Abstract
From November 2011 through March 2012, we surveyed 272 babies in our neonatal intensive care unit for rectal colonization with vancomycin-resistant enterococci (VRE). Using Spectra VRE medium (Remel Diagnostics, Lenexa, KS), we identified one neonate colonized with vancomycin-resistant Enterococcus faecium. In addition, 55 (13%) of the surveillance cultures yielded false-positive results with vancomycin-susceptible Enterococcus faecalis. During the same time period, 580 rectal swabs were collected from adult patients resulting in 20 (3%) false-positive cultures. The difference in false-positive rates between cultures from babies and adults was statistically significant (P < 0.001), prompting an investigation of factors that might influence the elevated false-positive rate in the neonates including patient demographics, nutrition, and topical ointments applied at the time of testing. Older neonates, with a median age of 6 weeks, were more likely to have false-positive cultures than younger neonates with a median age of 3 weeks (P < 0.001). The younger neonates receiving Similac Expert Care products were less likely to have false-positive surveillance cultures than those receiving other formulas (P < 0.001). Application of topical products was not associated with false-positive cultures. The false-positive E. faecalis strains were typed by Diversilab Rep-PCR (bioMérieux, Marcy l'Etoile, France) and found to represent eight different groups of isolates. The utility of the Spectra VRE media appeared to be significantly impacted by the age of the patients screened.
INTRODUCTION
Vancomycin-resistant enterococci (VRE) colonization of the gastrointestinal tract appears to be rare in neonates (1, 2). Most colonized neonates that develop infections have been associated with clusters and outbreaks (1, 2). Risk factors for VRE colonization in neonates include low birth weight, prematurity, and long-term antimicrobial therapy (2). Contact isolation precautions are usually implemented if VRE is detected in surveillance cultures in neonatal intensive care units (NNICU).
A variety of chromogenic agars have been developed for the qualitative detection of gastrointestinal colonization with vancomycin-resistant Enterococcus faecium and Enterococcus faecalis, including chromID VRE agar (bioMérieux Corp., Marcy l'Etoile, France), HardyChrom agar (Hardy Diagnostics, Santa Maria, CA), CHROMagar VanRE (BD, Baltimore, MD), and Spectra VRE agar (Remel Diagnostics, Lenexa, KS) (3). The purpose of the present study was to investigate false-positive Spectra VRE surveillance culture results in a NNICU population that initiated an outbreak investigation and contact isolation of a group of neonates.
MATERIALS AND METHODS
Spectra VRE surveillance cultures.
A total of 989 rectal swabs were collected from 676 adult and neonatal patients at Parkland Memorial Hospital, a 672-bed tertiary care medical center, from 9 November 2011 through 27 March 2012. The NNICU is a 90-bed unit that houses babies from birth until discharge or transfer to another Parkland unit. Weekly surveillance for VRE was initiated when vancomycin-resistant E. faecium was recovered from a neonate's urine. In January, the surveillance frequency was decreased to biweekly. Eswabs with liquid Amies transport medium (Becton Dickinson, Sparks, MD) were used to collect rectal samples. The swabs were then plated onto Spectra VRE agar (Remel) and incubated aerobically in ambient air at 35°C for 24 ± 5 h. After incubation, the plates were evaluated for suspected vancomycin-resistant E. faecium that appeared as navy blue or pink-purple colonies and vancomycin-resistant E. faecalis that appeared as light blue to blue colonies. Suspect VRE colonies were tested for pyrrolidonyl arylamidase production by disks (Key Scientific Products, Stamford, TX), and positive colonies were subcultured to sheep blood agar plates and saved frozen for later definitive identification and susceptibility testing. For the second phase of the study, suspect VRE colonies were prospectively inoculated into MicroScan Pos Gram combo panels (Siemens Healthcare Diagnostics, Inc., West Sacramento, CA) for definitive identification and susceptibility testing.
Interfering substances challenge.
Six products applied to neonates' buttocks—Sensi Care Protective Barrier, Aloe Vesta Skin Protectant, Boudreaux's Butt Paste, zinc oxide cream, nystatin powder, and nystatin cream—were evaluated for each product's ability to produce false-negative or false-positive results with Spectra VRE. Three vancomycin-resistant strains—one adult E. faecium, one adult E. faecalis, and one neonatal E. faecium—were tested. In addition, two vancomycin-susceptible strains, including a neonatal E. faecalis detected as a false positive, and E. faecalis ATCC 29212 strains were tested. Bacterial concentrations of 1.5 × 103 CFU/ml and 1.5 × 102 CFU/ml were prepared in 0.85% sterile saline from fresh subcultures; 0.1-ml portions of the diluted bacterial suspensions were added to the appropriate containers. The final concentrations in the Eswab transport media were approximately 1.5 × 102 CFU/ml and 1.5 × 101 CFU/ml. Each test was performed in duplicate. The products were applied to a sterile gloved hand or other clean disposable surface to simulate the amount used on a patient. Eswab (Becton Dickinson) were lightly rolled across the topical products and placed in the seeded transport containers. Spectra VRE plates were inoculated with the swabs and incubated overnight.
Repetitive sequence-based PCR (Rep-PCR).
Nineteen vancomycin-susceptible E. faecalis that demonstrated false-positive results on Spectra VRE agar, including 13 isolates from neonates and 6 from adults, were typed using a Diversilab Enterococcus DNA fingerprinting kit (bioMérieux). Briefly, DNA was extracted from colonies using an UltraClean microbial DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA). In accordance with the manufacturer's instructions, extracted DNA was amplified using a thermocycler and then separated, detected, and analyzed using a microfluidics DNA chip (bioMérieux) with an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA).
The relatedness was determined by cluster analysis and guidelines provided by the manufacturer. Isolates were categorized as indistinguishable, similar or different. In general, “different” was defined as <95% similarity and ≥2 band differences, “similar” was defined as <97% similarity and 1 band difference, and “indistinguishable” was defined as >95% similarity and no band differences.
Neonatal patient demographics.
Medical record review was performed to identify demographic characteristics, nutrition, and ointments in use at the time of testing. This study was exempt from the requirement for institutional review, and the privacy rule of the Health Insurance Portability and Accountability Act did not apply because the data were gathered as part of a hospital outbreak investigation.
Statistical analysis.
Group characteristics were compared between groups using a median two-sample test for continuous measures or test of proportion or Pearson chi-square test for categorical variables where appropriate. All P values in the analyses presented are two-sided and considered significant when P was ≤0.05 unless otherwise stated. The data were analyzed using the SAS version 9.2 (SAS, Inc., Cary, NC) statistical package and the WINKS Statistical Data Analysis software version 7.0 software (TexaSoft, Cedar Hill, TX).
RESULTS
Spectra VRE surveillance results.
A total of 580 fecal swabs were collected from 404 adult patients as shown in Table 1. Vancomycin-resistant E. faecium was recovered from 87 (15%) of the samples plated on Spectra VRE agar. Negative culture results were observed for 473 (82%) of the fecal swabs. False-positive results with blue or purple colonies growing on the media were seen for 20 (3%) of the cultures. The 20 isolates included 14 E. faecalis isolates with vancomycin MICs ranging from 0.5 to 4.0 μg/ml (median 2.0 μg/ml), 3 E. gallinarum isolates with vancomycin MICs of >16.0 μg/ml, and three E. durans isolates with vancomycin MICs of >16 μg/ml.
Table 1.
Comparison of the performance of Spectra VRE medium for surveillance cultures of adult and neonatal fecal samplesa
| Patient group | Total no. of tests | No. of results (%) |
||
|---|---|---|---|---|
| Positive | False positiveb | Negative | ||
| Adults | 580 | 87 (15) | 20 (3) | 473 (82) |
| Neonates | 409 | 1 (<1) | 55 (13) | 353 (86) |
Multiple lots of media were used during the study period.
Determined using a comparison of proportions for the false-positive adult and neonatal results (P < 0.001).
A total of 409 swabs from 272 neonates were plated on Spectra VRE. A single isolate of vancomycin-resistant E. faecium was recovered. Negative results were seen for 353 (86%) of the cultures. False-positive results were seen for 55 (13%) of the cultures collected from 45 patients; all isolates were identified as E. faecalis with vancomycin MICs ranging from 1.0 to 4.0 μg/ml (median, 2.0 μg/ml). False positives were observed throughout the study period on seven different lots of media.
Effect of topical products on the detection of VRE on Spectra VRE media.
Exposure of the vancomycin-susceptible strains to six topical products used on neonates did not produce false-positive results on the Spectra VRE media. However, low concentrations of vancomycin-resistant E. faecium and E. faecalis strains (1.5 × 102 CFU/ml) were inhibited by all of the products tested, with the exception of the E. faecium neonatal strain; this isolate was able to grow on Spectra VRE medium after exposure to nystatin powder. At the higher bacterial concentrations (1.5 × 103 CFU/ml), all of the VRE strains were inhibited by nystatin powder, and the adult E. faecalis strain was also inhibited by nystatin cream.
DNA fingerprinting of vancomycin-susceptible E. faecalis adult strains and neonatal E. faecalis strains demonstrating false-positive results on Spectra VRE.
The typing results are shown in Fig. 1. For the 14 isolates recovered from neonates, 8 different groups were identified: groups 1 through 5 and groups 7 through 9 with two or more band differences and <95% similarity between groups. Within group 1 (G1), G3, G5, and G8, there were multiple indistinguishable strains. Within G4, there were two similar but not indistinguishable strains.
Fig 1.
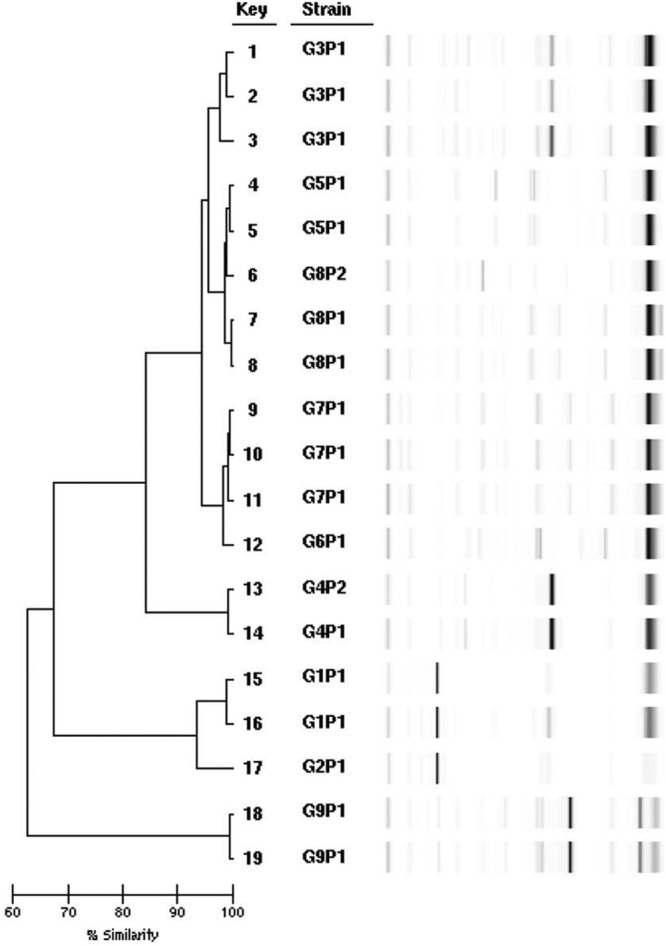
Rep-PCR dendrogram and simulated electrophoresis results for neonatal false-positive E. faecalis isolates corresponding to keys 1, 2, 4 to 8, 11, 13 to 17, and 19 and adult false-positive E. faecalis strains corresponding to keys 3, 9, 10, 12, and 18.
The five strains recovered from adults were assigned to groups 3, 6, 7, and 9. Two adult isolates placed in G7 were indistinguishable. Three adult strains were indistinguishable from neonatal isolates placed in the same groups, G3, G7, and G9.
Comparison of demographics, nutrition, and topical ointments in neonatal patients with false-positive and negative surveillance cultures.
The demographic characteristics and nutrition at the time of testing were compared between neonates with negative and false-positive surveillance cultures as shown in Table 2. The median age at testing was statistically significant between the two groups, P < 0.001; the false-positive group demonstrated a higher median age. When examining the nutrition received during the period in which testing occurred, only comparisons of the Similac Expert Care products showed statistical significance with P < 0.001; babies with false-positive results were less likely to have received these products. Babies with false-positive results were also less likely to have received breast milk. The number of chart-documented uses of topical products was too low to allow statistical comparisons.
Table 2.
Comparison of neonatal patients with false-positive and negative surveillance cultures for VRE
| Demographic characteristic | False positive (n = 55) |
Negative (n = 353) |
Pd | ||
|---|---|---|---|---|---|
| n or median | % or range | n or median | % or range | ||
| Female | 19 | 35 | 153 | 43 | 0.219 |
| Age at testing | 6 wks | 4 days to 20 wks | 3 wks | 0 days to 24 wks | <0.001* |
| Birth wt (g) | 0.683 | ||||
| < 1,000 | 5 | 9 | 47 | 13 | |
| 1,000–2000 | 22 | 40 | 134 | 38 | |
| >2,000 | 28 | 51 | 172 | 49 | |
| Preterm | 41 | 75 | 259 | 73 | 0.854 |
| Nutrition at testing | |||||
| Breast milk | 41 | 75 | 305 | 86 | 0.023* |
| Similac Special Carea | 16 | 29 | 119 | 34 | 0.498 |
| Similac Special Care 24 High Protein | 1 | 2 | 15 | 4 | 0.388 |
| Similac Expert Careb | 61 | 17 | 0.001* | ||
| Similac Expert Care Neosure | 16 | 29 | 27 | 7 | <0.001* |
| Similac Advance Early | 19 | 36 | 114 | 32 | 0.741 |
| Neocate Infant with DHA + ARAc | 1 | 2 | 3 | 1 | |
| Isomil soy | 1 | <1 | |||
| Pregestimil LIPIL | 1 | 2 | 3 | 1 | |
| Total parenteral nutrition + fat emulsion 20% infusion | 3 | 5 | 36 | 10 | 0.266 |
Includes Similac Special Care 20-cal/oz, 24-cal/oz, or 30-cal/oz formulations.
Includes Similac Expert Care 24 cal with iron or alimentum formulation.
Neocate Infant with (DHA) decosohexanoic acid plus (ARA) arachadonic acid.
*, P ≤ 0.05.
DISCUSSION
False-positive results for VRE fecal colonization were significantly higher in neonatal screening cultures than in adult cultures, 13% versus 3%, respectively, contributing to a pseudo-outbreak of colonized infants in the NNICU. As a result, the neonates were unnecessarily placed in isolation, increasing the labor and hospital costs in caring for these neonates. In addition, a recent review of the possible impacts of contact precautions on patients identified adverse outcomes, including reduced contact with clinical staff and changes in systems of care that produced delays and increases in preventable, noninfectious adverse events (4). These results suggested that there are differences between adult and neonatal fecal samples that influenced the results on the Spectra VRE agar. The published evaluations and manufacturer's published findings of the Spectra VRE agar did not segregate the results according to the ages of the patients tested (5, 6, 7, 8). This is the first report to describe differences in the performance of Spectra VRE based on the patient populations tested.
All of the isolates that produced false-positive results in neonates were vancomycin-susceptible E. faecalis. In Peterson's evaluation of Spectra VRE agar testing 399 fecal specimens, two vancomycin-sensitive E. faecalis produced light blue colonies on the media and were interpreted as false-positive results (7). The neonatal strains of E. faecalis in our study were typed to determine whether the vancomycin-susceptible strains demonstrating false-positive results were clonal. Using Rep-PCR, the DNA fingerprinting results showed that the strains were not clonal with 8 unrelated groups among 14 strains. Therefore, the false-positive results did not appear to be caused by an aberrant strain of E. faecalis.
In adults, 6 of 20 bacterial strains producing false-positive results were vancomycin-resistant E. gallinarum and E. durans. E. gallinarum has an intrinsic intermediate level of vancomycin resistance mediated by the vanC gene located on the chromosome and does not pose a risk for the dissemination of vancomycin resistance to other bacteria. The manufacturer indicated in the package insert that additional antibiotics, in combination with vancomycin, are present in the Spectra VRE medium to suppress the growth of E. gallinarum (8), although we had breakthrough colonies in our surveillance cultures. A single vancomycin-resistant E. durans strain was also recovered on Spectra VRE in the Peterson study (7). The false-positive rate for Spectra VRE at 24 h of incubation was <1% in three different studies (5, 6, 7). If the E. gallinarum and E. durans isolates were excluded from our study, the false-positive rate in adult cultures decreased to 2%, comparable to these studies.
Enterococci other than E. faecalis and E. faecium were also recovered on other chromogenic agars. In evaluations of CHROMagar VanRE, false-positive results were observed with E. gallinarum, E. casseliflavus, and E. raffinosus (9, 10). In another study, 40% of green colonies consistent with vancomycin-resistant E. faecalis on the CHROMagar VanRE were later identified as vancomycin-susceptible E. faecalis (11). Grabsch et al. also detected a high number of vancomycin-sensitive enterococci on another chromagar, chromID VRE, although the medium was incubated for 48 h (12).
The possible influence of the babies' ages at the time of testing on the surveillance cultures was examined. A statistically significant difference in the age at testing was noted between those with false-positive and negative surveillance cultures. The older infants with a median age of 6 weeks were more likely to have false-positive cultures than the younger neonates. Extended hospitalization in NNICUs with prolonged antibiotic therapy, parenteral nutrition, delayed oral feedings, and intubation seems to affect the composition of the intestinal flora (13).
Nutrition at the time of testing, which might have altered the composition of the feces, was also examined. Neonates receiving Similac Expert Care formulations were less likely to be associated with false-positive fecal cultures. The babies receiving these formulations were the youngest group of neonates. The older group of babies, with a median age of 6 weeks, who received other types of formula were more likely to have false-positive surveillance cultures. The Similac Expert Care and Advance products for older neonates were more complex, containing additional nutrients such as monoglycerides, carrageenan, galacto-oligosaccharides, and lycopene. Galacto-oligosaccharides have been shown to promote an increase in beneficial bacteria such as bifidobacteria in the gastrointestinal tracts of preterm infants (14) that could change the composition of the feces.
We also investigated the possible effects of topical products that were used on the neonates during collection of the fecal swabs. None of the ointments appeared to contribute to the false-positive culture results in the babies, although the number of documented applications was low. However, some products, such as nystatin, had an inhibitory effect on the recovery of VRE. The manufacturer evaluated miconazole as a possible interfering substance and indicated that the antifungal might reduce the recovery of VRE (8).
Findings from this pseudo-outbreak of VRE colonization in neonates suggested that verification studies from manufacturers and hospitals should include specimens from all age groups intended for inclusion in surveillance culturing to ensure that the screening medium will accurately detect the colonizing organisms in fecal specimens from each group. In addition, our findings suggested that additional identification and susceptibility testing should be performed to confirm that the pigmented colonies growing on Spectra VRE were vancomycin-resistant E. faecalis and E. faecium before reporting the results.
ACKNOWLEDGMENTS
We are grateful to the staff of the microbiology laboratory at Parkland Health and Hospital System, especially Susan Webb, for technical assistance in this investigation. We thank Cari Brown and Cheryl Lair for providing information concerning infant formulas. Finally, we thank Allan Elliot for help in the statistical analysis of the data.
Footnotes
Published ahead of print 19 December 2012
REFERENCES
- 1. Lee WG, Ahn SH, Jung MK, Jin HY, Park IJ. 2012. Characterization of a vancomycin-resistant Enterococcus faecium outbreak caused by two genetically different clones at a neonatal intensive care unit. Ann. Lab. Med. 32:82–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sherer CR, Sprague BM, Campos JM, Nambiar S, Temple R, Short B, Singh N. 2005. Characterizing vancomycin-resistant enterococci in neonatal intensive care unit. Emerg. Infect. Dis. 11:1470–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rank EL. 2012. Chromogenic agar media in the clinical, food, and environmental testing arenas, part 1. Clin. Microbiol. Newsl. 34:43–47 [Google Scholar]
- 4. Morgan DJ, Diekema DJ, Sephowitz K, Perenocevich EN. 2009. Adverse outcomes associated with contact precautions: a review of the literature. Am. J. Infect. Control. 37:85–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jenkins SG, Raskoshina L, Schuetz AN. 2011. Comparison of performance of the novel chromogenic Spectra VRE agar to that of bile esculin azide and Campylobacter agars for detection of vancomycin-resistant enterococci in fecal samples. J. Clin. Microbiol. 49:3947–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nguyen TDH, Evans KD, Goh RA, Tan GL, Peterson EM. 2012. Comparison of medium, temperature, and length of incubation for detection of vancomycin-resistant enterococcus. J. Clin. Microbiol. 50:2503–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peterson JF, Doern CD, Kallstrom G, Riebe KM, Sander T, Dunne WM, Jr, Ledeboer NA. 2010. Evaluation of Spectra VRE, a new chromogenic agar medium designed to screen for vancomycin-resistant Enterococcus faecalis and Enterococcus faecium. J. Clin. Microbiol. 48:4627–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Remel Diagnostics 2011. Spectra™ VRE package insert. Remel Diagnostics, Lenexa, KS [Google Scholar]
- 9. Kallstrom G, Doern CD, Dunn WM., Jr 2010. Evaluation of a chromogenic agar under development to screen VRE colonization. J. Clin. Microbiol. 48:999–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peltroche-Llacsahuanga H, Top J, Weber-Heynemann J, Lütticken R, Haase G. 2009. Comparison of two chromogenic media for selective isolation of vancomycin-resistant enterococci from stool specimens. J. Clin. Microbiol. 47:4113–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stamper PD, Shulder S, Bekalo P, Manandhar D, Ross TL, Speser S, Kingery J, Carroll KC. 2010. Evaluation of BBL CHROMagar VanRE for detection of vancomycin-resistant enterococci in rectal swab specimens. J. Clin. Microbiol. 48:4294–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grabsch EA, Ghaly-Derias S, Gao W, Howden BP. 2008. Comparative study of selective chromogenic (chromID VRE) and bile esculin agars for isolation and identification of vanB-containing vancomycin-resistant enterococci from feces and rectal swabs. J. Clin. Microbiol. 46:4034–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fanaro S, Chierici R, Guerrini P, Vigi V. 2003. Intestinal microflora in early infancy: composition and development. Acta Pediatr. Suppl. 91:48–55 [DOI] [PubMed] [Google Scholar]
- 14. Boehm G, Lidestri M, Casetta P, Jelinek J, Negretti F, Stahl S, Martini A. 2002. Supplementation of a bovine milk formula with an oligosaccharide mixture increases counts of faecal bifidobacteria in preterm infants. Arch. Dis. Child Fetal Neonatal ed. 86:F178–F181 [DOI] [PMC free article] [PubMed] [Google Scholar]


