D, Volman G, Ohana P, Calhoon RD, Wong HC, Emerick AW, Benziman M. 1991. Polypeptide composition of bacterial cyclic diguanylic acid-dependent cellulose synthase and the occurrence of immunologically crossreacting proteins in higher plants. Proc. Natl. Acad. Sci. U. S. A. 88:5472–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Weinhouse H, Sapir S, Amikam D, Shilo Y, Volman G, Ohana P, Benziman M. 1997. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett. 416:207–211 [DOI] [PubMed] [Google Scholar]
- 146. Alm RA, Bodero AJ, Free PD, Mattick JS. 1996. Identification of a novel gene, pilZ, essential for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 178:46–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Pratt JT, Tamayo R, Tischler AD, Camilli A. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 282:12860–12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Freedman JC, Rogers EA, Kostick JL, Zhang H, Iyer R, Schwartz I, Marconi RT. 2010. Identification and molecular characterization of a cyclic-di-GMP effector protein, PlzA (BB0733): additional evidence for the existence of a functional cyclic-di-GMP regulatory network in the Lyme disease spirochete, Borrelia burgdorferi. FEMS Immunol. Med. Microbiol. 58:285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65:876–895 [DOI] [PubMed] [Google Scholar]
- 150. Ramelot TA, Yee A, Cort JR, Semesi A, Arrowsmith CH, Kennedy MA. 2007. NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins 66:266–271 [DOI] [PubMed] [Google Scholar]
- 151. Shin JS, Ryu KS, Ko J, Lee A, Choi BS. 2011. Structural characterization reveals that a PilZ domain protein undergoes substantial conformational change upon binding to cyclic dimeric guanosine monophosphate. Protein Sci. 20:270–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Li TN, Chin KH, Fung KM, Yang MT, Wang AH, Chou SH. 2011. A novel tetrameric PilZ domain structure from xanthomonads. PLoS One 6:e22036 doi:10.1371/journal.pone.0022036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Hobley L, Fung RK, Lambert C, Harris MA, Dabhi JM, King SS, Basford SM, Uchida K, Till R, Ahmad R, Aizawa S, Gomelsky M, Sockett RE. 2012. Discrete cyclic di-GMP-dependent control of bacterial predation versus axenic growth in Bdellovibrio bacteriovorus. PLoS Pathog. 8:e1002493 doi:10.1371/journal.ppat.1002493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116 [DOI] [PubMed] [Google Scholar]
- 155. Fang X, Gomelsky M. 2010. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 76:1295–1305 [DOI] [PubMed] [Google Scholar]
- 156. Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38:128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Oglesby LL, Jain S, Ohman DE. 2008. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology 154:1605–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Wilksch JJ, Yang J, Clements A, Gabbe JL, Short KR, Cao H, Cavaliere R, James CE, Whitchurch CB, Schembri MA, Chuah ML, Liang ZX, Wijburg OL, Jenney AW, Lithgow T, Strugnell RA. 2011. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 7:e1002204 doi:10.1371/journal.ppat.1002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Guzzo CR, Salinas RK, Andrade MO, Farah CS. 2009. PILZ protein structure and interactions with PILB and the FIMX EAL domain: implications for control of type IV pilus biogenesis. J. Mol. Biol. 393:848–866 [DOI] [PubMed] [Google Scholar]
- 160. Li TN, Chin KH, Liu JH, Wang AH, Chou SH. 2009. XC1028 from Xanthomonas campestris adopts a PilZ domain-like structure without a c-di-GMP switch. Proteins 75:282–288 [DOI] [PubMed] [Google Scholar]
- 161. Beyhan S, Odell LS, Yildiz FH. 2008. Identification and characterization of cyclic diguanylate signaling systems controlling rugosity in Vibrio cholerae. J. Bacteriol. 190:7392–7405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 23:93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Petters T, Zhang X, Nesper J, Treuner-Lange A, Gomez-Santos N, Hoppert M, Jenal U, Søgaard-Andersen L. 2012. The orphan histidine protein kinase SgmT is a c-di-GMP receptor and regulates composition of the extracellular matrix together with the orphan DNA binding response regulator DigR in Myxococcus xanthus. Mol. Microbiol. 84:147–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65:1474–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Whitney JC, Colvin KM, Marmont LS, Robinson H, Parsek MR, Howell PL. 2012. Structure of the cytoplasmic region of PelD, a degenerate diguanylate cyclase receptor that regulates exopolysaccharide production in Pseudomonas aeruginosa. J. Biol. Chem. 287:23582–23593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3′,5′)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. U. S. A. 106:3461–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O'Toole GA. 2011. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. J. Bacteriol. 193:4685–4698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69:376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Srivastava D, Harris RC, Waters CM. 2011. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J. Bacteriol. 193:6331–6341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Chin KH, Lee YC, Tu ZL, Chen CH, Tseng YH, Yang JM, Ryan RP, McCarthy Y, Dow JM, Wang AH, Chou SH. 2009. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J. Mol. Biol. 396:646–662 [DOI] [PubMed] [Google Scholar]
- 171. Fazli M, O'Connell A, Nilsson M, Niehaus K, Dow JM, Givskov M, Ryan RP, Tolker-Nielsen T. 2011. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol. Microbiol. 82:327–341 [DOI] [PubMed] [Google Scholar]
- 172. Gomelsky M. 2009. Cyclic-di-GMP-binding CRP-like protein: a spectacular new role for a veteran signal transduction actor. J. Bacteriol. 191:6785–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Leduc JL, Roberts GP. 2009. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J. Bacteriol. 191:7121–7122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Ferreira RB, Chodur DM, Antunes LC, Trimble MJ, McCarter LL. 2012. Output targets and transcriptional regulation by a cyclic dimeric GMP-responsive circuit in the Vibrio parahaemolyticus Scr network. J. Bacteriol. 194:914–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175. Gomelsky M. 2012. Cyclic dimeric GMP-mediated decisions in surface-grown Vibrio parahaemolyticus: a different kind of motile-to-sessile transition. J. Bacteriol. 194:911–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Ma Q, Yang Z, Pu M, Peti W, Wood TK. 2011. Engineering a novel c-di-GMP-binding protein for biofilm dispersal. Environ. Microbiol. 13:631–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Düvel J, Bertinetti D, Möller S, Schwede F, Morr M, Wissing J, Radamm L, Zimmermann B, Genieser HG, Jänsch L, Herberg FW, Häussler S. 2012. A chemical proteomics approach to identify c-di-GMP binding proteins in Pseudomonas aeruginosa. J. Microbiol. Methods 88:229–236 [DOI] [PubMed] [Google Scholar]
- 178. Nesper J, Reinders A, Glatter T, Schmidt A, Jenal U. 2012. A novel capture compound for the identification and analysis of cyclic di-GMP binding proteins. J. Proteomics 75:4874–4878 [DOI] [PubMed] [Google Scholar]
- 179. Roelofs KG, Wang J, Sintim HO, Lee VT. 2011. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc. Natl. Acad. Sci. U. S. A. 108:15528–15533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Barrick JE, Breaker RR. 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 8:R239 doi:10.1186/gb-2007-8-11-r239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Bordeleau E, Fortier LC, Malouin F, Burrus V. 2011. c-di-GMP turn-over in Clostridium difficile is controlled by a plethora of diguanylate cyclases and phosphodiesterases. PLoS Genet. 7:e1002039 doi:10.1371/journal.pgen.1002039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ. 2010. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 78:361–379 [DOI] [PubMed] [Google Scholar]
- 183. Bobrov AG, Kirillina O, Ryjenkov DA, Waters CM, Price PA, Fetherston JD, Mack D, Goldman WE, Gomelsky M, Perry RD. 2011. Systematic analysis of cyclic di-GMP signalling enzymes and their role in biofilm formation and virulence in Yersinia pestis. Mol. Microbiol. 79:533–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183a. Chen ZH, Schaap P. 2012. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature 488:680–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Ho YS, Burden LM, Hurley JH. 2000. Structure of the GAF domain, a ubiquitous signaling motif and a new class of cyclic GMP receptor. EMBO J. 19:5288–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184a. Deng Y, Schmid N, Wang C, Wang J, Pessi G, Wu D, Lee J, Aguilar C, Ahrens CH, Chang C, Song H, Eberl L, Zhang LH. 2012. Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc. Natl. Acad. Sci. U. S. A. 109:15479–15484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Cashel M, Gallant J. 1969. Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221:838–841 [DOI] [PubMed] [Google Scholar]
- 186. Cao Z, Livoti E, Losi A, Gartner W. 2010. A blue light-inducible phosphodiesterase activity in the cyanobacterium Synechococcus elongatus. Photochem. Photobiol. 86:606–611 [DOI] [PubMed] [Google Scholar]
- 187. Savakis P, De Causmaecker S, Angerer V, Ruppert U, Anders K, Essen LO, Wilde A. 2012. Light-induced alteration of c-di-GMP level controls motility of Synechocystis sp. PCC 6803. Mol. Microbiol. 85:239–251 [DOI] [PubMed] [Google Scholar]
- 188. Liu N, Xu Y, Hossain S, Huang N, Coursolle D, Gralnick JA, Boon EM. 2012. Nitric oxide regulation of cyclic di-GMP synthesis and hydrolysis in Shewanella woodyi. Biochemistry 51:2087–2099 [DOI] [PubMed] [Google Scholar]
- 189. Plate L, Marletta MA. 2012. Nitric oxide modulates bacterial biofilm formation through a multicomponent cyclic-di-GMP signaling network. Mol. Cell 46:449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Galperin MY. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Zhulin IB, Nikolskaya AN, Galperin MY. 2003. Common extracellular sensory domains in transmembrane receptors for diverse signal transduction pathways in bacteria and archaea. J. Bacteriol. 185:285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Ryan RP, McCarthy Y, Andrade M, Farah CS, Armitage JP, Dow JM. 2010. Cell-cell signal-dependent dynamic interactions between HD-GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris. Proc. Natl. Acad. Sci. U. S. A. 107:5989–5994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Malone JG, Jaeger T, Manfredi P, Dotsch A, Blanka A, Bos R, Cornelis GR, Häussler S, Jenal U. 2012. The YfiBNR signal transduction mechanism reveals novel targets for the evolution of persistent Pseudomonas aeruginosa in cystic fibrosis airways. PLoS Pathog. 8:e1002760 doi:10.1371/journal.ppat.1002760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. He M, Ouyang Z, Troxell B, Xu H, Moh A, Piesman J, Norgard MV, Gomelsky M, Yang XF. 2011. Cyclic di-GMP is essential for the survival of the Lyme disease spirochete in ticks. PLoS Pathog. 7:e1002133 doi:10.1371/journal.ppat.1002133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Neunuebel MR, Golden JW. 2008. The Anabaena sp. strain PCC 7120 gene all2874 encodes a diguanylate cyclase and is required for normal heterocyst development under high-light growth conditions. J. Bacteriol. 190:6829–6836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Hull TD, Ryu MH, Sullivan MJ, Johnson RC, Klena NT, Geiger RM, Gomelsky M, Bennett JA. 2012. Cyclic di-GMP phosphodiesterases RmdA and RmdB are involved in regulating colony morphology and development in Streptomyces coelicolor. J. Bacteriol. 194:4642–4651 doi:10.1128/JB.00157-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196a. Li W, He ZG. 2012. LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Res. 40:11292–11307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Pesavento C, Becker G, Sommerfeldt N, Possling A, Tschowri N, Mehlis A, Hengge R. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22:2434–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Petrova OE, Sauer K. 2012. Sticky situations: key components that control bacterial surface attachment. J. Bacteriol. 194:2413–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Barken KB, Pamp SJ, Yang L, Gjermansen M, Bertrand JJ, Klausen M, Givskov M, Whitchurch CB, Engel JN, Tolker-Nielsen T. 2008. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 10:2331–2343 [DOI] [PubMed] [Google Scholar]
- 200. McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. 2012. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Microbiol. 10:39–50 [DOI] [PubMed] [Google Scholar]
- 201. Merritt JH, Brothers KM, Kuchma SL, O'Toole AG. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189:8154–8164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Purcell EB, McKee RW, McBride SM, Waters CM, Tamayo R. 2012. Cyclic diguanylate inversely regulates motility and aggregation in Clostridium difficile. J. Bacteriol. 194:3307–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Petersen E, Chaudhuri P, Gourley C, Harms J, Splitter G. 2011. Brucella melitensis cyclic di-GMP phosphodiesterase BpdA controls expression of flagellar genes. J. Bacteriol. 193:5683–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Kulesekara H, Lee V, Brencic A, Liberati N, Urbach J, Miyata S, Lee DG, Neely AN, Hyodo M, Hayakawa Y, Ausubel FM, Lory S. 2006. Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc. Natl. Acad. Sci. U. S. A. 103:2839–2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Ryan RP, Fouhy Y, Lucey JF, Jiang BL, He YQ, Feng JX, Tang JL, Dow JM. 2007. Cyclic di-GMP signalling in the virulence and environmental adaptation of Xanthomonas campestris. Mol. Microbiol. 63:429–442 [DOI] [PubMed] [Google Scholar]
- 206. Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD, Wozniak DJ, Harwood CS, Parsek MR. 2009. Pseudomonas aeruginosa rugose small colony variants have adaptations likely to promote persistence in the cystic fibrosis lung. J. Bacteriol. 191:3492–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Zhang LH. 2010. A novel c-di-GMP effector linking intracellular virulence regulon to quorum sensing and hypoxia sensing. Virulence 1:391–394 [DOI] [PubMed] [Google Scholar]
- 208. Monds RD, O'Toole GA. 2009. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17:73–87 [DOI] [PubMed] [Google Scholar]
- 209. O'Toole GA, Kolter R. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449–461 [DOI] [PubMed] [Google Scholar]
- 210. O'Toole GA, Kolter R. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295–304 [DOI] [PubMed] [Google Scholar]
- 211. Berg HC. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19–54 [DOI] [PubMed] [Google Scholar]
- 212. Minamino T, Imada K, Namba K. 2008. Molecular motors of the bacterial flagella. Curr. Opin. Struct. Biol. 18:693–701 [DOI] [PubMed] [Google Scholar]
- 213. Girgis HS, Liu Y, Ryu WS, Tavazoie S. 2007. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 3:1644–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Wolfe AJ, Berg HC. 1989. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. U. S. A. 86:6973–6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Ko M, Park C. 2000. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J. Mol. Biol. 303:371–382 [DOI] [PubMed] [Google Scholar]
- 216. Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30:285–293 [DOI] [PubMed] [Google Scholar]
- 216a. Zorraquino V, Garcia B, Latasa C, Echeverz M, Toledo-Arana A, Valle J, Lasa I, Solano C. 2013. Coordinated c-di-GMP repression of Salmonella motility through YcgR and cellulose. J. Bacteriol. 195:417–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Guttenplan SB, Blair KM, Kearns DB. 2010. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 6:e1001243 doi:10.1371/journal.pgen.1001243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217a. Chen Y, Chai Y, Guo JH, Losick R. 2012. Evidence for cyclic di-GMP-mediated signaling in Bacillus subtilis. J. Bacteriol. 194:5080–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218. Greer-Phillips SE, Stephens BB, Alexandre G. 2004. An energy taxis transducer promotes root colonization by Azospirillum brasilense. J. Bacteriol. 186:6595–6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218a. Russell MH, Bible AN, Fang X, Gooding J, Campagna S, Gomelsky M, Alexandre G. Integration of the second messenger c-di-GMP into the chemotactic signaling pathway promotes sensory adaptation. mBio, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Christen M, Kulasekara HD, Christen B, Kulasekara BR, Hoffman LR, Miller SI. 2010. Asymmetrical distribution of the second messenger c-di-GMP upon bacterial cell division. Science 328:1295–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220. Jyot J, Dasgupta N, Ramphal R. 2002. FleQ, the major flagellar gene regulator in Pseudomonas aeruginosa, binds to enhancer sites located either upstream or atypically downstream of the RpoN binding site. J. Bacteriol. 184:5251–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221. Baraquet C, Murakami K, Parsek MR, Harwood CS. 2012. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 40:7207–7218 doi:10.1093/nar/gks1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Tao F, He YW, Wu DH, Swarup S, Zhang LH. 2010. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J. Bacteriol. 192:1020–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Lee MC, Weng SF, Tseng YH. 2003. Flagellin gene fliC of Xanthomonas campestris is upregulated by transcription factor Clp. Biochem. Biophys. Res. Commun. 307:647–652 [DOI] [PubMed] [Google Scholar]
- 224. Choy WK, Zhou L, Syn CK, Zhang LH, Swarup S. 2004. MorA defines a new class of regulators affecting flagellar development and biofilm formation in diverse Pseudomonas species. J. Bacteriol. 186:7221–7228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225. Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH. 2006. Bacterial birth scar proteins mark future flagellum assembly site. Cell 124:1025–1037 [DOI] [PubMed] [Google Scholar]
- 226. Simm R, Remminghorst U, Ahmad I, Zakikhany K, Römling U. 2009. A role for the EAL-like protein STM1344 in regulation of CsgD expression and motility in Salmonella enterica serovar Typhimurium. J. Bacteriol. 191:3928–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Wada T, Morizane T, Abo T, Tominaga A, Inoue-Tanaka K, Kutsukake K. 2011. EAL-domain protein YdiV acts as an anti-FlhD4C2 factor responsible for nutritional control of the flagellar regulon in Salmonella enterica serovar Typhimurium. J. Bacteriol. 193:1600–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228. Caiazza NC, Merritt JH, Brothers KM, O'Toole GA. 2007. Inverse regulation of biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:3603–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Kuchma SL, Brothers KM, Merritt JH, Liberati NT, Ausubel FM, O'Toole GA. 2007. BifA, a cyclic-di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 189:8165–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230. Merritt JH, Ha DG, Cowles KN, Lu W, Morales DK, Rabinowitz J, Gitai Z, O'Toole GA. 2010. Specific control of Pseudomonas aeruginosa surface-associated behaviors by two c-di-GMP diguanylate cyclases. mBio 1:e00183–10 doi:10.1128/mBio.00183-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. McCarter L. 1999. The multiple identities of Vibrio parahaemolyticus. J. Mol. Microbiol. Biotechnol. 1:51–57 [PubMed] [Google Scholar]
- 232. Gotz F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367–1378 [DOI] [PubMed] [Google Scholar]
- 232a. Kuchma SL, Griffin EF, O'Toole GA. 2012. Minor pilins of the type IV pilus system participate in the negative regulation of swarming motility. J. Bacteriol. 194:5388–5403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233. Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175 [DOI] [PubMed] [Google Scholar]
- 234. Kader A, Simm R, Gerstel U, Morr M, Römling U. 2006. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 60:602–616 [DOI] [PubMed] [Google Scholar]
- 235. Mendez-Ortiz MM, Hyodo M, Hayakawa Y, Membrillo-Hernandez J. 2006. Genome wide transcriptional profile of Escherichia coli in response to high levels of the second messenger c-di-GMP. J. Biol. Chem. 281:8090–8099 [DOI] [PubMed] [Google Scholar]
- 236. Nakhamchik A, Wilde C, Rowe-Magnus DA. 2008. Cyclic-di-GMP regulates extracellular polysaccharide production, biofilm formation, and rugose colony development by Vibrio vulnificus. Appl. Environ. Microbiol. 74:4199–4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237. Rahman M, Simm R, Kader A, Basseres E, Römling U, Möllby R. 2007. The role of c-di-GMP signaling in an Aeromonas veronii biovar sobria strain. FEMS Microbiol. Lett. 273:172–179 [DOI] [PubMed] [Google Scholar]
- 238. Rashid MH, Rajanna C, Ali A, Karaolis DK. 2003. Identification of genes involved in the switch between the smooth and rugose phenotypes of Vibrio cholerae. FEMS Microbiol. Lett. 227:113–119 [DOI] [PubMed] [Google Scholar]
- 239. Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 188:2681–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240. Branda SS, Vik S, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends Microbiol. 13:20–26 [DOI] [PubMed] [Google Scholar]
- 241. Römling U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 62:1234–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Yildiz FH, Visick KL. 2009. Vibrio biofilms: so much the same yet so different. Trends Microbiol. 17:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243. Hay ID, Remminghorst U, Rehm BH. 2009. MucR, a novel membrane-associated regulator of alginate biosynthesis in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 75:1110–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Lee B, Schjerling CK, Kirkby N, Hoffmann N, Borup R, Molin S, Hoiby N, Ciofu O. 2011. Mucoid Pseudomonas aeruginosa isolates maintain the biofilm formation capacity and the gene expression profiles during the chronic lung infection of CF patients. APMIS 119:263–274 [DOI] [PubMed] [Google Scholar]
- 245. Malone JG, Jaeger T, Spangler C, Ritz D, Spang A, Arrieumerlou C, Kaever V, Landmann R, Jenal U. 2010. YfiBNR mediates cyclic di-GMP dependent small colony variant formation and persistence in Pseudomonas aeruginosa. PLoS Pathog. 6:e1000804 doi:10.1371/journal.ppat.1000804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246. Römling U. 2002. Molecular biology of cellulose production in bacteria. Res. Microbiol. 153:205–212 [DOI] [PubMed] [Google Scholar]
- 247. Kawano Y, Saotome T, Ochiai Y, Katayama M, Narikawa R, Ikeuchi M. 2011. Cellulose accumulation and a cellulose synthase gene are responsible for cell aggregation in the cyanobacterium Thermosynechococcus vulcanus RKN. Plant Cell Physiol. 52:957–966 [DOI] [PubMed] [Google Scholar]
- 248. Nobles DR, Romanovicz DK, Brown RM., Jr 2001. Cellulose in cyanobacteria. Origin of vascular plant cellulose synthase? Plant Physiol. 127:529–542 [PMC free article] [PubMed] [Google Scholar]
- 249. Ogawa K, Maki Y. 2003. Cellulose as extracellular polysaccharide of hot spring sulfur-turf bacterial mat. Biosci. Biotechnol. Biochem. 67:2652–2654 [DOI] [PubMed] [Google Scholar]
- 250. Zogaj X, Nimtz M, Rohde M, Bokranz W, Römling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452–1463 [DOI] [PubMed] [Google Scholar]
- 251. Brandl MT, Carter MQ, Parker CT, Chapman MR, Huynh S, Zhou Y. 2011. Salmonella biofilm formation on Aspergillus niger involves cellulose-chitin interactions. PLoS One 6:e25553 doi:10.1371/journal.pone.0025553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252. Matthysse AG, Holmes KV, Gurlitz RH. 1981. Elaboration of cellulose fibrils by Agrobacterium tumefaciens during attachment to carrot cells. J. Bacteriol. 145:583–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Monteiro C, Saxena I, Wang X, Kader A, Bokranz W, Simm R, Nobles D, Chromek M, Brauner A, Brown RM, Jr, Römling U. 2009. Characterization of cellulose production in Escherichia coli Nissle 1917 and its biological consequences. Environ. Microbiol. 11:1105–1116 [DOI] [PubMed] [Google Scholar]
- 254. Saldana Z, Xicohtencatl-Cortes J, Avelino F, Phillips AD, Kaper JB, Puente JL, Giron JA. 2009. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ. Microbiol. 11:992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Le Quere B, Ghigo JM. 2009. BcsQ is an essential component of the Escherichia coli cellulose biosynthesis apparatus that localizes at the bacterial cell pole. Mol. Microbiol. 72:724–740 [DOI] [PubMed] [Google Scholar]
- 256. Saxena IM, Brown RM., Jr 1995. Identification of a second cellulose synthase gene (acsAII) in Acetobacter xylinum. J. Bacteriol. 177:5276–5283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257. Bokranz W, Wang X, Tschäpe H, Römling U. 2005. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 54:1171–1182 [DOI] [PubMed] [Google Scholar]
- 258. Da Re S, Ghigo JM. 2006. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 188:3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259. Garcia B, Latasa C, Solano C, Portillo FG, Gamazo C, Lasa I. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 54:264–277 [DOI] [PubMed] [Google Scholar]
- 259a. Amarasinghe JJ, D'Hondt RE, Waters CM, Mantis NJ. 2013. Exposure of Salmonella enterica serovar Typhimurium to a protective monoclonal IgA triggers exopolysaccharide production via a diguanylate cyclase-dependent pathway. Infect. Immun. 81:653–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260. Gal M, Preston GM, Massey RC, Spiers AJ, Rainey PB. 2003. Genes encoding a cellulosic polymer contribute toward the ecological success of Pseudomonas fluorescens SBW25 on plant surfaces. Mol. Ecol. 12:3109–3121 [DOI] [PubMed] [Google Scholar]
- 261. Goymer P, Kahn SG, Malone JG, Gehrig SM, Spiers AJ, Rainey PB. 2006. Adaptive divergence in experimental populations of Pseudomonas fluorescens. II. Role of the GGDEF regulator WspR in evolution and development of the wrinkly spreader phenotype. Genetics 173:515–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Zogaj X, Bokranz W, Nimtz M, Römling U. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71:4151–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262a. Morgan JL, Strumillo J, Zimmer J. 2013. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493:181–186 doi:10.1038/nature11744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263. Bentancor LV, O'Malley JM, Bozkurt-Guzel C, Pier GB, Maira-Litran T. 2012. Poly-N-acetyl-β-(1–6)-glucosamine is a target for protective immunity against Acinetobacter baumannii infections. Infect. Immun. 80:651–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Itoh Y, Wang X, Hinnebusch BJ, Preston JF, 3rd, Romeo T. 2005. Depolymerization of beta-1,6-N-acetyl-d-glucosamine disrupts the integrity of diverse bacterial biofilms. J. Bacteriol. 187:382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Yakandawala N, Gawande PV, Lovetri K, Cardona ST, Romeo T, Nitz M, Madhyastha S. 2011. Characterization of the poly-β-1,6-N-acetylglucosamine polysaccharide component of Burkholderia biofilms. Appl. Environ. Microbiol. 77:8303–8309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266. Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U. 2009. Second messenger signalling governs Escherichia coli biofilm induction upon ribosomal stress. Mol. Microbiol. 72:1500–1516 [DOI] [PubMed] [Google Scholar]
- 267. Perez-Mendoza D, Coulthurst SJ, Sanjuan J, Salmond GP. 2011. N-Acetyl-glucosamine-dependent biofilm formation in Pectobacterium atrosepticum is cryptic and activated by elevated c-di-GMP levels. Microbiology 157:3340–3348 [DOI] [PubMed] [Google Scholar]
- 268. Bobrov AG, Kirillina O, Forman S, Mack D, Perry RD. 2008. Insights into Yersinia pestis biofilm development: topology and co-interaction of Hms inner membrane proteins involved in exopolysaccharide production. Environ. Microbiol. 10:1419–1432 [DOI] [PubMed] [Google Scholar]
- 269. Perry RD, Bobrov AG, Kirillina O, Jones HA, Pedersen L, Abney J, Fetherston JD. 2004. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J. Bacteriol. 186:1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Wang X, Preston JF, 3rd, Romeo T. 2004. The pgaABCD locus of Escherichia coli promotes the synthesis of a polysaccharide adhesin required for biofilm formation. J. Bacteriol. 186:2724–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Tagliabue L, Antoniani D, Maciag A, Bocci P, Raffaelli N, Landini P. 2010. The diguanylate cyclase YddV controls production of the exopolysaccharide poly-N-acetylglucosamine (PNAG) through regulation of the PNAG biosynthetic pgaABCD operon. Microbiology 156:2901–2911 [DOI] [PubMed] [Google Scholar]
- 271a. Steiner S, Lori C, Boehm A, Jenal U. 2013. Allosteric activation of exopolysaccharide synthesis through cyclic di-GMP-stimulated protein-protein interaction. EMBO J. 32:354–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272. Holland LM, O'Donnell ST, Ryjenkov DA, Gomelsky L, Slater SR, Fey PD, Gomelsky M, O'Gara JP. 2008. A staphylococcal GGDEF domain protein regulates biofilm formation independently of c-di-GMP. J. Bacteriol. 190:5178–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273. Güvener ZT, Harwood CS. 2007. Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 66:1459–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273a. Irie Y, Borlee BR, O'Connor JR, Hill PJ, Harwood CS, Wozniak DJ, Parsek MR. 2012. Self-produced exopolysaccharide is a signal that stimulates biofilm formation in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 109:20632–20636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274. Ueda A, Wood TK. 2009. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 5:e1000483 doi:10.1371/journal.ppat.1000483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275. Pu M, Wood TK. 2010. Tyrosine phosphatase TpbA controls rugose colony formation in Pseudomonas aeruginosa by dephosphorylating diguanylate cyclase TpbB. Biochem. Biophys. Res. Commun. 402:351–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Bassis CM, Visick KL. 2010. The cyclic-di-GMP phosphodiesterase BinA negatively regulates cellulose-containing biofilms in Vibrio fischeri. J. Bacteriol. 192:1269–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu. Rev. Microbiol. 60:131–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Ghigo JM. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442–445 [DOI] [PubMed] [Google Scholar]
- 279. Korea CG, Ghigo JM, Beloin C. 2011. The sweet connection: solving the riddle of multiple sugar-binding fimbrial adhesins in Escherichia coli: multiple E. coli fimbriae form a versatile arsenal of sugar-binding lectins potentially involved in surface-colonisation and tissue tropism. Bioessays 33:300–311 [DOI] [PubMed] [Google Scholar]
- 280. Mandlik A, Swierczynski A, Das A, Ton-That H. 2008. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16:33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281. Nudleman E, Kaiser D. 2004. Pulling together with type IV pili. J. Mol. Microbiol. Biotechnol. 7:52–62 [DOI] [PubMed] [Google Scholar]
- 282. Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55:368–380 [DOI] [PubMed] [Google Scholar]
- 283. Mikkelsen H, Ball G, Giraud C, Filloux A. 2009. Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS One 4:e6018 doi:10.1371/journal.pone.0006018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Jagnow J, Clegg S. 2003. Klebsiella pneumoniae MrkD-mediated biofilm formation on extracellular matrix- and collagen-coated surfaces. Microbiology 149:2397–2405 [DOI] [PubMed] [Google Scholar]
- 285. Langstraat J, Bohse M, Clegg S. 2001. Type 3 fimbrial shaft (MrkA) of Klebsiella pneumoniae, but not the fimbrial adhesin (MrkD), facilitates biofilm formation. Infect. Immun. 69:5805–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286. Johnson JG, Clegg S. 2010. Role of MrkJ, a phosphodiesterase, in type 3 fimbrial expression and biofilm formation in Klebsiella pneumoniae. J. Bacteriol. 192:3944–3950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287. Ong CL, Beatson SA, Totsika M, Forestier C, McEwan AG, Schembri MA. 2010. Molecular analysis of type 3 fimbrial genes from Escherichia coli, Klebsiella and Citrobacter species. BMC Microbiol. 10:183 doi:10.1186/1471-2180-10-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288. Sjöström AE, Sonden B, Muller C, Rydström A, Dobrindt U, Wai SN, Uhlin BE. 2009. Analysis of the sfaX(II) locus in the Escherichia coli meningitis isolate IHE3034 reveals two novel regulatory genes within the promoter-distal region of the main S fimbrial operon. Microb. Pathog. 46:150–158 [DOI] [PubMed] [Google Scholar]
- 288a. Spurbeck RR, Tarrien RJ, Mobley HL. 2012. Enzymatically active and inactive phosphodiesterases and diguanylate cyclases are involved in regulation of motility or sessility in Escherichia coli CFT073. mBio 3:e00307–00312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Giraud C, de Bentzmann S. 2012. Inside the complex regulation of Pseudomonas aeruginosa chaperone usher systems. Environ. Microbiol. 14:1805–1816 [DOI] [PubMed] [Google Scholar]
- 290. Klebensberger J, Birkenmaier A, Geffers R, Kjelleberg S, Philipp B. 2009. SiaA and SiaD are essential for inducing autoaggregation as a specific response to detergent stress in Pseudomonas aeruginosa. Environ. Microbiol. 11:3073–3086 [DOI] [PubMed] [Google Scholar]
- 291. Pelicic V. 2008. Type IV pili: e pluribus unum? Mol. Microbiol. 68:827–837 [DOI] [PubMed] [Google Scholar]
- 292. Merz AJ, So M, Sheetz MP. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98–102 [DOI] [PubMed] [Google Scholar]
- 293. Wall D, Kaiser D. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1–10 [DOI] [PubMed] [Google Scholar]
- 294. Jain R, Behrens AJ, Kaever V, Kazmierczak BI. 2012. Type IV pilus assembly in Pseudomonas aeruginosa over a broad range of c-di-GMP concentrations. J. Bacteriol. 194:4285–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. McCarthy Y, Ryan RP, O'Donovan K, He YQ, Jiang BL, Feng JX, Tang JL, Dow JM. 2008. The role of PilZ domain proteins in the virulence of Xanthomonas campestris pv. campestris. Mol. Plant Pathol. 9:819–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295a. Ryan RP, McCarthy Y, Kiely PA, O'Connor R, Farah CS, Armitage JP, Dow JM. 2012. Dynamic complex formation between HD-GYP, GGDEF and PilZ domain proteins regulates motility in Xanthomonas campestris. Mol. Microbiol. 86:557–567 [DOI] [PubMed] [Google Scholar]
- 296. Ryan RP, Dow JM. 2011. Intermolecular interactions between HD-GYP and GGDEF domain proteins mediate virulence-related signal transduction in Xanthomonas campestris. Virulence 1:404–408 [DOI] [PubMed] [Google Scholar]
- 297. Simm R, Lusch A, Kader A, Andersson M, Römling U. 2007. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189:3613–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298. Sommerfeldt N, Possling A, Becker G, Pesavento C, Tschowri N, Hengge R. 2009. Gene expression patterns and differential input into curli fimbriae regulation of all GGDEF/EAL domain proteins in Escherichia coli. Microbiology 155:1318–1331 [DOI] [PubMed] [Google Scholar]
- 299. Tagliabue L, Maciag A, Antoniani D, Landini P. 2010. The yddV-dos operon controls biofilm formation through the regulation of genes encoding curli fibers' subunits in aerobically growing Escherichia coli. FEMS Immunol. Med. Microbiol. 59:477–484 [DOI] [PubMed] [Google Scholar]
- 300. Gjermansen M, Ragas P, Sternberg C, Molin S, Tolker-Nielsen T. 2005. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 7:894–906 [DOI] [PubMed] [Google Scholar]
- 301. Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. 2003. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 49:905–918 [DOI] [PubMed] [Google Scholar]
- 302. Monds RD, Newell PD, Gross RH, O'Toole GA. 2007. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 63:656–679 [DOI] [PubMed] [Google Scholar]
- 303. Chatterjee D, Boyd CD, O'Toole GA, Sondermann H. 2012. Structural characterization of a conserved, calcium-dependent periplasmic protease from Legionella pneumophila. J. Bacteriol. 194:4415–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304. Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. 2010. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Mol. Microbiol. 75:827–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305. Beyhan S, Yildiz FH. 2007. Smooth to rugose phase variation in Vibrio cholerae can be mediated by a single nucleotide change that targets c-di-GMP signalling pathway. Mol. Microbiol. 63:995–1007 [DOI] [PubMed] [Google Scholar]
- 306. Lim B, Beyhan S, Meir J, Yildiz FH. 2006. Cyclic-diGMP signal transduction systems in Vibrio cholerae: modulation of rugosity and biofilm formation. Mol. Microbiol. 60:331–348 [DOI] [PubMed] [Google Scholar]
- 307. Shikuma NJ, Fong JC, Yildiz FH. 2012. Cellular levels and binding of c-di-GMP control subcellular localization and activity of the Vibrio cholerae transcriptional regulator VpsT. PLoS Pathog. 8:e1002719 doi:10.1371/journal.ppat.1002719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308. Fong JC, Yildiz FH. 2007. The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae. J. Bacteriol. 189:2319–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Zakikhany K, Harrington CR, Nimtz M, Hinton JC, Römling U. 2010. Unphosphorylated CsgD controls biofilm formation in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 77:771–786 [DOI] [PubMed] [Google Scholar]
- 310. Ogasawara H, Yamamoto K, Ishihama A. 2011. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J. Bacteriol. 193:2587–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Ahmad I, Lamprokostopoulou A, Le Guyon S, Streck E, Peters V, Barthel M, Hardt W-D, Römling U. 2011. Complex c-di-GMP signaling networks mediate the transition between virulence properties and biofilm formation in Salmonella enterica serovar Typhimurium. PLoS One 6:e28351 doi:10.1371/journal.pone.0028351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Güvener ZT, McCarter LL. 2003. Multiple regulators control capsular polysaccharide production in Vibrio parahaemolyticus. J. Bacteriol. 185:5431–5441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Ueda A, Wood TK. 2010. Tyrosine phosphatase TpbA of Pseudomonas aeruginosa controls extracellular DNA via cyclic diguanylic acid concentrations. Environ. Microbiol. Rep. 2:449–455 [DOI] [PubMed] [Google Scholar]
- 314. An S, Wu J, Zhang LH. 2010. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl. Environ. Microbiol. 76:8160–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Roy AB, Petrova OE, Sauer K. 2012. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol. 194:2904–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315a. Petrova OE, Sauer K. 2012. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc. Natl. Acad. Sci. U. S. A. 109:16690–16695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316. Curtis PD, Brun YV. 2010. Getting in the loop: regulation of development in Caulobacter crescentus. Microbiol. Mol. Biol. Rev. 74:13–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Paul R, Jaeger T, Abel S, Wiederkehr I, Folcher M, Biondi EG, Laub MT, Jenal U. 2008. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell 133:452–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, Baker TA, Jenal U. 2011. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol. Cell 43:550–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319. Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 3:e334 doi:10.1371/journal.pbio.0030334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320. Hecht GB, Lane T, Ohta N, Sommer JM, Newton A. 1995. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 14:3915–3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321. Sockett RE, Lambert C. 2004. Bdellovibrio as therapeutic agents: a predatory renaissance? Nat. Rev. Microbiol. 2:669–675 [DOI] [PubMed] [Google Scholar]
- 322. Flardh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7:36–49 [DOI] [PubMed] [Google Scholar]
- 323. Tran NT, Den Hengst CD, Gomez-Escribano JP, Buttner MJ. 2011. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. J. Bacteriol. 193:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324. Solano C, Garcia B, Latasa C, Toledo-Arana A, Zorraquino V, Valle J, Casals J, Pedroso E, Lasa I. 2009. Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc. Natl. Acad. Sci. U. S. A. 106:7997–8002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324a. Zogaj X, Wyatt GC, Klose KE. 2012. Cyclic di-GMP stimulates biofilm formation and inhibits virulence of Francisella novicida. Infect. Immun. 80:4239–4247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325. Schild S, Tamayo R, Nelson EJ, Qadri F, Calderwood SB, Camilli A. 2007. Genes induced late in infection increase fitness of Vibrio cholerae after release into the environment. Cell Host Microbe 2:264–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326. Tamayo R, Schild S, Pratt JT, Camilli A. 2008. Role of cyclic di-GMP during El Tor biotype Vibrio cholerae infection: characterization of the in vivo-induced cyclic di-GMP phosphodiesterase CdpA. Infect. Immun. 76:1617–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Römling U. 2009. Cyclic di-GMP (c-di-GMP) goes into host cells—c-di-GMP signaling in the obligate intracellular pathogen Anaplasma phagocytophilum. J. Bacteriol. 191:683–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328. Kumagai Y, Matsuo J, Cheng Z, Hayakawa Y, Rikihisa Y. 2011. c-di-GMP signaling regulates intracellular aggregation, sessility, and growth of Ehrlichia chaffeensis. Infect. Immun. 79:3905–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329. Lai TH, Kumagai Y, Hyodo M, Hayakawa Y, Rikihisa Y. 2008. Anaplasma phagocytophilum PleC histidine kinase and PleD diguanylate cyclase two-component system and role of cyclic di-GMP in host-cell infection. J. Bacteriol. 191:693–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Hisert KB, MacCoss M, Shiloh MU, Darwin KH, Singh S, Jones RA, Ehrt S, Zhang Z, Gaffney BL, Gandotra S, Holden DW, Murray D, Nathan C. 2005. A glutamate-alanine-leucine (EAL) domain protein of Salmonella controls bacterial survival in mice, antioxidant defence and killing of macrophages: role of cyclic diGMP. Mol. Microbiol. 56:1234–1245 [DOI] [PubMed] [Google Scholar]
- 331. Kumagai Y, Matsuo J, Hayakawa Y, Rikihisa Y. 2010. Cyclic di-GMP signaling regulates invasion by Ehrlichia chaffeensis of human monocytes. J. Bacteriol. 192:4122–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332. Lacey MM, Partridge JD, Green J. 2010. Escherichia coli K-12 YfgF is an anaerobic cyclic di-GMP phosphodiesterase with roles in cell surface remodelling and the oxidative stress response. Microbiology 156:2873–2886 [DOI] [PubMed] [Google Scholar]
- 333. Lamprokostopoulou A, Monteiro C, Rhen M, Römling U. 2010. Cyclic di-GMP signaling controls virulence properties of Salmonella enterica serovar Typhimurium at the mucosal lining. Environ. Microbiol. 12:40–53 [DOI] [PubMed] [Google Scholar]
- 334. McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, Ishii KJ, Akira S, Colonna M, Chen ZJ, Fitzgerald KA, Hayakawa Y, Vance RE. 2009. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J. Exp. Med. 206:1899–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335. Sauer J-D, Sotelo-Troha K, von Moltke J, Monroe KM, Rae CS, Brubaker SW, Hyodo M, Hayakawa Y, Woodward JJ, Portnoy DA, Vance RE. 2011. The N-ethyl-N-nitrosourea-induced Goldenticket mouse mutant reveals an essential function of Sting in the in vivo interferon response to Listeria monocytogenes and cyclic dinucleotides. Infect. Immun. 79:688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336. Claret L, Miquel S, Vieille N, Ryjenkov DA, Gomelsky M, Darfeuille Michaud A. 2007. The flagellar sigma factor FliA regulates adhesion and invasion of Crohn's disease-associated Escherichia coli via a c-di-GMP-dependent pathway. J. Biol. Chem. 282:33275–33283 [DOI] [PubMed] [Google Scholar]
- 337. Lee HS, Gu F, Ching SM, Lam Y, Chua KL. 2010. CdpA is a Burkholderia pseudomallei cyclic di-GMP phosphodiesterase involved in autoaggregation, flagellum synthesis, motility, biofilm formation, cell invasion, and cytotoxicity. Infect. Immun. 78:1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Levi A, Folcher M, Jenal U, Shuman HA. 2011. Cyclic diguanylate signaling proteins control intracellular growth of Legionella pneumophila. mBio 2:e00316–10 doi:10.1128/mBio.00316-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Yi X, Yamazaki A, Biddle E, Zeng Q, Yang CH. 2010. Genetic analysis of two phosphodiesterases reveals cyclic diguanylate regulation of virulence factors in Dickeya dadantii. Mol. Microbiol. 77:787–800 [DOI] [PubMed] [Google Scholar]
- 340. Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. 2011. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13:3128–3138 [DOI] [PubMed] [Google Scholar]
- 341. He YW, Ng AY, Xu M, Lin K, Wang LH, Dong YH, Zhang LH. 2007. Xanthomonas campestris cell-cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol. Microbiol. 64:281–292 [DOI] [PubMed] [Google Scholar]
- 342. Eichelberg K, Galan JE. 2000. The flagellar sigma factor FliA (σ28) regulates the expression of Salmonella genes associated with the centisome 63 type III secretion system. Infect. Immun. 68:2735–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343. Ramos HC, Rumbo M, Sirard JC. 2004. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 12:509–517 [DOI] [PubMed] [Google Scholar]
- 344. Chua KL, Chan YY, Gan YH. 2003. Flagella are virulence determinants of Burkholderia pseudomallei. Infect. Immun. 71:1622–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345. Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SL, Cookson BT, Aderem A. 2003. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 4:1247–1253 [DOI] [PubMed] [Google Scholar]
- 346. Boudeau J, Barnich N, Darfeuille-Michaud A. 2001. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol. Microbiol. 39:1272–1284 [DOI] [PubMed] [Google Scholar]
- 347. Kang PJ, Hauser AR, Apodaca G, Fleiszig SM, Wiener-Kronish J, Mostov K, Engel JN. 1997. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol. Microbiol. 24:1249–1262 [DOI] [PubMed] [Google Scholar]
- 348. Gerlach RG, Claudio N, Rohde M, Jackel D, Wagner C, Hensel M. 2008. Cooperation of Salmonella pathogenicity islands 1 and 4 is required to breach epithelial barriers. Cell. Microbiol. 10:2364–2376 [DOI] [PubMed] [Google Scholar]
- 349. Lara-Tejero M, Galán JE. 2009. Salmonella enterica serovar Typhimurium pathogenicity island 1-encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect. Immun. 77:2635–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350. Misselwitz B, Kreibich SK, Rout S, Stecher B, Periaswamy B, Hardt WD. 2011. Salmonella enterica serovar Typhimurium binds to HeLa cells via Fim-mediated reversible adhesion and irreversible type three secretion system 1-mediated docking. Infect. Immun. 79:330–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351. Sun YC, Koumoutsi A, Jarrett C, Lawrence K, Gherardini FC, Darby C, Hinnebusch BJ. 2011. Differential control of Yersinia pestis biofilm formation in vitro and in the flea vector by two c-di-GMP diguanylate cyclases. PLoS One 6:e19267 doi:10.1371/journal.pone.0019267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352. Häussler S, Tummler B, Weissbrodt H, Rohde M, Steinmetz I. 1999. Small-colony variants of Pseudomonas aeruginosa in cystic fibrosis. Clin. Infect. Dis. 29:621–625 [DOI] [PubMed] [Google Scholar]
- 353. Häussler S. 2004. Biofilm formation by the small colony variant phenotype of Pseudomonas aeruginosa. Environ. Microbiol. 6:546–551 [DOI] [PubMed] [Google Scholar]
- 354. Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355. Byrd MS, Pang B, Hong W, Waligora EA, Juneau RA, Armbruster CE, Weimer KE, Murrah K, Mann EE, Lu H, Sprinkle A, Parsek MR, Kock ND, Wozniak DJ, Swords WE. 2011. Direct evaluation of Pseudomonas aeruginosa biofilm mediators in a chronic infection model. Infect. Immun. 79:3087–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356. Jarrett CO, Deak E, Isherwood KE, Oyston PC, Fischer ER, Whitney AR, Kobayashi SD, DeLeo FR, Hinnebusch BJ. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190:783–792 [DOI] [PubMed] [Google Scholar]
- 357. Bruggemann H, Hagman A, Jules M, Sismeiro O, Dillies MA, Gouyette C, Kunst F, Steinert M, Heuner K, Coppee JY, Buchrieser C. 2006. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell. Microbiol. 8:1228–1240 [DOI] [PubMed] [Google Scholar]
- 358. Del Favero M, Mazzantini E, Briani F, Zangrossi S, Tortora P, Deho G. 2008. Regulation of Escherichia coli polynucleotide phosphorylase by ATP. J. Biol. Chem. 283:27355–27359 [DOI] [PubMed] [Google Scholar]
- 359. Nurmohamed S, Vincent HA, Titman CM, Chandran V, Pears MR, Du D, Griffin JL, Callaghan AJ, Luisi BF. 2011. Polynucleotide phosphorylase activity may be modulated by metabolites in Escherichia coli. J. Biol. Chem. 286:14315–14323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360. Mohanty BK, Kushner SR. 2006. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 34:5695–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361. De Lay N, Gottesman S. 2011. Role of polynucleotide phosphorylase in sRNA function in Escherichia coli. RNA 17:1172–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362. Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, Wang JX, Lee ER, Block KF, Sudarsan N, Neph S, Tompa M, Ruzzo WL, Breaker RR. 2007. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 35:4809–4819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363. Chen AG, Sudarsan N, Breaker RR. 2011. Mechanism for gene control by a natural allosteric group I ribozyme. RNA 17:1967–1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364. Matilla MA, Travieso ML, Ramos JL, Ramos-Gonzalez MI. 2011. Cyclic diguanylate turnover mediated by the sole GGDEF/EAL response regulator in Pseudomonas putida: its role in the rhizosphere and an analysis of its target processes. Environ. Microbiol. 13:1745–1766 [DOI] [PubMed] [Google Scholar]
- 365. Wang X, Kim Y, Hong SH, Ma Q, Brown BL, Pu M, Tarone AM, Benedik MJ, Peti W, Page R, Wood TK. 2011. Antitoxin MqsA helps mediate the bacterial general stress response. Nat. Chem. Biol. 7:359–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366. Maisonneuve E, Shakespeare LJ, Jorgensen MG, Gerdes K. 2011. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U. S. A. 108:13206–13211 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 367. Wang X, Wood TK. 2011. Toxin-antitoxin systems influence biofilm and persister cell formation and the general stress response. Appl. Environ. Microbiol. 77:5577–5583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368. Kanazawa T, Ren S, Maekawa M, Hasegawa K, Arisaka F, Hyodo M, Hayakawa Y, Ohta H, Masuda S. 2010. Biochemical and physiological characterization of a BLUF protein-EAL protein complex involved in blue light-dependent degradation of cyclic diguanylate in the purple bacterium Rhodopseudomonas palustris. Biochemistry 49:10647–10655 [DOI] [PubMed] [Google Scholar]
- 368a. Massie JP, Reynolds EL, Koestler BJ, Cong JP, Agostoni M, Waters CM. 2012. Quantification of high-specificity cyclic diguanylate signaling. Proc. Natl. Acad. Sci. U. S. A. 109:12746–12751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369. Grantcharova N, Peters V, Monteiro C, Zakikhany K, Römling U. 2010. Bistable expression of CsgD in biofilm development of Salmonella enterica serovar Typhimurium. J. Bacteriol. 192:456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369a. Pultz IS, Christen M, Kulasekara HD, Kennard A, Kulasekara B, Miller SI. 2012. The response threshold of Salmonella PilZ domain proteins is determined by their binding affinities for c-di-GMP. Mol. Microbiol. 86:1424–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370. Wei BL, Brun-Zinkernagel AM, Simecka JW, Pruss BM, Babitzke P, Romeo T. 2001. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol. Microbiol. 40:245–256 [DOI] [PubMed] [Google Scholar]
- 371. Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. 2005. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol. Microbiol. 56:1648–1663 [DOI] [PubMed] [Google Scholar]
- 372. Yakhnin H, Baker CS, Berezin I, Evangelista MA, Rassin A, Romeo T, Babitzke P. 2011. CsrA represses translation of sdiA, which encodes the N-acylhomoserine-l-lactone receptor of Escherichia coli, by binding exclusively within the coding region of sdiA mRNA. J. Bacteriol. 193:6162–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373. Takaya A, Erhardt M, Karata K, Winterberg K, Yamamoto T, Hughes KT. 2012. YdiV: a dual function protein that targets FlhDC for ClpXP-dependent degradation by promoting release of DNA-bound FlhDC complex. Mol. Microbiol. 83:1268–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374. Lawhon SD, Frye JG, Suyemoto M, Porwollik S, McClelland M, Altier C. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48:1633–1645 [DOI] [PubMed] [Google Scholar]
- 375. Lucchetti-Miganeh C, Burrowes E, Baysse C, Ermel G. 2008. The post-transcriptional regulator CsrA plays a central role in the adaptation of bacterial pathogens to different stages of infection in animal hosts. Microbiology 154:16–29 [DOI] [PubMed] [Google Scholar]
- 375a. Lu XH, An SQ, Tang DJ, McCarthy Y, Tang JL, Dow JM, Ryan RP. 2012. RsmA regulates biofilm formation in Xanthomonas campestris through a regulatory network involving cyclic di-GMP and the Clp transcription factor. PLoS One 7:e52646 doi:10.1371/journal.pone.0052646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376. Brencic A, Lory S. 2009. Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72:612–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377. Irie Y, Starkey M, Edwards AN, Wozniak DJ, Romeo T, Parsek MR. 2010. Pseudomonas aeruginosa biofilm matrix polysaccharide Psl is regulated transcriptionally by RpoS and post-transcriptionally by RsmA. Mol. Microbiol. 78:158–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378. O'Callaghan J, Reen FJ, Adams C, O'Gara F. 2011. Low oxygen induces the type III secretion system in Pseudomonas aeruginosa via modulation of the small RNAs rsmZ and rsmY. Microbiology 157:3417–3428 [DOI] [PubMed] [Google Scholar]
- 379. Camilli A, Bassler BL. 2006. Bacterial small-molecule signaling pathways. Science 311:1113–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380. Srivastava D, Waters CM. 2012. A tangled web: regulatory connections between quorum sensing and cyclic di-GMP. J. Bacteriol. 194:4485–4493 doi:10.1128/JB.00379-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381. Hammer BK, Bassler BL. 2008. Distinct sensory pathways in Vibrio cholerae El Tor and classical biotypes modulate c-di-GMP levels to control biofilm formation. J. Bacteriol. 191:169–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382. He YW, Boon C, Zhou L, Zhang LH. 2009. Co-regulation of Xanthomonas campestris virulence by quorum sensing and a novel two-component regulatory system RavS/RavR. Mol. Microbiol. 71:1464–1476 [DOI] [PubMed] [Google Scholar]
- 383. Pesavento C, Hengge R. 2009. Bacterial nucleotide-based second messengers. Curr. Opin. Microbiol. 12:170–176 [DOI] [PubMed] [Google Scholar]
- 384. McDonough KA, Rodriguez A. 2011. The myriad roles of cyclic AMP in microbial pathogens: from signal to sword. Nat. Rev. Microbiol. 10:27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385. Makman RS, Sutherland EW. 1965. Adenosine 3′,5′-phosphate in Escherichia coli. J. Biol. Chem. 240:1309–1314 [PubMed] [Google Scholar]
- 386. Fong JC, Yildiz FH. 2008. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J. Bacteriol. 190:6646–6659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387. Monds RD, Newell PD, Wagner JC, Schwartzman JA, Lu W, Rabinowitz JD, O'Toole GA. 2010. Di-adenosine tetraphosphate (Ap4A) metabolism impacts biofilm formation by Pseudomonas fluorescens via modulation of c-di-GMP-dependent pathways. J. Bacteriol. 192:3011–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388. Goldman SR, Sharp JS, Vvedenskaya IO, Livny J, Dove SL, Nickels BE. 2011. NanoRNAs prime transcription initiation in vivo. Mol. Cell 42:817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389. Ghosh S, Deutscher MP. 1999. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc. Natl. Acad. Sci. U. S. A. 96:4372–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390. Nickels BE, Dove SL. 2011. NanoRNAs: a class of small RNAs that can prime transcription initiation in bacteria. J. Mol. Biol. 412:772–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391. Hsu CY, Dennis D. 1982. RNA polymerase: linear competitive inhibition by bis-(3′ to 5′)-cyclic dinucleotides, NpNp. Nucleic Acids Res. 10:5637–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392. Shang F, Xue T, Sun H, Xing L, Zhang S, Yang Z, Zhang L, Sun B. 2009. The Staphylococcus aureus GGDEF domain-containing protein, GdpS, influences protein A gene expression in a cyclic diguanylic acid-independent manner. Infect. Immun. 77:2849–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393. Rao F, See RY, Zhang D, Toh DC, Ji Q, Liang ZX. 2010. YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 285:473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394. Tschowri N, Busse S, Hengge R. 2009. The BLUF-EAL protein YcgF acts as a direct anti-repressor in a blue-light response of Escherichia coli. Genes Dev. 23:522–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394a. Li B, Li N, Wang F, Guo L, Huang Y, Liu X, Wei T, Zhu D, Liu C, Pan H, Xu S, Wang HW, Gu L. 2012. Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility. Nucleic Acids Res. 40:11073–11085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395. Stewart MK, Cummings LA, Johnson ML, Berezow AB, Cookson BT. 2011. Regulation of phenotypic heterogeneity permits Salmonella evasion of the host caspase-1 inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 108:20742–20747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396. Zhou X, Meng X, Sun B. 2008. An EAL domain protein and cyclic AMP contribute to the interaction between the two quorum sensing systems in Escherichia coli. Cell Res. 18:937–948 [DOI] [PubMed] [Google Scholar]
- 397. Timmermans J, Van Melderen L. 2010. Post-transcriptional global regulation by CsrA in bacteria. Cell. Mol. Life Sci. 67:2897–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398. Jonas K, Tomenius H, Römling U, Georgellis D, Melefors O. 2006. Identification of YhdA as a regulator of the Escherichia coli carbon storage regulation system. FEMS Microbiol. Lett. 264:232–237 [DOI] [PubMed] [Google Scholar]
- 399. Suzuki K, Babitzke P, Kushner SR, Romeo T. 2006. Identification of a novel regulatory protein (CsrD) that targets the global regulatory RNAs CsrB and CsrC for degradation by RNase E. Genes Dev. 20:2605–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400. Hu DL, Narita K, Hyodo M, Hayakawa Y, Nakane A, Karaolis DK. 2009. c-di-GMP as a vaccine adjuvant enhances protection against systemic methicillin-resistant Staphylococcus aureus (MRSA) infection. Vaccine 27:4867–4873 [DOI] [PubMed] [Google Scholar]
- 401. Karaolis DK, Cheng K, Lipsky M, Elnabawi A, Catalano J, Hyodo M, Hayakawa Y, Raufman JP. 2005. 3′,5′-Cyclic diguanylic acid (c-di-GMP) inhibits basal and growth factor-stimulated human colon cancer cell proliferation. Biochem. Biophys. Res. Commun. 329:40–45 [DOI] [PubMed] [Google Scholar]
- 402. Chen W, Kuolee R, Yan H. 2010. The potential of 3′,5′-cyclic diguanylic acid (c-di-GMP) as an effective vaccine adjuvant. Vaccine 28:3080–3085 [DOI] [PubMed] [Google Scholar]
- 403. Kline T, Jackson SR, Deng W, Verlinde CL, Miller SI. 2008. Design and synthesis of bis-carbamate analogs of cyclic bis-(3′-5′)-diguanylic acid (c-di-GMP) and the acyclic dimer PGPG. Nucleosides Nucleotides Nucleic Acids 27:1282–1300 [DOI] [PubMed] [Google Scholar]
- 404. Sintim HO, Smith JA, Wang J, Nakayama S, Yan L. 2010. Paradigm shift in discovering next-generation anti-infective agents: targeting quorum sensing, c-di-GMP signaling and biofilm formation in bacteria with small molecules. Future Med. Chem. 2:1005–1035 [DOI] [PubMed] [Google Scholar]
- 405. Wood TK, Hong SH, Ma Q. 2011. Engineering biofilm formation and dispersal. Trends Biotechnol. 29:87–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405a. Römling U, Balsalobre C. 2012. Biofilm infections, their resilience to therapy and innovative treatment strategies. J. Intern. Med. 272:541–561 [DOI] [PubMed] [Google Scholar]
- 406. Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. 2010. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol. Microbiol. 75:815–826 [DOI] [PubMed] [Google Scholar]
- 407. Ma Q, Zhang G, Wood TK. 2011. Escherichia coli BdcA controls biofilm dispersal in Pseudomonas aeruginosa and Rhizobium meliloti. BMC Res. Notes 4:447 doi:10.1186/1756-0500-4-447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408. Barraud N, Storey MV, Moore ZP, Webb JS, Rice SA, Kjelleberg S. 2009. Nitric oxide-mediated dispersal in single- and multi-species biofilms of clinically and industrially relevant microorganisms. Microb. Biotechnol. 2:370–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409. Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. 2009. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J. Bacteriol. 191:7333–7342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410. Morgan R, Kohn S, Hwang SH, Hassett DJ, Sauer K. 2006. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J. Bacteriol. 188:7335–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411. Carlson HK, Vance RE, Marletta MA. 2010. H-NOX regulation of c-di-GMP metabolism and biofilm formation in Legionella pneumophila. Mol. Microbiol. 77:930–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412. Rao F, Ji Q, Soehano I, Liang ZX. 2011. Unusual heme-binding PAS domain from YybT family proteins. J. Bacteriol. 193:1543–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413. Kelsey I, Nakayama S, Sintim HO. 2011. Diamidinium and iminium aromatics as new aggregators of the bacterial signaling molecule, c-di-GMP. Bioorg. Med. Chem. Lett. 22:881–885 [DOI] [PubMed] [Google Scholar]
- 413a. Sambanthamoorthy K, Sloup RE, Parashar V, Smith JM, Kim EE, Semmelhack MF, Neiditch MB, Waters CM. 2012. Identification of small molecules that antagonize diguanylate cyclase enzymes to inhibit biofilm formation. Antimicrob. Agents Chemother. 56:5202–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414. Steinberger O, Lapidot Z, Ben-Ishai Z, Amikam D. 1999. Elevated expression of the CD4 receptor and cell cycle arrest are induced in Jurkat cells by treatment with the novel cyclic dinucleotide 3′,5′-cyclic diguanylic acid. FEBS Lett. 444:125–129 [DOI] [PubMed] [Google Scholar]
- 415. Brouillette E, Hyodo M, Hayakawa Y, Karaolis DK, Malouin F. 2005. 3′,5′-Cyclic diguanylic acid reduces the virulence of biofilm-forming Staphylococcus aureus strains in a mouse model of mastitis infection. Antimicrob. Agents Chemother. 49:3109–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416. Yan H, KuoLee R, Tram K, Qiu H, Zhang J, Patel GB, Chen W. 2009. 3′,5′-Cyclic diguanylic acid elicits mucosal immunity against bacterial infection. Biochem. Biophys. Res. Commun. 387:581–584 [DOI] [PubMed] [Google Scholar]
- 417. Zhao L, KuoLee R, Harris G, Tram K, Yan H, Chen W. 2011. c-di-GMP protects against intranasal Acinetobacter baumannii infection in mice by chemokine induction and enhanced neutrophil recruitment. Int. Immunopharmacol. 11:1378–1383 [DOI] [PubMed] [Google Scholar]
- 418. Karaolis DKR. 4 August 2009. Method for stimulating the immune, inflammatory or neuroprotective response. US patent 7,569,555. [Google Scholar]
- 419. Madhun AS, Haaheim LR, Nostbakken JK, Ebensen T, Chichester J, Yusibov V, Guzman CA, Cox RJ. 2011. Intranasal c-di-GMP-adjuvanted plant-derived H5 influenza vaccine induces multifunctional Th1 CD4+ cells and strong mucosal and systemic antibody responses in mice. Vaccine 29:4973–4982 [DOI] [PubMed] [Google Scholar]
- 420. Pedersen GK, Ebensen T, Gjeraker IH, Svindland S, Bredholt G, Guzman CA, Cox RJ. 2011. Evaluation of the sublingual route for administration of influenza H5N1 virosomes in combination with the bacterial second messenger c-di-GMP. PLoS One 6:e26973 doi:10.1371/journal.pone.0026973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421. Abdul-Sater AA, Grajkowski A, Erdjument-Bromage H, Plumlee C, Levi A, Schreiber MT, Lee C, Shuman H, Beaucage SL, Schindler C. 2012. The overlapping host responses to bacterial cyclic dinucleotides. Microbes Infect. 14:188–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421a. Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, Liu ZJ, Modlin RL, Liu YJ, Cheng G. 2012. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nat. Immunol. 13:1155–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422. Ross P, Mayer R, Weinhouse H, Amikam D, Huggirat Y, Benziman M, de Vroom E, Fidder A, de Paus P, Sliedregt LA. 1990. The cyclic diguanylic acid regulatory system of cellulose synthesis in Acetobacter xylinum. Chemical synthesis and biological activity of cyclic nucleotide dimer, trimer, and phosphothioate derivatives. J. Biol. Chem. 265:18933–18943 [PubMed] [Google Scholar]
- 423. Hayakawa Y, Nagata R, Hirata A, Hyodo M, Kawai R. 2003. A facile synthesis of cyclic bis(3′-5′)diguanylic acid. Tetrahedron 59:6465–6471 [Google Scholar]
- 424. Kawai R, Nagata R, Hirata A, Hayakawa Y. 2003. A new synthetic approach to cyclic bis(3′→5′)diguanylic acid. Nucleic Acids Res. Suppl. 2003:103–104 [DOI] [PubMed] [Google Scholar]
- 425. Zhang Z, Gaffney BL, Jones RA. 2004. c-di-GMP displays a monovalent metal ion-dependent polymorphism. J. Am. Chem. Soc. 126:16700–16701 [DOI] [PubMed] [Google Scholar]
- 426. Amiot N, Heintz K, Giese B. 2006. New approach for the synthesis of c-di-GMP and its analogues. Synthesis 2006:4230–4236 [Google Scholar]
- 427. Gaffney BL, Veliath E, Zhao J, Jones RA. 2010. One-flask syntheses of c-di-GMP and the [Rp,Rp] and [Rp,Sp] thiophosphate analogues. Org. Lett. 12:3269–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428. Kiburu I, Shurer A, Yan L, Sintim HO. 2008. A simple solid-phase synthesis of the ubiquitous bacterial signaling molecule, c-di-GMP and analogues. Mol. Biosyst. 4:518–520 [DOI] [PubMed] [Google Scholar]
- 429. Rao F, Pasunooti S, Ng Y, Zhuo W, Lim L, Liu AW, Liang ZX. 2009. Enzymatic synthesis of c-di-GMP using a thermophilic diguanylate cyclase. Anal. Biochem. 389:138–142 [DOI] [PubMed] [Google Scholar]
- 430. Spehr V, Warrass R, Hocherl K, Ilg T. 2011. Large-scale production of the immunomodulator c-di-GMP from GMP and ATP by an enzymatic cascade. Appl. Biochem. Biotechnol. 165:761–775 [DOI] [PubMed] [Google Scholar]
- 431. Simm R, Morr M, Remminghorst U, Andersson M, Römling U. 2009. Quantitative determination of cyclic diguanosine monophosphate concentrations in nucleotide extracts of bacteria by matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry. Anal. Biochem. 386:53–58 [DOI] [PubMed] [Google Scholar]
- 432. Spangler C, Bohm A, Jenal U, Seifert R, Kaever V. 2010. A liquid chromatography-coupled tandem mass spectrometry method for quantitation of cyclic di-guanosine monophosphate. J. Microbiol. Methods 81:226–231 [DOI] [PubMed] [Google Scholar]
- 433. Liaw YC, Gao YG, Robinson H, Sheldrick GM, Sliedregt LA, van der Marel GA, van Boom JH, Wang AH. 1990. Cyclic diguanylic acid behaves as a host molecule for planar intercalators. FEBS Lett. 264:223–227 [DOI] [PubMed] [Google Scholar]
- 434. Nakayama S, Kelsey I, Wang J, Sintim HO. 2010. c-di-GMP can form remarkably stable G-quadruplexes at physiological conditions in the presence of some planar intercalators. Chem. Commun. (Camb.) 47:4766–4768 [DOI] [PubMed] [Google Scholar]
- 435. Nakayama S, Roelofs K, Lee VT, Sintim HO. 2012. A c-di-GMP-proflavine-hemin supramolecular complex has peroxidase activity—implication for a simple colorimetric detection. Mol. Biosyst. 8:726–729 [DOI] [PubMed] [Google Scholar]
- 436. Nakayama S, Kelsey I, Wang J, Roelofs K, Stefane B, Luo Y, Lee VT, Sintim HO. 2011. Thiazole orange-induced c-di-GMP quadruplex formation facilitates a simple fluorescent detection of this ubiquitous biofilm regulating molecule. J. Am. Chem. Soc. 133:4856–4864 [DOI] [PubMed] [Google Scholar]
- 437. Gu H, Furukawa K, Breaker RR. 2012. Engineered allosteric ribozymes that sense the bacterial second messenger cyclic diguanosyl 5′-monophosphate. Anal. Chem. 84:4935–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438. Ho CL, Koh SL, Chuah ML, Luo Z, Tan WJ, Low DK, Liang ZX. 2011. Rational design of fluorescent biosensor for cyclic di-GMP. Chembiochem 12:2753–2758 [DOI] [PubMed] [Google Scholar]
- 438a. Rybtke MT, Borlee BR, Murakami K, Irie Y, Hentzer M, Nielsen TE, Givskov M, Parsek MR, Tolker-Nielsen T. 2012. Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 78:5060–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438b. Ho CL, Chong KS, Oppong JA, Chuah ML, Tan SM, Liang ZX. 2013. Visualizing the perturbation of cellular cyclic di-GMP levels in bacterial cells. J. Am. Chem. Soc. 135:566–569 [DOI] [PubMed] [Google Scholar]
- 439. Witte G, Hartung S, Buttner K, Hopfner KP. 2008. Structural biochemistry of a bacterial checkpoint protein reveals diadenylate cyclase activity regulated by DNA recombination intermediates. Mol. Cell 30:167–178 [DOI] [PubMed] [Google Scholar]
- 440. Hsu C-Y, Dennis D, Jones RA. 1985. Synthesis and physical characterization of bis 3′→5′ cyclic dinucleotides c(NpNp): RNA polymerase inhibitors. Nucleosides Nucleotides 4:377–389 [Google Scholar]
- 441. Römling U. 2008. Great times for small molecules: c-di-AMP, a second messenger candidate in Bacteria and Archaea. Sci. Signal. 1:pe39 doi:10.1126/scisignal.133pe39 [DOI] [PubMed] [Google Scholar]
- 442. Aravind L, Koonin EV. 1998. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem. Sci. 23:17–19 [DOI] [PubMed] [Google Scholar]
- 443. Corrigan RM, Abbott JC, Burhenne H, Kaever V, Gründling A. 2011. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS Pathog. 7:e1002217 doi:10.1371/journal.ppat.1002217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444. Kamegaya T, Kuroda K, Hayakawa Y. 2011. Identification of a Streptococcus pyogenes SF370 gene involved in production of c-di-AMP. Nagoya J. Med. Sci. 73:49–57 [PMC free article] [PubMed] [Google Scholar]
- 445. Luo Y, Helmann JD. 2012. Analysis of the role of Bacillus subtilis σM in β-lactam resistance reveals an essential role for c-di-AMP in peptidoglycan homeostasis. Mol. Microbiol. 83:623–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446. Oppenheimer-Shaanan Y, Wexselblatt E, Katzhendler J, Yavin E, Ben-Yehuda S. 2011. c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 12:594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447. Woodward JJ, Iavarone AT, Portnoy DA. 2010. c-di-AMP secreted by intracellular Listeria monocytogenes activates a host type I interferon response. Science 328:1703–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448. Bejerano-Sagie M, Oppenheimer-Shaanan Y, Berlatzky I, Rouvinski A, Meyerovich M, Ben-Yehuda S. 2006. A checkpoint protein that scans the chromosome for damage at the start of sporulation in Bacillus subtilis. Cell 125:679–690 [DOI] [PubMed] [Google Scholar]
- 449. Rallu F, Gruss A, Ehrlich SD, Maguin E. 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35:517–528 [DOI] [PubMed] [Google Scholar]
- 450. Thibessard A, Borges F, Fernandez A, Gintz B, Decaris B, Leblond-Bourget N. 2004. Identification of Streptococcus thermophilus CNRZ368 genes involved in defense against superoxide stress. Appl. Environ. Microbiol. 70:2220–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450a. Zhang L, Li W, He ZG. 2013. DarR, a TetR-like transcriptional factor, is a cyclic-di-AMP responsive repressor in Mycobacterium smegmatis. J. Biol. Chem. 288:3085–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451. Chaudhuri RR, Allen AG, Owen PJ, Shalom G, Stone K, Harrison M, Burgis TA, Lockyer M, Garcia-Lara J, Foster SJ, Pleasance SJ, Peters SE, Maskell DJ, Charles IG. 2009. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH). BMC Genomics 10:291 doi:10.1186/1471-2164-10-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452. French CT, Lao P, Loraine AE, Matthews BT, Yu H, Dybvig K. 2008. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol. Microbiol. 69:67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453. Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA, 3rd, Smith HO, Venter JC. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U. S. A. 103:425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454. Song JH, Ko KS, Lee JY, Baek JY, Oh WS, Yoon HS, Jeong JY, Chun J. 2005. Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol. Cells 19:365–374 [PubMed] [Google Scholar]
- 455. Davies BW, Bogard RW, Young TS, Mekalanos JJ. 2012. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149:358–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455a. Sun L, Wu J, Du F, Chen X, Chen ZJ. 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339:786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456. Gomelsky M. 2010. The core pathway: diguanylate cyclases, phosphodiesterases, and c-di-GMP-binding proteins, p 37–56 In Wolfe AJ, Visick KL. (ed), The second messenger cyclic di-GMP. ASM Press, Washington, DC [Google Scholar]
- 457. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458. Römling U. 2009. Rationalizing the evolution of EAL domain-based cyclic di-GMP-specific phosphodiesterases. J. Bacteriol. 191:4697–4700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459. Wang H, Wu JH, Ayala JC, Benitez JA, Silva AJ. 2011. Interplay among cyclic diguanylate, HapR, and the general stress response regulator (RpoS) in the regulation of Vibrio cholerae hemagglutinin/protease. J. Bacteriol. 193:6529–6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460. Li G, Brown PJ, Tang JX, Xu J, Quardokus EM, Fuqua C, Brun YV. 2011. Surface contact stimulates the just-in-time deployment of bacterial adhesins. Mol. Microbiol. 83:41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461. Wozniak DJ, Cram DC, Daniels CJ, Galloway DR. 1987. Nucleotide sequence and characterization of toxR: a gene involved in exotoxin A regulation in Pseudomonas aeruginosa. Nucleic Acids Res 15:2123–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]