Abstract
Determining the viral etiology of respiratory tract infections (RTI) has been limited for the most part to specific primer PCR-based methods due to their increased sensitivity and specificity compared to other methods, such as tissue culture. However, specific primer approaches have limited the ability to fully understand the diversity of infecting pathogens. A pathogen chip system (PathChip), developed at the Genome Institute of Singapore (GIS), using a random-tagged PCR coupled to a chip with over 170,000 probes, has the potential to recognize all known human viral pathogens. We tested 290 nasal wash specimens from Filipino children <2 years of age with respiratory tract infections using culture and 3 PCR methods—EraGen, Luminex, and the GIS PathChip. The PathChip had good diagnostic accuracy, ranging from 85.9% (95% confidence interval [CI], 81.3 to 89.7%) for rhinovirus/enteroviruses to 98.6% (95% CI, 96.5 to 99.6%) for PIV 2, compared to the other methods and additionally identified a number of viruses not detected by these methods.
INTRODUCTION
Respiratory infections are the single most important cause of death in childhood (1, 2), primarily due to bacterial and viral pathogens. Unfortunately, clinical features and current laboratory methods do not readily identify the etiologic agent. These laboratory methods have traditionally involved culture (3) and antibody-based approaches (1, 2, 4, 5), but in recent years, nucleic acid-based methods, such as PCR (3, 6), microarrays (7, 8), and next-generation sequencing (NGS) (9–11), have gradually gained acceptance and even preference over traditional methods for pathogen identification due to their higher sensitivities and specificities, decreasing cost, and multiplexing capabilities. Nevertheless, large-scale pathogen diagnostics (covering extensive pathogen diversity) for discovery and biosurveillance are still not in routine use. PCR, even though it is highly sensitive and fast, has limitations in detecting novel pathogens since it requires the selection of suitably specific primers from known sequences (12). Conversely, NGS approaches can provide sequence information of known pathogens in a sample but require complex postprocessing analysis, such as sequence assembly and alignment for diagnostic conclusions (13, 14).
The microarray technology has the potential to overcome both of these shortcomings and has thus begun to establish itself as an important diagnostic tool. It consists of thousands of fluorescence-labeled nucleic acid probes that bind with high specificity to complementary sequences of nucleic acid extracted from biological samples. While microarrays can detect multiple pathogens simultaneously, their clinical utility has been limited by their sensitivity in clinical specimens (11). Much effort has thus been spent on improving sample amplification techniques (15) and developing more-sophisticated algorithms to increase the sensitivity and accuracy of detection (15, 16). With a deeper understanding of probe hybridization properties, nonspecific hybridization noise (once considered a major drawback of microarrays) can now be used to detect or implicate novel pathogens in the specimen (17). In the last decade, many pathogen detection and discovery microarrays, such as the ViroChip (18), GreeneChip (19), PathChip (17), and Lawrence Livermore microbial detection array (LLMDA) (20), as well as resequencing microarrays (21–24), have been developed and some have been commercialized. LLMDA, comprised of 388,000 probes representing 38,000 virus sequences and 3,500 bacteria sequences, is the most comprehensive microarray to date (20). Recently, resequencing microarrays (24–26) and low-density arrays (27, 28) have also been used to detect multiple respiratory pathogens simultaneously in clinical samples.
The most prevalent PCR methods in clinical use are single-plex kits, which have been widely commercialized and FDA approved for specific pathogens. Multiplex PCR panels, which can detect 5 to 30 respiratory pathogens in a single assay, have been developed on a variety of formats, ranging from standard TaqMan quantitative PCR (qPCR) assays (29) and liquid bead array platforms (30–34) to lab-on-a-chip devices such as the BioFire FilmArray (35, 36). FilmArray incorporates on-board nucleic acid extraction together with automated nested multiplex PCR for detection of 25 pathogens. This device, as well as some of the multiplex panels, has since received FDA approval for diagnostic use (36, 37).
In this study, we demonstrate that the current version of the Genome Institute of Singapore (GIS) PathChip can detect at least 76 viruses, with sensitivity and specificity comparable to those of other molecular diagnosis methods.
MATERIALS AND METHODS
Study population.
From July 2000 to December 2004, a cohort of 12,194 Filipino children <2 years of age participated in a pneumococcal vaccine trial conducted in 6 barangays (villages) in Bohol, Philippines (38). The data on all hospital admissions and other important medical events were collected until a study termination visit at 23 months of age or the end of study follow-up at the end of December 2004. Concomitantly, all children from the 6 barangays who were ≤5 years of age, who were not in the trial, and who were admitted to the hospital or who visited the outpatient department of the Bohol Regional Hospital were enrolled in a separate epidemiologic study. Informed consent was obtained for all children for participation in the vaccine trial, and separate consent was obtained at admission for each child to obtain a history and specimens. The institutional review boards (IRB) at the Research Institute for Tropical Medicine (RITM) in Manila, Philippines, KTL Finland, and COMIRB, Aurora, CO, approved the study. In both of these groups of subjects, 5,570 clinical episodes of pneumonia were observed, at which time 2,066 nasopharyngeal aspirate samples were obtained to determine a viral etiology. We tested a random sample of 290 of these nasal wash specimens for this study.
Specimen collection and processing.
Nasal wash specimens were collected from all children admitted in the trial who were hospitalized at the Bohol Regional Hospital with acute lower respiratory tract infections (ALRI) and 1 in 5 outpatients with ALRI. The baby was restrained by being wrapped in a sheet and placed on the back with the neck extended. Five milliliters of normal saline was squirted into one nostril, and a 6 Fr suction catheter was placed into the nasopharynx using the opposite nostril, pointing the end of the tube directly backward, with the patient aspirating while the suction catheter was gently removed. Suction was applied using a pressure-regulated, trapped electrical suction pump or, when not available, a 10-ml sterile syringe. The procedure was repeated in the other naris if necessary. After we ensured that the specimen was cloudy and had mucus and cells, it was immediately placed on ice and transported to the laboratory, where it was then mixed with Remel M4 viral transport medium (Thermo Fisher Scientific, Lenexa, KS) in a 1:1 ratio, within about half an hour of collection. After being mixed and vortexed briefly, the materials were transferred into 3 to 5 cryovials and snap-frozen in liquid nitrogen, in which the specimens were stored for up to a month. Next, batches of specimens were transported to Denver, CO, on dry ice, where they were stored at −86°C.
Viral culture.
We utilized both routine respiratory tissue culture and conventional respiratory shell vial culture for detection of viruses, as described earlier (39). Briefly, respiratory tissue culture was performed by inoculating 0.3 ml of specimen into each of six tissue culture tubes: two rhesus monkey kidney (RhMK) tubes (BioWhittaker and ViroMed), one Hep-2 (ATCC), one A549 (ATCC), and two primary human embryonic lung fibroblasts (HELF) were prepared in the laboratory. Tubes were incubated at 37°C for up to 28 days. One HELF tube was incubated at 35°C on a roller drum to enhance rhinovirus isolation. Tubes were examined by light microscopy for up to 28 days, and hemadsorption of guinea pig red blood cells was performed weekly. Monolayers that showed cytopathic effect or positive hemadsorption were scraped and stained with specific monoclonal antibodies. Conventional respiratory shell vial cultures were performed by inoculation of 0.3 ml of the specimens into a shell vial containing either RhMK (Viromed) or locally made Hep-2 cell. Monolayers were scraped after 48 h of incubation at 37°C, spotted onto slides, fixed with acetone, stained with specific monoclonal antibodies (Dako for the first phase study and Bartel's for the second one) for influenza A and B, parainfluenza virus 1 (PIV 1), 2, and 3 (RhMK shell vial [SV]), respiratory syncytial virus (RSV), and adenovirus (Hep-2), and read under fluorescence microscopy. A positive result was defined as the presence of bright green fluorescence in the cytoplasm of ≥2 cells.
Nucleotide extraction and PCR.
RNA was extracted from 1 aliquot of nasal wash specimen stored in M4 transport medium using TRIzol LS (Invitrogen, Grand Island, NY) according to the manufacturer's instructions except as follows. The RNA pellet was dried at 55°C for 5 min. A total of 60 μl of RNA storage solution (Ambion, Grand Island, NY) was added to the dried pellet and heated to 55°C for 5 min to help resuspend the pellet. Three aliquots were made, each with 20 μl of the RNA, and then stored at −80°C. RNA samples were then processed according to manufacturers' instructions for the EraGen and Luminex platforms (PCR) and the GIS specific protocol for the PathChip. All the laboratory experiments were conducted at University of Colorado School of Medicine laboratories.
EraGen.
The EraGen MultiCode-PLs system assay has four steps and produces cDNA that has been labeled twice and then bound to a Luminex bead. All the steps were performed according to the manufacturer's protocol (40). Manufacturer-specified, assay-specific positive and negative controls were included on each plate of the run.
Luminex.
ID-Tag RVP (Tm Bioscience Corp., Toronto, Canada) has several steps, and it was combined with the Luminex X-Map technology for detection and analysis (41). All steps were done in accordance with the manufacturer's protocol. Manufacturer-specified, assay-specific positive and negative controls were included on each plate of the run.
GIS PathChip.
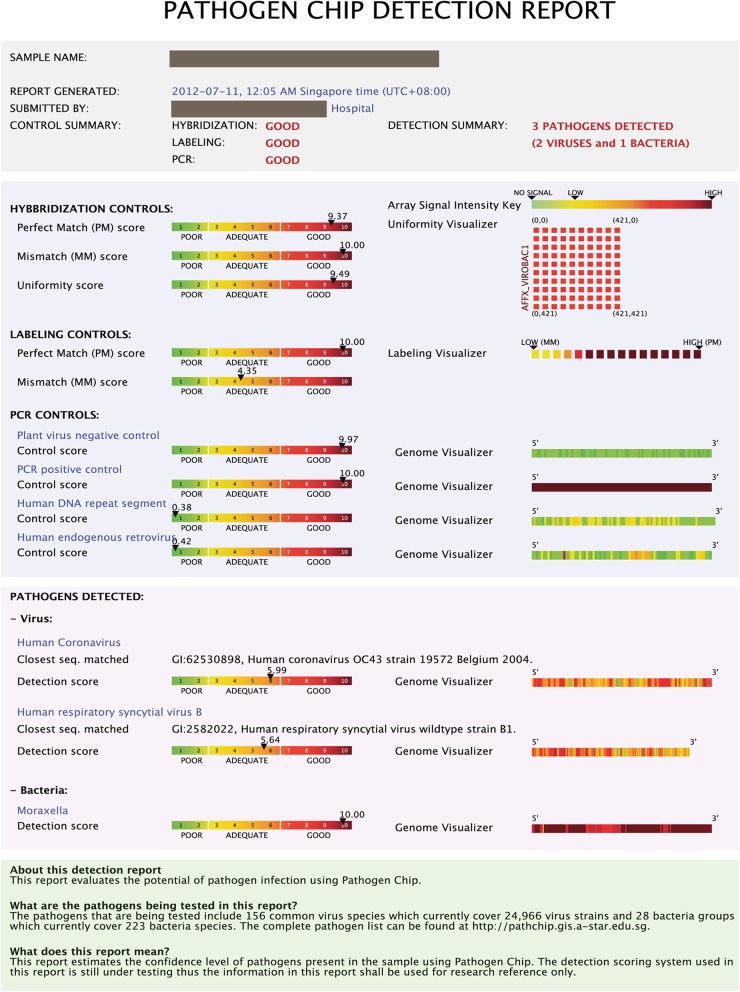
A total of 25,000 full-genome sequences of viruses clinically relevant to humans were downloaded from the NCBI Taxonomy database (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/) and organized into 59 genera and 154 clinically relevant groups. For each viral genome, a probe recognition signature consisting of probes uniformly spaced out across the entire genome was constructed as previously described (15, 17). The chip design was custom manufactured by Affymetrix, Inc. (Santa Clara, CA), in their GeneChip cartridge format. The cDNA was amplified from extracted nucleic acid samples using proprietary primers as described previously (15, 17) in a single-tube reaction (for each sample). The samples were then purified, fragmented, labeled, and hybridized onto the PathChip according to the manufacturer's protocol (PathGEN Dx, Singapore). After hybridization, the PathChip was washed, stained, and scanned using the Affymetrix GeneChip system. The Affymetrix image file (.CEL) containing all the raw signal intensities for each PathChip was uploaded into the GIS proprietary software described previously (17), which automatically detects which pathogen recognition signatures are present. A report which provides a summary of the assay, quality control metrics, and the pathogen genomes which were detected was then generated (Fig. 1). The entire process (starting from patient RNA) was completed within 20 h.
Fig 1.
Sample PathChip report. The 1st section contains a summary of the report. The 2nd section contains hybridization, labeling, and PCR controls. The 3rd section shows the heat maps of the detected pathogens and the closest matches to known strains found in the patient sample.
Development of a gold standard for viral diagnosis.
To test the sensitivity and specificity of the GIS PathChip compared to those of the multiplex panel tests, a gold standard was developed from results of culture and PCR for each specimen. The gold standard for comparison was considered to be positive for a virus if that virus was positive for culture or both PCR methods (EraGen and Luminex). The gold standard was then used to classify each specimen by type of viral infection, with the significance of any differences reported between individual assays determined using the Fisher exact test.
Sensitivity, specificity, and diagnostic accuracy.
The PathChip was compared to the gold standard to evaluate its sensitivity, specificity, and overall diagnostic accuracy. Sensitivity was computed as the proportion of positive specimens that tested positive; likewise, specificity was computed as the proportion of negative specimens that tested negative. The diagnostic accuracy was the proportion of all specimens that were correctly classified among all subjects.
Detection of novel pathogens by the PathChip.
For pathogens that were not covered by the viral culture or the EraGen and Luminex PCR approaches, we reported all positive samples based on the pathogen report, autogenerated by the PathChip software (and initially described as false positive compared to the gold standard).
Validation of viruses detected by PathChip but not detected by other methods (PathChip false-positive results).
This examination was done using 3 strategies. Since the gold standard included only viruses detected by both EraGen and Luminex methods, we considered a PathChip false-positive virus to be confirmed if it was also detected in one of the 2 PCR methods. For the poliovirus and the influenza viruses, we sequenced the VP1 region of poliovirus (42) and the complete hemagglutination and neuraminidase genes for influenza (43) using Sanger sequencing. Finally, for those samples with detected viruses that were not confirmed by these 2 methods, we performed NGS of the sample aliquots when available.
Validation of PathChip results by next-generation sequencing.
The 12 samples that were not confirmed by other PCR approaches or Sanger sequencing were selected for NGS validation. Each sample was amplified using PathChip's random-tagged PCR protocol. The amplified sample was then further processed for MiSeq multiplexing sequencing by following the Illumina TruSeq protocol (Illumina Inc., San Diego, CA). This generated 3.5 million paired-end reads (2 × 100 bp) covering the 12 samples. After the primer tag was removed from the paired-end reads, we used Bowtie 2 (44) to map the reads onto viral sequences downloaded from the NCBI Taxonomy database (http://www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html/). A read was considered mapped to a virus if at least one paired-end read matched the same specific sequence with a mismatch of <5 bp. For each sample, a virus was called if at least one paired-end read aligned to a distinct fragment covering at least 200 bp of the virus sequence with a minimum seed length of 20 bp and with an alignment score greater than 20 + 8.0 × ln(read length) in a multiple-seed heuristic local alignment.
RESULTS
Clinical features of the subjects.
This study was conducted in the context of a pneumococcal vaccine trial (38), and 47% of the patients included in this study had been in the vaccinated group (see Table S1 in the supplemental material). The mean (± standard deviation) age of the children was 11.2 ± 7.1 months. Nearly 60% of children had severe or very severe pneumonia and only 4 had upper respiratory tract infections, while the remainder had nonsevere pneumonia.
Laboratory results.
The gold standard panel identified respiratory syncytial virus (RSV) (culture only) or RSVA and RSVB (when culture was confirmed by PCR), parainfluenza viruses (PIV) 1, 2, and 3, influenza (INF) A and B, human rhinovirus (HRV) or enterovirus (EV), and adenoviruses (AD) (Table 1). PIV 4 was detected only by the PathChip, while the coronaviruses were detected only by PathChip and EraGen (as our tissue culture method was not suitable for coronavirus detection). Thus, PIV 4 was not included in the gold standard, and the gold standard for coronaviruses was considered inadequate. Furthermore, tissue culture was less sensitive in detecting enteroviruses than were the PCR platforms (P < 0.005).
Table 1.
Number of positive infections by virus type and platform
| Virus | No. (%) of positive infections bya: |
||||
|---|---|---|---|---|---|
| Gold standard | Culture | EraGen | Luminex | PathChip | |
| Adenovirus | 13 (4.5) | 13 (4.5) | 19 (6.6) | 5 (1.7) | 4 (1.4) |
| Coronavirusb | NAd | NA | 42 (14.5) | NA | 18 (6.2) |
| OC43 | NA | NA | 40 (13.8) | NA | 12 (4.1) |
| 229E | NA | NA | 0 (0) | NA | 4 (1.4) |
| NL63 | NA | NA | 2 (0.7) | NA | 2 (0.7) |
| Enterovirus/HRV | 83 (28.6) | 24 (8.3) | 113 (39) | 91 (31.4) | 90 (31) |
| HMPV | 20 (6.9) | NA | 27 (9.3) | 22 (7.6) | 23 (7.9) |
| INF A | 26 (9) | 23 (7.9) | 24 (8.3) | 24 (8.3) | 26 (9) |
| INF B | 11 (3.8) | 11 (3.8) | 11 (3.8) | 8 (2.8) | 11 (3.8) |
| PIV 1 | 15 (5.2) | 15 (5.2) | 11 (3.8) | 11 (3.8) | 11 (3.8) |
| PIV 2 | 12 (4.1) | 10 (3.4) | 11 (3.8) | 8 (2.8) | 8 (2.8) |
| PIV 3 | 19 (6.6) | 16 (5.5) | 24 (8.3) | 16 (5.5) | 18 (6.2) |
| RSVc | 88 (30.3) | 75 (25.9) | 94 (32.4) | 76 (26.2) | 72 (24.8) |
| RSV A | 46 (15.9) | NA | 64 (22.1) | 42 (14.5) | 37 (12.8) |
| RSV B | 34 (11.7) | NA | 39 (13.4) | 34 (11.7) | 37 (12.8) |
| No. (%) of virus-negative specimens | 36 (12.4) | 101 (34.8) | 37 (12.8) | 60 (20.7) | 49 (16.9) |
There were 290 samples tested by each platform.
Only EraGen was used to test for coronavirus; therefore, there was no possible positive coronavirus result for the gold standard.
Culture detected some that PCR did not, and two subjects had both RSV A and RSV B infections.
NA, not available.
Overall, there was no significant difference between the numbers of virus-negative specimens by any of the 3 PCR-based detection methods between themselves or with the gold standard (P > 0.15). However, all the PCR methods had significantly more viruses detected than did culture (P < 0.005). Compared to the gold standard (Table 2), the PathChip had a diagnostic accuracy that ranged between 85.9% for enterovirus/rhinovirus to 98.6% for PIV 2.
Table 2.
PathChip concordance with the gold standard, sensitivity, and specificity analysis by virus group
| Virusb | No. true positive | No. false positive | No. false negative | No. true negative | Sensitivity (%) (95% CI)a | Specificity (%) (95% CI) | Diagnostic accuracy (95% CI) |
|---|---|---|---|---|---|---|---|
| ADV | 4 | 0 | 9 | 277 | 30.8 (9.1, 61.4) | 100.0 (98.7, 100) | 96.9 (94.2, 98.6) |
| CoVc | 0 | 18 | 0 | 272 | NA | 93.8 (90.4, 96.3) | 93.8 (90.4, 96.3) |
| OC43 | 0 | 12 | 0 | 278 | NA | 95.9 (92.9, 97.8) | 95.9 (92.9, 97.8) |
| 229E | 0 | 4 | 0 | 286 | NA | 98.6 (96.5, 99.6) | 98.6 (96.5, 99.6) |
| NL63 | 0 | 2 | 0 | 288 | NA | 99.3 (97.5,99.9) | 99.3 (97.5,99.9) |
| EV/HRV | 66 | 24 | 17 | 183 | 79.5 (69.2, 87.6) | 88.4 (83.2, 92.4) | 85.9 (81.3, 89.7) |
| HMPV | 19 | 4 | 1 | 266 | 95.0 (75.1, 99.9) | 98.5 (96.3, 99.6) | 98.3 (96.0, 99.4) |
| INF A | 22 | 4 | 4 | 260 | 84.6 (65.1, 95.6) | 98.5 (96.2, 99.6) | 97.2 (94.6, 98.8) |
| INF B | 8 | 3 | 3 | 276 | 72.7 (39.0, 94.0) | 98.9 (96.9, 99.8) | 97.9 (95.6, 99.2) |
| PIV 1 | 10 | 1 | 5 | 274 | 66.7 (38.4, 88.2) | 99.6 (98.0, 100) | 97.9 (95.6, 99.2) |
| PIV 2 | 8 | 0 | 3 | 279 | 66.7 (34.9, 90.1) | 100.0 (98.7, 100) | 98.6 (96.5, 99.6) |
| PIV 3 | 16 | 2 | 3 | 269 | 84.2 (60.4, 96.6) | 99.3 (97.4, 99.9) | 98.3 (96.0, 99.4) |
| RSV A | 36 | 1 | 10 | 243 | 78.3 (63.6, 89.1) | 99.6 (97.7, 100) | 96.2 (93.3, 98.9) |
| RSV B | 31 | 6 | 3 | 250 | 91.2 (76.3, 98.1) | 97.7 (95.0, 99.1) | 96.9 (94.2, 98.6) |
| RSV | 68 | 4 | 20 | 198 | 77.3 (67.1, 85.5) | 98.0 (95.0, 99.5) | 91.7 (87.9, 94.6) |
Confidence intervals for the binomial proportion were computed with exact (Clopper-Pearson) confidence limits. NA, not applicable.
ADV, human adenovirus; CoV, human coronavirus; EV, human enterovirus; HRV, human rhinovirus; INF, influenza virus; PIV, human parainfluenza virus; RSV, respiratory syncytial virus.
Despite a clear gold standard for coronavirus, this reflects GIS false positives compared to EraGen only.
PathChip.
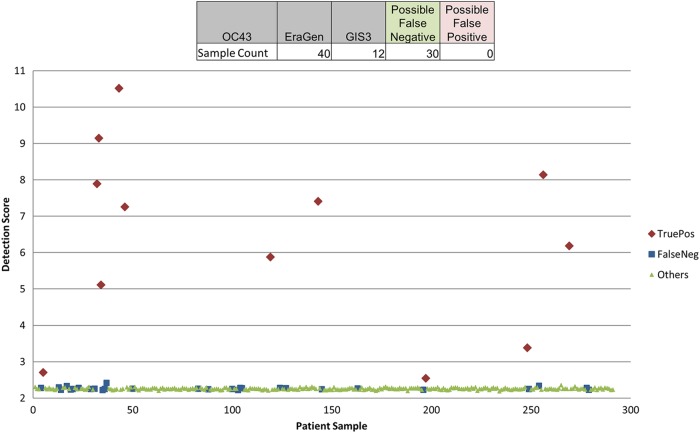
Figure 1 shows a sample report from the PathChip analysis, which is divided into 3 sections. The 1st section contains a summary including basic identification of the patient, technician, and laboratory, whether the test was successfully run, and the number of pathogens detected. The 2nd section contains a detailed description of various controls for the test, including PCR controls, labeling controls, and hybridization controls, that indicate the quality of the test. The final section shows heat maps of any detected pathogens, illustrating which regions of the pathogen genome were detected and identifying the known virus strains which are most similar to that found in the patient sample. The PathChip had >93% specificity for all of the gold standard viruses (Table 2), with the capacity to detect individual viral infections and describe the genome with the closest match. The PathChip had a lower sensitivity for adenovirus than did the other methods. For coronavirus OC43, there was a divergence between the PathChip and EraGen results that was difficult to interpret without reference to another method (Table 1). While EraGen detected coronavirus OC43 more often than did the PathChip, some samples clearly positive by PathChip were negative by EraGen (Fig. 2).
Fig 2.
OC43 detection score plot. EraGen identified 40 specimens with coronavirus OC43 infection (samples indicated by red and blue). The PathChip identified 12 specimens with coronavirus OC43 (red). The other 30 specimens (blue) were not identified by either PathChip or Luminex. The background detection scores of coronavirus OC43 in other specimens are also shown in green. TruePos, true positive; FalseNeg, false negative.
In total, the PathChip identified 76 different viruses, including a number of viruses not identified by any of the other methods, including poliovirus, bocavirus, parechovirus, and rotavirus (Table 3).
Table 3.
Viruses detected by GIS PathChip
| Virusa | No. detected | % of total |
|---|---|---|
| AD type 35 | 1 | 0.3 |
| AD type 5 | 1 | 0.3 |
| AD type 7 | 1 | 0.3 |
| AD type C | 1 | 0.3 |
| Bocavirus | 6 | 1.6 |
| CMV (HHV5) | 7 | 1.9 |
| HCoV 229E | 4 | 1.1 |
| HCoV NL63 | 2 | 0.5 |
| HCoV OC43 | 12 | 3.3 |
| Coxsackievirus A12 | 1 | 0.3 |
| Coxsackievirus A16 | 22 | 6.0 |
| Coxsackievirus A18 | 1 | 0.3 |
| Coxsackievirus A24 | 1 | 0.3 |
| Coxsackievirus A4 | 1 | 0.3 |
| Coxsackievirus B4 | 1 | 0.3 |
| Coxsackievirus B6 | 1 | 0.3 |
| Echovirus 11 | 2 | 0.5 |
| Echovirus 16 | 2 | 0.5 |
| Echovirus 19 | 1 | 0.3 |
| Echovirus 27 | 1 | 0.3 |
| Echovirus 9 | 2 | 0.5 |
| EV | 1 | 0.3 |
| EV 101 | 1 | 0.3 |
| EV 68 | 4 | 1.1 |
| EV 74 | 1 | 0.3 |
| EV 86 | 4 | 1.1 |
| EV 87 | 2 | 0.5 |
| EV Hangzhou | 3 | 0.8 |
| EV Ningbo | 1 | 0.3 |
| HMPV | 23 | 6.3 |
| HRV 10 | 2 | 0.5 |
| HRV 100 | 1 | 0.3 |
| HRV 12 | 4 | 1.1 |
| HRV 15 | 3 | 0.8 |
| HRV 1B | 1 | 0.3 |
| HRV 2 | 1 | 0.3 |
| HRV 20 | 1 | 0.3 |
| HRV 22 | 2 | 0.5 |
| HRV 24 | 4 | 1.1 |
| HRV 25 | 7 | 1.9 |
| HRV 26 | 1 | 0.3 |
| HRV 3 | 1 | 0.3 |
| HRV 31 | 1 | 0.3 |
| HRV 33 | 3 | 0.8 |
| HRV 38 | 2 | 0.5 |
| HRV 47 | 1 | 0.3 |
| HRV 49 | 1 | 0.3 |
| HRV 50 | 2 | 0.5 |
| HRV 60 | 3 | 0.8 |
| HRV 61 | 1 | 0.3 |
| HRV 65 | 1 | 0.3 |
| HRV 67 | 1 | 0.3 |
| HRV 7 | 3 | 0.8 |
| HRV 70 | 1 | 0.3 |
| HRV 76 | 2 | 0.5 |
| HRV 78 | 2 | 0.5 |
| HRV 80 | 5 | 1.4 |
| HRV 81 | 1 | 0.3 |
| HRV 85 | 2 | 0.5 |
| HRV 89 | 1 | 0.3 |
| HRV 9 | 1 | 0.3 |
| HRV 92 | 1 | 0.3 |
| HRV C | 5 | 1.4 |
| INF A H1N1 | 3 | 0.8 |
| INF A H3N2 | 23 | 6.3 |
| INF B | 11 | 3.0 |
| INF C | 1 | 0.3 |
| Parechovirus | 5 | 1.4 |
| PIV 1 | 11 | 3.0 |
| PIV 2 | 8 | 2.2 |
| PIV 3 | 18 | 4.9 |
| PIV 4 | 4 | 1.1 |
| Poliovirus | 1 | 0.3 |
| Rotavirus | 1 | 0.3 |
| RSV A | 37 | 10.1 |
| RSV B | 37 | 10.1 |
AD, human adenovirus; CMV (HHV5), cytomegalovirus (human herpesvirus 5); HCoV, human coronavirus; EV, human enterovirus; HMPV, human metapneumovirus; HRV, human rhinovirus; INF, influenza virus; PIV, human parainfluenza virus; RSV, respiratory syncytial virus.
Validation of PathChip false positives not detected by the gold standard.
There were 57 specimens in which one or more viruses that were detected by the PathChip were not confirmed by the gold standard (Table 2). For these samples, we investigated if there was supporting evidence for the presence of the PathChip-identified virus. For 12 of the 57 samples, no RNA remained for further testing and no alternative confirmation was available. For two samples, Sanger sequencing corroborated the identity, finding poliovirus in 1 specimen and influenza virus in another specimen. In 34 specimens, the virus identified by the PathChip was also identified by one of the other PCR methods (EraGen or Luminex). The remaining 12 samples were subjected to NGS, as described in Materials and Methods, with the results shown in Table 4. For nine of these 12 samples, the PathChip results correlated well with the NGS results, although for sample 702-0-1100, the PathChip reported the third coinfecting virus to be PIV 1 whereas the NGS and culture identified it as PIV 3. For the remaining three samples, discrepancies were observed (Table 4). They include PathChip-identified INF B virus in sample 701-0-1761 and INF A in sample 702-0-2174, findings which were not seen by any of the other methods, and EV 86 and RSV B in sample 701-0-1464, which were not seen by any other method. Overall, of the 17 viruses described as false positives in the 12 samples investigated by NGS, 12 were concordant with the NGS results (including two, bocavirus and influenza C virus, that were not covered by other technologies) while five were divergent.
Table 4.
Examination of PathChip false positivesa
| Sample | PathChip v.1.2a | False positive reported | MiSeq sequencing | Other platform result(s)b | Conclusion for the false positivesc |
|---|---|---|---|---|---|
| 701-0-0379 | HCoV OC43 | HCoV OC43 | HCoV OC43 | E+; others NT | HCoV OC43: TP |
| CMV/HHV5 | C−; others NT | ||||
| 701-0-1056 | HCoV 229E | HCoV 229E | HCoV 229E | E−; others NT | HCoV 229E: TP |
| HRV 7 | HRV 88/7 RSVA | C−, E+, L+ | |||
| 701-0-1464 | EV 86 | EV/HRV | C−, E−, L− | EV/HRV: FP | |
| RSV B, RSV A | RSV B | RSVA | RSV, C+; RSV A, E+, L+ | RSV B: FP | |
| CMV/HHV 5 | C−; others NT | ||||
| 701-0-1686 | HCoV 229E | HCoV 229E | HCoV 229E | E−; others NT | HCoV 229E: TP |
| Echovirus 27 | EV/HRV | C−, E+, L− | EV/HRV: TP | ||
| CMV/HHV 5 | CMV/HHV 5 | C−, others NT | |||
| 701-0-1747 | HCoV 229E | HCoV 229E | HCoV 229E | E−; others NT | HCoV 229E: TP |
| HRV 47 | HRV 12/31/47 | C+, E+, L+ | |||
| 701-0-1761 | INF B | INF B | C−, E−, L− | INF B: FP | |
| PIV 2 | PIV 2 | C+, E+, L+ | |||
| 701-0-1880 | RSV A, RSV B | RSV A, RSV B | RSV A, RSV B | C−, E−, L− | RSV A: TP; RSV B: TP |
| INF B | INF B | C+, E−, L− | |||
| HHV 7 | |||||
| HCoV 229E | E−; others NT | ||||
| 701-0-2422 | HCoV 229E | HCoV 229E | HCoV 229E | E−; others NT | HCoV 229E: TP |
| HRV 25 | C−, E+, L+ | ||||
| 702-0-1100 | Coxsackievirus A16 | EV/HRV | C−, E−, L+ | EV/HRV: TP | |
| HCoV OC43 | HCoV OC43 | E+, others NT | HCoV OC43: TP | ||
| PIV 1 | PIV 1 | PIV 3 | PIV1, C−, E−, L−; PIV3, C+, E−, L− | PIV 1: FP | |
| 702-0-1989 | INF A | INF A | C−, E−, L−, S+ | INF A: TP | |
| HRV 12 | HRV 12 | C−, E+, L+ | |||
| RSV A | |||||
| 702-0-2128 | HCoV OC43 | HCoV OC43 | HCoV OC43 | E+; others NT | HCoV OC43: TP |
| BV | BV | ||||
| 702-0-2174 | PIV 2, INF A | INF A | PIV 2 | C−, E−, L− | INF A: FP |
BV, bocavirus.
C, culture; E, EraGen; L, Luminex; S, capillary sequencing following specific PCR; NT, not tested.
FP, false positive; TP, true positive. One HCoV OC43 GIS FP, per the gold standard of requiring two PCR-positive results, was positive for EraGen but negative for MiSeq. This specimen has not been included in this table. For the final conclusion regarding the suspected false positives, the confirmed false positives are bolded.
Detection of coinfections with the PathChip.
The numbers of nasal wash specimens with single and dual infections are illustrated in Table S2 in the supplemental material, with the number of single infections on the diagonal for each respective virus. A single virus was identified in 171 (59%) of the specimens, and two viruses were identified in 59 (20.3%) of the specimens. Three or more viruses were detected in 11 (3.8%) of the specimens (see Table S3 in the supplemental material), and 49 (16.9%) specimens had no viruses detected. Most of those with multiple coinfections included EV and/or HRV. The most frequent types of dual coinfections were enterovirus with HRV (n = 16) and enterovirus with RSV A (n = 5).
DISCUSSION
In our analysis, we have demonstrated a similar broad agreement between the three PCR approaches and culture viral diagnosis, as has been previously reported (2, 4). Our data also show that these advanced PCR techniques were able to detect a wide range of viruses and coinfections in a fast and systematic way. However, the results also highlight the difficulty of virological diagnosis in these children, as the gold standard failed to identify a causative agent in 36 patients (Table 1). This was most likely because the agent was not a virus, it was not investigated by the gold standard assays, or the sample did not contain sufficient concentration of the virus for detection.
The PathChip was able to detect a wider variety of organisms than other approaches and, in particular, detected 5 virus types in 14 patients that were not found by culture, EraGen, or Luminex, consisting of human bocavirus (6 patients), human parechovirus (5 patients), rotavirus (1 patient), influenza C virus (1 patient), and poliovirus (1 patient), leading to a total of 76 viral types described (compared to culture [11], Luminex [15], and EraGen [18]).
However, we observed a reduction in sensitivity of the PathChip for some viruses (most notably adenovirus, with 4 detections, compared to 13 detections by the gold standard) that prevented a significant increase in diagnosis sensitivity compared to the other techniques.
The sensitivity differences between the PCR approaches are most likely a function of the PCR performance. The PathChip utilizes a semirandom PCR that makes it resilient to sequence diversity but intrinsically less efficient than the specific primers utilized by the other two PCR approaches. However, the PathChip approach does offer some routes to increase its sensitivity in the future, such as modeling the chip's individual probe hybridization characteristics in negative samples, which may allow for a reduction of the detection threshold.
We used NGS to estimate the pathogen identification accuracy of the PathChip when this assay reported false positives (Table 4) and found that 12 of 17 concurred. Of the 5 divergent results, one was the false finding of an RSV B virus in a sample that was positive for RSV A. In this case, the most likely explanation is that there was a false cross-hybridization from the RSV A array signal.
As the PathChip provided genomic information (the closest viral genome match in the PathChip database), it additionally allowed for potential molecular epidemiology investigations and was also able to provide evidence for novel viruses (those without whole-genome sequences in public records). Interestingly, several of the enteroviruses and rhinoviruses were not able to be matched well to a database genome, suggesting that these may be previously unsequenced viral types.
The PathChip also identified group A, B, and C rhinoviruses. The large number of probes on the PathChip allowed for the accurate classification of these enteroviruses, which is typically beyond the scope of typical specific PCR assays.
The PathChip detected the presence of coinfecting viruses in almost 30% of the samples, highlighting the complex nature of this disease. Traditionally, coinfections have been difficult to detect using array technology, but here the PathChip performed equally as well as the specific primer PCR approaches. The clinical importance of coinfections is only now being evaluated, with the advent of technology that allows their detection. However, it is clear that they can easily mislead a conventional approach to diagnosis (which would typically identify only one of the agents).
In conclusion, the high sensitivity and specificity, comparable to those of commercially available and FDA-licensed respiratory pathogen panels, and the ability to simultaneously species type enteroviruses, rhinoviruses, and influenza viruses suggest that use of this technology for detection and identification of viruses in the respiratory tract is now a viable option.
Supplementary Material
ACKNOWLEDGMENTS
This study is part of the research of the ARIVAC Consortium. We are indebted to the Consortium study team and the following collaborators: The Data Safety Monitoring Board, including Kim Mulholland (chair), Keith Klugman, Mary Ann Lansang (local safety monitor), David Sack, Pratap Singhashivanon, Peter Smith, and Chongsuphajaisiddhi Tan; the National Public Health Institute (KTL), including Tarja Kaijalainen and Kaisa Jousimies; the Research Institute for Tropical Medicine (RITM), including Vernoni Ermata Dulalia and Leilani T. Nillos; the University of Colorado, Denver, including Adriana Weinberg; and Sanofi Pasteur, including S. Arnoux, F. Bailleux, S. B'Chir, E. Boutry, J. M. Chapsal, Y. Couedel, V. Delore, H. DyTioco, E. Feroldi, J. Lang, J. R. Maleckar, M. Moreau, R. Ryall, D. Schulz, D. Teuwen, S. Vital, and C. Zocchetti.
The ARIVAC Consortium thanks and acknowledges the participation of the infants, parents, staff, Local Government of the Province of Bohol and Local Government Units (LGUs) of Baclayon, Balilihan, Cortes, Dauis, Panglao and Tagbilaran City, staff of the Pathology and Pediatric Departments of the Bohol Regional Hospital, and the private hospitals Tagbilaran Community Hospital, Borja Family Clinic, Medical Mission Group of Hospitals, Ramiro Community Hospital, St. Jude Hospital, Englewood Hospital, and Tagbilaran Puericulture Center.
Support for research was provided by the European Commission DG Research INCO program (contracts IC18-CY97-2019, ICA4-CT-1999-10008, and ICA4-CT-2002-10062), the Academy of Finland (contracts 206283, 106974, 108873, and 108878), the Finnish Ministry of Foreign Affairs (bilateral contracts 75502901 and 327/412/2000), Finnish Physicians for Social Responsibility, GAVI ADIP Pneumo, Sanofi Pasteur, the Research Institute for Tropical Medicine of the Philippines, National Public Health Institute Finland, the University of Queensland, the University of Colorado, the National Health and Medical Research Council of Australia, and PATH. This work was also supported by a grant from the Bill and Melinda Gates Foundation (OPP49865). Support was also received from the Department of Infectious Diseases and the Research Institute of Children's Hospital Colorado.
The views expressed by the authors do not necessarily reflect the views of PATH.
The GIS provided the proprietary gene chips and bioinformatic support for analysis of the PathChip data and receives funding through the Singapore Agency for Science, Technology and Research (A*STAR).
Eric A. F. Simões, Champa Patel, Marilla Lucero, Hanna Nohynek, Phyllis Carosone-Link, Geraldine Nai, Pei Ling Thien, Chee Wee Koh, Yang Sun Chan, Jianmin Ma, and Sebastian Maurer-Stroh declare that they have no conflict of interest. Four authors (Christopher W. Wong, Wing-Kin Sung, Martin L. Hibberd, and Charlie W. H. Lee) are cofounders and directors of PathGEN Dx Pte. Ltd., which has licensed the PathChip technology from A*STAR.
Footnotes
Published ahead of print 9 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02317-12.
REFERENCES
- 1. Adegbola RA. 2012. Childhood pneumonia as a global health priority and the strategic interest of the Bill & Melinda Gates Foundation. Clin. Infect. Dis. 54:S89–S92 [DOI] [PubMed] [Google Scholar]
- 2. Liu L, Johnson HL, Cousens S, Perin J, Scott S, Lawn JE, Rudan I, Campbell H, Cibulskis R, Li M, Mathers C, Black RE, Child Health Epidemiology Reference Group of WHO and UNICEF 2012. Global, regional, and national causes of child mortality: an updated systematic analysis for 2010 with time trends since 2000. Lancet 379:2151–2161 [DOI] [PubMed] [Google Scholar]
- 3. Leland DS, Ginocchio CC. 2007. Role of cell culture for virus detection in the age of technology. Clin. Microbiol. Rev. 20:49–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Metcalf JA, Davey RT. 1997. Acquired immunodeficiency syndrome: serologic and virologic tests, 4th ed Lippincott-Raven, Philadelphia, PA [Google Scholar]
- 5. Washington JA. 1996. Principles of diagnosis: serodiagnosis. In Baron S. (ed), Medical microbiology, 4th ed University of Texas Medical Branch at Galveston, Galveston, TX: [PubMed] [Google Scholar]
- 6. Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. 1986. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 51(Part 1):263–273 [DOI] [PubMed] [Google Scholar]
- 7. Schena M, Shalon D, Davis RW, Brown PO. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467–470 [DOI] [PubMed] [Google Scholar]
- 8. Liu Q, Bai Y, Ge Q, Zhou S, Wen T, Lu Z. 2007. Microarray-in-a-tube for detection of multiple viruses. Clin. Chem. 53:188–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sanger F, Nicklen S, Coulson AR. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Harris TD, Buzby PR, Babcock H, Beer E, Bowers J. 2008. Single-molecule DNA sequencing of a viral genome. Science 320:106–109 [DOI] [PubMed] [Google Scholar]
- 11. Yozwiak NL, Skewes-Cox P, Stenglein MD, Balmaseda A, Harris E, Derisi JL. 2012. Virus identification in unknown tropical febrile illness cases using deep sequencing. PLoS Negl Trop. Dis. 6:e1485 doi:10.1371/journal.pntd.0001485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nicholson TL, Kukielka D, Vincent AL, Brockmeier SL, Miller LC, Faaberg KS. 2011. Utility of a panviral microarray for detection of swine respiratory viruses in clinical samples. J. Clin. Microbiol. 49:1542–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stratton MR, Campbell PJ, Futreal PA. 2009. The cancer genome. Nature 458:719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quail M, Smith ME, Coupland P, Otto TD, Harris SR, Connor TR, Bertoni A, Swerdlow HP, Gu Y. 2012. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee WH, Wong CW, Leong WY, Miller LD, Sung WK. 2008. LOMA: a fast method to generate efficient tagged-random primers despite amplification bias of random PCR on pathogens. BMC Bioinformatics 9:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Urisman A, Fischer KF, Chiu CY, Kistler AL, Beck S, Wang D, Derisi JL. 2005. E-Predict. Genome Biol. 6:R78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wong CW, Heng CLW, Wan Yee L, Soh SWL, Kartasasmita CB, Simoes EAF, Hibberd ML, Sung W-K, Miller LD. 2007. Optimization and clinical validation of a pathogen detection microarray. Genome Biol. 8:R93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen EC, Miller SA, Derisi JL, Chiu CY. 2011. Using a pan-viral microarray assay (Virochip) to screen clinical samples for viral pathogens. J. Vis. Exp. 2011:pii=2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Quan P-L, Palacios G, Jabado OJ, Conlan S, Hirschberg DL, Pozo F, Jack PJM, Cisterna D, Renwick N, Hui J, Drysdale A, Amos-Ritchie R, Baumeister E, Savy V, Lager KM, Richt JA, Boyle DB, García-Sastre A, Casas I, Perez-Breña P, Briese T, Lipkin WI. 2007. Detection of respiratory viruses and subtype identification of influenza A viruses by GreeneChipResp oligonucleotide microarray. J. Clin. Microbiol. 45:2359–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gardner SN, Jaing CJ, McLoughlin KS, Slezak TR. 2010. A microbial detection array (MDA) for viral and bacterial detection. BMC Genomics 11:668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin B, Wang Z, Vora GJ, Thornton JA, Schnur JM, Thach DC, Blaney KM, Ligler AG, Malanoski AP, Santiago J, Walter EA, Agan BK, Metzgar D, Seto D, Daum LT, Kruzelock R, Rowley RK, Hanson EH, Tibbetts C, Stenger DA. 2006. Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays. Genome Res. 16:527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee CWH, Koh CW, Chan YS, Aw PPK, Loh KH, Han BL, Thien PL, Nai GYW, Hibberd ML, Wong CW, Sung W-K. 2010. Large-scale evolutionary surveillance of the 2009 H1N1 influenza A virus using resequencing arrays. Nucleic Acids Res. 38:e111 doi:10.1093/nar/gkq089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wong CW, Albert TJ, Vega VB, Norton JE, Cutler DJ, Richmond TA, Stanton LW, Liu ET, Miller LD. 2004. Tracking the evolution of the SARS coronavirus using high-throughput, high-density resequencing arrays. Genome Res. 14:398–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Z, Malanoski AP, Lin B, Kidd C, Long NC, Blaney KM, Thach DC, Tibbetts C, Stenger DA. 2008. Resequencing microarray probe design for typing genetically diverse viruses: human rhinoviruses and enteroviruses. BMC Genomics 9:577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Berthet N, Leclercq I, Dublineau A, Shigematsu S, Burguière AM, Filippone C, Gessain A, Manuguerra J-C. 2010. High-density resequencing DNA microarrays in public health emergencies. Nat. Biotechnol. 28:25–27 [DOI] [PubMed] [Google Scholar]
- 26. Dacheux L, Berthet N, Dissard G, Holmes EC, Delmas O, Larrous F, Guigon G, Dickinson P, Faye O, Sall AA, Old IG, Kong K, Kennedy GC, Manuguerra J-C, Cole ST, Caro V, Gessain A, Bourhy H. 2010. Application of broad-spectrum resequencing microarray for genotyping rhabdoviruses. J. Virol. 84:9557–9574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kodani M, Yang G, Conklin LM, Travis TC, Whitney CG, Anderson LJ, Schrag SJ, Taylor TH, Beall BW, Breiman RF, Feikin DR, Njenga MK, Mayer LW, Oberste MS, Tondella MLC, Winchell JM, Lindstrom SL, Erdman DD, Fields BS. 2011. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J. Clin. Microbiol. 49:2175–2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sakthivel SK, Whitaker B, Lu X, Oliveira DBL, Stockman LJ, Kamili S, Oberste MS, Erdman DD. 2012. Comparison of fast-track diagnostics respiratory pathogens multiplex real-time RT-PCR assay with in-house singleplex assays for comprehensive detection of human respiratory viruses. J. Virol. Methods 185:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chidlow GR, Harnett GB, Shellam GR, Smith DW. 2009. An economical tandem multiplex real-time PCR technique for the detection of a comprehensive range of respiratory pathogens. Viruses 1:42–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pabbaraju K, Tokaryk KL, Wong S, Fox JD. 2008. Comparison of the Luminex xTAG respiratory viral panel with in-house nucleic acid amplification tests for diagnosis of respiratory virus infections. J. Clin. Microbiol. 46:3056–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Merante F, Yaghoubian S, Janeczko R. 2007. Principles of the xTAG respiratory viral panel assay (RVP Assay). J. Clin. Virol. 40(Suppl 1):S31–S35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mahony J, Chong S, Merante F, Yaghoubian S, Sinha T, Lisle C, Janeczko R. 2007. Development of a respiratory virus panel test for detection of twenty human respiratory viruses by use of multiplex PCR and a fluid microbead-based assay. J. Clin. Microbiol. 45:2965–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brunstein J, Thomas E. 2006. Direct screening of clinical specimens for multiple respiratory pathogens using the Genaco Respiratory Panels 1 and 2. Diagn. Mol. Pathol. 15:169–173 [DOI] [PubMed] [Google Scholar]
- 34. Létant SE, Ortiz JI, Bentley Tammero LF, Birch JM, Derlet RW, Cohen S, Manning D, McBride MT. 2007. Multiplexed reverse transcriptase PCR assay for identification of viral respiratory pathogens at the point of care. J. Clin. Microbiol. 45:3498–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Poritz MA, Blaschke AJ, Byington CL, Meyers L, Nilsson K, Jones DE, Thatcher SA, Robbins T, Lingenfelter B, Amiott E, Herbener A, Daly J, Dobrowolski SF, Teng DHF, Ririe KM. 2011. FilmArray, an automated nested multiplex PCR system for multi-pathogen detection: development and application to respiratory tract infection. PLoS One 6:e26047 doi:10.1371/journal.pone.0026047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hayden RT, Gu Z, Rodriguez A, Tanioka L, Ying C, Morgenstern M, Bankowski MJ. 2012. Comparison of two broadly multiplexed PCR systems for viral detection in clinical respiratory tract specimens from immunocompromised children. J. Clin. Virol. 53:308–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Balada-Llasat J-M, Larue H, Kelly C, Rigali L, Pancholi P. 2011. Evaluation of commercial ResPlex II v2.0, MultiCode-PLx, and xTAG respiratory viral panels for the diagnosis of respiratory viral infections in adults. J. Clin. Virol. 50:42–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lucero MG, Nohynek H, Williams G, Tallo V, Simoes EAF, Lupisan S, Sanvictores D, Forsyth S, Puumalainen T, Ugpo J, Lechago M, de Campo M, Abucejo-Ladesma E, Sombrero L, Nissinen A, Soininen A, Ruutu P, Riley I, Mäkelä HP. 2009. Efficacy of an 11-valent pneumococcal conjugate vaccine against radiologically confirmed pneumonia among children less than 2 years of age in the Philippines: a randomized, double-blind, placebo-controlled trial. Pediatr. Infect. Dis. J. 28:455–462 [DOI] [PubMed] [Google Scholar]
- 39. Weinberg A, Brewster L, Clark J, Simoes E, ARIVAC Consortium 2004. Evaluation of R-Mix shell vials for the diagnosis of viral respiratory tract infections. J. Clin. Virol. 30:100–105 [DOI] [PubMed] [Google Scholar]
- 40. Nolte FS, Marshall DJ, Rasberry C, Schievelbein S, Banks GG, Storch GA, Arens MQ, Buller RS, Prudent JR. 2007. MultiCode-PLx system for multiplexed detection of seventeen respiratory viruses. J. Clin. Microbiol. 45:2779–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krunic N, Merante F, Yaghoubian S, Himsworth D, Janeczko R. 2011. Advances in the diagnosis of respiratory tract infections: role of the Luminex xTAG respiratory viral panel. Ann. N. Y. Acad. Sci. 1222:6–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kilpatrick DR, Iber JC, Chen Q, Ching K, Yang S-J, De L, Mandelbaum MD, Emery B, Campagnoli R, Burns CC, Kew O. 2011. Poliovirus serotype-specific VP1 sequencing primers. J. Virol. Methods 174:128–130 [DOI] [PubMed] [Google Scholar]
- 43. WHO 2009. WHO information for laboratory diagnosis of pandemic (H1N1) 2009 virus in humans—revised. WHO, Geneva, Switzerland: http://www.who.int/csr/resources/publications/swineflu/WHO_Diagnostic_RecommendationsH1N1_20090521.pdf [Google Scholar]
- 44. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.