Abstract
Necrotic enteritis (NE) is an economically important disease of poultry caused by certain Clostridium perfringens type A strains. NE pathogenesis involves the NetB toxin, which is encoded on a large conjugative plasmid within a 42-kb pathogenicity locus. Recent multilocus sequence type (MLST) studies have identified two predominant NE-associated clonal groups, suggesting that host genes are also involved in NE pathogenesis. We used microarray comparative genomic hybridization (CGH) to assess the gene content of 54 poultry isolates from birds that were healthy or that suffered from NE. A total of 400 genes were variably present among the poultry isolates and nine nonpoultry strains, many of which had putative functions related to nutrient uptake and metabolism and cell wall and capsule biosynthesis. The variable genes were organized into 142 genomic regions, 49 of which contained genes significantly associated with netB-positive isolates. These regions included three previously identified NE-associated loci as well as several apparent fitness-related loci, such as a carbohydrate ABC transporter, a ferric-iron siderophore uptake system, and an adhesion locus. Additional loci were related to plasmid maintenance. Cluster analysis of the CGH data grouped all of the netB-positive poultry isolates into two major groups, separated according to two prevalent clonal groups based on MLST analysis. This study identifies chromosomal loci associated with netB-positive poultry strains, suggesting that the chromosomal background can confer a selective advantage to NE-causing strains, possibly through mechanisms involving iron acquisition, carbohydrate metabolism, and plasmid maintenance.
INTRODUCTION
Clostridium perfringens is an important Gram-positive anaerobic pathogen of humans and animals and is found ubiquitously in soil and in the gastrointestinal tract of vertebrates. It causes a number of histotoxic and enterotoxemic diseases, including necrotic enteritis (NE), an economically important disease of poultry. NE is characterized by necrotic lesions in the small intestine and can occur in an acute form, which is often responsible for high flock mortality, and a subclinical form, which results in production losses (1). A novel toxin, NetB, is present in the majority of disease-associated isolates and plays a critical role in NE pathogenesis (2, 3).
As a species, C. perfringens produces an array of extracellular toxins, four of which (alpha, beta, epsilon, and iota) form the basis for a toxin-typing scheme (4). Several of these toxins, including beta2-toxin, C. perfringens enterotoxin (CPE) (in non-food-borne strains), and all of the typing toxins except for alpha-toxin, are encoded on a conserved family of large plasmids related to the pCW3 tetracycline resistance plasmid (5–7). These plasmids share a conserved core region that includes the transfer of the clostridial plasmid (tcp) locus required for conjugation (6, 8, 9). The gene encoding NetB was recently localized to a 42-kb pathogenicity locus, NELoc-1, which resides on an ∼85-kb plasmid in this family (10). Two additional NE-associated loci were also identified, one of which (NELoc-3) is located on a similar but distinct plasmid from NELoc-1, whereas the other locus (NELoc-2) is chromosomally located (10). A NetB-encoding plasmid (pNetB) was recently sequenced and found to contain a locus with 99% sequence identity to NELoc-1; furthermore, this plasmid was confirmed to undergo conjugative transfer (5). It is still not clear how these distinct but related large plasmids are maintained and coexist within a single bacterium.
The transmissible nature of key C. perfringens toxin and related virulence genes raises the prospect that strains might be converted to different pathotypes through plasmid acquisition or loss and suggests a need for further examination of the contribution of the bacterial host to pathogenesis. Previous pulsed-field gel electrophoresis (PFGE) analyses of NE-associated C. perfringens isolates failed to provide evidence for a relationship between subtypes from different flocks (11, 12), indicating that there were numerous clonal origins of virulent strains that resulted from horizontal transfer of virulence genes. However, two recent multilocus sequence typing (MLST) studies identified two prevalent clonal groups among NE-associated isolates, thus suggesting that specific core chromosomal genes might be important for NE pathogenesis (13, 14).
In this study, we used microarray comparative genomic hybridization (CGH) to examine the genome content of a set of 54 genetically diverse C. perfringens isolates derived from either healthy broiler chickens (intestinal tract or retail chicken meat) or from the intestine of broiler chickens with NE. We have determined the variable gene regions that represent, in part, the accessory genome among these isolates, and we have identified those regions that are significantly associated with the presence of the netB plasmid. The genotypic relatedness of these isolates was also determined based on CGH data, providing insight into the evolutionary basis of the prevalence of certain NE-associated clones (13, 14).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
All C. perfringens strains and genome sequences used in this study are described in Table S1 in the supplemental material. The virulence of several strains has been assessed in a chicken NE model, and some strains have been shown to be virulent (CP1, CP2, CP3, CP4, and JGS4143), whereas others are avirulent (CP5 and CP6) (15). As determined by PCR using netB- and cpb2-specific primers (see Table S2 in the supplemental material), of the 29 healthy bird origin isolates, six were netB positive, and of the 25 NE-diseased bird origin isolates, 15 were netB positive.
Pulsed-field gel electrophoresis.
PFGE was performed on the 54 C. perfringens poultry isolates essentially as described previously (10). Briefly, C. perfringens strains were grown overnight in TGY broth (3% tryptone, 2% glucose, 1% yeast extract), and the bacterial pellets were suspended in a final concentration of 1% PFGE-certified agarose (Bio-Rad Laboratories, Hercules, CA). Agarose plugs were incubated overnight with gentle shaking at 37°C in lysis buffer (0.5 M EDTA [pH 8.0], 2.5% of 20% Sarkosyl) (Fisher Scientific, Fair Lawn, NJ), 0.25% lysozyme (Sigma-Aldrich Co., St. Louis, MO) and subsequently incubated in 2% proteinase K (Roche Applied Science, Indianapolis, IN) buffer for 48 h at 55°C. For each isolate, a portion of a plug was equilibrated in 200 μl restriction buffer at room temperature for 20 min and then digested with 40 U of SmaI (New England BioLabs, Ipswich, MA) at 37°C overnight. Electrophoresis was performed in a 1% PFGE-certified agarose gel and separated with the CHEF-II PFGE system (Bio-Rad Laboratories) in 0.5× Tris-borate-EDTA buffer at 14°C at 6 V for 19 h with a ramped pulse time of 4 to 38 s. Gels were stained with ethidium bromide and visualized by UV light. Low-range PFG markers (New England BioLabs) were used as a molecular DNA ladder. PFGE gels were analyzed using BioNumerics version 6.6 software (Applied Maths, Austin, TX). Band matching was performed using a 1% position tolerance and a 0% optimization factor, and cluster analysis was performed using the Dice similarity coefficient and unweighted-pair group method with arithmetic means (UPGMA).
Microarray construction and quality assessment.
A total of 3,312 67- to 73-mer oligonucleotide probes were designed using OligoWiz version 2.1.3 software (Center for Biological Sequence Analysis) (16), representing 3,091 (90.4%) and 2,806 (80.3%) coding sequences (CDSs) from the draft genomes of NE-producing strains CP4 and JGS4143, respectively (15, 17). These draft genomes consist of both plasmid and chromosomal sequences, and in most cases, the origins of the sequences were determined based on their similarities to existing completely sequenced C. perfringens chromosomes and plasmids. The use of these two strains as templates for the design of the C. perfringens array is particularly relevant to NE, since they represent two phylogenetically distinct clonal groups most frequently isolated from birds suffering from NE (9, 18). The probes were also specific (>80% identity over >80% coverage) for 2,437 (91.6%), 2,589 (90%), and 2,178 (85.6%) CDSs of the C. perfringens strain 13 (Str13), ATCC 13124, and SM101 sequenced genomes, respectively, indicating that the array is suitable for surveying the vast majority of genes in diverse C. perfringens strains. An additional 23 probes were designed and represented various plasmid CDSs and the C. perfringens major and minor toxin genes. The optimal melting temperatures, potential for cross-hybridization, probe positions, secondary structures, and low sequence complexity were taken into account in the design of the probes. Oligonucleotides were synthesized by Ocimum Biosolutions (Gaithersburg, MD) and Operon Technologies (Huntsville, AL) and resuspended in 3× SSC (0.45 M sodium citrate, 45 mM sodium chloride) to a concentration of 50 pmol/μl in 384-well plates. Oligonucleotides were spotted in quadruplicate onto Corning GAPS II slides (Fisher Scientific, Ottawa, ON, Canada) with a Bio-Rad ChipWriter Pro microarray spotter. To assess the quality (i.e., specificity and sensitivity) of the probes, microarrays were hybridized separately with Cy-labeled genomic DNA (gDNA) from C. perfringens strains ATCC 13124 and Str13, for which complete genome sequences are available, and the present/absent calls from the hybridization results were evaluated against the predicted calls determined from BLAST alignment of the probe and genome sequences. Sensitivity was calculated as [true positives/(true positives + false negatives)] and specificity as [true negatives/(true negatives + false positives)].
DNA extraction and Cy labeling.
Genomic DNA was extracted from 1 ml of overnight culture in brain heart infusion (BHI) broth using the Gentra Puregene yeast/bacteria kit (Qiagen, Mississauga, ON, Canada). Genomic DNA from each test isolate was labeled with Cy3 using an indirect labeling method. Briefly, 3 μg of genomic DNA was labeled with aminoallyl dUTP (aa-dUTP) (Sigma, St. Louis, MO) in a 50-μl reaction mixture containing 15 μg random hexamers (Invitrogen, Grand Island, NY), deoxynucleotide-aminoallyl dUTP mix (0.2 mM each dATP, dCTP, and dGTP and 0.12 mM dTTP and aa-dUTP) and 50 U of Klenow (exo−) (New England BioLabs, Ipswich, MA) at 37°C for 2 h. Unincorporated deoxynucleoside triphosphates (dNTPs) were removed by passage through an Amicon Ultracell-30 filter unit, and the eluted aminoallyl-labeled genomic DNA (gDNA) was evaporated in a vacuum concentrator and resuspended in 4.5 μl sodium bicarbonate (pH 9.3). Cy3 monofunctional reactive dye (GE Healthcare, Piscataway, NJ) suspended in 4.5 μl dimethyl sulfoxide (DMSO) was added and allowed to incorporate for 1 h at room temperature in the dark. Unincorporated dye was removed by purification with the Qiagen PCR purification kit according to the manufacturer's instructions, and the incorporation of Cy was assessed by measuring the absorbances at 260, 550, and 650 nm with a NanoDrop 1000 spectrophotometer (Wilmington, DE).
Microarray hybridizations.
For each array, genomic DNA pooled from the two reference strains, CP4 and JGS4143 (3 μg each), was labeled with Cy5 as described above and cohybridized with a Cy3-labeled test sample. Before applying to the array, the Cy3- and Cy5-labeled samples were evaporated to several microliters, combined in 29 μl hybridization buffer (Ocimum), heat denatured for 3 min at 95°C, and snap-cooled on ice for 3 min. The arrays were hybridized overnight at 42°C in a humidified chamber, and the slides were washed once in 2× SSC, 0.1% SDS that was prewarmed to 42°C for 5 min, twice in 0.1× SSC, 0.1% SDS for 5 min, five times in 0.1× SSC for 2 min, and once in 0.01× SSC for 10 s to remove the unbound sample. The slides were dried by centrifugation and scanned with a ScanArray Express scanner (PerkinElmer, Waltham, MA).
Microarray data analysis.
Fluorescent intensities were extracted from the scanned images using ScanArray version 3.0 software (PerkinElmer). Normalization was performed with BRB ArrayTools version 4.1.0 software (see http://linus.nci.nih.gov/BRB-ArrayTools.html) using the median over arrays function, and spots with a Cy3 raw intensity of <2,000 were removed and designated undetectable. Genes were determined to be present or absent by GACK version 3.631 software (19) using the bi-output function with a 0% estimated probability of presence (EPP) cutoff, and heat maps were generated with GenePattern version 3.3.3 software (Broad Institute) (18). Clustering of binary CGH data was performed with BioNumerics version 6.6 software (Applied Maths) using the binary simple-matching and UPGMA methods. Fisher's exact test for independence (two-tailed) was used to test the null hypothesis that there was no difference in the numbers of netB-positive and netB-negative (or poultry and nonpoultry) strains testing positive for a given gene. Contingency tables were analyzed using the false discovery rate calculator for 2×2 contingency tables web-based tool (Microsoft), which performs P value adjustment to correct for multiple testing (20). The genomic context of the variable regions detected by CGH, including their boundaries and the presence of additional genes not represented on the array, was inferred based on nucleotide sequence comparison of the CP4 and JGS4143 draft genomes with the genomes of ATCC 13124, SM101, and Str13 using the Artemis Comparison Tool (ACT) version 8 software (Sanger Institute) (21). A probe sequence was considered to be of plasmid origin if it was 100% identical to a sequence from C. perfringens plasmid pCW3 (NCBI reference sequence NC_010937), pCPF5603 (NC_007773), pCPF4969 (NC_007772), pCP8533etx (NC_011412), pNetB (JN689219), pTet (JN689220), pBeta2 (JN689217), pSM101A (NC_008263), pSM101B (NC_008264), pCP13 (NC_003042), pIP404 (NC_001388), pBCNF5603 (NC_006872), or pCPPB-1 (NC_015712) based on BLAST nucleotide alignments.
PCR confirmation of variable genes.
To validate the microarray results, PCR primers were designed for nine variable genes and one conserved gene using Primer3 (22). Genomic DNA was prepared from overnight single colonies using InstaGene Matrix (Bio-Rad Laboratories). Each PCR mixture contained approximately 1 μl of template DNA, 0.5 U of Platinum Taq high-fidelity DNA polymerase (Invitrogen), 0.2 mM dNTPs, 1× PCR buffer, 2 mM MgSO4, and 0.2 μM each primer in a 25-μl reaction mixture. The reaction mixtures were subjected to the following amplification conditions: one cycle of 95°C for 5 min; 35 cycles of 95°C for 30 s, 53°C for 30 s, and 68°C for 5 min; and one cycle of 72°C for 10 min. All primers used are described in Table S2 in the supplemental material. PCR product sizes were determined by agarose gel electrophoresis, and visualization was accomplished by ethidium bromide staining.
RESULTS
PFGE typing of Clostridium perfringens poultry isolates.
Several of the 54 C. perfringens poultry isolates examined in the current study had been typed by disparate methods, including MLST, PFGE, and multilocus variable number of tandem repeats analysis (MLVA), in previous studies (14, 23, 24). To assess the genetic relatedness of the isolates using a common method, all were typed by PFGE, with the exception of one strain (JP77) that was not typeable (see Fig. S1 in the supplemental material). PFGE analysis revealed a wide range of genetic diversity among the 53 typeable isolates, which produced 53 different PFGE patterns that were distributed into 44 major types. A dendrogram generated from the PFGE profiles placed all except for four of the isolates into three main clusters (I through III).
Identification of variable genomic regions among 63 Clostridium perfringens strains.
The genomic content of 54 C. perfringens isolates of poultry origin, 29 from birds with NE and 25 from healthy chickens, was examined by microarray CGH (Fig. 1). In addition, nine sequenced nonpoultry strains were included via in silico hybridization. To validate the array, hybridizations with genomic DNA from C. perfringens Str13 and ATCC 13124, for which complete genome sequences are available (25, 26), were performed and compared against the in silico hybridization results, revealing an average specificity and sensitivity of 99.46% and 96.3%, respectively.
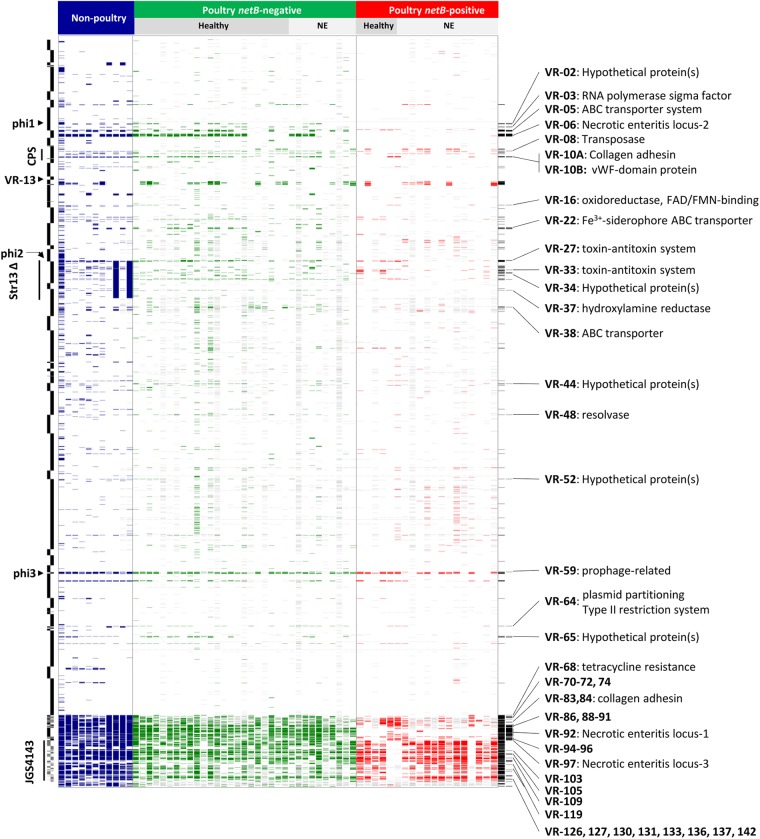
Fig 1.
Comparative genomic hybridization results of C. perfringens isolates from healthy and diseased chickens. Each column represents a different strain, and each row represents a different microarray probe. Gene probes called absent are colored blue, green, or red, and undetectable probes are colored gray. The colors indicate nonpoultry strains (blue), netB-negative isolates (green), and netB-positive isolates (red). Nonpoultry strains (from left to right) are SM101, NCTC 8239, F4969, ATCC 3626, JGS1495, JGS1721, JGS1987, ATCC 13124 (in silico), Str13 (in silico), ATCC 13124 (hybridization), and Str13 (hybridization). The host disease statuses of the isolate sources are given at the top. Probes are ordered according to the draft CP4 genome sequence, with the start of the genome at the top, and the CP4 contig boundaries are indicated on the left side. Probes specific for JGS4143 are placed at the bottom. On the left side, two regions identified as variable by Myers et al. (25), the capsular polysaccharide locus (CPS) and the large deletion found in Str13 (Str13Δ), are labeled, as are the locations of prophage. Variable probes and those significantly associated with netB-positive isolates (Pcorr < 0.05, Fisher's exact test) are indicated in the final two columns (black) on the right. The latter probes are annotated with the variable region and its putative function, where known (see Table S3 in the supplemental material for details on the variable regions). vWF, von Willebrand factor; FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide.
We initially sought to identify the genomic regions that exhibit variability among the set of isolates, as these regions are most likely to encode phenotypic differences between strains. Those genes present in 10% to 90% of the combined 63 poultry and nonpoultry strains were defined as variable, thus excluding both very low- and high-frequency genes. The vast majority of the gene probes (86.7%) were called present in 90% or more of the strains. The variable genes were grouped into regions if they were within 5 kb of each other and located on the same contig, based on the CP4 or JGS4143 draft genome sequence coordinates. A total of 400 variable genes were identified and grouped into 142 different regions, ranging in size from 116 bp to 42,019 bp (see Table S3 in the supplemental material). The variable genes were classified into 19 functional categories by the RAST annotation system (27), although for the majority of them (n = 322, 80.5%), a putative function could not be determined (see Fig. S2 in the supplemental material). Of the 78 that were classified, the most abundant categories were “carbohydrates” (n = 28) and “cell wall and capsule” (n = 19). A number of genes in the carbohydrates category were predicted to be involved in carbohydrate metabolism, including the metabolism of lactose (VR-01), fructooligosaccharides (FOS) and raffinose (VR-05), N-acetylglucosamine (VR-05, VR-92, and VR-122), and d-glucoronate (VR-100). Additionally, a C4-dicarboxylate transport system, involved in anaerobic energy metabolism, was found in VR-136 and VR-137 (28).
Variable genes with functions related to the cell wall and capsule included two sortases (VR-10B and VR-92), a dTDP-rhamnose biosynthesis operon (VR-13), and a capsular polysaccharide (CPS) biosynthesis locus (VR-9 and VR-109). The dTDP-rhamnose biosynthesis operon is closely linked to other variably present genes related to CPS biosynthesis (VR-13 and VR-14) and might represent a previously unidentified locus involved in the production of the C. perfringens CPS. The majority of the genes representing the core CPS biosynthesis locus, found in VR-9, VR-109, and VR-110, were variably present among the 63 poultry and nonpoultry isolates (see Table S3 in the supplemental material), consistent with the diversity observed in the C. perfringens CPS structure (25, 29, 30).
A previous study reported variability in the genes for several chromosomally encoded extracellular toxins and enzymes among three strains examined (25). Of the 18 minor toxins and extracellular enzymes examined in that study, only nagL, encoding a hyaluronidase, exhibited variability among the 63 strains (see Table S4 in the supplemental material).
To verify the CGH results, nine genes that were variable and one gene that was conserved among the poultry isolates were confirmed by PCR in 30 of the 54 poultry isolates. Of the 291 PCRs for which corresponding CGH data were available, 286 (98.3%) were in agreement with the microarray results (see Table S5 in the supplemental material), indicating a high degree of concordance between the PCR and CGH results.
Variable regions specific to poultry isolates.
To identify genetic loci that might play a role in host adaptation, the variable genes present among the poultry and nonpoultry strains were subjected to association analysis using Fisher's exact test for independence. A total of 241 variable genes were significantly associated (Pcorr < 0.05, two-tailed test) with poultry isolates, compared with the nonpoultry strains, with 99 of them absent from all nine of the nonpoultry strains (see Table S6 in the supplemental material). The majority of the latter group consisted of phage and plasmid-related genes and included the two previously identified plasmid-borne NE-associated loci (NELoc-1 and -3) (10). In addition, a number of nonprophage chromosomal genes were found exclusively in poultry isolates, including those found in VR-10B, VR-62, VR-101, VR-105, VR-109, and VR-110.
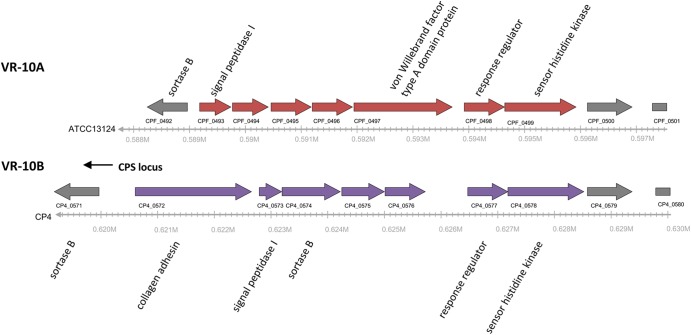
The VR-10 locus is found immediately downstream of the CPS locus and consists of two alleles, VR-10A and VR-10B, which appear to encode different cell surface proteins related to adherence (Fig. 2). A von Willebrand factor type A domain protein is present in VR-10A, and a collagen adhesin is in VR-10B (31). Although both alleles encode a two-component regulatory system, they are distantly related based on amino acid sequence identity (see Fig. S3 in the supplemental material). Four of seven genes within VR-10A were negatively associated with the poultry isolates, exhibiting an average prevalence of 44% among poultry strains and 94% among nonpoultry strains (see Table S6 in the supplemental material). In contrast, the genes constituting the alternative VR-10B allele exhibited an average prevalence of 64% among the poultry isolates and were absent from the nonpoultry strains. According to the array results, two of the poultry isolates appeared to carry both alleles; however, these anomalies have not yet been confirmed by an alternative method such as PCR. Given the association of VR-10B exclusively with poultry isolates, this locus might encode a poultry-specific colonization factor.
Fig 2.
Genetic map comparing two VR-10 adhesion-related variable region alleles. The genetic map shows the ATCC 13124 VR-10A region (which is also conserved in nine nonpoultry strains and JGS4143) and the corresponding CP4 VR-10B region. Genes conserved between the two alleles are shown in gray, and those unique to each allele are shown in red (VR-10A) or blue (VR-10B).
A cluster of six genes related to drug resistance (VR-62; two genes are not represented on the array) were present in, on average, 66% of the poultry isolates and absent from the nonpoultry strains. This locus contains a transcriptional regulator (CP4_2430), a VanZ family protein (CP4_2429), and two proteins with an ABC drug transporter domain (CP4_2433 and CP4_2436), a domain that is associated with drug resistance and lantibiotic immunity. Additional poultry-associated variable regions included a CPS locus (VR-109 and VR-110) and two regions encoding hypothetical proteins (VR-101 and VR-105).
Prevalence and conservation of NE loci among poultry isolates.
A previous study identified three loci highly conserved in NE-producing strains (NELoc-1 to -3), but only a limited set of virulent strains and just one healthy bird isolate were used in the comparison (10). To extend the previous findings, the prevalences of these three loci as determined by microarray CGH were examined in the present study among the larger set of isolates, which includes 29 obtained from healthy birds. PCR detection of netB revealed prevalences of 60% (15/25) in the NE bird isolates and 21% (6/29) in the healthy bird isolates (see Table S1 in the supplemental material). The presence of NELoc-1, as detected by CGH, was generally concordant with the netB PCR results and revealed that the locus, when present, was conserved essentially in its entirety (Fig. 3). Strain CP2 was previously shown to carry a small deletion in NELoc-1 (10), which was correctly detected by the array. A number of NELoc-1 genes were unexpectedly identified as present in netB-negative isolates. Spot testing of several of these probes by PCR, however, confirmed in each case a false-positive array result (data not shown), suggesting that the majority, if not entirety, of the NELoc-1 genes called present among the netB-negative isolates were in fact false positives. In some cases, the poor specificity of NELoc-1 probes can be explained by the presence of similar regions on other plasmids (e.g., CP4_3451-53, CP4_3470-74, CP4_3477), as evidenced by the in silico hybridization results with nonpoultry strains, which might result in cross-hybridization.
Fig 3.
Heat map of NELoc-1, -2, and -3 gene distribution among 54 poultry and nine nonpoultry C. perfringens isolates. Microarray CGH results for the genes found in the three NE-associated loci are shown. Each column represents a different strain, and each row represents a different microarray probe. Gene probes called absent are colored blue, green, or red, and undetectable probes are colored gray. The colors indicate nonpoultry strains (blue), netB-negative isolates (green), and netB-positive isolates (red). The nonpoultry strains (from left to right) are SM101, NCTC 8239, F4969, ATCC 3626, JGS1495, JGS1721, JGS1987, ATCC 13124, and Str13. The host disease statuses of the isolate sources are given at the top. Probes are ordered according to the draft CP4 genome sequence, with the start of the genome at the top. See Table S3 in the supplemental material for details on the variable regions.
NELoc-2, which is located on the chromosome, was present in 84% (21/25) of NE isolates and 41.4% (12/29) of non-NE isolates, and when NELoc-2 was present, it was found in its entirety (Fig. 3). This locus was present in all isolates that carried NELoc-1 and was the most closely associated with host disease status among the three loci.
The average prevalences of NELoc-3 genes were 71% in NE bird isolates and 52% in healthy bird isolates. As with NELoc-1, three NELoc-3 genes also exist on other plasmids (CP4_3567, CP4_3569, and CP4_3570), and these gene probes might cross-hybridize with sequences from other coexisting plasmids. Based on the remaining two genes specific to NELoc-3 (CP4_3472 and CP4_3568), this locus was found in 65% of NE bird isolates and 36% of healthy bird isolates. It was demonstrated previously that NELoc-3 and cpb2 are colocalized to the same plasmid (10). However, in the current study, only 55.2% (21/38) of the cpb2-positive isolates also carried NELoc-3, indicating that this linkage is not conserved (see Table S1 in the supplemental material). Similarly, there was not a strict linkage between NELoc-3 and NELoc-1, with at least two isolates possessing NELoc-1 but not NELoc-3. NELoc-2, on the other hand, was present in all strains that carried NELoc-1, supporting the hypothesis that this chromosomal locus plays an important role in NE pathogenesis.
Association of chromosomal “fitness” loci with netB-positive poultry isolates.
To find additional genes that might play a role in virulence, the 400 variable genes underwent association analysis, and those exhibiting significant linkage disequilibrium with netB (Pcorr < 0.05, Fisher's exact two-tailed test) were identified. The presence of the NetB toxin has been demonstrated to be a reliable predictor of NE-producing capability (3, 32) and was therefore chosen as a marker for virulence. Of the 400 variable genes identified, 128 were significantly associated with netB carriage, distributed into 48 variable regions (Fig. 4 and Table 1; also see Table S6 in the supplemental material for details). Ten genes exhibited a negative association with netB, and the remainder were positively associated. The 34 genes found on NELoc-1 were excluded from the analysis, given their physical linkage to netB.
Fig 4.
Heat map of genes significantly associated with netB-positive C. perfringens poultry isolates. Genes positively associated are shown at the top, and genes negatively associated are at the bottom. Each column represents a different strain, and each row represents a different microarray probe. Gene probes called absent are colored blue, green, or red, and undetectable probes are colored gray. The colors indicate nonpoultry strains (blue), netB-negative isolates (green), and netB-positive isolates (red). The nonpoultry strains (from left to right) are SM101, NCTC 8239, F4969, ATCC 3626, JGS1495, JGS1721, JGS1987, ATCC 13124, and Str13. The host disease statuses of the isolate sources are given at the top. Probes are ordered according to the draft CP4 genome sequence, with the start of the genome at the top. See Table S3 in the supplemental material for details on the variable regions.
Table 1.
Variable genomic regions significantly associated with netB-positive C. perfringens poultry isolates
| Variable | Strain associationa | Predicted functionb | Size (bp) | No. of genes | No. of significant genes | Average prevalence (%) |
Predicted locationc | |
|---|---|---|---|---|---|---|---|---|
| netB positive | netB negative | |||||||
| VR-02 | Both | Hypothetical protein(s) | 1,080 | 3 | 3 | 100 | 70 | Chromosome |
| VR-03 | Both | Alternative sigma factor | 1,794 | 2 | 2 | 100 | 72 | Chromosome |
| VR-05 | Both | Carbohydrate ABC transporter system | 6,393 | 5 | 5 | 100 | 70 | Chromosome |
| VR-06 | Both | NELoc-2 | 11,720 | 11 | 11 | 100 | 44 | Chromosome |
| VR-08 | Both | Phage/insertion sequence related | 3,445 | 2 | 1 | 67 | 24 | Chromosome |
| VR-10A | JGS4143 | Two-component system, vWF domain protein | 5,877 | 7 | 5 | 23 | 70 | Chromosome |
| VR-10B | CP4 | Alternative to VR-10A; two-component system, collagen adhesin | 5,743 | 4 | 3 | 87 | 42 | Chromosome |
| VR-16 | CP4 | Oxidoreductase, FAD/FMN-binding | 1,106 | 1 | 1 | 86 | 53 | Chromosome |
| VR-22 | Both | Fe3+-siderophore ABC transporter | 3,882 | 4 | 4 | 100 | 69 | Chromosome |
| VR-27 | Both | Phage/insertion sequence-related, toxin-antitoxin | 2,443 | 2 | 2 | 95 | 63 | Chromosome |
| VR-33 | Both | Phage/insertion sequence-related, toxin-antitoxin | 5,379 | 5 | 1 | 100 | 77 | Chromosome |
| VR-34 | Both | Hypothetical protein(s) | 14,946 | 8 | 3 | 98 | 71 | Chromosome |
| VR-37 | Both | Hydroxylamine reductase | 1,655 | 1 | 1 | 100 | 53 | Chromosome |
| VR-38 | Both | ABC transporter | 6,062 | 4 | 1 | 90 | 61 | Chromosome |
| VR-44 | CP4 | Hypothetical protein(s) | 770 | 1 | 1 | 95 | 55 | Chromosome |
| VR-48 | CP4 | Phage/insertion sequence-related | 152 | 1 | 1 | 90 | 58 | Chromosome |
| VR-52 | CP4 | Hypothetical protein(s) | 503 | 1 | 1 | 95 | 67 | Chromosome |
| VR-59 | CP4 | Phage/insertion sequence-related | 128 | 1 | 1 | 60 | 24 | Chromosome |
| VR-64 | CP4 | Plasmid partitioning, restriction modification system | 755 | 1 | 1 | 81 | 45 | Chromosome |
| VR-65 | Both | Hypothetical protein(s) | 6,351 | 3 | 3 | 100 | 65 | Chromosome |
| VR-68 | Both | AraC family transcriptional regulator | 4,994 | 3 | 1 | 95 | 61 | pTet |
| VR-70 | Both | Hypothetical protein(s) | 1,289 | 3 | 3 | 98 | 64 | Unknown plasmid |
| VR-71 | Both | LexA repressor | 559 | 1 | 1 | 100 | 56 | pTet |
| VR-72 | CP4 | Putative plasmid partitioning protein | 1,135 | 2 | 1 | 95 | 65 | pTet |
| VR-74 | Both | Putative ATPase of HSP70 class | 1,099 | 2 | 1 | 95 | 29 | pNetB |
| VR-83 | CP4 | Collagen adhesin (partial) | 4,754 | 2 | 1 | 81 | 29 | Multiple plasmids |
| VR-84 | CP4 | Collagen adhesin (partial) | 665 | 1 | 1 | 62 | 30 | Multiple plasmids |
| VR-86 | CP4 | FtsK/SpoI | 1,316 | 1 | 1 | 86 | 31 | Multiple plasmids |
| VR-88 | Both | LexA repressor | 404 | 1 | 1 | 100 | 24 | pBeta2 |
| VR-89 | CP4 | Radical SAM domain proteins | 3,737 | 4 | 4 | 69 | 19 | Unknown plasmid |
| VR-90 | Both | Putative ATPase | 1,561 | 3 | 3 | 93 | 41 | pBeta2 |
| VR-91 | CP4 | cpb2 | 1,776 | 2 | 1 | 100 | 79 | pBeta2 |
| VR-92 | Both | NELoc-1 | 42,019 | 34 | 34 | 96 | 28 | pNetB |
| VR-94 | Both | Hypothetical protein(s) | 1,114 | 2 | 1 | 53 | 4 | Multiple plasmids |
| VR-95 | Both | Putative ATPase | 1,503 | 3 | 2 | 92 | 40 | Multiple plasmids |
| VR-96 | CP4 | Hypothetical protein(s) | 2,466 | 2 | 1 | 76 | 29 | pBeta2 |
| VR-97 | CP4 | NELoc-3 | 6,487 | 5 | 5 | 88 | 41 | pBeta2 |
| VR-103 | Both | Hypothetical protein(s) | 371 | 1 | 1 | 100 | 58 | Chromosome |
| VR-105 | Both | Phage/insertion sequence-related | 8,069 | 8 | 3 | 91 | 53 | Chromosome |
| VR-109 | JGS4143 | Capsular polysaccharide | 16,185 | 12 | 1 | 19 | 48 | Chromosome |
| VR-119 | JGS4143 | Hypothetical protein(s) | 323 | 1 | 1 | 43 | 79 | Chromosome |
| VR-126 | Both | RNA polymerase sigma-70 factor | 1,875 | 2 | 1 | 63 | 93 | Chromosome |
| VR-127 | Both | Phage/insertion sequence-related | 8,901 | 15 | 1 | 95 | 71 | Chromosome |
| VR-130 | Both | Endo-alpha-N-acetylgalactosaminidase | 152 | 1 | 1 | 81 | 100 | Chromosome |
| VR-131 | Both | NELoc-1 fragment | 206 | 1 | 1 | 100 | 45 | pNetB |
| VR-133 | JGS4143 | Phage/insertion sequence-related, restriction system | 12,137 | 9 | 1 | 86 | 52 | Chromosome |
| VR-136 | JGS4143 | C4-dicarboxylate transport system | 2,699 | 2 | 1 | 19 | 48 | Chromosome |
| VR-137 | JGS4143 | C4-dicarboxylate transport system | 2,875 | 2 | 1 | 33 | 67 | Chromosome |
| VR-142 | Both | Conjugation | NAd | 6 | 2 | 93 | 60 | Multiple plasmids |
The locus is present in CP4, JGS4143, or both strains.
vWF, von Willebrand factor; FAD, flavin adenine dinucleotide; FMN, flavin mononucleotide.
Predicted location of variable regions based on sequence comparisons with Str13, ATCC 13124, SM101, and 13 plasmid sequences (see Materials and Methods). If an identical alignment to only one plasmid was found, the plasmid name is indicated. “Multiple plasmids” indicates that identical alignments to multiple different plasmids were found. “Unknown plasmid” indicates that only part of the variable region has alignments to a known plasmid.
NA, not available.
Thirty-one of the significant genes were predicted to be plasmid borne based on BLAST alignment of the oligonucleotide probe sequences with the complete sequences of 13 C. perfringens plasmids (Table 1). These included two genes localized to pNetB, but outside NELoc-1, and 11 specific to pBeta2, including those for NELoc-3. An additional three gene probes were specific for the tetracycline resistance plasmids, pTet and pCW3, which have not been shown to be associated with NE. For two netB-associated VRs (VR-70 and VR-89), only some of the probes exhibited sequence similarity to known plasmids; therefore, they appear to represent previously uncharacterized C. perfringens plasmids. The remaining plasmid-related probes represented genes present on multiple plasmids and therefore could not be localized to a specific plasmid.
The remaining 63 netB-associated genes were predicted to be chromosomally located. The most significantly associated were those in NELoc-2 (VR-06), which were present in all of the netB-positive isolates and, on average, in 44% of netB-negative isolates. Twelve of the significant chromosomally located genes were related to prophage or insertion sequences, and an additional 13 were hypothetical proteins for which a putative function could not be assigned; therefore, these genes were not considered further.
Several significant variable chromosomal regions encoded proteins putatively involved in nutrient acquisition or metabolism, including VR-05 and VR-38, both of which encode ABC transporter systems. VR-22 consists of four genes that encode a putative ferric iron siderophore ABC transporter system, all of which were significantly more prevalent in the netB-positive isolates. In addition to this novel iron acquisition locus, both CP4 and JGS4143 were found to carry two duplications of the feoAB ferrous iron transporter operon, based on genomic sequence comparisons with Str13 and SM101. It was not possible to determine how prevalent these additional feoAB copies were in the entire set of poultry isolates, because of the inability of the array to detect duplications.
Two chromosomal VRs encoding putative cell surface-related proteins, VR-10B and VR-109, were significantly associated with netB, though only one gene among the 12 found in VR-109 was found to be significant. VR-10B contains a poultry-specific putative adherence locus, as described above (Fig. 2). VR-10A, the alternate allele to VR-10B, accordingly exhibited a significant negative association with netB-positive isolates and in nearly all cases was detected only when VR-10B was absent (Fig. 4).
Association of plasmid maintenance loci with netB-positive poultry isolates.
Variable regions characteristic of these systems were found on several netB-associated chromosomal loci, including two toxin-antitoxin systems (VR-27 and VR-33) (Fig. 4). Additionally, VR-64 consists of a soj sporulation inhibitor protein (CP4_2645), part of the soj-spoOJ type I par locus involved in plasmid and chromosome segregation. Genome sequence comparison of the region surrounding CP4_2645 with the nonpoultry strains revealed that this gene is part of a larger variable locus consisting of 10 genes, nine of which are not represented on the array. These nine genes include a second component of the soj-spoOJ operon, four degenerate transposases, and two genes that encode a type IIS restriction-modification system, which is absent from the nine nonpoultry strains. A second soj-spoOJ operon located within the C. perfringens core genome shares only 35% amino acid identity with the VR-64 soj gene, whereas a soj gene (PCP01) on pCP13 is 86% identical, suggesting that VR-64 is of plasmid origin.
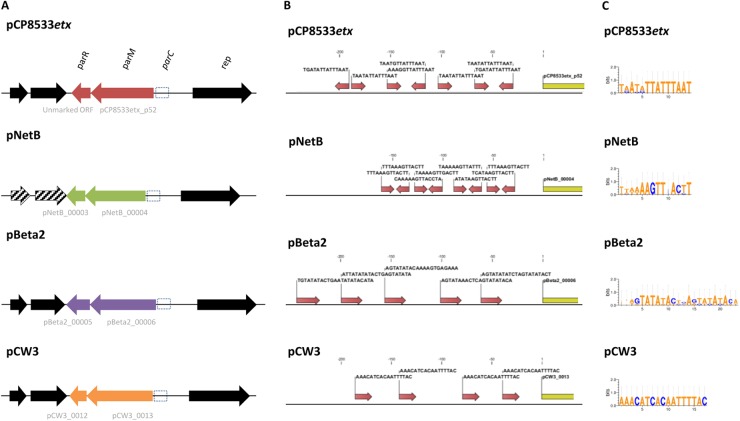
Several plasmid-borne netB-associated variable regions that carry putative plasmid partitioning systems were also identified. VR-72 consists of two genes that share 99% nucleotide sequence identity with a putative partitioning system located on pCW3 (pCW3_0012-13), based on the CP4 sequence. Sequence comparisons of several C. perfringens toxin plasmids revealed that each contains a similarly organized region located within the conserved core sequence. Despite exhibiting synteny, these regions share little sequence identity (Fig. 5A). Three additional netB-associated variable regions (VR-74, VR-90 and VR-95) were each subsequently found to contain two genes corresponding to this region in different plasmids. Specifically, the CP4 nucleotide sequences for VR-74, VR-90 and VR-95 are 99% identical to sequences in plasmids pNetB, pBeta2, and pCP8533etx, respectively, though nearly identical sequences were also found in other C. perfringens genomes. Pairwise amino acid alignments of the larger CDS present in each VR revealed only 22% to 34% sequence identity among them. The pCW3 CDS contains a provisional parM domain, suggesting that these regions might be related to the parMRC partitioning system. This system is often found in large, low-copy-number plasmids and consists of a ParM actin-like ATPase, which forms a dynamic filament that pushes ParR DNA-binding proteins to opposite poles of the cell, once bound to the parC cis-acting centromere element (33). Analogous to the Escherichia coli parC element, a series of direct repeats was located upstream from each putative ParM gene, with each upstream region containing a unique sequence motif (Fig. 5B and C).
Fig 5.
parMRC-like plasmid partitioning systems associated with netB-positive isolates. (A) Genetic maps of four putative parMRC-like systems associated with netB-positive C. perfringens poultry isolates. Genes conserved among the four loci are in black, and variable genes are colored. Two open reading frames (ORFs) in pNetB (hatched) were missing start codons and therefore not annotated in the sequence. (B) parC-like repeat elements identified upstream of putative parM orthologs. (C) Consensus sequences of different parC-like motifs found in different plasmids.
Conservation of netB-associated loci in prevalent NE clonal groups.
A recent MLST study examining NE and healthy bird C. perfringens isolates found that the majority of those associated with NE belonged to two phylogenetically distinct clonal groups: clonal cluster 4 (CC-4) and sequence type 31 (ST-31) (13). Based on in silico MLST analysis, CP4 and JGS4143 belong to CC-4 and ST-31, respectively (data not shown). Six additional poultry isolates had previously been typed by MLST (14) (see Table S1 in the supplemental material), of which two were closely related to CC-4 (MLST31 and MLST23) and another two to ST-31 (JGS4135, MLST11), based on previous analyses (13). All of these isolates were netB positive and were obtained from diseased birds. Since these two clonal groups appear to have a selective advantage over other NE-associated strains in the environment, we sought to identify those netB-associated variable regions common to both types. Of the 94 variable genes that were significantly associated with netB, not including those in NELoc-1, 48 were found in all six of the isolates (Table 2). These included genes in variable chromosomal regions associated with both nutrient acquisition (VR-05, VR-22, and VR-38) and plasmid maintenance (VR-27 and VR-33). Overall, the distribution of variable regions among the isolates of the same clonal group was highly concordant (Fig. 6).
Table 2.
Variable genomic regions significantly associated with netB-positive C. perfringens poultry isolates common to clonal groups CC-4 and ST-31
| Variable region | Locus tag | Product |
|---|---|---|
| VR-02 | CP4_0377 | Conserved hypothetical protein |
| CP4_0378 | Conserved hypothetical protein | |
| CP4_0379 | Conserved hypothetical protein | |
| VR-03 | CP4_0391 | Conserved hypothetical protein |
| CP4_0392 | RNA polymerase ECF-type sigma factor | |
| VR-05 | CP4_0444 | Transcriptional regulator |
| CP4_0445 | Extracellular solute-binding protein | |
| CP4_0446 | ABC transporter | |
| CP4_0447 | Sugar ABC transporter | |
| CP4_0448 | Alpha-galactosidase 1 | |
| VR-06 | CP4_0458 | Sigma factor SgiI |
| CP4_0459 | Conserved hypothetical protein | |
| CP4_0460 | Putative VTC domain superfamily | |
| CP4_0461 | Putative tubulin/FtsZ, GTPase | |
| CP4_0462 | Resolvase | |
| CP4_0463 | Putative VTC domain superfamily | |
| CP4_0464 | Tubulin/FtsZ, GTPase | |
| CP4_0465 | CotH protein | |
| CP4_0466 | Conserved hypothetical protein | |
| CP4_0467 | Putative heat repeat | |
| CP4_0468 | Chitin synthase | |
| VR-22 | CP4_0874 | Iron |
| CP4_0875 | Iron compound ABC transporter, permease protein | |
| CP4_0876 | ABC transport system permease protein | |
| CP4_0877 | Ferrichrome transport ATP-binding protein FhuC | |
| VR-27 | CP4_1010 | Putative toxin-antitoxin system, toxin component |
| VR-33 | CP4_1131 | Toxin-antitoxin system, antitoxin component, Xre family |
| VR-34 | CP4_1153 | Conserved hypothetical protein |
| CP4_1154 | Conserved hypothetical protein | |
| CP4_1158 | Conserved hypothetical protein | |
| VR-37 | CP4_1251 | Hydroxylamine reductase |
| VR-38 | CP4_1325 | Putative permease |
| VR-65 | CP4_2699 | Conserved hypothetical protein |
| CP4_2700 | Conserved hypothetical protein | |
| VR-70 | CP4_3125 | Hypothetical protein |
| CP4_3127 | Conserved hypothetical protein | |
| VR-71 | CP4_3126 | LexA repressor |
| VR-88 | CP4_3431 | LexA repressor |
| VR-90 | CP4_3437 | Conserved hypothetical protein |
| CP4_3438 | Putative ATPase | |
| VR-91 | CP4_3439 | Transcriptional regulator |
| VR-97 | CP4_3570 | Conserved hypothetical protein |
| VR-103 | JGS4143_0370 | Conserved hypothetical protein |
| VR-105 | JGS4143_0473 | Conserved hypothetical protein |
| JGS4143_0475 | Hypothetical protein | |
| JGS4143_0478 | Hypothetical protein | |
| VR-131 | JGS4143_1408 | Hypothetical protein |
| VR-142 | pCPF5603_70 | Putative Tn916 ORF16 protein |
Fig 6.
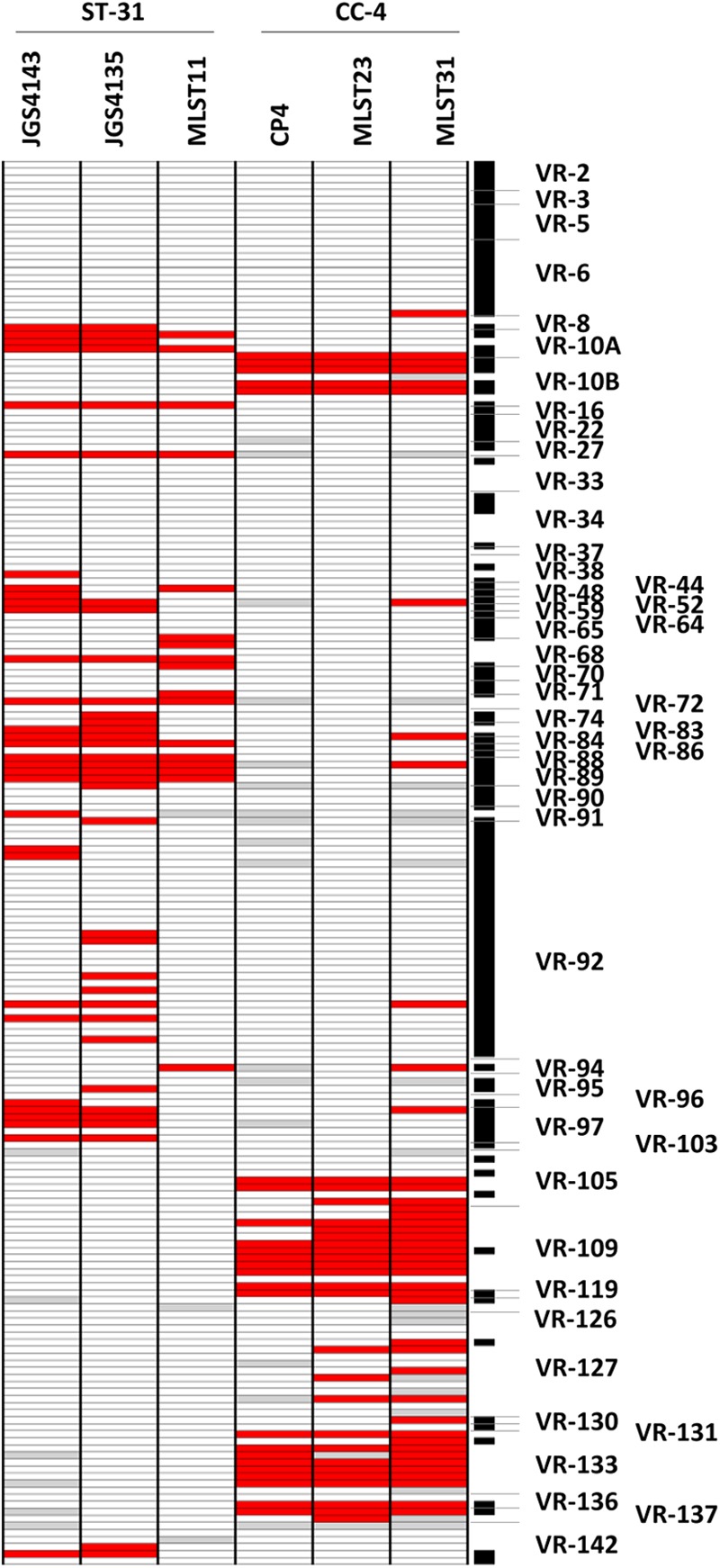
Heat map of variable regions significantly associated with netB-positive isolates common to two prevalent clonal groups. Each column represents a different strain, and each row represents a different microarray probe. Gene probes called absent are colored red, and undetectable probes are colored gray. CGH results from all of the variable regions containing one or more significantly associated genes are shown, and variable regions are labeled at the side. Those genes identified as significant are indicated on the right by a black bar. The clonal group (CC-4 or ST-31) to which each isolate belongs is indicated at the top.
Cluster analysis of microarray CGH data.
We performed hierarchical clustering on the binary CGH data to evaluate the relationships in the genomic content among the 63 C. perfringens strains. Forty-nine of the 54 poultry isolates distributed into three major clusters (I through III). Clusters II and III contained 76% (19/25) of the disease-associated isolates and all of the netB-positive isolates, whereas cluster I contained mostly healthy bird isolates (65%) and only netB-negative isolates (Fig. 7). Clusters II and III were composed of 61% (11/18) and 67% (10/15) netB-positive isolates, respectively, and were more closely related to each other than to cluster I. The dendrograms resulting from PFGE (see Fig. S1 in the supplemental material) and CGH (Fig. 7) exhibited broad concordance; in particular, cluster III from the CGH dendrogram completely overlapped the same cluster from PFGE analysis, although six strains in PFGE cluster III were not in CGH cluster III (CP1, CP5, JGS4120, JGS4141, JP148, and MLST19).
Fig 7.
Hierarchical cluster analysis of C. perfringens strains based on microarray CGH. Cluster analysis was performed on binary microarray CGH data or on in silico hybridization data where indicated (i). The netB and host NE statuses of each strain are indicated by + (positive) and − (negative). MLST types are shown where available, with type designations from Chalmers et al. (14) prefaced with a “C” and from Hibberd et al. (13) with an “H.” N/A, not available.
The six poultry isolates closely related to clonal groups CC-4 and ST-31 segregated into clusters II and III according to their clonal group. The three ST-31-related isolates assembled into cluster II, and the three CC-4 related isolates assembled into cluster III, indicating that these two predominant lineages carry distinct chromosomal backgrounds.
The nine nonpoultry C. perfringens strains for which genome sequences are available, representing the entire range of toxin types and a diversity of host and disease associations, were also included in the analysis using data from in silico hybridizations. All of these strains, including the five type A strains, clustered separately from the poultry isolates (Fig. 7), suggesting that poultry isolates share a core genome distinct from strains that occupy other niches.
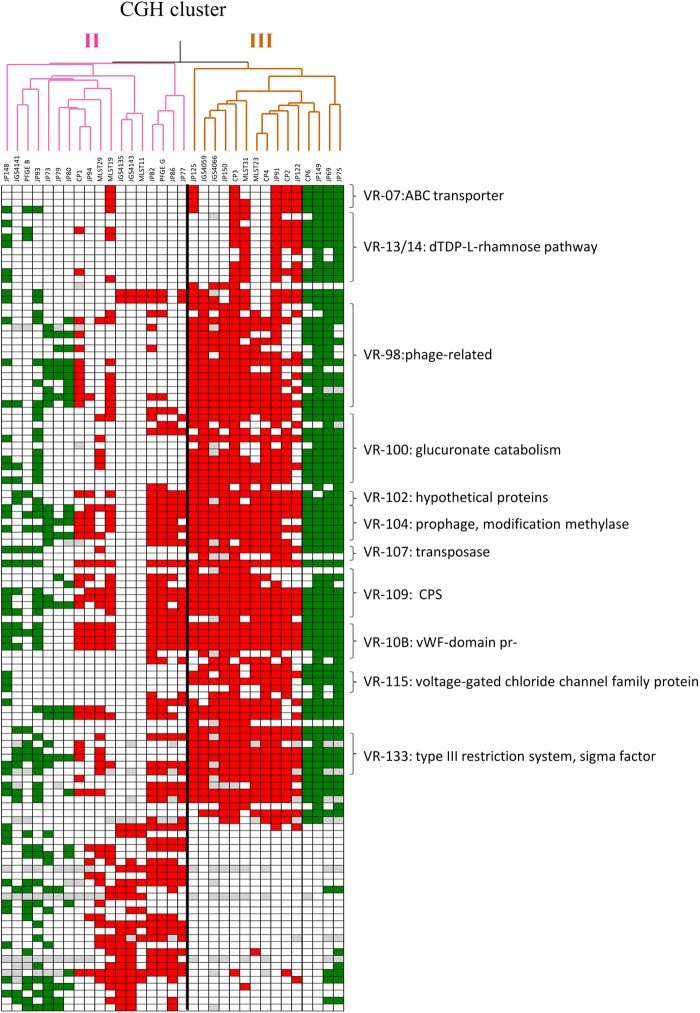
Variable genomic regions distinguishing NE-associated groups II and III.
To further examine the genetic basis of the netB-associated clusters II and III, and to identify putative distinguishing phenotypic and genotypic markers, we performed association analysis on this subset of isolates as described above. A total of 91 genes were significantly associated (Pcorr < 0.05) with cluster II and 27 genes with cluster III (Fig. 8). Several variable regions related to the cell wall and the capsule were predominantly present in cluster II, including VR-13 and VR-14, encoding a dTDP-l-rhamnose pathway, and VR-109, containing the JGS4143-specific CPS locus. A glucuronate catabolism operon (VR-100) and an ABC transporter system (VR-07) were also significantly represented in cluster II. Together, these additional variable regions found in many cluster II isolates predict phenotypic differences from isolates belonging to cluster III, possibly related to capsule structure and the ability to take up and utilize specific substrates, including glucuronate.
Fig 8.
Heat map of C. perfringens poultry isolates found in CGH clusters II and III showing genes significantly associated with each cluster. Each column represents a different strain, and each row represents a different microarray probe. Gene probes called absent are colored green or red, and undetectable probes are colored gray. The colors for absent genes indicate netB-negative isolates (green) and netB-positive isolates (red). Only the 33 poultry isolates that assembled into groups II and III based on hierarchical clustering of microarray CGH data are shown. See Table S3 in the supplemental material for details on the variable regions.
DISCUSSION
Clostridium perfringens is responsible for a diverse set of human and animal diseases, owing largely to the extensive pool of plasmid-encoded toxins carried by this species. Many of these toxin-encoding plasmids undergo conjugative transfer (6, 8, 9), so in principle, plasmid composition might be the only feature distinguishing a commensal strain from one of the many pathotypes. The identification of specific chromosomal regions associated with the netB plasmid in the present study suggests, however, that both plasmid and host genes contribute to NE pathogenesis. These results provide for the first time an explanation for the linkage between plasmid-mediated virulence in C. perfringens and the chromosomal background in which the plasmid resides, on the basis of the clonality recently identified in NE isolates (13). Results of phylogenetic studies of C. perfringens strains associated with other diseases suggest that the contribution of chromosomal background to virulence varies by pathotype. For example, C. perfringens non-food-borne disease isolates possessing a plasmid-borne cpe are phylogenetically distinct, indicating that cpe-carrying plasmids are acquired into unrelated chromosomal backgrounds and still confer virulence (34–36). In contrast, an MLST study examining C. perfringens isolates from 10 different host species found that the majority of bovine type E isolates assembled into the same clonal complex, as did porcine type C isolates, indicating an interrelationship between host and plasmid-encoded factors, the basis of which is not understood (37). The findings reported here for NE isolates represent a new paradigm in our understanding of C. perfringens as an enteric pathogen, and it might be applicable to these and other serious enteric infections caused by this organism.
Microarray CGH analysis revealed 400 variable genomic regions among the 54 poultry and nine nonpoultry isolates examined. Many of the variable genomic regions corresponded to cell surface structures, such as CPS, consistent with the intense selective pressure faced by these proteins through direct exposure to the host immune system. A number of the variable genomic regions were also putatively involved in the metabolism of various carbohydrates, which might be expected to impart fitness advantages via access to supplemental carbon sources.
There has been one previous comparative study of C. perfringens genomes, in which a number of variable regions were identified among three type A strains (Str13, ATCC 13124, and SM101). The largest of these was a 243-kb genomic island that was present in ATCC 13124 but absent from Str13 and only partially present in SM101 (25). This island was found to be largely intact in the 54 poultry and six nonpoultry isolates examined here and thus appears to be a deletion specific to Str13. A significant amount of variability was observed in this region, however, with 23 kb of sporadic insertions encoding 44 additional CDSs in the CP4 genome compared to the results from ATCC 13124. The 5′ end of this island contains one of three prophages found in the CP4 genome, all of which exhibit significant local variability and likely represent recombinational hotspots (38). This previous study also found that several minor extracellular toxins and enzymes were variably present (25); however, only nagL exhibited variability among our isolates and the six nonpoultry strains, suggesting that these genes are constituents of the C. perfringens core genome.
The NetB toxin gene was present in 60% of NE bird isolates and, in general, was found together with the other genes that comprise NELoc-1. Given the strong potential for recombination among C. perfringens plasmids, the strict conservation of this locus might indicate that NELoc-1 genes, in addition to netB, are important for pathogenesis. The relatively low prevalences of netB among the poultry NE isolates were similar to the findings from previous studies (2, 3, 39, 40). Assuming that netB is critical to NE pathogenesis, these low prevalences might reflect the occasional inadvertent isolation of a commensal strain, a propensity for the netB plasmid to be lost during isolation, or both. It is interesting that, of the three previously identified loci, NELoc-2 was most closely associated with NE bird isolates, which might be a consequence of its more stable location on the chromosome. Studies have demonstrated that netB-positive isolates from healthy birds will initiate disease in a model system, while netB-negative isolates from diseased birds will not (3, 32). For this reason, we chose to use netB carriage, as opposed to host disease status, as an indicator of virulence in our analyses. Additionally, all of our netB-positive isolates, regardless of host disease status, were cytotoxic against the chicken leghorn male hepatoma (LMH) cell line (data not shown), indicating that they produce functional NetB toxin and are therefore most likely virulent (32).
One of the most striking findings was the association of several chromosomal loci that appeared to be involved in enhancing fitness as it relates to virulence with netB. These included genes for iron acquisition (VR-22), carbohydrate utilization (VR-05 and VR-06), and adherence (VR-10B). It is hypothesized that some or all of these loci provide a selective advantage to strains that have acquired NELoc-1- and NELoc-3-containing plasmids by contributing to the initiation of infection.
A second hypothesis that might account for the association of specific chromosomal genes with plasmid-borne virulence genes is that the chromosomal genes play a role in maintenance of the virulence plasmid. It has been proposed that plasmid addiction, partitioning, and multimer resolution systems are all required for successful transmission of large low-copy-number virulence plasmids (41). Several chromosomal loci encoding putative plasmid maintenance systems, such as addiction modules and partitioning systems, were significantly associated with netB-positive isolates. These systems appear to be active in maintaining pNetB, since we were unable to cause any significant reduction in plasmid carriage even after approximately 80 generations (data not shown), despite anecdotal evidence to suggest that it can be readily lost during subculture. While these systems are typically encoded on the plasmid itself, examples of host-encoded plasmid maintenance systems do exist (42–45).
A number of plasmid-associated regions were also significantly associated with netB, including four different loci that appear to encode plasmid partitioning systems. The genetic organization of these loci and the presence of upstream parC-like sequence motifs (46) strongly suggest that they encode parMRC-like partition systems. The presence of different parMRC-like systems on different plasmids might explain how multiple large plasmids are able to successfully segregate within a single host and would likely form a basis for plasmid incompatibility (46). Two of these loci were localized to pNetB or pBeta2 (carrying NELoc-3), thus offering an explanation for their association; however, it is not clear why regions specific to other plasmids, or part of the conserved core region, were also significantly associated. It is possible that factors encoded by multiple different plasmids contribute to NE pathogenesis or, alternatively, that strains carrying pNetB are more likely to harbor other additional plasmids. An assessment of the plasmid complement among this set of isolates, in addition to functional studies, would help to elucidate the exact nature of this association.
The segregation of the netB-positive isolates into two major clusters based on gene content coincides well with recent MLST findings that most NE-associated isolates belong to two predominant clonal groups (13, 14) and suggests that at least two phenotypically distinct subpopulations of NE-causing strains exist. The lack of pathways for glucuronate catabolism (VR-100) and nutrient uptake (VR-07) by the majority of cluster III isolates does not necessarily preclude a selective advantage, as the loss of metabolic pathways can be beneficial if there is an unnecessary cost to their maintenance (47). The contribution of these differentiating loci to overall fitness might permit different subtypes to prevail in response to variations in host or environmental conditions. These distinguishing loci among the two clusters might be used, in combination with markers that differentiate these strains from healthy isolates, to identify specific NE subtypes.
In summary, these results suggest that netB-positive strains have arisen through clonal expansion, as opposed to acquisition of virulence plasmids by disparate hosts. The distribution and incomplete conservation of the chromosomally located loci associated with netB-positive isolates suggest that they provide subtle incremental effects on fitness as it relates to pathogenesis. It is also apparent that not all NE-causing strains are identical in terms of chromosomal backgrounds, and that at least two distinct subpopulations exist. Further studies targeting specific loci to examine their impacts on overall fitness and plasmid stability will help elucidate the mechanisms responsible for these chromosomal and extrachromosomal linkages.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Agriculture and Agri-Food Canada, the Canadian Poultry Research Council through the Poultry Cluster Agreement with Agriculture and Agri-Food Canada, the Ontario Ministry of Agriculture, Food and Rural Affairs, and the Natural Sciences and Engineering Research Council.
Footnotes
Published ahead of print 4 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01032-12.
REFERENCES
- 1. Cooper KK, Songer JG. 2009. Necrotic enteritis in chickens: a paradigm of enteric infection by Clostridium perfringens type A. Anaerobe 15:55–60 [DOI] [PubMed] [Google Scholar]
- 2. Keyburn AL, Boyce JD, Vaz P, Bannam TL, Ford ME, Parker D, Di Rubbo A, Rood JI, Moore RJ. 2008. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathog. 4:e26 doi:10.1371/journal.ppat.0040026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Keyburn AL, Yan XX, Bannam TL, Van Immerseel F, Rood JI, Moore RJ. 2010. Association between avian necrotic enteritis and Clostridium perfringens strains expressing NetB toxin. Vet. Res. 41:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Petit L, Gibert M, Popoff MR. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7:104–110 [DOI] [PubMed] [Google Scholar]
- 5. Bannam TL, Yan XX, Harrison PF, Seemann T, Keyburn AL, Stubenrauch C, Weeramantri LH, Cheung JK, McClane BA, Boyce JD, Moore RJ, Rood JI. 2011. Necrotic enteritis-derived Clostridium perfringens strain with three closely related independently conjugative toxin and antibiotic resistance plasmids. mBio 2(5):e00190-11 doi:10.1128/mBio.00190-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brynestad S, Sarker MR, McClane BA, Granum PE, Rood JI. 2001. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect. Immun. 69:3483–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Katayama S, Dupuy B, Daube G, China B, Cole ST. 1996. Genome mapping of Clostridium perfringens strains with I-CeuI shows many virulence genes to be plasmid-borne. Mol. Gen. Genet. 251:720–726 [DOI] [PubMed] [Google Scholar]
- 8. Hughes ML, Poon R, Adams V, Sayeed S, Saputo J, Uzal FA, McClane BA, Rood JI. 2007. Epsilon-toxin plasmids of Clostridium perfringens type D are conjugative. J. Bacteriol. 189:7531–7538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sayeed S, Li J, McClane BA. 2007. Virulence plasmid diversity in Clostridium perfringens type D isolates. Infect. Immun. 75:2391–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lepp D, Roxas B, Parreira VR, Marri PR, Rosey EL, Gong J, Songer JG, Vedantam G, Prescott JF. 2010. Identification of novel pathogenicity loci in Clostridium perfringens strains that cause avian necrotic enteritis. PLoS One 5:e10795 doi:10.1371/journal.pone.0010795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nauerby B, Pedersen K, Madsen M. 2003. Analysis by pulsed-field gel electrophoresis of the genetic diversity among Clostridium perfringens isolates from chickens. Vet. Microbiol. 94:257–266 [DOI] [PubMed] [Google Scholar]
- 12. Gholamiandekhordi AR, Ducatelle R, Heyndrickx M, Haesebrouck F, Van Immerseel F. 2006. Molecular and phenotypical characterization of Clostridium perfringens isolates from poultry flocks with different disease status. Vet. Microbiol. 113:143–152 [DOI] [PubMed] [Google Scholar]
- 13. Hibberd MC, Neumann AP, Rehberger TG, Siragusa GR. 2011. Multilocus sequence typing subtypes of poultry Clostridium perfringens isolates demonstrate disease niche partitioning. J. Clin. Microbiol. 49:1556–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chalmers G, Bruce HL, Hunter DB, Parreira VR, Kulkarni RR, Jiang YF, Prescott JF, Boerlin P. 2008. Multilocus sequence typing analysis of Clostridium perfringens isolates from necrotic enteritis outbreaks in broiler chicken populations. J. Clin. Microbiol. 46:3957–3964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thompson DR, Parreira VR, Kulkarni RR, Prescott JF. 2006. Live attenuated vaccine-based control of necrotic enteritis of broiler chickens. Vet. Microbiol. 113:25–34 [DOI] [PubMed] [Google Scholar]
- 16. Wernersson R. 2009. Probe design for expression arrays using OligoWiz. Methods Mol. Biol. 529:23–36 [DOI] [PubMed] [Google Scholar]
- 17. Barbara AJ, Trinh HT, Glock RD, Songer JG. 2008. Necrotic enteritis-producing strains of Clostridium perfringens displace non-necrotic enteritis strains from the gut of chicks. Vet. Microbiol. 126:377–382 [DOI] [PubMed] [Google Scholar]
- 18. Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. 2006. GenePattern 2.0. Nat. Genet. 38:500–501 [DOI] [PubMed] [Google Scholar]
- 19. Kim C, Joyce E, Chan K, Falkow S. 2002. Improved analytical methods for microarray-based genome-composition analysis. Genome Biol. 3:RESEARCH0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carlson J, Heckerman D, Shani G. 2009. False discovery rate for 2×2 contingency tables. Microsoft, Mountain View, CA. [Google Scholar]
- 21. Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 22. Rozen S, Skaletsky H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365–386 [DOI] [PubMed] [Google Scholar]
- 23. Chalmers G, Martin SW, Hunter DB, Prescott JF, Weber LJ, Boerlin P. 2008. Genetic diversity of Clostridium perfringens isolated from healthy broiler chickens at a commercial farm. Vet. Microbiol. 127:116–127 [DOI] [PubMed] [Google Scholar]
- 24. Sawires YS, Songer JG. 2006. Clostridium perfringens: insight into virulence evolution and population structure. Anaerobe 12:23–43 [DOI] [PubMed] [Google Scholar]
- 25. Myers GSA, Rasko DA, Cheung JK, Ravel J, Seshadri R, DeBoy RT, Ren Q, Varga J, Awad MM, Brinkac LM, Daugherty SC, Haft DH, Dodson RJ, Madupu R, Nelson WC, Rosovitz MJ, Sullivan SA, Khouri H, Dimitrov GI, Watkins KL, Mulligan S, Benton J, Radune D, Fisher DJ, Atkins HS, Hiscox T, Jost BH, Billington SJ, Songer JG, McClane BA, Titball RW, Rood JI, Melville SB, Paulsen IT. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aziz R, Bartels D, Best A, DeJongh M, Disz T, Edwards R, Formsma K, Gerdes S, Glass E, Kubal M, Meyer F, Olsen G, Olson R, Osterman A, Overbeek R, McNeil L, Paarmann D, Paczian T, Parrello B, Pusch G, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75 doi:10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Janausch IG, Zientz E, Tran QH, Kröger A, Unden G. 2002. C4-dicarboxylate carriers and sensors in bacteria. Biochim. Biophys. Acta 1553:39–56 [DOI] [PubMed] [Google Scholar]
- 29. Sheng S, Cherniak R. 1997. Structure of the capsular polysaccharide of Clostridium perfringens Hobbs 10 determined by NMR spectroscopy. Carbohydr. Res. 305:65–72 [DOI] [PubMed] [Google Scholar]
- 30. Kalelkar S, Glushka J, van Halbeek H, Morris LC, Cherniak R. 1997. Structure of the capsular polysaccharide of Clostridium perfringens Hobbs 5 as determined by NMR spectroscopy. Carbohydr. Res. 299:119–128 [DOI] [PubMed] [Google Scholar]
- 31. Konto-Ghiorghi Y, Mairey E, Mallet A, Duménil G, Caliot E, Trieu-Cuot P, Dramsi S. 2009. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 5:e1000422 doi:10.1371/journal.ppat.1000422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smyth JA, Martin TG. 2010. Disease producing capability of netB positive isolates of Clostridium perfringens recovered from normal chickens and a cow, and netB positive and negative isolates from chickens with necrotic enteritis. Vet. Microbiol. 146:76–84 [DOI] [PubMed] [Google Scholar]
- 33. Salje J, Gayathri P, Löwe J. 2010. The ParMRC system: molecular mechanisms of plasmid segregation by actin-like filaments. Nat. Rev. Microbiol. 8:683–692 [DOI] [PubMed] [Google Scholar]
- 34. Cornillot E, Saint-Joanis B, Daube G, Katayama S-i, Granum PE, Canard B, Cole ST. 1995. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 15:639–647 [DOI] [PubMed] [Google Scholar]
- 35. Lahti P, Heikinheimo A, Johansson T, Korkeala H. 2008. Clostridium perfringens type A strains carrying a plasmid-borne enterotoxin gene (genotype IS1151-cpe or IS1470-like-cpe) as a common cause of food poisoning. J. Clin. Microbiol. 46:371–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Deguchi A, Miyamoto K, Kuwahara T, Miki Y, Kaneko I, Li J, McClane BA, Akimoto S. 2009. Genetic characterization of type A enterotoxigenic Clostridium perfringens strains. PLoS One 4:e5598 doi:10.1371/journal.pone.0005598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jost BH, Trinh HT, Songer JG. 2006. Clonal relationships among Clostridium perfringens of porcine origin as determined by multilocus sequence typing. Vet. Microbiol. 116:158–165 [DOI] [PubMed] [Google Scholar]
- 38. Brüssow H, Canchaya C, Hardt W-D. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martin TG, Smyth JA. 2009. Prevalence of netB among some clinical isolates of Clostridium perfringens from animals in the United States. Vet. Microbiol. 136:202–205 [DOI] [PubMed] [Google Scholar]
- 40. Abildgaard L, Sondergaard TE, Engberg RM, Schramm A, Højberg O. 2010. In vitro production of necrotic enteritis toxin B, NetB, by netB-positive and netB-negative Clostridium perfringens originating from healthy and diseased broiler chickens. Vet. Microbiol. 144:231–235 [DOI] [PubMed] [Google Scholar]
- 41. Sengupta M, Austin S. 2011. Prevalence and significance of plasmid maintenance functions in the virulence plasmids of pathogenic bacteria. Infect. Immun. 79:2502–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sherratt DJ, Arciszewska LK, Blakely G, Colloms S, Grant K, Leslie N, McCulloch R. 1995. Site-specific recombination and circular chromosome segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 347:37–42 [DOI] [PubMed] [Google Scholar]
- 43. Lin DCH, Grossman AD. 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92:675–685 [DOI] [PubMed] [Google Scholar]
- 44. Hester CM, Lutkenhaus J. 2007. Soj (ParA) DNA binding is mediated by conserved arginines and is essential for plasmid segregation. Proc. Natl. Acad. Sci. U. S. A. 104:20326–20331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yamaichi Y, Niki H. 2000. Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:14656–14661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dam M, Gerdes K. 1994. Partitioning of plasmid R1 Ten direct repeats flanking the parA promoter constitute a centromere-like partition site parC, that expresses incompatibility. J. Mol. Biol. 236:1289–1298 [DOI] [PubMed] [Google Scholar]
- 47. Lawrence JG. 2005. Common themes in the genome strategies of pathogens. Curr. Opin. Genet. Dev. 15:584–588 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.