Abstract
Putative integrative and conjugative elements (ICEs), i.e., genomic islands which could excise, self-transfer by conjugation, and integrate into the chromosome of the bacterial host strain, were previously identified by in silico analysis in the sequenced genomes of Streptococcus agalactiae (M. Brochet et al., J. Bacteriol. 190:6913–6917, 2008). We investigated here the mobility of the elements integrated into the 3′ end of a tRNALys gene. Three of the four putative ICEs tested were found to excise but only one (ICE_515_tRNALys) was found to transfer by conjugation not only to S. agalactiae strains but also to a Streptococcus pyogenes strain. Transfer was observed even if recipient cell already carries a related resident ICE or a genomic island flanked by attL and attR recombination sites but devoid of conjugation or recombination genes (CIs-Mobilizable Element [CIME]). The incoming ICE preferentially integrates into the 3′ end of the tRNALys gene (i.e., the attR site of the resident element), leading to a CIME-ICE structure. Transfer of the whole composite element CIME-ICE was obtained, showing that the CIME is mobilizable in cis by the ICE. Therefore, genomic islands carrying putative virulence genes but lacking the mobility gene can be mobilized by a related ICE after site-specific accretion.
INTRODUCTION
Mobile genetic elements (MGEs) and genomic islands play a key role in bacterial genome evolution by disseminating new genes and phenotypes to the recipient cells (1). The multiplication of bacterial genome sequencing projects in the last few years provides a remarkable opportunity to explore the pool of bacterial genetic mobile elements or “mobilome” (2, 3).
Bacterial genome analyses pointed out a novel class of widespread MGEs, called integrative and conjugative elements (ICEs), and related genomic islands (4–7). These chromosomal elements can excise by site-specific recombination, transfer by conjugation to another bacterial cell and integrate into the chromosome of the recipient cell (4, 7). ICEs are characterized by a combination of modules that can either be involved in their dissemination and maintenance (recombination, conjugation, and regulation modules) or confer adaptive functions to their host (catabolic properties, virulence, and antibiotic resistance) (8, 9). They evolve by acquisition, deletion, and exchange of these modules between different ICEs or with other MGEs (5, 8). The excision of most ICEs relies on an integrase of the tyrosine recombinase family, which catalyzes the site-specific recombination between identical sequences carried by attL (left attachment site) and attR (right attachment site) recombination sites flanking the element. This leads to the excision of a circular form of the ICE harboring an attI site (attachment site of ICE) and to a chromosomal attB (bacterial attachment site) empty site. After transfer, the circular ICE generally integrates into the chromosome of the recipient cell by site-specific recombination between identical sequences carried by the attI site and the attB site, including the 3′ end of a tRNA gene, of a gene encoding ribosomal protein or another gene encoding a conserved protein (4).
In Gram-negative bacteria, the transfer of conjugative plasmids is initiated by a relaxase that nicks plasmid at the origin of transfer (oriT) and interacts with a coupling protein to convey DNA to a transport channel crossing the bacterial cell envelope (10). Little is known about the components of the conjugation machinery of conjugative plasmids and ICEs from Firmicutes (7). The conjugation machinery of Firmicutes has been studied extensively for two conjugative plasmids: pIP501 of Streptococcus agalactiae and pCW3 of Clostridium perfringens (10). Major proteins of these conjugative systems include a relaxase ensuring DNA processing, an ATPase that likely energizes the DNA transport process, a coupling protein that links the relaxosome with the transport apparatus, and a peptidoglycan hydrolase to facilitate the assembly of the transport channel in the membrane (10). For ICEBs1 of Bacillus subtilis, the ATPase ConE colocalized with excised ICEBs1 DNA at or near the cell poles, suggesting that the conjugation machinery assembles at the donor cell poles (11).
Genomic islands, which carry a recombination module but encode only some the proteins required for conjugation (relaxase, other proteins of the relaxosome, and sometimes the coupling protein), could excise and use the transport apparatus of unrelated ICEs and conjugative plasmids to transfer into a recipient cell and were thus called integrative mobilizable elements (IMEs) (5). Other elements that lack recombination and conjugation modules but are flanked by recombination sites and derived from ICEs have been reported (8, 12). We recently demonstrated in Streptococcus thermophilus that a related ICE can integrate in these recombination sites and mobilize in cis the element (thus called CIs-Mobilizable Element [CIME]) (13).
We have previously detected 35 different ICEs, IMEs, and related genomic islands in eight sequenced genomes of S. agalactiae (group B streptococci) (14), an opportunistic pathogen responsible for invasive bacterial diseases in human neonates (15) and causing infections in various animals (16, 17). In the present study, we examined the functionality of putative ICEs of S. agalactiae that are integrated at the 3′ end of a gene encoding a tRNA lysine (CTT anticodon) and carry a conjugation module distantly related to those of the ICEs RD2 from S. pyogenes and ICESt3 from S. thermophilus. We first test their excision as a circular form and then their conjugative transfer inside the S. agalactiae species and to other Firmicutes species. We also examine the ability of the ICEs to cis-mobilize a genomic island integrated into the same locus.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The original strains, spontaneous resistant strains, and plasmids used to obtain recipient or donor strains are listed in Table 1. Strains which were modified or mutated were named according to the modification. For example, the S. agalactiae Nem316 Rifr Strr strain refers to a spontaneous mutant selected from S. agalactiae Nem316 strain which is resistant to rifampin and spectinomycin.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype or genotypea | Source or reference |
|---|---|---|
| Strains | ||
| S. agalactiae | ||
| 515 | Wild-type strain, carrying a putative ICE (ICE_515_tRNALys) | 19 |
| 2603V/R | Wild-type strain, carrying a composite ICE-IME element (ICE_2603_tRNALys), encoding a truncated conjugal transfer protein OrfD (ATPase VirB4 homolog) | 20 |
| COH1 | Wild-type strain, carrying a putative ICE (ICE_COH1_tRNALys), encoding a truncated conjugal transfer protein OrfD (ATPase VirB4 homolog) | 19 |
| 18RS21 | Wild-type strain, carrying a putative ICE (ICE_18RS21_tRNALys), encoding three truncated conjugal proteins (OrfC, OrfD, and OrfL) | 19 |
| Nem316 | Wild-type strain, carrying a putative CIME (CIME_Nem_tRNALys) | 21 |
| A909 | Wild-type strain, carrying a putative IME (IME_A909_tRNALys) | 19 |
| 515 (ICE_515_tRNALysery) | Strain carrying ICE_515_tRNALys tagged by an Eryr cassette, Eryr | This study |
| 515 (ICE_515_tRNALyscat) | Strain carrying ICE_515_tRNALys tagged by a Cmr cassette, Cmr | This study |
| 2603V/R (ICE_2603V/R_tRNALysery) | Strain carrying ICE_2603V/R_tRNALys tagged by an Eryr cassette, Eryr | This study |
| COH1 (ICE_COH1_tRNALysery) | Strain carrying ICE_COH1_tRNALys tagged by an Eryr cassette, Eryr | This study |
| 18RS21 (ICE_18RS21_tRNALysery) | Strain carrying ICE_18RS21_tRNALys tagged by an Eryr cassette, Eryr | This study |
| Nem316 (CIME_Nem_tRNALyscat) | Strain carrying carrying CIME_Nem_tRNALys tagged by a Cmr cassette, Cmr | This study |
| COH1 Rifr Strr | Spontaneous rifampin- and streptomycin-resistant mutant | This study |
| Nem316 Rifr Strr | Spontaneous rifampin- and streptomycin-resistant mutant | This study |
| A909 Rifr Strr | Spontaneous rifampin- and streptomycin-resistant mutant | This study |
| S. mutans | ||
| UA159 | Wild-type strain, no element integrated in the tRNALys CTT gene | 22 |
| UA159 Rifr Strr | Spontaneous rifampin- and streptomycin-resistant mutant | This study |
| S. salivarius | ||
| CIP102503 | Wild-type strain | Institut Pasteur collection |
| JIM8777 | Wild-type strain, with a putative IME integrated in the tRNALys CTT gene | 23 |
| CIP102503 Rifr Strr | Spontaneous rifampin- and streptomycin-resistant mutant | This study |
| JIM8777 Rifr Strr | Spontaneous rifampin- and streptomycin-resistant mutant | This study |
| S. thermophilus | ||
| LMG18311 | Wild-type strain, with a putative CIME-IME composite element integrated in the tRNALys CTT gene | 24 |
| LMG18311 Rifr Strr | Spontaneous rifampin- and streptomycin-resistant mutant | This study |
| S. uberis | ||
| 20388 | Wild-type strain | 25 |
| 21458 | Wild-type strain | 25 |
| 20388 Rifr Strr | Spontaneous rifampin- and streptomycin-resistant mutant | This study |
| 21458 Rifr Strr | Spontaneous rifampin- and streptomycin-resistant | This study |
| S. dysgalactiae | ||
| 14998 | Wild-type strain | 25 |
| 16192 | Wild-type strain | 25 |
| 14998 Rifr Strr | Spontaneous rifampin- and streptomycin-resistant | This study |
| 16192 Rifr Strr | Spontaneous rifampin- and streptomycin-resistant mutant | This study |
| S. pyogenes | ||
| ATCC 12202 | Wild-type strain | ATCC |
| ATCC 12202 Rifr | Spontaneous rifampin-resistant mutant | 26 |
| E. faecalis | ||
| JH2-2 | Rifr, Fusr | 27 |
| JH2-2 Rifr Strr | Spontaneous streptomycin-resistant mutant | This study |
| E. coli | ||
| EC101 | supE hsd-5 thi (lac-proAB) F (traD6 proAB lacIq lacZ M15) repA, derivative of strain TG1 | 18 |
| DH5α | supE44 lacU169 (ϕ80 lacZ M15) hsdR17 endA1 gyrA96 thi-1 relA1 | 28 |
| Plasmids | ||
| pG+host9 | 3.8 kb, pWV01-type thermosensitive replication origin (Ts) from pVE6002, Eryr | 29 |
| pSET4s | 3.8 kb, pWV01-type Ts from pVE6002, lacZ′, Spcr | 30 |
| pSET5s | 3.8 kb, pWV01-type Ts from pVE6002, lacZ′, Cmr | 30 |
| pSL1180 | 3.4 kb, pMB1 origin, superpolylinker for vector construction, Ampr | 31 |
Abbreviations: Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Eryr, erythromycin resistance; Fusr, fusidic acid resistance; Rifr, rifampin resistance; Spcr, spectinomycin resistance; Strr, streptomycin resistance.
S. agalactiae, Streptococcus uberis, and Streptococcus dysgalactiae subsp. dysgalactiae strains were grown in brain heart infusion (BHI; Difco) broth at 37°C with shaking at 150 rpm. Streptococcus pyogenes and Enterococcus faecalis strains were grown in the same conditions but without shaking. Solid cultures of these species were made on tryptic soy plates supplemented with defibrinated horse blood (5%). S. thermophilus strains were grown in reconstituted skim milk (10% [wt/vol]), M17 broth supplemented with 0.5% lactose (LM17; Oxoid) at 42°C in anaerobic conditions (GENbox Anaer atmosphere generators and incubation jars from bioMérieux). Streptococcus salivarius strains were grown in M17 broth supplemented with 0.5% glucose (GM17; Oxoid) at 37°C in anaerobic conditions. Streptococcus mutans strains were grown in Todd-Hewitt broth supplemented with 0.1% of yeast extract (Oxoid) at 37°C in anaerobic conditions. Cultures were supplemented with the following antibiotic when required: chloramphenicol, 16 μg ml−1; erythromycin, 50 μg ml−1; rifampin, 75 μg ml−1; spectinomycin, 500 μg ml−1; or streptomycin, 250 μg ml−1.
Escherichia coli strains were grown in Luria-Bertani (LB) broth at 37°C with shaking at 200 rpm. Recombinant plasmids derived from the pG+host9 vector were transformed into E. coli EC101 strain, a strain which contains a chromosomal copy of the pWV01 repA gene (18), and selected at 37°C on LB medium containing 150 μg of erythromycin ml−1.
DNA manipulations, PCR, and pulsed-field gel electrophoresis.
Preparation of chromosomal and plasmid DNAs was performed according to standard protocols (28).
PCRs and high-fidelity PCRs were carried out according to the manufacturer's instructions using the ThermoPol PCR kit (New England BioLabs) and the Phusion high-fidelity DNA polymerase (Thermo Scientific), respectively. PCRs were performed with 4 μg of DNA template ml−1 and included 30 cycles of amplification. The annealing step was executed at 5°C below the melting temperature (Tm) of the primers for standard PCR (ThermoPol PCR kit) and at a 3°C above the Tm of the primers for high-fidelity PCR (Phusion high-fidelity DNA polymerase). The primers used in the present study were purchased from Eurogentec and are listed in Table S1 in the supplemental material.
Transconjugants were confirmed by PCR using primers located in the SAG2026 (corresponding to the SAL_2079 gene in strain 515, 100% identical to the SAG2026 gene) gene. Transconjugants of S. pyogenes were also confirmed by PCR using a primer located in the ICE integrase gene (intICESa03 Fwd) and a primer located in the tRNALys gene (tRNALys) (see Table S1 in the supplemental material).
The characterization of the CIME-ICE composite elements in the transconjugants of strain Nem316 was performed using three sets of primers: SAG2026-1 and intICESa03 Fwd (for integration in the attL site), intICESa03 Fwd and SAG1986 Rev (for integration in the attI site), and SAG1986 Fwd and SAG2026-1 (for integration in the attR site). Two pairs of primers (SAG2026-1/tRNAMet Rev and SAG2026-1/tRNA16S Rev) were also used to examine integration in the two other tRNALys genes.
Nested PCR were performed to determine the presence of an attI site and an attB site using two steps. The first step corresponds to a standard PCR except that 25 cycles of amplification were made instead of 30. In the second step, template corresponds to the PCR product obtained in the first step and primers used are internal to this fragment. Primers location and orientation are indicated in Fig. 2 and 5. Conditions used are those of the standard PCR described above. The attI and attB fragments obtained by PCR were sequenced to confirm their specificity.
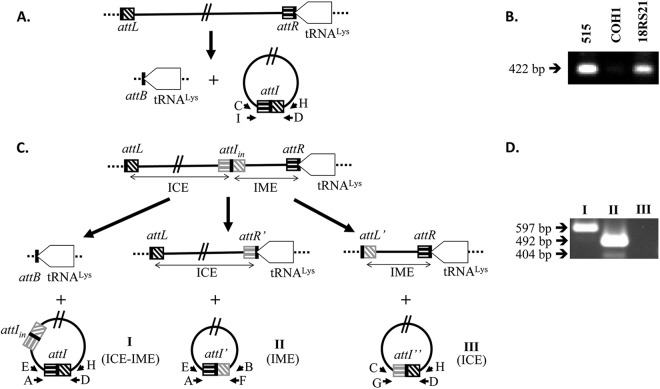
Fig 2.
Schematic representation of the site-specific excision of the ICEs integrated in the 3′ end of the S. agalactiae tRNALys gene. Primers used to detect DNA molecules resulting from site-specific recombination events are indicated by letters. (A) Model of ICE_515_tRNALys, ICE_COH1_tRNALys, and ICE_18RS21_tRNALys excision. (B) Electrophoresis of nested PCR product obtained for the 515, COH1, and 18RS21 strains of S. agalactiae. (C) Model of ICE_2603V/R_tRNALys excision where three different circular forms could be obtained. (D) Electrophoresis of nested PCR products obtained for the 2603V/R strain.
Fig 5.
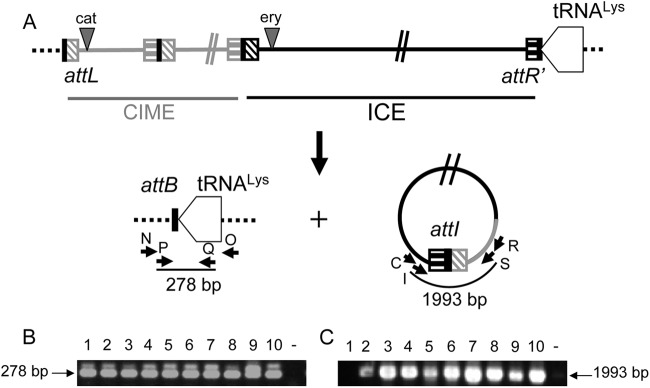
Analysis of the site-specific excision of the CIME cat-ICE ery composite element integrated in the 3′ end of the tRNALys gene of transconjugants of S. agalactiae Nem316. (A) Schematic representation of the circular form (with its attI site) and the empty attB integration site resulting from excision of the composite element. Primers used to detect DNA molecules resulting from site-specific recombination events are indicated by letters. (B) Amplification of an attB site in 10 transconjugants of S. agalactiae Nem316. (C) Amplification of an attI site in the same 10 clones. ICE_515_tRNALys appears in black, and CIME_Nem_tRNALys appears in gray.
Pulsed-field gel electrophoresis experiments were carried out as described previously using SmaI as the restriction enzyme (25).
ICE tagging.
ICE was tagged at its left hand side by a resistance gene using the pG+host9 spc vector. Vector pG+host9spc is a derivative of the pG+host9 plasmid carrying a spectinomycin resistance gene (X. Bellanger, unpublished data). The resistance cassette was obtained from the pSET4S plasmid by digestion with the SpeI restriction enzyme and ligated to XbaI-digested pG+host9 plasmid (Bellanger, unpublished).
The locus tagged is the SAG2026 (SAL_2079) gene encoding the ATPase subunit of an ABC transporter. This gene belongs to an operon homologous to the yydFGHIJ operon of from Bacillus subtilis (32), which likely encodes a system of synthesis, maturation, and export of a signaling and/or antibacterial peptide. This operon is complete only in strain COH1; two genes are missing in the three other strains.
To construct the mutant, the 5′ and 3′ ends of SAG2026 gene were independently amplified by PCR (using the primers SAG2026-1-HindIII, SAG2026-2-AvrII, SAG2026-3-AvrII, and SAG2026-4-EcoRI) and subcloned in the pSL1180 vector. An erythromycin resistance gene amplified from pG+host9 was inserted in the internal AvrII site, and the whole insert (containing the two SAG2026 fragments separated by the ery cassette) was then cloned into pG+host9spc to give pG+host9spc-SAG2026ery, which was used to transform S. agalactiae by electroporation (33). Two crossovers, upstream and downstream from the tagged region, were selected as described previously to obtain replacement of the gene by the ery resistance gene (26).
Attempts to obtain strains of S. agalactiae devoid of elements integrated in tRNALys.
In order to find a strain devoid of element integrated into the 3′ end the tRNALys CTT gene to be used as recipient strain during mating experiments, a large collection of strains of S. agalactiae (n = 70) was screened by PCR. The primers used are specific for SAG1986 (SAG1986 Fwd and SAG1986 Rev) and SAG1993 (intICESa03 Fwd and intICESa03 Rev) integrase genes of ICE_2603_tRNALys and specific for the SAG2026 gene (SAG2026-1 and SAG2026-4).
Two strategies were also tested for curing cells from their resident element. First, plasmids expressing the integrase and excisionase genes and carrying attR recombination site of ICE_515_tRNALys were constructed and electroporated in the Nem316 and 515 strains. The purpose was to increase the excision of the elements and to select cells which would have lost them after 100 bacterial generations, as previously obtained for ICESt3 (26). The second strategy relied on exposition of the cells to mitomycin C in order to increase the excision of the ICE, as described for the distantly related ICEs ICESt3 and RD2. This strategy was tested on strain 515. Since the loss of the element could be hampered due to a putative toxin-antitoxin system encoded by ICE_515_tRNALys (SAL_2044-SAL_2045 ORF), we also constructed a mutant deleted in the toxin gene to try to obtain cells cured of their ICEs.
Selection of spontaneous rifampin- and streptomycin-resistant mutants to be used as recipient cells in filter-mating experiments.
In a first step, rifampin-resistant mutants were selected by plating the parental strain on appropriate media containing rifampin (75 μg ml−1). In a second step, rifampin- and streptomycin-resistant mutants were selected by plating the rifampin-resistant mutants obtained in the first step on appropriate media containing streptomycin (250 μg ml−1). All of the selected mutants were confirmed by sequencing of the sodA gene (34) (see Table S1 in the supplemental material).
Filter-mating experiments.
The filter-mating protocol used was described previously (26). Briefly, both donor and recipient strains were grown overnight. A 15-ml culture in the relevant broth was inoculated with 150 μl of overnight culture of the recipient or the donor strain. The cultures were grown at the relevant temperature until mid-exponential phase (optical density at 600 nm of 0.4). Cultures of the donor and recipient were mixed and centrifuged for 15 min in a prewarmed centrifuge at 4,500 × g to form a cell pellet. The pellet was resuspended in 1 ml of BHI broth with or without 50 μg of DNase I ml−1 (35), and 150-μl aliquots were spread on 0.45-μm-pore-size nitrocellulose filters (Sartorius) on tryptic soy agar plates, which were then incubated for 15 h at 37°C. The filters were removed from the agar plates and placed in 50-ml tubes containing 10 ml of sterile BHI broth, and bacteria were recovered by vortexing for 30 s. Various dilutions were spread on agar plates supplemented with the appropriate antibiotics, and the plates were incubated for 24 h in order to count the CFUs of the donor, the recipient, and the transconjugants.
DNA sequencing and sequence analysis.
DNA sequencing was performed on recombinant plasmids and PCR products by Beckman Coulter genomics. BLASTN, BLASTX, BLASTP, and PSI-BLAST (36) were used to search similarities to sequences in the GenBank database. Sequences were aligned using Vector NTI advance 11 (Invitrogen).
Statistical analysis.
Statistical analysis was performed as described by Georgin and Mouet (37) and Cumming et al. (38). The means and standard errors from at least three independent experiments were determined.
RESULTS
Excision of the putative ICEs integrated in the 3′ end of a tRNA lysine gene in S. agalactiae.
We previously reported that eight sequenced genomes of S. agalactiae display a genomic island integrated into the 3′ end of the tRNALys CTT gene (14). Among these elements, four could be ICEs (in strains 18RS21, 515, COH1, and 2603V/R). The name that was given to the elements is composed of the nature of the element followed by the name of the strain and the integration locus. One of these putative ICEs, ICE_2603_tRNALys, is a composite element that carries an additional recombination module, an additional putative relaxase gene and an internal attI site that probably results from the accretion of a putative ICE (left side from attL to attI) and a putative IME (right side from attI to attR) (Fig. 1).
Fig 1.
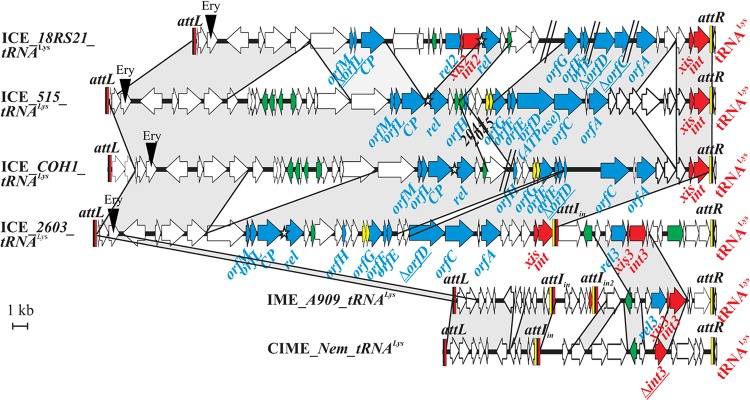
Open reading frame (ORF) organization and comparison of the elements integrated into the 3′ end of the tRNALys CTT gene in S. agalactiae 18RS21, 515, COH1, 2603V/R, A909, and Nem316. ORFs appear as arrows (truncated genes are indicated by a delta letter and underlined). The name of the gene where the element is integrated appears in red. Genes of the conjugation module are indicated with blue arrows, genes of the regulation module are indicated with green arrows, and genes of the recombination module are indicated with red arrows. When a putative domain has been found for the gene product or a putative function can be assigned to its product, the gene is named accordingly (rel for relaxase, CP for coupling protein, ATPase, xis for excisionase, and int for integrase). Other genes were named according to their similarity to ICESt1/St3 of S. thermophilus. The different genes encoding integrase, excisionase, and relaxase have been given different numbers. The putative toxin-antitoxin system (SAL2044-2045 and homologs in the other ICEs) appear as a yellow ORFs. The putative oriT is indicated by a star. Recombination sites are drawn as vertical rectangles. Black rectangles indicate identical sequences found in attL, attR, and attI sites. Yellow rectangles indicate the arm of attR sites and the related arm of attI sites, and red rectangles indicate the arm of attL sites and the related arms of attI sites. Protein identity higher than 80% is indicated in gray. Gaps in the genome due to missing contigs are indicated by a double slash.
We looked for excised circular forms of these four putative ICEs: one for ICE_515_tRNALys, ICE_COH1_tRNALys, and ICE_18RS21_tRNALys (Fig. 2A) and three different possible circular forms for ICE_2603V/R_tRNALys since this element carries an internal attI site (Fig. 2C). By nested PCR, we detected an excised circular form for ICE_515_tRNALys and ICE_18RS21_tRNALys but not for ICE_COH1_ tRNALys (Fig. 2B). An empty attB site was, however, detected for this latter strain (data not shown). Furthermore, two different excised forms were observed for ICE_2603_tRNALys (Fig. 2D), indicating that the IME integrated beside the ICE in this strain is able to excise alone or together with the ICE. All of the fragments obtained by nested PCR were confirmed by sequencing.
Test of intraspecies conjugative transfer of the putative ICEs integrated in the 3′ end of a tRNA lysine gene in S. agalactiae.
To test whether the four putative ICEs described above are able to transfer by conjugation, they were tagged at their left extremity by an erythromycin resistance cassette after allelic replacement of a gene encoding a membrane subunit of a putative ABC transporter (SAL_2079/SAG2026 gene) (Fig. 1).
The strains harboring these tagged elements were then used as donor cells in filter mating experiments with three different recipient strains that all harbor elements flanked by attL and attR sites: (i) strain COH1, which harbors a putative ICE as mentioned previously; (ii) strain A909, which harbors a putative IME (i.e., an element with a recombination module, a gene encoding the relaxase, but lacks genes required for mating pore assembly); and (iii) strain Nem316, which carries an element devoid of recombination and conjugation genes (CIME). We were not able to test a recipient strain devoid of element integrated in the tRNALys CTT gene since all our efforts to obtain such a strain (by screening of a large collection of strains or by artificial curing of a strain carrying an element) were unsuccessful (see Materials and Methods for more details).
ICE_515_tRNALys was found to transfer by conjugation to the Nem316 strain and to the COH1 strain at frequencies of 4.0 × 10−7 ± 0.8 × 10−7 and 0.7 × 10−7 ± 0.1 × 10−7 CFU of transconjugants per CFU of donor cells, respectively. As expected, PCR on the SAL_2079/SAG2026 gene gave two fragments for the transconjugants (one for the gene present on the resident element and another of higher size corresponding to the gene that is carried by the incoming ICE and is interrupted by the erythromycin cassette) (data not shown). No transconjugant was obtained when cell-cell contacts were prevented by placing a filter between donor and recipient cells. Furthermore, the addition of DNase I in the growth medium had no impact on the number of transconjugants, indicating that DNA transfer occurred by conjugation. No transconjugant was obtained when the A909 strain was used as recipient cells.
ICE was found to retransfer if transconjugants (carrying a CIME cat-ICE ery element) were used as donor cells and rifampin-streptomycin mutants of the Nem316 strain were used as recipient cells. The frequency of retransfer was higher (2.2 × 10−5 ± 0.9 × 10−5 CFU of transconjugants per CFU of donor cells) than the one obtained for transfer from strain 515 to strain Nem316. This indicates that ICE_515_tRNALys is able to self-transfer by conjugation and therefore is an ICE. In contrast, no conjugative transfer was observed for the three other putative ICEs tested (ICE_2603_ tRNALys, ICE_18RS21_ tRNALys, and ICE_COH1_tRNALys).
Test of interspecies conjugative transfer of ICE_515_tRNALys.
Since ICE_515_tRNALys was shown to transfer by conjugation to a S. agalactiae recipient strain, we also tested whether it can transfer to other bacterial species. Seven different species belonging to the Firmicutes were tested as recipient cells: S. salivarius (CIP102503 and JIM8777), S. thermophilus (LMG18311), S. pyogenes (ATCC 12202), S. uberis (20388 and 21458), S. dysgalactiae subsp. dysgalactiae (14998 and 16192), S. mutans (UA159), and E. faecalis (JH2-2). Spontaneous rifampin and streptomycin mutants were first selected for these strains in order to use them as recipient cells in the mating experiments. Transconjugants were obtained only when using S. pyogenes as recipient cells. However, even for this species, transconjugants were obtained only in three experiments out of nine experiments performed, and only 20 different clones of transconjugants were isolated. The acquired ICE is integrated into the 3′ end of the tRNALys CTT gene of the transconjugants, as shown by PCR using primers located in this tRNALys gene and in the integrase gene of ICE_515_tRNALys (Fig. 3A). In addition, they have the same genetic background than the recipient S. pyogenes strain, as shown by pulsed-field gel electrophoresis (Fig. 3B).
Fig 3.
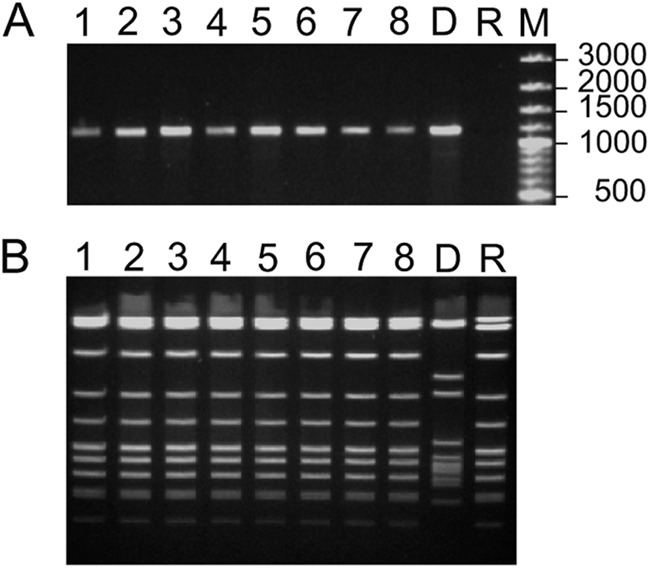
Analysis of eight clones of transconjugants obtained after filter-mating experiments between donor strain 515 (carrying ICE_515_tRNALys) and recipient strain S. pyogenes ATCC 12202. (A) Amplification of the right junction of the ICEs in these clones as for donor cells, using primers specific of the integrase genes and of the tRNALys gene (lane D). The negative control is S. pyogenes ATCC 12202, used as recipient cells (lane R). Lane M shows DNA molecular weight marker. (B) Analysis of the transconjugants by pulsed-field gel electrophoresis in parallel to the donor strain (lane D) and the recipient strain (lane R).
Site-specific accretion between ICE_515_tRNALys and related elements.
Since the Nem316 strain used as recipient cells for the mating experiments carries an internal recombination site in its element integrated in the tRNALys CTT gene, the incoming ICE could integrate into three different positions: the left (attL), the internal (attI), or the right (attR) recombination sites of the resident CIME (Fig. 4). Testing of 100 transconjugants by PCR indicated that integration occurs preferentially in the attR recombination site (96% versus 4% in the attL site and 0% in the attI site), giving rise to a CIME-ICE ery tandem (Fig. 4). Mating experiments were also performed with a Nem316 recipient strain carrying a CIME tagged with a chloramphenicol resistance gene, giving rise to CIME cat-ICE ery tandem. PCRs were performed to amplify the two other tRNALys genes to determine whether they can be used as secondary integration sites. None of the 100 transconjugants tested carried an ICE in the two other tRNALys genes, indicating that integration is specific to the tRNALys CTT gene.
Fig 4.
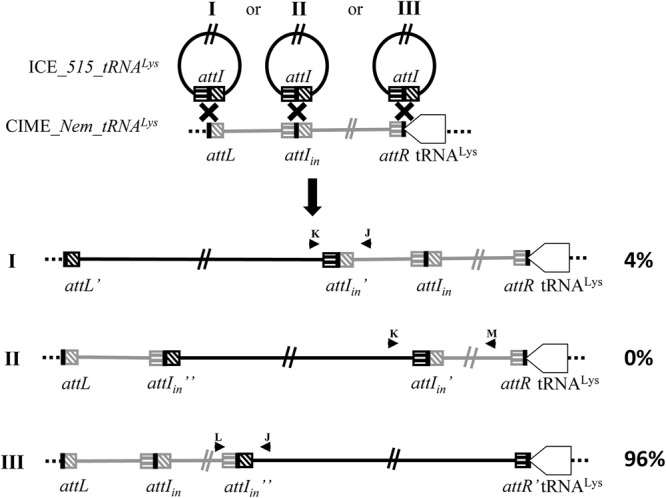
Schematic representation of the site-specific integration of ICE_515_tRNALys in the genome of strain Nem316 and schematic localization of primers used to detect DNA molecules resulting from the site-specific recombination events. ICE_515_tRNALys appears in black, and CIME_Nem_tRNALys appears in gray.
Strain COH1 also gave transconjugants, although it already carries a putative resident ICE that is integrated into the same site and harbors very closely related conjugation and recombination modules. The incoming ICE can thus theoretically integrate at two different positions: in the left (attL) or in the right (attR) recombination site of the resident ICE. Analysis of 12 transconjugants indicated that they all integrated in the attR recombination site giving rise to the formation of a tandem of ICEs.
Conjugative mobilization of CIME_316_ tRNALys in cis by ICE_515_tRNALys.
The empty attB site (Fig. 5B) and the attI site (Fig. 5C) resulting from the excision of the whole CIME cat-ICE ery tandem were detected by nested PCR in transconjugants, showing the intracellular mobilization in cis of the CIME by the ICE. The conjugative transfer of this composite element was then investigated by filter matings using, as donor cells, the transconjugant deriving from Nem316 strain carrying this tandem and, as recipient cells, rifampin-streptomycin spontaneous mutant of the Nem316 strain. Putative transconjugants deriving from Nem316 Rifr Smr were recovered on agar plates containing rifampin, streptomycin, and either erythromycin or chloramphenicol. Among 900 erythromycin-resistant transconjugants analyzed, only one carried both ICE and CIME; the others carried only the ICE alone. The only transconjugant recovered by plating on chloramphenicol-containing agar was shown to carry both ICE and CIME. These two transconjugants were analyzed by pulsed-field gel electrophoresis. Their electrophoresis pattern reflects the acquisition of a CIME-ICE tandem (one or two copies depending on the transconjugant) (Fig. 6). Therefore, the accretion has led not only to the intracellular mobilization in cis of the CIME by the ICE but also to its conjugative mobilization in cis.
Fig 6.
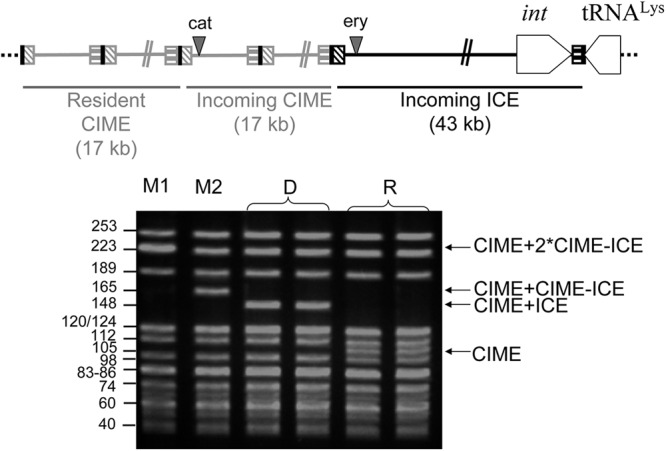
Pulsed-field gel electrophoresis analysis of the two transconjugants (M1 and M2) obtained after conjugative mobilization in cis of the CIME cat by ICE ery. A strain deriving from Nem316 and harboring a tandem CIME cat–ICE ery (indicated by “D” on the schema) and a mutant strain resistant to rifampin and streptomycin deriving from Nem316 already harboring a resident CIME (indicated as recipient “R”) were used as donor and recipient strains, respectively.
DISCUSSION
A circular form was detected for three out of the four ICEs integrated in a tRNALys gene of S. agalactiae tested. In strain 2603V/R, the whole composite ICE and the IME corresponding to its right end are able to excise. The lack of excision of the putative ICE corresponding to the left part of the composite ICE suggests that the integrase requires sequences present in the tRNALys gene which are absent in the internal attI site. Alternatively, it is possible that integrase binding to the attL site and to attR moiety of attIin is not optimal for recombination, explaining why the distal attR site is used for recombination instead of internal attR moiety of attIin. Since the attR and attL sites of the ICE and of the IME are very different, the excision of the whole composite element ICE-IME requires that the integrase (of the ICE or of the IME) recognizes the other type of sequence (attL of the ICE if excision is mediated by the integrase of the IME or attR of the IME if the excision is ensured by the integrase of the ICE). This suggests that this IME is functional and could thus be trans- or cis-mobilized by an ICE. In both cases, it would need a functional helper conjugation pore to be transferred to a recipient cell. Although putative IMEs are frequent in genomes in particular in S. agalactiae (14) and the presence of a high number of isolated relaxase genes in bacterial chromosomes suggests that these elements are very frequent in bacteria (6), very few IMEs have actually been characterized until now in bacteria (5, 39). No circular form was detected for the fourth ICE (ICE_COH1_tRNALys), but an empty integration site was detectable. This could indicate a loss of circular form in this strain in the extraction conditions tested due to less quantity or higher degradation of circular form than in the other strains. Alternatively, the absence of amplification can also be due to the longer expected size of the amplified fragment in this strain that carries two additional genes at the left-hand side of its ICE.
Screening of a large collection of strains of S. agalactiae did not enable us to find a strain that does not carry a genetic element integrated in the tRNALys CTT gene in order to be used as recipient in the mating experiments. Even a strain isolated from fish carries an element at this locus (40), which thus appears as a hot spot of integration in this species. All curing attempts failed, although we tested different strategies that proved to be efficient for distantly related ICEs (26, 41). This could be due to the presence of a toxin-antitoxin system that kills the cells that have lost the ICE. However, interruption of the toxin gene of the putative toxin-antitoxin system identified on ICE_515_tRNALys did not allow curing the strain even after a mitomycin C treatment. Our hypothesis is that the ICE replicates in the cell as reported for two ICEs belonging to the ICESt3 family (26, 41), leading to a high frequency of reintegration in the chromosome even if excision is artificially increased. This hypothesis would also explain why the numerous attempts to interrupt the excisionase, integrase, relaxase, and other genes of the ICE failed (data not shown). Since we did not find a S. agalactiae strain with an empty tRNALys gene site, we used in the mating experiments a recipient strain that already carries a genetic element (CIME, IME, or ICE) integrated at this locus.
In this study, we demonstrated the functionality of an ICE carried by a human isolate of S. agalactiae. This is only the second description of conjugative transfer of an ICE in this species (the first one is TnGBS2 which uses a DDE transposase for its integration instead of a tyrosine recombinase [42] and carries an unrelated conjugation module). This ICE is related to two other ICEs whose conjugative transfer has been demonstrated in other streptococcal species: (i) RD2 of S. pyogenes (67 to 96% of identity for the proteins involved in conjugation transfer but different recombination module enabling integration into the 3′end of tRNAThr gene) (41) and (ii) ICESt3 of S. thermophilus (35 to 67% of protein identity for the conjugation proteins, unrelated recombination module providing specific integration into the 3′end of fda gene) (26). ICE_515_tRNALys was successfully transferred by conjugation to two other S. agalactiae strains: Nem316 strain (carrying a CIME) and COH1 strain (carrying a defective ICE). This led in both cases to an accretion of two elements (CIME-ICE or ICE-ICE) with a preferential integration of the incoming ICE at the attR recombination site (that includes the end of the tRNALys gene). This attR preference was also observed for ICESt3 of S. thermophilus, although it encodes an unrelated integrase that catalyzes the integration in the fda gene instead of the tRNALys gene (13). This suggests that the integrase or a cofactor needs to bind sequences present in the tRNALys gene but absent in attL and attI sites for efficient integration of the ICE.
The frequency of conjugative transfer was low compared to the frequency of intraspecies transfer reported for ICEs of other Firmicutes (10-fold less than ICESt3 of S. thermophilus [26] and RD2 of S. pyogenes [41]). One hypothesis is that the occupation of the recipient attB site by an element flanked by attR and attL recombination sites reduces the frequency of ICE_515_tRNALys transfer, as was previously found for ICESt3 (13). The efficiency of ICE integration in recombination sites flanking the resident element is likely reduced compared to an empty integration site. In addition, the frequency of transfer is even lower when the resident element of the recipient strain is an ICE (i.e., in strain COH1). This could be due to an exclusion mechanism as described for other ICEs (7). Furthermore, the attR recombination site of IME_A909_tRNALys and CIME_Nem316_tRNALys resident elements greatly differs from the attR site of ICE_COH1_tRNALys.
The genetic background of the recipient cells also likely plays a role since the frequency of transfer was 100-fold higher when transferring the ICE between strains with the same genetic background (experiments of retransfer using Nem316 strain as donor and recipient strain). This could be due to restriction-modification systems that degrade foreign DNA, including incoming ICE. Furthermore, no conjugative transfer of ICE_515_tRNALys was obtained when the A909 strain (carrying an IME) was used. A CRISPR system has been described recently in this strain (43). Analysis of the spacers present in this CRISPR system indicate that three of them match a sequence found in ICE_515_tRNALys (the coupling protein gene, the intergenic region between coupling protein and relaxase genes, and the gene of ICE_515_tRNALys encoding a putative DNA adenine methylase). Strain A909 thus likely carry a functional CRISPR system which targets foreign DNA including ICEs belonging to the same family than ICE_515_tRNALys and protect cell from invasion by these MGEs.
No conjugative transfer was observed for the three other putative ICEs tested (ICE_COH1_tRNALys, ICE_2603_tRNALys, and ICE_18RS21_tRNALys). These three elements all carry an orfD pseudogene. This conjugation gene encodes a putative ATPase belonging to the FtsK superfamily with characteristic Walker A and Walker B domains. It is thus a pTi VirB4 homolog like ConE (YddE) of ICEBs1, TcpF of pCW3, or Orf16 of Tn916. This protein could play a role in energizing the conjugation machinery and as such is likely crucial for conjugative transfer of the element. This would explain why we did not observe conjugative transfer of ICE_COH1_tRNALys, ICE_2603_tRNALys, and ICE_18RS21_tRNALys.
Interspecies transfer was obtained only with S. pyogenes as recipient cells. In this species, the tRNALys CTT gene displays one difference in the 11 terminal base pairs compared to S. agalactiae. Other species tested in the present study (Streptococcus salivarius, Streptococcus uberis, and S. thermophilus) carry a tRNALys CTT gene with a 3′ end identical to the gene of S. agalactiae. Other factors thus impact the compatibility of donor-recipient cells. At least two of the strains tested as recipient already carry a putative IME integrated in the tRNALys CTT gene. This could limit the integration of the incoming ICE. Little is known about the cell-cell contacts that need to be established between donor and recipient cells during conjugative transfer. These contacts could be more difficult between bacteria of different species. Furthermore, restriction-modification systems probably interfere with the maintenance of ICE foreign DNA in the recipient cell. Even if we did not find in vitro conditions propitious to interspecies conjugative transfer of ICE_515_tRNALys, there are evidences of a spread of this type of ICE in other streptococcal species. For example, S. urinalis 2285-97 carries an almost identical ICE, and we recently identified an ICE integrated in a tRNALys gene and carrying a related conjugation module in a strain of S. uberis (unpublished results). We are currently characterizing the functions encoded by the accessory genes carried by ICE_515_tRNALys since they could confer an adaptive advantage to the recipient strains. We showed in particular that this ICE encodes a functional CAMP factor toxin (a gene also found on S. urinalis putative ICE) (44). Transfer of ICEs carrying such accessory gene to other bacterial species would thus disseminate this toxin.
Conjugative transfer of ICE_515_tRNALys to the Nem316 that carries a CIME led to a CIME-ICE tandem in the recipient strains, thus offering us the opportunity to examine if ICE_515_tRNALys is able to cis-mobilize genes of CIME_Nem_tRNALys. The attB and attI sites resulting from the coexcision of both elements were detected by PCR, showing the intracellular mobilization in cis of the CIME by the ICE. Although quite infrequently (0.1% of the events of retransfer), the ICE-CIME tandem was also shown to transfer by conjugation. Such phenomenon has been described recently for ICESt3 of S. thermophilus (13) and could be a shared characteristic of ICEs using site-specific recombinases to excise and integrate. ICE_COH1_tRNALys, ICE_2603_tRNALys, and ICE_18RS21_tRNALys, although not self-transferable, could also be transferred by conjugation after accretion of an ICE and cis-mobilization. Since ICE_2603_tRNALys is able to excise, it could also be transferred by conjugation by using the mating apparatus encoded by a related ICE (mobilization in trans).
ICEs likely play an even higher role in horizontal gene transfer since it was reported recently that ICEBs1 of B. subtilis is able to mobilize in trans plasmids lacking dedicated mobilization proteins (45). Furthermore, Tn916 was found to mobilize in trans MTnSag1, an element from S agalactiae which encodes a DDE transposase and carries its own oriT unrelated to the one of Tn916 but lacks conjugation or mobilization genes (46). Mobile genomic islands (MGI) carrying their own tyrosine integrase and an their own oriT but no conjugation or mobilization gene were also reported to be mobilized in trans by an ICE harboring a related oriT in Vibrio (47). Mobilization in cis can also lead to conjugative transfer of chromosomal sequences (up to 1 Mb) located at the 5′ side of the MGI (47). Conjugative transfer of chromosomal DNA (up to 334 kb) by an Hfr-type mobilization initiated from the oriT of a genomic island was also reported in S. agalactiae (48). cis- and trans-mobilization of DNA by ICEs are thus probably very common and likely contribute to the evolution of genomic islands and bacterial genomes.
Supplementary Material
ACKNOWLEDGMENTS
A.P. was supported by a grant from the Institut National de la Recherche Agronomique and a grant from Région Lorraine.
We thank C. Delorme, A. Roberts, J. Y. Madec, and Marisa Haenni for providing the strains used in this study. We also thank Mylène Maury for technical help and X. Bellanger for constructing the pG+host9spc vector.
We are grateful to P. Glaser for helpful discussions.
Footnotes
Published ahead of print 28 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.02199-12.
REFERENCES
- 1. Frost LS, Leplae R, Summers AO, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat. Rev. Microbiol. 3:722–732 [DOI] [PubMed] [Google Scholar]
- 2. Koonin EV, Wolf YI. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res. 36:6688–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Toussaint A, Chandler M. 2012. Prokaryote genome fluidity: toward a system approach of the mobilome, p 57–80 In van Helden J, Toussaint A, Thieffry D. (ed), Bacterial molecular networks: methods and protocols, vol 804 Humana Press, New York, NY: [DOI] [PubMed] [Google Scholar]
- 4. Burrus V, Pavlovic G, Decaris B, Guedon G. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601–610 [DOI] [PubMed] [Google Scholar]
- 5. Burrus V, Pavlovic G, Decaris B, Guédon G. 2002. The ICESt1 element of Streptococcus thermophilus belongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48:77–97 [DOI] [PubMed] [Google Scholar]
- 6. Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP. 2011. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 7:e1002222 http://dx.doi.org/10.1371/journal.pgen.1002222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8:552–563 [DOI] [PubMed] [Google Scholar]
- 8. Burrus V, Waldor MK. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376–386 [DOI] [PubMed] [Google Scholar]
- 9. Seth-Smith H, Croucher NJ. 2009. Genome watch: breaking the ICE. Nat. Rev. Microbiol. 7:328–329 [DOI] [PubMed] [Google Scholar]
- 10. Alvarez-Martinez CE, Christie PJ. 2009. Biological diversity of prokaryotic type IV secretion systems. Microbiol. Mol. Biol. Rev. 73:775–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Berkmen MB, Lee CA, Loveday EK, Grossman AD. 2010. Polar positioning of a conjugation protein from the integrative and conjugative element ICEBs1 of Bacillus subtilis. J. Bacteriol. 192:38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pavlovic G, Burrus V, Gintz B, Decaris B, Guédon G. 2004. Evolution of genomic islands by deletion and tandem accretion by site-specific recombination: ICESt1-related elements from Streptococcus thermophilus. Microbiology 150:759–774 [DOI] [PubMed] [Google Scholar]
- 13. Bellanger X, Morel C, Gonot F, Puymege A, Decaris B, Guedon G. 2011. Site-specific accretion of an integrative conjugative element together with a related genomic island leads to cis mobilization and gene capture. Mol. Microbiol. 81:912–925 [DOI] [PubMed] [Google Scholar]
- 14. Brochet M, Couve E, Glaser P, Guedon G, Payot S. 2008. Integrative conjugative elements and related elements are major contributors to the genome diversity of Streptococcus agalactiae. J. Bacteriol. 190:6913–6917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doran KS, Nizet V. 2004. Molecular pathogenesis of neonatal group B streptococcal infection: no longer in its infancy. Mol. Microbiol. 54:23–31 [DOI] [PubMed] [Google Scholar]
- 16. Keefe GP. 1997. Streptococcus agalactiae mastitis: a review. Can. Vet. J. 38:429–437 [PMC free article] [PubMed] [Google Scholar]
- 17. Mian GF, Godoy DT, Leal CA, Yuhara TY, Costa GM, Figueiredo HC. 2009. Aspects of the natural history and virulence of Streptococcus agalactiae infection in Nile tilapia. Vet. Microbiol. 136:180–183 [DOI] [PubMed] [Google Scholar]
- 18. Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, Deboy RT, Davidsen TM, Mora M, Scarselli M, Margarit y Ros I, Peterson JD, Hauser CR, Sundaram JP, Nelson WC, Madupu R, Brinkac LM, Dodson RJ, Rosovitz MJ, Sullivan SA, Daugherty SC, Haft DH, Selengut J, Gwinn ML, Zhou L, Zafar N, Khouri H, Radune D, Dimitrov G, Watkins K, O'Connor KJ, Smith S, Utterback TR, White O, Rubens CE, Grandi G, Madoff LC, Kasper DL, Telford JL, Wessels MR, Rappuoli R, Fraser CM. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc. Natl. Acad. Sci. U. S. A. 102:13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tettelin H, Masignani V, Cieslewicz MJ, Eisen JA, Peterson S, Wessels MR, Paulsen IT, Nelson KE, Margarit I, Read TD, Madoff LC, Wolf AM, Beanan MJ, Brinkac LM, Daugherty SC, DeBoy RT, Durkin AS, Kolonay JF, Madupu R, Lewis MR, Radune D, Fedorova NB, Scanlan D, Khouri H, Mulligan S, Carty HA, Cline RT, Van Aken SE, Gill J, Scarselli M, Mora M, Iacobini ET, Brettoni C, Galli G, Mariani M, Vegni F, Maione D, Rinaudo D, Rappuoli R, Telford JL, Kasper DL, Grandi G, Fraser CM. 2002. Complete genome sequence and comparative genomic analysis of an emerging human pathogen, serotype V Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 99:12391–12396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Glaser P, Rusniok C, Buchrieser C, Chevalier F, Frangeul L, Msadek T, Zouine M, Couve E, Lalioui L, Poyart C, Trieu-Cuot P, Kunst F. 2002. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 45:1499–1513 [DOI] [PubMed] [Google Scholar]
- 22. Ajdic D, McShan WM, McLaughlin RE, Savic G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. 2002. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. U. S. A. 99:14434–14439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guedon E, Delorme C, Pons N, Cruaud C, Loux V, Couloux A, Gautier C, Sanchez N, Layec S, Galleron N, Almeida M, van de Guchte M, Kennedy SP, Ehrlich SD, Gibrat JF, Wincker P, Renault P. 2011. Complete genome sequence of the commensal Streptococcus salivarius strain JIM8777. J. Bacteriol. 193:5024–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bolotin A, Quinquis B, Renault P, Sorokin A, Ehrlich SD, Kulakauskas S, Lapidus A, Goltsman E, Mazur M, Pusch GD, Fonstein M, Overbeek R, Kyprides N, Purnelle B, Prozzi D, Ngui K, Masuy D, Hancy F, Burteau S, Boutry M, Delcour J, Goffeau A, Hols P. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haenni M, Saras E, Bertin S, Leblond P, Madec J-Y, Payot S. 2010. Diversity and mobility of integrative and conjugative elements in S. agalactiae, S. dysgalactiae subsp. dysgalactiae, and S. uberis bovine isolates. Appl. Environ. Microbiol. 76:7957–7965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bellanger X, Roberts AP, Morel C, Choulet F, Pavlovic G, Mullany P, Decaris B, Guedon G. 2009. Conjugative transfer of the integrative conjugative elements ICESt1 and ICESt3 from Streptococcus thermophilus. J. Bacteriol. 191:2764–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jacob AE, Hobbs SJ. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 29. Maguin E, Duwat P, Hege T, Ehrlich D, Gruss A. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takamatsu D, Osaki M, Sekizaki T. 2001. Construction and characterization of Streptococcus suis-Escherichia coli shuttle cloning vectors. Plasmid 45:101–113 [DOI] [PubMed] [Google Scholar]
- 31. Brosius J. 1989. Superpolylinkers in cloning and expression vectors. DNA 8:759–777 [DOI] [PubMed] [Google Scholar]
- 32. Butcher BG, Lin YP, Helmann JD. 2007. The yydFGHIJ operon of Bacillus subtilis encodes a peptide that induces the LiaRS two-component system. J. Bacteriol. 189:8616–8625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Framson PE, Nittayajarn A, Merry J, Youngman P, Rubens CE. 1997. New genetic techniques for group B streptococci: high-efficiency transformation, maintenance of temperature-sensitive pWV01 plasmids, and mutagenesis with Tn917. Appl. Environ. Microbiol. 63:3539–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poyart C, Quesne G, Coulon S, Berche P, Trieu-Cuot P. 1998. Identification of streptococci to species level by sequencing the gene encoding the manganese-dependent superoxide dismutase. J. Clin. Microbiol. 36:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Christie PJ, Korman RZ, Zahler SA, Adsit JC, Dunny GM. 1987. Two conjugation systems associated with Streptococcus faecalis plasmid pCF10: identification of a conjugative transposon that transfers between S. faecalis and Bacillus subtilis. J. Bacteriol. 169:2529–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Georgin P, Mouet M. 2000. Statistiques avec Excel. Presses Universitaires de Rennes, Paris, France [Google Scholar]
- 38. Cumming G, Fidler F, Vaux DL. 2007. Error bars in experimental biology. J. Cell Biol. 177:7–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Doublet B, Boyd D, Mulvey MR, Cloeckaert A. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55:1911–1924 [DOI] [PubMed] [Google Scholar]
- 40. Wang B, Jian J, Lu Y, Cai S, Huang Y, Tang J, Wu Z. 2012. Complete genome sequence of Streptococcus agalactiae ZQ0910, a pathogen causing meningoencephalitis in the GIFT strain of Nile tilapia (Oreochromis niloticus). J. Bacteriol. 194:5132–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sitkiewicz I, Green NM, Guo N, Mereghetti L, Musser JM. 2011. Lateral gene transfer of streptococcal ICE element RD2 (region of difference 2) encoding secreted proteins. BMC Microbiol. 11:65 http://dx.doi.org/10.1186/1471-2180-11-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brochet M, Da Cunha V, Couve E, Rusniok C, Trieu-Cuot P, Glaser P. 2009. Atypical association of DDE transposition with conjugation specifies a new family of mobile elements. Mol. Microbiol. 71:948–959 [DOI] [PubMed] [Google Scholar]
- 43. Lopez-Sanchez MJ, Sauvage E, Da Cunha V, Clermont D, Ratsima Hariniaina E, Gonzalez-Zorn B, Poyart C, Rosinski-Chupin I, Glaser P. 2012. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol. Microbiol. 85:1057–1071 [DOI] [PubMed] [Google Scholar]
- 44. Chuzeville S, Puymege A, Madec JY, Haenni M, Payot S. 2012. Characterization of a new CAMP factor carried by an integrative and conjugative element in Streptococcus agalactiae and spreading in streptococci. PLoS One 7:e48918 http://dx.doi.org/10.1371/journal.pone.0048918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee CA, Thomas J, Grossman AD. 2012. The Bacillus subtilis conjugative transposon ICEBs1 mobilizes plasmids lacking dedicated mobilization functions. J. Bacteriol. 194:3165–3172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Achard A, Leclercq R. 2007. Characterization of a small mobilizable transposon, MTnSag1,in Streptococcus agalactiae. J. Bacteriol. 189:4328–4331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Daccord A, Ceccarelli D, Burrus V. 2010. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol. Microbiol. 78:576–588 [DOI] [PubMed] [Google Scholar]
- 48. Brochet M, Rusniok C, Couve E, Dramsi S, Poyart C, Trieu-Cuot P, Kunst F, Glaser P. 2008. Shaping a bacterial genome by large chromosomal replacements, the evolutionary history of Streptococcus agalactiae. Proc. Natl. Acad. Sci. U. S. A. 105:15961–15966 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.