Abstract
Y is the putative holin gene of the paradigm coliphage P2 and encodes a 93-amino-acid protein. Y is predicted to be an integral membrane protein that adopts an N-out C-in membrane topology with 3 transmembrane domains (TMDs) and a highly charged C-terminal cytoplasmic tail. The same features are observed in the canonical class I lambda holin, the S105 protein of phage lambda, which controls lysis by forming holes in the plasma membrane at a programmed time. S105 has been the subject of intensive genetic, cellular, and biochemical analyses. Although Y is not related to S105 in its primary structure, its characterization might prove useful in discerning the essential traits for holin function. Here, we used physiological and genetic approaches to show that Y exhibits the essential holin functional criteria, namely, allele-specific delayed-onset lethality and sensitivity to the energization of the membrane. Taken together, these results suggest that class I holins share a set of unusual features that are needed for their remarkable ability to program the end of the phage infection cycle with precise timing. However, Y holin function requires the integrity of its short cytoplasmic C-terminal domain, unlike for S105. Finally, instead of encoding a second translational product of Y as an antiholin, as shown for lambda S107, the P2 lysis cassette encodes another predicted membrane protein, LysA, which is shown here to have a Y-specific antiholin character.
INTRODUCTION
Holins are small phage-encoded membrane proteins that control the length of the bacteriophage infection cycle by determining the time of host lysis (1, 2). Almost all of the experimental work done on holin function at the physiological or molecular level has been focused on a few holin genes: lambda S, phage 21 S21, and T4 t. These genes encode the holins S105, S2168, and T, which are, respectively, the prototypes of three distinct holin classes, distinguished by their experimentally determined membrane topologies (Fig. 1A). S105 has class I topology, with three transmembrane domains (TMDs) arranged N-out C-in, whereas S2168 has class II topology, with two TMDs arranged N-in C-in. As of the last major review, ∼90% of the >100 genes from phages and prophages of eubacteria that have been assigned as putative holins fit into either class I or class II, based on predicted topology using widely accepted algorithms (1). These predicted holins are very diverse, with 12 and 14 unrelated sequence families grouped into classes I and II, respectively. Class III topology, with a single TMD and a large periplasmic domain, is restricted to the sequence homologs of protein T, the holin of T4. These proteins are found only in phages of the large myophage class of T4-like phages and in the large siphophage class of T5-like phages.
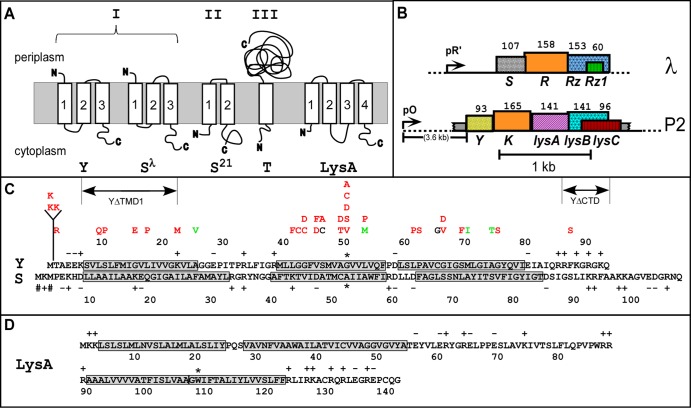
Fig 1.
Genes, sequences, and topologies. (A) Membrane topologies of P2 Y, λ S105, phage 21 S21, T4 T, and P2 LysA (see panel D). Three topology classes of the holins are indicated by Roman numerals. (B) The lysis cassettes of λ and P2. λ genes S, R, Rz, and Rz1 are shown along with late promoter pR′. P2 genes Y, K, lysA, lysB and lysC are shown along with late promoter pO. Genes are color coded according to their primary structure homology. Hatching pattern corresponds to homologous function. (C) Primary structures of Y (top) and S (bottom). Insertion, deletion, and missense changes relevant to the text are shown. Mutations are indicated above the Y sequence, color coded to phenotype: red = nonlytic; green = early lysis; black = normal lysis timing. Dual-start residues for S107 and S105 are indicated by # signs and the extent of the ΔTMD1 and ΔCTD deletions. Asterisks indicate the Ala52 residue in S105 and Gly52 in Y, chosen as analogous small residues in the central region of TMD2. (D) Primary structure and predicted topology of LysA. TMDs 1 and 2 were predicted by the algorithms TMHMM (27) and TMPRED (28). Both programs predicted a single TMD between 91 and 121. However, the homologous Y protein from a P2-like prophage in Enterobacter sp. 638 has a Lys residue at the position equivalent to Trp108 in Y, indicated by an asterisk, strongly suggesting that there are two TMDs in this hydrophobic stretch. This allows the highly basic N and C termini of LysA to be located in the cytoplasmic phase.
Despite this diversity, holins share some universal functional features, based mainly on extensive work that has been done with these three holin paradigms. Holin genes are turned on at the beginning of viral morphogenesis, so that the holins accumulate continuously and harmlessly in the cytoplasmic membrane while the viral structural proteins are synthesized and the virus particles are assembled. For the class I and II prototypes, S105 and S2168, the holins accumulate as harmless, mobile dimers in the membrane (3–6). There is no detectable effect on membrane integrity, respiration, proton motive force (PMF), or energy metabolism until there is a sudden irreversible holin-mediated halt of respiration, collapse of the PMF, release of small ions and small molecules, and loss of viability. This event is designated holin “triggering” (1, 2). For the class I and class II prototypes S105 and S2168, these effects have been shown to be caused by the sudden formation of lethal membrane lesions, or holes. Moreover, the triggering of these paradigm holins can be provoked prematurely by treatment with energy poisons, indicating that the timing of hole formation depends on the energization of the membrane. In the absence of holin function, lysis does not occur, so that virions and endolysins continue to accumulate indefinitely in the cytoplasm; the lysis defect is instantly relieved by the addition of chloroform, in effect constituting chemical complementation of the hole-forming function.
Despite these phenotypic commonalities, the prototype holins also exhibit unique features. The holes formed by S105 are known to be very large, capable of releasing molecules the size of the homotetrameric 0.5-MDa β-galactosidase enzyme in vivo (7). Purified S105 forms highly multimeric rings in detergent, estimated at >70 copies per ring (8). In contrast, the S2168 holin forms very small heptameric pores, estimated to be ca. ≤2 nm in diameter, both in vivo and, when purified, in detergent (9). The S105 protein is extremely sensitive to missense mutations within the TMDs, with single amino acid changes causing dramatic acceleration, retardation, or abolishment of the triggering time (10, 11). This has been interpreted as an indication that the timing of holin triggering involves packing of the TMD helices within the bilayer.
No other class I holin protein has been studied in detail, so it is unclear how many of the striking features of S105 are characteristic of other holins of this class, unrelated in terms of their sequence. Among the paradigm phages, coliphage P2 has a lysis cassette that includes a putative holin gene, Y, encoding a protein with predicted class I topology, adjacent to the gene for the endolysin, K (Fig. 1B). Ziermann et al. (12) constructed a P2 Yam mutant and showed that it exhibited a lysis defect that was suppressible by the addition of chloroform, indicating that Y functioned as the P2 holin. Here, we report the results of experiments designed to test this notion rigorously and to determine whether Y shares the extensively described physiological and biochemical features of the lambda S105 holin. The results are discussed in terms of a general model for holin topology and function.
MATERIALS AND METHODS
Materials, strains, bacteriophages, plasmids, and growth media.
The strains, phages, plasmids, and primers used in this study are listed and described in Table 1. The plasmids pYRRzRz1 (renamed pKT1-Y) and pYRamRzamRz1am (renamed pKT2-Y) are derivatives of the plasmid pS105, a medium-copy-number plasmid that has the entire λ lysis cassette under its native late promoter (13). The media, growth conditions, and thermal induction of the λ lysis genes from a prophage and/or plasmid have been described previously (13–15). All associated mutants were made by a commercially available error-prone PCR kit (Stratagene) and standard site-directed mutagenesis (14). Bacterial cultures were grown in standard LB medium supplemented with ampicillin (Amp) (100 μg/ml) and chloramphenicol (Cam) (10 μg/ml) (LBAmpCam) for the maintenance of plasmids and prophages, respectively.
Table 1.
Strains, phages, plasmids, and primers used in this study
| Phage, strain, plasmid, or primer | Genotypes and relevant features or sequence | Source or reference |
|---|---|---|
| Phages | ||
| λ Cmr Δ(SR) | stf::cat::tfa cI857 Δ(SR); replacement of stf and tfa genes (λ nt 19996–22220) with cat gene; Δ(SR)(29); deletion of λ nt 45136–45815 (10) | Laboratory stock |
| λ-S105 | cI857 SM1L; encodes S105 only | Laboratory stock |
| λ-Y | cI857; S105 deleted, replaced by Y (nt 6691–6997 of phage P2) | This study |
| Strains | ||
| MC4100 | E. coli K-12 F− araD139 Δ(argF-lac)U169 ΔfhuA rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 30 |
| XL1-Blue | E. coli K-12 recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15::Tn10 (Tetr)] | Stratagene |
| Plasmids | ||
| pS105 | λ lysis cassette under pR′ on pBR322 backbone; SM1L allele, encodes S105 only | 13 |
| pKT1-Y | Isogenic to pS105; S105 replaced by Y (nt 6691–6997 of phage P2) | This study |
| pKT2-Y | Isogenic to pKT1-Y; RT54am,K60am RzS100am RzlW38am | This study |
| pKT2-Yam | Isogenic to pKT2-Y; YS46am | This study |
| pKT2-S105 | Isogenic to pS105; RT54am,K60am RzS100am RzlW38am | This study |
| pKT1-YΔC-term | Isogenic to pKT1-Y; Arg86 to Gln93 deleted | This study |
| pKT1-YΔTMD1 | Isogenic to pKT1-Y; Ser7 to Lys22 deleted | This study |
| pKT1-Y+K | Isogenic to pKT1-Y; one Lys inserted after first Met | This study |
| pKT1-Y + 2K | Isogenic to pKT1-Y; two Lys inserted after first Met | This study |
| pLysA | Isogenic to pS105; S105RRzRz1 deleted and replaced by lysA (nt 7487 to 7934 of phage P2) | This study |
| pc-mycR::lacZ | mycR::lacZ replacement of luc in pZA32-luc | 7 |
| Primers | ||
| pKT1-Yfor | 5′GCGGTATTTCACACCGCACCTGGTGCACTCTCAG3′ | |
| pKT1-YRev | 5′CTGAGAGTGCACCAGGTGCGGTGTGAAATACCGC3′ |
YΔTMD1 or YΔC-term was expressed from pKT1-Y as mentioned above. The Y gene was replaced by the truncated YΔTMD1 gene (renamed pKT1-YΔTMD1) or truncated YΔC-term gene (renamed pKT1-YΔC-term). Either pKT1-YΔTMD1 or pKT1-YΔC-term was expressed in trans to full-length Y. Two lysogenic strains were used for the expression of Y: MC4100ΔfhuA(λCamΔSR) was used for the expression of Y and the lambda lysis genes R, Rz, and Rz1 from pKT1; and MC4100 (λ-Y) was used for the expression of Y and the lambda lysis genes R, Rz, and Rz1 from the integrated prophage. lysA was expressed from pS105 without the entire lysis cassette and under lambda native late promoter (renamed plysA). plysA was expressed in trans to MC4100 (λ-Y) and MC4100 (λ-S105). MC4100 (λ-S105) was used for the expression of S105 and the lambda lysis genes R, Rz, and Rz1 from the integrated prophage.
Mutagenesis of P2 Y.
Error-prone PCR with Mutazyme II DNA polymerase was performed using the GeneMorph II random mutagenesis kit (Stratagene) as recommended by the supplier. Each amplification reaction mixture (50 μl) contained 0.1 μM (each) of the primers pKT1-Yfor and pKT1-Yrev (Table 1), 40 μM (each) the deoxynucleoside triphosphates (dNTPs), 5 μl of 10× Mutazyme II buffer, 4.5 μg of pKT1-Y template, and 1 μl of Mutazyme II DNA polymerase. PCR was done with the following program: initial 5 min at 95°C, 20 cycles of amplification (30 s at 95°C, 30 s at 44°C, and 1 min at 72°C), and final 10 min hold at 72°C. PCR products were treated with DpnI, digested with NdeI and HindIII, which are the restriction sites flanking the Y gene, and then religated back into NdeI- and HindIII-digested pKT1-Y as described above. The ligated DNA was transformed into XL1-Blue, and the transformant colonies (∼500 colonies) were slurried and used to prepare a plasmid library. This DNA was transformed into the lysogenic host MC4100(λCmrΔ(SR)). The transformants were again slurried in 25 ml of LBAmpCam and aerated at 30°C. After standard thermal induction at A550 of ∼0.2 and lysis, the lysate was passed through a 0.45-μm filter to collect the cells that did not undergo lysis. The cells collected on the filter were resuspended in 5 ml LBAmpCam, and the resuspended cells were used to prepare plasmid DNA in a final volume of 50 μl. For each iteration of the screening process, 1 μl of this lysis-defective-enriched library was transformed into MC4100(λCmrΔ(SR)). Forty individual transformants were inoculated onto 5 ml of LBAmpCam and grown at 30°C in a roller drum until the cultures were in mid-logarithmic phase. Fifty microliters of each culture was reserved and then the tube cultures were transferred to a rack, shaken in a 42°C water bath for 15 min, and then shaken in a 37°C water bath for 65 min. Typically, ∼60% of the induced tube cultures underwent visible lysis. Nonlysed cultures were treated with 50 μl CHCl3 to test for endolysin accumulation. Typically, 90% of the nonlysing cultures lysed upon the addition of CHCl3, which is consistent with an absolute defect in the function of the Y holin. These presumptive Y mutants were grown to saturation from the reserved culture and were used for plasmid preparation and DNA sequencing. Of a total of 120 transformants, 72 exhibited an absolute lysis defect that was complemented by CHCl3. Of these, 64 transformants had at least one mutation in the Y gene. Of these, 43 had a single mutation, 15 had two mutations, and 6 had more than two mutations. Of the 72 mutations, two were −1 frameshifts, and the rest were single-base changes, approximately evenly distributed between transversions and transitions. All of the mutants that had a single nonsilent mutation are listed in Table 2. The mutant library is not saturated because only two of the 18 nonsense mutations accessible by a single base change were isolated.
Table 2.
Mutant alleles of Y isolated in this study
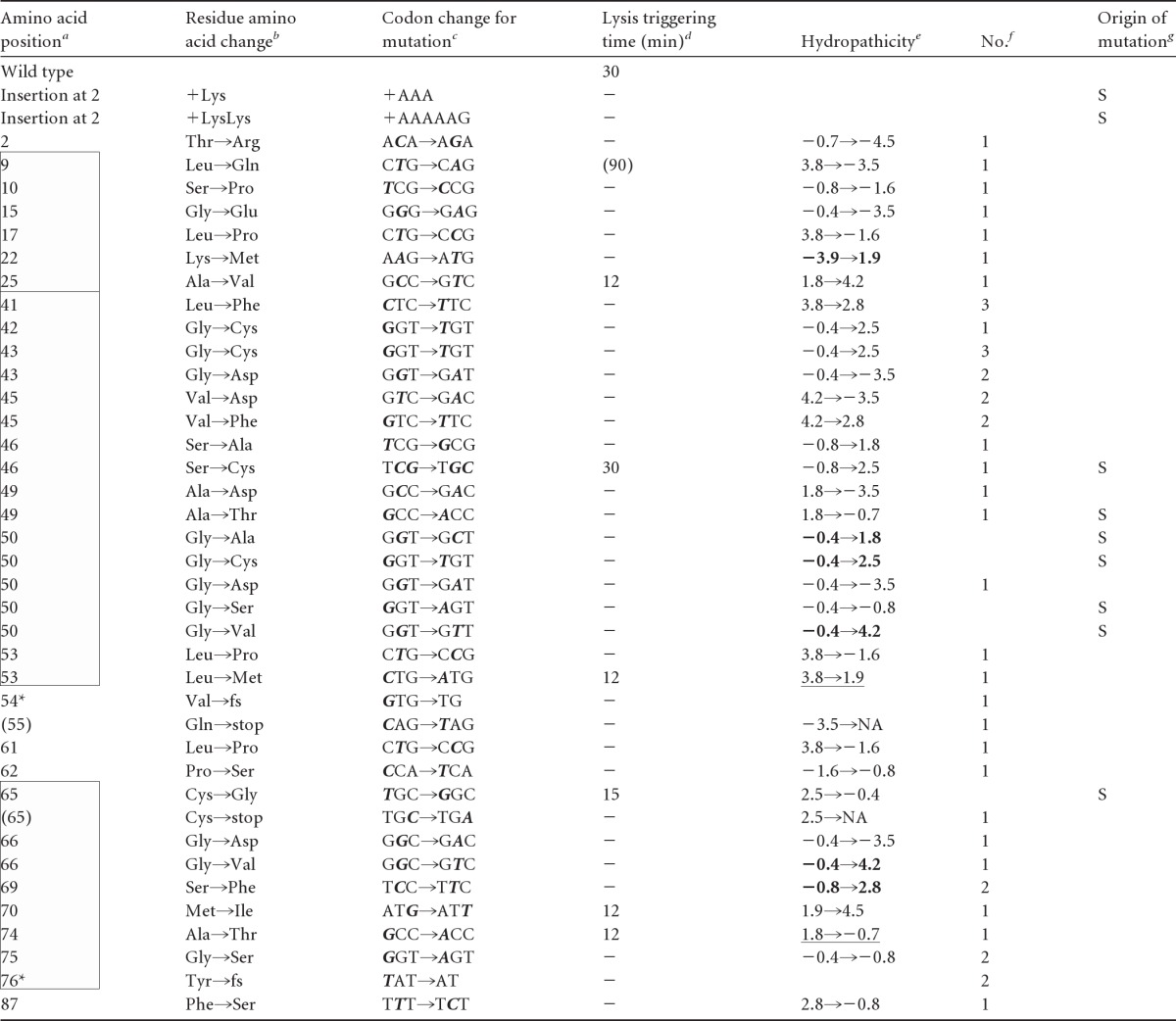
See Fig. 1C. All changes are missense changes except the two insertions (Lys and LysLys) at position 2, two frameshift mutants (denoted by asterisks), and two nonsense mutants (parentheses). Positions in TMDs 1, 2, and 3 (from top to bottom) are indicated by boxes.
fs, frameshift.
Altered bases are in bold italic type.
Triggering time in min after induction. Parentheses indicate gradual lysis. −, nonlytic.
Change in hydropathic character (31). Bold or underlined entries indicate increased hydrophobicity for lysis-defective alleles or decreased hydrophobicity for early-lysis alleles, respectively. NA, not applicable.
Number of times isolated for error-prone PCR mutants.
S, site-directed mutagenesis. All others were obtained by random error-prone PCR.
Growth conditions, inductions, and TCA precipitation.
For lysis curves and protein expression experiments, overnight cultures were used to inoculate 25-ml cultures of LB supplemented with the appropriate antibiotic. The cells were grown at 30°C to an A550 of ∼0.2 (for lysis curves) and an A550 of ∼0.4 (for protein expression experiments), thermally induced by aeration at 42°C for 15 min, and then returned to 37°C. A plaque-forming titer of progeny phage was determined as described previously (10). For protein expression experiments, 1-ml samples were removed from the growing cultures and added to 100% trichloroacetic acid (TCA) on ice. The samples were incubated for 30 min and centrifuged at maximum speed in a tabletop microcentrifuge at 4°C. The supernatant was removed and the pellet was resuspended in 1 ml cold acetone, vortexed, and spun at maximum speed in the tabletop microcentrifuge at 4°C. The acetone wash was repeated, and the supernatant was removed carefully after the final spin. The pellet was air-dried overnight and resuspended in 2× reducing sample loading buffer for SDS-PAGE. Western blotting was done with an antibody directed against the C terminus of Y (GenScript).
Antibodies.
Antibodies against the peptide CEIAIQRRFKGRGKQ corresponding to the C-terminal sequence of Y (Fig. 1C) were prepared and affinity purified by GenScript.
Site-specific photocrosslinking.
A 25-ml culture of MC4100ΔfhuA(λCamΔSR) carrying the desired plasmid was thermally induced and harvested at an A550 of ∼0.5 or 45 min after induction. One-milliliter samples of different Y alleles were collected by centrifugation, and the cell pellet was resuspended and washed twice in 1 ml phosphate-buffered saline (PBS) (pH 7.2), 2 mM EDTA (pH 8.0), and 1 mM phenylmethylsulfonyl fluoride (PMSF). The cell pellet was then resuspended in 200 μl PBS and pipetted into a sterile petri dish (15-mm diameter per well, 24 wells per dish), and 2 μl of 100 mM 4-maleimidobenzophenone (MBP) (Sigma) prepared in dimethylformamide, or 1 μl of dimethylformamide for the mock treatment, was added. The mixture was incubated at room temperature for 30 min in the dark, after which 2 μl 1 M dithiothreitol (DTT) was added to quench unreacted MBP. The samples were placed on ice and irradiated with 366-nm light using a handheld lamp for 30 min. Samples were mixed with 2× sample loading buffer and analyzed by SDS-PAGE and then by Western blotting using Y-specific antibody.
RESULTS
Y exhibits holin function.
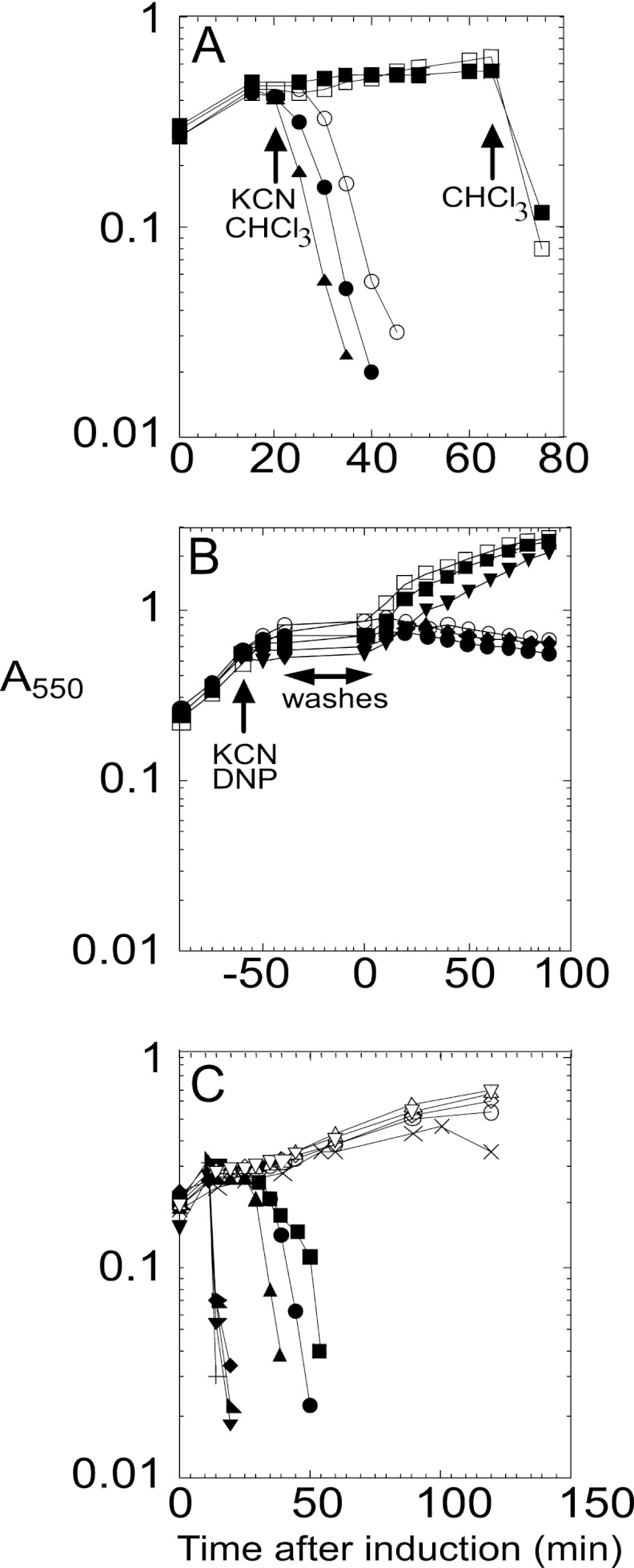
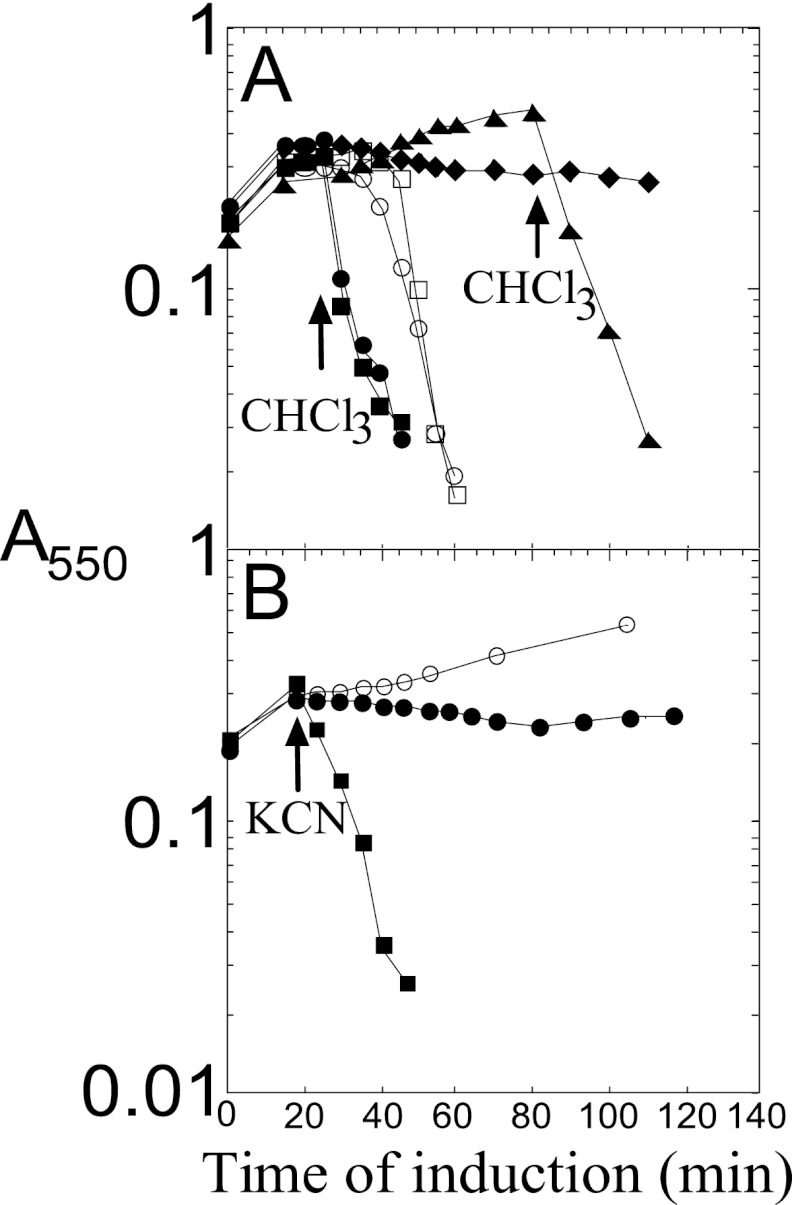
As a first step in characterizing the lytic function of P2 Y, we cloned the Y gene into plasmid pS105 (13), replacing the S105 gene in the lambda lysis cassette (Fig. 1B). Inserts in this plasmid are expressed in the proper temporal and transcriptional context by inducing a lysis-defective prophage, λcamΔSR, resulting in trans-activation of the plasmid-borne lambda late promoter, pR′, by the phage-encoded late gene activator, Q. The resulting hybrid lysis cassette, YRRzRz1, in plasmid pKT1-Y, was found to cause abrupt lysis ∼30 min after induction (Fig. 2A). Since R and RzRz1 are known to encode a soluble endolysin and a two-component spanin, respectively, this result strongly suggests that Y is a functional holin. However, inappropriate expression of a membrane protein or membrane-active toxins can suppress a holin lysis defect, presumably by nonspecific damage to the membrane, allowing the escape of endolysins that are sufficient to destroy the cell wall (12). To eliminate this possibility, cells induced for the Y-RRzRz1 chimera were tested for sensitivity to the addition of energy poisons; premature triggering was unambiguously demonstrated (Fig. 2A), as has been shown for the prototype holins of classes I, II, and III (16). Moreover, even in the absence of endolysin and spanin function, Y exhibits inducible lethality, and, like S105 and S2168, the addition of energy poisons triggers lethal function (Fig. 2B). Thus, Y exhibits all the physiological characteristics that are unique to the well-characterized holins.
Fig 2.
(A) P2 Y has lytic properties of a holin. Inductions: pKT1-Yam, with CHCl3 added at 65 min (□); pKT1-Yam, with cyanide added at 20 min and CHCl3 at 65 min (■); pKT1-Y, with CHCl3 added at 20 min (▲); pKT1-Y, with cyanide added at 20 min (●); pY (○). (B) Inducible lethality of P2 Y. Inductions: pKT2-Y (○); pKT2-Y, with 2,4-dinitrophenol (DNP) added at −60 min (●); pKT2-Y, with potassium cyanide (KCN) added at −60 min (◆); pKT2-Yam (□); pKT2-Yam, with DNP added at −60 min (■); pKT2-Yam, with KCN added at −60 min (▼). (C) Lysis phenotypes of P2 Y mutants. Inductions: pKT1-Y (▲), pKT1-YL9Q (×), pKT1-YL17P (○), pKT1-YK22M (△), pKT1-YA25V (+), pKT1-YS46C (●), pKT1-YA49T (♢), pKT1-YL53M (◆), pKT1-YC65G (■), pKT1-YM70I (▼), pKT1-YA74T (◣), and pKT1-YG75S (▽).
Single amino acid changes in Y have dramatic and unpredictable effects on lysis timing.
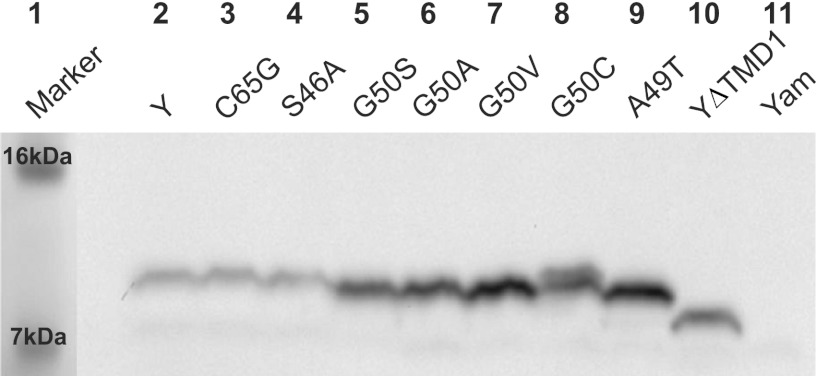
For the prototype holins of class I and II, a striking common feature has been that relatively conservative missense changes, especially in the TMD sequences, have led to dramatically altered lysis timing compared to that in the wild type (WT), either advancing or retarding the triggering time (4, 10, 11, 17, 18). To see if the same was true for Y, site-directed mutagenesis was used initially to probe key positions in the TMDs for timing phenotypes. In S105, alleles with missense changes at a central residue in TMD2, Ala52, block plaque formation for opposite reasons: S105A52V is completely nonlytic, whereas S105A52G causes lysis before the first virion is assembled (10, 11). An alignment of Y and S105 suggested that a residue analogous to Ala52 in S105 would be Gly50 in P2 Y (Fig. 1C, see asterisks). Indeed, the G50V mutation in Y eliminates lytic function without affecting accumulation, similar to the A52V mutation in S105; but, unlike in position 52 in S105, the substitution of small residues (Ala, Ser) in this position causes the loss of lytic function (Fig. 2C; Table 2). To address further the mutational sensitivity of the P2 holin timing function, a library of point mutations was generated by error-prone PCR and selected for either defective or early lysis. A summary of the timing phenotypes of all the site-directed mutants and mutants obtained from the PCR library is shown in Table 2 and Fig. 1C, and representative lysis phenotypes are illustrated in Fig. 2C. As in lambda S105, missense changes in all three TMDs were found to abolish lysis, including some relatively conservative mutations, like L41F, V45F, S46A, and G75S. Moreover, a number of missense changes led to early lysis phenotypes, including A25V, L53M, M70I, and A74T (Fig. 2C; Table 2). These timing phenotypes could not be explained by profound changes in the hydrophobic character of the substituted residues; both increases and decreases in hydrophobicity are associated with both early lysis and lysis-defective mutations (Table 2). The phenotypes also cannot be ascribed to a differential accumulation of Y protein; all the alleles supported normal accumulation of Y, as determined by Western blotting (Fig. 3). Thus, although the mutagenesis was not saturated, it was sufficient to show that P2 Y, like S105, appears to have the same extreme and unpredictable sensitivity to missense changes in the TMDs. Considering this with the detailed studies on lambda S105 and S2168, we conclude that the ability of conservative changes in TMD sequences to cause a profound alteration of the triggering time is a common feature of holin proteins.
Fig 3.
Y mutants accumulate normally. Lane 1, molecular mass marker; lane 2, pKT1-Y; lane 3, pKT1-YC65G; lane 4, pKT1-YS46A; lane 5, pKT1-YG50S; lane 6, pKT1-YG50A; lane 7, pKT1-YG50V; lane 8, pKT1-YG50C; lane 9, pKT1-YA49T; lane 10, pKT1-YΔTMD1; lane 11, pKT1-Yam.
Requirement for C-terminal cytoplasmic domain.
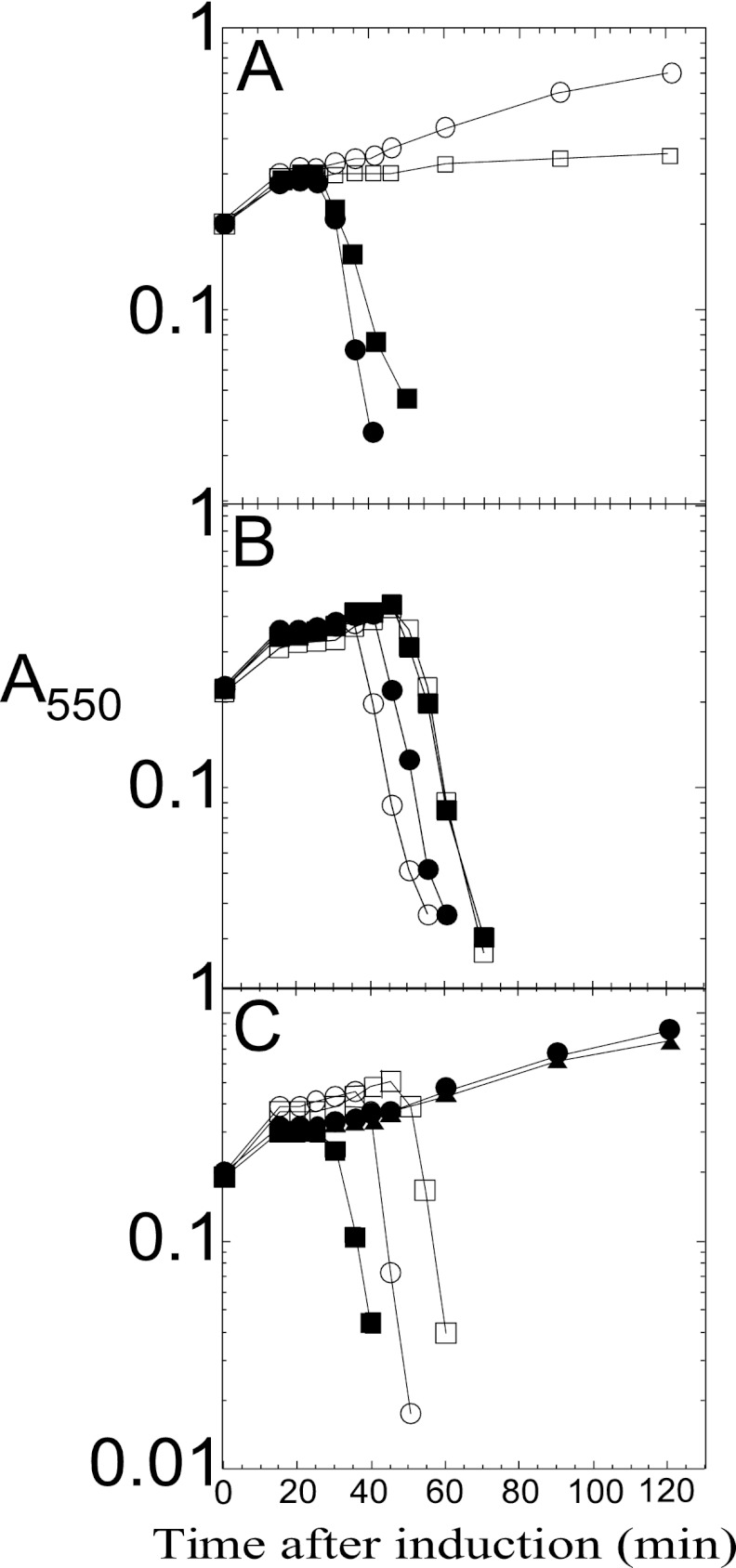
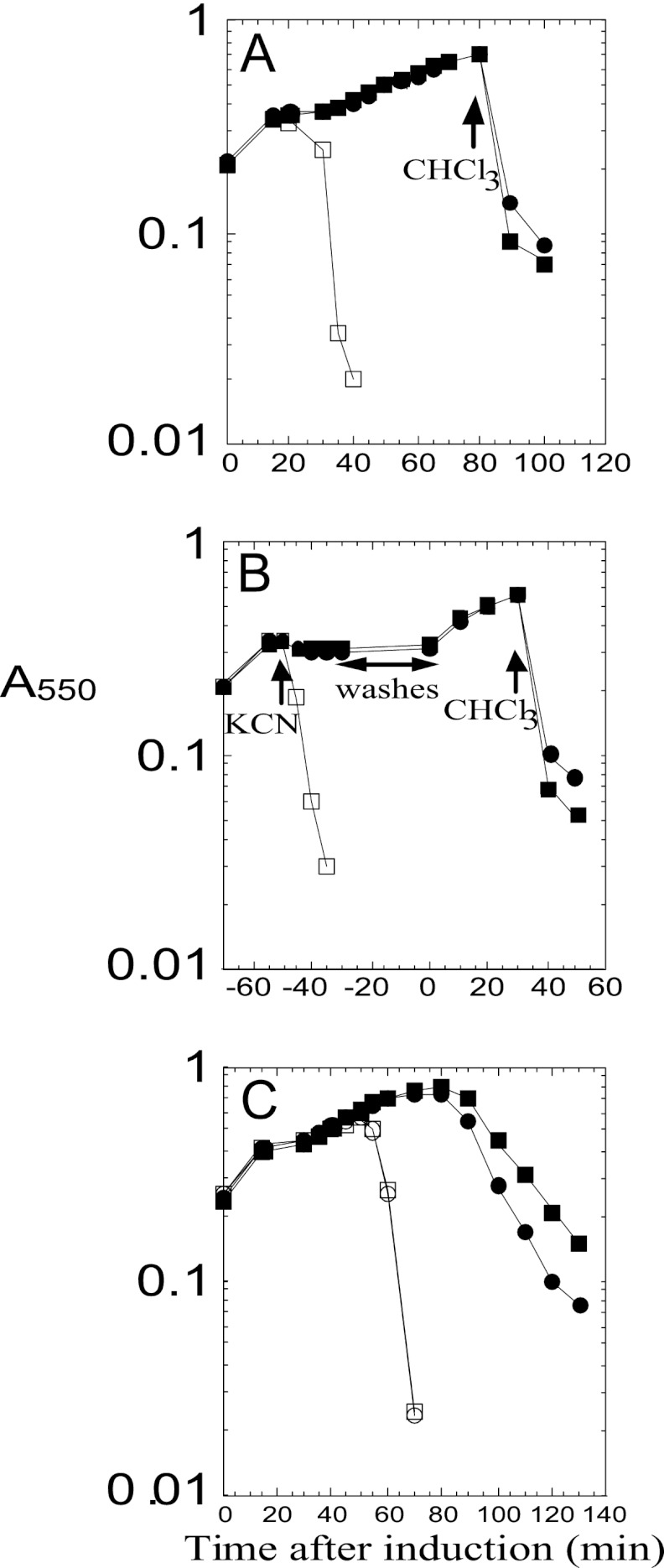
The most conserved feature of class I and II holins is that the most distal TMD is followed by a short C-terminal cytoplasmic tail enriched in basic residues (1). It has been reported that the cytoplasmic C-terminal domain (CTD) of lambda S105 is not necessary for host cell lysis, but instead plays a regulatory role in lysis timing (11, 19, 20). Site-directed mutagenesis experiments with S105 indicated that lytic function was retained as long as one positive-charge residue remained at the C terminus and that the timing of lysis was sensitive to the number of basic residues encoded in the C-terminal tail, with a more-positive charge being associated with delayed triggering time. We tested whether the highly charged C-terminal tail of Y served a similar function by constructing YΔCTD, in which the last 8 codons, Arg86 to Gln93, were deleted (Fig. 1C), so that the YΔCTD product has only a single basic residue at the C terminus, instead of five in the parental protein. Unlike the S105ΔCTD, which terminates with the single positively charged residue Arg93 and has an extreme early lysis phenotype (19), YΔCTD has an absolute lysis defect (Fig. 4A). Since the antibody against Y recognizes a C-terminal epitope, it was not possible to rule out the possibility that YΔCTD does not accumulate. However, in trans to the parental Y allele, YΔCTD is as functional as the WT gene (Fig. 4A). The simplest interpretation is that the C-terminal domain must be present in some n-mer state for Y to function but that not all protomers of the n-mer have to have the domain. In any case, we conclude that the C-terminal cytoplasmic domain of Y is essential for Y function in vivo, marking a distinct difference from the canonical class I holin, S105.
Fig 4.
(A) Deletion of the Y CTD inactivates holin function. Inductions in trans to λCamΔSR: no plasmid (○), pKT1-Y (●), pKT1-YΔC-term (□), and λ-Y lysogen harboring pKT1-YΔC-term (■). (B) LysA delays Y-mediated lysis. Inductions in trans to λ-Y: no plasmid (○) and plysA (●). Inductions in trans to λ-S105: no plasmid (□) and plysA (■). (C) YΔTMD1 delays Y-mediated lysis. Inductions in trans to λCamΔSR: no plasmid (●), pKT1-YΔTMD1 (▲), and pKT1-Y (■). Inductions in trans to λ-Y: no plasmid (○) and pKT1-YΔTMD1 (□).
Sizing the Y hole.
Early in the molecular characterization of the canonical class I holin S105, it was shown that the holes were lethal and nonspecific for endolysins. An indication of the size of these holes was obtained using fusion constructs in which the lambda R endolysin gene was fused to the full-length lacZ gene, thus creating a chimera of >1,100 codons encoding a hybrid R-βgal enzyme (7). In addition, the chimeric gene was modified to encode an N-terminal c-myc tag, to allow for monitoring of the production and stability of the hybrid protein. When this fusion gene was substituted for the parental R gene, the lytic and plating capacities of the phage were unaffected. Moreover, it was demonstrated not only that R-βgal was produced as an uncleaved stable 120-kDa polypeptide but also that the hybrid protein was fully active as an endolysin and also as a homotetrameric β-galactosidase of ∼0.5 MDa. To ask whether Y also formed holes of such capacity, plasmids encoding the c-mycR-lacZ fusion were induced in both S+ R− and Y+ R− contexts (Fig. 5A). The results show unambiguously that the Y holin is permissive for the R-βgal hybrid protein, suggesting that the nonspecificity of the Y hole is, like S105, due to a very large pore size in the membrane. However, when the Y holin was prematurely triggered by the addition of cyanide, lysis was observed with the WT R endolysin but not the R-βgal fusion (Fig. 5B). This indicates that the holes formed by premature triggering are much smaller than those formed at the spontaneous triggering time, which is consistent with similar observations with the lambda holin (7).
Fig 5.
(A) Sizing the Y hole. Inductions in trans to λKanΔ(SR); pKT2-Y and pc-mycR::lacZ (○); pKT2-Y and pc-mycR::lacZ, with CHCl3 added at 25 min (●); pKT2-S105 and pc-mycR::lacZ (□); pKT2-S105 and pc-mycR::lacZ, with CHCl3 added at 25 min (■); pKT2-Y, with CHCl3 added at 80 min (◆) and pc-mycR::lacZ (▲). (B) Premature Y holes are permissive for R endolysin but not the endolysin-β-Gal hybrid. pKT2-Y and pc-mycR, with KCN added at 15 min (■); pKT2-Y and pc-mycR::lacZ (●); pKT2-Yam (○).
Lysis deficiency of Y mutants is associated with an oligomerization defect.
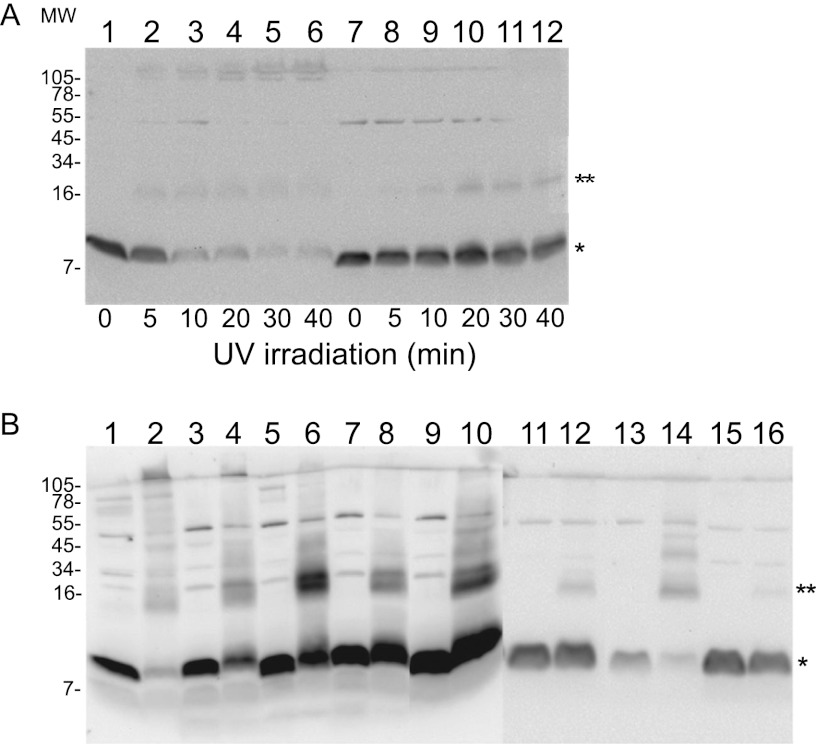
If membranes are treated with the membrane-permeable amine-specific cross-linker dithio-bis(succinimidyl propionate) (DSP), S105 can be trapped in covalent oligomeric states, up to at least n equal to 5; by this criterion, almost all lysis-defective S missense alleles are defective in oligomerization (14). To distinguish the molecular differences between the functional and nonfunctional alleles of Y, we attempted the same analysis, but treatment with DSP did not generate oligomeric species that were detectable by immunoblotting (data not shown). However, the presence of the single Cys residue at position 65 (Fig. 1C) made it possible to use the membrane-permeable photoactive heterobifunctional cross-linker MBP (21). When whole-cell samples that were treated with MBP and irradiated for increasing times were analyzed by immunoblotting, we observed that most of the Y protein became cross-linked in species that correspond to a dimer and also to high-order aggregates that are unable to enter the resolving gel, as is shown for the representative alleles in Fig. 6A. In contrast, when the same treatment was performed with cells expressing lysis-defective missense Y mutants, irradiation had a reduced or no effect on the amount of the monomeric species. The dimer species was more prevalent and the high-order aggregates were severely reduced or absent for the lysis-defective Y proteins (Fig. 6B). The Leu9Gln mutation exhibits the weakest molecular phenotype, showing only a reduced amount of the higher-order cross-linked species, which is consistent with the leaky lysis defect, with lysis beginning at about 90 min (Fig. 2C). The simplest interpretation is that Y undergoes dimerization and then higher-order oligomerization and that the mutants are blocked at different stages of oligomerization. Thus, for lethal function, Y shares with S105 the requirement for high-order oligomerization that is detectable by cross-linking (14).
Fig 6.
(A) Y, but not defective Y proteins, can be cross-linked into high-order oligomers. Photoactivated MBP cross-linking of whole-cell samples. Cells carrying either pKT2-Y (lanes 1 to 6) or pKT1-YL17P (lanes 7 to 12) were induced, labeled with MBP, and UV activated for the indicated lengths of time, in minutes, as described in Materials and Methods. (B) Lysis-defective Y proteins lose the ability to be cross-linked into high molecular weight (MW) species. Cultures carrying the indicated plasmids were induced, labeled with MBP, and UV-irradiated for 30 min (even-numbered lanes) or not (odd-numbered lanes). Lanes 1 and 2, Y; lanes 3 and 4, L9Q; lanes 5 and 6, L17P; lanes 7 and 8, A49T; lanes 9 and 10, G75S; lanes 11 and 12, K22M; lanes 13 and 14, L53P; lanes 15 and 16, G66D; monomer (asterisk), dimer (2 asterisks). Molecular weights are shown in thousands.
LysA exhibits Y-specific antiholin character.
For each of the prototype holins of classes I, II, and III, there is a specific antiholin. In the first two cases, the lambda S gene and the S21 gene of lambdoid phage 21 encode both the holin and antiholin, by virtue of alternate translational start codons (e.g., holin S105 and antiholin S107 from translational initiations at codons 3 and 1, respectively) (Fig. 1C). Such “dual-start” motifs are a very common feature in genes identified as encoding class I and class II holins (1, 22). In contrast, in phage T4, the holin T is inhibited by a specific antiholin encoded by gene rI. Moreover, genetic evidence also suggests that in phage P1, the holin LydA is inhibited by LydB, which is encoded by the adjacent gene (23–25). Since Y has only a single start codon, we wondered whether one of the genes in the P2 lysis cassette encoded an antiholin. The P2 lysis cassette is syntenic with the lambdoid lysis cassette, with holin, endolysin, and spanin genes arrayed in a cluster, except for a single gene, lysA, which is interposed between the endolysin gene K and the spanin gene lysBC (Fig. 1B). LysA is predicted to be an integral membrane protein with four TMDs (Fig. 1A and D). Indeed, Ziermann et al. (12) constructed P2lysAam and reported that cells infected with this construct underwent lysis ∼10 min earlier than cells infected with the isogenic WT phage. To test the hypothesis that LysA might serve as a specific antiholin for P2 Y, we constructed a plasmid that would express lysA from the λ late promoter in trans to either induced λ-Y or the isogenic parental λ. Induction of λ-Y lysogens transformed with the lysA construct resulted in lysis ∼10 min later than the same lysogen transformed with the isogenic vector (Fig. 4B), with an ∼50% increase in the yield of PFU (1.4 × 109 PFU/ml versus 9.3 × 108 PFU/ml) in the final lysate, without affecting the accumulation of Y (data not shown). These results indicate that LysA causes a delay in lysis by retarding the lethal action of Y and thus allows continued virion morphogenesis. In contrast, the lysA plasmid had no effect on lysis of the induced parental λ lysogen, demonstrating that the LysA-mediated inhibition of lysis is Y specific, supporting the hypothesis that LysA is the P2 antiholin.
TMD1 is essential for holin activity and can be converted into an inhibitory domain.
The S107 antiholin exerts its inhibitory function as a consequence of the ectopic localization of TMD1, which is prevented from entering the bilayer by virtue of the positively charged Lys residue at position two (Fig. 1C), which is missing in S107. Since Y does not have a dual-start motif and appears to have a cognate antiholin encoded by a distinct gene, lysA, we wondered if there was something intrinsic about the TMD1 of Y that prevents P2 from using the more genomically parsimonious dual-start motif for encoding antiholins. First, we tested whether TMD1 was essential for Y function by constructing an in-frame deletion of codons Ser7 to Lys22 (Fig. 1A and C). The YΔTMD1 allele proved to be nonlytic in the context of the phage induction (Fig. 4C), although the deletion product accumulated normally (Fig. 3). Thus, as in S105, TMD1 is essential for the lethal function of Y. Next, we tested whether Y might be converted to dual-start regulation by constructing Y alleles with one (Y+K) or two (Y+2K) extra Lys residues at the N terminus, making these the equivalent of the antiholin products of lambda S (S107, with one Lys residue at the N terminus) and the homologous P22 gene 13 (gp13108, with two Lys residues at the N terminus) (26). Both modified alleles exhibited an absolute lysis defect (Fig. 7A), although the gene products accumulated normally in the membrane (data not shown). Importantly, both modified Y alleles exhibited a specific antiholin phenotype, blocking the triggering of Y but not S105, when expressed in trans (Fig. 7C). As for S105, the simplest interpretation of these results is that an extra positively charged residue at the N terminus of Y prevents the entry of TMD1 into the membrane, creating a dominant negative mutant protein. Importantly, however, neither modified Y allele might be triggered by the depolarization of the membrane with cyanide, either to release endolysin or to form lethal lesions in the membrane (Fig. 7B), indicating that a dual-start motif might be necessary, but not sufficient, to generate a functional antiholin.
Fig 7.
(A) Y+K and Y+2K are absolute defective holins. Inductions: pKT1-Y (□); pKT1-Y+K, with CHCl3 added at 80 min (●); pKT1-Y+2K, with CHCl3 added at 80 min (■). (B) Y+K and Y+2K have no lethality. Inductions; pKT1-Y (□); pKT1-Y+K, with KCN added at −50 min and CHCl3 at 30 min (●); pKT1-Y+2K, with KCN added at −50 min and CHCl3 at 30 min (■). (C) Y+K and Y+2K are dominant negative and Y specific. Inductions: Lambda S105 lysogen harboring pKT1-Y+K (○) or pKT1-Y+2K (□). Lambda Y lysogen harboring pKT1-Y+K (●) or pKT1-Y+2K (■).
DISCUSSION
Prior to this work, only three holins have been studied in detail: S105, S2168, and T, the prototypes of topology classes I, II, and III, respectively. Of these, only the first two have been purified biochemically (8, 13), and only one, S105, is a canonical large-hole-forming holin that allows the release of a prefolded soluble endolysin. Class I holins are the most numerous in terms of holins that have been annotated in the database (1, 2), and thus, it was important to see if the many striking features of the well-studied lambda S105 were representative of this important topological class. The predicted membrane topology for Y is three TMDs with N-out C-in, as was demonstrated for S105. The results presented here show that some of these features are common to both S105 and Y and thus may be considered representative of type I holins as a whole:
Potential-sensitive lethality.
It was shown previously that the S105 holin exerted potential-sensitive lethality when induced, independent of the other proteins required for lysis, the endolysin and the spanin. The lethality is manifested as a sudden halt in culture growth after induction and is associated with a loss of viability, i.e., the triggering phenomenon. Y fully shares this characteristic (Fig. 2B). The potential sensitivity is reflected in the fact that the collapse of membrane energization with energy poisons causes premature triggering; Y also exhibits this feature (Fig. 2A). Since the accumulation of any membrane protein can lead to lethality and membrane dysfunction, the susceptibility of S105 and Y to premature lethal triggering by the addition of energy poisons can now be regarded as the definitive feature of holins, and especially the common class I holins.
Sensitivity of the timing function to conservative amino acid changes in the TMDs.
Perhaps the most striking aspect of S105 is its extreme sensitivity of the triggering time even to conservative missense substitutions, especially in the TMDs (10). For example, in S105, changes in a central residue in TMD2, Ala52, to Gly or Val both result in the loss of plaque-forming ability, but for opposite reasons (11). The Ala52Val protein accumulates in the membrane but never causes triggering, and thus plaque formation is abolished because lysis does not occur. In contrast, the Ala52Gly protein causes lysis at 20 min, at about the time that the first progeny virion appears in the cytoplasm; plaque formation is thus squelched because lysis is too early. Our initial attempts to see if Y exhibited similar phenotypic traits led us to do site-directed mutagenesis on the TMD2 residue predicted to be analogous to Ala52 (Fig. 1C). Although we were correct in judging Gly50 to be a residue important for lysis function, it turned out that all changes in this residue resulted in an absolute lysis-defective phenotype, including for Gly50Ala and Gly50Val, without affecting accumulation of the holin (Fig. 1C; Table 2). However, when we conducted random mutagenesis of Y, we were able to isolate both lysis-defective and early lysis alleles with rather conservative missense changes. For example, Leu41Phe, Val45Phe, Gly66Val, and Gly75Ser inactivate Y, whereas Ala25Val, Leu53Met, Cys65Gly, Ala75Thr, and Met79Ile cause extremely early lysis. Importantly, these changes in lysis timing are not associated with changes in protein accumulation or membrane localization for either S105 or Y (Fig. 3). It should be noted that early lysis phenotypes were also found for some missense and suppressed amber mutations in the TMDs of another putative class I holin, gpXXXV of the tectivirus phage PRD1, although no protein accumulation studies were done (18). Based on mutagenesis studies on the S gene, as well as studies with a green fluorescent protein (GFP)-tagged S105 holin, a model has been proposed in which the triggering time of S105 depends on the kinetics of formation of large two-dimensional arrays, or “rafts” of holin proteins. These rafts are proposed to exclude lipids as a result of intimate intermolecular helical packing interactions between the TMDs of the holin proteins. Conceptually, the exclusion of lipids from within the rafts would allow hole formation to occur by reorientation of the holin molecules, such that a relatively hydrophilic surface of one or more TMDs faces the lumen of the hole. In this scenario, the radical phenotypic variation observed for relatively conservative missense changes would derive from the disruption or enhancement of helix-helix packing in the raft state. The results reported that Y shares with S105 the same extreme sensitivity to missense changes within the TMDs; this provides additional support for this perspective and suggests that this model might be applicable to all class I holins.
Antiholin function and sensitivity of hole formation to the topology of TMD1.
The data presented here show that P2 LysA expressed in trans can retard Y triggering but not S105 triggering (Fig. 4B), indicating that LysA is the P2 antiholin. In this light, the rather weak lysis timing phenotype associated with P2lysAam, with only a 5- to 10-min early onset of lysis, is not unexpected. Similar small advances in triggering times were associated with the equivalent antiholin-defective mutants of S and S21, and T4 rI lysis timing is unaltered compared to the WT. It should be noted that in all three cases, overexpression of the antiholin resulted in a complete blockage of holin triggering. Repeated efforts to achieve a greater lysis delay using overexpression constructs for lysA in trans to the P2 lysis cassette were unsuccessful (data not shown). It is possible that a physiological signal is needed to activate LysA, as in the case of T4 RI during T4 lysis inhibition; efforts are under way to look for lysA missense mutants that are nonpermissive for lysis.
It is interesting that despite the identical topology, the fact that TMD1 is required for Y function and that the deletion of TMD1 creates a dominant negative allele, it was not possible to convert Y to a functional antiholin by adding one or two basic residues to the N terminus, as is the case in the S107 and gp13108 antiholins of λ and P22. Although the altered Y proteins had a Y-specific inhibitory character, collapsing the membrane potential did not rescue lytic function. Thus, the addition of a positively charged residue at the N terminus of TMD1 is not sufficient to generate a useful antiholin protein, which presumably must be both inhibitory during the latent period and activatable by membrane depolarization at the triggering time. The simplest interpretations are that the addition of the basic residues does not prevent the entry of TMD1 into the bilayer but abrogates Y function in another fashion or that the depolarization of the membrane does not restore proper TMD1 localization. Thus, it seems that Y would have to have more changes than just a dual start to encode a useful antiholin product, presumably accounting for the necessity of a distinct gene to encode an antiholin. The most striking difference between the TMD1s encoded by S and Y is that the former has the sequence KEQGIG, which is both hydrophilic and high in turn propensity, in the middle of the helix, where Y has the much more hydrophobic sequence GVLIVV. It will be of interest to select suppressors of the Y+K lysis defect to see if they confer intrinsic antiholin properties.
Involvement of the cytoplasmic C-terminal domain.
In S105, the cytoplasmic CTD is largely dispensable, requiring only that at least one positively charged residue, Lys92, be present, presumably to secure TMD3 in its proper topology. Indeed, shortening the C-terminal domain caused the lysis timing to become progressively earlier, suggesting that the highly positively charged cytoplasmic tail played a regulatory role, possibly by interacting with the anionic surface of the cytoplasmic membrane. However, in Y, similar deletions inactivate the holin function, suggesting that the C-terminal cytoplasmic domain of Y plays a direct role in hole formation, rather than a regulatory one. Taken together, this suggests that the dispensability and regulatory function of the cytoplasmic CTD cannot be taken as a general property of class I holins. In this light, it should be noted that the λ CTD (23 residues) is much longer than the Y CTD (14 residues); it will be interesting to see if the size difference is indicative of regulatory or hole-forming function.
Hole formation.
A key functional difference between holins and other proteins that are involved in the export of proteins across the bilayer is the size of the membrane channels involved. As noted above, in the case of S105, it was shown that a prefolded R-βgal chimera of nearly 0.5 MDa was fully functional as an endolysin. Here, we have shown that the same is true for P2 Y (Fig. 5A), indicating that this is a feature common to class I holins. Recently, cryo-electron microscopy (cryo-EM) and tomography have revealed that the S105 lesions might be visualized as interruptions in the cytoplasmic membrane. These lesions were massive and heterogeneous, with an average diameter of ca. ≥300 nm, with some extending to micron size. We presume that these interruptions constitute the large nonspecific holes required for the escape of the R-βgal chimera to the periplasm. Although the fine structure within the large interruptions is still unclear, the sizes observed are consistent with the notion that all of the holin proteins present at the time of triggering go into lining the holes. It will be important to conduct similar cryo-EM studies on Y and other holins to determine whether these lesions are a common feature of class I holins and, if so, how the holin molecules are arranged within them. In addition, the fact that both Y and S105 apparently form much smaller holes when triggered prematurely suggests that the surprising size of the holes, far bigger than what is needed to effect the release of the typical phage endolysin, is a consequence of the timing mechanism rather than an evolutionarily selected feature.
Oligomerization.
The results presented here show that the functional Y protein can be cross-linked into multimers in vivo, and that lysis-defective Y mutants show a defect in this property, which are features shared by S105 and S105 lysis-defective mutants, respectively. In addition, in both cases, a dimeric species can be generated by cross-linking, and this species can be detected irrespective of the lytic function of a Y or S allele. This suggests that one of the fundamental properties of class I holins is the propensity for homo-oligomerization. In addition, multiple lines of evidence indicate that for S105, a dimeric species appears to be the mobile intermediate in the lysis pathway that exists before triggering. The presence of the dimeric cross-linking product in both Y and Y mutants is at least consistent with the notion that this too is a common feature of class I holins.
ACKNOWLEDGMENTS
We thank the Young laboratory members, past and present, for their helpful criticisms and suggestions.
This work was supported by Public Health Service grant no. GM27099 to R.Y.
Footnotes
Published ahead of print 18 January 2013
REFERENCES
- 1. Wang IN, Smith DL, Young R. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799–825 [DOI] [PubMed] [Google Scholar]
- 2. Young R. 2002. Bacteriophage holins: deadly diversity. J. Mol. Microbiol. Biotechnol. 4:21–36 [PubMed] [Google Scholar]
- 3. Gründling A, Smith DL, Bläsi U, Young R. 2000. Dimerization between the holin and holin inhibitor of phage lambda. J. Bacteriol. 182:6075–6081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pang T, Park T, Young R. 2010. Mutational analysis of the S21 pinholin. Mol. Microbiol. 76:68–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pang T, Park T, Young R. 2010. Mapping the pinhole formation pathway of S21. Mol. Microbiol. 78:710–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. White R, Chiba S, Pang T, Dewey JS, Savva CG, Holzenburg A, Pogliano K, Young R. 2011. Holin triggering in real time. Proc. Natl. Acad. Sci. U. S. A. 108:798–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang IN, Deaton JF, Young R. 2003. Sizing the holin lesion with an endolysin–β-galactosidase fusion. J. Bacteriol. 185:779–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Savva CG, Dewey JS, Deaton JF, White RL, Struck DK, Holzenburg A, Young R. 2008. The holin of bacteriophage lambda forms rings with large diameter. Mol. Microbiol. 69:784–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pang T, Savva CG, Fleming KG, Struck DK, Young R. 2009. Structure of the lethal phage pinhole. Proc. Natl. Acad. Sci. U. S. A. 106:18966–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Raab R, Neal G, Sohaskey C, Smith J, Young R. 1988. Dominance in lambda S mutations and evidence for translational control. J. Mol. Biol. 199:95–105 [DOI] [PubMed] [Google Scholar]
- 11. Johnson-Boaz R, Chang CY, Young R. 1994. A dominant mutation in the bacteriophage lambda S gene causes premature lysis and an absolute defective plating phenotype. Mol. Microbiol. 13:495–504 [DOI] [PubMed] [Google Scholar]
- 12. Ziermann R, Bartlett B, Calendar R, Christie GE. 1994. Functions involved in bacteriophage P2-induced host cell lysis and identification of a new tail gene. J. Bacteriol. 176:4974–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith DL, Struck DK, Scholtz JM, Young R. 1998. Purification and biochemical characterization of the lambda holin. J. Bacteriol. 180:2531–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gründling A, Bläsi U, Young R. 2000. Genetic and biochemical analysis of dimer and oligomer interactions of the λ S holin. J. Bacteriol. 182:6082–6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chang CY, Nam K, Young R. 1995. S gene expression and the timing of lysis by bacteriophage lambda. J. Bacteriol. 177:3283–3294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Young R, Wang IN. 2006. Phage lysis, p 104–126 In Calendar R. (ed), The bacteriophages, 2nd ed Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 17. Gründling A, Bläsi U, Young R. 2000. Biochemical and genetic evidence for three transmembrane domains in the class I holin, lambda S. J. Biol. Chem. 275:769–776 [DOI] [PubMed] [Google Scholar]
- 18. Rydman PS, Bamford DH. 2003. Identification and mutational analysis of bacteriophage PRD1 holin protein P35. J. Bacteriol. 185:3795–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bläsi U, Fraisl P, Chang CY, Zhang N, Young R. 1999. The C-terminal sequence of the lambda holin constitutes a cytoplasmic regulatory domain. J. Bacteriol. 181:2922–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rietsch A, Fraisl P, Graschopf A, Bläsi U. 1997. The hydrophilic C-terminal part of the lambda S holin is non-essential for intermolecular interactions. FEMS Microbiol. Lett. 153:393–398 [DOI] [PubMed] [Google Scholar]
- 21. Tao T, Lamkin M, Scheiner CJ. 1985. The conformation of the C-terminal region of actin: a site-specific photocrosslinking study using benzophenone-4-maleimide. Arch. Biochem. Biophys. 240:627–634 [DOI] [PubMed] [Google Scholar]
- 22. Bläsi U, Young R. 1996. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol. Microbiol. 21:675–682 [DOI] [PubMed] [Google Scholar]
- 23. Walker JT, Walker DH., Jr 1980. Mutations in coliphage P1 affecting host cell lysis. J. Virol. 35:519–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yarmolinsky MB, Sternberg N, Calendar R. 1988. Bacteriophage P1, p 291–438 In Fraenkel-Conrat H, Wagner RR. (ed), The bacteriophages, vol 1 Plenum Press, New York, NY [Google Scholar]
- 25. Lobocka M, Rose DJ, Plunkett G, III, Rusin M, Samojedny A, Lehnherr H, Yarmolinsky MB, Blattner FR. 2004. Genome of bacteriophage P1. J. Bacteriol. 186:7032–7068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nam K, Bläsi U, Zagotta MT, Young R. 1990. Conservation of a dual-start motif in P22 lysis gene regulation. J. Bacteriol. 72:204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sonnhammer EL, von Heijne G, Krogh A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175–182 [PubMed] [Google Scholar]
- 28. Hofmann K, Stoffel W. 1993. TMbase — a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374:166 [Google Scholar]
- 29. Schweizer HP. 1990. The pUC18CM plasmids: a chloramphenicol resistance gene cassette for site-directed insertion and deletion mutagenesis in Escherichia coli. Biotechniques 8:612–613, 616 [PubMed] [Google Scholar]
- 30. Silhavy TJ, Berman ML, Enquist LW. 1984. Bacterial strains, p. xi–xiii In Silhavy TJ, Berman ML, Enquist LW. (ed), Experiments with gene fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31. Kyte J, Doolittle RF. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105–132 [DOI] [PubMed] [Google Scholar]