Abstract
Amplification of the universal 16S rRNA gene using PCR has improved the diagnostic yield of microbiological samples. However, no data have been reported on the reliability of this technique with venous access ports (VAPs). We assessed the utility of 16S rRNA PCR for the prediction of VAP-related bloodstream infection (VAP-RBSI). During a 2-year period, we prospectively received all VAPs removed by interventional radiologists. PCR and conventional cultures were performed using samples from the different VAP sites. We compared the results of PCR with those of conventional culture for patients with confirmed VAP-RBSI. We collected 219 VAPs from 219 patients. Conventional VAP culture revealed 15 episodes of VAP-RBSI. PCR revealed a further 4 episodes in patients undergoing antibiotic therapy which would have gone undetected using conventional culture. Moreover, it had a negative predictive value of 97.8% for the prediction of VAP-RBSI when it was performed using biofilm from the internal surface of the port. In conclusion, universal 16S rRNA PCR performed with samples from the inside of VAPs proved to be a useful tool for the diagnosis of VAP-RBSI. It increased detection of VAP-RBSI episodes by 21.1% in patients undergoing antibiotic therapy whose episodes would have gone undetected using conventional culture. Therefore, we propose a new application of 16S rRNA PCR as a useful tool for the diagnosis of VAP-RBSI in patients receiving antibiotic therapy.
INTRODUCTION
Confirmation of colonization of venous access ports (VAPs) requires culture of both port reservoir contents and the catheter tip (1). However, conventional culture of VAPs could prove negative when antibiotic therapy is prescribed before catheter withdrawal. Therefore, an episode of VAP-related bloodstream infection (VAP-RBSI) might not be confirmed, as diagnosis requires the isolation of the same microorganism in blood culture as in the colonized VAP (2–4).
Molecular techniques overcome some of the limitations of conventional culture under several clinical conditions (5–10). Nevertheless, the role of molecular techniques in the diagnosis of VAP-RBSI remains unclear, and available approaches have not been extensively tested in the routine of a clinical microbiology laboratory.
Our main objective was to assess the validity values of 16S rRNA PCR to analyze VAPs from patients with confirmed VAP-RBSI.
MATERIALS AND METHODS
Setting.
Ours was a prospective study performed between July 2009 and April 2011 at a large institution in Madrid, Spain.
We included all tunneled VAPs (Port-A-Caths) that were routinely removed in the Department of Vascular Interventional Radiology, irrespective of the reason for withdrawal. We also included those devices with suspicion of infection that were removed in surgery departments or emergency rooms.
We excluded VAPs that were colonized exclusively by fungi.
Laboratory procedures.
When the VAPs arrived at the microbiology laboratory, we performed culture and broad-range real-time 16S rRNA gene PCR followed by direct sequencing using port content aspirate before and after sonication, port sonication fluid, and biofilm from the internal surface of the port (Fig. 1).
Fig 1.
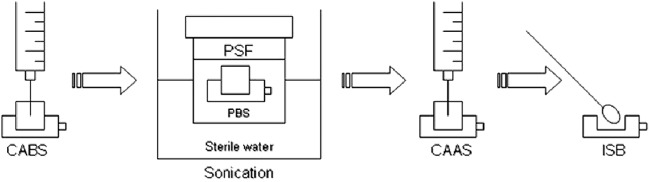
Laboratory procedures. PBS, sterile phosphate-buffered saline; CABS, port content aspirate before sonication; CAAS, port content aspirate after sonication; PSF, port sonication fluid; ISB, port internal surface biofilm.
Port content aspirate before sonication was obtained by aspirating all the liquid collected at the port with a 1-ml syringe. Port content aspirate after sonication was obtained by instilling and aspirating saline with a 1-ml syringe after sonication for 1 min (at 35,000 Hz and 125 W) and vortexing the whole port in 20 ml of sterile phosphate-buffered saline (port sonication fluid) for 15 s. Biofilm was obtained by opening the silicone membrane using a punch and rubbing a sterile swab on the inside. The swab was then inoculated into sterile water.
Conventional culture.
Samples from the different sites of VAPs were cultured on Columbia blood agar and brucella blood agar with hemin and vitamin K1 (Becton, Dickinson, Sparks, MD). The Columbia blood agar plates were incubated aerobically for 48 h at 37°C, and the brucella blood agar plates were incubated under anaerobic conditions for 5 days at 37°C. The colonies recovered were counted.
The microorganisms recovered from cultures were identified by standard microbiological methods. Aerobic microorganisms were identified using the automated MicroScan system with the POS Combo Panel Type 2S and NEG Combo Panel Type 1S (DADE Behring; Sacramento, CA), and anaerobic microorganisms were identified using the API ID 32A (bioMérieux, Marcy l'Etoile, France). Yeasts were identified using the API ID 32C (bioMérieux).
Molecular methods.
DNA was extracted from the port sites mentioned above using a QIAamp DNA Minikit (Qiagen Ltd., West Sussex, United Kingdom) according to the manufacturer's recommendations. In each run, a negative control with nuclease-free UV-treated water was included every 10 test samples.
The 16S rRNA gene was amplified using the methodology described by Marín et al. (8). PCR results were considered valid if all the controls were negative or positive, as appropriate, and the β-globin gene was detected in all the samples.
DNA sequencing reactions.
The first 500 bp of the 16S rRNA gene was sequenced with primers fD1 and E533R (5′-TTACCGCGGCTGCTGGCAC-3′) (11) in all the amplicons obtained using the BigDye Terminator method and detected in an AbiPrism 3100 DNA sequencer (Applied Biosystems Inc.). The sequences generated were compared with those stored in GenBank using BIBI software (http://pbil.univ-lyon1.fr/bibi). Sequence similarity was interpreted according to criteria reported elsewhere (12).
Data analysis.
We compared the validity values of the PCR results with those of conventional culture from patients with confirmed VAP-RBSI. We also compared PCR results with those of conventional VAP culture in order to calculate the validity values of the PCR for prediction of VAP colonization. The comparison was made using 2 different “gold standards” (routine and excellence) according to the incubation conditions of the VAP cultures.
In the case of patients undergoing antibiotic therapy, analysis of the PCR results was performed based only on those for whom therapy was prescribed within 7 days before VAP withdrawal.
Definitions. (i) Positive PCR.
We considered a PCR result to be positive when the microorganism identified had a percentage of similarity of ≥99%, even when there were mixed sequences. All PCRs were performed in parallel with culture and blind to culture results.
(ii) VAP colonization.
VAP colonization was defined as a positive quantitative culture (≥100 CFU/ml) of port content aspirate before or after sonication or of port sonication fluid and/or a positive qualitative culture of biofilm (13).
(iii) Routine gold standard.
For the routine gold standard, we used VAP colonization considering only VAP cultures incubated under aerobic conditions.
(iv) Excellence gold standard.
For the excellence gold standard, we used VAP colonization considering only VAP cultures incubated under both aerobic and anaerobic conditions.
(v) VAP-RBSI.
VAP-RBSI was defined as isolation of the same microorganism(s) both in the colonized VAP (by conventional culture) and in at least 1 peripheral blood culture obtained 7 days before or after catheter withdrawal (1).
Ethics.
The study was approved by the institutional ethics committee.
RESULTS
During the study period, we collected 219 VAPs from 219 patients. Median indwelling time was 435 days (interquartile range [IQR], 212 to 902 days). Colorectal cancer was the main underlying disease (22.8%), followed by breast cancer (21.9%) (Table 1). Most VAPs were removed because of end of use (61.6%), followed by suspicion of bloodstream infection (18.7%). Other patient and catheter characteristics are detailed in Table 1.
Table 1.
Characteristics of the 219 patients with a venous access port during the study perioda
| Characteristic | No. (%) |
P value | ||
|---|---|---|---|---|
| Total (n = 219) | Non-VAP-RBSI (n = 204) | VAP-RBSI (n = 15) | ||
| Age (yr), mean (SD) | 54.5 (17.80) | 54.26 (18.10) | 54.08 (18.64) | 0.970 |
| Male sex | 88 (40.2) | 77 (37.7) | 11 (73.3) | 0.015 |
| Underlying disease | ||||
| Colorectal cancer | 50 (22.8) | 48 (23.5) | 2 (13.3) | 0.556 |
| Breast cancer | 48 (21.9) | 47 (23.0) | 1 (11.1) | 0.248 |
| Lymphoma | 34 (15.5) | 34 (16.7) | 0 (0.0) | 0.177 |
| ENT cancer | 14 (6.4) | 10 (4.9) | 4 (26.7) | 0.006 |
| Leukemia | 13 (5.9) | 11 (5.4) | 2 (13.3) | 0.490 |
| Cervical cancer | 12 (5.5) | 12 (5.9) | 0 (0.0) | 0.705 |
| Digestive tract cancer | 8 (3.7) | 5 (2.5) | 3 (20.0) | 0.005 |
| Ovarian cancer | 3 (1.4) | 1 (0.5) | 2 (13.3) | 0.003 |
| Fibroma | 1 (0.5) | 0 (0.0) | 1 (6.7) | 0.087 |
| Other | 36 (16.4) | 36 (17.6) | 0 (0.0) | 0.156 |
| Charlson comorbidity index, median (IQR)b | 5 (2.00–7.00) | |||
| McCabe and Jackson score, median (IQR)b | 3 (2–3) | |||
| Cumulative catheter days | 136,356 | 125,198 | 11,158 | |
| Catheter days, median (IQR) | 435 (212–902) | 427 (214.50–896.50) | 613 (172.00–1,020.00) | 0.001 |
| No. of times of catheter use, median (IQR) | 20 (2–50) | 20 (3.00–48.00) | 25 (0.00–52.00) | 0.054 |
| Site of catheter insertion | ||||
| Jugular vein | 198 (90.4) | 187 (91.7) | 11 (73.3) | 0.061 |
| Subclavian vein | 21 (9.6) | 17 (8.3) | 4 (26.7) | 0.061 |
| Time of blood draw for cultureb | ||||
| Before catheter withdrawal | 14 (93.3) | |||
| At the time of catheter withdrawal | 0 (0.0) | |||
| After catheter withdrawal | 1 (6.7) | |||
| Reason for catheter withdrawal | ||||
| End of use | 135 (61.6) | 135 (66.2) | 0 (0.0) | <0.001 |
| Suspicion of bloodstream infection | 41 (18.7) | 30 (14.7) | 11 (73.3) | <0.001 |
| Suspicion of local infection | 21 (9.6) | 17 (8.3) | 4 (26.7) | 0.061 |
| Obstruction-malfunction | 15 (6.8) | 15 (7.4) | 0 (0.0) | 0.577 |
| Thrombosis | 4 (1.8) | 4 (2.0) | 0 (0.0) | 0.652 |
| Other | 3 (1.4) | 3 (1.5) | 0 (0.0) | 0.498 |
| Appearance of insertion site | ||||
| Intact | 190 (86.8) | 186 (91.2) | 4 (26.7) | <0.001 |
| Swollen | 16 (7.3) | 13 (6.4) | 3 (20.0) | 0.149 |
| Ulcerated | 10 (4.6) | 4 (2.0) | 6 (40.0) | <0.001 |
| Suppurative | 3 (1.4) | 1 (0.5) | 2 (13.3) | 0.003 |
| DDDs, median (IQR)b | 24 (17.80–70.00) | |||
| Total days of therapy, median (IQR)b | 19 (14.00–28.00) | |||
| Antibiotic use before catheter withdrawalb | 11 (73.3) | |||
Values are numbers with percentages in parentheses unless otherwise indicated. ENT, ear, nose, and throat; IQR, interquartile range; DDDs, defined daily doses. Boldface indicates statistical significance (P < 0.05).
Recorded only for patients with VAP-RBSI.
VAP colonization.
The overall VAP colonization rate by conventional culture under aerobic plus anaerobic conditions (excellence gold standard) was 37.9% (83/219); colonization was polymicrobial in 27.7% (23/83) of the colonized VAPs. The distribution of the isolated microorganisms was as follows: Gram-positive bacteria, 85.6%; Gram-negative bacteria, 5.9%; and yeasts, 8.5% (Table 2). The most common microorganism was Propionibacterium acnes (40.7%), which was isolated from 48 patients and was mainly unaccompanied by aerobic microorganisms (37/48 [77.1%]). No blood cultures were performed for the 37 patients with P. acnes as the only isolated microorganism colonizing VAPs, since 94.6% had their VAPs removed because of end of use. Of the remaining 11 patients in whom P. acnes was isolated together with aerobic microorganisms, 40.5% had bacteremia, which was caused only by aerobic microorganisms.
Table 2.
Microorganisms isolated in colonized venous access ports detected by conventional culture and PCR
| Microorganism(s) | No. (%) identified by: |
|
|---|---|---|
| Conventional culture | PCRa | |
| Gram positive | 101 (85.6) | 37 (82.2) |
| Propionibacterium acnes | 48 (40.7) | 6 (13.3) |
| Staphylococcus epidermidis | 24 (20.3) | 19 (42.2) |
| Staphylococcus aureus | 7 (5.9) | 6 (13.3) |
| CoNSc | 6 (5.1) | 2 (4.4) |
| Enterococcus faecium | 6 (5.1) | |
| Enterococcus faecalis | 3 (2.5) | 1 (2.2) |
| Corynebacterium amycolatum | 2 (1.7) | 1 (2.2) |
| Corynebacterium spp. | 1 (0.8) | 1 (2.2) |
| Streptomyces spp. | 1 (0.8) | |
| Propionibacterium granulosum | 1 (0.8) | |
| Anaerobic cocci | 1 (0.8) | |
| Clostridium spp. | 1 (0.8) | 1 (2.2) |
| Gram negative | 7 (5.9) | 8 (17.8) |
| Enterobacter cloacae | 2 (1.7) | 2 (4.4) |
| Proteus mirabilis | 1 (0.8) | 1 (2.2) |
| Escherichia coli | 1 (0.8) | 1 (2.2) |
| Ochrobactrum anthropi | 1 (0.8) | 1 (2.2) |
| Serratia marcescens | 1 (0.8) | 1 (2.2) |
| Chryseobacterium indologenes | 1 (0.8) | 1 (2.2) |
| Pseudomonas aeruginosa | 1 (2.2) | |
| Fungib | 10 (8.5) | |
| Candida albicans | 7 (5.9) | |
| Candida glabrata | 1 (0.8) | |
| Candida dubliniensis | 1 (0.8) | |
| Rhodotorula mucilaginosa | 1 (0.8) | |
| Total | 118 | 45 |
We excluded from the table those PCR results with a mixed sequence.
All fungi isolated in conventional cultures were accompanied by either Gram-positive or Gram-negative bacteria.
CoNS, coagulase-negative staphylococci.
In contrast, the overall VAP colonization rate by conventional culture under aerobic conditions only (routine gold standard) was 21.0% (46/219), and the distribution of the isolated microorganisms was as follows: Gram-positive bacteria, 74.6%; Gram-negative bacteria, 10.4%; and yeasts, 16.4%.
Validity of the 16S universal PCR for detection of VAP colonization.
PCR revealed that 29.7% (65/219) of VAPs were colonized. In 45 cases (69.2%), PCR identified a single microorganism, and in the remaining 20 cases (30.8%), it detected mixed sequences. Gram-positive microorganisms accounted for 82.2% of the total and Gram-negative organisms for 17.8% (Table 2). Of the 20 VAPs in which PCR detected mixed sequences, 14 (70.0%) had negative cultures, 5 (20.0%) had positive cultures with a single microorganism (4 with Propionibacterium acnes and 1 with Staphylococcus epidermidis), and 1 (5.0%) had a positive culture with 3 microorganisms (S. epidermidis, Enterococcus faecalis, and Corynebacterium amycolatum).
Compared to conventional culture, PCR with biofilm was highly specific for the prediction of VAP colonization using either the routine or the excellence gold standard (91.9% and 94.1%, respectively) (Table 3).
Table 3.
Validity values of the universal 16S rRNA gene PCR for the detection of colonizationa
| Test | No. of cases/total (%) (95% confidence interval) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Routine GSb |
Excellence GSc |
|||||||
| S | SP | PPV | NPV | S | SP | PPV | NPV | |
| PCR with any sample | 34/46 (73.9) (60.1–87.7) | 142/173 (81.5) (76.1–88.1) | 34/65 (52.3) (39.4–65.2) | 142/154 (92.2) (87.7–96.8) | 44/83 (53.0) (41.7–64.4) | 115/136 (84.6) (78.1–91.0) | 44/65 (67.7) (55.6–79.8) | 115/154 (74.7) (67.5–81.9) |
| PCR with CABS | 21/46 (45.7) (30.2–61.1) | 157/173 (90.6) (86.2–95.4) | 21/37 (56.8) (39.4–74.1) | 157/182 (86.3) (81.0–91.5) | 23/83 (27.7) (17.5–37.9) | 122/136 (89.7) (84.2–95.2) | 23/37 (62.2) (45.2–79.1) | 122/182 (67.0) (59.9–74.1) |
| PCR with CAAS | 24/46 (52.2) (36.7–67.7) | 155/173 (89.6) (84.8–94.4) | 24/42 (57.1) (41.0–73.3) | 155/177 (87.6) (82.4–92.7) | 29/83 (34.9) (24.1–45.8) | 123/136 (90.4) (85.1–95.8) | 29/42 (69.0) (53.9–84.2) | 123/177 (69.5) (62.4–76.6) |
| PCR with PSF | 18/46 (39.1) (23.9–54.3) | 157/173 (90.6) (86.2–95.4) | 18/34 (52.9) (34.7–71.2) | 157/185 (84.8) (79.4–90.3) | 25/83 (30.1) (19.7–40.6) | 127/136 (93.4) (88.8–97.9) | 25/34 (73.5) (57.2–89.8) | 127/185 (68.6) (61.7–75.6) |
| PCR with ISB | 23/46 (50.0) (34.5–65.5) | 159/173 (91.9) (87.6–96.3) | 23/37 (62.2) (45.2–79.1) | 159/182 (87.4) (82.3–92.5) | 29/83 (34.9) (24.1–45.8) | 128/136 (94.1) (89.8–98.4) | 29/37 (78.4) (63.8–93.0) | 128/182 (70.3) (63.4–77.2) |
GS, gold standard; S, sensitivity; SP, specificity; PPV, positive predictive value; NPV, negative predictive value; CABS, port content aspirate before sonication; CAAS, port content aspirate after sonication; PSF, port sonication fluid; ISB, port internal surface biofilm.
Incubation of VAP cultures under aerobic conditions.
Incubation of VAP cultures under aerobic plus anaerobic conditions.
VAP-RBSI episodes.
We detected 15 episodes of VAP-RBSI (incidence density, 0.11 episode/1,000 catheter days). The most frequent causative microorganism was Staphylococcus aureus (37.5%), followed by S. epidermidis (18.8%). The remaining microorganisms were Enterococcus faecalis (6.3%), Enterococcus faecium (6.3%), Enterobacter cloacae (6.3%), Escherichia coli (6.3%), and Proteus mirabilis (6.3%). Only 1 polymicrobial VAP-RBSI episode was detected (caused by Serratia marcescens and Candida glabrata).
Compared to the remaining 204 patients with non-VAP-RBSI, we found that patients with VAP-RBSI were predominantly men, had different underlying malignancies, had a long indwelling period, and showed more clinical signs of infection (Table 1).
Validity of 16S universal PCR for detection of VAP-RBSI.
The validity values of PCR performed at any site on the port for the prediction of VAP-RBSI were as follows: sensitivity, 80.0%; specificity, 74.0%; positive predictive value, 18.5%; and negative predictive value, 98.1%. PCR performed on biofilm from the internal surface had better validity values than PCR performed at the remaining port sites, as follows: sensitivity, 73.3%; specificity, 87.3%; positive predictive value, 29.7%; and negative predictive value, 97.8% (Table 4).
Table 4.
Validity values of the universal 16S rRNA gene PCR for the detection of venous access port-related bloodstream infectiona
| Test | No. of cases/total (%) (95% confidence interval) |
|||
|---|---|---|---|---|
| S | SP | PPV | NPV | |
| PCR with any sample | 12/15 (80.0) (51.9–95.7) | 151/204 (74.0) (67.8–80.3) | 12/65 (18.5) (8.3–28.7) | 151/154 (98.1) (94.4–99.6) |
| PCR with CABS | 8/15 (53.3) (26.6–78.7) | 175/204 (85.8) (80.7–90.8) | 8/37 (21.6) (7.0–36.2) | 175/182 (96.2) (93.1–99.2) |
| PCR with CAAS | 9/15 (60.0) (32.3–83.7) | 171/204 (83.8) (78.5–89.1) | 9/42 (21.4) (7.8–35.0) | 171/177 (96.6) (93.7–99.6) |
| PCR with PSF | 7/15 (46.7) (21.3–73.4) | 177/204 (86.8) (81.9–91.7) | 7/34 (20.6) (5.5–35.7) | 177/185 (95.7) (92.5–98.9) |
| PCR with ISB | 11/15 (73.3) (44.9–92.2) | 178/204 (87.3) (82.4–92.1) | 11/37 (29.7) (13.7–45.8) | 178/182 (97.8) (94.5–99.4) |
S, sensitivity; SP, specificity; PPV, positive predictive value; NPV, negative predictive value; CABS, port content aspirate before sonication; CAAS, port content aspirate after sonication; PSF, port sonication fluid; ISB, port internal surface biofilm.
Figure 2 shows the yield of PCR in bacteremic patients receiving systemic antibiotic therapy. PCR performed at any site on the port identified the same microorganism as that isolated in blood cultures for 4 (26.7%) of the 15 bacteremic patients whose VAP cultures were negative and who were receiving antibiotic therapy before VAP withdrawal, with no other source of infection confirmed. In other words, if we consider these episodes as confirmed (according to the standard definition by conventional culture), we would increase the number of detected VAP-RBSIs to 19. Hence, the detection of VAP-RBSI would have been increased by broad-range PCR by 21.1% (4/19) (Fig. 2). Moreover, in 3 of these episodes, the PCR of biofilm was always positive, whereas in the remaining case, the only positive PCR result was for the sonication fluid.
Fig 2.
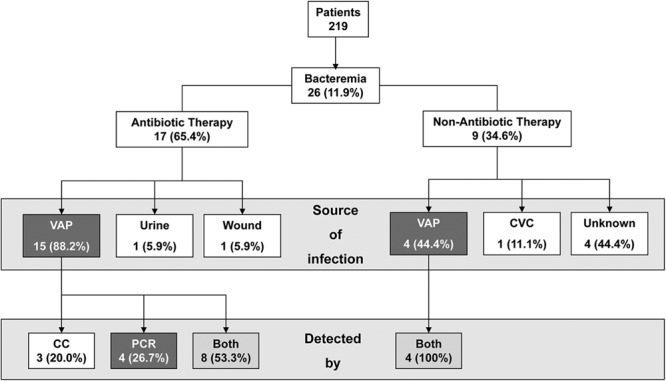
Flowchart describing results for samples obtained from bacteremic patients. CVC, central venous catheter; CC, conventional culture.
DISCUSSION
Universal 16S rRNA PCR is complementary to conventional culture in the microbiological diagnosis of colonized VAPs. For patients with bacteremia, a negative PCR result makes it possible to rule out the origin of VAP-RBSI. The technique is superior to conventional culture, mainly for patients undergoing antibiotic therapy.
PCR performed on VAPs cannot replace conventional culture. As a PCR requires a certain amount of DNA, it has high specificity but low sensitivity.
Confirmation of VAP colonization requires culture of both the catheter tip and the port reservoir contents (1), because it has been demonstrated that culturing the catheter tip alone could leave almost half of all VAP-RBSI episodes undiagnosed (14–16). However, conventional culture is subject to limitations, such as implementation of antibiotic therapy before catheter withdrawal or the impossibility of detecting fastidious microorganisms. Therefore, molecular methods could be useful to solve these problems.
16S rRNA PCR has proven useful for the identification of live or dead bacteria in cases of suspected infection in patients undergoing antibiotic therapy (2–4). The technique can be performed with normally sterile biological specimens and also on artificial devices (5–10). However, current studies based on molecular detection of catheter-related bloodstream infection mainly measure levels of bacterial DNA in blood samples drawn through the catheter using commercially available real-time PCR (17–19). Therefore, at present, no data are available on the yield of universal 16S rRNA PCR performed on the catheter in situ, irrespective of the type of catheter.
Our results showed that in addition to its good specificity for predicting colonization, 16S rRNA PCR performed on biofilm from the internal surface of the port reservoir proved to be very useful for ruling out or confirming an episode of VAP-RBSI (sensitivity, 73.3%; negative predictive value, 97.8%), particularly for patients undergoing antibiotic therapy.
Another important issue in our research was the incubation of additional plates under anaerobic conditions, which allowed us to recover 43.2% of anaerobic microorganisms, the most common being P. acnes (94.1%). However, we could not find any relationship between the presence of this microorganism colonizing VAPs and the consequent emergence of bloodstream infection.
The main limitation of our study was that we did not routinely obtain blood cultures before catheter withdrawal; therefore, some VAP-RBSIs could have gone undetected. Moreover, as we only included those VAP-RBSI episodes that were confirmed by VAP culture, episodes that could have been detected without catheter withdrawal (differential time to positivity) would have been missed.
Future studies should focus on the use of 16S rRNA PCR performed with samples from both the port reservoir and the catheter tip. Furthermore, it would be appropriate to perform a panfungal PCR assay to detect fungal DNA in port reservoirs and to perform an exhaustive analysis of those patients with P. acnes colonizing VAPs.
In conclusion, we present a new application of universal 16S PCR performed on port reservoir contents in order to rule out VAP-RBSI and to diagnose VAP-RBSI episodes in patients whose antibiotic therapy was prescribed before catheter withdrawal.
ACKNOWLEDGMENTS
This study was supported by Ministerio de Sanidad y Consumo (Instituto de Salud Carlos III) and Fundación para la Investigación Biomédica del Hospital General Gregorio Marañón (FIBHGM) (CM09/00028). This study was partially financed by grants from Fundación Mutua Madrileña de Madrid.
We thank Thomas O'Boyle for his help in the preparation of the manuscript.
We thank the members of the GEIDI Study Group for their contribution to this work: José Eugenio Guerrero, Milagros Sancho, Braulio de la Calle, Carlos Sotillo, Guiomar Sánchez, Esther Bermejo López, Lorenzo Fernández Quero, Ana Lajara, Isabel Frías, Carmen Heras, María Jesús Pérez, José Maria Barrio, Alejandro Garrido Sanchez, Patricia Muñoz, Marta Rodríguez-Créixems, Mar Santos, Eduardo Verde, Fernando González García, Emilia Bastida, Maite López Gil, Teresa Blanco, Cristina Cuerda, Laura Frías, José María Tellado, Antonio Echenagusia, Fernando Camúñez, Gracia Rodríguez Rosales, Gonzalo Simó, Mikel Echenagusia, Sonia Zamorano Caballero, Ana Barrientos Guerrero, Abilio Calderón Martín, Carmen Flores Sánchez, M. Jesús Ruano Sta. Engracia, Esperanza Arranz García, M. Ana Luna Caballero, Mar San Segundo Sánchez, Amelia V. Fernández Alonso, and M. Nieves Moro Tejedor.
We declare no conflicts of interest.
Footnotes
Published ahead of print 19 December 2012
REFERENCES
- 1. Mermel LA, Allon M, Bouza E, Craven DE, Flynn P, O'Grady NP, Raad II, Rijnders BJ, Sherertz RJ, Warren DK. 2009. Clinical practice guidelines for the diagnosis and management of intravascular catheter-related infection: 2009 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 49:1–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bauer TW, Parvizi J, Kobayashi N, Krebs V. 2006. Diagnosis of periprosthetic infection. J. Bone Joint Surg. Am. 88:869–882 [DOI] [PubMed] [Google Scholar]
- 3. Del Pozo JL, Patel R. 2009. Clinical practice. Infection associated with prosthetic joints. N. Engl. J. Med. 361:787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Trampuz A, Piper KE, Jacobson MJ, Hanssen AD, Unni KK, Osmon DR, Mandrekar JN, Cockerill FR, Steckelberg JM, Greenleaf JF, Patel R. 2007. Sonication of removed hip and knee prostheses for diagnosis of infection. N. Engl. J. Med. 357:654–663 [DOI] [PubMed] [Google Scholar]
- 5. Drancourt M, Berger P, Terrada C, Bodaghi B, Conrath J, Raoult D, LeHoang P. 2008. High prevalence of fastidious bacteria in 1520 cases of uveitis of unknown etiology. Medicine (Baltimore) 87:167–176 [DOI] [PubMed] [Google Scholar]
- 6. Goldenberger D, Kunzli A, Vogt P, Zbinden R, Altwegg M. 1997. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J. Clin. Microbiol. 35:2733–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Insa R, Martín MMA, Martín-Rabadán P, Alcalá L, Cercenado E, Calatayud L, Liñares J, Bouza E. 2012. Systematic use of universal 16s rRNA gene polymerase chain reaction (PCR) and sequencing for processing pleural effusions improves conventional culture techniques. Medicine (Baltimore) 91:103–110 [DOI] [PubMed] [Google Scholar]
- 8. Marín M, Garcia-Lechuz JM, Alonso P, Villanueva M, Alcala L, Gimeno M, Cercenado E, Sanchez-Somolinos M, Radice C, Bouza E. 2012. Role of universal 16S rRNA gene PCR and sequencing in diagnosis of prosthetic joint infection. J. Clin. Microbiol. 50:583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marin M, Munoz P, Sanchez M, del Rosal M, Alcala L, Rodriguez-Creixems M, Bouza E. 2007. Molecular diagnosis of infective endocarditis by real-time broad-range polymerase chain reaction (PCR) and sequencing directly from heart valve tissue. Medicine (Baltimore) 86:195–202 [DOI] [PubMed] [Google Scholar]
- 10. Xu J, Millar BC, Moore JE, Murphy K, Webb H, Fox AJ, Cafferkey M, Crowe MJ. 2003. Employment of broad-range 16S rRNA PCR to detect aetiological agents of infection from clinical specimens in patients with acute meningitis—rapid separation of 16S rRNA PCR amplicons without the need for cloning. J. Appl. Microbiol. 94:197–206 [DOI] [PubMed] [Google Scholar]
- 11. Baker GC, Smith JJ, Cowan DA. 2003. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 55:541–555 [DOI] [PubMed] [Google Scholar]
- 12. Drancourt M, Berger P, Raoult D. 2004. Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans. J. Clin. Microbiol. 42:2197–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sherertz RJ, Raad II, Belani A, Koo LC, Rand KH, Pickett DL, Straub SA, Fauerbach LL. 1990. Three-year experience with sonicated vascular catheter cultures in a clinical microbiology laboratory. J. Clin. Microbiol. 28:76–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Douard MC, Arlet G, Longuet P, Troje C, Rouveau M, Ponscarme D, Eurin B. 1999. Diagnosis of venous access port-related infections. Clin. Infect. Dis. 29:1197–1202 [DOI] [PubMed] [Google Scholar]
- 15. Longuet P, Douard MC, Arlet G, Molina JM, Benoit C, Leport C. 2001. Venous access port-related bacteremia in patients with acquired immunodeficiency syndrome or cancer: the reservoir as a diagnostic and therapeutic tool. Clin. Infect. Dis. 32:1776–1783 [DOI] [PubMed] [Google Scholar]
- 16. Whitman ED, Boatman AM. 1995. Comparison of diagnostic specimens and methods to evaluate infected venous access ports. Am. J. Surg. 170:665–669; discussion, 669–670 [DOI] [PubMed] [Google Scholar]
- 17. Ley BE, Linton CJ, Bennett DM, Jalal H, Foot AB, Millar MR. 1998. Detection of bacteraemia in patients with fever and neutropenia using 16S rRNA gene amplification by polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 17:247–253 [DOI] [PubMed] [Google Scholar]
- 18. Millar M, Zhou W, Skinner R, Pizer B, Hennessy E, Wilks M, Gilbert RE. 2011. Accuracy of bacterial DNA testing for central venous catheter-associated bloodstream infection in children with cancer. Health Technol. Assess. 15:1–114 [DOI] [PubMed] [Google Scholar]
- 19. Warwick S, Wilks M, Hennessy E, Powell-Tuck J, Small M, Sharp J, Millar MR. 2004. Use of quantitative 16S ribosomal DNA detection for diagnosis of central vascular catheter-associated bacterial infection. J. Clin. Microbiol. 42:1402–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]


