Abstract
Acetylation of α-tubulin at lysine 40 (K40) is a well-conserved posttranslational modification that marks long-lived microtubules but has poorly understood functional significance. Recently, αTAT1, a member of the Gcn5-related N-acetyltransferase superfamily, has been identified as an α-tubulin acetyltransferase in ciliated organisms. Here, we explored the function of αTAT1 with the aim of understanding the consequences of αTAT1-mediated microtubule acetylation. We demonstrate that α-tubulin is the major target of αTAT1 but that αTAT1 also acetylates itself in a regulatory mechanism that is required for effective modification of tubulin. We further show that in mammalian cells, αTAT1 promotes microtubule destabilization and accelerates microtubule dynamics. Intriguingly, this effect persists in an αTAT1 mutant with no acetyltransferase activity, suggesting that interaction of αTAT1 with microtubules, rather than acetylation per se, is the critical factor regulating microtubule stability. Our data demonstrate that αTAT1 has cellular functions that extend beyond its classical enzymatic activity as an α-tubulin acetyltransferase.
INTRODUCTION
Posttranslation modification of tubulin is a key regulatory mechanism for adapting subsets of microtubules to specialized functions in eukaryotic cells (1). Acetylation of the ε-amino group of K40 in α-tubulin is one such modification, and since its first description in the flagella of Chlamydomonas (2, 3), a number of cellular events have been shown to be influenced by this process. They include accelerated kinesin-mediated transport along acetylated microtubule tracks (4); increased sensitivity of acetylated microtubules to severing by the protein katanin (5); and numerous biological processes, such as primary cilium disassembly, cell migration, and the maturation of cortical neurons (6–9). Intriguingly, α-tubulin acetylation accumulates in long-lived microtubules, such as those found in neuronal axons and cilia. Whether acetylation is also responsible for enhanced microtubule stability has attracted much debate.
Enzymes implicated in the regulation of microtubule acetylation include the deacetylases histone deacetylase 6 (HDAC6) and SirT2 (7, 10) and the acetyltransferases ARD1-NAT1, ELP3, San, and αTAT1 (8, 11–15). Of these, recent evidence suggests that αTAT1 is likely to function as the major α-tubulin acetyltransferase in Tetrahymena and Caenorhabditis elegans. Genetic ablation of αTAT1 genes abolishes α-tubulin K40 acetylation in these organisms and leads to reduced mechanosensitivity in C. elegans (13, 14, 16, 17). Furthermore, in mammalian cells, depletion of αTAT1 delays assembly of the primary cilium (14). It has recently been shown that some of these phenotypes do not depend on the α-tubulin acetyltransferase activity of αTAT1, although it is still not clear whether αTAT1 has further unknown functions on other proteins, different than α-tubulin (15).
In the present study, we sought to determine the specificity of αTAT1 for microtubules and investigated the relative contributions of αTAT1 and α-tubulin acetylation to microtubule stability. We found that, in addition to α-tubulin, αTAT1 acetylates itself and that this forms an important regulatory mechanism for its action at microtubules. We also examined the expression pattern of αTAT1 in mice and showed that it is particularly enriched in neuronal tissues. Moreover, in mammalian cells, we observed that overexpression of αTAT1 destabilizes microtubules and that this effect persists in a catalytically inactive mutant. Thus, αTAT1 has a conserved role as a microtubule-associated protein with enzymatic and nonenzymatic actions that may determine its function in vivo.
MATERIALS AND METHODS
Antibodies, plasmids, short hairpin RNA (shRNA), and recombinant protein.
Mouse monoclonal antibodies to α-tubulin (T5168; Sigma), acetylated α-tubulin (T7451; Sigma), and actin (MAB1501; Chemicon) were used. Rabbit monoclonal antibody was used for α-tubulin (2125; Cell Signaling). Rabbit polyclonal antibodies were used for panacetyllysine (AB3879 [Chemicon] and ICP0380 [Immunechem]), C6orf134/αTAT1 (ab58742; Abcam), and detyrosinated tubulin (AB3201; Millipore), as well as chicken polyclonal anti-green fluorescent protein (anti-GFP) (ab13970; Abcam). Secondary antibodies used for immunofluorescence were anti-rabbit, -chicken, or -mouse IgG–Alexa Fluor 488 (Invitrogen), anti-rabbit and -mouse IgG–Alexa Fluor 555 and 546 (Invitrogen), and anti-rabbit and -mouse IgG–Alexa Fluor 633 and 647 (Invitrogen). Secondary antibodies used for immunoblotting were horseradish peroxidase (HRP)-conjugated anti-rabbit and -mouse antibodies (NA931V and NA934V; GE Healthcare).
Mouse αTAT1 cDNA (ENSMUST00000056034) was used for all subcloning purposes. All αTAT1 variants (full length and mutants) were subcloned into the pEYFP vector and verified by sequencing. Point mutagenesis was carried out with the QuikChange II XL site-directed mutagenesis kit (Stratagene). Multiple shRNA plasmids (HuSH shRNA Plasmid Panels; Origene) were tested in knockdown experiments, and their effectiveness was quantified by reverse transcription (RT)-PCR and Western blotting against acetylated α-tubulin at various times after transfection. The best conditions were obtained with a combination of four shRNAs giving the strongest reduction of αTAT1 expression 3.5 days posttransfection. For recombinant protein production, αTAT1 was subcloned into the pET-14 vector, and subsequently, His-tagged αTAT1 was purified as previously described (18). For the glutathione S-transferase (GST) pulldown assays, αTAT1 was cloned into the pGEX-4T1 vector. Expression and extraction of proteins were performed as previously described (19).
Quantitative real-time PCR.
RNA from mouse tissues and NIH 3T3 cells was isolated using an RNeasy Micro kit (Qiagen) according to the manufacturer's instructions. mRNA was reverse transcribed by Superscript II reverse transcription (Invitrogen) using poly(T) primers. Quantitative PCR was performed on a LightCycler 480 PCR instrument (Roche) using SYBR green I master (Roche). The expression data were normalized to ubiquitin as a reference gene.
In situ hybridization.
Mouse tissues isolated from embryonic day 16.5 (E16.5) and adult wild-type C57BL/6 mice were fixed in 4% paraformaldehyde and embedded in paraffin. In situ hybridization was performed according to procedures previously described (20) and using a probe that targets the first seven exons that are common to all splice variants. The probe was cloned using the following primers: 5′ATGGAGTTCCCGTTCG and 5′GCTGATGGGCAAAG.
In vitro acetylation reaction.
Reactions were performed in 10 μl ADE buffer (14) with 1 μg bovine brain tubulin (TL238; Cytoskeleton), 5 μg recombinant αTAT1, and 8 μM acetyl coenzyme A (AcCoA) (A2056; Sigma) or [14C]AcCoA (NEC313N010UC; PerkinElmer) unless otherwise indicated. Reaction mixtures were incubated at 37°C for the time indicated and stopped by addition of SDS loading buffer. Gels were fixed, dried, and exposed for 3 days. The signal was quantified with ImageJ.
GST pulldown assays.
GST pulldown was performed as previously described (19). Specifically, GST fusion proteins and polymerized microtubules (TL238; Cytoskeleton) were allowed to form complexes for 1 h at 37°C. Pulldowns and washes were performed in the ADE buffer supplemented with 0.5% NP-40 and 200 mM KCl. After the elution, proteins were separated by SDS-PAGE and visualized by Western blotting with anti α-tubulin (T5168; Sigma) antibody.
Mass spectrometry.
The standard in vitro reaction (described above) was scaled up 40 times using [13C]AcCoA. In-solution digestion, enrichment of acetylated peptides, and mass spectrometry (MS) analysis were performed according to procedures previously described (21), with trypsin and Glu-C used for α-tubulin digestion and trypsin and chymotrypsin for αTAT1 digestion.
Transfection and immunocytochemistry.
Primary mouse dorsal root ganglion (DRG) neurons and CHO and NIH 3T3 cells were transfected using the Nucleofector system (Amaxa Biosystems) following the manufacturer's instructions. Unless otherwise indicated, experiments were conducted on cells 24 to 30 h after transfection. For the nocodazole-induced microtubule depolymerization test, cells were incubated in 10 μM nocodazole (M1404; Sigma) for the time indicated and then fixed. For trichostatin A (TSA) treatment, cells were incubated for 4 h with 5 μM trichostatin A (9950; Cell Signaling) before harvesting. For immunocytochemistry, cells were fixed in 4% paraformaldehyde in PBS for 15 min at room temperature, washed in PBS, and permeabilized with 0.05% Triton X. Nonspecific binding was blocked by incubation in 3% goat serum for 30 min. Incubations with primary antibodies were carried out overnight in 4% bovine serum albumin (BSA). Subsequently, coverslips were washed three times with PBS and incubated with a fluorescent secondary antibody for 2 h at room temperature. After three washes, the coverslips were mounted and examined on a Leica SP5 confocal microscope at room temperature using a 63× objective. The images were processed, and the fluorescent signal was quantified using ImageJ.
Live-cell imaging.
To record microtubule plus-end growth in live cells, NIH 3T3 cells were transfected with TagRFP-EB3 and yellow fluorescent protein (YFP)-αTAT1, YFP-αTAT1-GGL, αTAT1 shRNA, scrambled shRNA, or mock YFP using the Nucleofector system (Amaxa Biosystems). The cells were imaged 80 to 85 h after transfection on a Nikon Eclipse Ti system with a 100× objective for 1 min at 1 frame per second in a chamber at 37°C, as previously described (22). To process and track EB3 movement, the software package plusTipTracker was used, as described previously (22). For live imaging of TagRFP–α-tubulin, NIH 3T3 cells were transfected in the same way as for the plus-end growth assay. Cells were imaged 80 to 85 h after transfection on a Nikon Eclipse Ti system with a 64× objective for 5 min at 1 frame per 2 s in a chamber at 37°C. The analysis of microtubule dynamic instability was performed using MT-LHAP software as described previously (23).
Statistical analysis.
For all figures, the Student t test was used for statistical analysis.
RESULTS
Tubulin is the major substrate of αTAT1.
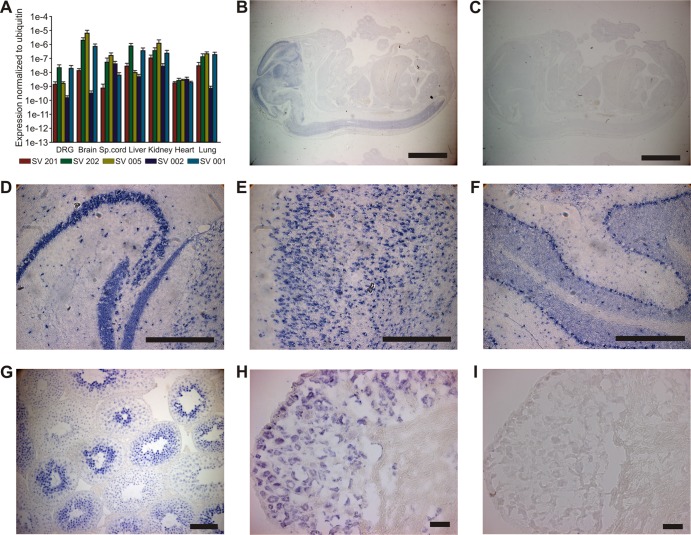
As a starting point in the current study, we examined the expression pattern of αTAT1 at the whole-organism and cellular levels in order to determine whether αTAT1 distribution mirrors α-tubulin acetylation levels. Acetylation of α-tubulin at K40 is present at low levels in most cell types, but with increased prevalence in specialized cells, such as neurons (24). Since multiple alternatively spliced transcripts are predicted for the αTAT1 gene (see Fig. S1 in the supplemental material), we first analyzed the expression patterns of five representative transcripts using quantitative RT-PCR. All αTAT1 transcripts were expressed at low and relatively comparable levels in each tissue, suggesting that alternative splicing does not regulate αTAT1 expression at the gross tissue level (Fig. 1A). We examined expression in more detail by performing in situ hybridization using a probe directed against the domain region of αTAT1 that is common to all splice variants. The strongest signal was detected in the nervous systems of embryonic and adult mice (Fig. 1B to I), and neuronal labeling was clearly evident in the cerebellum, hippocampus, spinal cord, and DRG.
Fig 1.
Expression pattern of αTAT1 mRNA. (A) Five different αTAT1 transcripts (see Fig. S1 in the supplemental material) were analyzed by quantitative real-time PCR (n = 3). All transcripts were expressed at low levels in all tested tissues (DRG, brain, spinal [Sp.] cord, heart, kidney, lungs, and liver). The error bars indicate standard errors. (B to I) In situ hybridization for αTAT1. (B) E16.5 mice show strong αTAT1 mRNA expression signal in the central nervous system. (D to H) Adult mice display expression in the nervous system, especially in the hippocampus (D), cortex (E), cerebellum (F) and DRG (H), and also in the testis (G). (C and I) Control hybridizations with sense probe. Bars, 100 μm (B and C), 500 μm (D, E, and F), 50 μm (G), and 40 μm (H and I).
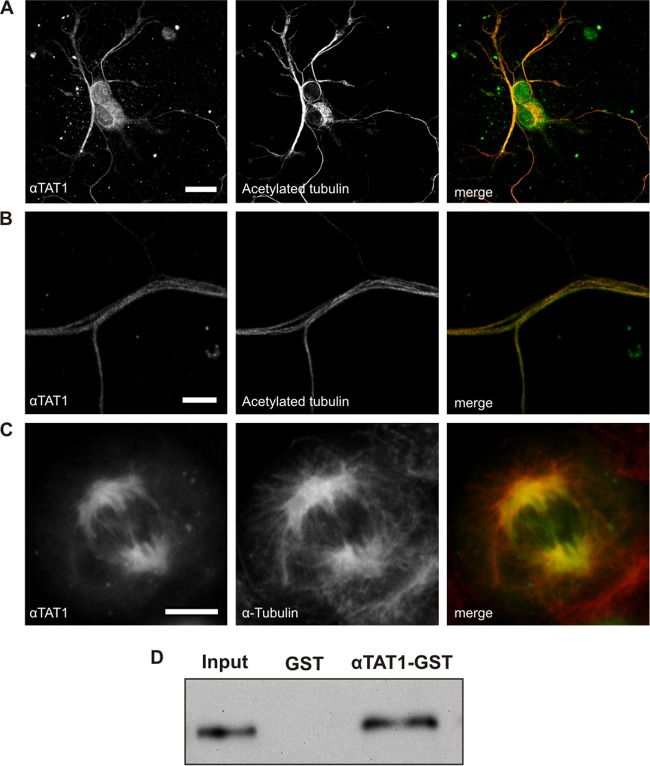
To examine the subcellular distribution of αTAT1, we assessed its colocalization with microtubules using immunocytochemistry and fluorescence microscopy. In primary DRG neurons, we detected native αTAT1 immunoreactivity enriched in the axons of neurons and overlapping with acetylated α-tubulin (Fig. 2A and B). Similarly, in transfected CHO cells, we observed pronounced colocalization of YFP-αTAT1 fusion protein with α-tubulin (Fig. 2C). Finally, we performed a GST pulldown assay and demonstrated direct interaction between αTAT1 protein and purified microtubules by staining with an anti-α-tubulin antibody (Fig. 2D). These data imply that αTAT1 is closely associated with microtubules in a variety of cells and that it can interact directly with microtubules in vitro.
Fig 2.
αTAT1 colocalizes and interacts with microtubules. (A and B) Endogenously expressed αTAT1 in DRG neurons colocalizes with acetylated α-tubulin, especially in axons (B). (C) Transfected CHO cells show that αTAT1-YFP is recruited to the mitotic spindle during cell division. The cells were costained with an antibody against α-tubulin. Endogenous levels of αTAT1 were too low to be detected with the anti-αTAT1 antibody. (D) GST pulldown assay showing interaction between bacterially expressed αTAT1-GST and purified bovine microtubules. Ten percent of the input was loaded, and the membrane was blotted against α-tubulin. Scale bars, 20 μm (A) and 5 μm (B and C).
We next examined the acetyltransferase activity of mouse αTAT1 in mammalian cells and whole-cell extracts. We initially used a well-characterized monoclonal antibody directed against K40-acetylated α-tubulin (6-11b-1 [25]) to assess the levels of tubulin acetylation. In agreement with previous reports (13, 14), overexpression of αTAT1, or transfection of cells with αTAT1 shRNA, increased or decreased the levels of α-tubulin acetylation as described previously (see Fig. S2A to F in the supplemental material). We also generated a functionally inactive form of αTAT1 by mutating 3 residues (G134W, G136W, and L139P, termed αTAT1-GGL), which we reasoned would disrupt the structure of the putative AcCoA binding domain (26). Overexpression of αTAT1-GGL, however, did not alter the levels of endogenous α-tubulin acetylation in CHO or NIH 3T3 cells (see Fig. S2C and F in the supplemental material).
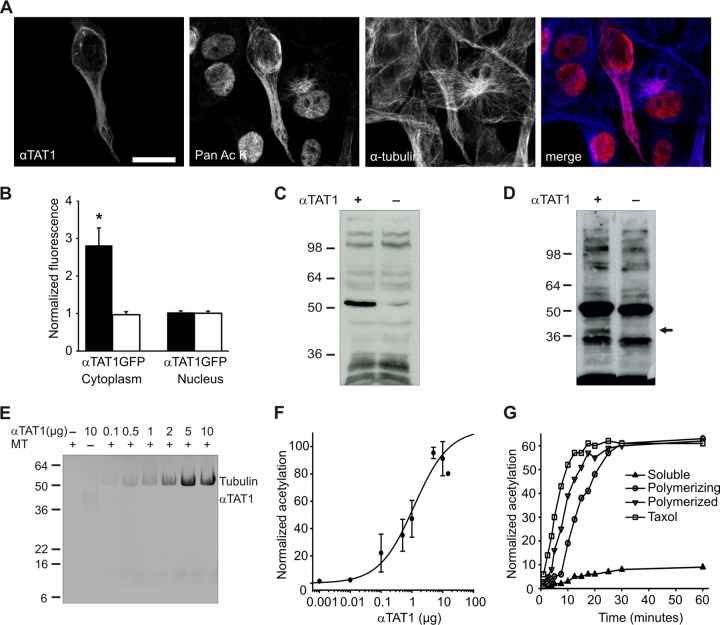
To determine whether αTAT1 has additional targets in mammalian cells besides its action at α-tubulin, we used a pan-anti-acetyl lysine antibody to screen for substrates upon overexpression of αTAT1. Immunostaining of control CHO cells revealed high levels of acetylation in the cell nucleus with less acetylation in the cytoplasm (see Fig. S2G in the supplemental material). Transfection of cells with αTAT1 induced a substantial increase in cytoplasmic acetylation levels, with no change in nuclear acetylation (Fig. 3A and B; see Fig. S2G in the supplemental material). Furthermore, cytoplasmic acetylation coincided entirely with α-tubulin localization (Fig. 3A), implying that spatially, αTAT1-mediated acetylation is restricted to microtubule-associated targets.
Fig 3.
Tubulin and αTAT1 itself are the primary substrates of αTAT1. (A) αTAT1-mediated acetylation predominates at microtubules. CHO cells were transfected with YFP-αTAT1 and stained with panspecific antiacetyllysine antibody and α-tubulin antibody. Scale bar, 20 μm. (B) αTAT1-YFP transfection increases cytoplasmic acetylation, as determined from fluorescence intensity measurements of acetyllysine staining (P < 0.01; n > 10). (C) Transfected CHO whole-cell extracts immunoblotted with panspecific antiacetyllysine antibody showing that tubulin is the major target of αTAT1. (D) Acetyllysine immunoblot of CHO cells treated with the histone deacetylase inhibitor TSA (5 μM) for 4 h. αTAT1-overexpressing cells display increased tubulin acetylation and an additional band (arrow) that corresponds to αTAT1. (E) Autoradiography of αTAT1-mediated [14C]acetyl incorporation into polymerized microtubules (MT). In the absence of tubulin, autoacetylation is more evident. (F) Concentration dependence of αTAT1-mediated acetylation of microtubules (n = 3). (G) αTAT1 preferentially acetylates polymerized tubulin. Paclitaxel and Polymerized, microtubules treated with 3 μM paclitaxel or 1 mM GTP plus 15% glycerol, respectively, for 30 min before the acetylation reaction; Polymerizing, microtubules polymerized with 1 mM GTP and 15% glycerol during the acetylation reaction; Soluble, microtubules incubated with 1 mM GDP and without glycerol during acetylation. The error bars indicate standard errors.
We tested this assumption further by immunoblotting whole-cell lysates with the panspecific acetylated lysine antibody. Transfection of αTAT1 increased acetylation in only a single band corresponding to tubulin (Fig. 3C). To increase the sensitivity of this assay and preclude the possibility of rapid deacetylation by endogenous deacetylases, we also treated cells with the general histone deacetylase inhibitor TSA. Again, the predominant increase in acetylation was most evident for tubulin, but we also detected an extra band with the same molecular weight as αTAT1 itself (Fig. 3D).
αTAT1 acetylates K40 on α-tubulin and multiple lysine residues on itself.
To determine whether αTAT1 acetylates itself in addition to α-tubulin, we established an in vitro acetylation assay utilizing the transfer of [14C]acetyl from acetyl-CoA. As reported previously (13, 14), increasing concentrations of recombinant mouse αTAT1 augmented incorporation of [14C]acetyl into tubulin (Fig. 3E and F), and acetylation proceeded more efficiently in the presence of paclitaxel (Taxol)-stabilized microtubules than in the presence of soluble tubulin (Fig. 3G; see Fig. S3A in the supplemental material) (13, 14, 16, 17). In our assay, αTAT1-mediated acetylation saturated within 30 min, contrasting with a previous report that the reaction required more than 12 h to reach maximum (14). While we performed experiments at 37°C compared to 24°C, it is unlikely that αTAT1 turnover has such high temperature dependence (Q10, approximately 12, where the temperature coefficient Q10 is a measure of the rate of change of a biological or chemical system as a consequence of increasing the temperature by 10°C); instead subtle differences in reaction conditions or recombinant protein purity may be important in determining the kinetics of αTAT1 activity. We also investigated the specificity of the in vitro assay by substituting recombinant histone H3 or the histone acetyltransferase P300/CPB as an alternative substrate or enzyme. αTAT1 was not able to acetylate H3, nor was P300/CPB able to acetylate tubulin (see Fig. S3B in the supplemental material), confirming that microtubules are the preferred specific target of αTAT1 (14).
In agreement with our cellular data, we also detected incorporation of [14C]acetyl into αTAT1 itself (Fig. 3E). This band was consistently weaker than tubulin and more readily observed in the absence of tubulin, suggesting that tubulin has higher affinity for αTAT1 than αTAT1 itself and explaining the relatively low levels of αTAT1 autoacetylation in cells. To explore the specificity of αTAT1-mediated acetylation further, we subjected in vitro reaction samples to mass spectrometry analysis. As expected, we detected incorporation of [13C]acetyl onto K40 of α-tubulin (see Table S1 in the supplemental material). Intriguingly, we also identified four separate acetylated lysine residues on αTAT1 itself, spanning the putative acetyltransferase domain (K56 in motif C and K146 in motif A) (26) and the unstructured C terminus (K210 and K221) (see Table S1 in the supplemental material).
Autoacetylation of αTAT1 increases its catalytic activity at microtubules.
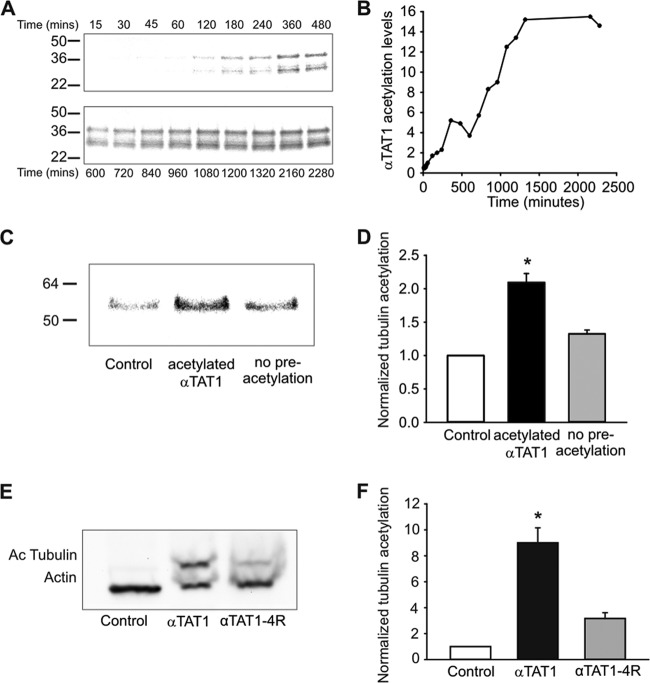
We investigated αTAT1 autoacetylation in more detail to determine whether it might function as a regulatory mechanism for acetylation of microtubules. We first examined the kinetics of the in vitro reaction and observed that autoacetylation took 24 h to saturate (Fig. 4A and B). This was approximately 50 times slower than tubulin acetylation, and [14C]acetyl incorporation never attained levels equivalent to that of tubulin. We next investigated the effects of αTAT1 autoacetylation on its catalytic activity at tubulin. We preincubated αTAT1 with or without [14C]acetyl-CoA for 24 h and then added tubulin and measured [14C]acetyl incorporation (Fig. 4C). Autoacetylation of αTAT1 strongly increased tubulin acetylation, indicating that this may be an important regulatory loop for enzymatic activity or αTAT1 protein stability (Fig. 4D).
Fig 4.
Autoacetylation increases the catalytic activity of αTAT1. (A and B) Autoradiography of [14C]acetyl incorporation into αTAT1. Recombinant αTAT1 is present as two bands (37 kDa and 30 kDa). For both bands, saturation of the autoacetylation reaction is achieved after 24 h (n = 3). (C and D) Preincubation of αTAT1 with [14C]acetyl-CoA for 24 h increases its subsequent catalytic activity at tubulin (n = 3; P < 0.02). (E and F) αTAT1 autoacetylation regulates its catalytic activity in vivo. (E) Immunoblot of mock-, αTAT1-, and αTAT1-4R-transfected NIH 3T3 cell extracts against acetylated tubulin and actin (n = 3). (F) Quantification of acetylated tubulin (n = 3; P < 0.02). The error bars indicate standard errors.
To explore this further at the cellular level, we mutated four of the lysine acetylation targets identified in mass spectrometry experiments to arginine to eliminate acetylation at these sites (K56, K146, K210, and K221: αTAT1-4R). We observed a dramatic reduction in tubulin acetylation in the presence of αTAT1-4R compared to wild-type αTAT1 in NIH 3T3 cells (Fig. 4E and F). Although other acetylation sites on αTAT1 might still be present, we believe that these sites account for the majority of the αTAT1 acetylation, since we could not detect a 37-kDa band on the Western blot after TSA treatment (see Fig. S3C in the supplemental material). Furthermore, αTAT1-4R still interacted with microtubules in GST pulldown assays (see Fig. S3D in the supplemental material), suggesting that although autoacetylation is important for acetylation of tubulin, it is not needed for the in vitro association of αTAT1 with microtubules.
αTAT1 regulates microtubule dynamics.
We next examined the effects of αTAT1-mediated acetylation on microtubule function in mammalian cells. The relationship between acetylation and microtubule stability has attracted much attention. It is generally agreed that posttranslational modifications, such as acetylation, accumulate in stable microtubules (1); however, the question as to whether acetylation actively stabilizes microtubules remains largely unresolved (27–31). Armed with our tools and findings on the function of αTAT1, we sought to determine whether αTAT1-mediated acetylation influences microtubule stability.
As a measure of microtubule stability, we first investigated their resistance to nocodazole, a microtubule-depolymerizing drug. Unexpectedly, we found that shRNA-mediated knockdown of αTAT1 in NIH 3T3 cells led to increased resistance of microtubules to nocodazole (Fig. 5). We analyzed cells 5 and 10 min after the application of nocodazole and observed significantly more cells with intact microtubules (50% in the knockdown compared to 30% in the scrambled control after 5 min of nocodazole application). To evaluate this more systematically, we divided cells with microtubules that were not intact into two groups: cells with sporadic depolymerization (which we termed fragmented) and cells with full depolymerization of microtubules (termed depolymerized). The image in Fig. 5A shows that after 10 min of nocodazole application some cells transfected with shRNA against αTAT1 (transfected cells are shown in the insets) have a fully polymerized microtubule cytoskeleton, while the untransfected and scrambled control cells display an accumulation of diffuse tubulin with occasional short, fragmented microtubules. In contrast, in NIH 3T3 cells overexpressing αTAT1, most microtubules were fully depolymerized following 10 min of nocodazole application (Fig. 5A, compare the two αTAT1-transfected cells to the untransfected cells). Quantification of these data revealed that only 25% of the shRNA-transfected cells show depolymerized microtubules after 10 min compared to approximately 50% in the scrambled control and more than 80% of the αTAT1-transfected cells (Fig. 5B). These experiments imply that αTAT1 renders microtubules more prone to nocodazole-mediated depolymerization.
Fig 5.
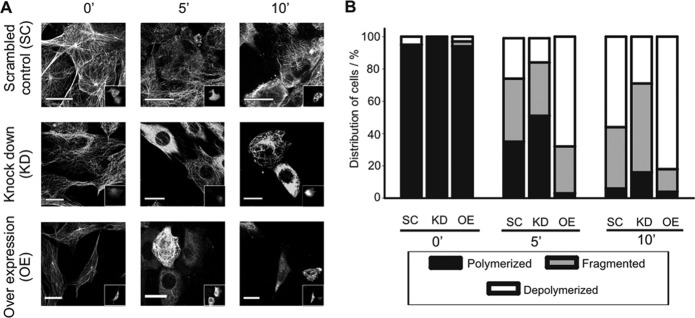
αTAT1 decreases resistance to nocodazole. (A) NIH 3T3 cells were transfected with αTAT1 shRNA (KD), scrambled shRNA (SC), or αTAT1-YFP (OE). The insets show transfected cells. Twenty-four hours posttransfection, nocodazole was applied for the indicated time. Cells with αTAT1 knockdown show increased resistance on nocodazole. (B) Quantification of data from panel A. Polymerized, cells with an intact MT network; Fragmented and Depolymerized, partial and widespread depolymerization, respectively.
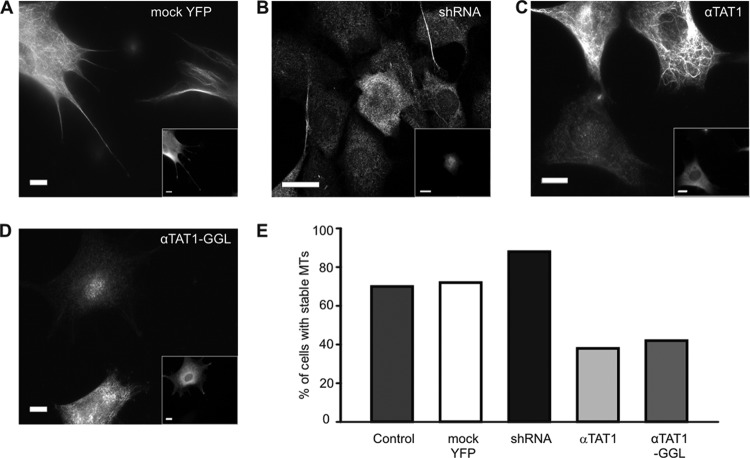
To explore these findings further, we used an assay in which we distinguished stable from short-lived microtubules by immunostaining cells with an antibody directed against detyrosinated tubulin that accumulates in stable microtubules. In agreement with our nocodazole experiments, knockdown of αTAT1 in NIH 3T3 cells led to an increased intensity of detyrosinated tubulin staining while overexpression had the reverse effect and significantly decreased the number of cells containing detyrosinated microtubules (Fig. 6A to C and E). We also examined the effects of the catalytically inactive mutant αTAT1-GGL in these cells and, surprisingly, found that the mutant had the same effect as overexpression of wild-type αTAT1 (Fig. 6D and E). These observations indicate that increased acetylation per se is not sufficient to stabilize microtubules. Levels of tubulin acetylation were approximately 4-fold higher in cells transfected with αTAT1 (see Fig. S2B in the supplemental material), yet these cells displayed fewer stable microtubules and accumulated less detyrosinated α-tubulin. Furthermore, since decreased microtubule stability persisted in cells expressing the acetyltransferase inactive mutant, our data suggest that αTAT1 might exert its influence on microtubule stability independently of its acetyltransferase activity.
Fig 6.
αTAT1 reduces levels of detyrosinated tubulin. (A to D) NIH 3T3 cells, transfected with mock YFP (A), αTAT1 shRNA (B), αTAT1-YFP (C), and catalytically inactive αTAT1-GGL-YFP (D), were stained with anti-detyrosinated tubulin antibody. The αTAT1- and αTAT1-GGL-transfected cells (the insets show transfected cells) show reduced detyrosination. (E) The proportion of cells with stable microtubules is increased upon transfection with αTAT1 shRNA and reduced after transfection with αTAT1 and the catalytically inactive mutant αTAT1-GGL (n > 100). Scale bars, 20 μm.
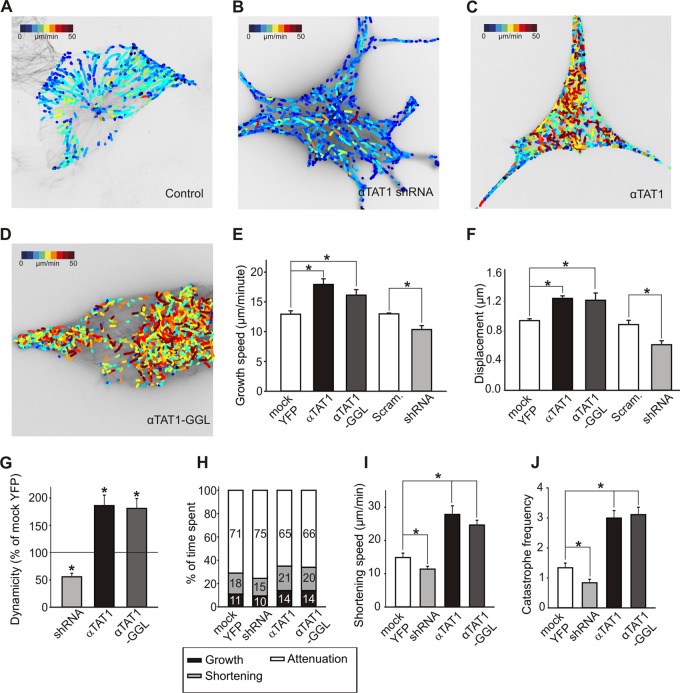
To explore this in detail and to obtain a more dynamic readout of microtubule behavior in the presence of αTAT1, we assayed microtubule growth using the plus-end marker EB3. We used a recently developed computational-tracking package (plusTipTracker [22, 32, 33]) to measure microtubule growth events from live-cell images of EB3-TagRFP (see Movie S1 in the supplemental material). We quantified the speed, displacement, and lifetime of EB3 particles during the growth phase of microtubule dynamics and evaluated the effects of αTAT1 depletion and overexpression on these parameters. shRNA targeted against αTAT1 reduced the speed and displacement of microtubules undergoing growth (Fig. 7A, B, E, and F; see Fig. S4 in the supplemental material). In contrast, overexpression of αTAT1 induced substantial increases in the speed and displacement of EB3 particles (Fig. 7C, E, and F; see Fig. S4 in the supplemental material). Finally, we examined the effects of the catalytically inactive mutant αTAT1-GGL. This mutant closely copied the behavior of wild-type αTAT1 and displayed similar effects on the speed and displacement of EB3 particles during the growth phase (Fig. 7D, E, and F; see Fig. S4 in the supplemental material). Thus, acetylation is not required for αTAT1-mediated acceleration of microtubule plus-end growth.
Fig 7.
αTAT1 increases microtubule dynamics. (A to F) Live-cell images with a color-coded overlay representing EB3 particle growth speed. NIH 3T3 cells were transfected with TagRFP-EB3 and mock YFP (A), αTAT1 shRNA (B), YFP-αTAT1 (C), and catalytically inactive YFP-αTAT1-GGL (D). Video microscopy data were analyzed using the plusTipTracker software package. (E and F) Data for the speed of plus-tip growth (E) and for particle displacement (F) (n = 20). (G to J) Live-cell imaging with TagRFP–α-tubulin. Microtubule dynamicity (the sum of all growth events and all shortening events divided by total time) (G) and the percentage of time microtubules spend in growth or shortening (H) are increased upon YFP-αTAT1 and YFP-αTAT1-GGL overexpression and decreased upon αTAT1 knockdown (the numbers indicate percentages). (I and J) Data for the speed of shortening and frequency of catastrophe (the number of transitions from growth and pause to shortening per minute). The error bars indicate standard errors.
To understand how αTAT1 increases microtubule growth, we performed an additional set of experiments in which we quantified microtubule growth, shortening, and time spent in pause by directly monitoring microtubule dynamics using a red fluorescent protein (RFP)-tagged α-tubulin construct. Live imaging of TagRFP–α-tubulin in NIH 3T3 cells showed that microtubule dynamicity (the rate of total tubulin exchanged at a microtubule end during all growing and shortening events) was increased upon αTAT1 or αTAT1-GGL overexpression and reduced when αTAT1 was knocked down (Fig. 7G). Detailed analysis revealed that αTAT1- or αTAT1-GGL-overexpressing cells spent marginally more time in shortening and growth phases and less time at pause, while αTAT1-shRNA had opposite effects (Fig. 7H). Moreover, microtubules from overexpressing cells also displayed greater growth and shortening speeds and higher catastrophe frequency, which were again reversed by αTAT1-shRNA (Fig. 7I and J). Together, these data argue that αTAT1 has a role in regulating microtubule dynamic instability and that this role is independent of its acetylation activity.
DISCUSSION
Here, we have examined the function of mouse αTAT1 and report that in mammalian cells, α-tubulin is the major target of αTAT1. αTAT1 is also able to acetylate itself, and this acts as a regulatory mechanism for its action at α-tubulin. Moreover, we establish that, in addition to its role as an acetyltransferase, αTAT1 regulates microtubule stability through a nonenzymatic effect on microtubule dynamics. This mechanism could contribute to the cellular function of αTAT1 and underlie some of the phenotypes associated with its depletion.
Our experiments on the expression and acetyltransferase activity of αTAT1 argue for a constrained role of αTAT1 at microtubules. The endogenous localization of αTAT1 mRNA and protein overlaps with the distribution of acetylated tubulin, and YFP-fused αTAT1 colocalizes with microtubules. Moreover, αTAT1 is able to interact directly with microtubules in vitro. Similarly, overexpression of αTAT1 induces detectable acetylation only on microtubules, and α-tubulin is the principal acetylated protein in whole-cell extracts. Intriguingly, we also observed that αTAT1 itself becomes acetylated in vitro and upon pharmacological inhibition of the endogenous α-tubulin deacetylase HDAC6. These data imply that while α-tubulin is the major substrate of αTAT1, self-acetylation could function to regulate activity at microtubules.
We explored this idea further in vitro and demonstrated that autoacetylation of αTAT1 is functionally significant, albeit at acetylation levels that are substantially lower than those of tubulin. From our in vitro analysis, it is difficult to gauge whether this process is an essential requirement for catalytic activity at tubulin, since presumably, αTAT1 is undergoing acetylation continually. However, cellular experiments using nonacetylatable mutants suggest that self-acetylation is critical for activity at tubulin and that levels of αTAT1 acetylation may serve as a means to regulate enzymatic activity. In support of this hypothesis, autoacetylation of acetyltransferases has been described previously and shown to be essential for several different classes of histone acetyltransferase (34–37). Mechanistically, autoacetylation could regulate enzymatic activity either by acting as an intermediate for transfer of the acetyl group to its substrate (38) or via conformational effects on the enzyme and subsequent alterations in complex formation (39). Recently, studies describing the crystal structure of αTAT1 (40, 41) suggest that residues 196 to 236 of human αTAT1 (where acetylated lysines K210 and K221 are located) are disordered and do not contribute significantly to catalytic activity. In contrast, acetylated residues K56 and K146 are both within the catalytic domain (α1 and α3 helices, respectively) and close to the AcCoA binding site, which suggests that these residues might act as an intermediate for the transfer of the acetyl group (40, 41). However, further structural data with autoacetylation mutants are needed to fully understand this mechanism and to test the possibility of conformational changes caused by αTAT1 autoacetylation.
The role of α-tubulin acetylation in microtubule stability has been the subject of extensive debate. Early in vitro studies showed that acetylation does not affect the polymerization rate of tubulin (42) and that Tetrahymena mutants lacking detectable microtubule acetylation have no impairment of microtubule function (43). Since the identification of HDAC6 as an α-tubulin deacetylase enzyme, several studies have reported that acetylation of α-tubulin increases the stability of microtubules (27, 28), while others have observed no change (29–31). Many of these studies relied upon pharmacological or genetic approaches to manipulate the activity or expression of HDAC6. Since HDAC6 has numerous other targets in addition to α-tubulin, some of which regulate microtubule dynamics themselves (44), interpretation of these findings is complicated by difficulties in discriminating between direct HDAC6-mediated deacetylation of microtubules and indirect effects via other molecules. We reasoned that the identification of αTAT1 as the primary α-tubulin acetyltransferase should allow us to address these issues and determine whether acetylation influences microtubule dynamics.
We used four distinct approaches to assess the influence of αTAT1 and α-tubulin acetylation on microtubule stability: resistance of microtubules to depolymerization, accumulation of detyrosinated tubulin, microtubule plus-end growth, and direct monitoring of microtubule dynamics with fluorescently tagged α-tubulin. In all of these measures, overexpression of αTAT1 (and increased α-tubulin acetylation) led to a decrease in microtubule stability, while depletion resulted in more stable microtubules, contrary to previous reports (27, 28). This was most evident in those experiments where we measured microtubule dynamics directly using RFP-tagged α-tubulin. We observed an increase in catastrophe frequency and dynamicity (the rate of tubulin exchanged at a microtubule end) upon overexpression of αTAT1 and a decrease in these parameters after treatment with αTAT1 shRNA. Thus, the majority of microtubules recorded in these cells display either an increase in dynamics in the presence of aTAT1 or a decrease when it is depleted.
Importantly, the influence of αTAT1 on microtubule stability does not appear to be dependent upon levels of α-tubulin acetylation. Overexpression of a catalytically inactive mutant of αTAT1 also destabilized microtubules in the absence of any change in α-tubulin acetylation. In support of this finding, catalytically inactive mutants of the tubulin deacetylase HDAC6 have also been demonstrated to modulate microtubule dynamics without altering acetylation (44). Moreover, it has recently been demonstrated that MEC-17, the C. elegans orthologue of αTAT1, does not require catalytic activity for normal neuronal development and touch sensitivity (15). These data, together with ours, suggest that αTAT1 has a more complex effect on microtubule dynamics than would be expected from its function as an acetyltransferase, and it is the physical presence of αTAT1 and its interactions with tubulin or other as-yet-unidentified regulatory proteins that are the key factors in regulating microtubule stability. Accordingly, we suggest that microtubule acetylation per se does not overtly influence microtubule stability but that αTAT1 and HDAC6 have additional roles at microtubules that extend beyond their classical enzymatic activities.
Our study demonstrates that αTAT1 is functionally more sophisticated than its original description as a K40 α-tubulin acetyltransferase (13). We identified a self-regulatory mechanism for catalytic activity involving autoacetylation of αTAT1 that has functional equivalents among many other acetyltransferases. Most importantly, we also demonstrated that, in addition to its acetyltransferase activity, αTAT1 accelerates microtubule dynamics via a mechanism distinct from acetylation. Further in vitro and in vivo studies are required to reveal the details of these processes and to determine the importance of αTAT1 for mammalian physiology and microtubule-associated diseases.
Supplementary Material
ACKNOWLEDGMENTS
This study was technically supported by the EMBL Protein Expression and Purification Core Facility, the EMBL Proteomics Core Facility, and the EMBL Monterotondo Microscopy Facility. We thank Cinzia Magagnotti and Angela Bachi at the San Raffaele Institute, Milan, Italy, for assistance with mass spectrometry.
Footnotes
Published ahead of print 28 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01044-12.
REFERENCES
- 1. Hammond JW, Cai D, Verhey KJ. 2008. Tubulin modifications and their cellular functions. Curr. Opin. Cell Biol. 20: 71–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. L'Hernault SW, Rosenbaum JL. 1983. Chlamydomonas alpha-tubulin is posttranslationally modified in the flagella during flagellar assembly. J. Cell Biol. 97: 258–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. LeDizet M, Piperno G. 1987. Identification of an acetylation site of Chlamydomonas alpha-tubulin. Proc. Natl. Acad. Sci. U. S. A. 84: 5720–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Reed NA, Cai D, Blasius TL, Jih GT, Meyhofer E, Gaertig J, Verhey KJ. 2006. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 16: 2166–2172 [DOI] [PubMed] [Google Scholar]
- 5. Sudo H, Baas PW. 2010. Acetylation of microtubules influences their sensitivity to severing by katanin in neurons and fibroblasts. J. Neurosci. 30: 7215–7226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. 2007. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129: 1351–1363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. 2002. HDAC6 is a microtubule-associated deacetylase. Nature 417: 455–458 [DOI] [PubMed] [Google Scholar]
- 8. Creppe C, Malinouskaya L, Volvert ML, Gillard M, Close P, Malaise O, Laguesse S, Cornez I, Rahmouni S, Ormenese S, Belachew S, Malgrange B, Chapelle JP, Siebenlist U, Moonen G, Chariot A, Nguyen L. 2009. Elongator controls the migration and differentiation of cortical neurons through acetylation of alpha-tubulin. Cell 136: 551–564 [DOI] [PubMed] [Google Scholar]
- 9. Li L, Wei D, Wang Q, Pan J, Liu R, Zhang X, Bao L. 2012. MEC-17 deficiency leads to reduced alpha-tubulin acetylation and impaired migration of cortical neurons. J. Neurosci. 32: 12673–12683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. 2003. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell 11: 437–444 [DOI] [PubMed] [Google Scholar]
- 11. Ohkawa N, Sugisaki S, Tokunaga E, Fujitani K, Hayasaka T, Setou M, Inokuchi K. 2008. N-acetyltransferase ARD1-NAT1 regulates neuronal dendritic development. Genes Cells 13: 1171–1183 [DOI] [PubMed] [Google Scholar]
- 12. Chu CW, Hou F, Zhang J, Phu L, Loktev AV, Kirkpatrick DS, Jackson PK, Zhao Y, Zou H. 2011. A novel acetylation of beta-tubulin by San modulates microtubule polymerization via down-regulating tubulin incorporation. Mol. Biol. Cell 22: 448–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Akella JS, Wloga D, Kim J, Starostina NG, Lyons-Abbott S, Morrissette NS, Dougan ST, Kipreos ET, Gaertig J. 2010. MEC-17 is an alpha-tubulin acetyltransferase. Nature 467: 218–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shida T, Cueva JG, Xu Z, Goodman MB, Nachury MV. 2010. The major alpha-tubulin K40 acetyltransferase alphaTAT1 promotes rapid ciliogenesis and efficient mechanosensation. Proc. Natl. Acad. Sci. U. S. A. 107: 21517–21522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Topalidou I, Keller C, Kalebic N, Nguyen KC, Somhegyi H, Politi KA, Heppenstall P, Hall DH, Chalfie M. 2012. Genetically separable functions of the MEC-17 tubulin acetyltransferase affect microtubule organization. Curr. Biol. 22: 1057–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang Y, Ma C, Delohery T, Nasipak B, Foat BC, Bounoutas A, Bussemaker HJ, Kim SK, Chalfie M. 2002. Identification of genes expressed in C. elegans touch receptor neurons. Nature 418: 331–335 [DOI] [PubMed] [Google Scholar]
- 17. Chalfie M, Au M. 1989. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science 243: 1027–1033 [DOI] [PubMed] [Google Scholar]
- 18. Grimm C, Matos R, Ly-Hartig N, Steuerwald U, Lindner D, Rybin V, Muller J, Muller CW. 2009. Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J. 28: 1965–1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hublitz P, Kunowska N, Mayer UP, Muller JM, Heyne K, Yin N, Fritzsche C, Poli C, Miguet L, Schupp IW, van Grunsven LA, Potiers N, van Dorsselaer A, Metzger E, Roemer K, Schule R. 2005. NIR is a novel INHAT repressor that modulates the transcriptional activity of p53. Genes Dev. 19: 2912–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mirabeau O, Perlas E, Severini C, Audero E, Gascuel O, Possenti R, Birney E, Rosenthal N, Gross C. 2007. Identification of novel peptide hormones in the human proteome by hidden Markov model screening. Genome Res. 17: 320–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. 2009. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325: 834–840 [DOI] [PubMed] [Google Scholar]
- 22. Matov A, Applegate K, Kumar P, Thoma C, Krek W, Danuser G, Wittmann T. 2010. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat. Methods 7: 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yenjerla M, Lopus M, Wilson L. 2010. Analysis of dynamic instability of steady-state microtubules in vitro by video-enhanced differential interference contrast microscopy with an appendix by Emin Oroudjev. Methods Cell Biol. 95: 189–206 [DOI] [PubMed] [Google Scholar]
- 24. Fukushima N, Furuta D, Hidaka Y, Moriyama R, Tsujiuchi T. 2009. Post-translational modifications of tubulin in the nervous system. J. Neurochem. 109: 683–693 [DOI] [PubMed] [Google Scholar]
- 25. Piperno G, Fuller MT. 1985. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell Biol. 101: 2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Steczkiewicz K, Kinch L, Grishin NV, Rychlewski L, Ginalski K. 2006. Eukaryotic domain of unknown function DUF738 belongs to Gcn5-related N-acetyltransferase superfamily. Cell Cycle 5: 2927–2930 [DOI] [PubMed] [Google Scholar]
- 27. Matsuyama A, Shimazu T, Sumida Y, Saito A, Yoshimatsu Y, Seigneurin-Berny D, Osada H, Komatsu Y, Nishino N, Khochbin S, Horinouchi S, Yoshida M. 2002. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO J. 21: 6820–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tran AD, Marmo TP, Salam AA, Che S, Finkelstein E, Kabarriti R, Xenias HS, Mazitschek R, Hubbert C, Kawaguchi Y, Sheetz MP, Yao TP, Bulinski JC. 2007. HDAC6 deacetylation of tubulin modulates dynamics of cellular adhesions. J. Cell Sci. 120: 1469–1479 [DOI] [PubMed] [Google Scholar]
- 29. Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. 2003. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl. Acad. Sci. U. S. A. 100: 4389–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Palazzo A, Ackerman B, Gundersen GG. 2003. Cell biology: tubulin acetylation and cell motility. Nature 421: 230. [DOI] [PubMed] [Google Scholar]
- 31. Zhang Y, Kwon S, Yamaguchi T, Cubizolles F, Rousseaux S, Kneissel M, Cao C, Li N, Cheng HL, Chua K, Lombard D, Mizeracki A, Matthias G, Alt FW, Khochbin S, Matthias P. 2008. Mice lacking histone deacetylase 6 have hyperacetylated tubulin but are viable and develop normally. Mol. Cell. Biol. 28: 1688–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thoma CR, Matov A, Gutbrodt KL, Hoerner CR, Smole Z, Krek W, Danuser G. 2010. Quantitative image analysis identifies pVHL as a key regulator of microtubule dynamic instability. J. Cell Biol. 190: 991–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Myers KA, Applegate KT, Danuser G, Fischer RS, Waterman CM. 2011. Distinct ECM mechanosensing pathways regulate microtubule dynamics to control endothelial cell branching morphogenesis. J. Cell Biol. 192: 321–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thompson PR, Wang D, Wang L, Fulco M, Pediconi N, Zhang D, An W, Ge Q, Roeder RG, Wong J, Levrero M, Sartorelli V, Cotter RJ, Cole PA. 2004. Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 11: 308–315 [DOI] [PubMed] [Google Scholar]
- 35. Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, Ding H, Xu H, Han J, Ingvarsdottir K, Cheng B, Andrews B, Boone C, Berger SL, Hieter P, Zhang Z, Brown GW, Ingles CJ, Emili A, Allis CD, Toczyski DP, Weissman JS, Greenblatt JF, Krogan NJ. 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- 36. Berndsen CE, Albaugh BN, Tan S, Denu JM. 2007. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry 46: 623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Santos-Rosa H, Valls E, Kouzarides T, Martinez-Balbas M. 2003. Mechanisms of P/CAF auto-acetylation. Nucleic Acids Res. 31: 4285–4292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yan Y, Harper S, Speicher DW, Marmorstein R. 2002. The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nat. Struct. Biol. 9: 862–869 [DOI] [PubMed] [Google Scholar]
- 39. Wang L, Tang Y, Cole PA, Marmorstein R. 2008. Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr. Opin. Struct. Biol. 18: 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Taschner M, Vetter M, Lorentzen E. 2012. Atomic resolution structure of human alpha-tubulin acetyltransferase bound to acetyl-CoA. Proc. Natl. Acad. Sci. U. S. A. 109: 19649–19654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Friedmann DR, Aguilar A, Fan J, Nachury MV, Marmorstein R. 2012. Structure of the alpha-tubulin acetyltransferase, alphaTAT1, and implications for tubulin-specific acetylation. Proc. Natl. Acad. Sci. U. S. A. 109: 19655–19660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Maruta H, Greer K, Rosenbaum JL. 1986. The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. J. Cell Biol. 103: 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gaertig J, Cruz MA, Bowen J, Gu L, Pennock DG, Gorovsky MA. 1995. Acetylation of lysine 40 in alpha-tubulin is not essential in Tetrahymena thermophila. J. Cell Biol. 129: 1301–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zilberman Y, Ballestrem C, Carramusa L, Mazitschek R, Khochbin S, Bershadsky A. 2009. Regulation of microtubule dynamics by inhibition of the tubulin deacetylase HDAC6. J. Cell Sci. 122: 3531–3541 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.