Abstract
RVB1/RVB2 (RuvBL1/RuvBL2 or pontin/reptin) are enigmatic AAA+ ATPase proteins that are present in multiple cellular complexes. Although they have been implicated in many cellular functions, the exact molecular function of RVB proteins in the various complexes is not clear. TIP60 complex (TIP60.com) is a tumor suppressor chromatin-remodeling complex containing RVB proteins. RVBs are required for the lysine acetyltransferase activity of TIP60.com but not for that of the pure recombinant TIP60 polypeptide. Here we describe two molecular functions of RVBs in TIP60.com. First, RVBs negate the repression of catalytic activity of TIP60 by another protein in TIP60.com, p400. RVBs competitively displace the SNF2 domain of p400 from the TIP60 polypeptide. In addition RVBs are also required for heat stability of TIP60.com by a p400-independent pathway. RVB1 and RVB2 are redundant with each other for these functions and do not require their ATPase activities. Thus, RVB proteins act as molecular adaptors that can substitute for one another to facilitate the optimal assembly, heat stability, and function of the TIP60 complex.
INTRODUCTION
RVB1 and RVB2 (also known as RuvBL1/RuvBL2 or pontin/reptin), two closely related proteins, belong to the AAA+ ATPase (ATPase associated with multiple activities) family, have an ATPase core domain with Walker A and Walker B motifs, and are conserved in different species (1, 2). We and others have identified RVB1 in various chromatin-remodeling complexes in humans and yeasts, such as TIP60.com, SWR1.com, and INO80.com (3, 4, 5, 6, 7, 8, 9, 10, 11). Besides the chromatin-remodeling complexes mentioned above, RVBs have also been identified in complex with other transcription activators, such as the RNA polymerase II holoenzyme, c-Myc, and β-catenin. RVBs have also been purified as part of the small nucleolar RNA (snoRNA) biogenesis machinery, annealing helicases such as ANH2 that are involved in DNA damage control, phosphatidylinositol 3-kinase-related kinase (PIKK) (e.g., ATM, ATR, mTOR, and DNA dependent protein kinase)-containing complexes, and the telomerase complex (reviewed in references 1, 12, and 13). Yeast RVBs have independently been discovered as part of an R2TP complex that interacts with the HSP90 chaperone and is involved in snoRNA biogenesis (14). Although they are widely present, the molecular function of RVBs in a complex is usually unclear even in cases where they are required for the activity of the complex. In one clear example, yeast RVBs are required for the proper assembly of functional INO80.com (7), as the lack of RVBs results in depletion of ARP5, an essential subunit of INO80.com, from the complex. The importance of RVBs in cancer has been highlighted by recent discoveries showing overexpression of the proteins in hepatocellular carcinomas (15) and an important role in the regulation of metastasis (16, 17).
TIP60.com is a multisubunit chromatin-remodeling complex that contains a histone (lysine) acetyltransferase activity (6, 18). It acts in transcription as a coactivator (19), as a repressor of viral and cellular genes (20, 21), and in the DNA damage response (22). TIP60 acetylates histones H2A and H4 and makes the chromatin environment favorable for transcription (18, 23, 24). TIP60 can also act as a repressor, but in this case acetylation of histones is recognized by a repressor complex (20, 21). TIP60.com has been implicated at two different steps of the DNA damage response pathway. Members of the phosphatidylinositol 3-kinase family such as ATR and ATM are at the apex of a cell's response to DNA damage. These kinases are activated upon damage to phosphorylate proteins that amplify the damage signal, leading to a cell cycle arrest that gives the cells time to recover from the damage or to cell death in case of irreparable DNA damage. TIP60.com interacts with ATM and acetylates it on lysine 3016, promoting the kinase activity of ATM (25, 26). TIP60.com is also involved in attenuating this damage response signal. Once the histone H2AX is phosphorylated on its C-terminal tail, TIP60.com is recruited to these sites of damage (25, 27, 28, 29, 30). This recruitment results in local acetylation of histones H2AX (H2A in yeast and H2A.v in Drosophila) and H4, leading to the local decondensation of chromatin, dephosphorylation of phospho-H2AX, and attenuation of the DNA damage signal (29, 30).
The activity of TIP60.com is modulated by many cellular factors. Sun et al. showed that trimethylation of histone H3 on lysine 9 activates TIP60.com both in vitro and in vivo (31). In addition, multiple subunits of the TIP60 complex are required for its optimal activity (29, 30). We had identified RVB1 as a positive cofactor of TIP60.com (29). Mammalian cells depleted for RVB1 show increased levels of phosphorylated H2AX, and this is due to the decreased activity of TIP60.com (29). p400, another subunit of TIP60.com, is known to both facilitate and antagonize the complex's function (32, 33, 34, 35). p400 is an SWI2/SNF2-related ATPase and has been implicated in the remodeling of chromatin by nucleosomal exchange (32).
Here we show that RVBs have at least two independent molecular roles in TIP60.com. They are involved in negating the inhibitory effect of p400 on TIP60.com and, independent of p400, are required for sustaining the activity of TIP60.com upon exposure to heat stress in vitro. Together with their molecular role in INO80.com (7), we now have two instances where RVBs act as molecular adaptors necessary for the proper assembly and functional stability of a multiprotein chromatin-remodeling complex. Intriguingly, although there are examples where RVB1 or RVB2 have antagonistic functions in vivo and examples where the Walker B motifs of RVB proteins are essential, RVB1 and RVB2 can substitute for each other in TIP60.com, and the ATPase activity of RVB1 or RVB2 is dispensable for supporting the catalytic activity of TIP60.com.
MATERIALS AND METHODS
Cell culture and siRNA transfection.
293T were cultured in Dulbecco modified Eagle medium (DMEM) containing 10% donor calf serum, whereas HCT116 p53−/− cells was cultured in McCoy's 5A medium containing 10% fetal bovine serum. Small interfering RNA (siRNA) transfection was carried out using 30 nM annealed siRNA duplex (Invitrogen) with Lipofectamine-2000 RNAi-max (Invitrogen) reagent following the manufacturer's instructions. Cells were split 1:2 after 48 h of transfection to maintain them in log phase and harvested after 72 h of transfection. The target sequences of the oligonucleotides used are as follows: siControl (GL2), CGTACGCGGAATACTTCGA; siRVB1-A, AATGGCTGGAAGAGCTGTC; siRVB2-B, ACACTGTGACTCTGTATAA; and sip400-A, GGATTCGCTTTCATTTAAT.
Plasmid constructs and transfections.
Construction of the human TIP60-expressing plasmid is described elsewhere (29). For expressing recombinant proteins, the gene of interest was cloned either in pET28a (TIP60) (29), pET30b (RVB1 and RVB2) (29), or pGEX-2T (p400 fragments) and transformed into BL21(De) cells. Short hairpin RNA (shRNA) expression vectors were purchased from Open Biosystems and modified by expressing the shRNA cassette from a cytomegalovirus (CMV)-driven promoter. Details about the shRNA construct can be provided upon request. 293T cells were transfected either with vector or plasmid expressing FLAG-TIP60 together with a plasmid expressing a short hairpin against RVB1 (shRVB1-A), p400 (shp400-A), or control hairpin (shControl). Puromycin was added 24 h after transfection, and cells were harvested after 48 h of drug selection.
Protein purification and glutathione S-transferase (GST) pulldown assay.
To purify His-tagged proteins, cells were grown at 37°C until the culture reached an optical density (OD) of 0.4. The expression of the recombinant protein was induced by treatment with isopropyl-β-d-thiogalactopyranoside (IPTG) (0.25 mM final concentration) for 16 h at 20°C. Pellets were resuspended in buffer H (50 mM Tris [pH 8.0], 150 mM NaCl, 10 mM imidazole, 10% glycerol, and protease inhibitor cocktail). Lysozyme (10 mg/ml) was added and incubated at 37°C for 30 min, followed by three 30-s sonication cycles at 30% output of a Branson microtip (3.2 mm) and Fisher model 500 Sonic Dismembrator. Triton X-100 (final concentration, 0.1%) was then added, and the slurry was rotated for an hour at 4°C. The soluble fraction was separated by spinning at 13,000 rpm for 15 min at 4°C and then incubated with a preequilibrated Ni-nitrilotriacetic acid (NTA) 50% slurry. Ni-NTA-bound proteins were washed 3 times with wash buffer (buffer H containing 40 mM imidazole) by rotating for 5 min at 4°C. The proteins were eluted in buffer H containing 250 mM imidazole by rotating at 4°C for 30 min and dialyzed in histone acetyltransferase (HAT) assay buffer for 4 h at 4°C. Proteins were aliquoted and stored at −80°C until further use.
For GST-tagged protein purification, lysates were prepared similarly to those for His-tagged protein, except that cells were resuspended in phosphate-buffered saline (PBS) and the soluble fraction was incubated with glutathione-agarose beads and eluted by incubating the beads with elution buffer (10 mM glutathione prepared in 50 mM Tris, pH 8.0) for 30 min at room temperature.
For GST pulldown assays, bacterial lysates were incubated at 4°C for an hour alone or in combination with indicated proteins, e.g., with or without recombinant TIP60 (rTIP60), rRVB1, or rRVB2. Where necessary, different amounts of bacterial lysate (without recombinant protein) were included to keep the total protein amount equal within each sample. The soluble fraction was separated by centrifugation at 13,000 rpm for 10 min at 4°C and incubated with glutathione-agarose beads for an hour. Proteins bound to glutathione-agarose beads were washed three times, first with 1 ml of PBS plus 0.2% Triton X-100, then with 1 ml of PBS plus 0.2% Triton X-100 plus 300 mM NaCl, and finally with 1 ml of PBS plus 0.2% Triton X-100 plus 450 mM NaCl. Proteins bound to the beads were eluted by adding 200 μl 2× SDS sample loading buffer.
Western blotting, immunoprecipitation, and antibodies.
For Western blotting, cells were lysed in IPH buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM EDTA, 0.5% vol/vol NP-40, 1 mM dithiothreitol, 20 mM NaF, and protease inhibitor cocktail [Sigma]) and sonicated three times for 15 s at 25% output using a Branson microtip (3.2 mm) and Fisher model 500 Sonic Dismembrator. For immunoprecipitation, anti-Flag M2 affinity gel (Sigma) was incubated with cell lysate overnight at 4°C. The beads were washed 3 times with 1 ml lysis buffer, and the bound proteins were eluted either by boiling in 2× SDS-PAGE sample buffer (for Western blotting) or by incubating with 100 μl of 3× Flag peptide (dissolved in HAT assay buffer at 100 ng/μl) for 30 min at 4°C.
The antibodies used were as follows: anti-His from Qiagen diluted 1:1,000 in 3% bovine serum albumin (BSA) prepared in PBS-Tween 20 (PBST), anti-AcH4 and anti-AcK from Millipore diluted 1:1,000 in 3% BSA prepared in PBST, anti-GST from Santa Cruz diluted 1:500 in 5% milk prepared in PBST, anti-Flag from Sigma diluted 1:500 in 5% milk prepared in PBST, anti-H2AX and anti-phospho-S139-H2AX from Cell Signaling diluted 1:1,000 in 3% BSA prepared in TBST, anti-p400 from Bethyl diluted 1:1,000 in 3% milk prepared in PBST, and anti-RVB1 (36) and anti-TIP60 (20) diluted 1:500 in 5% milk prepared in PBST.
HAT assay.
TIP60.com was immunoprecipitated as described above, washed twice with 1 ml HAT assay buffer (50 mM Tris [pH 8.0], 10% glycerol, 0.1 mM EDTA, 1 mM dithiothreitol [DTT]), and incubated in 50 μl of HAT assay buffer containing acetyl coenzyme A (acetyl-CoA) (100 μM) and core histone (0.5 μg) for 5, 10, or 20 min at 30°C before addition of 10 μl of 6× SDS sample loading buffer. Twenty microliters of the reaction mixture was separated by 15% SDS-PAGE, and acetylated proteins were visualized by Western blotting using either antiacetyllysine or anti-pan-acetylated histone H4 antibody.
For enzyme linked immunosorbent assay (ELISA)-based HAT assay, each well of a streptavidin-coated plate was precoated by adding 100 μl of 1 μg/ml reconstituted biotinylated histone H4 peptide. The plate was incubated for 30 min at room temperature and then washed 5 times with 200 μl of Tris-buffered saline (TBS). Blocking was performed by adding 200 μl of 3% BSA to each well and incubating for 30 min at 30°C, following which the plates were washed 5 times with TBS. HAT assay was performed by adding 50 μl HAT reaction cocktail per well at 30°C. After 30 min of incubation, the wells were washed 5 times with 200 μl TBS. One hundred microliters of a 1:250 dilution of ELISA-grade antiacetyllysine in 3% BSA-containing TBS was added and incubated for 1 h 30 min at room temperature. The wells were washed 5 times with TBS, and 100 μl of a 1:10,000 dilution of goat anti-rabbit IgG–horseradish peroxidase (HRP) conjugate in TBS containing 3% BSA was incubated for 30 min at room temperature. The wells were washed 3 times with 200 μl of TBS containing 0.1% (vol/vol) Tween 20 to enhance the signal-to-background ratio. One hundred microliters of tetramethylbenzidine (TMB) substrate mixture was added to each well and incubated in the dark for 10 min at room temperature, and then 50 μl of fresh 1 M sulfuric acid was added to each well to stop the HRP reaction. The plate was then read at wavelengths of 450 nm and 570 nm. The values at 570 nm were subtracted from those at 450 nm to remove any well-to-well variation.
Real-time PCR.
Total RNA was isolated with TRIzol reagent (Invitrogen) according to the manufacturer's instructions and used for cDNA synthesis with Superscript III (Invitrogen). The cDNAs were used as templates for real-time PCR using SYBR green PCR master mix (Applied Biosystems). The primers used for PCR were as follows: p400-F, AAAACCATCACACCTGCACA; p400-R, AGAGGTCGTCGTCGTCTGTT; ACTIN-F, CCAGATCATGTTTGAGACCTTCAAC; and ACTIN-R, CCAGAGGCGTACAGGGATAGC.
RESULTS
Recombinant RVBs rescue the HAT activity of TIP60.com isolated from RVB-depleted cells.
TIP60.com purified from RVB1-depleted cells has reduced HAT activity, and addition of rRVB1 in vitro rescued the HAT activity (29). We first tested whether RVB1 and RVB2 were redundant with each other in this function. In order to address this, TIP60.com was purified from either RVB1- or RVB2-depleted cell lysate. Knockdown of RVB1 or RVB2 decreased TIP60.com's activity (Fig. 1A and B). This is not surprising, because depletion of either of the RVB proteins destabilizes the other RVB protein in cells (37). Interestingly, addition of either rRVB1 or rRVB2 was able to restore the activity of TIP60.com extracted from RVB-depleted cells (Fig. 1C to F). These data suggest that RVB1 and RVB2 are redundant with each other in their ability to support the activity of TIP60.com. rRVB1 did not, however, stimulate the HAT activity of recombinant TIP60 (29), leading us to conclude that RVB proteins are required for the activity of TIP60 when it is associated with other cellular factors present in TIP60.com.
Fig 1.
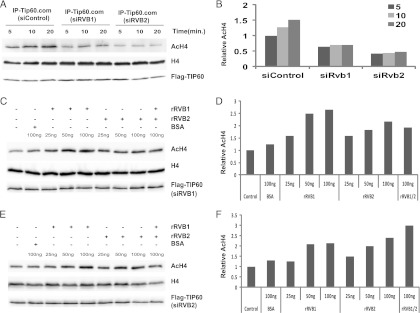
Recombinant RVBs (rRVBs) are able to restore TIP60.com's HAT activity. (A) Both RVB1 and RVB2 are required for TIP60.com's activity. In vitro HAT assay was performed on TIP60.com purified from either siControl-, siRVB1-, or siRVB2-treated cells. Immunoblotting of reaction products with the indicated antibodies is shown. (B) The graph shows the relative amount of acetylated H4 (AcH4) to total H4 as in panel A. (C and E) rRVB1 and rRVB2 can restore TIP60.com's HAT activity. TIP60.com was purified from RVB1 (C)- or RVB2 (E)-depleted cells. Equal amounts of TIP60.com were incubated with the indicated amount of recombinant proteins or BSA (negative control) and HAT assay performed for 20 min. The rest is as for panel A. (D and F) Relative AcH4 levels in panels C and E, respectively.
Depletion of p400 bypasses the need for RVB1 in the activity of TIP60.com.
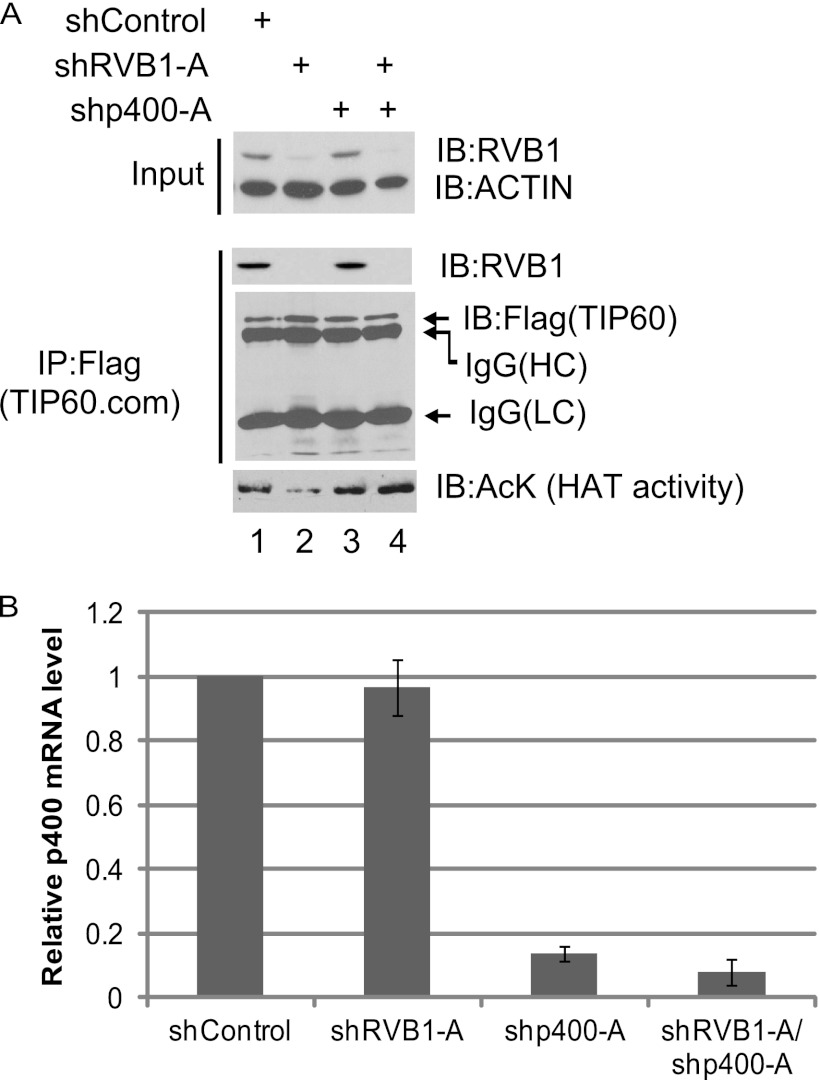
Of the many subunits in TIP60.com, p400 is known to antagonize TIP60.com's function both in vitro and in vivo (34, 35). In view of our data above, suggesting that RVB1 is required to antagonize the repression of TIP60 by a component of TIP60.com, we tested whether p400 was the relevant repressor. As expected, TIP60.com purified from cells that were depleted for RVB1 showed a decrease in HAT activity (Fig. 2A, compare lanes 1 and 2). Although depletion of p400 alone (Fig. 2A and B) does not have any effect on TIP60.com's HAT activity (Fig. 2A, compare lanes 1 and 3), it effectively bypasses the requirement of RVB1 for TIP60.com's HAT activity (Fig. 2A, compare lanes 2 and 4).
Fig 2.
RVB1 relieves p400-mediated inhibition of TIP60's activity in vivo. (A) Codepletion of p400 bypasses the requirement of RVB1 for TIP60.com's HAT activity. 293T cells were cotransfected with Flag-TIP60 or shControl/shRVB1-A/shp400-A-expressing plasmids. Flag-TIP60 complex was immunoprecipitated (IP) from the lysate and assayed for HAT activity as described in Materials and Methods. Depletion of RVB1 was detected using anti-RVB1 antibody. Immunoblotting (IB) with anti-Flag antibody shows the amounts of TIP60 in the lanes. Actin and IgG served as loading controls for input and immunoprecipitation, respectively. (B) p400 is depleted in cells expressing shp400-A plasmid. Total RNA was isolated, and the relative p400 mRNA measured by quantitative reverse transcription-PCR (Q-RT-PCR) and normalized to actin mRNA is shown. Results are means ± standard deviations (SD) from three experiments.
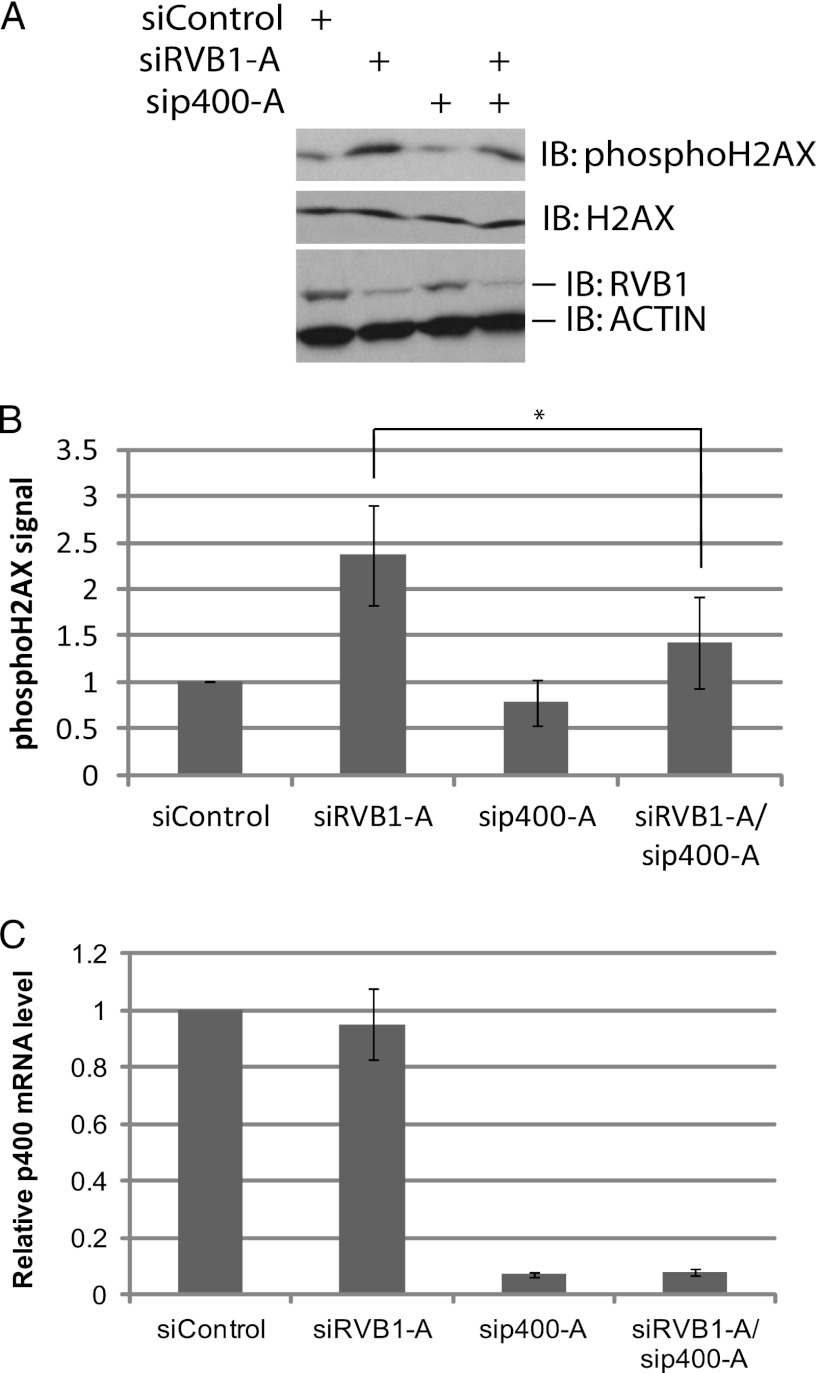
We and others have shown the requirement of TIP60.com in the DNA damage response (25, 28, 29, 30, 31, 38). TIP60.com downregulates phospho-H2AX, and RVB1 is required for execution of this function of the complex (29). We therefore tested whether the requirement of RVB1 for this function of TIP60.com in vivo can also be relieved by depleting p400. RVB1 was depleted from cells using siRNA, and the levels of phosphorylated H2AX were investigated as an in vivo read-out of functional TIP60.com (29, 30) (Fig. 3A and B). Depletion of RVB1 resulted in increased phospho-H2AX levels (Fig. 3A, lanes 1 and 2). Depletion of p400 alone did not affect phospho-H2AX levels (Fig. 3A, lanes 1 and 3, and C), but suppressed the increase of phospho-H2AX seen after depletion of RVB1 (Fig. 3A, lanes 2 and 4, and B). These results suggest that the decrease in catalytic activity of TIP60 in vivo seen upon depletion of RVB1 is significantly due to the presence of p400 in the TIP60.com. p400 has been identified as both an activator and a repressor of TIP60 in different contexts (33, 34), but in the absence of RVB, p400 appears to be mainly a repressor of the acetyltransferase.
Fig 3.
The increase in phospho-H2AX level after RVB1 knockdown is rescued by codepletion of p400. (A) Lysates from HCT116 p53−/− cells transfected with the indicated siRNA were harvested at 60 min after UV treatment. Cell lysates were resolved on a 15% SDS-polyacrylamide gel and immunoblotted with the indicated antibodies. (B) Quantitation of data in panel A. The phospho-H2AX signal was quantitated using ImageJ software. Means ± SD from three independent experiments are shown. *, P < 0.05 by Student's t test. (C) p400 mRNA is depleted after si-p400-A transfection. Total RNA was isolated, and relative p400 mRNA levels measured by Q-RT-PCR, and normalized p400 mRNA levels are shown. The rest is as for panel A.
Identifying the domain of p400 that interacts with TIP60.
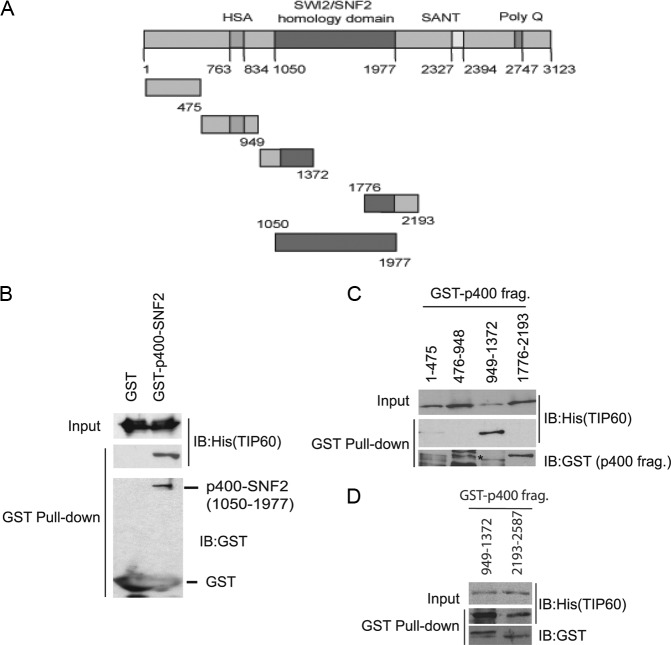
We next sought evidence for whether TIP60 can be inhibited in vitro by p400 and whether this inhibition can be reversed by RVBs. Since a 400-kDa protein is difficult to make in bacteria, we decided to make the regions of p400 that would specifically bind to TIP60 in vitro (Fig. 4A) (34). Among these, we decided to focus on the N-terminal region that was also shown to interact with RVB in a yeast two-hybrid screen (39). p400 fragments were cloned and expressed as GST-tagged proteins in bacteria. Interestingly, p400 interacted with TIP60 through its SNF2 domain (positions 1050 to 1977) (Fig. 4B). Further deletions identified the fragment from position 949 to 1372 of p400 [p400(949-1372)] as interacting specifically with TIP60, compared to GST alone or other p400 fragments (positions 1 to 475, 476 to 948, 1776 to 2193) (Fig. 4C). The interaction of TIP60 with p400(949-1372) appears to be stronger than that with a fragment containing the SANT domain [p400(2193-2587)] (Fig. 4D). These experiments suggest that TIP60 can bind directly to the SNF2 domain of p400 through the latter's 1050 to 1372 region and with lesser avidity with a fragment containing the SANT domain of p400. Because RVBs have been suggested to interact with the p400(951-1365) (39), we focused our subsequent experiments on p400(949-1372).
Fig 4.
A fragment of the SNF2 domain of p400 binds directly to recombinant TIP60. (A) Schematic representing different domains of p400. The indicated fragments of p400 were expressed as GST-tagged proteins. (B and C) p400 fragment(949-1372) interacts with rTIP60. The indicated GST proteins were incubated with rTIP60 in bacterial lysate for an hour at 4°C. The glutathione-agarose-bound proteins were immunoblotted with the indicated antibodies. Five percent of input is shown for each sample. The asterisk in the second lane of panel C indicates the size of the p400(476-948) fragment. (D) GST pulldown similar to that for panel B using p400(949-1372) or p400(2193-2587).
p400(949-1372) inhibits TIP60 activity in vitro.
We next tested whether the p400(949-1372), which interacts with TIP60, can also inhibit rTIP60's HAT activity. The in vitro HAT assay was performed in the presence of increasing amounts of purified p400(949-1372). rTIP60 plus GST or rTIP60 plus GST-p400 fragment were preincubated for 30 min at 30°C before the HAT-ELISA. The p400(1-475), which does not interact with TIP60, was used as a negative control (Fig. 5A). The p400(949-1372) but not p400(1-475) inhibited rTIP60's activity (Fig. 5B). These data suggest that p400(949-1372) associates with and inhibits TIP60's activity.
Fig 5.
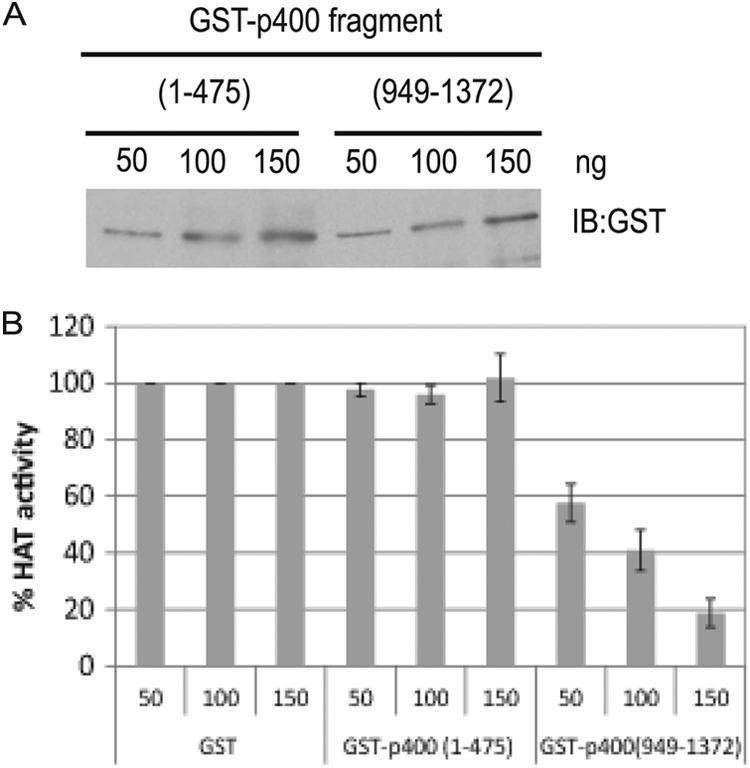
rTIP60's activity is inhibited by p400(949-1372). (A) Estimation of purified p400 fragments. The indicated amounts of purified p400 fragments [p400(1-475) or p400(949-1372)] were resolved and immunoblotted with anti-GST antibody. (B) p400(949-1372) inhibits rTIP60's HAT activity. An ELISA-based HAT assay was performed with rTIP60 in the presence of 100 ng of GST, GST-p400(1-475), or GST-p400(949-1372). The HAT activity is normalized to that seen in the presence of GST alone. Means ± SD from three independent experiments are shown.
RVBs bind to p400(949-1372) and antagonize the inhibitory effect on TIP60.
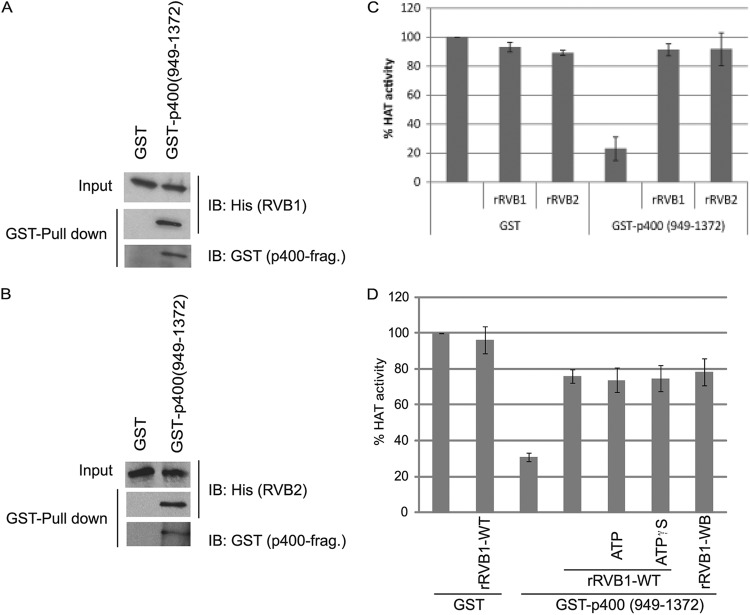
p400(951-1365) was found to coimmunoprecipitate with RVB from cell extracts (39), leading us to test whether recombinant RVB directly binds to the part consisting of positions 949 to 1372 of p400 (also made in bacteria), which interacts with and inhibits TIP60. In GST pulldown experiments, p400(949-1372) interacted with rRVBs (rRVB1 or rRVB2) (Fig. 6A and B). Notably, the RVBs also neutralized the inhibitory effect of p400(949-1372) on TIP60: addition of 100 ng of rGST-p400 fragment inhibited the HAT activity of rTIP60, and the presence of 100 ng of rRVB1 or rRVB2 attenuated this inhibition (Fig. 6C).
Fig 6.
Recombinant RVB interacts with p400 fragment and neutralizes its inhibitory effect on rTIP60. (A and B) p400(949-1372) interacts with rRVB1 and rRVB2 in a GST pulldown assay. Glutathione-agarose-bound proteins were immunoblotted with the indicated antibodies. (C) p400-mediated inhibition of rTIP60's HAT activity (as measured by an ELISA) can be reversed by both rRVB1 and rRVB2. Means ± SD of six measurements are shown. (D) GST-p400 fragment inhibits rTIP60's HAT activity and rRVB1 antagonizes its effect in the presence of nonhydrolyzable ATP or with a Walker B mutation. The indicated proteins [GST, GST-p400(949-1372), RVB1-WT (wild type), or RVB1-WB (Walker B mutant)] were incubated in an in vitro ELISA-HAT assay. A 10 μM concentration of either ATP or ATPγS was added where indicated. Means ± SD of four measurements are shown.
RVBs have Walker A and B motifs and are ATP binding ATPases, though the requirement of these motifs for RVB function appears to be context dependent (7, 40). There is at least one example where the ATPase activity of RVB was not required for its molecular function (7, 40). In the current study, we were curious to see whether the relief of TIP60 from inhibition by p400 required the ATPase function of RVB1. Addition of ATP did not stimulate, while addition of ATPγS (nonhydrolyzable ATP) did not inhibit, the derepression by RVB1 (Fig. 6D). Consistent with this, the Walker B mutant of RVB1 was also able to relieve the repression of TIP60 by p400 (Fig. 6D).
RVB1 competitively inhibits the interaction of p400(949-1372) with TIP60.
Next, we wanted to know whether this relief of inhibition was due to RVB1 competitively inhibiting the interaction of p400(949-1372) with rTIP60. To ensure that the interaction of p400(949-1372) with rTIP60 was near the top end of the linear range, we titrated the amount of rTIP60 lysate input into a reaction mixture with 450 ng of GST-p400 fragment (Fig. 7A). The results suggest that the 4× amount of rTIP60 gives binding near the top end of the linear range (Fig. 7A). Using these optimized conditions, we tested whether RVB1 can compete with TIP60 for binding with the p400 fragment. When added to the p400 fragment along with rTIP60, rRVB1 was able to displace the rTIP60 in a dose-dependent manner while itself binding to the p400 fragment (Fig. 7B and C). These data suggest that the portion of p400 consisting of positions 949 to 1372 binds to TIP60 to inhibit the latter's activity and that RVB1 relieves this inhibition by interfering with this particular p400-TIP60 interaction.
Fig 7.
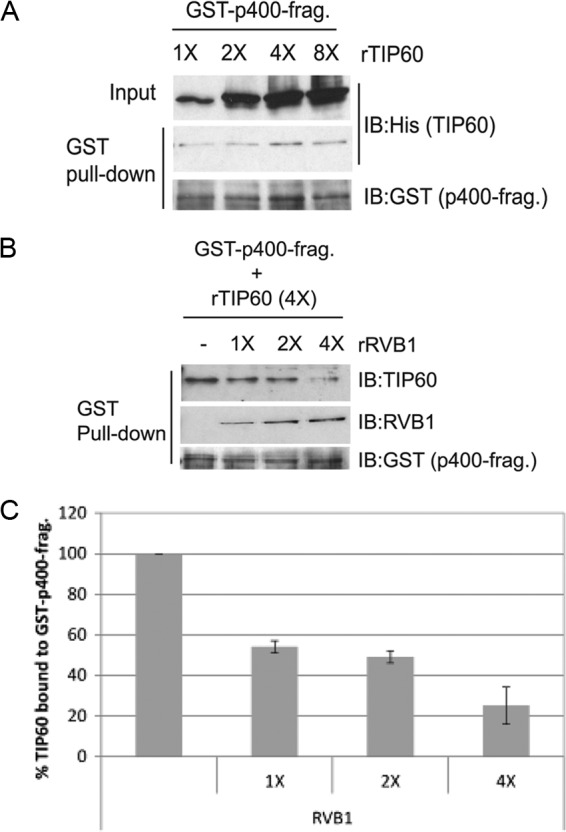
RVB1 competitively inhibits the binding of rTIP60 to GST-p400(949-1372). (A) rTIP60 binding to 450 ng GST-p400 fragment saturates when 4× TIP60-containing bacterial lysate (containing about 400 ng of TIP60) is added to the reaction mixture. Increasing amounts of rTIP60 lysate were incubated, and TIP60 binding to GST-p400 fragment was estimated by GST pulldown and immunoblotting. (B) Competition between rTIP60 and rRVB1 for binding to p400 fragment. Lysates containing 450 ng GST-p400 fragment, 400 ng rTIP60, and the indicated amounts of rRVB1 (1× = 50 ng) were incubated for an hour before pulldown on glutathione-agarose beads and immunoblotting with the indicated antibodies. (C) Quantitation of TIP60 bound to GST-p400 fragment to show competition by rRVB1. The level of TIP60 bound to GST-p400 fragment in the absence of RVB1 is 100%. Means ± SD from three independent experiments are shown.
RVB is required to stabilize TIP60.com activity following heat stress in vitro, and this activity is independent of p400.
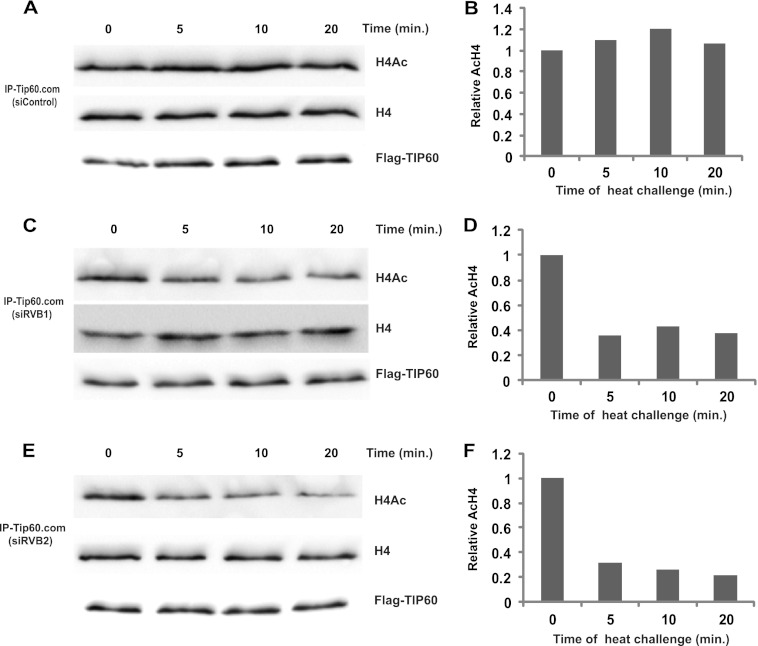
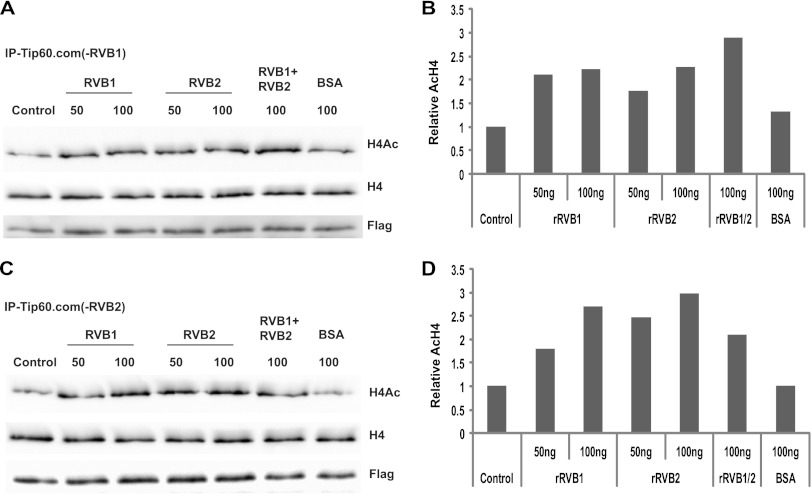
If RVB was required to facilitate the proper assembly of TIP60.com, we expected that loss of RVB would produce multiple deficits in the complex. Indeed, during our experiments we observed that TIP60.com depleted of RVB not only was less active at the first time point measured but also did not show any time-dependent increase in acetylation activity at subsequent time points compared to control TIP60.com (Fig. 1A and B). We hypothesized that RVB might be required for the stability of TIP60.com activity even under the reaction conditions in vitro. To seek evidence for such heat lability TIP60.com was immunoprecipitated from control (GL2) or RVB1- or RVB2-depleted cells and was preincubated at 30°C for various periods of time before addition of histone H4 to measure the acetylation activity over 20 min. While control TIP60.com retained its HAT activity despite 20 min of heat stress (Fig. 8A and B), knockdown of RVB1 (Fig. 8C and D) or RVB2 (Fig. 8E and F) made the HAT activity of TIP60.com labile to even 5 min of preincubation at 30°C. The TIP60 protein itself was not degraded by the preincubation, nor was such heat inactivation seen upon preincubation of purified recombinant TIP60 polypeptide (data not shown). Addition of rRVB1 or rRVB2, but not bovine serum albumin (BSA), to the RVB-depleted TIP60.com before preincubation stabilized the HAT activity of TIP60.com against heat stress (Fig. 9A to D).
Fig 8.
Depletion of RVBs makes TIP60 complex heat labile. Flag-TIP60.com purified from cells transfected with either siControl (GL2) (A and B), siRVB1 (C and D), or siRVB2 (E and F) was preincubated at 30°C for indicated minutes prior to an in vitro HAT assay on histone H4 for 20 min. Immunoblotting of reaction products with the indicated antibodies is shown. (A) Western blots show that the HAT activity of TIP60.com from siControl-transfected cells is stable to heat challenge. (B) The graph shows the relative amount of AcH4 relative to total H4 as in panel A, with 1 being the acetylation seen with 0 min of heat challenge. The x axis shows time of heat challenge in minutes as indicated. (C and E) Western blots for an experiment similar to that for panel A, except cells RVB1 and RVB2 depleted. (D and F) Graphs for AcH4 relative to total H4 in panels C and E, respectively.
Fig 9.
Recombinant RVB1 and RVB2 can stabilize TIP60.com in the face of heat challenge. Flag-TIP60.com was purified from RVB1- or RVB2-depleted cells. (A) rRVB1, rRVB2, or BSA was incubated with RVB1-depleted TIP60.com prior to 20 min of heat challenge at 30°C. In vitro acetylation assay was performed following this for 20 min at 30°C. Western blotting was performed with the indicated antibodies. (B) Amount of AcH4 relative to total H4 in panel A, normalized to that in the control (heat-inactivated but unrescued) lane. (C) Same as panel A except with RVB2-depleted TIP60 complex. (D) Quantitation of results in panel C as done for panel B.
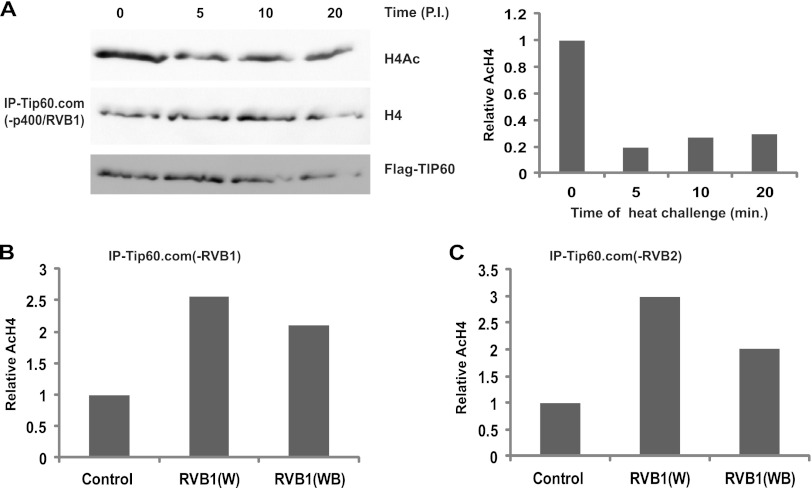
Since RVB proteins rescued TIP60.com from inhibition by p400, we were interested to check whether the heat lability of RVB-depleted TIP60.com is also due to the unfettered action of p400. RVB-depleted TIP60.com continued to be heat labile even when p400 was codepleted from the complex by siRNA against p400 (Fig. 10A), suggesting that the heat lability was not due to the inhibitory interaction of p400 with TIP60.com.
Fig 10.
Heat lability of RVB-depleted TIP60.com is not due to p400. (A) TIP60.com was purified from p400- and RVB1-depleted cells and preheated for the indicated times (minutes) prior to in vitro acetylation assay. Immunoblotting of reaction products with the indicated antibodies is shown. The graph shows the relative acetylation of H4 in each lane. (B) Restoration of heat stability by RVB1 regardless of whether RVB1 or RVB2 is depleted and independent of RVB1's ATP hydrolysis activity. TIP60.com was purified from RVB1-depleted cells and complemented with either wild-type RVB1 or Walker B-mutated RVB1 prior to heat inactivation. Acetylation of H4 in the subsequent in vitro acetylation assay is shown. (C) Same as panel B except with TIP60.com from RVB2-depleted cells.
To ascertain whether the ATPase activity of RVB was necessary for stabilizing the activity of TIP60.com against heat stress, we tested whether RVB1 without a functional Walker B motif could stabilize the HAT activity. Preincubation with the Walker B mutant of RVB1 successfully stabilized the HAT activity of TIP60.com from heat inactivation (Fig. 10B and C).
These results show that RVB proteins stabilize the activity of TIP60.com when challenged with heat stress in vitro and that this function of RVB is independent of p400 and does not require RVB's ATPase activity.
DISCUSSION
RVBs, members of AAA+ ATPase family, have been implicated in different cellular processes, such as the regulation of chromatin-remodeling complexes such as INO80.com and TIP60.com (1, 7, 29), small nucleolar RNA (snoRNA) biogenesis, Myc- and β-catenin-dependent pathways, and telomerase maturation (1). While RVBs have been implicated in many cellular processes, their exact molecular function is unclear. In the best example where the molecular function of RVBs has been elucidated, i.e., their role in yeast INO80.com, the RVBs were found to be essential for the recruitment of an essential component, ARP5, into the chromatin-remodeling complex. In this study, we demonstrate that RVBs have a similar role in the proper assembly of the TIP60 chromatin-remodeling and lysine acetyltransferase complex.
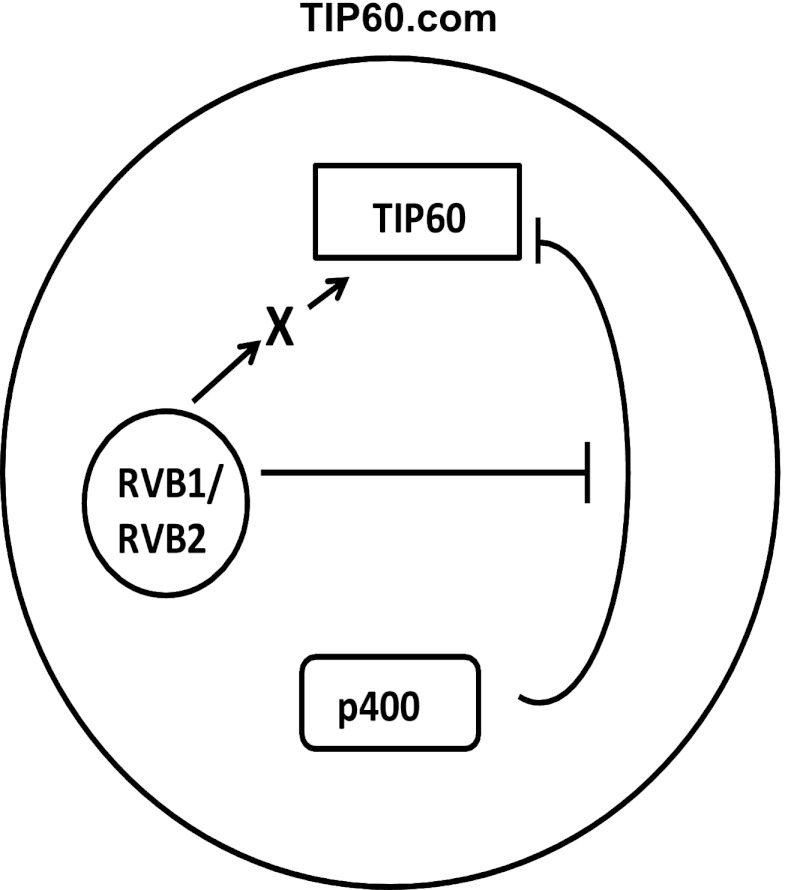
TIP60, a tumor suppressor and a lysine acetyltransferase (or histone acetyltransferase [HAT]), has been implicated in a number of biological processes (19, 22). Recently, two different cofactors have been identified for TIP60's HAT function. Sun et al. discovered that association of TIP60 with methylated histones activates TIP60, both in vitro and in vivo (31). Using a peptide library, the authors showed that addition of H3K9Me3 and H3K36Me3 peptide in the reaction mixture increases TIP60's HAT activity. In another study, Charvet et al. and Lin et al. showed that glycogen synthase kinase 3 (GSK-3) phosphorylates TIP60 on serine 86 and that this modification is required for TIP60's activity (41, 42). Our data show that a constitutive component of the TIP60.com, RVBs, is required to promote TIP60 activity by at least two mechanisms (Fig. 11). First, the SNF2 domain of p400, another constitutive component of TIP60.com, interacts with and represses the catalytic activity of the TIP60 polypeptide in the absence of RVBs. This repression is relieved by the addition of RVBs, at least partly because the RVBs interact with the SNF2 domain of p400 and interferes with the inhibitory interaction of p400 with TIP60. It is not clear that this constitutive function of RVB is actually used to regulate TIP60 activity under conditions when TIP60 activity is required, for example, after DNA damage, when TIP60 activity is required to activate checkpoint and proapoptotic pathways and later to limit the action of the checkpoint enzymes by promoting the dephosphorylation of phospho-H2AX. It is worth noting that p400 and RVBs coexist in TIP60.com and that there is at least another part of p400, i.e., the SANT domain, that interacts with TIP60 (34). Thus, we do not expect RVBs to completely displace p400 from TIP60.com or even from the TIP60 polypeptide. RVBs can fulfill their function by selectively interfering with the repressive interaction of the SNF2 domain of p400 with TIP60 while permitting other interactions. Second, consistent with the notion that RVBs act as a general adaptor or chaperone that is required for the proper assembly of the TIP60 complex, we discovered that RVBs are also required to stabilize the catalytic activity of TIP60.com when the complex is challenged with heat stress in vitro. This stabilization activity is not dependent on interfering with the action of p400 or on maintaining the physical integrity of the TIP60 polypeptide, but it is also not necessary when recombinant TIP60 polypeptide is challenged with heat. We conclude, therefore, that this second function of RVB in TIP60.com reflects a role of RVB in organizing the complex such that yet another (non-p400) component of the complex does not acquire an inhibitory conformation when the complex is preheated to 30°C for as short a time as 5 min. Both these molecular activities of RVBs are consistent with a role of these proteins as molecular adaptors or chaperones involved in proper assembly of TIP60.com.
Fig 11.
Putative model for the two roles of RVBs in TIP60.com. RVBs negate the inhibitory effect of p400 on TIP60 activity and are required to directly or indirectly stabilize the activity of TIP60.com when it is challenged by heat.
RVBs (RVB1 and RVB2) have a typical ATPase core domain with well-characterized Walker A and Walker B motifs that are essential for growth in yeast (36). However, there have been examples where the ATPase activity of RVB is dispensable for its function. For instance, a functional INO80.com was not dependent on the ATPase activity of RVBs (7). Loss of RVBs in yeast depleted ARP5 from INO80.com, but the Walker B mutant of RVB2 effectively recruited ARP5 into INO80.com (7). In contrast to this, the Walker motifs of RVBs are required for c-Myc-dependent transformation (40). It appears that RVB's requirement as an ATPase is context dependent. Our results suggest that hydrolysis of ATP is not necessary for RVB1 to relieve the repression of TIP60 by p400 (Fig. 6D) or to stabilize TIP60.com when it is challenged by heat in vitro (Fig. 10B and C). In our experience, the mutation in the Walker A motif of RVB1 decreased the yield of the protein by 10-fold or more, most likely because ATP binding is necessary for the proper folding of the protein. Because of the poor yield and insolubility of the Walker A mutant of RVB, we could not experimentally evaluate whether RVBs need to bind ATP to be functional in the two in vitro assays reported here.
Although RVB1 and RVB2 are complexed with each other in cells, there are several examples where the two proteins act independently of each other (43, 44, 45, 46). The rescue of TIP60.com's HAT activity by either rRVB1 or rRVB2, however, suggests that the two proteins can also function alone and are redundant with each other.
To summarize, we propose that TIP60's HAT activity in TIP60.com is dependent on RVBs, because RVBs are required to prevent the inhibition of the TIP60 polypeptide by p400. In addition, RVBs prevent the inhibition of TIP60 by other components of TIP60.com when the complex is challenged by heat in vitro. These molecular functions of RVBs are consistent with a role of these proteins as adaptors or chaperones that facilitate the proper assembly of TIP60.com.
ACKNOWLEDGMENTS
We thank the members of the Dutta laboratory for helpful comments.
This work was supported by grant RO1 GM084465 (to A.D.). This work was also supported by a start-up grant from the Cancer Science Institute of Singapore and the School of Medicine at the National University of Singapore (to S.J.). A.G. was supported by U.S. Army postdoctoral fellowship for breast cancer research DOD-W81XWH1110687.
Footnotes
Published ahead of print 7 January 2013
REFERENCES
- 1. Jha S, Dutta A. 2009. RVB1/RVB2: running rings around molecular biology. Mol. Cell 34: 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Torreira E, Jha S, López-Blanco JR, Arias-Palomo E, Chacón P, Cañas C, Ayora S, Dutta A, Llorca O. 2008. Architecture of the pontin/reptin complex, essential in the assembly of several macromolecular complexes. Structure 16: 1511–1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, Conaway JW. 2005. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 280: 13665–13670 [DOI] [PubMed] [Google Scholar]
- 4. Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. 2010. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J. Biol. Chem. 285: 4268–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cai Y, Jin J, Tomomori-Sato C, Sato S, Sorokina I, Parmely TJ, Conaway RC, Conaway JW. 2003. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J. Biol. Chem. 278: 42733–42736 [DOI] [PubMed] [Google Scholar]
- 6. Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102: 463–473 [DOI] [PubMed] [Google Scholar]
- 7. Jonsson ZO, Jha S, Wohlschlegel JA, Dutta A. 2004. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol. Cell 16: 465–477 [DOI] [PubMed] [Google Scholar]
- 8. Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348 [DOI] [PubMed] [Google Scholar]
- 9. Shen X, Mizuguchi G, Hamiche A, Wu C. 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406: 541–544 [DOI] [PubMed] [Google Scholar]
- 10. Shen X, Ranallo R, Choi E, Wu C. 2003. Involvement of actin-related proteins in ATP-dependent chromatin remodeling. Mol. Cell 12: 147–155 [DOI] [PubMed] [Google Scholar]
- 11. Shen X, Xiao H, Ranallo R, Wu WH, Wu C. 2003. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science 299: 112–114 [DOI] [PubMed] [Google Scholar]
- 12. Yuan J, Ghosal G, Chen J. 2012. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol. Cell 47: 410–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Izumi N, Yamashita A, Ohno S. 2012. Integrated regulation of PIKK-mediated stress responses by AAA+ proteins RUVBL1 and RUVBL2. Nucleus 3: 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao R, Kakihara Y, Gribun A, Huen J, Yang G, Khanna M, Costanzo M, Brost RL, Boone C, Hughes TR, Yip CM, Houry WA. 2008. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 180: 563–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haurie V, Ménard L, Nicou A, Touriol C, Metzler P, Fernandez J, Taras D, Lestienne P, Balabaud C, Bioulac-Sage P, Prats H, Zucman-Rossi J, Rosenbaum J. 2009. Adenosine triphosphatase pontin is overexpressed in hepatocellular carcinoma and coregulated with reptin through a new posttranslational mechanism. Hepatology 50: 1871–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kim JH, Choi HJ, Kim B, Kim MH, Lee JM, Kim IS, Lee MH, Choi SJ, Kim KI, Kim SI, Chung CH, Baek SH. 2006. Roles of sumoylation of a reptin chromatin-remodelling complex in cancer metastasis. Nat. Cell Biol. 8: 631–639 [DOI] [PubMed] [Google Scholar]
- 17. Huber O, Ménard L, Haurie V, Nicou A, Taras D, Rosenbaum J. 2008. Pontin and reptin, two related ATPases with multiple roles in cancer. Cancer Res. 68: 6873–6876 [DOI] [PubMed] [Google Scholar]
- 18. Doyon Y, Selleck W, Lane WS, Tan S, Cote J. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell. Biol. 24: 1884–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sapountzi V, Logan IR, Robson CN. 2006. Cellular functions of TIP60. Int. J. Biochem. Cell Biol. 38: 1496–1509 [DOI] [PubMed] [Google Scholar]
- 20. Jha S, Vande Pol S, Banerjee NS, Dutta AB, Chow LT, Dutta A. 2010. Destabilization of TIP60 by human papillomavirus E6 results in attenuation of TIP60-dependent transcriptional regulation and apoptotic pathway. Mol. Cell 38: 700–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gupta A, Jha S, Engel DA, Ornelles DA, Dutta A. 2012. TIP60 degradation by adenovirus relieves transcriptional repression of viral transcription activator EIA. Oncogene doi:10.1038/onc.2012.534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Squatrito M, Gorrini C, Amati B. 2006. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol. 16: 433–442 [DOI] [PubMed] [Google Scholar]
- 23. Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B. 2004. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol. Cell. Biol. 24: 4546–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4: 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun Y, Jiang X, Chen S, Fernandes N, Price BD. 2005. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc. Natl. Acad. Sci. U. S. A. 102: 13182–13187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sun Y, Xu Y, Roy K, Price BD. 2007. DNA damage-induced acetylation of lysine 3016 of ATM activates ATM kinase activity. Mol. Cell. Biol. 27: 8502–8509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419: 411–415 [DOI] [PubMed] [Google Scholar]
- 28. Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, Kimura H, Ikura M, Nishikubo S, Ito T, Muto A, Miyagawa K, Takeda S, Fishel R, Igarashi K, Kamiya K. 2007. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 27: 7028–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jha S, Shibata E, Dutta A. 2008. Human Rvb1/Tip49 is required for the histone acetyltransferase activity of Tip60/NuA4 and for the downregulation of phosphorylation on H2AX after DNA damage. Mol. Cell. Biol. 28: 2690–2700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, III, Abmayr SM, Washburn MP, Workman JL. 2004. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science 306: 2084–2087 [DOI] [PubMed] [Google Scholar]
- 31. Sun Y, Jiang X, Xu Y, Ayrapetov MK, Moreau LA, Whetstine JR, Price BD. 2009. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat. Cell Biol. 11: 1376–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. 2007. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 21: 1869–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD. 2010. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. J. Cell Biol. 191: 31–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Park JH, Sun XJ, Roeder RG. 2010. The SANT domain of p400 ATPase represses acetyltransferase activity and coactivator function of TIP60 in basal p21 gene expression. Mol. Cell. Biol. 30: 2750–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tyteca S, Vandromme M, Legube G, Chevillard-Briet M, Trouche D. 2006. Tip60 and p400 are both required for UV-induced apoptosis but play antagonistic roles in cell cycle progression. EMBO J. 25: 1680–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qiu XB, Lin YL, Thome KC, Pian P, Schlegel BP, Weremowicz S, Parvin JD, Dutta A. 1998. An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J. Biol. Chem. 273: 27786–27793 [DOI] [PubMed] [Google Scholar]
- 37. Venteicher AS, Meng Z, Mason PJ, Veenstra TD, Artandi SE. 2008. Identification of ATPases pontin and reptin as telomerase components essential for holoenzyme assembly. Cell 132: 945–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun Y, Jiang X, Price BD. 2010. Tip60: connecting chromatin to DNA damage signaling. Cell Cycle 9: 930–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fuchs M, Gerber J, Drapkin R, Sif S, Ikura T, Ogryzko V, Lane WS, Nakatani Y, Livingston DM. 2001. The p400 complex is an essential E1A transformation target. Cell 106: 297–307 [DOI] [PubMed] [Google Scholar]
- 40. Wood MA, McMahon SB, Cole MD. 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5: 321–330 [DOI] [PubMed] [Google Scholar]
- 41. Charvet C, Wissler M, Brauns-Schubert P, Wang SJ, Tang Y, Sigloch FC, Mellert H, Brandenburg M, Lindner SE, Breit B, Green DR, McMahon SB, Borner C, Gu W, Maurer U. 2011. Phosphorylation of Tip60 by GSK-3 determines the induction of PUMA and apoptosis by p53. Mol. Cell 42: 584–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, Zhang SM, Lian G, Liu Q, Ruan K, Wang Z, Zhang CS, Chien KY, Wu J, Li Q, Han J, Lin SC. 2012. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science 336: 477–481 [DOI] [PubMed] [Google Scholar]
- 43. Bauer A, Chauvet S, Huber O, Usseglio F, Rothbächer U, Aragnol D, Kemler R, Pradel J. 2000. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J. 19: 6121–6130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Diop SB, Bertaux K, Vasanthi D, Sarkeshik A, Goirand B, Aragnol D, Tolwinski NS, Cole MD, Pradel J, Yates JR, III, Mishra RK, Graba Y, Saurin AJ. 2008. Reptin and Pontin function antagonistically with PcG and TrxG complexes to mediate Hox gene control. EMBO Rep. 9: 260–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim JH, Kim B, Cai L, Choi HJ, Ohgi KA, Tran C, Chen C, Chung CH, Huber O, Rose DW, Sawyers CL, Rosenfeld MG, Baek SH. 2005. Transcriptional regulation of a metastasis suppressor gene by Tip60 and beta-catenin complexes. Nature 434: 921–926 [DOI] [PubMed] [Google Scholar]
- 46. Rottbauer W, Saurin AJ, Lickert H, Shen X, Burns CG, Wo ZG, Kemler R, Kingston R, Wu C, Fishman M. 2002. Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell 111: 661–672 [DOI] [PubMed] [Google Scholar]