Abstract
Human enteroviruses (HEVs) are endemic worldwide and among the most common viruses infecting humans. Nevertheless, there are very limited data on the circulation and genetic diversity of HEVs in developing countries and sub-Saharan Africa in particular. We investigated the circulation and genetic diversity of HEVs among 436 healthy children in a limited area of the far north region of Cameroon in 2008 and 2009. We also characterized the genetic biodiversity of 146 nonpolio enterovirus (NPEV) isolates obtained throughout the year 2008 from stool specimens of patients with acute flaccid paralysis (AFP) in Cameroon, Chad, and Gabon. We found a high rate of NPEV infections (36.9%) among healthy children in the far north region of Cameroon. Overall, 45 different HEV types were found among healthy children and AFP patients. Interestingly, this study uncovered a high rate of HEVs of species C (HEV-C) among all typed NPEVs: 63.1% (94/149) and 39.5% (49/124) in healthy children and AFP cases, respectively. Besides extensive circulation, the most prevalent HEV-C type, coxsackievirus A-13, featured a tremendous intratypic diversity. Africa-specific HEV lineages were discovered, including HEV-C lineages and the recently reported EV-A71 “genogroup E.” Virtually all pathogenic circulating vaccine-derived polioviruses (cVDPVs) that have been fully characterized were recombinants between oral poliovaccine (OPV) strains and cocirculating HEV-C strains. The extensive circulation of diverse HEV-C types and lineages in countries where OPV is massively used constitutes a major viral factor that could promote the emergence of recombinant cVDPVs in the Central African subregion.
INTRODUCTION
Enteroviruses (EVs) are members of the genus Enterovirus in the family Picornaviridae. This genus comprises at least 10 different species, including 7 species of human EVs (HEVs): 3 rhinovirus species (species A to C) and 4 EV species (species A to D). In particular, HEV-C includes 3 types of poliovirus (PV) (PV-1, -2, and -3), 9 types of coxsackievirus A (e.g., CVA-11, -13, and -24), and 11 types of EVs (e.g., EV-C95, -C99, and -C116) (http://www.picornaviridae.com/). Further EV species infect pigs and cows (porcine and bovine EVs) and nonhuman primates (NHPs) (simian EVs) (1).
EVs are small nonenveloped viruses having a capsid with icosahedral symmetry. Their genome is made up of a single polyadenylated positive-strand RNA of about 7.5 kb with a single open reading frame flanked by two noncoding regions. The large polyprotein translated from the genomic RNA strand is processed to yield four structural proteins, VP1 to VP4, and nonstructural proteins implicated in the multiplication of the virus, including the viral RNA-dependent RNA polymerase.
HEVs are among the most common viruses infecting humans. Most HEV infections remain subclinical. However, they can cause a wide spectrum of diseases, with symptoms ranging from mild febrile illness to severe forms, such as the common cold, upper respiratory illness, acute hemorrhagic conjunctivitis, aseptic meningitis, myocarditis, encephalitis, and acute flaccid paralysis (AFP) (2).
One of the most worrying HEV-induced diseases is paralytic poliomyelitis, specifically caused by PVs. Poliomyelitis has been one of the greatest human pandemics of the last century. The Global Polio Eradication Initiative (http://www.polioeradication.org/) is based on massive immunization campaigns with the live attenuated oral poliovaccine (OPV). This strategy has led to a drastic decrease in the poliomyelitis incidence worldwide. However, OPV-related PVs can circulate for a long time in underimmunized communities, thereby maintaining a reservoir of neurovirulent circulating vaccine-derived polioviruses (cVDPVs) that can cause poliomyelitis outbreaks. These cVDPVs have been implicated in several poliomyelitis outbreaks, primarily in resource-limited countries (3–8). The largest outbreak of cVDPV infection, in terms of the number of cases and geographic distribution, has been reported in northern Nigeria. Actually, cVDPVs have been circulating in northern Nigeria from 2005 to 2012 and have occasionally been involved in poliomyelitis cases in neighboring countries, including Chad and Niger (9, 10). With few exceptions (11), all analyzed cVDPVs were shown to be recombinant strains having capsid coding regions derived from OPV-related PVs and certain or all nonstructural regions of the genome originating from other HEV-C strains (3, 4, 12, 13). The fact that virtually all cVDPVs were PV/non-PV HEV-C recombinants suggests that a high rate of HEV-C strains and their cocirculation with OPV-related PVs may constitute a major viral factor favoring the emergence and circulation of these pathogenic viruses (3, 5, 13–15).
The Centre Pasteur du Cameroun (CPC) in Yaoundé, Cameroon, has been involved in poliomyelitis surveillance since 1993. As the national and intercountry reference laboratory for poliomyelitis, its activities include the reception of stool specimens from AFP patients and virus isolation and typing of PVs. During investigations for PV, nonpolio enteroviruses (NPEVs) are also detected, either because they are possibly involved in the etiology of AFP or because they are shed with feces without any association with AFP. Routinely obtained NPEV isolates are added to the laboratory biological collection without further investigations.
Despite the worldwide endemicity of HEVs, only very limited data are available on the circulation and biodiversity of NPEVs in sub-Saharan Africa. In particular, there are no data about the rate and genetic diversity of HEVs in Cameroon and other neighboring countries. The last indigenous wild poliovirus strain from Cameroon was reported in 1999. However, the far northern region of Cameroon has remained at high risk of wild PV importation from countries where PV is endemic. This region is a narrow region flanked by the Nigerian northern state (Borno) and Chad (see Fig. S1 in the supplemental material). These two countries bordering Cameroon are still facing the transmission of wild PVs and cVDPVs.
In order to investigate the circulation and genetic biodiversity of HEVs in the far north region of Cameroon, we characterized HEVs infecting healthy children in that particular region. In addition, to have an idea of the nationwide viral diversity in Cameroon and neighboring countries, we also performed molecular characterization of all NPEV strains isolated within the frame of poliomyelitis surveillance at the CPC in 2008.
We found an extensive circulation of diverse NPEVs belonging to as many as 45 different NPEV types, including recently reported and hitherto-unknown lineages. There was a high rate of isolation of HEV-C strains among all isolates. Furthermore, the most prevalent HEV-C types featured a tremendous intratypic variability.
MATERIALS AND METHODS
Field investigations.
Stool specimens were collected from apparently healthy children in three urban districts in Maroua and four neighboring rural villages (Djinglya, Kolofata, Koza, and Tokombere) (see Fig. S1 in the supplemental material). Two rounds of specimen collection were carried out in November 2008 and November 2009. The entire protocol of the study was reviewed and approved by the National Research Ethics Committee, and an administrative approval was obtained from the Cameroonian Ministry of Public Health. A questionnaire including data on date of birth, sex, site of enrollment, previous routine immunization (based on health card), and immunization campaign (based on parent's declarations) with oral poliovaccine was completed for each child. Written informed consent was obtained from the parents or legal guardians of the children enrolled in the study.
Cell lines and virus isolation.
Human rhabdomyosarcoma (RD), human larynx epidermoid carcinoma (HEp-2c), and murine L20B (a derivative of murine L cells expressing the PV human receptor) cell lines were used in this study. Chloroform-treated and clarified stool suspensions were inoculated onto monolayered RD and HEp-2c cells maintained in Dulbecco's modified Eagle's medium (D-MEM) supplemented with 2% fetal calf serum and 2 mM l-glutamine at 36°C. Infected tubes were microscopically checked for 5 days to detect the appearance of cytopathogenic effects (CPE). In order to increase the sensitivity of virus isolation, a blind passage (inoculation of fresh cells) was carried out using the supernatant of the inoculated cultures which had remained negative, and newly inoculated cells were then checked for the next 5 days. The isolates harvested from infected RD and HEp-2c cell lines were systematically inoculated onto L20B cells, and the resulting PV isolates were further typed, as described below. The isolates showing CPE only on RD or HEp-2c and not on L20B cells were considered to be nonpolioviruses and were characterized by molecular analyses, as described below. Throughout the text, the names of strains from field investigations in healthy children are given in the following format: a three-letter code standing for the local district where the stool was sampled (DJA, Djarengol; DJI, Djinglya; DOU, Dougoi; FOU, Founangue; KOL, Kolofata; KOZ, Koza; TOK, Tokombere) followed by the enrollment number of the child. For example, strain TOK-230 was isolated from the stool of the 230th healthy child enrolled during field investigations in the district of Tokombere.
Molecular characterization of PVs.
In order to determine whether PVs isolated from healthy children originated from the vaccine strain or the wild type, they were analyzed by two intratypic differentiation (ITD) methods according to the protocol recommended by the WHO. The molecular ITD method was carried out by conventional reverse transcription-PCR (RT-PCR) using PV group- and serotype-specific and Sabin strain-specific primers sets (16, 17). The second ITD method was performed by an enzyme-linked immunosorbent assay (ELISA) using intratype-specific cross-absorbed antisera (16, 18). Besides ITD tests, the full-length VP1 sequences of Sabin-like PVs were determined by RT-PCR and sequencing using the following type-specific primers, named by numbering according to the position in the corresponding PV Sabin strain and orientation (forward [F]/reverse [R]) (in the 5′-to-3′ direction): Sab1-2359-F (5′-AGT CGT CCC TCT TTC GAC A-3′) and Sab1-3619-R (5′-TGG GCC AAC GAA GGA T-3′) for Sabin 1, Sab2-2355-F (5′-TAG GGT TGT TGT CCC GTT G-3′) and Sab2-3462-R (5′-GTC TTC TTG TGT AGC TAG G-3′) for Sabin 2, and Sab3-2356-F (5′-TGT GGT GCC ACT GTC CAC C-3′) and Sab3-3462-R (5′-GTC CCA CAA ACG ACA CAG-3′) for Sabin 3 (M.-L. Joffret, unpublished data).
Virus isolates from patients with AFP.
A total of 146 NPEV isolates obtained from the stool specimens of 502 AFP patients during the year 2008 were available for the present study. The original stool specimens investigated by the CPC in 2008 originated from 222, 235, 35, 7, and 3 AFP patients from Cameroon, Chad, Gabon, Equatorial Guinea, and Sao Tomé and Principe, respectively (see Fig. S1 in the supplemental material). The processing of stool samples, virus isolation, and storage of virus isolates were carried out according to the instructions described in the Polio Laboratory Manual (World Health Organization) (16). In addition, all clarified stool suspensions were inoculated onto HEp-2c cell cultures. The isolates showing CPE only on RD or HEp-2c cells, but not on L20B cells, were considered to be NPEVs and were characterized by molecular techniques, as described below. Two stool specimens were processed for most patients. When two isolates originated from the two stool specimens of the same patient, only one isolate was analyzed. Throughout the text, the names of strains from AFP patients are given in the following format: a one-letter code standing for the country of origin of AFP patient (C for Cameroon, T for Chad, and G for Gabon), followed by the year of isolation (08 for 2008) and the serial number of the AFP case. For instance, strain C08-146 was isolated from the 146th case of AFP patients from Cameroon in 2008.
RNA extraction and gene amplification.
Viral RNAs were extracted and purified from 140 μl of infected cell culture supernatants using a QIAamp viral RNA minikit according to the manufacturer's instructions (Qiagen, France).
Initially, a 1,452-bp DNA fragment encompassing the 3′ one-third of the VP1 capsid region and the 2A, the 2B, and part of the 2C coding regions were amplified by using RT-PCR, as described previously (19). Alternatively, amplification techniques targeting the partial or complete VP1 capsid coding region using degenerate primers or strain-specific primers were also used, as previously described (20, 21).
Virus isolates that were refractory to all RT-PCR amplifications targeting the VP1 gene were tested with a RT-PCR technique amplifying a portion of the most conserved 5′-noncoding region of the HEV genome, using the UG52-UC53 primer pair, as described previously (22). Total DNA was then purified from the isolates that were negative by enterovirus-specific RT-PCR using a QIAamp DNA extraction kit (Qiagen, France). The purified DNA was subjected to a nested PCR for the detection of adenovirus, as previously reported (23).
Amplicons were analyzed by agarose gel electrophoresis. Depending on the presence of a single or multiple bands in the gel, amplicons were either directly purified with a QIAquick PCR purification kit or gel isolated and purified by means of the QIAquick gel extraction kit (Qiagen, France), according to the manufacturer's protocol.
Sequencing.
Amplicons were subjected to direct sequencing using the BigDye Terminator v3.1 kit (Applied Biosystems) and an ABI Prism 3140 automated sequencer (Applied Biosystems). The sequencing of the VP1-2C DNA fragment was performed in one direction by using the genomic primer EUG3abc (19). Other amplicons were sequenced in both directions by using PCR primers.
Molecular typing of NPEVs.
Nucleotide sequences corresponding to the partial (3′ one-third or 5′ one-half) or complete VP1 capsid gene of each NPEV isolate were pairwise compared with the homologous sequences of prototype strains retrieved from databases, as previously reported (19, 24). Scores were established for each strain according to nucleotide and amino acid identities with homotypic and heterotypic strains. Field isolates were considered to belong to the same serotype as the closest prototype strain according to the results of pairwise comparisons of nucleotide and amino acid sequences, as previously reported (19). In most cases, nucleotide identities with the corresponding prototype strains were equal to or higher than the type assignment nucleotide and amino acid thresholds (75 and 85%, respectively). In a few cases in which nucleotide identity scores related to the homotypic prototype strain fell between 71 and 75%, databases were screened for similar sequences by using the NCBI Basic Local Alignment Search Tool (BLAST) implemented directly from the CLC Main Workbench interface. The type assigned to the studied isolates was supported by the serotype associated with the most similar HEV sequences present in databases giving higher nucleotide and amino acid identities.
Apart from type assignment based on partial VP1 sequences, we performed type confirmation based on the complete VP1 sequence of all HEV-C lineages by using both pairwise nucleotide and amino acid comparisons with all available HEV-C prototype strains. Alternatively, the automated Enterovirus Genotyping Tool version 0.1 (25) was used for the typing of strains representing all the genetic lineages identified among the studied HEV-C strains.
Sequence analyses.
Multiple-sequence alignments were performed with CLC Main Workbench 5.7.2 software (CLC bio, Aarhus, Denmark). From the resulting nucleotide sequence alignments, phylograms were inferred by both the distance and maximum likelihood (ML) methods.
Distance-based phylogenetic trees were reconstructed by the neighbor-joining (NJ) method using MEGA, version 5.05 (26), with the Jukes-Cantor algorithm for genetic distance determination and pairwise deletion for gaps. The reliability of tree topology was estimated by using 1,000 bootstrap replicates.
The ML algorithm was implemented in PhyML 3.0 software (27) under the HKY85 model of substitutions (28) with a transition/transversion ratio of 8.0. The reliability of the maximum likelihood phylogenies was assessed by 1,000 bootstrap resamplings. Trees were drawn with the NJ Plot program (29).
Nucleotide sequence accession numbers.
Sequences were submitted to the GenBank database under accession numbers JX307648 to JX307652 for the full-length VP1 sequence of HEV-A, JX417821 to JX417887 for the full-length VP1 sequence of HEV-C, JX431302 for the full-length VP1 sequence of the unique HEV-D, JX417717 to JX417820 and JX437641 to JX437661 for partial VP1 sequences of HEV-B, and JX426619 to JX426694 for partial VP1 sequences of HEV-C.
RESULTS
Virus isolation and differentiation of polioviruses.
Stool specimens collected from healthy children in the northern region of Cameroon were systematically tested for HEV isolation by using the human RD and HEp2-c cell lines. Among 436 stool specimens, 186 induced CPE in cell cultures. Among the 186 harvested isolates, only 17 could be propagated on murine L20B cells (expressing the human PV receptor) and were identified as PVs. Overall, only 3.9% (17/436) of children were infected by PV (Table 1). All 19 PV strains identified from the 17 samples (2 samples containing a mixture of PV type 1 and type 2 isolates) were shown to be closely related to the original Sabin strains (Sabin-like strains) by the ITD RT-PCR and ELISA methods. Sequence analyses also showed that all PV strains had accumulated ≤4 mutations in the full-length VP1 genomic region (data not shown). These results were consistent with the fact that these PV strains originated from recently immunized children.
Table 1.
Virus isolation from stool specimens of healthy children with respect to the round of stool collection in the far north region of Cameroon
| Isolatea | 2008 (n = 233) |
2009 (n = 203) |
Total (n = 436) |
|||
|---|---|---|---|---|---|---|
| No. of isolates | Isolation rate (%) | No. of isolates | Isolation rate (%) | No. of isolates | Isolation rate (%) | |
| Enteroviruses | ||||||
| Polioviruses | 10 | 4.3 | 7 | 3.4 | 17 | 3.9 |
| Typed NPEVs | 78 | 71 | 149 | |||
| Mixed strains of NPEVs | 8 | 4 | 12 | |||
| All NPEVs | 86 | 36.9 | 75 | 36.9 | 161 | 36.9 |
| All enteroviruses | 96 | 41.2 | 82 | 40.4 | 178 | 40.8 |
| Adenoviruses | 1 | 0.4 | 7 | 3.4 | 8 | 1.8 |
Among the remaining 169 nonpolio isolates, only 8 were not molecularly identified as HEVs (generic RT-PCR test) and were shown to be adenoviruses based on PCR evidence. This means that 161 NPEV isolates were obtained, with an overall isolation rate of 36.9% (161/436) (Table 1).
Molecular typing of NPEV isolates.
Comparison of partial or complete VP1 sequences of EV field strains with those of prototype strains has became an effective method for molecular typing of field strains (19, 20, 24, 30). In this study, NPEVs were initially typed by using sequences of the 3′ one-third and/or the 5′ one-half of the VP1 gene. The partial or full-length VP1 sequences of the field isolates were compared with the homologous sequences of prototype strains.
Among the 307 isolated NPEVs, including 161 from healthy children and 146 from AFP patients, 273 were successfully sequenced. The remaining 34 isolates (12 from healthy children and 22 from AFP patients) could not be typed due to unexploitable sequencing results associated with the presence of mixed isolates in the studied samples and were not further analyzed.
It was recently shown that the 3′ one-third (∼300 nucleotides [nt]) of the VP1 gene is in some cases insufficient for the accurate typing of certain HEV-C types (31). In order to refine the molecular typing of HEV-C strains, full-length VP1 sequences were analyzed for 67 HEV-C isolates. These HEV-C isolates included all representatives of the circulating genetic lineages identified through phylogenetic analyses of the 3′ one-third of VP1 sequences (data not shown). A comprehensive summary of nucleotide and deduced amino acid sequence similarity scores between the full-length VP1 sequences of the 67 studied HEV-C strains and both homotypic and heterotypic prototype strains is shown in Table S1 in the supplemental material. A few complete VP1 sequences did not reach the cutoff values of 75.0% nucleotide and 88.0% amino acid identities, which are considered to be the cutoff values allowing an unambiguous typing of HEV-C field isolates (see Table S1 in the supplemental material). However, some of them met the cutoff value at either the nucleotide or amino acid level. Among them, strain DJI-346 showed a similar nucleotide sequence identity (73%) to the heterotypic prototype strain CVA-24 Joseph and the prototype EV-C99 strain BAN00-10461. However, it displayed a significantly higher nucleotide identity (79.5%) to the EV-C99 variant USA/GA84, thus supporting its typing as EV-C99. Accordingly, strain DJI-346 displayed 88.5% and 84.3% amino acid similarities with the EV-C99 and CVA-24 prototype strains, respectively.
For all but two sequenced isolates, a search of the databases found homotypic strains featuring a nucleotide or amino acid similarity above the cutoff values of 75.0% and 88.0%, respectively. Indeed, field isolates T08-083 and T08-234 displayed low sequence similarity scores of 70% (nucleotide) and 84% (amino acid) compared to the closest prototype strain, CVA-21 Kuykendall (see Table S1 in the supplemental material). However, these strains were unambiguously typed as EV-C95, whose prototype sequence is no yet available (Picornavirus Study Group, ICTV) (J. Ayukekbong, J.-C. Kabayiza, M. Lindh, T. Nkuo-Akenji, F. Tah, T. Bergström, and H. Norder, unpublished data). Actually, the full-length VP1 sequence of strains T08-083 and T08-234 displayed similarity scores of 88.4 and 88.9% (nucleotide) and 98.0 and 98.3% (amino acid), respectively, with the prototype EV-C95 strain (Picornavirus Study Group, ICTV).
In addition, the type of all but three HEV-C strains was confirmed by using a Web-based molecular genotyping tool for enteroviruses (see Table S1 in the supplemental material) (25).
Diversity of HEV types in healthy children and patients with AFP.
Among the NPEVs isolated from healthy children, 28 different HEV types were identified. A summary of strain distribution into HEV types, with respect to the round of stool collection, is shown in Table 2. No isolate belonging to HEV-A was found, while the unique strain of the HEV-D species was EV-D111, a recently reported type. Overall, 36.2% (54/149) of isolates belonged to HEV-B. These isolates were split into 21 different types with a quite heterogeneous distribution between the two rounds of stool collection in 2008 and 2009. Interestingly, a high rate of 63.1% (94/149) HEV-C isolates was found among all typed isolates (see Fig. S2 in the supplemental material). Furthermore, CVA-13 was strikingly the most frequent type, accounting for 26.8% (40/149) of all typed isolates from healthy children (Table 2). The CVA-13 type was followed, in proportion, by other HEV-C types, including EV-C99 and CVA-20, -17, -24, and -11.
Table 2.
Distribution of NPEV types and species among healthy children with respect to the round of stool collection in the far north region of Cameroona
| Species and type | 2008 (n = 233) |
2009 (n = 203) |
Total (n = 436) |
|||
|---|---|---|---|---|---|---|
| No. of strains | % of strains among all typed isolates | No. of strains | % of strains among all typed isolates | No. of strains | % of strains among all typed isolates | |
| HEV-B | ||||||
| CVB-1 | 3 | 4.2 | 3 | 2.0 | ||
| CVB-5 | 2 | 2.6 | 2 | 1.3 | ||
| E-2 | 1 | 1.3 | 1 | 0.7 | ||
| E-3 | 1 | 1.3 | 1 | 0.7 | ||
| E-6 | 1 | 1.3 | 3 | 4.2 | 4 | 2.7 |
| E-7 | 4 | 5.1 | 4 | 2.7 | ||
| E-11 | 3 | 3.8 | 3 | 2.0 | ||
| E-12 | 1 | 1.3 | 1 | 1.4 | 2 | 1.3 |
| E-13 | 3 | 4.2 | 3 | 2.0 | ||
| E-14 | 1 | 1.4 | 1 | 0.7 | ||
| E-17 | 2 | 2.8 | 2 | 1.3 | ||
| E-19 | 3 | 3.8 | 5 | 7.0 | 8 | 5.4 |
| E-25 | 1 | 1.3 | 1 | 1.4 | 2 | 1.3 |
| E-26 | 2 | 2.6 | 2 | 1.3 | ||
| E-29 | 4 | 5.1 | 1 | 1.4 | 5 | 3.4 |
| E-30 | 1 | 1.3 | 1 | 1.4 | 2 | 1.3 |
| E-31 | 1 | 1.3 | 1 | 0.7 | ||
| E-32 | 1 | 1.3 | 1 | 0.7 | ||
| E-33 | 1 | 1.3 | 4 | 5.6 | 5 | 3.4 |
| EV-B75 | 1 | 1.4 | 1 | 0.7 | ||
| EV-B87 | 1 | 1.4 | 1 | 0.7 | ||
| All HEV-B isolates | 27 | 34.6 | 27 | 38.0 | 54 | 36.2 |
| HEV-C | ||||||
| CVA-11 | 1 | 1.3 | 1 | 0.7 | ||
| CVA-13 | 21 | 26.9 | 19 | 26.8 | 40 | 26.8 |
| CVA-17 | 5 | 6.4 | 6 | 8.5 | 11 | 7.4 |
| CVA-20 | 10 | 12.8 | 6 | 8.5 | 16 | 10.7 |
| CVA-24 | 6 | 7.7 | 3 | 4.2 | 9 | 6.0 |
| EV-C99 | 7 | 9.0 | 10 | 14.1 | 17 | 11.4 |
| All HEV-C isolates | 50 | 64.1 | 44 | 62.0 | 94 | 63.1 |
| HEV-D | ||||||
| EV-D111 | 1 | 1.3 | 1 | 0.7 | ||
| All HEV-D isolates | 1 | 1.3 | 1 | 0.7 | ||
| All typed isolates | 78 | 100.0 | 71 | 100.0 | 149 | 100.0 |
n, number of stool samples collected.
Concerning isolates derived from AFP patients, 124 NPEVs belonging to 39 different types of the species HEV-A, -B, and -C were identified. The largest proportion of isolates, 57.3% (71/124), belonged to HEV-B species (29 different types). The distribution of NPEVs from AFP patients, with respect to viral species, types, and countries of origin, is summarized in Table 3. As for healthy children, we found a high rate of 39.5% (49/124) HEV-C strains in AFP patients (Table 3). Indeed, similar high rates of HEV-C were found in patients from Cameroon, Chad, and Gabon: 41.5% (27/61), 35.8% (19/53), and 50% (3/6), respectively. CVA-13 isolates were also the most prevalent among all typed isolates from AFP patients (16.9%), followed in proportion by CVA-20, -17, or -24, EV-C95, and CVA-21 or -11. Surprisingly, no EV-C99 strains were isolated from AFP patients, while they accounted for 11.4% of all NPEVs identified in healthy children.
Table 3.
Distribution of NPEV types and species among acute flaccid paralysis patients with respect to country of origina
| Species and serotype | Cameroon (n = 71) |
Chad (n = 67) |
Gabon (n = 7) |
Total |
||||
|---|---|---|---|---|---|---|---|---|
| No. of strains | % of strains among all typed isolates | No. of strains | % of strains among all typed isolates | No. of strains | % of strains among all typed isolates | No. of strains | % of strains among all typed isolates | |
| HEV-A | ||||||||
| CVA-10 | 1 | 1.9 | 1 | 0.8 | ||||
| EV-A71 | 2 | 3.1 | 2 | 1.6 | ||||
| EV-A76 | 1 | 1.5 | 1 | 0.8 | ||||
| All HEV-A isolates | 3 | 4.6 | 1 | 1.9 | 0 | 0.0 | 4 | 3.2 |
| HEV-B | ||||||||
| CVB-1 | 4 | 7.5 | 4 | 3.2 | ||||
| CVB-2 | 1 | 1.5 | 1 | 0.8 | ||||
| CVB-4 | 1 | 1.5 | 1 | 0.8 | ||||
| CVB-5 | 2 | 3.1 | 1 | 1.9 | 3 | 2.4 | ||
| CVB-6 | 1 | 1.9 | 1 | 0.8 | ||||
| E-1 | 1 | 1.5 | 2 | 3.8 | 3 | 2.4 | ||
| E-2 | 1 | 1.5 | 1 | 1.9 | 2 | 1.6 | ||
| E-3 | 1 | 1.5 | 2 | 3.8 | 3 | 2.4 | ||
| E-6 | 7 | 10.8 | 1 | 1.9 | 8 | 6.5 | ||
| E-7 | 3 | 4.6 | 2 | 3.8 | 5 | 4.0 | ||
| E-12 | 3 | 4.6 | 1 | 1.9 | 4 | 3.2 | ||
| E-13 | 2 | 3.1 | 4 | 7.5 | 6 | 4.8 | ||
| E-14 | 1 | 1.5 | 1 | 1.9 | 2 | 1.6 | ||
| E-17 | 2 | 3.1 | 2 | 1.6 | ||||
| E-19 | 1 | 1.5 | 1 | 1.9 | 2 | 1.6 | ||
| E-20 | 1 | 1.9 | 1 | 0.8 | ||||
| E-21 | 1 | 1.5 | 1 | 0.8 | ||||
| E-24 | 1 | 1.5 | 1 | 1.9 | 2 | 1.6 | ||
| E-26 | 1 | 1.9 | 1 | 0.8 | ||||
| E-27 | 1 | 1.9 | 1 | 0.8 | ||||
| E-30 | 2 | 3.1 | 2 | 1.9 | 1 | 16.7 | 5 | 4.0 |
| E-31 | 1 | 1.9 | 1 | 0.8 | ||||
| E-33 | 1 | 1.5 | 2 | 3.8 | 1 | 16.7 | 4 | 3.2 |
| EV-B69 | 1 | 1.5 | 2 | 3.8 | 3 | 2.4 | ||
| EV-B75 | 1 | 1.5 | 1 | 0.8 | ||||
| EV-B80 | 1 | 1.9 | 1 | 0.8 | ||||
| EV-B81 | 1 | 16.7 | 1 | 0.8 | ||||
| EV-B85 | 1 | 1.5 | 1 | 0.8 | ||||
| EV-B97 | 1 | 1.5 | 1 | 0.8 | ||||
| All HEV-B isolates | 35 | 53.8 | 33 | 62.3 | 3 | 50.0 | 71 | 57.3 |
| HEV-C | ||||||||
| CVA-11 | 1 | 16.7 | 1 | 0.8 | ||||
| CVA-13 | 14 | 21.5 | 7 | 13.2 | 21 | 16.9 | ||
| CVA-17 | 3 | 4.6 | 2 | 3.8 | 1 | 16.7 | 6 | 4.8 |
| CVA-20 | 6 | 9.2 | 6 | 11.3 | 12 | 9.7 | ||
| CVA-21 | 1 | 1.9 | 1 | 0.8 | ||||
| CVA-24 | 4 | 6.2 | 1 | 1.9 | 1 | 16.7 | 6 | 4.8 |
| EV-C95 | 2 | 3.8 | 2 | 1.6 | ||||
| All HEV-C isolates | 27 | 41.5 | 19 | 35.8 | 3 | 50.0 | 49 | 39.5 |
| All serotypes | 65 | 100.0 | 53 | 100.0 | 6 | 100.0 | 124 | 100.0 |
n, number of isolates analyzed.
Genetic diversity of HEV-A and -D strains.
We analyzed the phylogenetic relatedness of the HEV isolates circulating in Cameroon, Chad, and Gabon with those reported in other contexts. Phylogenetic relationships were inferred by using both ML and NJ algorithms. Since tree topologies from both methods displayed very similar patterns, only NJ trees are presented.
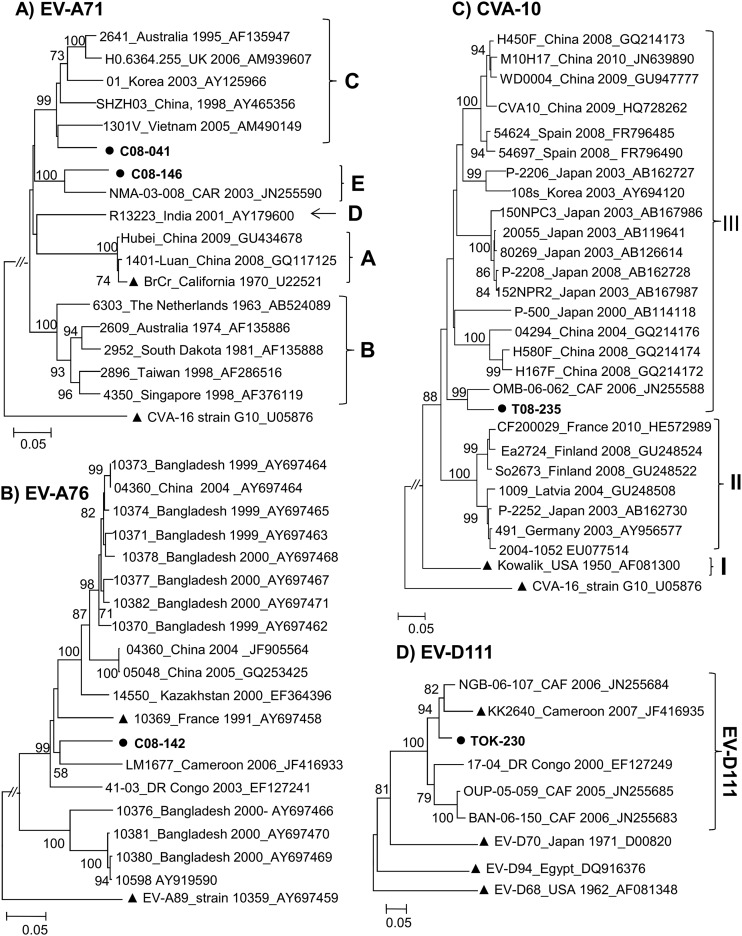
Among the 273 strains identified in this study, only 4 HEV-A strains, 2 EV-A71 strains, and an EV-A76 strain from Cameroon and a CVA-10 strain from Chad were isolated. Phylogenetic analysis showed that the VP1 sequences of the two EV-A71 strains fell into distinct subclusters (Fig. 1A). Strain C08-041, from Douala, was a member of genogroup C, while the second EV-A71 strain, C08-146, from Yaoundé grouped consistently with the recently reported Central African strain NMA-03-008. The latter strain, showing a high sequence similarity score of 89.6% and 97% at the nucleotide and amino acid levels, respectively, with strain C08-146, was previously shown to belong to a new EV-A71 genogroup tentatively called “genogroup E” (32). The identification of strains of this genogroup in the Central African Republic, Nigeria (32), and Cameroon indicates that it was circulating without known disease outbreaks in Africa.
Fig 1.
Phylogenetic relationships of the VP1 sequences of the studied HEV-A and HEV-D strains. (A) Phylogram based on partial VP1 sequences of EV-A71 strains (nt 1 to 855 according to EV-A71 prototype strain BrCr VP1 numbering). EV-A71 genogroups A to E are indicated by the corresponding letters. (B) Phylogram based on partial VP1 sequences of EV-A76 strains (nt 13 to 782 according to EV-A76 prototype strain FRA91-10369 VP1 numbering). (C) Phylogram based on partial VP1 sequences of CVA-10 strains (nt 478 to 862 according to CVA-10 prototype strain Kowalik VP1 numbering). (D) Phylogram based on partial VP1 sequences of EV-D111 strains (nt 117 to 471 according to EV-D111 prototype strain KK2640 VP1 numbering). The newly sequenced strains are in boldface type and highlighted by circles. For the reference sequences, the location and year of isolation and GenBank accession number are indicated in trees, if known. Triangles indicate prototype strains. For clarity, most bootstrap values of less than 70% have been omitted. The scale bars indicate nucleotide distance as substitutions per site.
The unique EV-A76 strain C08-142 isolated in this study was the third EV-A76 strain reported so far in sub-Saharan Africa (33). The three African strains fell into a subcluster with different strains (bootstrap value of 99%), including prototype strain 10369 from France (Fig. 1B). Interestingly, strain C08-142, which was isolated from a patient with AFP in Cameroon, was particularly related to EV-A76 strain LM1677, detected in a wild chimpanzee in this country (33).
Chadian strain T08-235 was a member of CVA-10 lineage III (Fig. 1C). Lineage III includes CVA-10 strains that had been circulating in China, Japan, South Korea, and Spain from 2000 to 2010. Within this lineage, the Chadian strain showed high sequence similarity scores (93.0% and 96.6% at the nucleotide and amino acid levels, respectively) and grouped consistently with CVA-10 strain OMB-06-062, a recently reported strain from the Central African Republic (32).
The single HEV-D strain, TOK-230, belonged to the EV-D111 type, whose prototype strain was recently isolated from a Cameroonian wild chimpanzee (33). Overall, five isolates of EV-D111 have been reported so far, and all of them originated from sub-Saharan Africa (32–34). They segregated into two reliable subclusters (Fig. 1D). Interestingly, the EV-D111 strain, isolated from the stool specimen of a healthy child in northern Cameroon, featured a close phylogenetic relatedness to strain KK2640, isolated from a chimpanzee in the same country.
Genetic diversity of HEV-B strains.
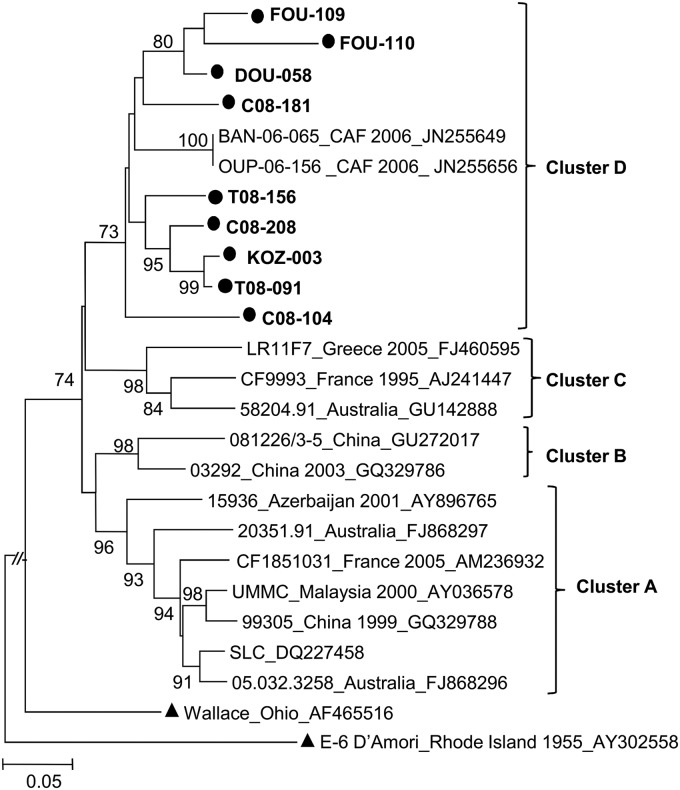
As many as 34 out of the 60 currently known HEV-B types were found among all isolates analyzed in this study, thus revealing the huge diversity of HEV-B isolates in Cameroon and neighboring countries. Individual HEV-B types seemed to be circulating in a sporadic pattern. Accordingly, a number of types detected in healthy children in 2008 were not found in 2009. All echovirus 7 (E-7) VP1 sequences formed a sub-Saharan cluster along with E-7 strains recently reported in the Central African Republic (32) (Fig. 2). Several recently described new HEV-B types, EV-B75, EV-B80, EV-B81, EV-B85, EV-B87, and EV-B97, were also found. These types were first isolated in Bangladesh and the United States (35, 36).
Fig 2.
Phylogenetic relationships of the VP1 sequences of the studied E-7 isolates. The phylogram is based on an alignment of the partial VP1 sequences (nt 571 to 876 according to the E-7 prototype strain Wallace VP1 numbering). The newly sequenced strains are in boldface type and highlighted by circles. The location and year of isolation and GenBank accession number of the reference strains are indicated in the tree, if known (CAF, Central African Republic). Prototype strains are highlighted by triangles. Bootstrap values are indicated if higher than 70%.
Genetic diversity of HEV-C strains.
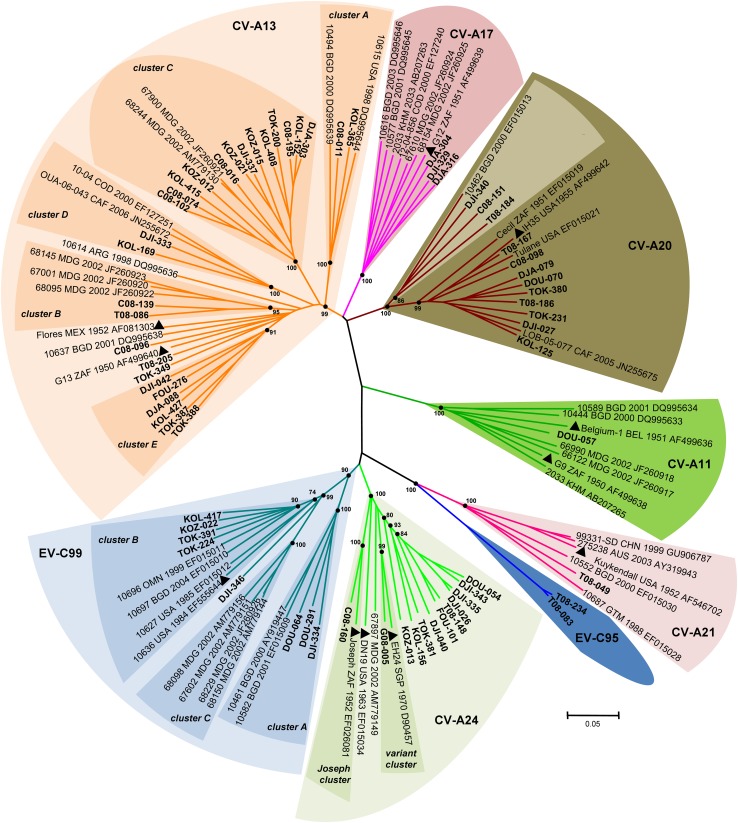
Representatives of all HEV-C genetic lineages were analyzed by using complete VP1 sequences (Fig. 3). As expected, individual nucleotide VP1 sequences of the study strains segregated into type-specific clusters (with strong bootstrap support). Within their type-related clusters, individual strains belonging to types CVA-13, -20, and -24 and EV-C99 featured a wide range of genetic variability, expressed through segregation into consistent subclusters.
Fig 3.
Phylogenetic relationships based on the full-length VP1 sequences of newly sequenced HEV-C isolates from Cameroon, Chad, and Gabon. Studied isolates are indicated in boldface type. The year and country of isolation and GenBank accession number of each reference isolate are indicated, if known (ARG, Argentina; AUS, Australia; BEL, Belgium; BGD, Bangladesh; CAF, Central African Republic; CHN, China; COD, Democratic Republic of the Congo; GTM, Guatemala; KHM, Cambodia; MEX, Mexico; MDG, Madagascar; OMN, Oman; SGP, Singapore; USA, United States; ZAF, South Africa). Prototype strains are highlighted by triangles. For clarity, most bootstrap values of less than 70% have been omitted. The scale is shown at the bottom as substitutions per site.
Interestingly, the highest intratypic genetic variability was observed among CVA-13 strains, which constituted the most prevalent NPEV type. Indeed, CVA-13 sequences fell into all previously reported subclusters A to D (32, 37) and an apparently new cluster, tentatively called “cluster E” (Fig. 3). Besides these five major subclusters, isolates TOK-349 from northern Cameroon and T08-205 and C08-096 from Chad may form additional subclusters.
Within the CVA-20-related group, three strains formed a bootstrap-supported subcluster with strain 10462 from Bangladesh. The remaining CVA-20 strain fell in another subcluster along with CVA-20 prototype strain IH-35 and CVA20-Tulane (Fig. 3).
Concerning CVA-24 isolates, one grouped with CVA-24 prototype strain Joseph, while another one clustered with the CVA-24 variant EH24 (Fig. 3). The remaining strains did not group with any of the prototype strains, constituting a third subcluster (bootstrap value of 90%) of related strains with sequence identities from 80.6 to 96.7% and 91.5 to 99.3% at the nucleotide and amino acid levels, respectively.
Isolates belonging to the CVA-11, -17, and -21 types formed relatively compact clusters in terms of sequence diversity. However, CVA-17 strains isolated in this study formed a distinct subgroup (bootstrap value of 71%) that was quite distinct from that formed by the other CVA-17 strains, including Madagascan ones (37).
As expected, Chadian strains T08-083 and T08-234, identified by pairwise nucleotide sequence comparison as EV-C95 strains, grouped separately from all of the other type-defined clusters.
Within-type segregation was also found for the eight EV-C99 strains isolated in this study. Most of them fell into two out of the three previously defined EV-C99 clusters (37): three strains grouped into subcluster A along with prototype strain 10461 from Bangladesh, while four strains formed a well-defined group within subcluster B along with two strains isolated from Bangladesh and Oman (31). The eighth isolate, DJI-346, branched separately from all previously reported EV-C99 clusters, defining a putative new genetic lineage within this HEV type.
Interestingly, the VP1 sequences of the EV-C99 isolates of cluster A shared particular features compared to those of EV-C99 strains of other clusters. The nucleotide sequences of the 5′ one-third of the VP1 region of cluster A were more closely related to those of CVA-24 strains than to those of other EV-C99 strains (see Fig. S3A in the supplemental material). In addition, the VP1 peptide sequences of cluster A, including that of strain DJI-346, fell into a consistent cluster apart from that formed by the other EV-C99 isolates (see Fig. S3B in the supplemental material). These results indicated that EV-C99 isolates of cluster A were distantly related to homotypic strains belonging to other clusters and suggested that they possibly originated from intertypic recombination events.
DISCUSSION
This study aimed to investigate the occurrence and genetic diversity of HEV in healthy children in the far north region of Cameroon as well as in AFP patients from several Central African countries. We found a low rate of PV strains among healthy children in the far north region of Cameroon, and all of them were OPV strains. Only 3.9% of the stool specimens were positive for PV, while a rate of at least 10% was expected from other studies of healthy children in other developing countries where OPV is routinely used (6, 38, 39). This low rate of PV circulation can be explained by the high poliovaccine coverage rate of about 90% among the healthy children enrolled in the study. The coverage rate was above the national average of about 80% in 2008 and 2009 (40). Accordingly, almost all PVs were isolated from recently immunized children, and all appeared to be Sabin-like, with ≤4 nt substitutions in their full-length VP1 sequences.
In contrast to PVs, we found an extensive circulation of NPEV among healthy children, with an overall isolation rate of 36.9%. The high rate of EV infection among healthy children enrolled in this study can be explained by the fact that the children were between the ages of 0 and 5 years. During this early period of life, such young children are still building up their immunity against each of the numerous circulating HEV types and are thus highly susceptible to NPEV infections. Accordingly, the isolation rate seemed to be higher in children aged 1 to 3 years than in children under 1 and over 3 years of age (data not shown). Before the age 1 year, children are still breastfed and may be minimally exposed to infections, while from 3 years of age, they have possibly mounted efficient immunity against most circulating NPEV lineages. A high rate of EV infection among 54 healthy children between the ages of 5 and 15 years was also reported in a recent study conducted in the southwestern region of Cameroon. In that previous study, in which HEVs were not typed (generic RT-PCR test), HEV infection was found in 31.5% of healthy children, while a lower rate of 13.9% was reported among 93 HIV-infected adults (41).
High rate of HEV-C.
Overall, 45 different NPEV types were identified in this study, including 9 types recently reported and considered to be new types, such as EV-B75, -A76, -B80, -B81, -B85, -B87, -B97, and -D111 (33, 35, 36, 42). In addition, a strikingly high rate of HEV-C isolates was uncovered for both healthy children from northern Cameroon and AFP patients from Cameroon, Chad, and Gabon. In particular, a rate of 63.1% HEV-C was found among NPEV-positive healthy children. There was not a striking change in the rate and the genetic diversity of circulating HEV-C types between 2008 and 2009 (Table 2), highly suggesting that the high rate of HEV-C infection among the children enrolled was not due to a monomorphic and short-lived outbreak. As in healthy children, the rates of HEV-C infection were also high in AFP patients from all three countries (Table 3). Overall, the rate of HEV-C infections in Cameroon and neighboring countries was huge compared to rates reported by most studies of EV-infected patients in temperate countries, including North Africa (43–45). In our study, HEp-2c cells were systematically used for virus isolation, in addition to the primary RD and L20B cells recommended by the WHO for poliomyelitis surveillance (16). HEp-2c cells are known to be particularly suitable for the efficient isolation of coxsackie A viruses belonging to the HEV-C species (15, 46, 47). Accordingly, 88.1% (126/143) of HEV-C isolates were propagated exclusively on HEp-2c cell cultures, while this cell line was conducive to only 48.8% (61/125) of HEV-B isolates. A low rate of HEV-C isolation was found in a recent study of HEVs infecting healthy children originating from the northeastern Nigeria (48), despite the fact that tropical conditions and socioeconomic factors are similar to those of the northern Cameroon. The absence of the HEp-2c cell line in the isolation technique used in that study possibly led to an underestimation of the isolation rate of NPEV and HEV-C in particular (48). Indeed, high rates of HEV-C isolation were repeatedly reported when isolation techniques made use of both the HEp-2c and RD cell lines in some tropical countries, including Cambodia (3), the Central African Republic (32), Madagascar (15), and China (49). In contrast to tropical countries, several previous epidemiological studies using the HEp2-c cell line in industrialized countries recurrently reported HEV-C isolation rates of less than 5% (43, 45, 50–52). Overall, it appears that the high rate of HEV-C is linked to tropical conditions, where the use of the HEp2-c cell line for the NPEV surveillance would be of great interest.
Some contrasting results were observed between healthy children and AFP patients in terms of HEV species, ratios, and types. The rate of HEV-C seemed to be relatively higher in healthy children than in AFP patients (see Fig. S2 in the supplemental material). Moreover, no EV-C99 isolate was identified in AFP patients despite high rates of isolation and wide diversity in healthy children. Conversely, no HEV-A strain was found in healthy children. These contrasting type distribution patterns of NPEVs could constitute differential factors between healthy children and AFP patients. However, these two populations are not strictly comparable. AFP isolates were obtained from specimens collected year-round from patients from different geographic and climate origins (from dry tropical to equatorial climates). This could have an influence on the circulation of HEV types and species. Furthermore, in contrast to the stool specimens from healthy children, which were collected and transported under uniform conditions, samples from AFP patients usually face different storage and transport conditions (53). Possibly, some HEV types in feces may be more sensitive than others, depending on physical conditions (53). Therefore, additional studies are required before drawing a definitive conclusion about the differences of HEV species and types found in healthy children and AFP patients.
Genetic diversity and phylogenetic relationships among HEVs.
As expected from previous studies in sub-Saharan Africa (15, 32, 34, 39), isolation of HEV-A strains was very uncommon. Only 4 HEV-A strains, including 2 EV-A71 strains, were identified in AFP patients. One strain of EV-A71 belonged to genogroup C, which has been associated with disease outbreaks worldwide, including Africa (54–56). Two novel genogroups were recently identified in Africa (32) and India (57), respectively. Our study identified an isolate belonging to the novel Central African EV-A71 genogroup (32). The identification of this new EV-A71 genogroup in three distinct countries, at different times, indicates that it has been circulating in sub-Saharan Africa for several years. Since NPEVs are poorly sampled in sub-Saharan Africa, the extent of the circulation of this novel EV-A71 genogroup in African populations is probably underestimated. The major EV-A71 genogroups A, B, and C have been involved in several epidemics of hand-foot-and-mouth disease, severe neurological disease, and other health concerns primarily in Asia but also in Australia, Europe, and the United States (55, 58, 59). It has been demonstrated that diverse lineages of genogroup B and C have each silently circulated in the human population for several years before causing large outbreaks (56). Thus, the virtually quiescent circulation of a new EV-A71 genogroup in Africa and its high potential to cause severe disease outbreaks emphasize the need for additional surveillance and detailed characterization.
Most of the regions where EV-A76 has been reported, including Bangladesh, Cameroon, the Democratic Republic of Congo, and Southeast Asia (34, 42, 49, 60), are home to a variety of nonhuman primate (NHP) species. We identified an EV-A76 isolate that was related to human- and chimpanzee-derived EV-A76 strains (Fig. 1B). As for EV-A76, the unique HEV-D strain EV-D111 was recently shown to infect both humans and chimpanzees in Central Africa (32–34). Together with previous reports, this study suggests that cross-species transmission of EVs may play a role in the diversity and evolution of HEVs in certain regions, including sub-Saharan Africa (60, 61). Contacts between humans and nonhuman primates have significantly increased in Central Africa during the last decades (62), and this could promote the cross-species transmission of new viral pathogens to humans.
For the most prevalent type, CVA-13, a tremendous genetic diversity was found. We identified at least five different clusters, including all previously reported clusters A to D (31, 32, 37) along with a new sub-Saharan “cluster E.” A similar clustering pattern was observed for other prevalent types, including CVA-20, CVA-24, and EV-C99. Their high rates coupled with their high genetic diversity indicate that they have been circulating with high endemicity in Cameroon and neighboring countries for many years. Interestingly, the genetic diversity reported here covered virtually all genetic lineages reported elsewhere, including variants that were previously considered Malagasy topotypes (37).
High risk of cVDPV emergence.
Recombination events are known to play a key role in the high plasticity of EV genomes and have been shown to be ubiquitous in the nonstructural regions of the viral genome (63, 64). Most cVDPVs have been shown to be recombinants emerging through recombination between PVs and other non-PV HEV-C strains (12). This includes the cVDPVs that were recently isolated in Chad and Cameroon (S. A. Sadeuh-Mba and F. Delpeyroux, unpublished results) and most of those circulating in the neighboring Nigeria and Niger (C. Burns, personal communication). As discussed above, there was a high rate and a tremendous genetic diversity of HEV-C strains circulating in Cameroon and neighboring countries. Given that OPV is massively used in this region, the frequency of PV/non-PV HEV-C coinfections could be particularly high. This provides an ideal setting for recombination between cocirculating PVs and non-PV HEV-C strains. The data from cVDPV outbreak studies in Cambodia and Madagascar have shown that CVA-13 and -17 are efficient recombination partners for PVs (3, 5, 13). CVA-13 strains have been shown to be the non-PV HEV-C strains most closely related to PVs in the capsid coding region, followed by CVA-20 and -17 (65). The particular viral ecosystem, characterized by high rate and high variability of CVA-13, -17, and -20 in Cameroon and neighboring countries, offers ideal viral conditions for the emergence of pathogenic cVDPVs. cVDPVs have been continuously reported in the Nigerian northern states, where they have been causing poliomyelitis cases for the past 7 years and occasionally spread to Chad and Niger (66). To avoid the emergence and circulation of recombinant cVDPVs in other countries of the Central African subregion, especially where indigenous wild-type polioviruses have been eradicated, the achievement and maintenance of high poliovaccine coverage remain essential.
In conclusion, this study showed an extensive circulation of HEVs among healthy children and AFP patients in Cameroon and neighboring countries. Most of the circulating NPEV types and variants were previously reported in many other parts of the world and may be distributed worldwide. Moreover, a number of HEV strains belonging to genetic lineages that seemed to be specific to Africa were identified. These Africa-specific strains included the new EV-A71 genogroup and EV-D111 as well as new lineages of HEV-C types. This study demonstrated that the genetic variability among some prevalent HEV-C types, including CVA-13, -20, and -24 and EV-C99, was higher than previously known. The particular enteroviral ecosystem characterized by a high rate of HEV-C isolation coupled with a wide genetic diversity constitutes a major viral factor rendering Cameroon and neighboring countries at high risk of emergence and circulation of pathogenic recombinant cVDPVs.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Cressence Essamba and Kamga Daniel for virus isolation; Coralie Tran, Jean-Michel Thiberge, Laure Diancourt, and Valérie Caro (Plateforme de Génotypage des Pathogènes et Santé Publique, Institut Pasteur, Paris, France) for virus sequencing; Florence Colbère-Garapin for advice; Nick J. Knowles for information about new HEV types; and Cara Burns for personal communication. We also thank the local health districts in northern Cameroon for their collaboration and the parents of the children enrolled in this study for their participation.
We are grateful for the financial support of the Institut Pasteur (PTR-276), the Société de Pathologie Exotique, the Agence Nationale pour la Recherche (ANR 09 MIEN 019), the Fondation pour la Recherche Médicale (FRM DMI20091117313), the French Ministry of Foreign and European Affairs, and the World Health Organization (HQPOL1206310).
Footnotes
Published ahead of print 19 December 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02119-12.
REFERENCES
- 1. Hyypia T, Hovi T, Knowles NJ, Stanway G. 1997. Classification of enteroviruses based on molecular and biological properties. J. Gen. Virol. 78:1–11 [DOI] [PubMed] [Google Scholar]
- 2. Pallansch MA, Roos R. 2007. Enteroviruses: polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p 839–894 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3. Arita M, Zhu SL, Yoshida H, Yoneyama T, Miyamura T, Shimizu H. 2005. A Sabin 3-derived poliovirus recombinant contained a sequence homologous with indigenous human enterovirus species C in the viral polymerase coding region. J. Virol. 79:12650–12657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kew O, Morris-Glasgow V, Landaverde M, Burns C, Shaw J, Garib Z, Andre J, Blackman E, Freeman CJ, Jorba J, Sutter R, Tambini G, Venczel L, Pedreira C, Laender F, Shimizu H, Yoneyama T, Miyamura T, van Der Avoort H, Oberste MS, Kilpatrick D, Cochi S, Pallansch M, de Quadros C. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356–359 [DOI] [PubMed] [Google Scholar]
- 5. Rakoto-Andrianarivelo M, Guillot S, Iber J, Balanant J, Blondel B, Riquet F, Martin J, Kew O, Randriamanalina B, Razafinimpiasa L, Rousset D, Delpeyroux F. 2007. Co-circulation and evolution of polioviruses and species C enteroviruses in a district of Madagascar. PLoS Pathog. 3:e191 doi:10.1371/journal.ppat.0030191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rakoto-Andrianarivelo M, Gumede N, Jegouic S, Balanant J, Andriamamonjy SN, Rabemanantsoa S, Birmingham M, Randriamanalina B, Nkolomoni L, Venter M, Schoub BD, Delpeyroux F, Reynes JM. 2008. Reemergence of recombinant vaccine-derived poliovirus outbreak in Madagascar. J. Infect. Dis. 197:1427–1435 [DOI] [PubMed] [Google Scholar]
- 7. Shimizu H, Thorley B, Paladin FJ, Brussen KA, Stambos V, Yuen L, Utama A, Tano Y, Arita M, Yoshida H, Yoneyama T, Benegas A, Roesel S, Pallansch M, Kew O, Miyamura T. 2004. Circulation of type 1 vaccine-derived poliovirus in the Philippines in 2001. J. Virol. 78:13512–13521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang CF, Naguib T, Yang SJ, Nasr E, Jorba J, Ahmed N, Campagnoli R, van der Avoort H, Shimizu H, Yoneyama T, Miyamura T, Pallansch M, Kew O. 2003. Circulation of endemic type 2 vaccine-derived poliovirus in Egypt from 1983 to 1993. J. Virol. 77:8366–8377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jenkins HE, Aylward RB, Gasasira A, Donnelly CA, Mwanza M, Corander J, Garnier S, Chauvin C, Abanida E, Pate MA, Adu F, Baba M, Grassly NC. 2010. Implications of a circulating vaccine-derived poliovirus in Nigeria. N. Engl. J. Med. 362:2360–2369 [DOI] [PubMed] [Google Scholar]
- 10. Wassilak S, Pate MA, Wannemuehler K, Jenks J, Burns C, Chenoweth P, Abanida EA, Adu F, Baba M, Gasasira A, Iber J, Mkanda P, Williams AJ, Shaw J, Pallansch M, Kew O. 2011. Outbreak of type 2 vaccine-derived poliovirus in Nigeria: emergence and widespread circulation in an underimmunized population. J. Infect. Dis. 203:898–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang X, Zhang Y, Xu W, Wen N, Zuo S, Lee LA, Yu J. 2006. An outbreak of poliomyelitis caused by type 1 vaccine-derived poliovirus in China. J. Infect. Dis. 194:545–551 [DOI] [PubMed] [Google Scholar]
- 12. Combelas N, Holmblat B, Joffret ML, Colbere-Garapin F, Delpeyroux F. 2011. Recombination between poliovirus and coxsackie A viruses of species C: a model of viral genetic plasticity and emergence. Viruses 3:1460–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joffret ML, Jegouic S, Bessaud M, Balanant J, Tran C, Caro V, Holmblat B, Razafindratsimandresy R, Reynes JM, Rakoto-Andrianarivelo M, Delpeyroux F. 2012. Common and diverse features of cocirculating type 2 and 3 recombinant vaccine-derived polioviruses isolated from patients with poliomyelitis and healthy children. J. Infect. Dis. 205:1363–1373 [DOI] [PubMed] [Google Scholar]
- 14. Jegouic S, Joffret ML, Blanchard C, Riquet FB, Perret C, Pelletier I, Colbere-Garapin F, Rakoto-Andrianarivelo M, Delpeyroux F. 2009. Recombination between polioviruses and co-circulating coxsackie A viruses: role in the emergence of pathogenic vaccine-derived polioviruses. PLoS Pathog. 5:e1000412 doi:10.1371/journal.ppat.1000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rakoto-Andrianarivelo M, Rousset D, Razafindratsimandresy R, Chevaliez S, Guillot S, Balanant J, Delpeyroux F. 2005. High frequency of human enterovirus species C circulation in Madagascar. J. Clin. Microbiol. 43:242–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anonymous 2004. Isolation and identification of polioviruses. In World Health Organization (ed), Polio laboratory manual. World Health Organization, Geneva, Switzerland [Google Scholar]
- 17. Kilpatrick D, Nottay B, Yang C, Yang S, Mulders M, Holloway B, Pallansch M, Kew O. 1996. Group-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residue at positions of codon degeneracy. J. Clin. Microbiol. 34:2990–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van der Avoort HG, Hull BP, Hovi T, Pallansch MA, Kew OM, Crainic R, Wood DJ, Mulders MN, van Loon AM. 1995. Comparative study of five methods for intratypic differentiation of polioviruses. J. Clin. Microbiol. 33:2562–2566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Caro V, Guillot S, Delpeyroux F, Crainic R. 2001. Molecular strategy for ‘serotyping’ of human enteroviruses. J. Gen. Virol. 82:79–91 [DOI] [PubMed] [Google Scholar]
- 20. Bessaud M, Jegouic S, Joffret ML, Barge C, Balanant J, Gouandjika-Vasilache I, Delpeyroux F. 2008. Characterization of the genome of human enteroviruses: design of generic primers for amplification and sequencing of different regions of the viral genome. J. Virol. Methods 149:277–284 [DOI] [PubMed] [Google Scholar]
- 21. Oberste MS, Maher K, Williams AJ, Dybdahl-Sissoko N, Brown BA, Gookin MS, Penaranda S, Mishrik N, Uddin M, Pallansch MA. 2006. Species-specific RT-PCR amplification of human enteroviruses: a tool for rapid species identification of uncharacterized enteroviruses. J. Gen. Virol. 87:119–128 [DOI] [PubMed] [Google Scholar]
- 22. Guillot S, Caro V, Cuervo N, Korotkova E, Combiescu M, Persu A, Aubert-Combiescu A, Delpeyroux F, Crainic R. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434–8443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Allard A, Albinsson B, Wadell G. 2001. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J. Clin. Microbiol. 39:498–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oberste MS, Maher K, Kilpatrick DR, Flemister MR, Brown BA, Pallansch MA. 1999. Typing of human enteroviruses by partial sequencing of VP1. J. Clin. Microbiol. 37:1288–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kroneman A, Vennema H, Deforche K, Avoort HVD, Penaranda S, Oberste MS, Vinje J, Koopmans M. 2011. An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 51:121–125 [DOI] [PubMed] [Google Scholar]
- 26. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 28. Hasegawa M, Kishino H, Yano T. 1985. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22:160–174 [DOI] [PubMed] [Google Scholar]
- 29. Perriere G, Gouy M. 1996. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78:364–369 [DOI] [PubMed] [Google Scholar]
- 30. Nix WA, Oberste MS, Pallansch MA. 2006. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J. Clin. Microbiol. 44:2698–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brown BA, Maher K, Flemister MR, Naraghi-Arani P, Uddin M, Oberste MS, Pallansch MA. 2009. Resolving ambiguities in genetic typing of human enterovirus species C clinical isolates and identification of enterovirus 96, 99 and 102. J. Gen. Virol. 90:1713–1723 [DOI] [PubMed] [Google Scholar]
- 32. Bessaud M, Pillet S, Ibrahim W, Joffret ML, Pozzetto B, Delpeyroux F, Gouandjika-Vasilache I. 2012. Molecular characterization of human enteroviruses in the Central African Republic: uncovering wide diversity and identification of a new human enterovirus A71 genogroup. J. Clin. Microbiol. 50:1650–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harvala H, Sharp CP, Ngole EM, Delaporte E, Peeters M, Simmonds P. 2011. Detection and genetic characterization of enteroviruses circulating among wild populations of chimpanzees in Cameroon: relationship with human and simian enteroviruses. J. Virol. 85:4480–4486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Junttila N, Leveque N, Kabue JP, Cartet G, Mushiya F, Muyembe-Tamfum JJ, Trompette A, Lina B, Magnius LO, Chomel JJ, Norder H. 2007. New enteroviruses, EV-93 and EV-94, associated with acute flaccid paralysis in the Democratic Republic of the Congo. J. Med. Virol. 79:393–400 [DOI] [PubMed] [Google Scholar]
- 35. Oberste MS, Maher K, Nix WA, Michele SM, Uddin M, Schnurr D, al-Busaidy S, Akoua-Koffi C, Pallansch MA. 2007. Molecular identification of 13 new enterovirus types, EV79-88, EV97, and EV100-101, members of the species Human enterovirus B. Virus Res. 128:34–42 [DOI] [PubMed] [Google Scholar]
- 36. Oberste MS, Michele SM, Maher K, Schnurr D, Cisterna D, Junttila N, Uddin M, Chomel J-J, Lau C-S, Ridha W, al-Busaidy S, Norder H, Magnius LO, Pallansch MA. 2004. Molecular identification and characterization of two proposed new enterovirus serotypes, EV74 and EV75. J. Gen. Virol. 85:3205–3212 [DOI] [PubMed] [Google Scholar]
- 37. Bessaud M, Joffret ML, Holmblat B, Razafindratsimandresy R, Delpeyroux F. 2011. Genetic relationship between cocirculating human enteroviruses species C. PLoS One 6:e24823 doi:10.1371/journal.pone.0024823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kuramitsu M, Kuroiwa C, Yoshida H, Miyoshi M, Okumura J, Shimizu H, Narantuya L, Bat-Ochir D. 2005. Non-polio enterovirus isolation among families in Ulaanbaatar and Tov province, Mongolia: prevalence, intrafamilial spread, and risk factors for infection. Epidemiol. Infect. 133:1131–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Silva P, Stark K, Mockenhaupt F, Reither K, Weitzel T, Ignatius R, Saad E, Seidu-Korkor A, Bienzle U, Schreier E. 2008. Molecular characterization of enteric viral agents from children in northern region of Ghana. J. Med. Virol. 80:1790–1798 [DOI] [PubMed] [Google Scholar]
- 40. Anonymous 2012. WHO-UNICEF immunization summary. World Health Organization, Geneva, Switzerland: http://www.childinfo.org/files/immunization_summary_en.pdf [Google Scholar]
- 41. Ayukekbong J, Lindh M, Nenonen N, Tah F, Nkuo-Akenji T, Bergstrom T. 2011. Enteric viruses in healthy children in Cameroon: viral load and genotyping of norovirus strains. J. Med. Virol. 83:2135–2142 [DOI] [PubMed] [Google Scholar]
- 42. Oberste MS, Maher K, Michele SM, Belliot G, Uddin M, Pallansch MA. 2005. Enteroviruses 76, 89, 90 and 91 represent a novel group within the species Human enterovirus A. J. Gen. Virol. 86:445–451 [DOI] [PubMed] [Google Scholar]
- 43. Bahri O, Rezig D, Nejma-Oueslati BB, Yahia AB, Sassi JB, Hogga N, Sadraoui A, Triki H. 2005. Enteroviruses in Tunisia: virological surveillance over 12 years (1992-2003). J. Med. Microbiol. 54:63–69 [DOI] [PubMed] [Google Scholar]
- 44. Khetsuriani N, Lamonte A, Oberste M, Pallansch M. 2006. Neonatal enterovirus infections reported to the national enterovirus surveillance system in the United States, 1983-2003. Pediatr. Infect. Dis. J. 25:889–893 [DOI] [PubMed] [Google Scholar]
- 45. Tan C, Ninove L, Gaudart J, Nougairede A, Zandotti C, Thirion-Perrier L, Charrel R, de Lamballerie X. 2011. A retrospective overview of enterovirus infection diagnosis and molecular epidemiology in the public hospitals of Marseille, France (1985-2005). PLoS One 6:e18022 doi:10.1371/journal.pone.0018022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bryden A. 1992. Isolation of enteroviruses and adenoviruses in continuous simian cell lines. Med. Lab. Sci. 49:60–65 [PubMed] [Google Scholar]
- 47. Heim A. 2005. From poliovirus surveillance to enterovirus surveillance: a complete picture? J. Med. Microbiol. 54:1–2 [DOI] [PubMed] [Google Scholar]
- 48. Baba MM, Oderinde BS, Patrick PZ, Jarmai MM. 2012. Sabin and wild polioviruses from apparently healthy primary school children in northeastern Nigeria. J. Med. Virol. 84:358–364 [DOI] [PubMed] [Google Scholar]
- 49. Bingjun T, Yoshida H, Yan W, Lin L, Tsuji T, Shimizu H, Miyamura T. 2008. Molecular typing and epidemiology of non-polio enteroviruses isolated from Yunnan Province, the People's Republic of China. J. Med. Virol. 80:670–679 [DOI] [PubMed] [Google Scholar]
- 50. Antona D, Leveque N, Chomel J, Dubrou S, Levy-Bruhl D, Lina B. 2007. Surveillance of enteroviruses in France, 2000-2004. Eur. J. Clin. Microbiol. Infect. Dis. 26:403–412 [DOI] [PubMed] [Google Scholar]
- 51. Roth B, Enders M, Arents A, Pfitzner A, Terletskaia-Ladwig E. 2007. Epidemiologic aspects and laboratory features of enterovirus infections in Western Germany, 2000-2005. J. Med. Virol. 79:956–962 [DOI] [PubMed] [Google Scholar]
- 52. Tseng F, Huang H, Chi C, Lin T, Liu C, Jian J, Hsu L, Wu H, Yang J, Chang Y, Wang H, Hsu Y, Su I, Wang J. 2007. Epidemiological survey of enterovirus infections occurring in Taiwan between 2000 and 2005: analysis of sentinel physician surveillance data. J. Med. Virol. 79:1850–1860 [DOI] [PubMed] [Google Scholar]
- 53. Adu F, Idowu A, Akintokun A, Adeniji J, Ajuwon B. 2012. Effect of temperature and distance on the viral outcome of clinical specimens from acute flaccid paralysis cases in Nigeria. J. Clin. Lab. Anal. 26:254–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chakraborty R, Iturriza-Gomara M, Musoke R, Palakudy T, D'Agostino A, Gray J. 2004. An epidemic of enterovirus 71 infection among HIV-1-infected orphans in Nairobi. AIDS 18:1968–1970 [DOI] [PubMed] [Google Scholar]
- 55. Mirand A, Schuffenecker I, Henquell C, Billaud G, Jugie G, Falcon D, Mahul A, Archimbaud C, Terletskaia-Ladwig E, Diedrich S, Huemer HP, Enders M, Lina B, Peigue-Lafeuille H, Bailly J-L. 2010. Phylogenetic evidence for a recent spread of two populations of human enterovirus 71 in European countries. J. Gen. Virol. 91:2263–2277 [DOI] [PubMed] [Google Scholar]
- 56. Tee KK, Lam TT, Chan YF, Bible JM, Kamarulzaman A, Tong CY, Takebe Y, Pybus OG. 2010. Evolutionary genetics of human enterovirus 71: origin, population dynamics, natural selection, and seasonal periodicity of the VP1 gene. J. Virol. 84:3339–3350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Deshpande JM, Nadkarni SS, Francis PP. 2003. Enterovirus 71 isolated from a case of acute flaccid paralysis in India represents a new genotype. Curr. Sci. 84:1350–1353 [Google Scholar]
- 58. McMinn P, Lindsay K, Perera D, Chan HM, Chan KP, Cardosa MJ. 2001. Phylogenetic analysis of enterovirus 71 strains isolated during linked epidemics in Malaysia, Singapore, and Western Australia. J. Virol. 75:7732–7738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. van der Sanden S, van der Avoort H, Lemey P, Uslu G, Koopmans M. 2010. Evolutionary trajectory of the VP1 gene of human enterovirus 71 genogroup B and C viruses. J. Gen. Virol. 91:1949–1958 [DOI] [PubMed] [Google Scholar]
- 60. Smura T, Blomqvist S, Paananen A, Vuorinen T, Sobotova Z, Bubovica V, Ivanova O, Hovi T, Roivainen M. 2007. Enterovirus surveillance reveals proposed new serotypes and provides new insight into enterovirus 5′-untranslated region evolution. J. Gen. Virol. 88:2520–2526 [DOI] [PubMed] [Google Scholar]
- 61. Harvala H, McIntyre C, Imai N, Clasper L, Djoko C, LeBreton M, Vermeulen M, Saville A, Mutapi F, Tamoufe U, Kiyang J, Biblia T, Midzi N, Mduluza T, Pepin J, Njoum R, Smura T, Fair J, Wolfe N, Roivainen M, Simmonds P. 2012. High seroprevalence of enterovirus infections in apes and Old World monkeys. Emerg. Infect. Dis. 18:283–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wolfe N. 2005. Bushmeat hunting, deforestation, and prediction of zoonotic disease emergence. Emerg. Infect. Dis. 11:1822–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lukashev AN, Lashkevich VA, Ivanova OE, Koroleva GA, Hinkkanen AE, Ilonen J. 2003. Recombination in circulating enteroviruses. J. Virol. 77:10423–10431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Simmonds P, Welch J. 2006. Frequency and dynamics of recombination within different species of human enteroviruses. J. Virol. 80:483–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jiang P, Faase JA, Toyoda H, Paul A, Wimmer E, Gorbalenya AE. 2007. Evidence for emergence of diverse polioviruses from C-cluster coxsackie A viruses and implications for global poliovirus eradication. Proc. Natl. Acad. Sci. U. S. A. 104:9457–9462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Anonymous 2011. Vaccine-derived polioviruses detected worldwide, July 2009-March 2011. Wkly. Epidemiol. Rec. 86:277–288 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.