Abstract
Members of the ROK family of proteins are mostly transcriptional regulators and kinases that generally relate to the control of primary metabolism, whereby its member glucose kinase acts as the central control protein in carbon control in Streptomyces. Here, we show that deletion of SCO6008 (rok7B7) strongly affects carbon catabolite repression (CCR), growth, and antibiotic production in Streptomyces coelicolor. Deletion of SCO7543 also affected antibiotic production, while no major changes were observed after deletion of the rok family genes SCO0794, SCO1060, SCO2846, SCO6566, or SCO6600. Global expression profiling of the rok7B7 mutant by proteomics and microarray analysis revealed strong upregulation of the xylose transporter operon xylFGH, which lies immediately downstream of rok7B7, consistent with the improved growth and delayed development of the mutant on xylose. The enhanced CCR, which was especially obvious on rich or xylose-containing media, correlated with elevated expression of glucose kinase and of the glucose transporter GlcP. In liquid-grown cultures, expression of the biosynthetic enzymes for production of prodigionines, siderophores, and calcium-dependent antibiotic (CDA) was enhanced in the mutant, and overproduction of prodigionines was corroborated by matrix-assisted laser desorption ionization–time-of-flight analysis. These data present Rok7B7 as a pleiotropic regulator of growth, CCR, and antibiotic production in Streptomyces.
INTRODUCTION
Streptomycetes are high-G+C soil bacteria that are well known as major producers of antibiotics and natural products. Under nutrient-limited conditions they undergo complex morphological changes, which result in formation of aerial mycelium and chains of spores. The developmental program coincides with production of antibiotics (1). Sequencing of the genome of the model organism Streptomyces coelicolor A3(2) identified around 7,825 genes and more than 20 secondary metabolite gene clusters (2). Over 12% of the genome encodes predicted regulatory proteins, underlining the complexity of the regulatory networks that govern the transcriptional control of cell differentiation, secondary metabolite production, and signaling cascades.
Carbon source regulation is a major factor in the control of morphological differentiation and secondary metabolism (recently reviewed in references 3 and 4). We previously showed that changes in N-acetylglucosamine metabolism have a major effect on the production of pigmented antibiotics in S. coelicolor (5, 6). This is in part explained by the fact that glucosamine-6P is an allosteric effector of the global regulator DasR, which represses antibiotic production (7, 8), but it is likely that other factors play a role, including global carbon control mechanisms. Carbon catabolite repression (CCR) allows bacteria to utilize a preferred carbon source (such as glucose) when exposed to multiple carbohydrates. In Gram-negative bacteria and low-G+C Gram-positive bacteria, such as Escherichia coli and Bacillus subtilis, the phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) plays a major role in carbon regulation (9, 10). During PTS-mediated carbon source internalization, a phosphoryl group is transferred from the glycolytic intermediate phosphoenolpyruvate, via the PTS enzymes phosphotransferase enzyme I (EI), the histidine phosphor-carrier protein (HPr), and enzyme IIA (EIIA), onto the incoming carbon source. The PTS-mediated transport triggers the cyclic AMP (cAMP)-receptor protein (Crp) in Gram-negative bacteria and the catabolite control protein CcpA in low-GC Gram-positive bacteria (9). In Streptomyces, GlcNAc and fructose transport depend on the PTS, but glucose transport is mediated via the major facilitator superfamily (MFS) transporter GlcP (11). In terms of global regulation, streptomycetes lack a CcpA orthologue, while Crp is present but does not play a role in CCR (12). In contrast, in streptomycetes CCR largely depends on the enzyme glucose kinase (Glk; SCO2126), and deletion of the glkA gene results in global loss of CCR (13). Interestingly, CCR is independent of the Glk enzymatic activity and the flux of glucose, and replacing Glk of S. coelicolor with that of Zymomonas mobilis restores normal glycolysis but not CCR (13).
The crystal structure of Glk of Streptomyces griseus was recently resolved (14). The Streptomyces Glk protein belongs to group III of the glucokinases, which contain a ROK signature (Pfam PF00480) that is found in a diverse family of proteins (ROK stands for repressors, open reading frames, and kinases) (15). Some 5,000 ROK-family proteins have been identified so far, mostly in prokaryotes, but family members are found in all kingdoms of life (16). ROK kinases contain a conserved N-terminal ATP binding motif (DxGxT), while ROK regulators contain a canonical helix-turn-helix DNA binding motif in their N-terminal domains. In E. coli, the ROK-family proteins NagC and Mlc are involved in the control of GlcNAc and glucose-PTS genes, respectively (17, 18). The S. coelicolor genome contains 25 genes encoding putative ROK-family proteins, 12 of which contain a DNA binding motif. Additionally, 16 of the ROK proteins contain a zinc binding motif, which conforms to the consensus GHX(9-11)CXCGX2G(C/H)XE and was shown to be essential for function of E. coli Mlc and B. subtilis Glk (19, 20). Interestingly, SCO6008 (Rok7B7) was identified as an activator of actinorhodin biosynthesis in a mass spectrometry (MS)-based assay for proteins that bind upstream of the regulatory gene actII-ORF4 (21), and expression of a metagenomic library in Streptomyces lividans revealed that the rok7B7 homolog rep accelerates antibiotic production and sporulation (22). Additionally, Rok7B7 was recently implicated in CCR in S. coelicolor (23).
In search of new regulatory genes that may play a role in carbon-dependent control of antibiotic production, we describe here the mutational analysis of seven of the rok genes of S. coelicolor. These seven genes all contain a DNA binding domain and a cysteine motif involved in zinc binding (24) and include the three closest relatives of rep. Deletion of rok7B7 (SCO6008) resulted in global changes in antibiotic production and morphological differentiation under various growth conditions. Detailed phenotypic and genome-wide transcriptome and proteome analyses of the rok7B7 null mutant established a relationship between Rok7B7 and CCR depending on the nutritional status of the environment.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All bacterial strains are listed in Table S1 in the supplemental material. Escherichia coli JM109 (25) and ET12567 (26) were used for routine cloning procedures and for extracting nonmethylated DNA, respectively. Streptomyces coelicolor A3(2) M145 was the parent for all mutants described in this work. S. coelicolor M512 (M145 ΔactII-ORF4 ΔredD) (27) lacks the pathway-specific activator genes for actinorhodin and undecylprodigiosin biosynthetic clusters and therefore fails to produce these antibiotics, although the strain has an intact biosynthetic machinery (28). All media and routine Streptomyces techniques are described in the Streptomyces manual (26). E. coli cells were grown in Luria-Bertani broth (LB) at 37°C. Phenotypic characterization of Streptomyces mutants was carried out on minimal medium (MM) and on R2YE− (i.e., lacking glucose) agar plates (26) with 1% carbon sources as indicated. SFM (soy flour mannitol) agar plates were used to prepare spore suspensions. For growth curves, liquid NNMP (26) was used with a low (0.1%, wt/vol) concentration of Casamino Acids. An adapted NMMP medium was used for 14N/15N labeling, containing (per liter) 2 g NH4Cl, 1 g algal amino acids, 0.6 g MgSO4 · 7 H2O, 50 g polyethylene glycol 6000 (NBS Biologicals, Huntingdon, United Kingdom), 0.1 mg ZnSO4 · 7 H2O, 0.1 mg FeSO4 · 7 H2O, 0.1 mg MnCl2 · 7 H2O, 0.1 mg CaCl2, 15 mM Na/K phosphate buffer (pH 6.8), and 5 g mannitol. For 15N labeling, NH4Cl (>99% 15N) and algal amino acids (>98% 15N) were added as 15N-labeled compounds (Cambridge Stable Isotopes, Andover, MA).
Plasmids and constructs.
All constructs described in this work are listed in Table S2 in the supplemental material.
Constructs for gene replacement and deletion mutants.
The strategy for creating knockout mutants is based on the unstable multicopy vector pWHM3 (29) as described previously (11). For each knockout construct, roughly 1.5-kb stretches of upstream and downstream regions of the respective genes were amplified by PCR from the S. coelicolor M145 genome. The upstream region was thereby cloned as an EcoRI-XbaI fragment and the downstream part as an XbaI-HindIII fragment, and these were ligated into EcoRI-HindIII-digested pWHM3 (for the exact location of the oligonucleotides, see Table S3 in the supplemental material). In this way, an XbaI site was engineered between the flanking regions of the gene of interest. This was then used to insert the apramycin (Apr) resistance cassette aacC4 flanked by loxP sites, using engineered XbaI sites. The presence of the loxP recognition sites allows the efficient removal of the apramycin resistance cassette following the introduction of plasmid pUWLcre (expressing the Cre recombinase) (30, 31). We constructed the knockout plasmids pGAM8 to pGAM14 for the single gene replacement of SCO0794, SCO1060, SCO2846, SCO6008, SCO6566, SCO6600, SCO7543, respectively. To analyze correctness of the mutants, PCRs were done on mycelium from liquid-grown MM cultures (oligonucleotide pairs are described in Table S3).
For complementation of the GAM33 mutant (Δrok7B7), the integrative vector pSET152 (32) harboring a recombinant rok7B7 gene, along with a 250-bp promoter region and the stop codon replaced by the sequence 5′-ATGGACTACAAGGACCACGACGGCGACTACAAGGACCACGACATCGACTACAAGGACGACGACGACAAGTAGAGCACGGATGGCACCGTTGTCATCTCGTTAAGGATTTACTTCTT, was used. The recombinant rok7B7 gene was synthesized that allows production of Rok7B7 containing a C-terminal triple FLAG-tag epitope (GenScript, La Jolla, CA).
Agarase assays.
Spores of S. coelicolor M145, its rok7B7 null mutant GAM33, and the complemented mutant were spotted onto MM and R2YE− agar plates (26) with 1% (wt/vol) different carbon sources as indicated. After 3 days of growth, the agarase production was assessed by measuring the zones of clearing of the agar around the mycelial spot and related to the biomass. Experiments were done several times, with highly similar results.
Proteomics analysis.
S. coelicolor M145 and its rok7B7 null mutant, GAM33, were grown in liquid minimal media containing either 14N or 15N as the sole nitrogen source, as described above, until late logarithmic phase, when antibiotic production became apparent due to color changes of the growth medium. This was repeated for a label swap experiment in which the nitrogen sources were exchanged between strains.
At the indicated time points, mycelium was harvested by centrifugation, washed, and sonicated for 5 min at 12-W output power using 5-s-on/5-s-off intervals in 100 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 5 mM dithiothreitol (DTT). Debris was removed by centrifugation at 16,000 × g for 10 min at 4°C. Protein concentration of the extracts was determined with a Bradford protein assay, using bovine serum albumin (BSA) as a standard. Secreted proteins were precipitated from the growth medium in chloroform-methanol as described previously (33). Protein pellets were dissolved in 1% (wt/vol) SDS, 50 mM Tris-HCl, pH 7.5, at 70°C for 10 min, and the amount of protein was measured using a protein DC assay (Bio-Rad, Hercules, CA), using BSA as a standard.
To perform quantitative proteomics, 14N- and 15N-labeled media or mycelial extracts were mixed 1:1 for protein content, and proteins were precipitated using chloroform-methanol (33). Protein pellets were heated at 95°C for 5 min in SDS-PAGE sample buffer, and proteins were separated on mini-Protean TGX anyKD gels (Bio-Rad). Gels were stained with Coomassie brilliant blue, and gel lanes were cut into approximately 15 pieces. Cysteines were reduced with DTT and modified with iodoacetamide, proteins were in-gel digested overnight with trypsin, and peptides were extracted, all as described previously (34). Acidified peptides were purified and concentrated using C18 stage tips (35) as described previously. Peptides were analyzed on a Surveyor nanoLC system (Thermo, Waltham, MA) connected to an LTQ-Orbitrap mass spectrometer (Thermo) as described previously (36).
Peak lists were extracted and charge states determined using DTASuperCharge (version 2.0b1) (37). Spectra were searched against the Streptomyces coelicolor genome database (Sanger Institute, Hinxton, United Kingdom) using Mascot (version 2.2.03; Matrix Science, London, United Kingdom) with a parent ion tolerance of 10 ppm and a fragment ion mass tolerance of 0.9 Da. 15N-metabolic labeling was specified as the quantification method. Fixed and variable modifications were the iodoacetamide derivative of cysteine and oxidation of methionine, respectively. Extracted ion currents (XIC) of identified peptides and their relative ratios were derived using MSQuant (version 2.0a81) (37), and a correction algorithm for incomplete 15N incorporation rates was applied (38). StatQuant (version 1.2.3) (39) was used for peptide ratio outlier removal based on Z-values (n > 3), after which relative protein ratios were calculated by averaging peptide ratios. To compensate for errors in the mixing of samples, protein ratios were normalized based on the median ratio or, in case of the time series experiment, on the elongation factor G (SCO4661; fusA) ratio.
StatQuant was also used to determine whether the observed difference in the amount of protein between the two samples was significant using a one-sample Student's t test. Protein ratios with P < 0.1 were accepted and compared to protein ratios of the label swap experiment using an additional one-sample Student's t test. Proteins that demonstrated opposing ratios between the original and label swap experiment (P > 0.5) or were present in only one of the experiments were eliminated.
MALDI-TOF-MS analysis.
For matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF)-MS, the same mycelial samples as those used for proteomics were also extracted with methanol, and the extract was mixed 1:1 (vol/vol) with a saturated α-cyano-4-hydroxycinnamic acid solution in 50% (vol/vol) acetonitrile–0.05% (vol/vol) trifluoroacetic acid. One μl was spotted on a MALDI target plate, and samples were measured on a Brucker microflex LRF mass spectrometer in the positive-ion reflection mode using delayed extraction. For each spectrum, at least 1,000 shots were acquired at 60 Hz.
RNA isolation and DNA microarray analysis and validation.
For transcript analysis, total RNA was isolated as described previously (40) from surface-grown mycelia of S. coelicolor M145 and its rok7B7 null mutant, cultivated on MM agar plates with mannitol (1%, wt/vol) as the sole carbon source (plates were covered with cellophane discs). Two independent biological replicate experiments were performed. Samples were taken at 14 h (early vegetative growth), 24 h (vegetative growth), 30 h (early aerial growth), 36 h (aerial growth), 42 h (early sporulation), and 54 h (sporulation). Total RNA was purified using the Kirby-mix protocol (26). DNase I treatment was used to fully remove any traces of DNA. Before use, the RNA samples were checked for integrity on an Agilent 2100 Bioanalyzer (Agilent Technologies), and lack of DNA was checked by reverse transcription-PCR (RT-PCR) without reverse transcriptase. cDNA preparation, labeling, and hybridization was performed as previously described (41, 42). For the microarray analysis, 4×44K whole-genome Streptomyces arrays (Agilent Technologies) were used (42). The filtered data sets were analyzed using rank product analysis (43) via the Rank-Prod package in R (version 2.5.0) (44). Differentially expressed genes were identified as demonstrating a >2-fold change in expression between the parental strain M145 (wild type) and the rok7B7 null mutant. To verify the output from the microarray analysis, semiquantitative RT-PCR analysis was carried out as described previously (45). For each RT-PCR, 200 ng of RNA was used (concentration was assessed using a Nanodrop spectrophotometer), and amplification without reverse transcriptase was used as the control. Oligonucleotides used for RT-PCR are presented in Table S3 in the supplemental material.
Computer analysis.
Alignments of ROK-family proteins were made using ClustalW (46). This alignment was then used as the input for the Genebee website (http://www.genebee.msu.su/clustal/basic.html) to produce a phylogram. Shaded protein alignments were visualized using the BoxShade program at http://www.ch.embnet.org/software/BOX_form.html.
RESULTS
The rok-family genes in S. coelicolor.
In total, 25 genes encoding proteins with a ROK-family signature are found on the S. coelicolor genome (2). A phylogram is shown in Fig. 1A. The family includes glucose kinase (Glk; SCO2126), which catalyzes the phosphorylation of glucose and plays a key role in carbon catabolite repression (47), and N-acetylglucosamine kinase NagK (SCO4285) (5). Sixteen of the ROK-family proteins contain a conserved zinc binding cysteine motif (Fig. 1B). Introduction of rep, an ROK-family regulatory gene obtained from a metagenomic library, accelerated sporulation and enhanced antibiotic production in S. lividans (22). BLASTP analysis of the ROK-family proteins of S. coelicolor proteins with the Rep protein sequence as the input revealed SCO6008 as the nearest orthologue in S. coelicolor, with 48% amino acid identity, while the next strongest hits show 30 to 35% amino acid identity (Table 1). However, the gene synteny of rep does not correspond to that of SCO6008 or to that of any of the other ROK genes. To analyze the possible role of the ROK-family proteins of S. coelicolor, we performed mutational analysis of seven rok genes (SCO0794, SCO1060, SCO2846, SCO6008, SCO6566, SCO6600, and SCO7543), whereby the predicted gene products, similar to the Rep protein, contain both a DNA binding and a zinc binding motif and have not been studied previously.
Fig 1.
Phylogeny of the predicted ROK family proteins of S. coelicolor M145 and alignment of Cys motifs. (A) Phylogram of the ROK proteins of S. coelicolor based on ClustalW analysis (46). ROK-family proteins containing a conserved zinc binding motif are marked by asterisks. The names of proteins along with known or suspected functions are given. (B) Alignment of the partial amino acid sequences of the ROK-family proteins that contain the zinc binding motif, with B. subtilis XylR and E. coli NagC included for reference purposes. Asterisks refer to the conserved Cys and His residues that coordinate the zinc atom. For shading, at least 50% of the aligned proteins should share the same or similar amino acid residues. Identical residues are shaded black, similar residues are shaded gray.
Table 1.
ROK regulators and their knockout strains studied in this worka
| Gene | Name of the null mutant | Length of gene product (aa) | % Amino acid identity to: |
|
|---|---|---|---|---|
| NagC | Rep | |||
| SCO0794 | GAM30 | 429 | 28 | 28 |
| SCO1060 | GAM31 | 413 | 27 | 35 |
| SCO2846 | GAM32 | 406 | 29 | 29 |
| SCO6008 (rok7B7) | GAM33 | 403 | 30 | 48 |
| SCO6566 | GAM34 | 584 | 28 | 33 |
| SCO6600 | GAM35 | 441 | 26 | 30 |
| SCO7543 | GAM36 | 424 | 27 | 34 |
The amino acid identities of S. coelicolor A3(2) ROK-family proteins to NagC of E. coli and Rep, which was expressed from a metagenomic library, are presented. Protein lengths and amino acid identities are indicated.
Gene synteny for the rok genes.
The genetic context of the rok genes studied in this work is presented in Fig. S1 in the supplemental material. Suggestively, five out of seven genes are located in direct proximity of one or more ABC transporter systems. SCO6008, previously named rok7B7 (23, 24, 48), is highly conserved in streptomycetes and is flanked by two sugar transport operons, namely, SCO6005 to SCO6007 (upstream) and the xylose transporter operon xylFGH (SCO6009 to SCO6011; downstream), with its gene synteny highly conserved in streptomycetes. Rok7B7 exhibits 27% amino acid identity to the xylose repressor XylR of Bacillus subtilis. Rok7B7 itself shares 92% amino acid identity with NgcR of Streptomyces olivaceoviridis, which controls the upstream-located N-acetylglucosamine and chitobiose ABC transporter operon ngcEFG (49). However, in S. coelicolor N-acetylglucosamine is transported via the PTS, with NagE2 and NagF as the membrane components EIIC and EIIB, respectively (50), while chitobiose is transported via DasABC (51). Since SCO6005 to SCO6007 shows limited sequence similarity to NgcEFG (around 35% amino acid identity), it likely transports a molecule other than N-acetylglucosamine (5, 6).
Several of the other rok genes are also connected to ABC transporter operons. Like rok7B7, SCO6566 is connected to the utilization of C5 sugars, as it is located upstream of a likely ribose transporter operon, with conserved gene synteny in many streptomycetes. In the direct environment of SCO1060 are two sugar transport operons, with SCO1062 to SCO1065 possibly encoding a galactose transporter based on the phylogenetic linkage to galactose utilization genes (galK, galT, and genes for β-galactosidases) and SCO1056 to SCO105 having similarity to the maltose operon malEFG (SCO2231-2234) (52). SCO6600 is divergently transcribed from sugar transport operons SCO6601 to SCO6604, which are involved in glucoside transport (48). SCO7543 lies adjacent to the operons SCO7539 to SCO7542 and SCO7544 to SCO7548. SCO7547 to SCO7544 encode a likely sulfonate ABC transporter with similarity (37.5, 43.4, and 23.3% protein identity, respectively) to TauABC of E. coli (53).
Phenotypic characterization of the rok mutants.
As a first step toward the characterization of the rok genes, null mutants were constructed in S. coelicolor M145. For this, we first replaced the entire coding region of each gene of interest with the apramycin cassette (aacC4) flanked by loxP sequences. For each experiment, five transformants exhibiting the desired double-crossover phenotype (apramycin resistant and thiostrepton sensitive) were selected and verified by PCR as described in Materials and Methods. Following removal of the aacC4 cassette by Cre recombinase, deletion mutants were obtained and designated GAM30 (M145ΔSCO0794), GAM31 (M145ΔSCO1060), GAM32 (M145ΔSCO2846), GAM33 (M145ΔSCO6008), GAM34 (M145ΔSCO6566), GAM35 (M145ΔSCO6600), and GAM36 (M145ΔSCO7543). For all experiments, both the apramycin-resistant and the deletion mutants were analyzed. Considering that there were no phenotypic differences between them in terms of antibiotic production and development, from this point on we will only describe the results obtained for the deletion mutants unless stated otherwise.
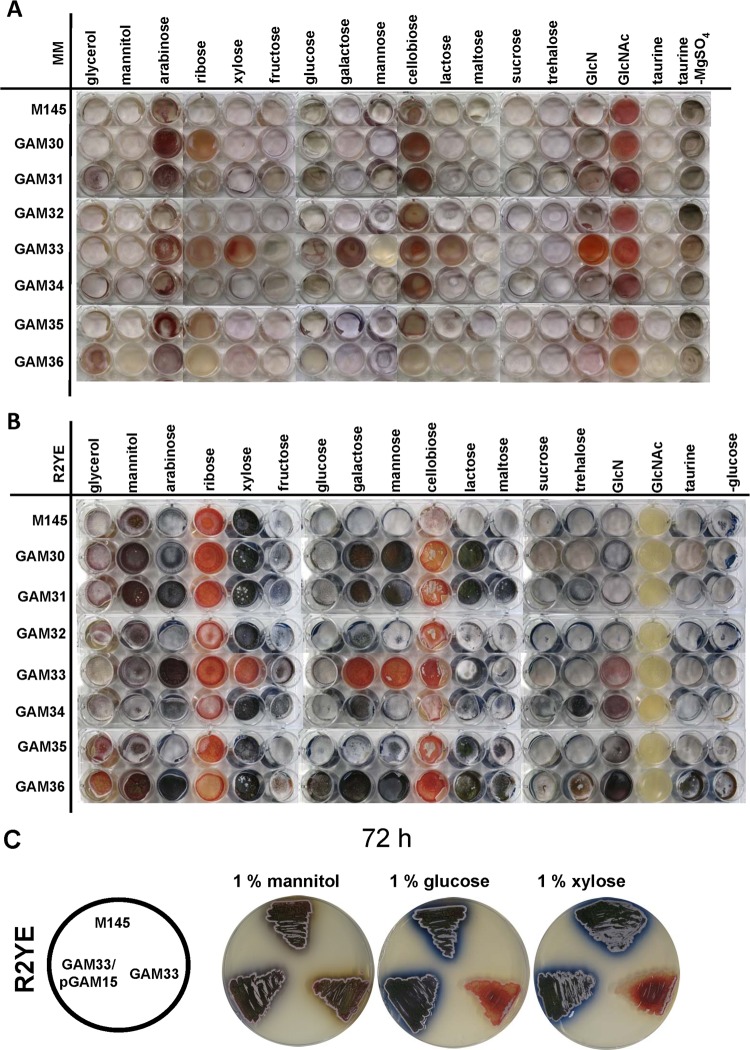
The phenotype of all mutants was tested on MM (minimal medium) and R2YE (rich medium) agar plates supplemented with glycerol, mannitol, arabinose, ribose, xylose, fructose, glucose, galactose, mannose, cellobiose, lactose, maltose, sucrose, trehalose, glucosamine (GlcN), or N-acetylglucosamine (GlcNAc) as the sole carbon source, while taurine was tested as a sulfur source considering the possible relationship between SCO7543 and sulfur utilization. The mutants were phenotypically similar to the parental strain M145 in terms of development when grown on R2YE− or MM agar plates supplemented with fructose, sucrose, trehalose, or taurine. The same was true for MM agar plates supplemented with mannitol or maltose and on R2YE− plates supplemented with GlcNAc.
GlcNAc activated antibiotic production of all strains under poor nutritional conditions (MM), while under rich growth conditions this process and aerial mycelium formation were blocked in all strains (Fig. 2A and B). This is in accordance with previous reports (8). Furthermore, cellobiose also inhibited development and antibiotic production on both MM- and R2YE-grown cultures, which may be due to the repressing effect of its monomer, glucose. Interestingly, actinorhodin production (blue pigment) and development was blocked for all strains grown on R2YE− agar plates with the C5 carbon source ribose and also for several of the strains when grown on MM with ribose (Fig. 2A and B). Development of all strains was also inhibited on MM supplemented with arabinose, which is a C′-2 carbon epimer of ribose. Carbon source dependence of antibiotic production (such as repression by glucose) is a rather well-known phenomenon (3, 54), but the effect of C5 sugars is far less well established.
Fig 2.
Phenotypic analysis of the rok mutants. Phenotypes of S. coelicolor M145 and its ΔSCO0794, ΔSCO1060, ΔSCO2846, ΔSCO6008 (Δrok7B7), ΔSCO6566, ΔSCO6600, and ΔSCO7543 mutants (designated GAM30 to GAM36, respectively) were tested on MM (A) and R2YE (B) microtiter plates supplemented with 1% (wt/vol) carbon sources as indicated. Cultures were grown for 120 h. (C) Complementation of the rok7B7 mutant GAM33 with pGAM15 harboring the rok7B7 gene and its native promoter. Phenotypes were analyzed on R2YE agar plates supplemented with 1% (wt/vol) mannitol, glucose, or xylose. Cultures were grown for 72 h. Note that development of the rok7B7 mutant is delayed on R2YE glucose and not completely blocked, and after 120 h normal sporulation occurred (see panel B).
The most pleiotropic changes were observed in the rok7B7 null mutant and, to a lesser extent, the ΔSCO7543 (GAM36) mutant (Fig. 2A and B). Growth on MM and R2YE agar medium supplemented with GlcNAc did not show significant differences between M145 and the rok7B7 deletion mutant, suggesting that GlcNAc transport still takes place. The rok7B7 null mutant showed delayed development and actinorhodin production on R2YE− with xylose, glucose, galactose, mannose, cellobiose, or GlcN (Fig. 2B and C). Furthermore, development of the rok7B7 mutant was delayed on MM with arabinose and blocked on MM with ribose, xylose, galactose, mannose, cellobiose, lactose, or GlcN (Fig. 2A). Development of the ΔSCO7543 null mutant GAM36 was delayed under most growth conditions (Fig. 2A and B). The other five rok gene null mutants were similar to the parent under most growth conditions (Fig. 2A and B). Considering the pleiotropic effect of the deletion of rok7B7, we subjected its null mutant, GAM33, to more extensive analysis. In a control experiment, GAM33 was complemented with pGAM15 harboring rok7B7 and its native promoter, which restored the wild-type phenotype (Fig. 2C).
Rok7B7 controls xylose utilization and carbon catabolite repression.
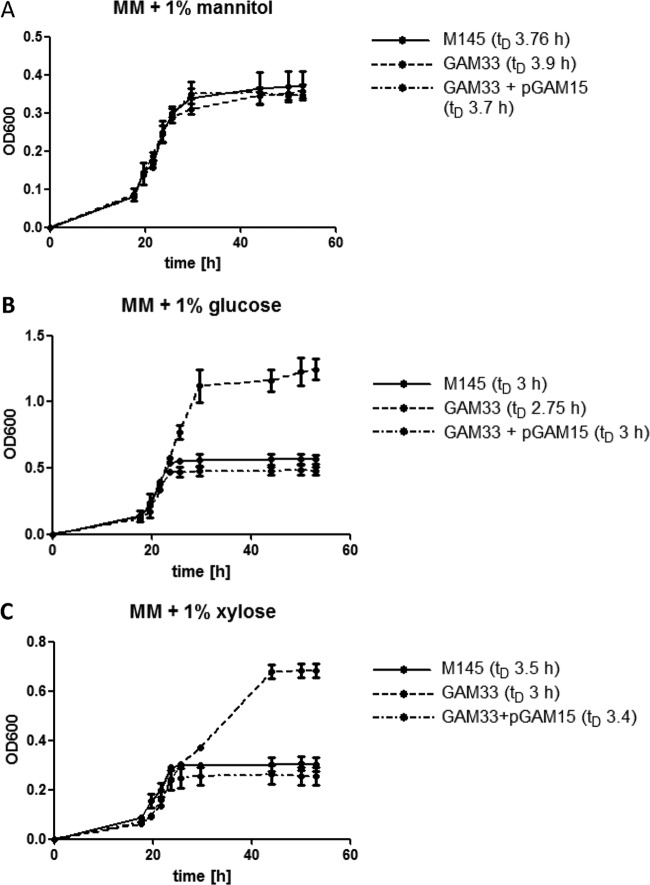
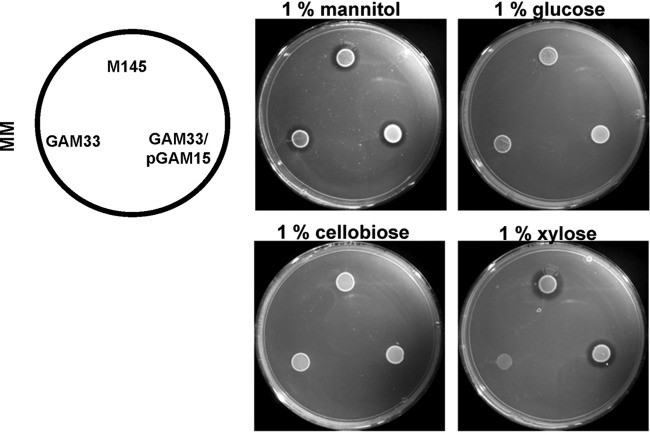
Considering the in silico predictions and the possible involvement of Rok7B7 in control of xylose utilization, we monitored growth of the rok7B7 mutant GAM33 in defined liquid MM with mannitol (1%, wt/vol), xylose (1%, wt/vol), or glucose (1%, wt/vol) as the sole carbon source. The rok7B7 mutant grew with a doubling time (∼4 h) similar to that of the parent strain M145 in the presence of mannitol (Fig. 3A). Interestingly, in MM with glucose or xylose, GAM33 had a slightly higher growth rate than the parent M145 and reached significantly higher biomass levels due to postponed entry into the stationary phase (around 6 or 20 h later for glucose and xylose, respectively), suggesting enhanced glucose and xylose utilization (Fig. 3B and C). Indeed, complementation of GAM33 with plasmid pGAM15 harboring rok7B7 and its native promoter restored the wild-type phenotype with reduced growth on glucose and xylose (Fig. 3B and C). This strongly suggests that the improved sugar utilization by GAM33 was due solely to the deletion of rok7B7. The prolonged vegetative growth phase corresponds well to the observed delay in development and antibiotic production on MM and R2YE agar plates supplemented with either glucose or xylose. The previously reported upregulation in glucose-grown cultures of the Rok7B7 protein in the glkA mutant, which is pleiotropically defective in CCR, prompted analysis of the role of Rok7B7 in carbon control (23). The secreted agarase produced by S. coelicolor is a classical CCR target and provides a simple assay (13). Therefore, agarase production by M145, its rok7B7 null mutant GAM33, and the complemented mutant (harboring pGAM15) was compared on solid media containing mannitol, glucose, cellobiose, or xylose as the sole carbon source. The same amount of agarase was produced by all three strains on MM agar plates supplemented with mannitol. On MM agar plates with glucose or its dimer, cellobiose, neither strain produced significant amounts of agarase due to efficient glucose repression (Fig. 4 and Table 2). Interestingly, while the parental strain M145 produced normal levels of agarase on MM with xylose, agarase production was completely repressed in the rok7B7 mutant (Fig. 4 and Table 2). Agarase production was restored following complementation of GAM33 with pGAM15 (Fig. 4), confirming that repression resulted solely from the inactivation of rok7B7. The rok7B7 mutant also showed enhanced CCR on MM agar plates with maltose or glucosamine as the sole carbon source, although not as strong as that with xylose (Table 2). CCR was strongly enhanced in the rok7B7 mutant grown on R2YE agar plates with various carbon sources (except glucose and cellobiose), with the complete block of agarase production on R2YE with GlcNAc. These data suggest that Rok7B7 acts as a global regulator of CCR.
Fig 3.
Growth curves in minimal media with various carbon sources. The wild-type strain M145 (solid line), GAM33 (lacking rok7B7) (dashed line), and the complemented mutant GAM33/pGAM15 (dashed/dotted line) were grown in MM supplemented with mannitol (A), glucose (B), or xylose (C). tD, doubling time. All growth analyses were performed in duplicate. Note that the mutant grows significantly better on glucose or xylose than the parental strain or the complemented mutant. The standard deviations for each data point are indicated. OD600, optical density at 600 nm.
Fig 4.
Effects of mannitol, glucose, cellobiose, and xylose on agarase production. Strains were M145 (parental strain), its rok7B7 null mutant GAM33, and GAM33 complemented with pGAM15 (harboring the rok7B7 gene). Five μl of spores of either strain (109 CFU/ml) was spotted onto MM plates. After 3 days of growth, the agarase production was detected as a zone of clearing of the agar around the mycelial spot and was related to the biomass (for details, see Table 2 and reference 63).
Table 2.
Rok7B7 and carbon catabolite repression of agarase production
| Agar plate and supplement | Value for straina: |
Fold change | |||||
|---|---|---|---|---|---|---|---|
| M145 |
Δrok7B7 (GAM33) |
||||||
| Agarase (mm) | Total (mm) | Ratio | Agarase (mm) | Total (mm) | Ratio | ||
| MM | |||||||
| Agar | 6 | 26 | 0.23 | 6 | 26 | 0.23 | 1 |
| Mannitol | 5 | 25 | 0.2 | 5 | 25 | 0.2 | 1 |
| GlcN | 5 | 23 | 0.22 | 3 | 19 | 0.16 | 0.73 |
| Fructose | 6 | 26 | 0.23 | 6 | 25 | 0.24 | 1.04 |
| Maltose | 6 | 26 | 0.23 | 5 | 26 | 0.19 | 0.83 |
| GlcNAc | 5 | 26 | 0.19 | 5 | 26 | 0.19 | 1 |
| Glucose | 1 | 17 | 0.06 | 1 | 17 | 0.06 | 1 |
| Xylose | 5 | 25 | 0.2 | 0 | 15 | 0 | 0 |
| Chitin | 6 | 26 | 0.23 | 6 | 27 | 0.22 | 0.96 |
| Cellobiose | 1 | 17 | 0.06 | 1 | 17 | 0.06 | 1 |
| R2YE | |||||||
| Agar | 5 | 22 | 0.23 | 1 | 15 | 0.07 | 0.29 |
| Mannitol | 5 | 22 | 0.23 | 1.5 | 16 | 0.09 | 0.41 |
| GlcN | 5 | 22 | 0.23 | 1.5 | 16 | 0.09 | 0.41 |
| Fructose | 5 | 23 | 0.22 | 2 | 17 | 0.12 | 0.54 |
| Maltose | 5 | 22 | 0.23 | 3 | 20 | 0.15 | 0.66 |
| GlcNAc | 2 | 17 | 0.12 | 0 | 14 | 0 | 0 |
| Glucose | 1 | 18 | 0.06 | 1 | 18 | 0.06 | 1 |
| Xylose | 5 | 25 | 0.2 | 2 | 17 | 0.12 | 0.59 |
| Chitin | 5 | 22 | 0.23 | 2 | 17 | 0.12 | 0.52 |
| Cellobiose | 1 | 18 | 0.06 | 1 | 18 | 0.06 | 1 |
Agarase indicates agarase zone, measured from the outside of the colony to the edge of the zone of clearing. Total indicates the combined length of the agarase zone and the colony. Ratio indicates the length of the agarase zone to the combined length of the agarase zone and the colony. Fold change indicates the ratio of the expression level of Δrok7B7 to that of M145.
Global transcriptional profiling of the rok7B7 null mutant by microarray analysis.
To obtain insight into the global transcriptional changes as a result of the deletion of rok7B7 DNA microarray analysis was performed on RNA isolated from solid-grown MM mannitol cultures of S. coelicolor M145 and its rok7B7 null mutant, GAM33. For this, plates were overlaid with cellophane discs and RNA was harvested after 14 h (early vegetative growth), 24 h (corresponding to vegetative growth), 30 h (vegetative/aerial growth), 36 h (aerial growth), 42 h (aerial growth/sporulation), and 54 h (sporulation). Under these conditions, which have been employed for a variety of microarray experiments with S. coelicolor, the mutant showed a timing of development similar to that of its parent.
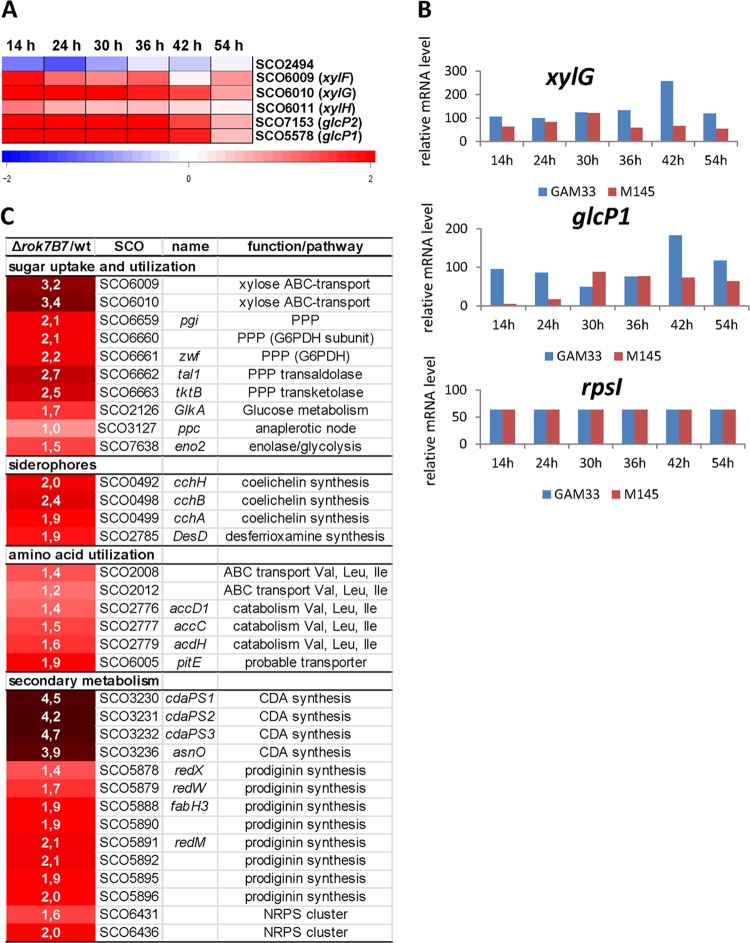
The transcription of over 160 genes was more than 2-fold up- or downregulated at one or more time points in the rok7B7 mutant. These genes are listed in Table S4 in the supplemental material. Only a few targets showed over 2-fold differential expression between GAM33 and the wild-type strain at several time points, including genes of the xylFGH operon (xylF and xylG) (Fig. 5A). Interestingly, the target that was most strongly downregulated in the rok7B7 mutant was SCO2494, encoding pyruvate phosphate dikinase (PPDK; SCO2494), which generates PEP from pyruvate. This enzyme was recently implicated to play a key role in glucose kinase-independent carbon control in streptomycetes (23). In support of the enhanced expression of xylFGH observed in the microarray data, quantitative proteomics experiments also showed that the components of the XylFGH transporter are about 10-fold increased at the protein level in the rok7B7 mutant compared to the parental strain M145 (see below). Interestingly, transcription of glcP for the glucose permease GlcP (which occurs in two identical copies, glcP1 and glcP2) was strongly upregulated at several time points (Fig. 5A). Most of the signal observed in the microarray experiments probably stems from glcP1, as glcP2 is hardly expressed under any of the conditions tested, including during growth on glucose (11). The genes for glucose kinase and for the glycolytic enzymes were not changed significantly. Besides the primary metabolic genes discussed above, another class of genes that was deregulated in the rok7B7 null mutant was that of the chpABCDEFGH, rdlAB, and ramS genes, which encode the surfactant proteins chaplins, rodlins, and the spore-associated protein SapB, respectively (55, 56). All of these genes showed the same trend, but transcription of chpDEH and rdlAB was most strongly downregulated, namely, over 2-fold at 42 h compared to the parental strain M145 (see Table S4). Conversely, transcription was upregulated at 54 h.
Fig 5.
Transcriptomic and proteomic analysis of the S. coelicolor rok7B7 null mutant. (A) Genes whose transcription was significantly differentially expressed between the rok7B7 null mutant (GAM33) and its parent, S. coelicolor M145. RNA was isolated from S. coelicolor M145 or its rok7B7 null mutant, GAM33, after 14 h (early vegetative growth), 24 h (vegetative growth), 30 h (vegetative/aerial growth), 36 h (aerial growth), 42 h (aerial growth/sporulation), and 54 h (sporulation) of growth on MM agar plates supplemented with 1% mannitol. The heat map indicates the fold changes. (B) Validation of microarray data by semiquantitative RT-PCR. RT-PCR was performed on RNA isolated from the parental strain M145 and the rok7B7 null mutant, using oligonucleotide pairs for a selection of targets (xylG and glcP1). rpsI (for ribosomal protein S9) was used as a control. (C) Differentially expressed proteins between the rok7B7 null mutant and the wild type. Quantitative proteomic comparison of the rok7B7 deletion strain and parent strain (M145) is shown. Strains were grown in liquid MM supplemented with mannitol. The comparison between strains is indicated as ratios of the Δrok7B7 strain to M145. Results are given as log2 ratios, therefore positive numbers indicate upregulation.
In a control experiment, the microarray data were further validated by semiquantitative RT-PCR performed on the same RNA, using oligonucleotide pairs for xylG and glcP1, while rpsI was the control (Fig. 5B). The relative transcript levels of xylG and glcP1 corresponded well to the microarray analysis, confirming their strong upregulation in the rok7B7 null mutant.
Global protein expression profiling of the rok7B7 mutant by proteomics.
To further study the involvement of Rok7B7 in primary metabolism, carbon catabolite repression, and secondary metabolites biosynthesis, proteomics analysis was performed. For this, S. coelicolor M145 and its rok7B7 null mutant, GAM33, were grown in liquid minimal media containing either 14N or 15N as the sole nitrogen source and mannitol as the sole carbon source until late logarithmic phase, when antibiotic production became apparent due to color changes of the growth medium. 14N- and 15N-labeled protein extracts were obtained from both the mycelium itself and the spent media. These growth conditions are very similar to those used in the study of the glkA mutant (23), allowing direct comparison of the outcome.
Quantitative proteomics identified 59 proteins (53 proteins in the mycelial fraction and 7 in the secretome, with the xylose binding protein SCO6009 in both lists) whose expression changed with statistical significance in the rok7B7 null mutant (Fig. 5C; also see Table S5 in the supplemental material). Classes of proteins that were overrepresented were those related to (i) primary metabolism (sugar and amino acid transport, metabolic enzymes), (ii) siderophore biosynthesis, and (iii) secondary metabolism (antibiotic biosynthesis) (Fig. 5C).
Control of primary metabolism and CCR.
In line with the notion that Rok7B7 controls the downstream-located xylose operon SCO6009 to SCO6011 (xylFGH), SCO6009 and SCO6010 were strongly upregulated (9- to 11-fold) in the rok7B7 null mutant. Interestingly, glucose kinase (Glk; SCO2126) was 3-fold upregulated in the rok7B7 null mutant, while phosphoenolpyruvate carboxylase (PPC; SCO3127) was 2-fold upregulated. These results are remarkable, since no glucose is present in the growth medium, and this again links Rok7B7 to glucose utilization and carbon catabolite repression.
Enzymes of the pentose phosphate pathway (PPP; SCO6659 to SCO6663) were also significantly more abundant (4- to 6-fold) in the rok7B7 mutant. Considering the chromosomal location, SCO6658 to SCO6663 is considered the secondary PPP cluster and may be involved in providing NADPH for secondary metabolite production (57). Abundance of proteins for glutamate import (SCO5775 to SCO5777) was 2- to 3-fold lower in the mutant, while those for import and catabolism of branched-chain amino acids (SCO2008 to SCO2012 and SCO2776 to SCO2779) and an enolase (SCO7638) were 2- to 3-fold enhanced in the mutant. Furthermore, the solute binding protein SCO6005, which is a distant homologue of the NgcE protein that is involved in N-acetylglucosamine transport in Streptomyces olivaceoviridis (49), was 4-fold more abundant in the rok7B7 mutant.
Control of siderophore and antibiotic biosynthesis.
The rok7B7 mutant shows strongly increased expression of biosynthetic proteins for the siderophores coelichelin (SCO0492, SCO0494, SCO0498, and SCO0499; 3- to 5-fold enhanced) and desferrioxamine (SCO2782 and SCO2785; 2- to 4-fold enhanced). These compounds bind extracellular iron, after which they can be reimported through ABC transporters (58–60). A significant number of proteins that showed differential expression in the rok7B7 mutant compared to M145 related to antibiotic production, namely, calcium-dependent antibiotic (CDA; SCO3230 to SCO3232 and SCO3236), prodiginine (SCO5878 to SCO5896, eight proteins detected), and an uncharacterized nonribosomal peptide (SCO6431 and SCO6436) all demonstrated increased expression levels in the rok7B7 mutant. In particular, the biosynthetic proteins for CDA production were massively upregulated (Fig. 5C).
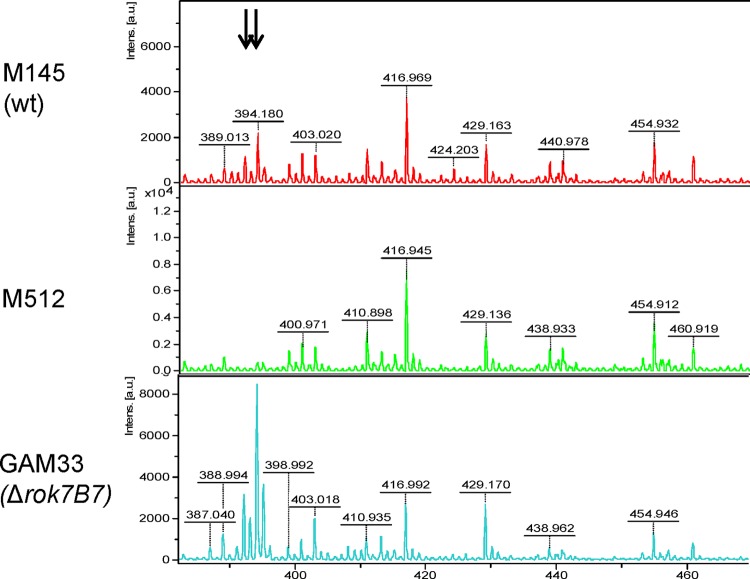
To validate these data and assess the effect of the enhanced expression of antibiotic-related proteins to secondary metabolite production, we subjected mycelial samples corresponding to the samples used for proteomics to MALDI-TOF-MS analysis. S. coelicolor M512, which fails to produce prodigionines, was used as a negative control. Interestingly, prodiginines (m/z 392 and 394) were readily detected in the rok7B7 mutant and approximately 4-fold enhanced compared to the level in the parent M145 (Fig. 6). This validates the proteomics data, demonstrating that the deletion of rok7B7 does result in upregulation of antibiotic production in minimal medium cultures.
Fig 6.
MALDI-TOF analysis of prodiginine production. Production of prodigionines was analyzed in mycelia of strains M145, GAM33, and M512. Arrows correspond to the 392- and 394-Da peaks for prodigionines. Horizontal axis, m/z (Da); vertical axis, intensity (arbitrary units [a.u.]).
DISCUSSION
ROK-family proteins are widespread in actinomycetes, which are generally associated with the control of carbon metabolism. Except for glucose kinase (13, 23, 47, 61), study of this promising family of regulators and (sugar) kinases is relatively scarce. Analysis of a series of novel ROK null mutants identified rok7B7 as a likely new pleiotropic regulatory gene in S. coelicolor. rok7B7 null mutants showed a delay in aerial mycelium formation under most growth conditions as well as deregulation of antibiotic production, and they showed enhanced carbon catabolite repression. Deletion of the other rok family genes, SCO0794, SCO1060, SCO2846, SCO6566, SCO6600, and SCO7543, had less obvious phenotypic consequences, although the ΔSCO7543 null mutant was somewhat affected in antibiotic production and aerial mycelium formation.
The rok7B7 mutant showed strongly enhanced growth in liquid-grown cultures with glucose or xylose as the sole carbon source, while at the same time development and antibiotic production were delayed in solid-grown cultures with the same carbon sources. This suggests that in the absence of rok7B7, the switch to development is postponed, as the onset of development and antibiotic production roughly correspond to the transition phase in liquid-grown cultures (62). This corresponds well to previous data showing that Rok7B7 likely activates actinorhodin biosynthesis (21). Furthermore, rok7B7 is the nearest orthologue of rep isolated from a metagenomic library, the introduction of which into S. lividans accelerated both development and antibiotic production (22). The control of development by Rok7B7 is likely mediated at least in part via the control of carbon utilization, and it is upregulated during CCR in S. coelicolor (23). A model for the Rok7B7 regulon is presented in Fig. 7.
Fig 7.
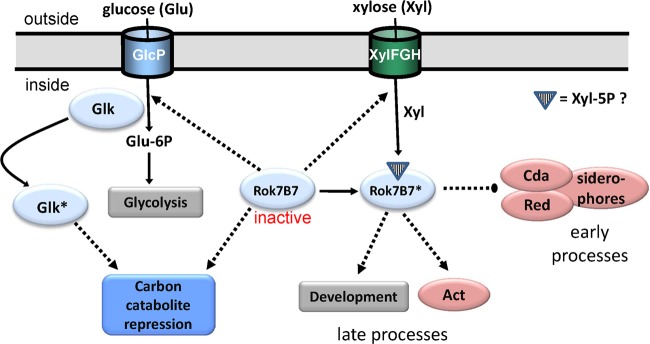
Model for the Rok7B7 regulatory network. Our data show that active Rok7B7 (indicated with an asterisk) activates actinorhodin (Act) production and development (as shown previously for its orthologue, Rep [22]), which explains the delayed development and Act production in the rok7B7 null mutant. Conversely, the production of the secondary metabolites Red, CDA, and siderophores is upregulated. Deletion of rok7B7 (presented in the model as “inactive Rok7B7”) results in enhanced expression of GlcP, Glk, and XylFGH as well as induction of CCR (including Glk-independent CCR). During glucose utilization, Glk is likely posttranslationally activated (shown as Glk*), resulting in CCR (11). Solid lines, conversion or pathway; dotted lines, regulation (arrows represent activation, large dots indicate repression). We speculate that a metabolic derivative of xylose, such as xylose-5P, acts as a ligand for Rok7B7, thus activating the protein.
Combined transcriptomic and proteomics analyses suggested that Rok7B7 acts as a repressor of the adjacent xylose operon xylFGH (SCO6009 to SCO6011), since expression of the operon was strongly enhanced in the mutant. Furthermore, in a model in which Rok7B7 acts as a repressor, one would expect that expression of the components of the xylose transporter are downregulated following enhanced expression of Rok7B7, but recent proteomics data revealed the opposite, namely, that Rok7B7 and XylFGH are both strongly induced in glkA deletion mutants (23). Such a direct correlation of expression is not consistent with a simple model of Rok7B7 acting as a specific repressor of xylFGH. In fact, quantitative proteomics experiments identified XylFGH as the only sugar transport system that was strongly activated by glucose in a Glk-independent manner, along with its putative ROK-family regulator, Rok7B7. We propose that the upregulation of the xylose transporter operon is an attempt to compensate for the absence of Rok7B7, which would be consistent with a role of xylose-5P or a related metabolite as an inducer molecule of Rok7B7.
In terms of the control of xylose utilization, the xylose regulator XylR of S. coelicolor is likely encoded by SCO1171, with the gene located adjacent to, and its gene product likely controlling the transcription of, xylA (SCO1169; for xylose isomerase) and xylB (SCO1170; for xylulose kinase). XylA and XylB together catalyze the conversion from xylose to xylulose-5P. Since there is no phylogenetic linkage between the xylABR genes and rok7B7 or xylFGH and deletion of rok7B7 does not affect any of the genes among SCO1169 to SCO1171, it is less likely that these genes belong to the rok7B7 regulon. However, to obtain full insight into the relationship between the regulons controlled by SCO1171-XylR and Rok7B7, the control of xylose and xylulose metabolism needs to be investigated in more detail. It should be noted, however, that deletion of rok7B7 alone is sufficient to obtain strong CCR of agarase on xylose-containing MM agar plates, suggesting that xylose transport and utilization was completely derepressed in the rok7B7 mutant. A similar observation was made for the control of maltose utilization by MalR in S. coelicolor, where malR deletion mutants showed complete catabolite control of agarase when maltose was used as the sole carbon source, while the wild-type strain hardly grows on maltose (63).
Interestingly, the combined data suggest that deletion of rok7B7 induces glucose kinase-independent CCR. In other words, Rok7B7 may repress this poorly studied CCR pathway (23). The glucose transporter gene glcP was strongly upregulated in the rok7B7 null mutant, suggesting direct control of glucose metabolism by Rok7B7. The expression of GlcP is strongly induced by glucose in a Glk-independent manner (23). The transmembrane GlcP binds to Glk, and its release during sugar transport may be a key mechanism in the activation of CCR (61). The target that was most strongly downregulated at the transcriptional level in the rok7B7 null mutant was SCO2494, encoding pyruvate phosphate dikinase (PPDK; SCO2494), which generates PEP from pyruvate, the opposite of the reaction catalyzed by pyruvate kinase. PPDK also belongs to those proteins that are most strongly repressed by glucose in a Glk-independent manner (23). In fact, PPDK is some 10- to 30-fold downregulated by glucose in a Glk-independent manner in both mannitol- and fructose-grown cultures, and it may play a key role in Glk-independent carbon control in streptomycetes. The change in expression of GlcP (upregulated) and SCO2494 (downregulated) by glucose in a Glk-independent manner is consistent with cells exerting glucose repression, as was indeed observed for the rok7B7 mutant. This is further corroborated by the enhanced expression of Glk in rok7B7 null mutants, as shown by the proteomics experiments. The fact that no significant change was found in transcription of the gene for glucose kinase (glkA) is consistent with the constitutive transcription of glkA and confirms the earlier concept that glucose kinase expression is controlled at the posttranscriptional level (61). All in all, our data suggest that Rok7B7 negatively controls CCR. However, despite our efforts using purified Rok7B7, we failed to detect direct binding of Rok7B7 to the promoter regions of any of the genes whose expression was most significantly affected by the deletion of rok7B7 (data not illustrated). We therefore performed global in vivo DNA binding studies using chromatin immunoprecipitation (ChIP)-chip technology on cultures grown under standard conditions of minimal media agar plates with mannitol as the sole carbon source, but this also failed to identify significant binding to specific sites. Therefore, we expect that a ligand or cofactor is required to facilitate binding of Rok7B7 to the DNA, with derivatives of xylose, cellobiose, or glucose as candidate molecules. ChIP-chip experiments under different growth conditions should provide more insight into the direct Rok7B7 regulon and its binding site.
In terms of antibiotic production, both the red-pigmented prodiginines and their biosynthetic proteins were enhanced in the rok7B7 mutant in liquid-grown MM cultures supplemented with mannitol, as shown by proteomics and subsequent MALDI-TOF-MS analysis of secondary metabolites. Conversely, in solid-grown cultures on rich media (R2YE), the production of the blue-pigmented antibiotic actinorhodin was strongly delayed in the rok7B7 mutant, which was independent of the carbon source and consistent with earlier reports that Rok7B7 activates actinorhodin production (21, 22). Following a DNA-affinity capture assay (DACA), Rok7B7 was isolated as one of the proteins that was bound to the promoter regions of actII-ORF4 and redD, the pathway-specific transcriptional regulators of the act and red gene clusters, respectively (21). All data point to control of antibiotic production by Rok7B7, with activation of Act and repression of Red and CDA. The proteomics experiments also showed upregulation of the degradation pathway for branched-chain amino acids (leucine, isoleucine, and valine), which are precursors for biosynthesis of polyketides such as actinorhodin (64), suggesting that besides antibiotic production, Rok7B7 also controls precursor supply.
While the C5 sugar xylose had a stimulatory effect on actinorhodin production in wild-type cells, perhaps acting via Rok7B7, the C5 sugars arabinose and ribose, which are closely related compounds, suppress actinorhodin production. To the best of our knowledge, the stimulatory effect of xylose on antibiotic production was not reported previously, but the inhibitory effect of the C5 sugars ribose and arabinose on antibiotic production was established in Streptomyces hygroscopicus (65) and Streptomyces olivocinereus (66). Actinorhodin production is strongly delayed in the rok7B7 mutant when grown on media containing xylose. As argued above, xylose or a derivative thereof (e.g., xylose-5P) may act as an effector molecule of Rok7B7, leading to its activation and linking nutrient sensing to antibiotic production and development. Antibiotic production was also repressed by cellobiose. Carbon source dependence of antibiotic production is widely reported in the literature (reviewed in references 3 and 54). Since cellobiose is a dimer of glucose (β-1,4), its repressing activity regarding antibiotic production may be explained by the accumulation of glucose produced from its extracellular hydrolysis, thus effecting glucose repression.
In conclusion, our work underlines the complexity of the regulatory networks involved in the control of antibiotic production and highlights Rok7B7 as a novel pleiotropic transcriptional regulator of CCR that links the control of primary and secondary metabolism in S. coelicolor (Fig. 7). Rok7B7 therefore qualifies as a possible target for genetic engineering approaches toward the improvement of antibiotic production.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Fritz Titgemeyer and Sébastien Rigali for stimulating discussions and to Bobby Florea and Hermen Overkleeft for help with mass spectrometry.
The work was supported by a VENI grant and a VICI grant from the Netherlands Technology Foundation (STW) to J.G. and G.P.W., respectively.
Footnotes
Published ahead of print 4 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02191-12.
REFERENCES
- 1. Hopwood DA. 1999. Forty years of genetics with Streptomyces: from in vivo through in vitro to in silico. Microbiology 145:2183–2202 [DOI] [PubMed] [Google Scholar]
- 2. Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O'Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147 [DOI] [PubMed] [Google Scholar]
- 3. Sanchez S, Chavez A, Forero A, Garcia-Huante Y, Romero A, Sanchez M, Rocha D, Sanchez B, Avalos M, Guzman-Trampe S, Rodriguez-Sanoja R, Langley E, Ruiz B. 2010. Carbon source regulation of antibiotic production. J. Antibiot. (Tokyo) 63:442–459 [DOI] [PubMed] [Google Scholar]
- 4. van Wezel GP, McKenzie NL, Nodwell JR. 2009. Chapter 5. Applying the genetics of secondary metabolism in model actinomycetes to the discovery of new antibiotics. Methods Enzymol. 458:117–141 [DOI] [PubMed] [Google Scholar]
- 5. Swiatek MA, Tenconi E, Rigali S, van Wezel GP. 2012. Functional analysis of the N-acetylglucosamine metabolic genes of Streptomyces coelicolor and role in the control of development and antibiotic production. J. Bacteriol. 194:1136–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Swiatek MA, Urem M, Tenconi E, Rigali S, van Wezel GP. 2012. Engineering of N-acetylglucosamine metabolism for improved antibiotic production in Streptomyces coelicolor A3(2) and an unsuspected role of NagA in glucosamine metabolism. Bioengineered 3:280–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rigali S, Nothaft H, Noens EE, Schlicht M, Colson S, Muller M, Joris B, Koerten HK, Hopwood DA, Titgemeyer F, van Wezel GP. 2006. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 61:1237–1251 [DOI] [PubMed] [Google Scholar]
- 8. Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, van Wezel GP. 2008. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9:670–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brückner R, Titgemeyer F. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141–148 [DOI] [PubMed] [Google Scholar]
- 10. Deutscher J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11:87–93 [DOI] [PubMed] [Google Scholar]
- 11. van Wezel GP, Mahr K, Konig M, Traag BA, Pimentel-Schmitt EF, Willimek A, Titgemeyer F. 2005. GlcP constitutes the major glucose uptake system of Streptomyces coelicolor A3(2). Mol. Microbiol. 55:624–636 [DOI] [PubMed] [Google Scholar]
- 12. Piette A, Derouaux A, Gerkens P, Noens EE, Mazzucchelli G, Vion S, Koerten HK, Titgemeyer F, De Pauw E, Leprince P, van Wezel GP, Galleni M, Rigali S. 2005. From dormant to germinating spores of Streptomyces coelicolor A3(2): new perspectives from the crp null mutant. J. Proteome Res. 4:1699–1708 [DOI] [PubMed] [Google Scholar]
- 13. Angell S, Lewis CG, Buttner MJ, Bibb MJ. 1994. Glucose repression in Streptomyces coelicolor A3(2): a likely regulatory role for glucose kinase. Mol. Gen. Genet. 244:135–143 [DOI] [PubMed] [Google Scholar]
- 14. Miyazono K, Tabei N, Morita S, Ohnishi Y, Horinouchi S, Tanokura M. 2012. Substrate recognition mechanism and substrate-dependent conformational changes of an ROK family glucokinase from Streptomyces griseus. J. Bacteriol. 194:607–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Titgemeyer F, Reizer J, Reizer A, Saier MH., Jr 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140:2349–2354 [DOI] [PubMed] [Google Scholar]
- 16. Conejo MS, Thompson SM, Miller BG. 2010. Evolutionary bases of carbohydrate recognition and substrate discrimination in the ROK protein family. J. Mol. Evol. 70:545–556 [DOI] [PubMed] [Google Scholar]
- 17. Nam TW, Cho SH, Shin D, Kim JH, Jeong JY, Lee JH, Roe JH, Peterkofsky A, Kang SO, Ryu S, Seok YJ. 2001. The Escherichia coli glucose transporter enzyme IICB(Glc) recruits the global repressor Mlc. EMBO J. 20:491–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Plumbridge J. 1995. Co-ordinated regulation of amino sugar biosynthesis and degradation: the NagC repressor acts as both an activator and a repressor for the transcription of the glmUS operon and requires two separated NagC binding sites. EMBO J. 14:3958–3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mesak LR, Mesak FM, Dahl MK. 2004. Bacillus subtilis GlcK activity requires cysteines within a motif that discriminates microbial glucokinases into two lineages. BMC Microbiol. 4:6 doi:10.1186/1471-2180-4-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schiefner A, Gerber K, Seitz S, Welte W, Diederichs K, Boos W. 2005. The crystal structure of Mlc, a global regulator of sugar metabolism in Escherichia coli. J. Biol. Chem. 280:29073–29079 [DOI] [PubMed] [Google Scholar]
- 21. Park SS, Yang YH, Song E, Kim EJ, Kim WS, Sohng JK, Lee HC, Liou KK, Kim BG. 2009. Mass spectrometric screening of transcriptional regulators involved in antibiotic biosynthesis in Streptomyces coelicolor A3(2). J. Ind. Microbiol. Biotechnol. 36:1073–1083 [DOI] [PubMed] [Google Scholar]
- 22. Martinez A, Kolvek SJ, Hopke J, Yip CL, Osburne MS. 2005. Environmental DNA fragment conferring early and increased sporulation and antibiotic production in Streptomyces species. Appl. Environ. Microbiol. 71:1638–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gubbens J, Janus M, Florea BI, Overkleeft HS, van Wezel GP. 2012. Identification of glucose kinase dependent and independent pathways for carbon control of primary metabolism, development and antibiotic production in Streptomyces coelicolor by quantitative proteomics. Mol. Microbiol. 86:1490–1507 [DOI] [PubMed] [Google Scholar]
- 24. Nothaft H. 2003. Carbon metabolism in Streptomyces coelicolor. Ph.D. thesis, Friedrich Alexander University, Erlangen, Germany [Google Scholar]
- 25. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 26. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 27. Floriano B, Bibb M. 1996. afsR is a pleiotropic but conditionally required regulatory gene for antibiotic production in Streptomyces coelicolor A3(2). Mol. Microbiol. 21:385–396 [DOI] [PubMed] [Google Scholar]
- 28. van Wezel GP, White J, Hoogvliet G, Bibb MJ. 2000. Application of redD, the transcriptional activator gene of the undecylprodigiosin biosynthetic pathway, as a reporter for transcriptional activity in Streptomyces coelicolor A3(2) and Streptomyces lividans. J. Mol. Microbiol. Biotechnol. 2:551–556 [PubMed] [Google Scholar]
- 29. Vara J, Lewandowska-Skarbek M, Wang YG, Donadio S, Hutchinson CR. 1989. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J. Bacteriol. 171:5872–5881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fedoryshyn M, Welle E, Bechthold A, Luzhetskyy A. 2008. Functional expression of the Cre recombinase in actinomycetes. Appl. Microbiol. Biotechnol. 78:1065–1070 [DOI] [PubMed] [Google Scholar]
- 31. Khodakaramian G, Lissenden S, Gust B, Moir L, Hoskisson PA, Chater KF, Smith MC. 2006. Expression of Cre recombinase during transient phage infection permits efficient marker removal in Streptomyces. Nucleic Acids Res. 34:e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43–49 [DOI] [PubMed] [Google Scholar]
- 33. Wessel D, Flugge UI. 1984. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem. 138:141–143 [DOI] [PubMed] [Google Scholar]
- 34. Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1:2856–2860 [DOI] [PubMed] [Google Scholar]
- 35. Rappsilber J, Mann M, Ishihama Y. 2007. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2:1896–1906 [DOI] [PubMed] [Google Scholar]
- 36. Florea BI, Verdoes M, Li N, van der Linden WA, Geurink PP, van den Elst H, Hofmann T, de Ru A, van Veelen PA, Tanaka K, Sasaki K, Murata S, den Dulk H, Brouwer J, Ossendorp FA, Kisselev AF, Overkleeft HS. 2010. Activity-based profiling reveals reactivity of the murine thymoproteasome-specific subunit beta5t. Chem. Biol. 17:795–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mortensen P, Gouw JW, Olsen JV, Ong SE, Rigbolt KT, Bunkenborg J, Cox J, Foster LJ, Heck AJ, Blagoev B, Andersen JS, Mann M. 2010. MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J. Proteome Res. 9:393–403 [DOI] [PubMed] [Google Scholar]
- 38. Gouw JW, Tops BB, Mortensen P, Heck AJ, Krijgsveld J. 2008. Optimizing identification and quantitation of 15N-labeled proteins in comparative proteomics. Anal. Chem. 80:7796–7803 [DOI] [PubMed] [Google Scholar]
- 39. van Breukelen B, van den Toorn HW, Drugan MM, Heck AJ. 2009. StatQuant: a post-quantification analysis toolbox for improving quantitative mass spectrometry. Bioinformatics 25:1472–1473 [DOI] [PubMed] [Google Scholar]
- 40. Noens EE, Mersinias V, Willemse J, Traag BA, Laing E, Chater KF, Smith CP, Koerten HK, van Wezel GP. 2007. Loss of the controlled localization of growth stage-specific cell-wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor. Mol. Microbiol. 64:1244–1259 [DOI] [PubMed] [Google Scholar]
- 41. Bucca G, Hindle Z, Smith CP. 1997. Regulation of the dnaK operon of Streptomyces coelicolor A3(2) is governed by HspR, an autoregulatory repressor protein. J. Bacteriol. 179:5999–6004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bucca G, Laing E, Mersinias V, Allenby N, Hurd D, Holdstock J, Brenner V, Harrison M, Smith CP. 2009. Development and application of versatile high density microarrays for genome-wide analysis of Streptomyces coelicolor: characterization of the HspR regulon. Genome Biol. 10:R5.doi:10.1186/gb-2009-10-1-r5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Breitling R, Armengaud P, Amtmann A, Herzyk P. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573:83–92 [DOI] [PubMed] [Google Scholar]
- 44. Hong F, Breitling R, McEntee CW, Wittner BS, Nemhauser JL, Chory J. 2006. RankProd: a bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 22:2825–2827 [DOI] [PubMed] [Google Scholar]
- 45. Colson S, Stephan J, Hertrich T, Saito A, van Wezel GP, Titgemeyer F, Rigali S. 2007. Conserved cis-acting elements upstream of genes composing the chitinolytic system of streptomycetes are DasR-responsive elements. J. Mol. Microbiol. Biotechnol. 12:60–66 [DOI] [PubMed] [Google Scholar]
- 46. Nguyen KD, Pan Y. 2011. Multiple sequence alignment based on dynamic weighted guidance tree. Int. J. Bioinformatics Res. Appl. 7:168–182 [DOI] [PubMed] [Google Scholar]
- 47. Angell S, Schwarz E, Bibb MJ. 1992. The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol. Microbiol. 6:2833–2844 [DOI] [PubMed] [Google Scholar]
- 48. Bertram R, Schlicht M, Mahr K, Nothaft H, Saier MH, Jr, Titgemeyer F. 2004. In silico and transcriptional analysis of carbohydrate uptake systems of Streptomyces coelicolor A3(2). J. Bacteriol. 186:1362–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xiao X, Wang F, Saito A, Majka J, Schlosser A, Schrempf H. 2002. The novel Streptomyces olivaceoviridis ABC transporter Ngc mediates uptake of N-acetylglucosamine and N,N′-diacetylchitobiose. Mol. Genet. Genomics 267:429–439 [DOI] [PubMed] [Google Scholar]
- 50. Nothaft H, Rigali S, Boomsma B, Swiatek M, McDowall KJ, van Wezel GP, Titgemeyer F. 2010. The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol. Microbiol. 75:1133–1144 [DOI] [PubMed] [Google Scholar]
- 51. Colson S, van Wezel GP, Craig M, Noens EE, Nothaft H, Mommaas AM, Titgemeyer F, Joris B, Rigali S. 2008. The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor. Microbiology 154:373–382 [DOI] [PubMed] [Google Scholar]
- 52. van Wezel GP, White J, Bibb MJ, Postma PW. 1997. The malEFG gene cluster of Streptomyces coelicolor A3(2): characterization, disruption and transcriptional analysis. Mol. Gen. Genet. 254:604–608 [DOI] [PubMed] [Google Scholar]
- 53. van der Ploeg JR, Eichhorn E, Leisinger T. 2001. Sulfonate-sulfur metabolism and its regulation in Escherichia coli. Arch. Microbiol. 176:1–8 [DOI] [PubMed] [Google Scholar]
- 54. van Wezel GP, McDowall KJ. 2011. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Nat. Prod. Rep. 28:1311–1333 [DOI] [PubMed] [Google Scholar]
- 55. Claessen D, de Jong W, Dijkhuizen L, Wösten HA. 2006. Regulation of Streptomyces development: reach for the sky! Trends Microbiol. 14:313–319 [DOI] [PubMed] [Google Scholar]
- 56. Willey JM, Gaskell AA. 2011. Morphogenetic signaling molecules of the streptomycetes. Chem. Rev. 111:174–187 [DOI] [PubMed] [Google Scholar]
- 57. Challis GL, Hopwood DA. 2003. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. U. S. A. 100:14555–14561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barona-Gomez F, Lautru S, Francou FX, Leblond P, Pernodet JL, Challis GL. 2006. Multiple biosynthetic and uptake systems mediate siderophore-dependent iron acquisition in Streptomyces coelicolor A3(2) and Streptomyces ambofaciens ATCC 23877. Microbiology 152:3355–3366 [DOI] [PubMed] [Google Scholar]
- 59. Craig M, Lambert S, Jourdan S, Tenconi E, Colson S, Maciejewska M, Martin JF, Ongena M, Van Wezel G, Rigali S. 2012. Unsuspected control of siderophore production by N-acetylglucosamine in streptomycetes. Env. Microbiol. Rep. [DOI] [PubMed] [Google Scholar]
- 60. Patel P, Song L, Challis GL. 2010. Distinct extracytoplasmic siderophore binding proteins recognize ferrioxamines and ferricoelichelin in Streptomyces coelicolor A3(2). Biochemistry 49:8033–8042 [DOI] [PubMed] [Google Scholar]
- 61. van Wezel GP, Konig M, Mahr K, Nothaft H, Thomae AW, Bibb M, Titgemeyer F. 2007. A new piece of an old jigsaw: glucose kinase is activated posttranslationally in a glucose transport-dependent manner in Streptomyces coelicolor A3(2). J. Mol. Microbiol. Biotechnol. 12:67–74 [DOI] [PubMed] [Google Scholar]
- 62. Huang J, Lih CJ, Pan KH, Cohen SN. 2001. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 15:3183–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. van Wezel GP, White J, Young P, Postma PW, Bibb MJ. 1997. Substrate induction and glucose repression of maltose utilization by Streptomyces coelicolor A3(2) is controlled by malR, a member of the lacl-galR family of regulatory genes. Mol. Microbiol. 23:537–549 [DOI] [PubMed] [Google Scholar]
- 64. Stirrett K, Denoya C, Westpheling J. 2009. Branched-chain amino acid catabolism provides precursors for the type II polyketide antibiotic, actinorhodin, via pathways that are nutrient dependent. J. Ind. Microbiol. Biotechnol. 36:129–137 [DOI] [PubMed] [Google Scholar]
- 65. Ilić S, Veljković KSVB, Savić DS, Gojgić-Cvijović GD. 1991. The impact of different carbon and nitrogen sources on antibiotic production by Streptomyces hygroscopicus CH-7. Antibiot. Khimioter. 36:5–8 [Google Scholar]
- 66. Deianova OA, Vinogradova KNKA, Korolev PN, Polin AN. 1988. Effect of L-arabinose and sucrose on the biosynthesis of heliomycin by its producer Streptomyces olivocinereus 11-98. Antibiot. Khimioter. 33:248–252 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.