Abstract
The cost of whole-genome sequencing (WGS) is decreasing rapidly as next-generation sequencing technology continues to advance, and the prospect of making WGS available for public health applications is becoming a reality. So far, a number of studies have demonstrated the use of WGS as an epidemiological tool for typing and controlling outbreaks of microbial pathogens. Success of these applications is hugely dependent on efficient generation of clean genetic material that is free from host DNA contamination for rapid preparation of sequencing libraries. The presence of large amounts of host DNA severely affects the efficiency of characterizing pathogens using WGS and is therefore a serious impediment to clinical and epidemiological sequencing for health care and public health applications. We have developed a simple enzymatic treatment method that takes advantage of the methylation of human DNA to selectively deplete host contamination from clinical samples prior to sequencing. Using malaria clinical samples with over 80% human host DNA contamination, we show that the enzymatic treatment enriches Plasmodium falciparum DNA up to ∼9-fold and generates high-quality, nonbiased sequence reads covering >98% of 86,158 catalogued typeable single-nucleotide polymorphism loci.
INTRODUCTION
Whole-genome sequencing (WGS) is beginning to provide new and efficient opportunities for the diagnosis and control of many clinically relevant pathogenic infections (1–5). High-resolution WGS is promising to be the most informative typing method (6) and is likely to be more sensitive than the traditional culture-based diagnostic procedures in clinical microbiology (1, 2, 4, 7). As the technology is moving from bench to bedside (8–10), sample quality and quantity have become a major technical bottleneck in clinical sequencing. A large proportion of field-derived pathogen specimens are heavily contaminated with host material. Attempts to sequence these samples without adequate removal of the host DNA negate some of the cost benefit realized by the current advances in sequencing technology (11). Whereas problems associated with the small amounts of starting material have been addressed extensively through the development of alternative library preparation methods as well as the discovery of novel amplification technologies (12–14), host contamination has remained a big challenge, especially for pathogens that are very difficult to culture in vitro.
Due to the small size of pathogen genomes in comparison to that of the genome of their host, the presence of just a few nucleated human cells in a pathogen clinical specimen can completely inundate that sample with host DNA contamination (11). Direct sequencing of host-contaminated samples is inefficient and therefore not cost-effective. For example, one lane of an Illumina HiSeq system run has the capacity to sequence more than 50 multiplexed pathogen genomes that are pure; however, this capacity is reduced to only a single or a very few samples—with maintenance of adequate coverage depth—when contaminated pathogen DNA is used. Similarly, smaller benchtop systems—like the Ion Torrent PGM (Life Technologies), MiSeq (Illumina), and 454 GS Junior (Roche) systems—are likely to produce very little pathogen sequence from contaminated samples, reporting data mainly from the overly abundant host material. It is therefore beneficial and cost-effective to remove host DNA contamination prior to sequencing of pathogen clinical samples.
Here, we report an enzyme-based DNA degradation method that selectively digests and depletes human DNA contamination from malaria clinical samples, thereby enriching the parasite material for high-throughput next-generation sequencing (NGS). This approach takes advantage of the differential methylation patterns that exist between host DNA and most pathogen genomes. We have identified the recently characterized modification-dependent restriction endonucleases (MD-REs) and harnessed their unique activities to selectively degrade human host DNA in clinical samples. MspJI (from Mycobacterium spp.), LpnPI (from Legionella pneumophila), and FspEI (from Frankia spp.) are homologous enzymes belonging to the Mrr superfamily of endonucleases that bind and cut near methylated DNA base pairs (15–17). Although these enzymes display different sequence recognition preferences, they all cleave at position N-12/N-16 on the 3′ side of methylated cytosines (15). DNA methylation in higher eukaryotes is dynamic and is affected by cellular processes that include differentiation, genome regulation, and disease conditions (18). In humans, methylated cytosine is known to occur predominantly in the context of methyl-CpGs and at an estimated frequency of 60% to 90% in that context (19, 20). Complete digestion of a contaminated clinical sample with methylation-dependent endonucleases is therefore expected to result in significant degradation of the host DNA and, consequently, enrich pathogen DNA. To test this hypothesis, we first assessed the effect of the enzymes on pure Plasmodium falciparum DNA and in mock host-contaminated samples. We then assessed the ability of the enzymes to deplete human host DNA in authentic highly contaminated malaria-containing clinical samples. Here we report our findings and recommendations on the use of MD-REs in degrading human DNA from malaria clinical samples to enrich for parasite DNA prior to NGS.
MATERIALS AND METHODS
DNA samples.
P. falciparum 3D7 genomic DNA was obtained from Chris Newbold's laboratory at the University of Oxford, Oxford, United Kingdom. Other P. falciparum genomic DNA samples were samples of clinical isolates containing ∼80 to 90% human DNA contamination. The clinical isolates were obtained from the Malaria Genetics Group's Sequencing Sample Repository at the Wellcome Trust Sanger Institute. All clinical samples were positive for P. falciparum by microscopic analysis. Human genomic DNA was purchased from Promega (Madison, WI). Mock samples were manually reconstituted by mixing 100 ng of pure P. falciparum 3D7 genomic DNA with 900 ng of pure human genomic DNA to obtain 1 μg of a simulated clinical genomic DNA sample.
Methylation-dependent digestion.
Digestion with methylation-dependent restriction endonucleases was performed in 0.2-ml PCR tubes. The reaction mixture (total volume, 30 μl), containing 1× NEB buffer 4, 10 μg of bovine serum albumin, and 6 units of MD-RE (New England BioLabs, MA), was placed in a thermocycler programmed to incubate at 37°C for 16 h, followed by heating at 65°C for 20 min to inactivate the enzymes, prior to cooling to 4°C. It has been shown that the cleavage activity of MD-REs is enhanced by inclusion of an activator sequence in the digestion reaction (17). The activator is a short double-stranded oligonucleotide designed to form a stem-loop structure and contains two methylation (m) sites (CTGCmCAGGATCTTTTTTGATCmCTGGCAG). The activator oligonucleotide was purchased from Integrated DNA Technologies (Coralville, IA). For reactions involving the activator, 0.05 μM oligonucleotide was used.
Gel-based sample preparation.
For procedures involving gel separation, genomic DNA (1 to 2 μg) was digested with MspJI as described above under “Methylation-dependent digestion.” After digestion, the entire reaction mixture was loaded on a 1% agarose-TBE (Tris-borate-EDTA) gel containing SYBR green and resolved at 60 V for 45 min. High-molecular-weight bands containing undigested genomic DNA (Fig. 1i and ii) were excised and weighed before extracting the DNA using a QIAEX II gel extraction kit (Qiagen, Germany) following the manufacturer's instructions. Briefly, weighed gel slices were put in 2-ml Eppendorf tubes and buffer QX 1 was added (3 volumes of buffer to 1 g of gel). The gel was allowed to dissolve by incubating at room temperature with agitation for 20 min or until complete dissolution. To bind the DNA, 10 μl of QIAEX II silica gel particles was added, and incubation was continued for a further 10 min with agitation. Samples were centrifuged for 30 s at 10,000 × g, and the supernatant was carefully removed. The pellet was washed once with 500 μl of buffer QX 1 and twice with 500 μl of wash buffer (PE). The pellet was air dried for 15 min, and the DNA was eluted in elution buffer (EB). Eluted DNA was sheared to ∼350 bp with a Covaris S2 sonicator (Covaris Inc., Woburn, MA) using the settings described below under “Illumina sequencing library preparation.” Sheared fragments were processed to generate Illumina sequencing libraries.
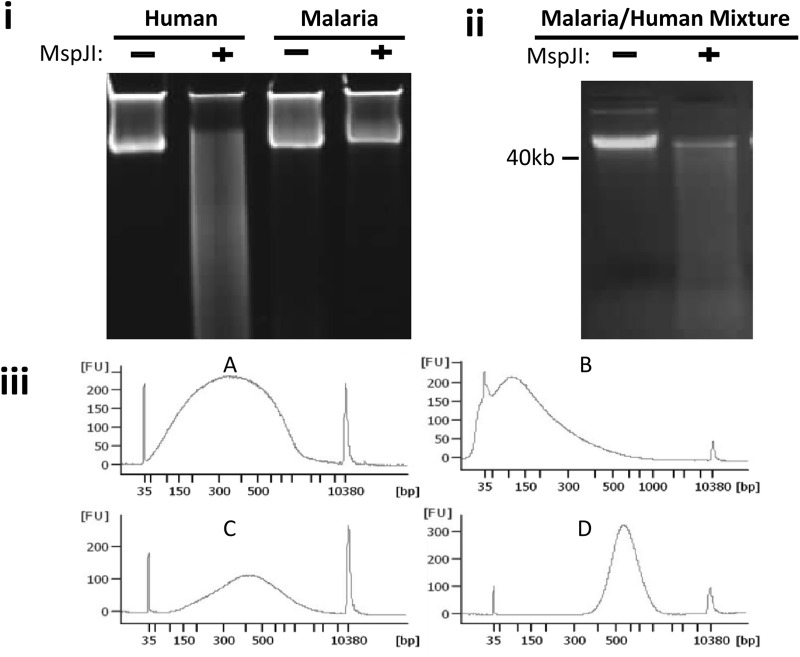
Fig 1.
Differential human DNA degradation analysis and separation procedures. (i) Agarose gel electrophoresis analysis of pure human and P. falciparum genomic DNA incubated in the absence (−) or presence (+) of MspJI enzyme. (ii) A mixture comprising 90% human and 10% P. falciparum genomic DNA incubated in the absence (−) or presence (+) of MspJI enzyme. (iii) Agilent bioanalyzer fragment analysis of mixtures of human and P. falciparum DNA. (A) Fragment sizes of a Covaris-treated randomly sheared mixture of human and P. falciparum genomic DNA before MspJI digestion; (B) fragment size distribution of the sheared DNA after MspJI digestion; (C) fragment size distribution of the MspJI-digested DNA after Ampure XP size selection that removes degraded human DNA and enriches for the larger undigested parasite DNA fragments; (D) fragment size distribution of a PCR enriched adapter-ligated DNA library prior to sequencing. FU, fragment units (i.e., number of fragments).
Gel-free sample preparation.
Genomic DNA (0.1 to 2 μg) was sheared to obtain random fragments with an average size of ∼350 bp using a Covaris S2 sonicator. Fragments (Fig. 1iiiA) were end repaired as described below under “Illumina sequencing library preparation.” End-repaired samples were digested with MspJI as described above under “Methylation-dependent digestion.” Digested samples (Fig. 1iiiB) were size selected using Agencourt Ampure XP beads (Beckman Coulter Inc., Beverly, MA) following the manufacturer's instructions with slight modifications. Briefly, equal volumes of beads and the digested sample were mixed and incubated for 5 min at room temperature. After incubation, the tube containing the bead-DNA mixture was placed on a magnetic rack to capture the DNA-bound beads and the unbound solution was discarded. Beads were washed twice with 80% ethanol, and the bound DNA was eluted with EB. Two rounds of this purification step ensure removal of smaller fragments generated by MspJI digestion. Purified samples (Fig. 1iiiC) were processed to generate Illumina paired-end libraries through A tailing, PE-adapter ligation and enrichment by 12 cycles of PCR (Fig. 1iiiD), as described below under “Illumina sequencing library preparation.”
Illumina sequencing library preparation.
Genomic DNA (0.05 to 2 μg) in 75 μl TE (Tris-EDTA) buffer was sheared for 70 s using a Covaris S2 device. To obtain an average fragment size of ∼350 bp, the settings used were 10% duty cycle, intensity 4, and 200 cycles per burst. Illumina paired-end sequencing libraries were constructed using an NEBNext DNA sample preparation kit (New England BioLabs) following the standard Illumina sample preparation protocol (13). All PCR library amplifications, after adapter ligation, were performed with an MJ Research PTC-225 thermocycler. Illumina PE 1.0 and 2.0 primers or PE 1.0- and 2.0-derived indexing primers were used to amplify 10 ng adapter-ligated library fragments by PCR. Libraries were amplified using optimized PCR conditions (13). Briefly, the PCR mixture with KAPA HiFi (KAPA Biosystems, South Africa) contained 1× KAPA HiFi master mix, 0.4 μM each primer pair, and 10 ng of DNA in a 50-μl volume. Amplification reactions were performed using the following thermocycling conditions: 1 min at 98°C for the initial denaturation, followed by 12 cycles of 10 s at 98°C and 1 min at 65°C and a final extension for 5 min at 65°C.
Sequencing.
DNA libraries were sequenced at the Wellcome Trust Sanger Institute using Illumina MiSeq and HiSeq 2000 instruments. Samples were sequenced using Illumina (version 3) chemistry; paired-end sequencing was done with 75-base reads and an 8-base index read.
Data analysis.
Sequence data obtained from each sample were subjected to standard Illumina quality control procedures before detailed analysis for enrichment and quality of coverage. For enrichment analysis, we counted the number of reads mapping to either human or P. falciparum reference genome sequences. Each data set was analyzed independently by mapping sequence reads to the reference genome using Burrows-Wheeler Aligner (BWA) (21). SAMtools (21) was used to generate coverage statistics from the BWA mapping output.
Nucleotide sequence accession number.
All data sets used in this study have been deposited in the European Nucleotide Archive read archive under accession number ERP000832.
RESULTS
Differential genomic endonuclease activity assay.
Genomic DNA methylation has been extensively studied in bacteria and higher eukaryotic organisms, but little is known about the extent and form of such modifications in eukaryotic pathogens. To determine whether MD-REs have endonuclease activity on the genomic DNA of an important eukaryotic pathogen, we performed an in vitro digestion assay on DNA extracted from the malaria parasite P. falciparum. As a positive control, a parallel assay was performed on human DNA. To control against any nonenzymatic degradation, P. falciparum genomic DNA was incubated in the presence of all the reagents present in the digestion reaction except the enzyme. As shown in Fig. 1i, positive endonuclease activity, which results in shorter fragments (seen as a smear below the main band), was observed only in the human genomic DNA sample that was incubated in the presence of MspJI enzyme. P. falciparum DNA samples incubated in the presence or absence of the enzyme remained intact without visible degradation. A mixture of human and parasite DNA (90% human DNA by mass) was also digested and analyzed by gel electrophoresis. As shown in Fig. 1ii, degradation of the human DNA was also observed. Presumably, the remaining DNA band contained the malaria component. This indicates a selective endonuclease activity exhibited by the MD-RE on human DNA. To further confirm that MD-RE digestion had no degradation activity on P. falciparum DNA, we prepared MspJI-treated and nontreated DNA for sequencing using Illumina technologies. Nondegraded parasite DNA was separated from degraded host DNA using either agarose gel electrophoresis (Fig. 1i and ii) or magnetic bead-based size selection (Fig. 1iii).
Effect of activator sequence on MD-RE activity.
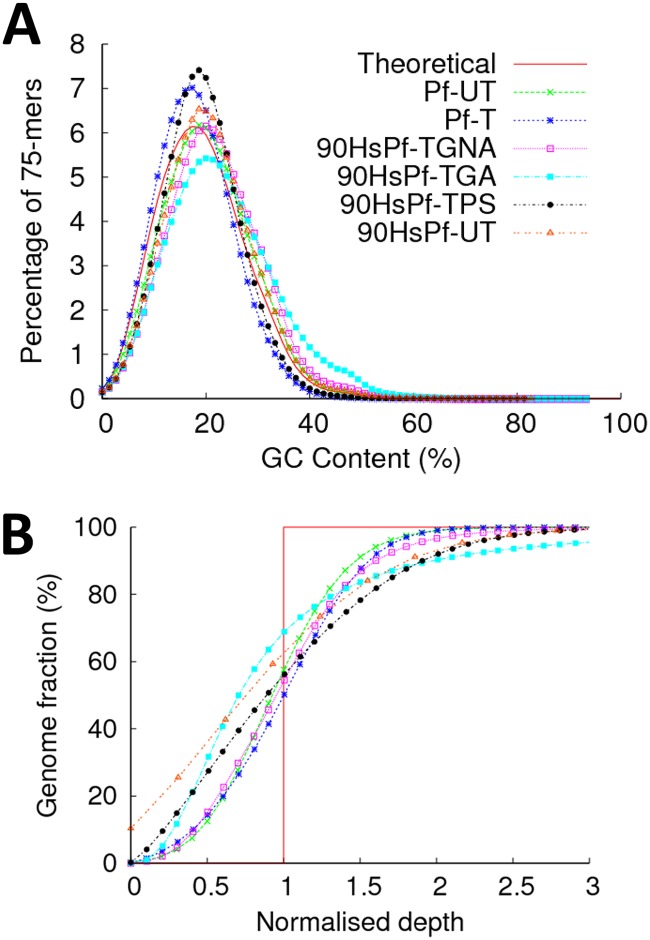
Cleavage activity of MD-REs has been shown to be enhanced by inclusion of an activator sequence in the digestion reaction (17). We performed an assay to detect the effect of this activator using the host DNA depletion reaction. Data analysis indicates no significant improvement to the overall enrichment of parasite DNA under the reaction conditions tested (Fig. 2A). However, a detailed analysis of the enriched sequence data (Fig. 3A and B) indicates a possible nonspecific degradation induced by the presence of the activator. We compared the quality of sequence data generated from samples treated in the absence (Fig. 3, sample 90HsPf-TGNA) or presence (Fig. 3, sample 90HsPf-TGA) of the activator sequence and observed a slightly biased base composition (higher G+C content; Fig. 3A; Table 1) and uneven genome coverage (Fig. 3B) in samples where digestion was supplemented with activator. Due to these observations, the activator was excluded in all subsequent assays.
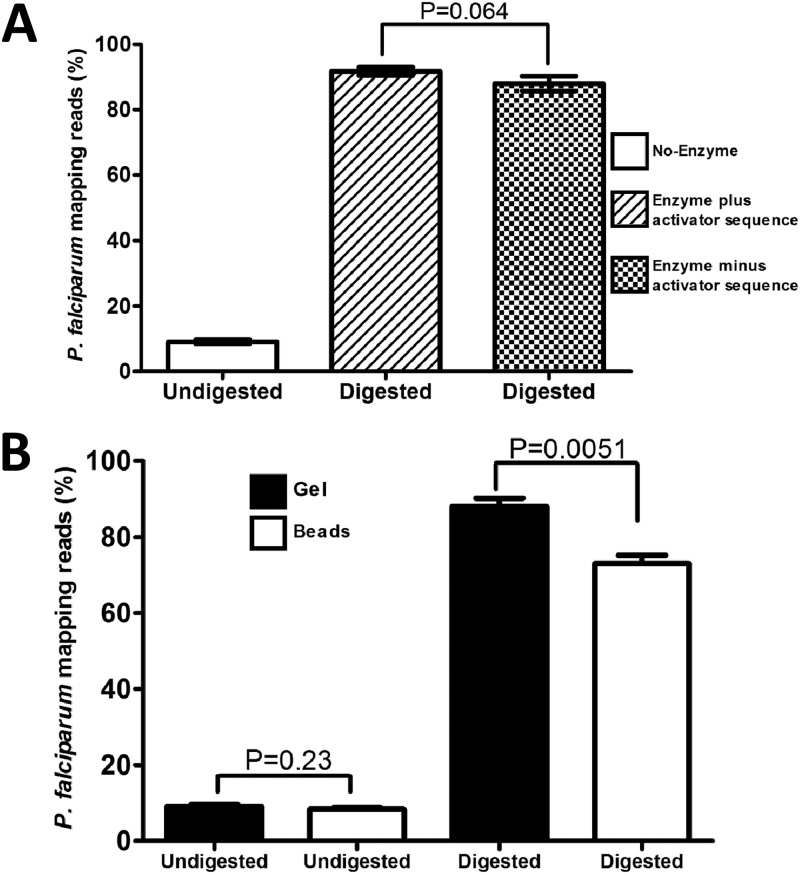
Fig 2.
Analysis of sequence reads generated from mock samples (90% human DNA contaminated) treated with or without MspJI enzyme. (A) Plots comparing the proportion of sequence reads mapping to the P. falciparum genome in an untreated sample and treated sample with or without activator supplement. (B) Plots comparing the proportion of sequence reads mapping to the P. falciparum genome in untreated and treated samples; degraded human DNA fragments were removed either following gel electrophoresis or with magnetic beads. In both panels, error bars illustrate the deviation observed in 3 independent replicates. Statistical differences were calculated using the unpaired Student's t test, with a P value of <0.05 considered significant.
Fig 3.
G+C bias and whole-genome coverage analysis of mock samples treated with or without MspJI. (A) G+C content distribution profile for samples treated and processed using different procedures. Libraries with G+C contents above or below 19.4% (theoretical) indicate base bias with respect to the P. falciparum 3D7 reference genome. (B) A plot of whole-genome coverage against average depth. Variance in coverage depth above and below the normalized average depth (red vertical line) across the genome is shown. Increased deviation of the sample curves from the average depth line indicates increased levels of nonuniformity in coverage depth distribution across the genome. The y-axis intercept indicates the percentage of the genome left uncovered. Pf-T, uncontaminated P. falciparum genomic DNA treated with MspJI and processed with the gel electrophoresis procedure; Pf-UT, uncontaminated P. falciparum genomic DNA processed with the gel electrophoresis procedure without MspJI treatment (positive control); 90HsPf-UT, human DNA contaminated (90%) with P. falciparum genomic DNA processed with the gel electrophoresis procedure without MspJI treatment; 90HsPf-TGA, P. falciparum genomic DNA contaminated with human DNA (90%) treated with MspJI in the presence of activator sequence and processed with the gel electrophoresis procedure; 90HsPf-TGNA, P. falciparum genomic DNA contaminated with human DNA (90%) treated with MspJI in the absence of activator sequence and processed with the gel electrophoresis procedure; 90HsPf-TPS, P. falciparum genomic DNA contaminated with human DNA (90%) treated with MspJI in the absence of activator sequence and processed with the magnetic bead size-selection procedure.
Table 1.
Average GC contenta
| Sample | Avg % G+C content |
|---|---|
| Pf-UT | 20.40 ± 0.75 |
| Pf-T | 19.70 ± 0.57 |
| 90HsPf-TGNA | 21.58 ± 0.45 |
| 90HsPf-TGA | 22.06 ± 0.45 |
| 90HsPf-TPS | 19.99 ± 1 |
| 90HsPf-UT | 21.00 ± 0.57 |
Calculated values of average G+C content corresponding to each library preparation. For the P. falciparum 3D7 reference genome, a G+C content above or below 19.4% indicates bias. Figure 3A also shows a graphical view of the G+C content distribution. Sample name abbreviations are given in the legend to Fig. 3.
Gel and bead size selection.
To separate degraded host fragments from the nondegraded pathogen DNA, we used either gel electrophoresis or magnetic bead-based size-selection procedures. The magnetic bead-based size selection allows high-throughput automation of the entire sample preparation through to the final library creation. As shown in Fig. 1iiiA, randomly sheared fragments are subjected to MD-RE digestion after end repair, during which host DNA fragments that are methylated are degraded to smaller fragments of about 150 bp (Fig. 1iiiB). The digestion product is subjected to magnetic bead-based size selection, where parasite DNA fragments are enriched relative to the degraded host DNA (Fig. 1iiiC). Purified adapter-ligated product is then amplified by PCR to generate a final library, which is shown in Fig. 1iiiD. Comparative analysis of samples processed through either gel- or bead-based procedures indicates slightly higher enrichment efficiency in gel-separated samples (Fig. 2B); however, the quality of data (in terms of uniformity of coverage) generated via both procedures was largely similar (Fig. 3).
Enrichment of pathogen DNA in human host-contaminated samples.
MspJI treatment selectively degrades human host DNA with no detectable effect on pathogen DNA. This makes it suitable for applications requiring removal of host contamination during microbial clinical sequencing. To test this application, we prepared mock samples (1 μg total DNA) consisting of 10% parasite and 90% human DNA by mass. We treated these samples with MspJI and purified nondegraded DNA using either gel- or magnetic bead-based size-selection procedures (Fig. 2). Posttreatment analysis indicates a recovery of ∼70 ng of undigested DNA material (presumably parasite DNA), representing ∼70% of the total parasite DNA input that is available for NGS. Treated and nontreated samples were both sequenced using Illumina sequencing. The number of reads from untreated samples confirmed the original proportions of parasite to human DNA to be 1:10. Analysis of reads from treated samples indicated up to a 9-fold enrichment of the parasite DNA, from 10% to ∼92% of reads (Fig. 2A and B). Mapping of reads obtained from treated samples to the P. falciparum reference genome showed 100% coverage (Fig. 3B).
Enrichment assay on malaria clinical samples.
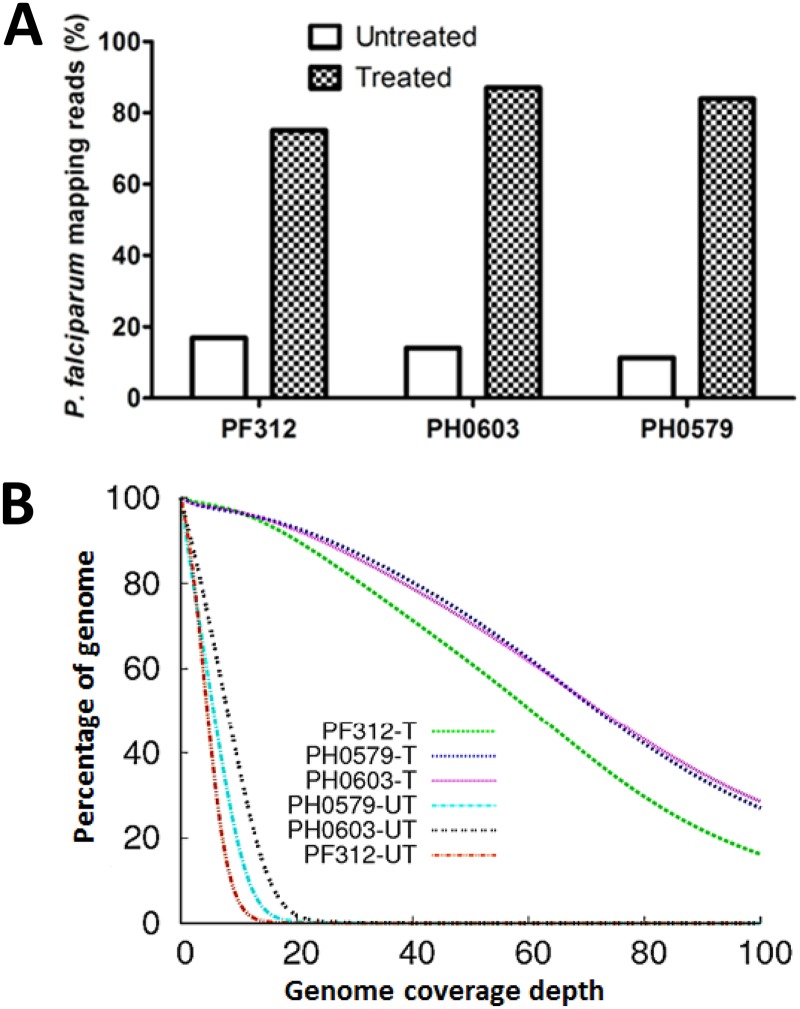
After optimization of the enrichment assay using mock samples, we extended these experiments to authentic malaria clinical samples containing human DNA contamination at levels ranging from ∼80 to 90%, as estimated by quantitative PCR. For the assay, each clinical sample was divided into two parts. One part was sequenced without treatment, and the other was treated with the MspJI MD-RE prior to sequencing. Degraded DNA was removed using gel separation. We analyzed the sequence data generated and compared treated versus untreated samples to determine the level of enrichment as well as the quality of sequence data generated. About 80% of the reads in the treated samples mapped to P. falciparum. We down-sampled reads from treated samples—to match their corresponding untreated data sets—for enrichment analysis. We show that between 4- and 7-fold enrichment of parasite DNA was achieved, depending on the extent of host contamination in the original sample (Table 2; Fig. 4A). We also assessed cumulative genome coverage and compared the depth and breadth of coverage for treated and untreated samples. As shown in Fig. 4B, >95% of the genome was covered 20 times or more in data from the treated samples. In contrast, in the untreated samples, none (0%) of the genome was covered at a depth greater than 20 times. In addition to the 3 clinical samples that contained sufficient DNA to perform both treated and untreated analyses, we also present data for 14 other clinical samples that were subjected only to MD-RE treatment followed by sequencing. As shown in Table S1 in the supplemental material, we obtained enrichment values ranging from 3- to 13-fold; data were of high quality, and coverage of the P. falciparum reference genome was 100%.
Table 2.
Summary of sequence data information for host depletion samplesa
| Sample | No. of reads (106) |
% reads mapping P. falciparum | Fold enrichment | |
|---|---|---|---|---|
| Avg | Down-sampled | |||
| Pf-UTm | 139.11 | 18.48 | 99.7 | NA |
| Pf-Tm | 37.96 | 18.48 | 99.6 ± 0.5 | NA |
| 90HsPf-TGNAm | 40.20 | 18.48 | 88.9 ± 2 | 10 |
| 90HsPf-TGAm | 50.94 | 18.48 | 88.9 ± 1 | 10 |
| 90HsPf-TPSm | 46.30 | 18.48 | 74.0 ± 4 | 8 |
| 90HsPf-UTm | 18.48 | 18.48 | 8.9 ± 1 | NA |
| PF312-UTc | 10.32 | 10.32 | 16.9 | NA |
| PF312-Tc | 37.62 | 10.32 | 75.0 | 4.4 |
| PH0579-UTc | 16.39 | 16.39 | 11.7 | NA |
| PH0579-Tc | 42.37 | 16.39 | 83.9 | 7.2 |
| PH0603-UTc | 18.55 | 18.55 | 14.0 | NA |
| PH0603-Tc | 40.43 | 18.55 | 87.0 | 6.2 |
Before filtering out human mapping reads (average number of reads), original data sets were randomly down-sampled to generate a matching number of reads for comparative analysis. Mock samples were analyzed in triplicate, and the data shown represent average numbers of reads.
, mock samples;
Fig 4.
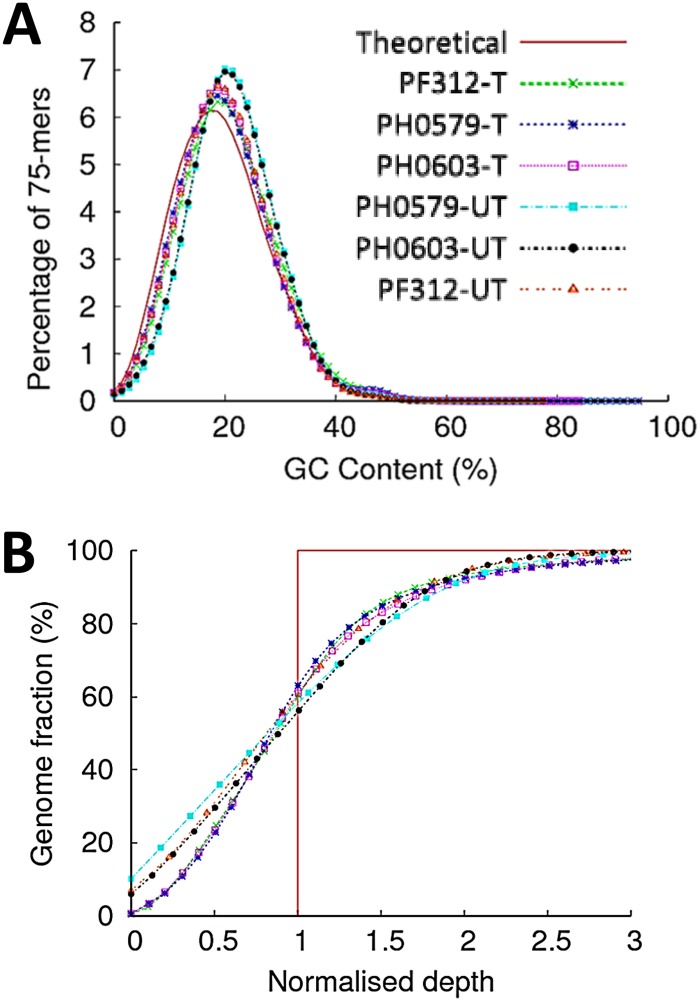
Analysis of sequence reads generated from clinical samples treated with or without MspJI enzyme. (A) Plots showing the proportion of sequence reads mapping to the P. falciparum genome in untreated samples and treated samples. (B) A plot showing the depth and breadth of the genome covered at various read depths by treated and untreated samples. Sample names have a T appended for MspJI treated and a UT appended for untreated. Host contamination was as follows: 86% for PF312, 92% for PH0579, and 90% for PH0603. All samples were processed using the gel separation procedure.
Sequence analysis.
Owing to the poor understanding of DNA methylation status in many pathogenic organisms, we performed extensive high-resolution comparative analysis of sequence data generated from treated and untreated samples. To compare corresponding treated and untreated samples, total sequence read data sets (before filtering host contamination) were normalized by random down-sampling to generate matching numbers of reads for analysis (Table 2). In this way, we assessed the quality of data generated posttreatment. Base composition was assessed using G+C content analysis. Our results (Fig. 3A and 5A) indicate no bias in base composition in treated samples that could be attributed to the effect of digestion. We also used evenness-of-coverage metrics (13, 14) to analyze the distribution of coverage across the entire genome for each sample. We did not detect any unintended loss of sequence or uneven coverage as a result of treatment (Fig. 3B, samples Pf-T and Pf-UT). However, as expected, untreated clinical samples with host contamination generated very few parasite-specific sequence reads, resulting in a very low depth of coverage and with up to 10.3% of the parasite genome not being covered at all (Fig. 3B, 4B, and 5B).
Fig 5.
G+C bias and whole-genome coverage analysis of authentic clinical samples treated or untreated with MspJI. (A) G+C content distribution profiles. Libraries with G+C content above or below 19.4% (theoretical) indicate base bias with respect to the P. falciparum 3D7 reference genome. (B) A plot showing uniformity of genome coverage against normalized average depth. Variance in coverage depth above and below the normalized average depth (red vertical line) across the genome is shown. Clinical isolates are PF312, PH0579, and PH0603. Sample names have a T appended for MspJI treated and a UT appended for untreated. Host contamination was as follows: 86% for PF312, 92% for PH0579, and 90% for PH0603. All samples were processed using the gel separation procedure.
Coverage of typeable SNPs.
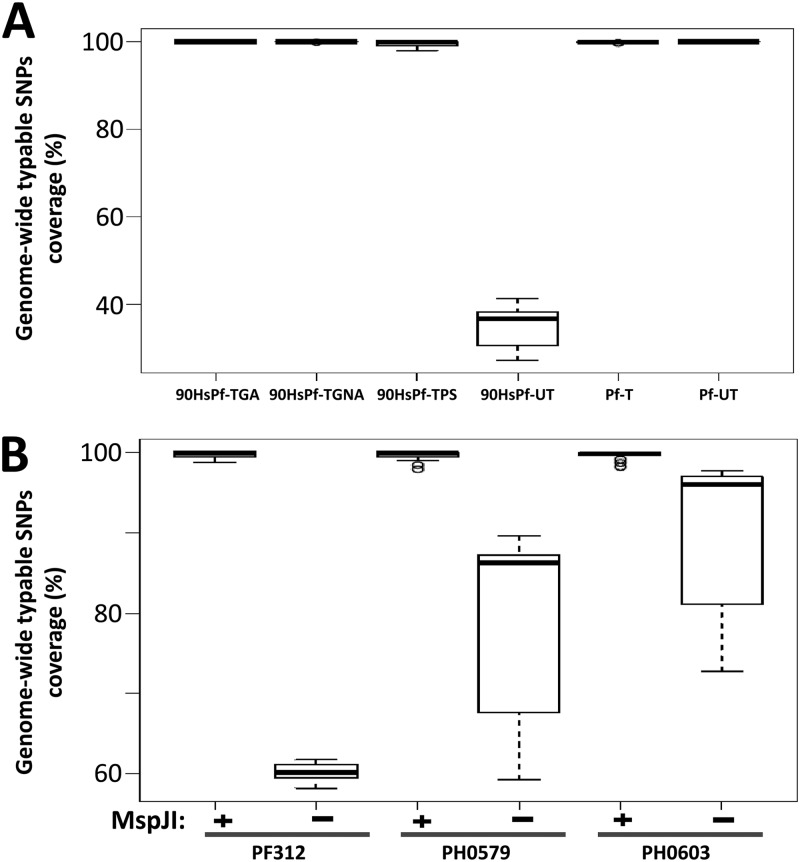
To evaluate the quality and complexity of sequence data generated following the host depletion method, we analyzed the coverage of 86,158 high-quality single-nucleotide polymorphisms (SNPs; typeable SNPs) that had previously been identified and used in determining P. falciparum population diversity (6). We analyzed the genome-wide coverage of typeable SNPs in both untreated and treated samples from mock (containing 90% human DNA by mass) and clinical samples. As shown in Fig. 6A (mock samples) and Fig. 6B (clinical samples), all treated samples (including 14 additional clinical samples shown in Table S1 in the supplemental material) produced >98% coverage of typeable SNPs at a depth of ≥5 times in all of the 14 P. falciparum chromosomes. On the other hand, none of the untreated samples with host contamination achieved >80% coverage at 5 times in all chromosomes. Most importantly, coverage in the treated samples was in concordance with the pure P. falciparum DNA that was processed untreated (Pf-UT, positive control). This indicated that host depletion treatment significantly enriched pathogen material and improved coverage without bias toward specific regions. The treatment maintained the complexity of the genome, thereby producing quality parasite genome data, meeting the required threshold for accurate SNP genotyping.
Fig 6.
Typeable SNP loci covered 5 times or more by treated and untreated samples. A total of 86,158 typeable SNP positions across the entire genome were analyzed for coverage at depths of 5 times or more. The mean percentage of SNP positions covered per chromosome was computed for each sample. The box plots show the mean and the variation of the percent coverage in the 14 chromosomes. (A) Mean percent coverage of the mock samples described in Fig. 3; (B) mean percent coverage of the clinical samples described in Fig. 4. Samples were treated without (−) or with (+) MspJI enzyme. Host contamination was as follows: 86% for PF312, 92% for PH0579, and 90% for PH0603. All samples were processed using the gel separation procedure.
DISCUSSION
Current solutions to host contamination include enrichment by hybrid selection techniques in which DNA probes complementary to the pathogen nucleic acid are hybridized to the contaminated sample. The captured pathogen DNA is pulled down and sequenced, while the nonhybridizing host contaminants are washed away. Although these techniques improve sample quality (22–24), they have numerous limitations and drawbacks. First, the oligonucleotide baits (probes) are strain specific and expensive and only partially remove the contaminating genome. Second, optimal hybridization conditions for complete genome pulldown that avoid nonspecific capture and bias are difficult to achieve. Third, the current techniques require specialized skills and relatively large quantities (at least 2 μg) of starting DNA material to complete the purification process (23). A traditional method for separating human and P. falciparum DNA involves the use of Hoechst dye and ultracentrifugation in a CsCl gradient (25). This method utilizes the inherent density differences between two DNA samples (e.g., human and P. falciparum) when their G+C content is significantly different. Although studies have reported successful separation, the method is inefficient and suffers from poor sensitivity. Furthermore, it is impractical if the quantity of starting material is limited, as is the case with many clinical samples.
Here we describe a novel approach to solve host contamination problems using an enzymatic method that has the potential for application to a wider range of pathogen samples. Unlike hybridization enrichment methods, the current technique is not limited by the scope of oligonucleotide probes and therefore suitable for WGS applications. Our tests show that MD-REs selectively degrade human DNA, thereby enriching malaria DNA in clinical samples. We have used various analysis metrics and show that MspJI treatment does not affect the quality and integrity of the Plasmodium genome. Whereas other methods have been associated with base bias problems (22), our analysis has detected no base bias associated with MspJI treatment. Evenness of genome coverage by the treated samples was similar to that by pure untreated samples. A further evaluation of the quality of data generated by MspJI treatment involved testing the coverage of 86,158 high-quality typeable SNPs previously used in parasite diversity studies (6). All treated samples were able to confidently call >98% of the typeable SNPs, including clinical samples with host contamination of up to ∼90%.
We further explored ways of making the treatment procedure compatible with high-throughput automation. To this end, we describe a gel-free option that makes the technique automation friendly. The gel-free option uses magnetic beads for size selection and is particularly suitable for samples where the starting DNA material is of limited quantity. However, the gel-free method suffers from poorer yield (Fig. 2B), mainly due to the suboptimal size separation by magnetic beads technology. The current enzyme digestion time (16 h) can be shortened depending on the level of contamination. Future optimization of the enzyme purity and buffer conditions may improve digestion efficiency.
The robustness and simplicity of the host depletion procedure make it suitable for routine applications. The method is practical even for poorly resourced clinical laboratories near the field, which process clinical samples before sending the DNA to larger sequencing centers. The technology is relatively rapid and cost-effective, with an estimated treatment cost of only $4 per sample.
Conclusion.
Host contamination in pathogen clinical sequencing is currently a major problem. The current development has the potential to alleviate human DNA contamination problems in clinical sequencing. Implementation will not only reduce the time and cost of pathogen sequencing for health care but also increase the sampling density of culture-free field isolates for control, research, and public health sequencing applications.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Abdoulaye Djimde (Malaria Research and Training Centre, Bamako, Mali), Jean-Bosco Ouedraogo (Institut de Recherche en Sciences de la Santé, Burkina Faso), Lucas Amenga-Etego (Navrongo Health Centre, Navrongo, Ghana), and Rick Fairhurst (National Institute of Allergy and Infectious Diseases, MD) for providing field samples for analysis.
This work was supported by the Wellcome Trust (grant number 079355/Z/06/Z). T.D.O. is supported by the European Union 7th framework EVIMalaR.
We declare no competing financial interests.
Footnotes
Published ahead of print 5 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02507-12.
REFERENCES
- 1. Chin CS, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, Bullard J, Webster DR, Kasarskis A, Peluso P, Paxinos EE, Yamaichi Y, Calderwood SB, Mekalanos JJ, Schadt EE, Waldor MK. 2011. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364:33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gardy JL, Johnston JC, Ho Sui SJ, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, Varhol R, Birol I, Lem M, Sharma MK, Elwood K, Jones SJ, Brinkman FS, Brunham RC, Tang P. 2011. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N. Engl. J. Med. 364:730–739 [DOI] [PubMed] [Google Scholar]
- 3. Harris SR, Cartwright EJP, Török ME, Holden MTG, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 9 November 2012. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect. Dis. [Epub ahead of print.] doi:10.1016/S1473-3099(12)70268-2 [DOI] [PMC free article] [PubMed]
- 4. Lienau EK, Strain E, Wang C, Zheng J, Ottesen AR, Keys CE, Hammack TS, Musser SM, Brown EW, Allard MW, Cao G, Meng J, Stones R. 2011. Identification of a salmonellosis outbreak by means of molecular sequencing. N. Engl. J. Med. 364:981–982 [DOI] [PubMed] [Google Scholar]
- 5. Pallen MJ, Loman NJ, Penn CW. 2010. High-throughput sequencing and clinical microbiology: progress, opportunities and challenges. Curr. Opin. Microbiol. 13:625–631 [DOI] [PubMed] [Google Scholar]
- 6. Manske M, Miotto O, Campino S, Auburn S, Almagro-Garcia J, Maslen G, O'Brien J, Djimde A, Doumbo O, Zongo I, Ouedraogo J-B, Michon P, Mueller I, Siba P, Nzila A, Borrmann S, Kiara SM, Marsh K, Jiang H, Su X-Z, Amaratunga C, Fairhurst R, Socheat D, Nosten F, Imwong M, White NJ, Sanders M, Anastasi E, Alcock D, Drury E, Oyola S, Quail MA, Turner DJ, Ruano-Rubio V, Jyothi D, Amenga-Etego L, Hubbart C, Jeffreys A, Rowlands K, Sutherland C, Roper C, Mangano V, Modiano D, Tan JC, Ferdig MT, Amambua-Ngwa A, Conway DJ, Takala-Harrison S, Plowe CV, Rayner JC, Rockett KA, Clark TG, Newbold CI, Berriman M, MacInnis B, Kwiatkowski DP. 2012. Analysis of Plasmodium falciparum diversity in natural infections by deep sequencing. Nature 487:375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pallen MJ, Loman NJ. 2011. Are diagnostic and public health bacteriology ready to become branches of genomic medicine? Genome Med. 3:53 doi:10.1186/gm269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N. Engl. J. Med. 366:2267–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Macdermed DM, Uribe-Bruce L. 2011. Diverse perspectives on the current state of genomic medicine: has the revolution begun? Genome Med. 3:27 doi:10.1186/gm241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Otto TD. 2011. Real-time sequencing. Nat. Rev. Microbiol. 9:633 doi:10.1038/nrmicro2638 [DOI] [PubMed] [Google Scholar]
- 11. Auburn S, Campino S, Clark TG, Djimde AA, Zongo I, Pinches R, Manske M, Mangano V, Alcock D, Anastasi E, Maslen G, Macinnis B, Rockett K, Modiano D, Newbold CI, Doumbo OK, Ouedraogo JB, Kwiatkowski DP. 2011. An effective method to purify Plasmodium falciparum DNA directly from clinical blood samples for whole genome high-throughput sequencing. PLoS One 6:e22213 doi:10.1371/journal.pone.0022213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aird D, Ross MG, Chen WS, Danielsson M, Fennell T, Russ C, Jaffe DB, Nusbaum C, Gnirke A. 2011. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 12:R18 doi:10.1186/gb-2011-12-2-r18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oyola SO, Otto TD, Gu Y, Maslen G, Manske M, Campino S, Turner DJ, Maclnnis B, Kwiatkowski DP, Swerdlow HP, Quail MA. 2012. Optimizing Illumina next-generation sequencing library preparation for extremely AT-biased genomes. BMC Genomics 13:1 doi:10.1186/1471-2164-13-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Quail MA, Otto TD, Gu Y, Harris SR, Skelly TF, McQuillan JA, Swerdlow HP, Oyola SO. 2012. Optimal enzymes for amplifying sequencing libraries. Nat. Methods 9:10–11 [DOI] [PubMed] [Google Scholar]
- 15. Cohen-Karni D, Xu D, Apone L, Fomenkov A, Sun Z, Davis PJ, Kinney SR, Yamada-Mabuchi M, Xu SY, Davis T, Pradhan S, Roberts RJ, Zheng Y. 2011. The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. Proc. Natl. Acad. Sci. U. S. A. 108:11040–11045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horton JR, Mabuchi MY, Cohen-Karni D, Zhang X, Griggs RM, Samaranayake M, Roberts RJ, Zheng Y, Cheng X. 2012. Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease. Nucleic Acids Res. 40:9763–9773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng Y, Cohen-Karni D, Xu D, Chin HG, Wilson G, Pradhan S, Roberts RJ. 2010. A unique family of Mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res. 38:5527–5534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bird AP. 1986. CpG-rich islands and the function of DNA methylation. Nature 321:209–213 [DOI] [PubMed] [Google Scholar]
- 20. Kricker MC, Drake JW, Radman M. 1992. Duplication-targeted DNA methylation and mutagenesis in the evolution of eukaryotic chromosomes. Proc. Natl. Acad. Sci. U. S. A. 89:1075–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bright AT, Tewhey R, Abeles S, Chuquiyauri R, Llanos-Cuentas A, Ferreira MU, Schork NJ, Vinetz JM, Winzeler EA. 2012. Whole genome sequencing analysis of Plasmodium vivax using whole genome capture. BMC Genomics 13:262 doi:10.1186/1471-2164-13-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Melnikov A, Galinsky K, Rogov P, Fennell T, Van Tyne D, Russ C, Daniels R, Barnes KG, Bochicchio J, Ndiaye D, Sene PD, Wirth DF, Nusbaum C, Volkman SK, Birren BW, Gnirke A, Neafsey DE. 2011. Hybrid selection for sequencing pathogen genomes from clinical samples. Genome Biol. 12:R73 doi:10.1186/gb-2011-12-8-r73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith M, Campino S, Gu Y, Clark TG, Otto TD, Maslen G, Manske M, Imwong M, Dondorp AM, Kwiatowski DP, Quail MA, Swerdlow H. 2012. An in-solution hybridisation method for the isolation of pathogen DNA from human DNA-rich clinical samples for analysis by NGS. Open Genomics J. 5:18–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dame JB, McCutchan TF. 1987. Plasmodium falciparum: Hoechst dye 33258-CsCl ultracentrifugation for separating parasite and host DNAs. Exp. Parasitol. 64:264–266 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.