Abstract
Helicobacter suis is a Gram-negative bacterium colonizing the majority of pigs, in which it causes gastritis and decreased daily weight gain. H. suis is also the most prevalent gastric non-Helicobacter pylori Helicobacter species in humans, capable of causing gastric disorders. To gain insight into the genetic diversity of porcine and human H. suis strains, a multilocus sequence typing (MLST) method was developed. In a preliminary study, 7 housekeeping genes (atpA, efp, mutY, ppa, trpC, ureI, and yphC) of 10 H. suis isolates cultured in vitro were investigated as MLST candidates. All genes, except the ureI gene, which was replaced by part of the ureAB gene cluster of H. suis, displayed several variable nucleotide sites. Subsequently, internal gene fragments, ranging from 379 to 732 bp and comprising several variable nucleotide sites, were selected. For validation of the developed MLST technique, gastric tissue from 17 H. suis-positive pigs from 4 different herds and from 1 H. suis-infected human patient was used for direct, culture-independent strain typing of H. suis. In addition to the 10 unique sequence types (STs) among the 10 isolates grown in vitro, 15 additional STs could be assigned. Individual animals were colonized by only 1 H. suis strain, whereas multiple H. suis strains were present in all herds tested, revealing that H. suis is a genetically diverse bacterial species. The human H. suis strain showed a very close relationship to porcine strains. In conclusion, the developed MLST scheme may prove useful for direct, culture-independent typing of porcine and human H. suis strains.
INTRODUCTION
Helicobacter suis is a Gram-negative, motile, tightly coiled, spiral-shaped, and microaerophilic bacterium that colonizes the gastric mucosa of the majority of pigs worldwide (1–3). Its reported prevalence depends on the study. Mostly, however, this bacterium is detected in more than 60% of pigs at slaughter age (1–4). H. suis infection in pigs is associated with chronic gastritis and decreased daily weight gain (5). In addition, associations have been made between a natural H. suis infection and the presence of ulcers in the pars oesophagea of the stomach (6–8). H. suis is also of zoonotic importance, as it is the most prevalent gastric non-Helicobacter pylori Helicobacter (NHPH) species in humans (4, 9). Pigs are considered to be an important source of infection for humans. Besides direct contact with animals, the consumption of raw or undercooked pig meat may also be a source of human infection (10, 11).
The first isolation of H. suis was described only in 2008 (12). In subsequent years, we gathered a total of 10 isolates, cultured in vitro, of this extremely fastidious microorganism. All isolates originate from pig stomachs, and until now, no isolates have been obtained from infected humans. To our knowledge, the above-mentioned in vitro isolates are the only ones available worldwide, and still now, H. suis isolation remains difficult and time-consuming. Often, mucus scrapings from half of a porcine stomach are needed to successfully isolate a new H. suis strain.
In order to gain insight into the strain diversity of both human and porcine H. suis strains, several typing methods can be used. However, given the extremely fastidious nature of this microorganism, a culture-independent method should be used, allowing typing of H. suis directly in stomach samples. Multilocus sequence typing (MLST), introduced in 1998, has been widely used in molecular epidemiology and population biology of bacterial species (13–16) and has proven its usefulness for typing strains of other Helicobacter species (17–19). In addition, this technique uses the unambiguousness and portability of nucleotide sequence data, which allows results from different laboratories to be compared without exchanging strains (20–22). Although it also has some drawbacks, including a relatively high cost (21), MLST is considered a gold standard for strain typing of bacterial species.
Our aim was thus to develop a robust H. suis MLST technique which can be applied to biological tissue without the need for cultivation.
MATERIALS AND METHODS
H. suis isolates cultured in vitro and DNA extraction.
H. suis strains HS1 to HS10 were isolated from the gastric mucosa of sows as described previously (12). Bacteria were grown microaerobically (85% N2, 10% CO2, 5% O2; 37°C; 48 to 96 h) on biphasic brucella (Becton Dickinson, Franklin Lakes, NJ) culture plates (with a pH adjusted to 5) supplemented with 20% fetal calf serum (HyClone, Logan, UT) and Vitox supplement (Oxoid), as described previously (23). Bacterial genomic DNA of all H. suis strains was extracted as described by Wilson (24).
Development of a Helicobacter suis multilocus sequence typing method.
PCR assays were performed on a Mastercycler thermal cycler (Eppendorf, Hamburg, Germany). Different DNA polymerases (Taq, Pwo, and Accuzyme) were used in this study depending on the application. PCR products were first purified using a MSB Spin PCRapace kit (STRATEC Molecular GmbH, Berlin, Germany). Purified PCR products were sequenced using the BigDye Terminator sequencing kit (Applied Biosystems, Foster City, CA), and sequences were determined on an automatic DNA sequencer (ABI Prism 3100 genetic analyzer; Applied Biosystems). The electropherograms were exported and converted to Kodon software (Applied Maths, Sint-Martens-Latem, Belgium).
Based on the whole genomic DNA sequence of H. suis strains 1 and 5 (25), seven housekeeping genes used for H. pylori multilocus sequence typing (atpA, efp, mutY, ppa, trpC, ureI, and yphC) were initially selected for the design of a multilocus sequence typing method for Helicobacter suis. The complete coding regions of these genes were amplified from 10 H. suis isolates cultured in vitro using Pwo polymerase with 3′-to-5′ proofreading exonuclease activity. Final reaction mixtures containing a total of 25 μl were prepared in 2 parts. Part 1 contained 6.25 μl deoxynucleotide triphosphates (dNTPs) (1 mM each), 0.3 μl of each primer at a concentration of 0.6 pmol/μl, 5.15 μl Aq HPLC, and 0.5 μl DNA sample. Part 2 included 0.25 μl Pwo DNA polymerase (5-U/μl stock solution), 2.5 μl 10× buffer containing MgSO4, and 9.75 μl Aq HPLC. Briefly, following an initial 3-min denaturation at 94°C, amplification was done by 10 cycles of 94°C for 30 s, 53°C to 58°C for 30 s, and 72°C for 90 s, followed by 25 or 30 cycles of 94°C for 30 s, 53°C to 58°C for 30 s, and 72°C for 90 s, with an additional 5-s elongation time for each cycle. A final extension step was performed at 72°C for 10 min. Alternatively, amplification was done using Accuzyme DNA polymerase. Final reaction mixtures (10 μl) included 5 μl Accuzyme mix, 0.1 μl of each primer at a concentration of 0.5 pmol/μl, 3.8 μl Aq HPLC, and 1.0 μl DNA sample. Briefly, following an initial denaturation for 3 min at 94°C, amplification was done by 10 cycles of 94°C for 30 s, 53°C to 58°C for 30 s, and 72°C for 90 s, followed by 25 or 30 cycles of 94°C for 30 s, 53°C to 58°C for 30 s, and 72°C for 90 s, with an additional 5-s elongation time for each cycle. A final extension step was performed at 72°C for 20 min. Purification of PCR products and sequencing were performed as described above. Primers used for amplification and sequencing, as well as annealing temperatures, are shown in Table 1.
Table 1.
Oligonucleotide primers used for amplification and sequencing of complete coding sequences
| Primer | Sequence (5′ to 3′) | Polarity | Amplicon size (bp) | Position (bp) | Annealing temp (°C) | Use of primera |
|---|---|---|---|---|---|---|
| atpA JunF1 | TTGTTAAAATTAAAAGCAGAAGAAATTAGCG | Forward | 1,512 | 1–31 | 54 | A, S |
| atpA JunR1 | CTAGAGAGTTAGACTTAGTTTAAATTCTTCAATCGC | Reverse | 1477–1512 | A, S | ||
| ATPA-F | CCCCCCTATTCTCCCTTGCCC | Forward | 1270–1290 | S | ||
| ATPA-R | CAAGCCCCTATCGCCGGTATC | Reverse | 169–189 | S | ||
| AtpA_fw_in | TACCCGTAGGCGATGCGGTC | Forward | 293–312 | S | ||
| efp JunF1 | ATGGCCATTGGAATGGGTGAAC | Forward | 567 | 1–22 | 53 | A, S |
| efp JunR1 | TTAAACTTTGAGCTTTTCTAAATACTCC | Reverse | 540–567 | A, S | ||
| EFP-F | GCCTAATTTTAGAGGGGATAC | Forward | 414–434 | S | ||
| EFP-R bis | ATTAGGCTCTTCGCACTTATCC | Reverse | 174–195 | S | ||
| mutY JunF1 | ATGTCCCAGCAAACCCAAATTGAGGTTG | Forward | 873 | 1–28 | 58 | A, S |
| mutY JunR1 | CTACAAAATCCCCGTCTTTTCTAGTAATACC | Reverse | 843–873 | A, S | ||
| MUTY-F | AGTACCGCGTGCAAGTTTGGC | Forward | 719–739 | S | ||
| MUTY-R | CACGGGCATAATAACCTAGAC | Reverse | 125–145 | S | ||
| ppa JunF1 | ATGAACATCTCAAAAATTCCAGTTAGTAACGC | Forward | 528 | 1–32 | 56 | A, S |
| ppa JunR1 | TCAACCTTGGGCTTGTTGGTAATTTGCAAGTGC | Reverse | 496–528 | A, S | ||
| PPA-F2 | CATTGTGCTACAAAAGATCAGAC | Forward | 378–400 | S | ||
| PPA-R2 | TAGCTGGATATACCATAGATG | Reverse | 134–154 | S | ||
| trpC JunF1 | ATGCATGATTTTTTAACAACCATGTTAGAA C | Forward | 1,359 | 1–31 | 57 | A, S |
| trpC JunR1 | TCAATACTCCCGTAACATTTTTGCAATTTGTGC | Reverse | 1327–1359 | A, S | ||
| TRPC-F | GCTAGCTGGAGGGTTAAATGC | Forward | 1209–1229 | S | ||
| TRPC-R | GAGGGGGAGGCTTGTTTGCAC | Reverse | 150–170 | S | ||
| TRPC_rv_in | TTTAAGCGCAGTTGGCATGCA | Reverse | 960–980 | S | ||
| ureI JunF1 | ATGCTAGGACTTGTGTTATTGTATGTTGCGATCG | Forward | 588 | 1–34 | 55 | A, S |
| ureI JunR1 | TTACACCCAGTGTTCGATAAAGAGAAGCCAAGCAGG | Reverse | 553–588 | A, S | ||
| UREI-F | ACTGGTTGGATTGAGGAAGTGG | Forward | 463–484 | S | ||
| UREI-R | CAAACAATAGAGATCCCACCTAC | Reverse | 106–128 | S | ||
| yphC JunF1 | ATGCTTAAAATTGCTATTTTGGGCAAGCC | Forward | 1,317 | 1–29 | 57 | A, S |
| yphC JunR1 | TCATGTTTGCAAAGTTTCTTCTTTGCCTCTGGC | Reverse | 1285–1317 | A, S | ||
| YPHC-F2 | TTGCTTTGGTTATGAATCGCCCTAAG | Forward | 1175–1200 | S | ||
| YPHC-R | AAAGCTCCACCCCATGGCCTGCTAG | Reverse | 136–160 | S |
A, primer used for amplification; S, primer used for sequencing.
For all genes except the ureI gene, variable sites were detected. Based on the position of these sites, internal gene fragments (450 to 732 bp) were selected (Table 2). Amplification was done using Pwo or Accuzyme DNA polymerase with 3′-to-5′ proofreading activity. Primers used for amplification and sequencing of these variable regions are shown in Table 2. Briefly, following an initial denaturation for 3 min at 94°C, amplification was done by 10 cycles of 94°C for 30 s, 59°C to 69°C for 30 s, and 72°C for 90 s, followed by 25 or 30 cycles of 94°C for 30 s, 59°C to 69°C for 30 s, and 72°C for 90 s, with an additional 5-s elongation time for each cycle. A final extension step was performed at 72°C for 20 min. Purification of PCR products and sequencing were performed as described above. Based on the multiple alignment, using Kodon software, of the ureAB gene cluster of different H. suis strains (HS1, HS5, and NCBI accession numbers AF507995, AF508001, AF508008, AF508009, and AF508013), variable sites were detected in this gene cluster. Therefore, primers were designed for amplification and sequencing of the variable region of this gene cluster (Table 2).
Table 2.
Oligonucleotide primers used for H. suis MLST
| Primer | Sequence (5′ to 3′) | Polarity | Amplicon size (bp) | Position (bp) | Annealing temp (°C) | Use of primera |
|---|---|---|---|---|---|---|
| atpA-MLST-A | TTATGAGGTGGTTGAATTTGATACCGGC | Forward | 790 (732)b | 150–177 | 63 | A, S |
| atpA-MLST-B | AGAGCCTGCCCCTTTCTTATCACTCATTT | Reverse | 911–939 | A, S | ||
| atpA-MLST-C | ATGATTGCATCAATGGCAACAGTGG | Reverse | 530–554 | S | ||
| efp-MLST-A | TACAAGGCGTTCCTTATCGCATTGT | Forward | 470 (379)b | 47–71 | 61 | A, S |
| efp-MLST-B | CACCTCCCCCTCTAGCACATGG | Reverse | 495–516 | A, S | ||
| efp_mlstAquintoc | GGCCTTTGTACGGGCTAAA | Forward | 379 | 105–123 | 58 | A, S |
| efp_mlstBbisc | CACCACTGCCCCGGT | Reverse | 469–483 | A, S | ||
| mutY-MLST-A | CGCCCCTTTAGACCGGGTTTTACTT | Forward | 650 | 90–114 | 61 | A, S |
| mutY-MLST-B | GCCAAACTTGCACGCGGTACTTG | Reverse | 717–739 | A, S | ||
| mutY-MLST-C | TTAGGCAAAATGTGGGCGTGCTAGA | Forward | 278–302 | S | ||
| ppa-MLST-A | TGCCGTTATTGAAATCCCGTATGGA | Forward | 480 | 45–69 | 60 | A, S |
| ppa-MLST-B | CCTTGGGCTTGTTGGTAATTTGCAA | Reverse | 500–524 | A, S | ||
| trpC-MLST-A | TGTGGCCTTAAGCGGGTTAAAGATG | Forward | 450 | 769–793 | 60 | A, S |
| trpC-MLST-B | TCCAGCTAGCATAAAGCGATGGGAT | Reverse | 1194–1218 | A, S | ||
| ureAB_mlstA | GTGCGCTTTGAACCTGGCG | Forward | 688 (676)b | 523–541 | 69 | A |
| ureB_mlstB | CCTGTTCCGCCTCCAAGCAT | Reverse | 1191–1210 | A, S | ||
| ureB_mlstA | ATGTATGGCCCCACTACAGGCG | Forward | 759–780 | S | ||
| yphC-MLST-A | GGATACAGGCGGGTTTGATGCAG | Forward | 850 (717)b | 162–184 | 59 | A, S |
| yphC-MLST-B | TTTGATTTGGAGGATATGGCGCTTAGA | Reverse | 985–1011 | A, S | ||
| yphc-MLST-C | AAATGCCCTGATAGAGCAAGAACGC | Forward | 579–603 | S | ||
| yphC_mlstAtrisc | AAAATCCCCCCACAAGATGAGGATAA | Forward | 717 | 268–293 | 62 | A, S |
| yphC_mlstBtrisc | GATAGCACTTGTTGTAAGAAGCG | Reverse | 962–984 | A, S |
A, primer used for amplification; S, primer used for sequencing.
The number in parentheses represents the actual length, after trimming, of nucleotide fragments used for multiple alignment, determination of ST, and generation of concatenated sequences.
Primer pair used for amplification and sequencing of internal efp and yphC genes from the human H. suis strain described in this study.
Based on the nucleotide sequence of the internal gene fragments, allele numbers for each gene were assigned for isolates HS1 to HS10, cultured in vitro. The sequence type (ST) was defined by the combined allelic profile of the 7 alleles.
Validation of Helicobacter suis MLST.
For validation of the developed MLST on tissue samples, gastric biopsy specimens were collected from slaughter pigs from 4 different herds. For each stomach, three pieces of tissue (1 cm2; 1 from the corpus, 2 from the antrum) were collected using sterile biopsy punches. In addition, an antral gastric biopsy specimen from a pig veterinarian, suffering from gastric complaints and diagnosed with H. suis infection (M. Joosten, B. Flahou, T. Meyns, A. Smet, J. Arts, L. De Cooman, F. Pasmans, R. Ducatelle, and F. Haesebrouck, submitted for publication), was included as well. DNA was extracted using the Isolate genomic DNA minikit (Bioline, London, United Kingdom) according to the manufacturer's instructions. All DNA samples were screened for the presence of H. suis using a Taq polymerase-based species-specific PCR, as described by De Groote et al. (7). Three to six H. suis-positive samples/herd, as well as the human sample, were selected for multilocus sequence typing of colonizing H. suis strains, as described above.
Amplification of the internal gene fragments for the efp and yphC genes from the human H. suis strain, however, yielded no pure PCR products. Therefore, new primer pairs were designed, resulting in the amplification of a single PCR product of the expected size (Table 2). In order to enable comparison, internal efp and yphC gene fragments from in vitro-cultured H. suis strains and the porcine and human gastric tissue samples, obtained as described above, were all trimmed to this new size (379 bp and 717 bp for efp and yphC, respectively). Contig assembly of sequences obtained using different primers was done using Kodon software. The obtained sequences of internal gene fragments (harboring variable nucleotide sites) for individual genes were aligned using the CLUSTAL W program.
In order to assess the stability of allelic polymorphisms during in vitro culture of H. suis, strains HS1 and HS5 were cultured and passed onto new plates continuously for another 6 weeks. These long-term in vitro-cultured strains were designated HS1p17 and HS5p21, respectively. DNA was extracted and used for sequencing of the internal gene fragments as described above.
Based on the nucleotide sequence of the internal gene fragments, allele numbers for each gene were assigned for H. suis strains detected in porcine and human stomach tissues. The sequence type (ST) was defined by the combined allelic profile of the 7 alleles.
The relationship among the different H. suis strains was examined using the concatenated internal gene fragment sequences of the 7 MLST loci. A phylogenetic tree was constructed using the neighbor-joining method via the PHYLIP 3.69 package, using DNADIST for distance analysis (26).
RESULTS
Selection of genes for multilocus sequence typing.
A total of 6,744 bp, representing the complete coding sequences of 7 housekeeping genes (atpA, efp, mutY, ppa, trpC, ureI, and yphC), were amplified and sequenced for all in vitro H. suis isolates available (HS1 to HS10), revealing a unique concatenated sequence for all isolates. Fifty-six nucleotide sites (56/6,744; 0.83%) were found to be polymorphic. The mutY gene was shown to have the highest discriminatory power, with 32 variable nucleotide sites. The ppa gene showed 9 variable nucleotide sites, whereas the yphC gene revealed 5, the atpA gene revealed 4, and the efp and trpC genes revealed 3 variable nucleotide sites. Based on the complete ureI coding sequence (strains HS1 to HS10), this gene was shown to lack discriminatory power (0 variable nucleotide sites).
MLST analysis of in vitro-isolated H. suis strains and H. suis strains present in porcine and human gastric tissue.
Based on the complete gene sequences, as described above, internal gene fragments/variability regions (379 to 732 bp) were selected for all 7 loci except the ureI gene. Instead, the latter was replaced by part of the ureAB gene cluster. For porcine H. suis strains HS1 to HS10, allele numbers and sequence types (STs) were assigned, as shown in Table 3. Determination of allele numbers for long-term in vitro-cultured HS1p17 and HS5p21 showed no differences for HS1p17 compared to its parental strain, HS1. For HS5p21, however, a new allele type was attributed for mutY and trpC, indicating that long-term in vitro culture, and possibly also in vivo colonization, may result in limited genetic diversification.
Table 3.
Determination of allele numbers and sequence types for 10 H. suis strains isolated in vitro and H. suis bacteria present in 1 human and 17 porcine gastric tissue samples
| Samplea | ST | Sample origin | Allele no. |
||||||
|---|---|---|---|---|---|---|---|---|---|
| atpA | efp | mutY | ppa | trpC | ureAB | yphC | |||
| HS1 | 1 | In vitro culture | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| HS1p17 | 1 | In vitro culture | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| HS2 | 2 | In vitro culture | 2 | 1 | 2 | 1 | 2 | 1 | 2 |
| HS3 | 3 | In vitro culture | 1 | 2 | 2 | 1 | 2 | 1 | 2 |
| HS4 | 4 | In vitro culture | 3 | 2 | 3 | 1 | 3 | 1 | 3 |
| HS5 | 5 | In vitro culture | 4 | 2 | 4 | 1 | 1 | 1 | 1 |
| HS5p21 | 6 | In vitro culture | 4 | 2 | 5 | 1 | 2 | 1 | 1 |
| HS6 | 7 | In vitro culture | 1 | 3 | 6 | 1 | 1 | 1 | 3 |
| HS7 | 8 | In vitro culture | 1 | 3 | 7 | 1 | 1 | 1 | 3 |
| HS8 | 9 | In vitro culture | 1 | 4 | 8 | 1 | 1 | 1 | 1 |
| HS9 | 10 | In vitro culture | 4 | 2 | 9 | 1 | 1 | 1 | 1 |
| HS10 | 11 | In vitro culture | 1 | 4 | 3 | 2 | 1 | 1 | 3 |
| A-P29 | 12 | Herd A | 4 | 2 | 9 | 1 | 2 | 1 | 1 |
| A-P49 | 13 | Herd A | 1 | 4 | 10 | 1 | 1 | 1 | 3 |
| A-P63 | 12 | Herd A | 4 | 2 | 9 | 1 | 2 | 1 | 1 |
| B-V16 | 11 | Herd B | 1 | 4 | 3 | 2 | 1 | 1 | 3 |
| B-V37 | 14 | Herd B | 1 | 2 | 3 | 1 | 1 | 2 | 4 |
| B-V57 | 15 | Herd B | 1 | 2 | 3 | 1 | 1 | 1 | 1 |
| B-V85 | 16 | Herd B | 1 | 2 | 3 | 1 | 1 | 2 | 3 |
| C-P1A1 | 17 | Herd C | 1 | 2 | 11 | 1 | 1 | 1 | 1 |
| C-P3A1 | 17 | Herd C | 1 | 2 | 11 | 1 | 1 | 1 | 1 |
| C-P8A1 | 17 | Herd C | 1 | 2 | 11 | 1 | 1 | 1 | 1 |
| C-P9A1 | 18 | Herd C | 1 | 2 | 12 | 1 | 1 | 1 | 1 |
| D-P21A1 | 19 | Herd D | 1 | 2 | 13 | 1 | 2 | 1 | 5 |
| D-P26A1 | 20 | Herd D | 1 | 2 | 14 | 1 | 1 | 1 | 2 |
| D-P27A1 | 21 | Herd D | 1 | 4 | 15 | 1 | 1 | 1 | 1 |
| D-P28A1 | 22 | Herd D | 1 | 3 | 16 | 1 | 2 | 1 | 2 |
| D-P29A1 | 23 | Herd D | 1 | 2 | 16 | 1 | 2 | 1 | 2 |
| D-P30A1 | 24 | Herd D | 1 | 2 | 14 | 1 | 1 | 1 | 6 |
| HA | 25 | Human | 1 | 2 | 17 | 1 | 1 | 1 | 1 |
HS1 to HS10, H. suis isolates cultured in vitro; HS1p17 and HS5p21, long-term (6-week) in vitro cultures originating from HS1 and HS5, respectively; A-P29, for porcine gastric tissue samples, the number of the stomach (P29 to P63; V16 to V85; P1A1 to P30A1) is preceded by a letter designating the herd (A to D); HA, H. suis strain colonizing the antrum of a human patient suffering from reflux esophagitis and dyspepsia.
In addition, direct H. suis MLST strain typing was performed on gastric tissue of 1 H. suis-infected human and 17 H. suis-positive pigs at slaughter age from 4 different herds, as determined by H. suis-specific PCR. Three stomachs from herd A were tested, 4 each from herds B and C were tested, and 6 stomachs from herd D were tested. For MLST analysis, selected variability regions were amplified and sequenced. Close examination of the electropherograms indicated that only 1 ST colonized each animal. For the human sample, as well as all 17 porcine gastric tissue samples, a total of 4,084 bp were aligned to the same gene regions of H. suis strains HS1 to HS10. In total, 48 nucleotide sites (48/4,084; 1.18%) were shown to be polymorphic. The mutY gene revealed a total of 27 variable nucleotide sites, whereas 7, 4, 3, 3, 2, and 2 variable nucleotide sites were detected in the ppa gene, yphC gene, atpA gene, efp gene, trpC gene, and ureAB gene, respectively. Twenty-five sequence types (STs) were assigned based on unique allele combinations (Table 3). In herd A, 2 out of 3 strains shared the same ST (ST11), while a third strain displayed an unrelated ST, belonging to another group (Fig. 1). In herd C, three out of four strains shared the same ST. For herds B and D, however, all strains showed a different ST. All data have been made available at http://pubmlst.org/hsuis/, developed and hosted at the University of Oxford (26a).
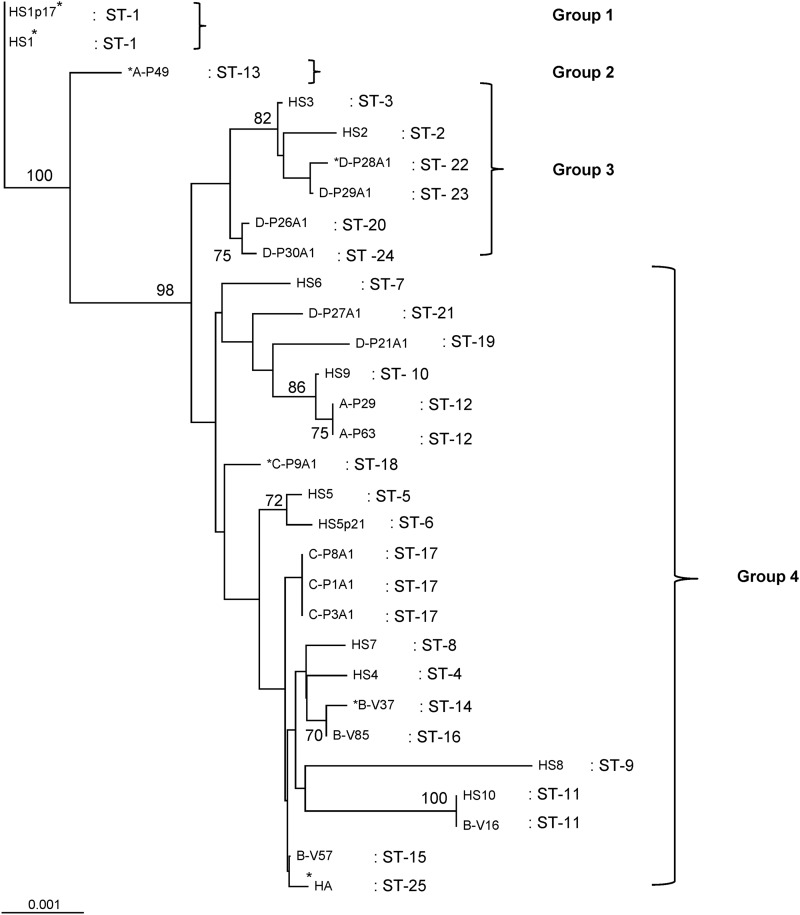
Fig 1.
Neighbor-joining phylogenetic tree (unrooted), constructed using the concatenated sequences (4,084 nucleotides) of 7 H. suis gene fragments from 27 H. suis strains tested. Bootstrap values equal to or greater than 70 are presented at the nodes of the phylogenetic tree. The topology shows that all sequence types cluster into four groups. HS1* to HS10, H. suis isolates cultured in vitro; HS1p17* and HS5p21, long-term (6-week) in vitro cultures originating from HS1 and HS5, respectively; *A-P49, for gastric tissue samples, the number of the stomach (P29 to P63; V16 to V85; P1A1 to P30A1) is preceded by a letter designating the herd (A to D); *HA, H. suis strain colonizing the antrum of a human patient suffering from reflux esophagitis and dyspepsia.
Relationships between the isolates.
A neighbor-joining tree (unrooted) was constructed based on the concatenated sequences of all 7 MLST gene fragments (4,084 bp), indicating the genetic relationship between all strains tested (Fig. 1). In general, calculated bootstrap values were low (<70), resulting in the assignment of 4 groups, of which groups 1 and 2 were singletons. Although most strains detected in porcine gastric tissue belonged to the same group with low internal bootstrap values (group 4), strains belonging to a particular herd tended to cluster, indicating a lower genetic distance between strains present in the same herd. Figure 1 clearly shows that the human H. suis strain is closely related to porcine H. suis strains.
DISCUSSION
Up to now, very little is known about transmission of H. suis between pigs and from pigs to humans. It is thought that pigs are the most important source of infection for humans through direct contact with the animals or through the consumption of raw or undercooked pig meat contaminated with H. suis (10, 11). In addition, few data are available on strain-related differences in virulence or the immune response evoked in its animal or human host. So undoubtedly, there is a strong need for a robust H. suis strain typing method, which is essential to provide answers to some of the above-mentioned questions.
Baele et al. (12), describing the first successful in vitro isolation of H. suis, performed genomic fingerprinting of isolates HS1, HS2, and HS3 by amplified fragment length polymorphism (AFLP), revealing distinct AFLP patterns for these 3 isolates. Although this typing method has proven its value for strain typing of isolates cultured in vitro of several bacterial species (27, 28), this cannot be used for typing of H. suis strains in a complex matrix, such as biological tissue samples. Keeping in mind that H. suis isolation remains extremely difficult and time-consuming, we decided to develop an MLST technique which can be applied on samples without the need for in vitro cultivation. As a genotyping method and technique, MLST has often been used in strain typing studies and can be considered a gold standard (15, 18, 20–22).
For MLST analysis of strains of H. pylori, a closely related major human gastric pathogen, 7 housekeeping genes are used (atpA, efp, mutY, ppa, trpC, yphC, and ureI) (17, 19). The whole-genome sequences of H. suis strains 1 and 5 revealed the presence of a homologue for all of these genes (25). Therefore, these genes were also investigated as MLST candidates in our preliminary study. Each gene was successfully amplified and sequenced from all H. suis strains. The mutY gene was found to have, by far, the most discriminatory power, revealing 17 alleles for 30 samples tested. The remaining 6 genes revealed a significantly lower number of alleles than the mutY gene. The ureI gene was even found to have no variable nucleotide sites, and this locus was therefore replaced by part of the ureAB gene cluster in the final MLST scheme.
For H. pylori, all 7 loci used for multilocus sequence typing show a similar discriminatory ability, in contrast to what we found for H. suis (http://pubmlst.org/helicobacter/). Nevertheless, assessment of the allelic profiles for the 10 available H. suis isolates cultured in vitro enabled us to discriminate 10 distinct sequence types (STs), underlining the suitability of the proposed MLST scheme to identify different H. suis strains. In addition, this scheme was applied to gastric biopsy specimens from 1 human patient and 17 slaughter pigs originating from 4 different herds. We were able to distinguish 14 additional STs, and all tissue samples revealed the presence of only 1 ST, based on the analysis of electropherograms. Because isolation and purification of colonizing H. suis strains were not attempted on these samples, we cannot exclude the presence of additional sequence types in 1 individual animal at a lower colonization rate than that of the dominant strain. For H. pylori, it has been shown that mixed infections with more than 1 strain in single individuals are indeed common in certain geographic regions. These mixed infections are, in fact, important for genetic diversification, since H. pylori genomes seem more stable in the absence of a mixed infection in one individual (29). On the other hand, long-term in vitro culture of H. suis strain HS5 resulted in the appearance of new alleles for a number of genes, indicating that changes or mutations do occur during in vitro culture and most likely also during in vivo colonization, as has been described for H. pylori in the absence of mixed infection (30). Recombination events in the presence of other H. suis strains can be excluded, since H. suis strains HS1 and HS5 were purified before the onset of the present study from a single colony on 1% brucella agar plates after initial isolation from slaughterhouse pigs by biphasic culture methods (12).
Based on the neighbor-joining tree and calculated bootstraps of concatenated sequences from all 7 housekeeping genes, four groups were distinguished among the isolates tested in this study, of which 2 (groups 1 and 2) were singletons. Most H. suis strains belonged to a large group (group 4) with low internal bootstrap values, although strains belonging to a particular pig herd showed a tendency to cluster. For 2 out of 4 herds (B and D) tested in the present study, all samples revealed the presence of a different ST, indicating a substantial genetic heterogeneity among H. suis bacteria present at the herd level. The human H. suis strain clearly showed a close relationship to porcine H. suis strains, indicating that the patient, a pig veterinarian, most likely contracted the infection through its close contact with pigs (Joosten et al., submitted).
At this point, no conclusions can be drawn on possible correlations between STs and pathologies in pigs and humans. For the slaughterhouse samples, no detailed histopathology was performed. H. suis strains HS1 to HS9 have all been shown to generate an immune response and gastritis in vivo (4, 23, 31). Strain HS1 generates an immune response that is somewhat different from that of most other in vitro-cultured H. suis strains with regard to the expression of interleukin 4 (IL-4) and IL-6 (31), and interestingly, this strain constitutes a distinct singleton in the present study. Future studies should try to elucidate whether additional correlations can be made between the ST and the clinical significance of the strain.
In conclusion, we developed a multilocus sequence typing scheme and method for identification of H. suis strain differences, both in pure in vitro cultures and in biological samples. Most likely, this technique will further prove its usefulness, since until today, H. suis isolation and cultivation remain a challenge. In the future, this scheme can be used for strain typing, not only of porcine H. suis strains but also of human strains and strains present in contaminated pork (11). In the end, this should provide better insights into the epidemiology of H. suis infections worldwide.
ACKNOWLEDGMENTS
This work was supported by the Research Fund of Ghent University, Belgium (grants BOF08/GOA/004 and 01W00310).
We are very grateful to Sofie De Bruyckere for her invaluable help with performing PCRs and sequencing analysis. We thank Sophie Callens for her help with collecting porcine stomachs and Venessa Eeckhaut for her help with phylogenetic analysis.
Footnotes
Published ahead of print 9 January 2013
REFERENCES
- 1. Grasso GM, Ripabelli G, Sammarco ML, Ruberto A, Iannitto G. 1996. Prevalence of Helicobacter-like organisms in porcine gastric mucosa: a study of swine slaughtered in Italy. Comp. Immunol. Microbiol. Infect. Dis. 19(3):213–217 [DOI] [PubMed] [Google Scholar]
- 2. Park JH, Seok SH, Cho SA, Baek MW, Lee HY, Kim DJ, Park JH. 2004. The high prevalence of Helicobacter sp. in porcine pyloric mucosa and its histopathological and molecular characteristics. Vet. Microbiol. 104:219–225 [DOI] [PubMed] [Google Scholar]
- 3. Hellemans A, Chiers K, De Bock M, Decostere A, Haesebrouck F, Ducatelle R, Maes D. 2007. Prevalence of ‘Candidatus Helicobacter suis’ in pigs of different ages. Vet. Rec. 161(6):189–192 [DOI] [PubMed] [Google Scholar]
- 4. Haesebrouck F, Pasmans F, Flahou B, Chiers K, Baele M, Meyns T, Decostere A, Ducatelle R. 2009. Gastric helicobacters in domestic animals and nonhuman primates and their significance for human health. Clin. Microbiol. Rev. 22:202–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Bruyne E, Flahou B, Chiers K, Meyns T, Kumar S, Vermoote M, Pasmans F, Millet S, Dewulf J, Haesebrouck F, Ducatelle R. 2012. An experimental Helicobacter suis infection causes gastritis and reduced daily weight gain in pigs. Vet. Microbiol. 160:449–454 [DOI] [PubMed] [Google Scholar]
- 6. Barbosa AJ, Silva JC, Nogueira AM, Paulino Junior E, Miranda CR. 1995. Higher incidence of Gastrospirillum sp. in swine with gastric ulcer of the pars oesophagea. Vet. Pathol. 32(2):134–139 [DOI] [PubMed] [Google Scholar]
- 7. De Groote D, Ducatelle R, van Doorn LJ, Tilmant K, Verschuuren A, Haesebrouck F. 2000. Detection of “Candidatus Helicobacter suis” in gastric samples of pigs by PCR: comparison with other invasive diagnostic techniques. J. Clin. Microbiol. 38:1131–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Roosendaal R, Vos JH, Roumen T, van Vugt R, Cattoli G, Bart A, Klaasen HL, Kuipers EJ, Vandenbroucke-Grauls CM, Kusters JG. 2000. Slaughter pigs are commonly infected by closely related but distinct gastric ulcerative lesion-inducing gastrospirilla. J. Clin. Microbiol. 38:2661–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Van den Bulck K, Decostere A, Baele M, Driessen A, Debongnie JC, Burette A, Stolte M, Ducatelle R, Haesebrouck F. 2005. Identification of non-Helicobacter pylori spiral organisms in gastric samples from humans, dogs, and cats. J. Clin. Microbiol. 43:2256–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Meining A, Kroher G, Stolte M. 1998. Animal reservoirs in the transmission of Helicobacter heilmannii. Results of a questionnaire-based study. Scand. J. Gastroenterol. 33:795–798 [DOI] [PubMed] [Google Scholar]
- 11. De Cooman LM, Pasmans F, Flahou B, Smet A, Houf K, Ducatelle R, Haesebrouck F. 2011. Detection of viable Helicobacter suis in pork by a combination of ethidium monoazide (EMA) and realtime-PCR. Helicobacter 16:142 [Google Scholar]
- 12. Baele M, Decostere A, Vandamme P, Ceelen L, Hellemans A, Mast J, Chiers K, Ducatelle R, Haesebrouck F. 2008. Isolation and characterization of Helicobacter suis sp. nov. from pig stomachs. Int. J. Syst. Evol. Microbiol. 58:1350–1358 [DOI] [PubMed] [Google Scholar]
- 13. Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95:3140–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Woo PC, Tsang AK, Wong AY, Chen H, Chu J, Lau SK, Yuen KY. 2011. Analysis of multilocus sequence typing schemes for 35 different bacteria revealed that gene loci of 10 bacteria could be replaced to improve cost-effectiveness. Diagn. Microbiol. Infect. Dis. 70:316–323 [DOI] [PubMed] [Google Scholar]
- 15. Chen KW, Chen YC, Lo HJ, Odds FC, Wang TH, Lin CY, Li SY. 2006. Multilocus sequence typing for analysis of clonality of Candida albicans strains in Taiwan. J. Clin. Microbiol. 44:2171–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diancourt L, Passet V, Verhoef J, Grimont PA, Brisse S. 2005. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43:4178–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Devi SM, Ahmed I, Francalacci P, Hussain MA, Akhter Y, Alvi A, Sechi LA, Megraud F, Ahmed N. 2007. Ancestral European roots of Helicobacter pylori in India. BMC Genomics 8:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rimbara E, Mori S, Matsui M, Suzuki S, Wachino J, Kawamura Y, Shen Z, Fox JG, Shibayama K. 2012. Molecular epidemiology analysis and antimicrobial resistance of Helicobacter cinaedi isolated from 7 hospitals in Japan. J. Clin. Microbiol. 50:2553–2560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamaoka Y. 2009. Helicobacter pylori typing as a tool for tracking human migration. Clin. Microbiol. Infect. 15:829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, Golubchik T, Harding RM, Jeffery KJ, Jolley KA, Kirton R, Peto TE, Rees G, Stoesser N, Vaughan A, Walker AS, Young BC, Wilcox M, Dingle KE. 2010. Multilocus sequence typing of Clostridium difficile. J. Clin. Microbiol. 48:770–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kilian M, Scholz CF, Lomholt HB. 2012. Multilocus sequence typing and phylogenetic analysis of Propionibacterium acnes. J. Clin. Microbiol. 50:1158–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laukkanen-Ninios R, Didelot X, Jolley KA, Morelli G, Sangal V, Kristo P, Brehony C, Imori PF, Fukushima H, Siitonen A, Tseneva G, Voskressenskaya E, Falcao JP, Korkeala H, Maiden MC, Mazzoni C, Carniel E, Skurnik M, Achtman M. 2011. Population structure of the Yersinia pseudotuberculosis complex according to multilocus sequence typing. Environ. Microbiol. 13:3114–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flahou B, Haesebrouck F, Pasmans F, D'Herde K, Driessen A, Van Deun K, Smet A, Duchateau L, Chiers K, Ducatelle R. 2010. Helicobacter suis causes severe gastric pathology in mouse and Mongolian gerbil models of human gastric disease. PLoS One 5:e14083 doi:10.1371/journal.pone.0014083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wilson K. 1994. Preparation of genomic DNA from bacteria, suppl 27 CPMB. In Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Struhl K. (ed), Current protocols in molecular biology, vol 1 John Wiley & Sons, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 25. Vermoote M, Vandekerckhove TT, Flahou B, Pasmans F, Smet A, De Groote D, Van Criekinge W, Ducatelle R, Haesebrouck F. 2011. Genome sequence of Helicobacter suis supports its role in gastric pathology. Vet. Res. 42:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eeckhaut V, Van Immerseel F, Teirlynck E, Pasmans F, Fievez V, Snauwaert C, Haesebrouck F, Ducatelle R, Louis P, Vandamme P. 2008. Butyricicoccus pullicaecorum gen. nov., sp. nov., an anaerobic, butyrate-producing bacterium isolated from the caecal content of a broiler chicken. Int. J. Syst. Evol. Microbiol. 58:2799–2802 [DOI] [PubMed] [Google Scholar]
- 26a. Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hagen F, Illnait-Zaragozi MT, Meis JF, Chew WH, Curfs-Breuker I, Mouton JW, Hoepelman AI, Spanjaard L, Verweij PE, Kampinga GA, Kuijper EJ, Boekhout T, Klaassen CH. 2012. Extensive genetic diversity within the Dutch clinical Cryptococcus neoformans population. J. Clin. Microbiol. 50:1918–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Overdevest IT, Willemsen I, Elberts S, Verhulst C, Rijnsburger M, Savelkoul P, Kluytmans JA. 2011. Evaluation of the DiversiLab typing method in a multicenter study assessing horizontal spread of highly resistant Gram-negative rods. J. Clin. Microbiol. 49:3551–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kennemann L, Didelot X, Aebischer T, Kuhn S, Drescher B, Droege M, Reinhardt R, Correa P, Meyer TF, Josenhans C, Falush D, Suerbaum S. 2011. Helicobacter pylori genome evolution during human infection. Proc. Natl. Acad. Sci. U. S. A. 108:5033–5038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Farnbacher M, Jahns T, Willrodt D, Daniel R, Haas R, Goesmann A, Kurtz S, Rieder G. 2010. Sequencing, annotation, and comparative genome analysis of the gerbil-adapted Helicobacter pylori strain B8. BMC Genomics 11:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Flahou B, Van Deun K, Pasmans F, Smet A, Volf J, Rychlik I, Ducatelle R, Haesebrouck F. 2012. The local immune response of mice after Helicobacter suis infection: strain differences and distinction with Helicobacter pylori. Vet. Res. 43:75. [DOI] [PMC free article] [PubMed] [Google Scholar]