Abstract
Prospective studies addressing the clinical value of broad-range PCR using the internal transcribed spacer region (ITS) for diagnosis of microscopy-negative fungal infections in nonselected patient populations are lacking. We first assessed the diagnostic performance of ITS rRNA gene PCR compared with that of routine microscopic immunofluorescence examination. Second, we addressed prospectively the impact and clinical value of broad-range PCR for the diagnosis of infections using samples that tested negative by routine microscopy; the corresponding patients' data were evaluated by detailed medical record reviews. Results from 371 specimens showed a high concordance of >80% for broad-range PCR and routine conventional methods, indicating that the diagnostic performance of PCR for fungal infections is comparable to that of microscopy, which is currently considered part of the “gold standard.” In this prospective study, 206 specimens with a negative result on routine microscopy were analyzed with PCR, and patients' clinical data were reviewed according to the criteria of the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group. We found that broad-range PCR showed a sensitivity, specificity, positive predictive value, and negative predictive value of 57.1%, 97.0%, 80%, and 91.7%, respectively, for microscopy-negative fungal infections. This study defines a possible helpful role of broad-range PCR for diagnosis of microscopy-negative fungal infections in conjunction with other tests.
INTRODUCTION
Invasive fungal infections (IFIs) remain a leading cause of death (1) and represent a massive financial burden to the health care system. Pulmonary invasive aspergillosis (IA) is the most common invasive mold infection (IMI) in immunocompromised patients (2); however, a shift to non-Aspergillus infections has become evident in the last few years (3). The crude mortality rate of IMIs is considerably high (4), though it is largely influenced by early diagnosis and adequate treatment (5–7). However, securing a firm diagnosis is difficult, as patients may not exhibit specific signs and symptoms related to IFIs. The traditional microbiological workup of clinical specimens is based on microscopic examination (8, 9), culture on various media (8, 9), and serological tests, such as the Platelia Aspergillus galactomannan enzyme immunoassay (GM-EIA; Bio-Rad) (9). No method has proven sufficiently sensitive and specific to allow adequate diagnosis, and the “gold standard” consists of microscopy and culture. Microscopic examination allows the cheap and rapid detection of fungal elements in clinical specimens. Despite this advantage of providing an early presumptive or definitive diagnosis of IFI, fungal classification is not possible. Hence, a differentiation within, e.g., Aspergillus and mucormycetes is desirable since the clinical management might be different (10, 11). Therefore, additional tests which overcome these drawbacks are highly warranted. Over the past 2 decades, molecular techniques have been implemented for accurate pathogen identification in diagnostic microbiology (12–14). Broad-range internal transcribed spacer (ITS) rRNA gene PCR is used to detect and successfully identify fungal pathogens predominantly in immunosuppressed patients (15). Despite the wide implementation of panfungal PCR (12, 15–17), there are no evidence-based studies systematically addressing its diagnostic impact in nonselected (random) populations of patients suspected of having an infectious disease not limited to particular disease entities (e.g., acute leukemia). In addition, little information on effective implementation of broad-range PCR with microscopy-negative samples is available. In case microscopic examination is negative, one has to go back to culture and antigen testing. However, both tests are associated with low or varying sensitivity, resting on the patient population and effective antifungal treatment (18).
Here, we performed both a prospective laboratory study to compare the diagnostic performance of fungal PCR with that of microscopy and a prospective clinical study to assess the impact of broad-range PCR with microscopy-negative specimens.
MATERIALS AND METHODS
The study design was composed of a laboratory study and a clinical study. In the laboratory study, specimens from primary sterile body sites and bronchoalveolar lavage (BAL) fluid specimens were subjected (in parallel) to fungal microscopy and broad-range PCR, to compare the diagnostic performance of PCR and conventional methods. We applied microscopic immunofluorescence examinations to all relevant clinical specimens (other than blood cultures) obtained from patients suspicious of having IFIs.
In the clinical study, an algorithm integrating the broad-range PCR into the diagnostic sample workup was used. In this algorithm, samples submitted to the laboratory were subjected to broad-range PCR. Tests to detect fungal pathogens were performed on specific request by the clinicians and for patients for whom any IFI was highly suspicious or were performed in cases where an IFI could not be excluded.
This study was approved by the ethical committee of the Innsbruck Medical University and was done according to good clinical practice.
Clinical specimens.
Routine patient samples, such as BAL fluid, biopsy, tissue, autopsy, and sterile fluid (e.g., cerebrospinal fluid, ascites) specimens, were obtained from 8 tertiary care hospitals of Tirol, Austria. Clinical specimens were collected at the Division of Hygiene and Medical Microbiology, Innsbruck Medical University, and samples were aseptically divided into three fractions for microscopy, culture, panfungal PCR, and the GM-EIA, if appropriate. GM-EIA is available for routine diagnosis in our laboratory, and the application is recommended for either serum or BAL fluid. If a BAL fluid volume and/or a tissue amount allows such additional investigation, we apply the GM-EIA in patients suspicious of suffering from IA. Two consecutive samples with a cutoff of >0.5 for serum and a cutoff of >1 for BAL fluid are suggestive of IA.
Medical record review.
Clinical data for the patients enrolled in the clinical study were obtained by medical record review and analyzed for the likelihood of a fungal infection according to the criteria of the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) (19). In the final analysis, patients were categorized as suffering from proven, probable, possible, or no IFI. The definition for proven IFI required microscopic documentation of an infection (existence of hyphae or yeast-like forms) or recovery of a fungus from a specimen from a normally sterile site. The definition of probable IFI required the fulfillment of criteria within host factors, clinical manifestations (symptoms and radiological features), and microbiological evidence. All probable cases fulfilled the diagnostic criteria with a surrogate non-culture-based method (a positive galactomannan assay [i.e., GM-EIA] result), radiologically compatible computed tomography (CT) findings, and recovery of a fungus.
Microscopy and cultures.
Fungi-Fluor (calcofluor white staining solution; Polysciences) stains of clinical specimens and fungal cultures were prepared as described previously (8). In brief, solid and tissue samples were placed in sterile 0.9% sodium chloride solution following retrieval, minced upon receipt in the laboratory, prepared for microscopy, inoculated for culture, and incubated on Sabouraud agar and broth. Agar plates were examined for growth after 72 h and 5 and 10 days. Cultures were considered negative if no fungal growth was visible after 14 days of incubation.
DNA extraction and PCR.
DNA extraction was done using a modified cetyltrimethylammonium bromide (CTAB) protocol with the addition of proteinase K (Fermentas, St. Leon-Rot, Germany) and chloroform-isoamyl alcohol (1:24; Sigma-Aldrich Corporation, St. Louis, MO) (20, 21). Extracted DNA was detected with panfungal PCR using ITS3 forward and ITS4 reverse primers (Metabion, Martinsried, Germany), which amplify the ITS2 region of fungal ribosomal DNA genes (15). The PCR mixture (50 μl) contained 2 μl of DNA, 16 mM (NH4)2SO4, 67 mM Tris-HCl (pH 8.8 at 25°C), 1.5 mM MgCl2, 0.01% Tween 20, 0.1 mM each deoxynucleoside triphosphate, 1.25 U of Taq polymerase (genXpress; Wiener Neudorf, Austria), as well as 0.1 μM each primer. The PCR cycling conditions consisted of an initial denaturation for 2 min at 94°C and then 35 cycles of 30 s at 94°C, 30 s at 58°C, and 1 min at 72°C and a final extension at 72°C for 5 min. All PCRs were run on a Biometra personal cycler (Biometra, Goettingen, Germany). Aspergillus DNA was used as a PCR positive control in each run. As a negative control, buffers and PCR reagents were routinely tested for fungal DNA contamination. In order to overcome the PCR-inhibitory effects of various compounds (e.g., hemoglobin), different amounts of the routine patient samples were extracted. Furthermore, DNA extracts used for PCR were diluted 1:2, 1:5, and 1:10. The PCR products were visualized on a gel imager (Gel Doc EZ Imager; Bio-Rad Laboratories, Austria) after electrophoresis on 1% agarose gels using GelRed (Biotum, Hayward, CA) for staining. All PCR products were cleaned with ExoSap-It (USB Corporation, Cleveland, OH) following the manufacturer's instructions. Aspergillus DNA was used as a positive control. As a negative control, buffers and PCR reagents were routinely tested for fungal DNA contamination. Sequencing was done with a BigDye Terminator (version 3.1) cycle sequencing kit and a capillary sequencer (3500 genetic analyzer; Applied Biosystems, Life Technologies Corporation, Carlsbad, CA). Suspicion of polymicrobial infection occurred once two or more bands were detected on the gel. Separated bands were cut out, and DNA was extracted using a QIAquick gel extraction kit (Qiagen, Hilden, Germany). Sequencing of these DNA eluates failed, and patient samples (n = 2) were excluded due to technical difficulties.
Statistical methods.
Statistical calculations were done using GraphPad Prism software, version 5.02 (GraphPad Software). The statistical methods related to the data set analyzed are described in the text or in the figure legend. Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) are given.
RESULTS
Sensitivity and specificity of broad-range PCR compared with microscopy (laboratory study).
We prospectively assessed broad-range PCR compared with fungal microscopy; in addition, positive cultures and GM-EIA data were adducted, if available. A total of 371 samples from a nonselected patient population (n = 306 patients) were investigated, and clinical data were not taken into account. There was a high concordance of both microscopy and PCR in 357 of 371 (96.1%) analyses (Tables 1 and 2). Discordant results were observed in 14 cases, with 10 (2.6%) microscopy-negative, PCR-positive results and 4 (1.0%) culture-positive, PCR-negative results.
Table 1.
Broad-range PCR compared with direct microscopy and conventional culturea
| Assay and result | No. (%) of specimens with the following result by microscopy: |
|
|---|---|---|
| + | − | |
| PCR | ||
| + | 89 (23.9) | 10 (2.6) |
| − | 4 (1.0) | 268 (72.2) |
| Culture | ||
| + | 83 (22.3) | 12 (2.6) |
| − | 16 (4.3) | 260 (70) |
A total of 371 clinical specimens were included in this laboratory study. For fungal identification, see Table 2. −, negative; +, positive.
Table 2.
Pathogens identified by broad-range sequencing and microscopy in the laboratory studya
| Specimen and species | No. of isolates |
|---|---|
| Microscopy-positive, PCR-positive specimens | |
| A. fumigatus | 27 |
| A. terreus | 12 |
| A. flavus | 10 |
| C. albicans | 19 |
| C. glabrata | 9 |
| C. tropicalis | 4 |
| Cryptococcus neoformans | 1 |
| Lichtheimia corymbifera | 3 |
| Rhizomucor spp. | 2 |
| Rhizopus spp. | 1 |
| Scedosporium apiospermum | 1 |
| Microscopy-negative, PCR-positive specimens | |
| A. fumigatus | 2 |
| A. terreus | 1 |
| C. albicans | 2 |
| Cladosporium spp. | 2 |
| Penicillium chrysogenum | 2 |
| Lichtheimia corymbifera | 1 |
| Microscopy-positive, PCR-negative specimens | |
| Aspergillus-like elementsb | 2 |
| Mucormycete-like elementsc | 2 |
The laboratory study included 371 specimens from 306 patients.
One sample (BAL fluid) was GM-EIA positive.
In one patient, a Mucor species was cultured from the specimen.
Species assignments in 10 microscopy-negative, PCR-positive samples (Table 2) included Aspergillus spp. (n = 3), Candida albicans (n = 2), Lichtheimia corymbifera (n = 1), Cladosporium spp. (n = 2), and Penicillium chrysosporium (n = 2). Of them, cultures were positive in 4 samples for an Aspergillus species, Candida albicans, a Cladosporium species, and P. chrysosporium. The isolates identified in microscopy-positive, PCR-negative results included Aspergillus spp. (n = 2) and mucormycetes (n = 2).
Broad-range PCR for diagnosis of infections in microscopy-negative samples (clinical study).
In the clinical study, we prospectively enrolled all patient specimens received from January 2009 to December 2011 (n = 206 samples, 190 patients) for which fungal tests had remained negative for microscopic examination. The likelihood of infection was retrospectively judged on the basis of EORTC/MSG criteria (19). Data were obtained by detailed patient medical record review. Proven IFI required microscopic documentation of hyphae or yeast-like forms or recovery of a fungus from specimens from a normally sterile site (Tables 3 and 4). Probable IFI required host factors, clinical manifestations (symptoms and radiological features), and microbiological evidence of a fungus. All cases enrolled either had a positive GM-EIA result from serum or BALs and/or recovery of fungi. Possible cases lacked positive GM-EIA and fungal culture results.
Table 3.
Summary of microscopy-negative, PCR-positive specimens in the clinical studya
| Patient no. (n = 16) | Clinical specimen | Fungal species identified by broad-range PCR | Fungal culture result | GM-EIAb result from serum/BAL fluid | Primary antifungal treatment | Fungus observed by culture in various other body sites | EORTC/MSG criteria |
|---|---|---|---|---|---|---|---|
| 1 | BAL fluid | A. flavus | A. flavus | + | Voriconazole | Probable IFI | |
| 2 | Biopsy | A. fumigatus | Neg | +/++ | L-AMB | Probable IFI | |
| 3 | Biopsy | A. terreus | Neg | Neg | Voriconazole | A. terreus (nose, throat) | Probable IFI |
| 4 | Biopsy | Rhizomucor spp. | Rhizomucor sp. | Neg | L-AMB, voriconazole | Rhizopus sp. (wound) | Proven IFI |
| 5 | BAL fluid | A. terreus | Neg | + | Voriconazole | Probable IFI | |
| 6 | BAL fluid | L. corymbifera | Neg | Neg | Voriconazole | Possible IFI | |
| 7 | BAL fluid | A. fumigatus | A. fumigatus (previous BAL fluid samples) | Not done | L-AMB | Probable IFI | |
| 8 | BAL fluid | Penicillium sp. | Penicillium sp. | Neg | Caspofungin, voriconazole | Penicillium sp. (subsequent 2nd and 3rd BAL fluid samples) | Probable IFI |
| 9 | Autopsy (lung) | A. fumigatus | A. fumigatus, A. terreus | +/++ | Voriconazole | A. fumigatus, A. terreus | Proven IFI |
| 10 | Autopsy (muscle) | Mucor sp. | Neg | Not done | Voriconazole, L-AMB | Mucor sp. (wound, lung) | Proven IFI |
| 11 | Tissue | L. corymbifera | Neg | Not done | L-AMB | Possible IFI | |
| 12 | Tissue | C. albicans | Neg | Neg | Anidulafungin | Possible IFI | |
| 13 | Tissue | C. albicans | Neg | Neg | Voriconazole | Possible IFI | |
| 14 | Fluid | A. fumigatus | Neg | +/++ | Caspofungin | A. fumigatus (sputum, 3 times) | Probable IFI |
| 15 | CNS | C. albicans | Neg | Not done | Caspofungin | C. albicans | Possible IFI |
| 16 | CNS | A. flavus | Neg | +/++c | L-AMB | A. fumigatus (previous samples) | Probable IFI |
CT, computed tomography; GM-EIA, galactomannan enzyme immunoassay; L-AMB, liposomal amphotericin B; IFI, invasive fungal infections; Neg, negative.
For several patients, GM-EIA was performed with either serum (+) or BAL fluid (++), or both serum/BAL fluid, unless indicated otherwise.
The results are for serum (+)/CNS (++).
Table 4.
Summary of microscopy-negative and PCR-negative specimens in proven and probable cases according to EORTC/MSG criteriaa
| Patient no. (n = 12) | Clinical specimen | Fungal culture result | GM-EIAb result from serum/BAL fluid | Secondary antifungal prophylaxisc | EORTC/MSG criteria |
|---|---|---|---|---|---|
| 1 | Lung biopsy | A. flavus | + | Proven IFI | |
| 2 | Lung biopsy | Neg | −/++ | Probable IFI | |
| 3 | Lung biopsy | Neg | Neg | Voriconazole | Proven IFI (histopathology-confirmed diagnosis) |
| 4 | BAL fluid | Neg | +/++ | Probable IFI | |
| 5 | BAL fluid | Neg | + | Probable IFI | |
| 6 | BAL fluid | Neg | −/+ | Probable IFI | |
| 7 | BAL fluid | A. fumigatus | Not done | Probable IFI | |
| 8 | BAL fluid | A. terreus | + | Voriconazole | Probable IFI |
| 9 | Autopsy | A. terreus | +/++ | Proven IFI | |
| 10 | Autopsy | Mucor spp. | Not done | L-AMB | Proven IFI |
| 11 | Autopsy | Neg | Not done | L-AMB | Proven IFI |
| 12 | CNS | Candida glabrata (blood) | Neg | Proven IFI |
GM-EIA, galactomannan enzyme immunoassay; L-AMB, liposomal amphotericin B; IFI, invasive fungal infections; Neg, negative.
GM-EIA was performed in several patients from either serum or BAL fluid, or both serum/BAL fluid.
These patients received secondary antifungal prophylaxis due to the proof of IFI during the previous hospitalization; clinical signs and symptoms showed deterioration.
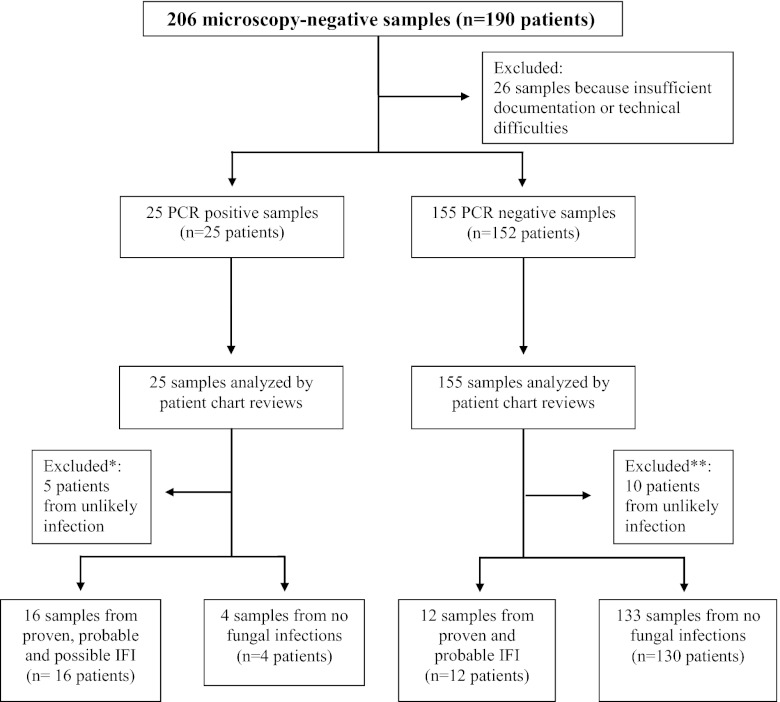
Twenty-six of 206 specimens were immediately excluded because of unavailable or insufficient patient documentation or due to technical difficulties. Of the remaining 180 microscopy-negative specimens, 25 and 155 samples were PCR positive and PCR negative, respectively. Five of 25 microscopy-negative, PCR-positive samples and 10 of 155 microscopy-negative, PCR-negative samples were further excluded because they were from patients unlikely to have infections (Fig. 1). The suspicions of mycoses were revised before broad-range PCR was interpreted and clinicians withdrew their request to examine the specimens for fungal pathogens. It became apparent that cancer (n = 6) and noninfectious processes (n = 9) were responsible for underlying diseases in these particular cases. So, in total, 165 specimens from 162 patients were included in the analysis: BAL fluid specimens (n = 78); CT-guided percutaneous lung biopsy specimens (n = 61); specimens of various other tissues (n = 15); sterile fluids, including cerebrospinal fluid (n = 4) and ascites (n = 2); and autopsy samples (n = 5).
Fig 1.
Enrollment of patients (specimens) from the clinical study on broad-range PCR for diagnosis of microscopy-negative fungal infections. *, cancer (n = 3) and noninfectious agents (n = 2) were documented as underlying causes of disease, and clinicians withdrew their request to examine the specimens for fungal pathogens; **, malignant and nonmalignant tumors (n = 3) and noninfectious agents (n = 7) were documented as underlying causes of disease, and clinicians withdrew their request to examine the specimens for fungal pathogens.
Twenty of 165 samples were PCR positive and included Aspergillus spp. (n = 8), mucormycetes (n = 4), a Penicillium species (n = 1), and Candida species (n = 3) (Table 3). Sixteen of the 20 PCR-positive samples were from patients categorized as having proven (n = 3), probable (n = 8), and possible (n = 5) fungal infections (Table 3). Clinical data obtained showed various underlying risk factor for IFIs, and all of the patients received antifungal therapy. Six of these patients showed multiple high GM-EIA values, and seven of them were positive by fungal culture of specimens from various body sites at a point in time during the study or a fungal pathogen had been identified in previous analyses; four cases were considered contaminants, as bacterial infections emerged.
Of 145 samples, 133 were negative by broad-range PCR, including samples from 12 patients with proven and probable fungal infection, according to EORTC/MSG criteria (Fig. 1). These patients had received secondary antifungal prophylaxis (n = 4), fungal pathogens had been detected during previous hospitalization (n = 6), the patients had had a positive result by a fungal test other than microscopy and PCR, or the patients died and autopsy tissues showed the presence of fungal elements (Table 4).
The sensitivity of broad-range PCR in diagnosing fungal infection for microscopy-negative samples was 57.1%, the specificity was 97.0%, the PPV was 80.0%, and the NPV was 91.7%. If we reintegrate for further analyses the excluded samples (n = 15) into the evaluation (Fig. 1), specificity, PPV, and NPV decrease to 94.0%, 64.0%, and 92.2%, respectively.
Overall, according to EORTC/MSG criteria, 28 patients showed disease from proven (n = 9), probable (n = 14), and possible (n = 5) IFIs. Of these, 18 and 4 cases suffered from hematological malignancies or underwent solid organ transplantation, respectively; another 3 patients received intensive medical treatment for other reasons. All of them met either host factor, clinical, or mycological criteria. If we extract only proven and probable cases (n = 23), broad-range PCR was positive in 50% of patients (11 cases) (Tables 3 and 4).
DISCUSSION
Clinicians are frequently challenged with the suspicion of fungal infectious diseases even when a conventional microbiological workup remains negative. The primary goal of this study was to assess the value of broad-range PCR for the diagnosis of fungal infections in a nonselected patient population, in particular, to provide evidence-based data to the clinician for a targeted use of panfungal PCR.
In the laboratory part of the study, we analyzed the diagnostic sensitivity and specificity of broad-range PCR with 371 specimens compared with those of routine fungal microscopic examination. We observed that the concordance between broad-range PCR and microscopy is >80%. The sensitivity of broad-range PCR in our data set was 95.6%, and the specificity was 96.4%. A recent meta-analysis reported similar data: sensitivity and specificity were 91% and 92%, respectively, by BAL fluid PCR assays for proven and probable IA (22). A subgroup analysis showed that the performance of the PCR assay was influenced by the PCR assay methodology, primer design, and the methods used for cell wall disruption and DNA extraction. Hammond et al. showed that Mucorales PCR is useful for confirmation of the diagnosis of mucormycosis and for further characterization of the infection in cases where cultures are negative (23).
In general, species-specific PCR is based on the amplification of species-specific genes, and its usage is restricted to a limited number of pathogens from defined clinical samples (24, 25). Previously, we tested the MycAssay Aspergillus real-time PCR kit (26). For all samples, the sensitivity of the MycAssay Aspergillus test was 82% and the specificity was 79% relative to the microscopy results and 90% and 64%, respectively, compared with Aspergillus culture results. The MycAssay Aspergillus test detected tissue-invasive infections with Aspergillus fumigatus, A. flavus, and A. terreus. However, the coverage of a wide range of fungal pathogens is important, as fungal epidemiology is changing dramatically (27). We face a shift from C. albicans to non-C. albicans and from Aspergillus to non-Aspergillus pathogens. The performance with CT-guided percutaneous lung biopsy specimens identified that more than 30% of our invasive fungal pathogens were mucormycetes (8). Mucor spp., Rhizopus spp., and Lichtheimia corymbifera are the main representatives. Such a change is of major clinical importance, as the fungal pathogens listed require different patient and treatment procedures. Aspergillus spp. are generally susceptible to voriconazole, but mucormycetes are not (5, 28, 29). Such a diversity of fungal pathogens requires a broad-range PCR for fast and sensitive detection.
In the clinical part of this study, we assessed the diagnostic power of broad-range PCR in the diagnosis of fungal infections in which microscopy remained negative. Patients with these results frequently pose a diagnostic dilemma, as one has to rely on cultures and antigen tests, which in turn display various sensitivities and specificities. Such a situation raises the question of whether a molecular-based diagnosis can provide valid data, especially in cases where cultures remain negative, too. However, this basically means a lack of proof of infection, as a positive PCR result might simply indicate contamination. We therefore concentrated on this issue and assessed the value of broad-range PCR with microscopy-negative samples. In our study, PCR identified fungal pathogens in 57.1% of microscopy-negative specimens; clinical analysis revealed that several of these patients suffered from proven and probable infection (Table 3). The PPV was 80.0%, indicating that a positive result might identify an infection. However, if for further analyses we also reintegrated the samples which were excluded (n = 15 specimens), the specificity, PPV, and NPV decreased to 94.0%, 64.0%, and 92.2%, respectively. Therefore, broad-range PCR is partly valuable and may add key diagnostic findings, especially in conjunction with other tests. The majority of pathogens identified in the microscopy-negative, PCR-positive group of the clinical study were common fungal pathogens. Conversely, in 12 cases broad-range PCR failed to detect IFI, as cultures and the antigen tests performed displayed the existence of an underlying infection. The discrepancies observed between microscopy, culture, and PCR might have occurred for multiple reasons (30). Fungal contamination (30, 31), loss of fungal viability (30, 32), aggressive specimen processing (33), and examination of samples from two different areas (26) are some of the potential explanations. However, regardless of the reason for the lack of fungal growth or for a negative fungal PCR result, fungal elements in tissues causing pathology should be treated immediately (32).
Our study reflects the difficulty of accurately diagnosing fungal infection at the bedside, emphasizing the need for an interdisciplinary approach toward accurate diagnosis, in which clinical presentation, laboratory findings, and imaging all contribute to the final diagnosis. Carrying out broad-range PCR with microscopy-negative samples in parallel with other tests will aid substantially in diagnosing fungal infections. However, it is also clear that our study has several limitations, starting with the use of a set of patients with microscopy-negative and PCR-positive results; herein we lacked hard facts for proof of IFI in 5 patients; on the basis of EORTC/MSG criteria, these cases suffered from a possible IFI. All of them received antifungal treatment, and the clinical course supported no other diseases. Further drawbacks are followed by the general diagnostic limitations of broad-range PCR, such as a lack of standardization of reagents and methods, the absence of optimal contamination controls, and uncertainty over which fungal targets result in high sensitivity and specificity (12, 15, 21). The existence of a double infection is also extremely difficult, if not impossible, to demonstrate. Whether patients 4 and 16 suffered from a double infection or whether species identification failed remains unknown. Patient 4 suffered from invasive mucormycoses, as Rhizomucor was identified in the lung biopsy specimen by culture and PCR, but in the surgical wound, a Rhizopus species was identified by morphology. In patient 16, A. fumigatus had been identified by morphology in a previous central nervous system (CNS) sample, whereas A. flavus detection was recorded by sequencing.
PCR has been used as an aid in the diagnosis of IA for almost 2 decades. A lack of standardization has limited both its acceptance as a diagnostic tool and multicenter clinical evaluations, preventing its inclusion in disease-defining criteria (18). In 2006, the European Aspergillus PCR Initiative was formed (34). The aim of the initiative is to provide optimal standardized protocols for the widespread clinical evaluation of the Aspergillus PCR to determine its diagnostic role and allow inclusion in disease diagnosis criteria.
On the basis of our data, we propose that broad-range PCR of samples from primary sterile body sites be done for patients with a high clinical suspicion of infection and negative microscopy results. Broad-range PCR may be helpful for diagnosis of microscopy-negative fungal infections in conjunction with other tests. The suspicion of infections with difficult-to-culture pathogens justifies a broad-range PCR on its own, in particular, in centers with emerging fungal pathogens involved in IFIs.
ACKNOWLEDGMENTS
We thank D. Moroder for technical support.
We declare that we have no conflict of interest. We all have submitted the International Committee of Medical Journal Editors form for disclosure of potential conflicts of interest.
Footnotes
Published ahead of print 26 December 2012
REFERENCES
- 1. Richardson M, Lass-Florl C. 2008. Changing epidemiology of systemic fungal infections. Clin. Microbiol. Infect. 14(Suppl 4):5–24 [DOI] [PubMed] [Google Scholar]
- 2. Segal BH. 2009. Aspergillosis. N. Engl. J. Med. 360:1870–1884 [DOI] [PubMed] [Google Scholar]
- 3. Binder U, Lass-Flörl C. 2011. Epidemiology of invasive fungal infections in the Mediterranean area. Mediterr. J. Hematol. Infect. Dis. 3:e20110016 doi:10.4084/MJHID.2011.0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Walsh T, Annaissie E, Dennind DW, Herbrecht R, Kontoyiannis DP, Marr AK, Morrison VA, Segal BH, Steinbach W, Stevens D, Burik JA, Wingard JR, Patterson J. 2009. Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin. Infect. Dis. 46:327–360 [DOI] [PubMed] [Google Scholar]
- 5. Kubak BM, Huprikar SS, AST Infectious Diseases Community of Practice 2009. Emerging & rare fungal infections in solid organ transplant recipients. Am. J. Transplant. 9(Suppl 4):208–226 [DOI] [PubMed] [Google Scholar]
- 6. Lass-Flörl C, Mayr A. 2009. Diagnosing invasive fungal diseases—limitations of microbiological diagnostic methods. Expert Opin. Med. Diagn. 3:1–10 [DOI] [PubMed] [Google Scholar]
- 7. Leventakos K, Russell EL, Kontoyiannis DP. 2010. Fungal infections in leukemia patients: how do we prevent and treat them? Clin. Infect. Dis. 50:405–415 [DOI] [PubMed] [Google Scholar]
- 8. Lass-Flörl C, Resch G, Nachbaur D, Mayr A, Gastl G, Auberger J, Bialek R, Freund M. 2007. The value of computed tomography-guided percutaneous lung biopsy for diagnosis of invasive fungal infection in immunocompromised patients. Clin. Infect. Dis. 45:e101–e104 doi:10.1086/521245 [DOI] [PubMed] [Google Scholar]
- 9. Rex JH. 2006. Galactomannan and the diagnosis of invasive aspergillosis. Clin. Infect. Dis. 42:1428–1430 [DOI] [PubMed] [Google Scholar]
- 10. Pound MW, Townsend ML, Drew RH. 2010. Echinocandin pharmacodynamics: review and clinical implications. J. Antimicrob. Chemother. 65:1108–1118 [DOI] [PubMed] [Google Scholar]
- 11. Steinbach WJ, Juvvadi PR, Fortwendel JR, Rogg LE. 2011. Newer combination antifungal therapies for invasive aspergillosis. Med. Mycol. 49:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bretagne S. 2011. Primary diagnostic approaches of invasive aspergillosis—molecular testing. Med. Mycol. 49:S48–S53 [DOI] [PubMed] [Google Scholar]
- 13. Hage CA, Knox KS, Davis TE, Wheat LJ. 2011. Antigen detection in bronchoalveolar lavage fluid for diagnosis of fungal pneumonia. Curr. Opin. Pulm. Med. 17:167–171 [DOI] [PubMed] [Google Scholar]
- 14. Preuner S, Lion T. 2009. Towards molecular diagnostics of invasive fungal infections. Expert Rev. Mol. Diagn. 9:397–401 [DOI] [PubMed] [Google Scholar]
- 15. Ciardo DE, Lucke K, Imhof A, Bloemberg GV, Böttger EC. 2010. Systematic internal transcribed spacer sequence analysis for identification of clinical mold isolates in diagnostic mycology: a 5-year study. J. Clin. Microbiol. 48:2809–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mengoli C, Cruciani M, Barnes RA, Loeffler J, Donnelly JP. 2009. Use of PCR for diagnosis of invasive aspergillosis: systematic review and meta-analysis. Lancet Infect. Dis. 9:89–96 [DOI] [PubMed] [Google Scholar]
- 17. Munoz-Cadavid C, Rudd S, Zaki SR, Patel M, Moser SA, Brandt ME, Gomez BL. 2010. Improving molecular detection of fungal DNA in formalin-fixed paraffin-embedded tissues: comparison of five tissue DNA extraction methods using panfungal PCR. J. Clin. Microbiol. 48:2147–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Loeffler J, Barnes R, Donnelly JP, European Aspergillus PCR Initiative 2012. Standardization of Aspergillus PCR diagnosis. Bone Marrow Transplant. 42:299–300 [DOI] [PubMed] [Google Scholar]
- 19. De Pauw B, Walsh T, Donnelly JP, Stevens DA, Edwards JE, Calandra T, Pappas P, Maertens J, Lortholary O, Kauffman C, Denning D, Patterson T, Maschmeyer G, Bille J, Dismukes W, Herbrecht R, Hope W, Kibbler C, Kullberg BJ, Marr KA, Munoz P, Odds FC, Perfect JR, Ruhnke M, Segal BH, Sobel J, Sorrell TC, Viscoli C, Wingard J, Zaoutis T, Bennett J. 2008. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin. Infect. Dis. 46:1813–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Demeke T, Jenkins GR. 2010. Influence of DNA extraction methods, PCR inhibitors and quantification methods on real-time PCR assay of biotechnology-derived traits. Anal. Bioanal. Chem. 396:1977–1990 [DOI] [PubMed] [Google Scholar]
- 21. Khot PD, Ko DL, Fredricks DN. 2009. Sequencing and analysis of fungal rRNA operons for development of broad-range fungal PCR assays. Appl. Environ. Microbiol. 75:1559–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun W, Wang K, Gao W, Su X, Qian Q, Lu X, Song Y, Guo Y, Shi Y. 2011. Evaluation of PCR on bronchoalveolar lavage fluid for diagnosis of invasive aspergillosis: a bivariate metaanalysis and systematic review. PLoS One 6:e28467 doi:10.1371/journal.pone.0028467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hammond SP, Bialek R, Milner DA, Petschnigg EM, Baden LR, Marty FM. 2011. Molecular methods to improve diagnosis and identification of mucormycosis. J. Clin. Microbiol. 49:2151–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamakami Y, Hashimoto A, Tokimatsu I, Nasu M. 1996. PCR detection of DNA specific for Aspergillus species in serum of patients with invasive aspergillosis. J. Clin. Microbiol. 34:2464–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamakami Y, Hashimoto A, Yamagata E, Kamberi P, Karashima R, Nagai H, Nasu M. 2000. Evaluation of PCR for detection of DNA specific for Aspergillus species in sera of patients with various forms of pulmonary aspergillosis. J. Clin. Microbiol. 36:3619–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lass-Flörl C, Follett SA, Moody A, Denning DW. 2011. Detection of Aspergillus in lung and other tissue samples using the MycAssay Aspergillus real-time PCR kit. Can. J. Microbiol. 57:765–768 [DOI] [PubMed] [Google Scholar]
- 27. Lass-Flörl C. 2009. The changing face of epidemiology of invasive fungal diseases in Europe. Mycoses 52:197–205 [DOI] [PubMed] [Google Scholar]
- 28. Maschmeyer G, Haas A, Cornely OA. 2007. Invasive aspergillosis: epidemiology, diagnosis and management in immunocompromised patients. Drugs 67:1567–1601 [DOI] [PubMed] [Google Scholar]
- 29. Varkey JB, Perfect JR. 2008. Rare and emerging fungal pulmonary infections. Semin. Respir. Crit. Care Med. 29:121–131 [DOI] [PubMed] [Google Scholar]
- 30. Sangoi AR, Rogers WM, Longacre TA, Montoya JG, Baron EJ, Banaei N. 2009. Challenges and pitfalls of morphologic identification of fungal infections in histologic and cytologic specimens: a ten-year retrospective review at a single institution. Am. J. Clin. Pathol. 131:364–375 [DOI] [PubMed] [Google Scholar]
- 31. Löffler J, Hebart H, Bialek R, Hagmeyer L, Schmidt D, Serey FP, Hartmann M, Eucker J, Einsele H. 2000. Contaminations occurring in fungal PCR assays. J. Clin. Microbiol. 37:1200–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guarner J, Brandt ME. 2011. Histopathologic diagnosis of fungal infections in the 21st century. Clin. Microbiol. Rev. 24:247–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sibley CD, Peirano G, Church DL. 2012. Molecular methods for pathogen and microbial community detection and characterization: current and potential application in diagnostic microbiology. Infect. Genet. Evol. 12:505–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. White LP, Bretagne S, Klingspor L, Melchers WJG, McCulloch E, Schulz B, Finnstrom N, Mengoli C, Barnes R, Donnelly JP, Loeffler J. 2010. Aspergillus PCR: one step closer to standardization. J. Clin. Microbiol. 48:1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]