Abstract
Candida africana was previously proposed as a new species within the Candida albicans species complex, together with C. albicans and C. dubliniensis, although further phylogenetic analyses better support its status as an unusual variant within C. albicans. Here we show that C. africana can be distinguished from C. albicans and C. dubliniensis by pyrosequencing of a short region of ITS2, and we have evaluated its occurrence in clinical samples by pyrosequencing all presumptive isolates of C. albicans submitted to the Mycology Reference Laboratory over a 9-month period. The C. albicans complex constituted 826/1,839 (44.9%) of yeast isolates received over the study period and included 783 isolates of C. albicans, 28 isolates of C. dubliniensis, and 15 isolates of C. africana. In agreement with previous reports, C. africana was isolated exclusively from genital specimens, in women in the 18-to-35-year age group. Indeed, C. africana constituted 15/251 (6%) of “C. albicans” isolates from female genital specimens during the study period. C. africana isolates were germ tube positive, grew significantly more slowly than C. albicans and C. dubliniensis on conventional mycological media, could be distinguished from the other members of the C. albicans complex by appearance on chromogenic agar, and were incapable of forming chlamydospores. Here we present the detailed evaluation of epidemiological, phenotypic, and clinical features and antifungal susceptibility profiles of United Kingdom isolates of C. africana. Furthermore, we demonstrate that C. africana is significantly less pathogenic than C. albicans and C. dubliniensis in the Galleria mellonella insect systemic infection model.
INTRODUCTION
The incidence of invasive fungal infections caused by unusual Candida spp. continues to rise, in part due to increased populations of immunocompromised patients and those undergoing invasive procedures (1–8). Nevertheless, Candida albicans remains the most frequently isolated Candida species in the clinical setting, is the principal agent of nosocomial yeast infections (1, 4–6), and is widely accepted as being the most pathogenic Candida species (reviewed in reference 9). In addition to C. albicans sensu stricto, two closely related species have been described and added to the C. albicans species complex. In 1995, Candida dubliniensis, predominantly associated with cases of oral candidiasis in human immunodeficiency virus-infected patients, was described (10). While C. dubliniensis is phylogenetically closely related to C. albicans and shares the ability to produce germ tubes, true hyphae, and chlamydospores, epidemiological analyses have revealed that C. dubliniensis is far less prevalent than C. albicans (11–14), is comparatively rarely associated with systemic infection (11, 14), and is less pathogenic than C. albicans in a variety of infection models (13–16).
Similarly, Candida africana was initially described as an atypical, chlamydospore-negative variant of C. albicans (17, 18) but was proposed as a new species on the basis of morphological, biochemical, and physiological differences (18, 19). While subsequent molecular analyses supported a varietal distinction (C. albicans var. africana) (20), the taxonomic status of C. africana remains controversial. Although they are germ tube positive like C. albicans and C. dubliniensis, C. africana isolates studied to date have reportedly grown and produced hyphae more slowly than either C. albicans or C. dubliniensis and could be distinguished from both by an inability to assimilate several sugars or to produce chlamydospores and by appearance on chromogenic agars (18, 19, 21–23). Moreover, despite an almost worldwide distribution, the overwhelming majority of C. africana isolates have been recovered from female genital specimens (reviewed in reference 16). Epidemiological analyses of C. africana have been hampered by the failure of the commonly employed commercially available identification methods to distinguish it from C. albicans (reviewed in references 21 and 16). However, studies employing PCR amplicon length analyses of the Hwp-1 gene (24) revealed that C. africana constituted 7.2% of C. albicans complex isolates in three different hospitals in southern Italy, a prevalence 3 times that of C. dubliniensis in samples from principally nonsterile sites from the same cohort of hospitalized patients (25).
Here we have shown that C. africana can be distinguished from C. albicans and C. dubliniensis by pyrosequencing of a short fragment of the internal transcribed spacer region 2 (ITS2) and have subsequently used pyrosequencing to assess the prevalence of C. africana among Candida isolates from clinical specimens submitted to the UK National Mycology Reference Laboratory (MRL) over a 9-month period spanning 2011 to 2012. Isolates identified as C. africana by pyrosequencing were subjected to phenotypic and biochemical analyses, and their antifungal susceptibility profiles were established. Finally, we compared the pathogenicity of C. africana, C. albicans, and C. dubliniensis clinical isolates in vivo in the Galleria mellonella systemic infection model.
MATERIALS AND METHODS
Antifungal agents.
Antifungal drugs were obtained from their respective manufacturers as standard powders. Amphotericin B, nystatin, and clotrimazole (all from Sigma Chemical Co, St. Louis, MO) were dissolved in dimethyl sulfoxide (DMSO) (amphotericin and nystatin) and polyethylene glycol (PEG) 400 with heating to 70°C (clotrimazole). Itraconazole, miconazole, econazole, and ketoconazole (all from Janssen Research Foundation, Beerse, Belgium) were prepared in PEG 400 with heating to 70°C (itraconazole and miconazole) or in DMSO (econazole and ketoconazole). Fluconazole (Pfizer Central Research, Sandwich, United Kingdom) was resuspended in sterile water. Serial 2-fold dilutions of the various drugs were prepared in RPMI 1640 medium (with l-glutamine, without bicarbonate; Sigma Chemical Co, St. Louis, MO), and buffered to pH 7.0 using a 0.165 M solution of MOPS (morpholinepropanesulfonic acid) (Sigma Chemical Co, St. Louis, MO).
Yeast isolates and conventional phenotypic identification approaches.
A total of 1,839 clinical isolates of yeast were included in the current study. Isolates had been submitted to the UK National Mycology Reference Laboratory (MRL) for identification, susceptibility testing, or both. Isolates were from both sterile and nonsterile sites (female genitals, n = 381; male genitals, n = 10; blood and line tips, n = 600; sputum and bronchoalveolar lavage [BAL] fluid, n = 199; feces, n = 16; drain fluids, n = 144; tissue, n = 136; eyes, n = 37; cerebrospinal fluid [CSF], n = 14; urine, n = 71; ear, n = 12; and site not stated, n = 219). All isolates were subcultured on plates of Oxoid Sabouraud dextrose agar containing 0.5% (wt/vol) chloramphenicol (Unipath Limited, Basingstoke, England). Cultures were incubated for 24 h at 35°C prior to testing. Isolates of C. africana have been stored in sterile water in a metabolically inactive state in the National Collection of Pathogenic Fungi (NCPF).
The clinical isolates included in this study were all subjected to initial germ tube testing (inoculation into sterile horse serum and incubation at 37°C for 3 h). Isolates were also cultured on Dalmau plates (Oxoid cornmeal agar, supplemented with 1% Tween 80, with a sterile coverslip over a single streak of the organism to establish the additional morphological characteristics required for complete AUXACOLOR2 profiles) and MAST-ID CHROMagar Candida plates to establish the purity of isolates. Strains formally identified as Candida africana by molecular approaches were also subjected to the AUXACOLOR2 identification kit (Bio-Rad Laboratories Ltd., Hemel Hempstead, United Kingdom) exactly as described previously (26).
Molecular identification.
All 1,839 test isolates were subjected to pyrosequencing analysis of a portion of ITS2 exactly as described previously (27–29). For isolates identified by pyrosequencing as C. africana, identity was confirmed by PCR amplification and sequencing of a region of the large subunit gene (LSU) and the internal transcribed spacer 1 (ITS1) region using the primers described previously (30, 31) and direct inoculation of PCR products with yeast suspensions as described in reference 32. Organisms were identified using BLAST searches against fungal sequences in existing public DNA databases (EMBL) and multiple sequence alignments using ClustalW and a database of formally identified organisms compiled by the MRL.
Broth microdilution determination of yeast MICs.
MICs were determined according to CLSI methodologies (CLSI M27-A3 [33]) in round-bottomed 96-well plates with yeast blastospore suspensions prepared in saline and then diluted into RPMI 1640 and adjusted to final concentrations of 2.5 × 103 CFU/ml. Inoculated plates were incubated for 24 to 48 h at 35°C. MICs were read at 24 and 48 h as the concentration of drug that elicited 100% inhibition of growth (amphotericin B, nystatin) or significant (approximately 50%) inhibition of growth compared with a drug-free control (itraconazole, fluconazole, miconazole, ketoconazole, econazole, and clotrimazole).
Galleria mellonella virulence assays.
Killing assays were performed in Galleria mellonella essentially as described previously (34–37). Briefly, final (sixth) instar larvae weighing approximately 300 mg each (Livefood UK Ltd., Rooks Bridge, Somerset, United Kingdom) that were absent of gray markings were maintained at room temperature in the dark and inoculated within 1 week of receipt. Suspensions of individual Candida isolates that had been grown on Sabouraud agar for 24 h at 37°C were harvested by gentle scraping of colony surfaces with sterile plastic loops, washed twice in sterile phosphate-buffered saline (PBS), counted in hemocytometers, and adjusted to 105 cells per microliter in sterile PBS. Individual larvae were inoculated in the left rear proleg with 1 × 106 yeast cells in PBS (final inoculum volume, 10 μl) using a 10-μl Hamilton syringe fitted with a 26-gauge blunt needle. Fifteen larvae were inoculated per isolate per experiment (experiments employed 4 independent isolates of each Candida test species). Control groups of larvae received 10 μl of sterile PBS in exactly the same manner. Inoculated larvae were incubated at 30°C, 37°C, or 42°C and scored for viability at 8-h intervals as described previously (34–37). Differences in resulting Kaplan-Meier survival plots were evaluated using the Mann-Whitney (two-sample Wilcoxon) test. In experiments designed to assess differences in fungal cell filamentation postinfection, representative larvae from each inoculum group were cut open at 24 h postinfection and the fat body/solid internal structures and hemolymph were collected aseptically as described previously (37) and examined microscopically.
RESULTS
We have previously demonstrated that pyrosequencing of 35 bp of a portion of the internal transcribed spacer region 2 (ITS2) is sufficient to robustly identify the vast majority of pathogenic yeasts (27), including genetically closely related isolates within species complexes (28, 29). This is also the case for the key species within the C. albicans species complex. To date, we have identified 4 C. albicans sequence variants within the 35-bp ITS2 signature sequence we employ for yeast identification, 2 C. dubliniensis variants, and a single signature sequence shared by all C. africana isolates encountered to date (Table 1). All C. dubliniensis and C. africana isolates differ from C. albicans at 10 or 11 and 1 or 2 positions, respectively, within the 35-base pyrosequencing region (Table 1). Conversely, the ITS2 sequence for C. stellatoidea (based on published ITS2 sequences for the type strain, isolate CBS 1905; EMBL AJ853768) is indistinguishable from that of C. albicans variant 1 (Table 1), which is the most common ITS2 sequence variant encountered in United Kingdom isolates.
Table 1.
Pyrosequencing signature sequences of members of the C. albicans species complexa
| Species/sequence variant | Pyrosequencing signature sequenceb | Identityc | Percentaged |
|---|---|---|---|
| Candida albicans 1 | GTCAA AGTTT GAAGA TATAC GTGGT GGACG TTACC | 35 | 65.3 |
| Candida albicans 2 | GTCAA AGTTT GAAGA TATAC GTGGT aGACG TTACC | 34 | 24.9 |
| Candida albicans 3 | GTCAA AGTTT GAAGA TATAC GTGGT aGACG cTACC | 33 | 7.0 |
| Candida albicans 4 | GTCAA AGTTT GAAGA TATAC GTGGT aGACG TTgCC | 33 | 2.8 |
| Candida stellatoidea (CBS1905) | GTCAA AGTTT GAAGA TATAC GTGGT GGACG TTACC | 35 | ND |
| Candida africana | GTCAA AGTTT GAAGA TATAC GTaGT aGACG TTACC | 33 | NA |
| Candida dubliniensis 1 | GTCAA AGTTT GAAGA ataAa aTGGg cGACG ccAga | 24 | 88.0 |
| Candida dubliniensis 2 | GTCAA AGTTT GAAGA ataAa aTGGc –GACG ccAga | 24 | 12.0 |
Sequences are aligned against the 35-bp signature of the most common C. albicans variant (C. albicans 1). A total of 213 isolates (C. albicans), 25 isolates (C. dubliniensis), and 15 isolates (C. africana) were subjected to pyrosequencing.
Bold type indicates totally conserved nucleotides. Lowercase letters indicate positions in which the sequence differs from that of C. albicans 1.
Identity is given as the number of identical bases compared to the signature sequence of C. albicans 1.
Percentage of isolates of a given species with that particular signature sequence. ND, not determined; NA, not applicable.
In order to evaluate the prevalence and potential clinical significance of C. africana in clinical samples in the United Kingdom, between September 2011 and July 2012, the UK National Mycology Reference Laboratory identified 1,839 yeast isolates to species level using pyrosequencing. C. albicans species complex constituted 44.9% (826/1,839) of all isolates identified and comprised 783 isolates of C. albicans (42.6% of all isolates), 28 isolates of C. dubliniensis (1.5%), and 15 isolates of C. africana (0.8%). Thus, when considering isolates from all clinical sites, C. dubliniensis and C. africana represented 3.4% and 1.8%, respectively, of all C. albicans complex isolates (Table 2). In agreement with the published reports concerning the occurrence of C. africana (22), all 15 isolates identified in this study had been isolated from genital specimens (Table 2) in young female adults (age range, 18 to 31 years; mean age, 24.7 years). Indeed, when only the C. albicans complex isolates from female genital specimens were considered (251 isolates in total), C. africana (15 isolates) was much more prevalent than C. dubliniensis (3 isolates) and represented 6% of C. albicans complex isolates from such specimens.
Table 2.
Strain collection data and MICs for C. africana isolates
| Straina | Patient siteb | Age (yr) | Location | MIC (mg/liter)c |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AMB | FLZ | ITR | MIC | NYS | KET | CLO | ECON | ||||
| NCPF 8945 | HVS | 22 | Wrexham | 0.125 | <0.125 | <0.03 | <0.125 | 2.0 | <0.125 | <0.125 | <0.125 |
| NCPF 8946 | Genital | 22 | Ipswich | 0.125 | <0.125 | <0.03 | <0.125 | 1.0 | <0.125 | <0.125 | <0.125 |
| NCPF 8947 | Vaginal swab | 25 | Bath | 0.125 | <0.125 | <0.03 | <0.125 | 1.0 | <0.125 | <0.125 | <0.125 |
| NCPF 8948 | Genital | 31 | Portsmouth | 0.25 | 0.25 | <0.03 | <0.125 | 2.0 | <0.125 | <0.125 | <0.125 |
| NCPF 8949 | HVS | 22 | Crewe | 0.125 | <0.125 | <0.03 | <0.125 | 1.0 | <0.125 | <0.125 | <0.125 |
| NCPF 8950 | HVS | 28 | Crewe | 0.125 | <0.125 | <0.03 | <0.125 | 1.0 | <0.125 | <0.125 | <0.125 |
| NCPF 8951 | HVS | 26 | Bangor | 0.125 | <0.125 | <0.03 | <0.125 | 2.0 | <0.125 | <0.125 | <0.125 |
| NCPF 8952 | Vaginal swab | 20 | Epsom | ND | <0.125 | <0.03 | <0.125 | 0.5 | <0.125 | 0.25 | ND |
| NCPF 8953 | HVS | 25 | London | ND | <0.125 | <0.03 | <0.125 | 0.5 | <0.125 | <0.125 | ND |
| NCPF 8954 | HVS | 22 | Coventry | ND | ND | ND | ND | ND | ND | ND | ND |
| NCPF 8955 | HVS | 22 | Stoke | ND | <0.125 | <0.03 | <0.125 | 0.5 | ND | ND | ND |
| NCPF 8956 | HVS | 31 | Bristol | ND | 0.25 | <0.03 | <0.125 | 1.0 | <0.125 | <0.125 | <0.125 |
| NCPF 8957 | LVS | 25 | Bristol | 0.25 | <0.125 | <0.03 | <0.125 | 0.5 | <0.125 | <0.125 | <0.125 |
| NCPF 8958 | HVS | 21 | Bristol | ND | <0.125 | ND | <0.125 | 1.0 | <0.125 | <0.125 | ND |
| Isolate F111 (dead) | Vaginal swab | 18 | Northampton | 0.125 | 0.25 | <0.03 | <0.125 | 1.0 | <0.125 | <0.125 | <0.125 |
NCPF, National Collection of Pathogenic Fungi, UK Mycology Reference Laboratory.
All patients were female. HVS, high vaginal swab; LVS, low vaginal swab.
Drug abbreviations: AMB, amphotericin B; FLZ, fluconazole; ITR, itraconazole; MIC, miconazole; NYS, nystatin; KET, ketoconazole; CLO, clotrimazole; ECON, econazole. ND, not determined.
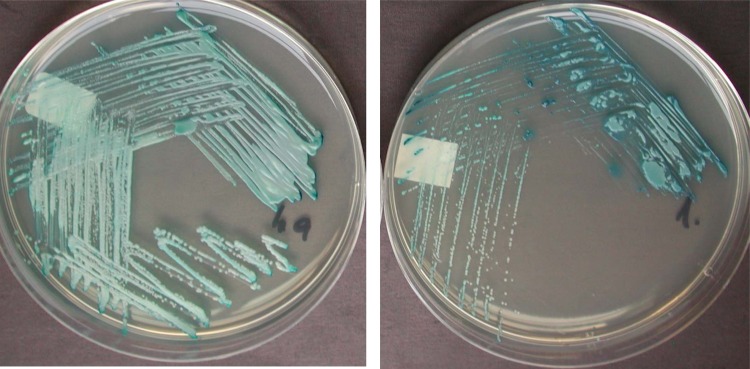
Candida africana isolates had been submitted to the MRL from hospitals across central and southern England and Wales. In agreement with one published evaluation (38), all isolates that were tested were found to be susceptible (with very low MICs) to all antifungal agents that would be appropriate for treating vaginal candidiasis (Table 2). Examination of clinical details submitted with the isolates did not reveal a significant or unusual proportion of recalcitrant or recurrent infections associated with C. africana, compared to those associated with isolates of C. albicans sensu stricto, although generally the majority of yeast isolates from genital sites received by the MRL are from recurrent or recalcitrant infections. In agreement with previous reports (18, 22, 23), all 15 C. africana isolates were capable of producing germ tubes upon incubation in horse serum for 3 h at 37°C (Table 3). Similarly, C. africana isolates formed true hyphae in Dalmau cultures, although the quantity of hyphae produced and the speed of hyphal formation were much reduced compared to those for C. albicans and C. dubliniensis isolates (Table 3; data not shown). All isolates of C. africana failed to produce chlamydospores in culture, even after prolonged incubation, were incapable of assimilating trehalose, and lacked N-acetyl-galactosaminidase (hexosaminidase) activity (18, 22, 23) (Table 3). Thus, C. africana could be distinguished from C. albicans and C. dubliniensis by AUXACOLOR2, although the former yielded unique profiles that are not currently in the databases provided with the kit. C. africana could also be distinguished from C. albicans on the basis of its appearance on CHROMagar, with the former producing smaller and deeper turquoise-green colonies than C. albicans (Fig. 1). However, in contrast to previously published descriptions (22, 39), all 15 United Kingdom isolates of C. africana were capable of growth at 30°C, 37°C, and 42°C, although growth was restricted at the latter temperature (Table 3). Importantly, current matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF) approaches are apparently unable to robustly distinguish C. africana and C. albicans (see, for example, Bruker Daltonik TechNote 1.0; Bruker Daltonik GmbH, Bremen, Germany).
Table 3.
Phenotypic characteristics of C. albicans, C. dubliniensis, and C. africana isolatesa
| Species | Germ tube result | Dalmau culture result |
Growthb at: |
Trehalose assimilationc | NAG actd | Color on CHROMagar | Growth on actidione | Growth speede | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hyphae | Chlamydospores | 30°C | 37°C | 42°C | |||||||
| Candida albicans | + | Abundant | Present | +++ | ++++ | +++ | + | + | Apple green | + | 100 |
| Candida dubliniensis | + | Abundant | Present | ++ | +++ | +/− | + | + | Apple green | + | 85 |
| Candida africana | + | Scant | Absent | ++ | ++ | + | − | − | Turquoise-green | + | 50 |
+, positive; −, negative (except as described in footnote b).
Growth: −, undetectable; +/−, barely detectable; +, detectable; ++, good; +++, abundant; ++++, prolific.
Ability to assimilate trehalose.
N-acetyl-galactosaminidase (hexosaminidase) activity.
Growth speed expressed as a percentage of that observed with C. albicans isolates.
Fig 1.
Appearance of C. albicans (left panel) and C. africana (right panel) after 48 h of incubation at 35°C on CHROMagar.
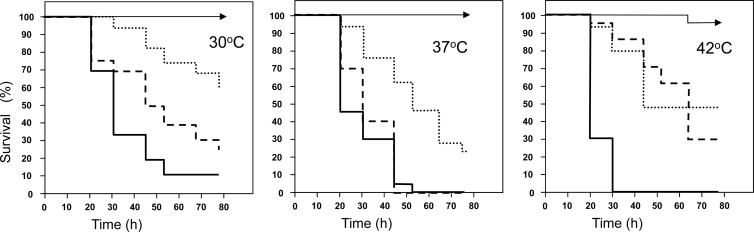
Given the reduced formation of hyphae and slower growth of C. africana compared to C. albicans, as well as previous reports demonstrating that C. dubliniensis exhibits reduced pathogenicity compared to C. albicans in a variety of infection models (reviewed in references 15 and 14), we compared the virulence of isolates of C. albicans, C. africana, and C. dubliniensis in a systemic infection model using the insect larval model Galleria mellonella (34–37). Although recent studies have failed to reveal differences in pathogenicity between mucosal and systemic isolates of key pathogenic yeast species in Galleria larvae (40), any such potential bias in the current study was avoided by comparing C. africana strains with C. albicans and C. dubliniensis isolates that had also been recovered from female genital samples. In preliminary experiments, and in agreement with previous reports (34), no significant larval killing was observed with any isolate when larvae were inoculated with 105 CFU/larva at any temperature (data not shown). However, when 106 CFU per larva were employed, species-specific and temperature-dependent larval killing was observed (Fig. 2). Candida albicans was the most virulent species studied, and larval killing increased in a temperature-dependent manner. C. dubliniensis isolates exhibited virulence (in terms of mortality rates and time to kill) that was indistinguishable from that of C. albicans in larvae maintained at 37°C, but they were significantly less virulent than C. albicans in larvae incubated at 42°C (P < 0.015), a temperature at which most C. dubliniensis isolates show extremely restricted growth in vitro (39). Conversely, larval killing by C. africana isolates was significantly slower than that observed with C. albicans in larvae incubated at all three temperatures (P < 0.030, P < 0.065, and P < 0.007 for 30°C, 37°C, and 42°C, respectively) or C. dubliniensis in larvae incubated at 30°C or 37°C (P < 0.08 and < 0.015, respectively) (Fig. 2). Finally, analysis of hemolymph from G. mellonella larvae inoculated with the various strains and incubated at 37°C for 8 h revealed significant hyphal and pseudohyphal proliferation in larvae inoculated with C. albicans and C. dubliniensis but not those inoculated with C. africana (data not shown). Indeed, no significant filamentation was observed in larvae infected with C. africana at any time postinfection.
Fig 2.
Virulence of C. albicans, C. dubliniensis, and C. africana in Galleria mellonella larvae is species specific and temperature dependent. Kaplan-Meier plots of G. mellonella survival after injection with 106 CFU/larva of C. albicans (solid lines), C. dubliniensis (dashed lines), or C. africana (dotted lines) or an equivalent volume of sterile PBS (arrowed lines) and incubation at 30°C (left panel), 37°C (middle panel), and 42°C (right panel). Four strains were tested per species, with 15 larvae per strain (60 larvae per species). Experiments were performed in duplicate; curves represent the combined (additive) data from all strains and all experiments.
DISCUSSION
In the current study, we have employed pyrosequencing to evaluate the prevalence of Candida africana among isolates of pathogenic Candida species submitted to the UK National Mycology Reference Laboratory. In agreement with the existing literature (reviewed in reference 22), C. africana was isolated exclusively in this study from female genital specimens. Indeed, C. africana constituted 6% of C. albicans species complex isolates from female genital specimens and was much more prevalent than the related C. dubliniensis in such samples, as has been reported previously from both Europe and the African subcontinent (18, 41). Phenotypically, C. africana differed from C. albicans and C. dubliniensis by the delayed formation of hyphae and absence of chlamydospores in Dalmau cultures, the inability to assimilate trehalose, the lack of N-acetyl-galactosaminidase activity, and the development of smaller, more intensely colored turquoise-green colonies on CHROMagar. Despite slower growth and reduced formation of hyphae, all isolates of C. africana were found to be germ tube positive under standard incubation conditions (3 h at 37°C). However, in contrast to previous reports (22, 39), the United Kingdom isolates of C. africana evaluated here were capable of growth, albeit restricted, at 42°C.
The current study is the first to have evaluated and compared the pathogenicity of C. africana isolates with that of its close relatives C. albicans and C. dubliniensis. As assessed by the ability to kill Galleria mellonella larvae in a systemic infection model, C. africana isolates were significantly less pathogenic than their C. albicans counterparts at 30°C, 37°C, and 42°C and less pathogenic than C. dubliniensis isolates at both 30°C and 37°C (Fig. 2). In agreement with many previous studies in a wide range of infection models (reviewed in reference 15), C. dubliniensis was less pathogenic than C. albicans in G. mellonella at all incubation temperatures evaluated, although this difference was not significant in larvae incubated at 37°C. Thus, at least in Galleria larvae, the members of the C. albicans complex can be ordered C. albicans > C. dubliniensis > C. africana in terms of virulence. Virulence factors have been extensively studied in C. albicans and include adhesins, kinases, secreted extracellular enzymes, and, importantly, the ability to alternate between filamentous (hyphal) and budding yeast forms (reviewed in references 15, 42, and 43). Indeed, hyphal formation has been proposed to play pivotal roles in adhesion to host cell surfaces and subsequent biofilm formation, tissue invasion, and immune evasion (44). In this respect, it is noteworthy that mutations in C. albicans genes associated with hyphal formation and regulation resulted in reduced virulence in Galleria larvae (36). Previously, the reduced virulence of C. dubliniensis compared to C. albicans has been attributed to lower filamentation rates in the former (45, 46), the loss of several virulence factor genes that appear to be preferentially expressed by the hyphal form (reviewed in reference 15), and a reduced ability to tolerate a wide range of environmental stresses (46).
Here we have reported that C. africana grows more slowly than both C. albicans and C. dubliniensis in vitro and has a much reduced ability to form hyphal filaments on Dalmau culture. This lower filamentation rate was also apparent in G. mellonella larvae. In larvae sacrificed 8 h after inoculation with the various Candida species, C. albicans and to a lesser degree C. dubliniensis were present throughout both the insect hemolymph and fat body as a mixture of budding yeasts, true mycelium, and pseudohyphae. In larvae inoculated with C. africana, only replicating yeast forms could be detected (data not shown). Thus, it is plausible that this lower filamentation rate contributes to the reduced virulence of C. africana in this infection model. For C. dubliniensis, it has been proposed that reductive evolution involving the loss of key virulence genes has resulted in the narrowing of environmental conditions conducive to hyphal formation (15), perhaps as an adaptive change to a particular anatomical niche in which filamentation is less required for colonization. In future comparative genomic studies, it will be important to establish whether the genetic repertoire of C. africana is similarly reduced and whether the inability of C. africana to form hyphae accounts for the apparent restriction of this organism in humans to colonization of the female genital tract. Further studies will be required to compare the abilities of C. africana, C. albicans, and C. dubliniensis to colonize this particular anatomical niche.
ACKNOWLEDGMENT
We are grateful to the other members of the United Kingdom MRL for their assistance with data collation and phenotypic and molecular analyses of isolates.
Footnotes
Published ahead of print 9 January 2013
REFERENCES
- 1. Marr KA. 2004. Invasive Candida infections: the changing epidemiology. Oncology 18(14 Suppl 13):9–14 [PubMed] [Google Scholar]
- 2. Nucci M, Marr KA. 2005. Emerging fungal diseases. Clin. Infect. Dis. 41:521–526 [DOI] [PubMed] [Google Scholar]
- 3. Pfaller MA, Jones RN, Doern GV, Fluit AC, Verhoef J, Sader HS, Messer SA, Houston A, Coffman S, Hollis RJ. 1999. International surveillance of blood stream infections due to Candida species in the European SENTRY program: species distribution and antifungal susceptibility including the investigational triazole and echinocandin agents. SENTRY Participaant Group (Europe). Diagn. Microbiol. Infect. Dis. 35:19–25 [DOI] [PubMed] [Google Scholar]
- 4. Pfaller MA, Jones RN, Messer SA, Edmond MB, Wenzel RP. 1998. National surveillance of nosocomial blood stream infection due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE Program. Diagn. Microbiol. Infect. Dis. 31:327–332 [DOI] [PubMed] [Google Scholar]
- 5. Ruhnke M. 2006. Epidemiology of Candida albicans infections and role of non-Candida albicans yeasts. Curr. Drug Targets 7:495–504 [DOI] [PubMed] [Google Scholar]
- 6. Snydman DR. 2003. Shifting patterns in the epidemiology of nosocomial Candida infections. Chest 123:500S–503S [DOI] [PubMed] [Google Scholar]
- 7. Wingard JR. 1994. Infections due to resistant Candida species in patients with cancer who are receiving chemotherapy. Clin. Infect. Dis. 19(Suppl 1):49–53 [DOI] [PubMed] [Google Scholar]
- 8. Wright WL, Wenzel RP. 1997. Nosocomial Candida. Epidemiology, transmission and prevention. Infect. Dis. Clin. North Am. 11:411–425 [DOI] [PubMed] [Google Scholar]
- 9. Moran GP, Coleman DC, Sullivan DJ. 2011. Comparative genomics and evolution of pathogenicity in human pathogenic fungi. Eukaryot. Cell 10:34–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sullivan DJ, Westerneng TJ, Haynes KA, Bennett DE, Coleman DC. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141:1507–1521 [DOI] [PubMed] [Google Scholar]
- 11. Khan Z, Ahmad S, Joseph L, Chandy R. 2012. Candida dubliniensis: an appraisal of its clinical significance as a bloodstream pathogen. PLoS One 7:e32952 doi:10.1371/journal.pone.0032952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sullivan DJ, Coleman DC. 1997. Candida dubliniensis: an emerging opportunistic pathogen. Curr. Top. Med. Mycol. 8:15–25 [PubMed] [Google Scholar]
- 13. Sullivan DJ, Moran GP, Coleman DC. 2005. Candida dubliniensis: ten years on. FEMS Microbiol. Lett. 253:9–17 [DOI] [PubMed] [Google Scholar]
- 14. Sullivan DJ, Moran GP, Pinjon E, Al-Mosaid A, Stokes C, Vaughan C, Coleman DC. 2004. Comparison of the epidemiology, drug resistance mechanisms, and virulence of Candida dubliniensis and Candida albicans. FEMS Yeast Res. 4:369–376 [DOI] [PubMed] [Google Scholar]
- 15. Moran GP, Coleman DC, Sullivan DJ. 2012. Candida albicans versus Candida dubliniensis: why is C. albicans more pathogenic? Int. J. Microbiol. 2012:205291 doi:10.1155/2012/205921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Romeo O, de Leo F, Criseo G. 2011. Adherence ability of Candida africana: a comparative study with Candida albicans and Candida dubliniensis. Mycoses 54:e57–e61 [DOI] [PubMed] [Google Scholar]
- 17. Forche A, Schönian G, Gräser Y, Vilgalys R, Mitchell TG. 1999. Genetic structure of typical and atypical populations of Candida albicans from Africa. Fungal Genet. Biol. 28:107–125 [DOI] [PubMed] [Google Scholar]
- 18. Tietz HJ, Küssner A, Thanos M, De Andrade MP, Presber W, Schönian G. 1995. Phenotypic and genotypic characterization of unusual vaginal isolates of Candida albicans from Africa. J. Clin. Microbiol. 33:2462–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Alonso-Vargas R, Elorduy L, Eraso E, Cano FJ, Guarro J, Ponton J, Quindos G. 2008. Isolation of Candida africana, probable atypical strains of Candida albicans, from a patient with vaginitis. Med. Mycol. 46:167–170 [DOI] [PubMed] [Google Scholar]
- 20. Jacobsen MD, Boekhout T, Odds FC. 2008. Multilocus sequence typing confirms synonymy but highlights differences between Candida albicans and Candida stellatoidea. FEMS Yeast Res. 8:764–770 [DOI] [PubMed] [Google Scholar]
- 21. Romeo O, Criseo G. 2009. Morphological, biochemical and molecular characterization of the first Italian Candida africana isolate. Mycoses 52:454–457 [DOI] [PubMed] [Google Scholar]
- 22. Romeo O, Criseo G. 2011. Candida africana and its closest relatives. Mycoses 54:475–486 [DOI] [PubMed] [Google Scholar]
- 23. Tietz HJ, Hopp M, Schmalreck A, Sterry W, Czaika V. 2001. Candida africana sp. nov., a new human pathogen or a variant of Candida albicans? Mycoses 44:437–445 [DOI] [PubMed] [Google Scholar]
- 24. Romeo O, Criseo G. 2008. First molecular method for discriminating between Candida africana, Candida albicans and Candida dubliniensis by using hwp1 gene. Diagn. Microbiol. Infect. Dis. 62:230–233 [DOI] [PubMed] [Google Scholar]
- 25. Romeo O, Criseo G. 2009. Molecular epidemiology of Candida albicans and its closely related yeasts Candida dubliniensis and Candida africana. J. Clin. Microbiol. 47:212–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Campbell CK, Davey KG, Holmes AD, Szekely A, Warnock DW. 1999. Comparison of the API Candida system with the AUXACOLOR2 system for identification of common yeast pathogens. J. Clin. Microbiol. 37:821–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Borman AM, Linton CJ, Oliver D, Palmer MD, Szekely A, Johnson EM. 2010. Rapid molecular identification of pathogenic yeasts by pyrosequencing analysis of 35 nucleotides of internal transcribed spacer 2. J. Clin. Microbiol. 48:3648–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Borman AM, Linton CJ, Oliver D, Palmer MD, Szekely A, Odds FC, Johnson EM. 2009. Pyrosequencing analysis of 20 nucleotides of internal transcribed spacer 2 discriminates Candida parapsilosis, Candida metapsilosis, and Candida orthopsilosis. J. Clin. Microbiol. 47:2307–2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Borman AM, Petch R, Linton CJ, Palmer MD, Bridge PD, Johnson EM. 2008. Candida nivariensis, an emerging pathogenic fungus with multidrug resistance to antifungal agents. J. Clin. Microbiol. 46:933–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Borman AM, Campbell CK, Linton CJ, Johnson EM. 2006. Polycytella hominis is a mutated form of Scedosporium apiospermum. Med. Mycol. 44:33–39 [DOI] [PubMed] [Google Scholar]
- 31. Linton CJ, Borman AM, Cheung G, Holmes AD, Szekely A, Palmer MD, Bridge PD, Campbell CK, Johnson EM. 2007. Molecular identification of unusual pathogenic yeast isolates by large ribosomal subunit gene sequencing: 2 years of experience at the United Kingdom Mycology Reference Laboratory. J. Clin. Microbiol. 45:1152–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Borman AM, Fraser M, Linton CJ, Palmer MD, Johnson EM. 2010. An improved method for the preparation of total genomic DNA from isolates of yeast and mould using Whatman FTA filter papers. Mycopathologia 169:445–449 [DOI] [PubMed] [Google Scholar]
- 33. Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution susceptibility testing of yeasts: approved standard, 3rd ed Document M27-A3 Clinical and Laboratory Standards Institute, Wayne, PA [Google Scholar]
- 34. Cotter G, Doyle S, Kavanagh K. 2000. Development of an insect model for the in vivo pathogenicity testing of yeasts. FEMS Immunol. Med. Microbiol. 27:163–169 [DOI] [PubMed] [Google Scholar]
- 35. Fallon J, Kelly J, Kavanagh K. 2012. Galleria mellonella as a model for fungal pathogenicity testing. Methods Mol. Biol. 845:469–485 [DOI] [PubMed] [Google Scholar]
- 36. Fuchs BB, Eby J, Nobile CJ, El Khoury JB, Mitchell AP, Mylonakis E. 2010. Role of filamentation in Galleria mellonella killing by Candida albicans. Microbes Infect. 12:488–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fuchs BB, O'Brien E, Khoury JB, Mylonakis E. 2010. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence 1:475–482 [DOI] [PubMed] [Google Scholar]
- 38. Odds FC, Bougnoux ME, Shaw DJ, Bain JM, Davidson AD, Diogo D, Jacobsen MD, Lecomte M, Li SY, Tavanti A, Maiden MC, Gow NA, d'Enfert C. 2007. Molecular phylogenetics of Candida albicans. Eukaryot. Cell. 6:1041–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tintelnot K, Haase G, Siebold M, Bergmann F, Staemmler M, Franz T, Naumann D. 2000. Evaluation of phenotypic markers for selection and identification of Candida dubliniensis. J. Clin. Microbiol. 38:1599–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Junqueira JC, Fuchs BB, Muhammed M, Coleman JJ, Suleiman JMAH, Vilela SFG, Costa ACBP, Rasteiro VMC, Jorge AOC, Mylonakis E. 2011. Oral Candida albicans isolates from HIV-positive individuals have similar in vitro biofilm-forming ability and pathogenicity as invasive Candida isolates. BMC Microbiol. 11:247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nnadi NE, Ayanbimpe GM, Scordina F, Okolo MO, Enweani IB, Criseo G, Romeo O. 2012. Isolation and molecular characterisation of Candida africana from Jos, Nigeria. Med. Mycol. 50:765–767 [DOI] [PubMed] [Google Scholar]
- 42. Sudbery P, Gow N, Berman J. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317–324 [DOI] [PubMed] [Google Scholar]
- 43. Sullivan DJ, Moran GP. 2011. Differential virulence of Candida albicans and C. dubliniensis: a role for Tor1 kinase? Virulence 2:77–81 [DOI] [PubMed] [Google Scholar]
- 44. Finkel JS, Mitchell AP. 2010. Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 9:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stokes C, Moran GP, Spiering MJ, Cole GT, Coleman DC, Sullivan DJ. 2007. Lower filamentation rates of Candida dubliniensis contribute to its lower virulence in comparison with Candida albicans. Fungal Genet. Biol. 44:920–931 [DOI] [PubMed] [Google Scholar]
- 46. Vilela MMS, Kamei K, Sano A, Tanaka R, Uno J, Takahashi I, Ito J, Yarita K, Miyaji M. 2002. Pathogenicity and virulence of Candida dubliniensis: comparison with C. albicans. Med. Mycol. 40:249–257 [DOI] [PubMed] [Google Scholar]