Abstract
We describe biophysical and ultrastructural differences in genome release from adeno-associated virus (AAV) capsids packaging wild-type DNA, recombinant single-stranded DNA (ssDNA), or dimeric, self-complementary DNA (scDNA) genomes. Atomic force microscopy and electron microscopy (EM) revealed that AAV particles release packaged genomes and undergo marked changes in capsid morphology upon heating in physiological buffer (pH 7.2). When different AAV capsids packaging ss/scDNA varying in length from 72 to 123% of wild-type DNA (3.4 to 5.8 kb) were incrementally heated, the proportion of uncoated AAV capsids decreased with genome length as observed by EM. Genome release was further characterized by a fluorimetric assay, which demonstrated that acidic pH and high osmotic pressure suppress genome release from AAV particles. In addition, fluorimetric analysis corroborated an inverse correlation between packaged genome length and the temperature needed to induce uncoating. Surprisingly, scAAV vectors required significantly higher temperatures to uncoat than their ssDNA-packaging counterparts. However, externalization of VP1 N termini appears to be unaffected by packaged genome length or self-complementarity. Further analysis by tungsten-shadowing EM revealed striking differences in the morphologies of ssDNA and scDNA genomes upon release from intact capsids. Computational modeling and molecular dynamics simulations suggest that the unusual thermal stability of scAAV vectors might arise from partial base pairing and optimal organization of packaged scDNA. Our work further defines the biophysical mechanisms underlying adeno-associated virus uncoating and genome release.
INTRODUCTION
Adeno-associated virus (AAV) is a small (25 nm) nonenveloped virus belonging to the family Parvoviridae and genus Dependovirus. The AAV capsid packages a single-stranded (ssDNA) genome approximately 4.7 kb in length (1). The wild-type genome consists of two open reading frames flanked by two inverted terminal hairpin repeats (ITRs). The ITRs, which are 145 nucleotides each, are the only cis element in the AAV genome required for successful packaging (2, 3). The AAV capsid is composed of 60 (T = 1) viral protein subunits VP1, VP2, and VP3, in approximately the ratio 1:1:10. The three different subunits are generated from overlapping reading frames and interact within the capsid through the common VP3 subunit region. The largest VP1 subunit is known to possess a phospholipase A2 domain required for infectivity (4). Because of its broad tropism, lack of pathogenicity, and flexibility in genome content, AAV has become a promising candidate for therapeutic gene transfer applications. In the past 2 decades, AAV has been utilized as a gene transfer vector in a number of phase I and phase II clinical trials treating various genetic diseases (5).
Different AAV serotypes infect cells by engaging a variety of cell surface glycans and coreceptors, followed by endocytic uptake (4, 6, 7). Viral particles are then thought to escape from the endosome and translocate to the nucleus, where the ssDNA genome is released and undergoes second-strand synthesis. Engineered AAV genomes containing a mutant 3′ ITR have been shown to package dimeric, self-complementary DNA (scDNA) (8). Such scAAV vectors have the advantage of escaping ssDNA degradation (9) and bypassing second-strand synthesis, which is a rate-limiting step preceding transgene expression by AAV vectors (10, 11). These features have been shown to enable rapid onset of transgene expression by scAAV vectors.
The packaging capacity of ssAAV and scAAV vectors has been extensively studied (12–17). However, little is known about the consequences of packaging subgenomic-length DNA or self-complementary genomes for AAV capsid uncoating. Previously, studies with minute virus of mice (MVM) packaging subgenomic-length DNA have demonstrated that such defective particles do not release their genomes in vitro (18). Along with the observation that the MVM genome reinforces and increases the stiffness of MVM capsids (19), these studies suggest that the viral genome exerts an internal pressure on the walls of the capsid, which is critical for proper capsid uncoating. Thermal analysis has previously been used to understand large conformational changes in AAV capsids such as exposure of the buried phospholipase A2 domain during infection (18, 20–22). In the current study, we utilized atomic force microscopy (AFM), electron microscopy (EM), and fluorescence-based assays to characterize the biophysical and ultrastructural properties of different AAV vectors and the impact of thermal stimuli on capsid uncoating/genome release. In addition, we utilized computational modeling and molecular dynamics (MD) to further understand the potential organization and architecture of DNA packaged within ssAAV and scAAV vectors.
MATERIALS AND METHODS
Chemicals and reagents.
Mica was purchased (Ted Pella Inc.) and cleaved immediately prior to use. Four-hundred-mesh copper Formvar carbon-coated TEM grids (Ted Pella Inc.) were made hydrophilic by glow discharge immediately before use. Uranyl acetate (Ted Pella Inc.) was used as received. Uranyl acetate solutions were centrifuged for several minutes prior to use to remove any precipitate or aggregates. SYBR gold (Invitrogen Life Sciences) was first diluted in water immediately prior to use. Tris-HCl and spermidine (ThermoFisher) were utilized as received.
Virus production and purification.
All viral vectors were generated at the UNC Vector Core by iodixanol gradient ultracentrifugation followed by ion-exchange chromatography and vector genome titers determined by dot blot assay as well as verified by quantitative PCR (qPCR) as described previously (23). Single-stranded AAV serotype 2 vectors packaging different transgene cassettes were as follows: wild-type AAV genome containing rep and cap genes (4.7 kb) (24), chicken β-actin (CBA) promoter-driven firefly luciferase (4.1 kb) (25), CBA promoter-driven tdTomato (3.8 kb) (25), cytomegalovirus (CMV) promoter-driven firefly luciferase (3.6 kb) (26), CMV promoter-driven green fluorescent protein (GFP) (3.4 kb) (23), and an EF1a promoter-driven mCherry with an internal ribosome entry site (IRES) signal followed by WGA-Cre (5.3 kb) (27). Self-complementary AAV2 vectors utilized in the study were as follows: CMV promoter-driven green fluorescent protein with lambda phage genome stuffer DNA (5.0 kb) (23), CMV promoter-driven green fluorescent protein (4.6 kb) (28), and CBA hybrid promoter-driven green fluorescent protein (4.1 kb) (28).
AFM.
Freshly cleaved mica was treated with 50 μl of polylysine (50 μg/ml) for 2 h at 37°C in a humid chamber. Mica was then washed three times with distilled water (dH2O) and dried. Viral particles (1 × 1010 vector genomes [vg]) in 1× phosphate-buffered saline (PBS) were heated in a thermocycler for 30 min to 37 or 65°C, followed by immediate chilling to 4°C. AAV was then deposited on the treated mica surface (10 μl) in a humid chamber for 45 min at room temperature. Mica pieces were then washed three times with dH2O and gently air dried. Atomic force microscopy (AFM) images were acquired using an Asylum MFP3D atomic force microscope. All images were collected in air using the tapping mode. Images were then processed using the MFP3D software.
TEM.
Viral particles (2 × 1010 vg) in 1× PBS were heated in a thermocycler for 30 min to 37, 55, or 65°C, followed by immediate chilling to 4°C. Vectors were then adsorbed onto 400-μm mesh carbon-coated copper transmission electron microscopy (TEM) grids for 2 min. After washing twice with 0.2-μm-filtered dH2O, the grids were stained with freshly prepared 2% uranyl acetate for 30 s. After drying, grids were imaged using a Zeiss LEO 910 transmission electron microscope. Ten to 20 images of each grid were captured in random locations to get an accurate sampling of the viral particles. Each image was then analyzed using ImageJ by counting the number of full particles based on their distinct morphologies and intensity of uranyl acetate staining. Particle counts for each virus and experiment ranged from 395 to 5,317 total particles. In all cases, the average and standard deviation for each temperature were determined using the percentage of full particles from each image. Empty particles determined by their differential staining (29) were excluded from counting due to the presence of fragmented capsids and diverse morphology.
Fluorometric assay for detection of thermally induced genome release.
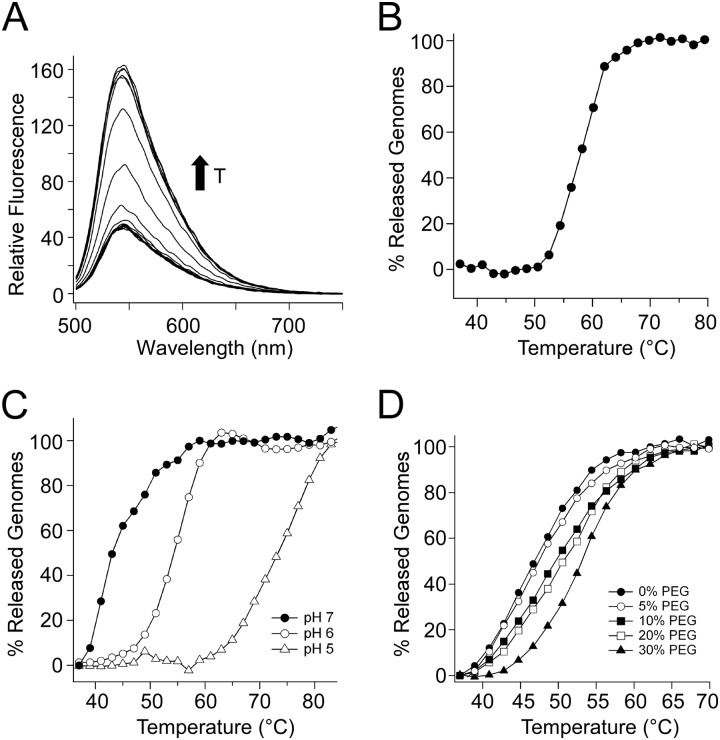
Viral vectors (1 × 1010vg) in 1× PBS with 25 μM SYBR gold were heated using a Roche LightCycler 480. Samples were heated in a stepwise manner (2°C steps) from 37°C to 95°C. Viruses were held at each temperature for 5 min prior to measuring fluorescence. Each sample was subjected to thermal treatment in triplicate. In addition to a dye-only control, each virus was heated to 95°C prior to addition of SYBR gold to determine whether any reversible fluorescence changes originated from melting of DNA secondary structures. Data were normalized to baseline spectra obtained prior to and after complete thermal transition to obtain the ratio of uncoated particles to intact particles. Melting temperature (Tm) values were defined as the temperature at which uncoated and intact viral particles were at 50% each (uncoated particle ratio = 0.5). For subjecting samples to increasing osmotic pressure, wild-type AAV in PBS (pH 7.2) was supplemented with 5%, 10%, 20%, or 30% polyethylene glycol (PEG) 8000 (Sigma). For pH studies, samples at pH 5 and pH 6 were prepared by dialyzing wild-type AAV in citrate buffer (pH 5 or pH 6) supplemented with 137 mM NaCl. Samples at pH 7.2 were maintained in PBS. Error bars represent the standard deviation (n = 3). All AAV vectors were obtained from the UNC Vector Core, and the size/nature of packaged genomes (ssDNA or scDNA) was blinded to the authors during these experiments.
Immunoblotting (dot blotting) of heat-treated AAV particles.
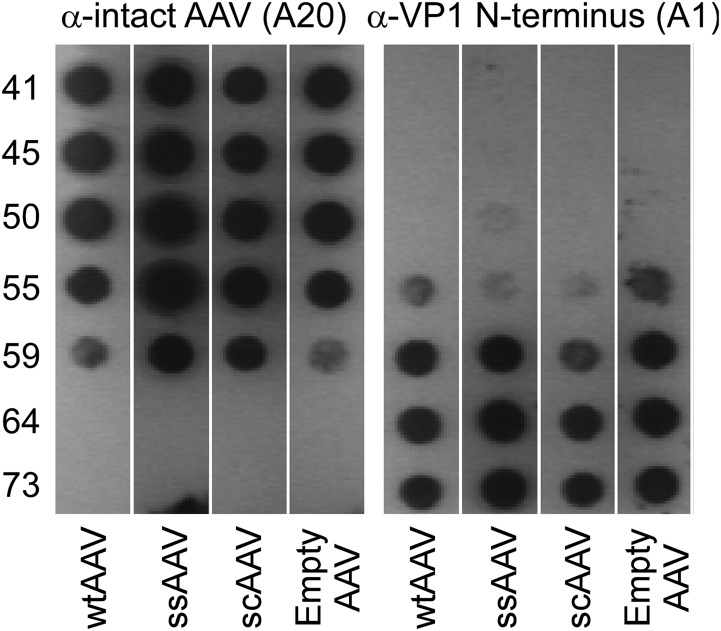
Using a thermocycler, AAV packaging different transgenes was subjected to heat treatment. Wild-type AAV, empty AAV capsids, and AAV packaging single-stranded and self-complementary CMV-GFP genomes were heated to temperatures ranging from 40 to 73°C for 30 min prior to rapid cooling to 4°C (5 × 109 vg in 200 μl per well). Each virus was heated in duplicate and applied to two separate dot blots. Samples were loaded onto two nitrocellulose membranes in a dot blot apparatus. Membranes were blocked using 5% dehydrated milk in 1× Tris-buffered saline (TBS)–0.1% Tween 20 (TBS-T) for 1 h. The monoclonal mouse antibody A20 recognizes a conformational epitope on the intact capsid, while the monoclonal mouse antibody A1 recognizes a linear epitope in the N terminus of the VP1 capsid protein (30). Membranes were soaked with primary antibodies (1:30 in 2% dehydrated milk in TBS-T) for 1 h. Horseradish peroxidase (HRP)-conjugated goat anti-mouse polyclonal antibody (1:5,000) was then bound to the primary antibody for 1 h, followed by 4 washes using TBS-T and detection of signal using the West Femto chemiluminescence detection kit (ThermoFisher Scientific).
Tungsten-shadowing electron microscopy.
AAV vectors packaging ssDNA and scDNA cassettes containing cytomegalovirus (CMV) promoter-driven green fluorescent protein (GFP) (3 × 109 vg) were diluted in 10 mM Tris-HCl (pH 7.5) to 1 × 1012 vg/ml. Dilutions were heated to 65°C for 5 min and then rapidly cooled on ice. Ten-microliter aliquots of these dilutions were then further diluted 1:3 in 10 mM Tris-HCl (pH 7.5), followed by 1:1 dilution with a buffer containing 4 mM spermidine, and mounted to charged-carbon-foil grids as published previously (31, 32). Carbon grids were washed in water, dehydrated in a series of ethanol washes, air dried, and rotary shadow cast with tungsten. Samples were visualized on a Tecnai 12 transmission electron microscope at 40 kV. All microscopy images were captured using a Gatan Ultrascan 4000 charge-coupled device (CCD) camera and supporting software (Gatan Inc.).
DNA and capsid modeling.
Following the report by Locker and Harvey (33), the AAV genome was coarsely modeled by an elastic bead-spring polymer chain model, where each bead represented six nucleotide pairs. The molecular mechanics potential function used in this model was of the form
| (1) |
The stretching (Eri) and (Eθi) bending terms were modeled as harmonic oscillators of the form
| (2) |
where the equilibrium bond distances (α = r, ri being the distance between monomers i and i + 1) and angles (α = θ, θi being the angle between monomers i, i + 1, and i + 2) and their spring constants were chosen to match the structure and properties of ssDNA and double-stranded DNA (dsDNA) under physiological conditions. Specifically, as described previously (33), we used statistical distributions of bond lengths in experimentally observed structures of DNA to derive stretching parameters of kr = 3.49 kcal/mol/Å2 and r0 = 20 Å. Parameters for the bending term were chosen to match the persistence lengths and elastic properties of dsDNA and ssDNA observed experimentally, yielding values of θ0 = 180°, kθ = 14.6 kcal/mol/rad2 for stiff chains representing double-stranded DNA, and kθ = 0.75 kcal/mol/rad2 for flexible chains representing single-stranded DNA. The exclusion term (Edij) gives the chain volume and prevents self-crossing using a shifted, repulsive-only Lennard-Jones function (equation 3):
| (3) |
where dij is the distance between monomers i and j, ε = 15 kcal/mol, σ = 22.27 Å, and d0, the effective chain diameter, was chosen to be 25 Å, consistent with observed values of interaction distances in packed dsDNA viral genomes (34). No torsional restraints were included in the force field, and electrostatic and other long-range nonbonded interactions were also excluded in this simplified model. Chains of 350 monomers (representing 2,100 bp) were generated through a Monte Carlo random walk, with initial angles assigned from a Boltzmann distribution using the bending energy function for flexible chains given above and torsion angles randomly chosen from a uniform distribution.
In this model, the icosahedral capsid was represented by a smooth sphere with a diameter of 216 Å (Rc = 108 Å), chosen to match the internal volume of the actual virus based on inspection of the X-ray crystal structure of the AAV-2 capsid (Protein Data Bank [PDB] no. 1LP3) (35). The capsid restraint term was modeled as in equation 4 as a purely repulsive semiharmonic potential:
| (4) |
where Ri is the distance of monomer i from the center of the capsid and kc = 8.8 kcal/mol/Å2.
MD simulations.
Molecular dynamics (MD) simulations were used to predict the organization and energies of encapsidated single-stranded and self-cDNA inside the AAV capsid. All simulations were carried out using the LAMMPS MD simulation package (36) with a time step of 500 fs and at a temperature of 300 K maintained by a Nosé-Hoover thermostat (37).
Models of virus particles encapsidating ssDNA genomes were created by placing randomly generated flexible chains representing ssDNA inside spherical semiharmonic potential boundaries of the form given in equation 4 and gradually reducing the confining radius, Rc, from an initial value of 2,000 Å to 254 Å in 1-Å decrements, equilibrating the DNA chain for 5 ns at each step. At a bounding radius of 254 Å, the DNA chain occupied 5% of the volume of the confining sphere, and it was allowed at this point to equilibrate over 50 ns of MD. The confining sphere was then shrunk further in 1-Å steps, following the same protocol of equilibrating for 5 ns after each 1-Å decrement and subsequently stopping to equilibrate for 50 ns at steps where the volume fraction of the DNA chain was a multiple of 5%. This was continued until the target Rc of 108 Å (containing 65% DNA by volume fraction) was reached.
Following confinement, the encapsidated ssDNA chains were converted step by step into dsDNA chains in order to simulate in situ base pairing of two adjacent ssDNA chains nucleated at the central ITR, as might occur in a self-complementary AAV vector. To accomplish this, the small spring constants of the bending terms in the flexible ssDNA chains (kθ = 0.75 kcal/mol/rad2) were converted, one angle at a time, to the larger value (kθ = 14.6 kcal/mol/rad2) characteristic of the stiffer dsDNA chains. The chains were allowed to equilibrate for 50 ns after each angle was stiffened, and the pressure (defined as the average total force exerted by the genome on the capsid walls divided by the surface area of the capsid) was calculated at intervals of 10% DNA base paired. Capsid pressure data were collected by using the radial positions of the monomers at each time step during the final 10 ns of the MD run to compute the average force exerted by the capsid restraining term (equation 4) and dividing by the surface area of the spherical capsid.
RESULTS
Thermally induced DNA release is dependent on genome length.
AAV vectors have been previously shown to undergo structural transitions in response to limited heating (20–22). We adapted this approach to determine the impact of thermal stimuli on encapsidated DNA. Atomic force microscopy (AFM) images of scAAV vectors heated to 65°C demonstrated release of AAV genomes from intact capsids (Fig. 1A), similar to the case for other parvoviruses such as MVM (18). Further, we utilized transmission electron microscopy (TEM) to investigate whether the heated capsids contained DNA (full) or not (empty). Capsids which are intact and contain an encapsidated genome appear as 25-nm opaque/white spheres (Fig. 1B, panel i). In contrast, capsids which have released their packaged DNA are permeable to uranyl acetate and therefore appear as 25-nm rings (Fig. 1B, panels ii to v). In addition, as shown in Fig. 1B (panels vi to x), several capsid fragments and other morphologies were observed upon heating.
Fig 1.
Ultrastructural characterization of AAV capsid uncoating. (A) Atomic force microscopy images of scAAV (sc-CMV-GFP) vectors heated to 37°C and 65°C. Viral particles are observed as 25-nm spheres. After heating to 65°C, viral genomes are detected both associated with viral particles and as free DNA (arrowheads). (B) Observed morphologies of heat treated AAV. Genome-containing AAV particles are impermeable to uranyl acetate (i), while empty AAV particles that have released DNA exhibit a ring-like architecture (ii). Genome release resulted in diverse morphologies characterized by small gaps in the capsid wall (iii to v) as well as capsid fragments forming multimeric aggregates (vi to x). The scale bar represents 20 nm.
Both ssDNA- and scDNA-packaging AAV capsids incubated at three different temperatures (37, 55, and 65°C) were then visualized by TEM to quantify the populations of full and empty particles (Fig. 2). At 55°C and 65°C, we observed an increase in the number of empty particles compared to that at 37°C. Three different ssAAV vectors with genomes ranging from 72% (3.4 kb) to 100% (4.7 kb) of the wild-type genome length were characterized (Fig. 2A). After counting the numbers of full and empty particles, we observed a trend wherein AAV capsid stability appears to increase as genome size is reduced (Fig. 3A). Notably, when heated to 55°C, wild-type AAV (4.7 kb) has only 40% intact capsids remaining, while the 3.4-kb ssAAV vector is unaffected, with nearly 100% intact capsids. Similarly, at 65°C, the wild-type AAV sample has ∼16% intact capsids remaining, while the shorter, 3.4-kb ssAAV vector still has 75% intact capsids. This trend is also seen in case of scAAV vectors, where the smallest of the three tested genomes (4.1 kb) has 79% intact particles at 65°C, while the largest of the three (5.0 kb) has only 9% intact capsids remaining at 65°C (Fig. 2B and 3B).
Fig 2.
Effect of packaged genome length on AAV capsid uncoating. ssAAV (A) and scAAV (B) vectors were heated to different temperatures for 30 min prior to TEM imaging. Full AAV virions are viewed as 25-nm solid spheres, and empty virions are 25-nm donut-like structures. Differences in the relative amount of empty particles are apparent for different vector genome lengths. Genome lengths shown represent single-stranded DNA lengths (e.g., sc CMV-GFP is 2 × 2,058 bases + 3 × ITRs = 4,551 bases). Images are representative of 10 to 20 captured images. Scale bars are 50 nm.
Fig 3.
Quantitative analysis of TEM images. Thermally induced uncoating efficiency was assessed for intact-genome-containing (full) AAV particles packaging ssDNA (A) and scDNA (B) genomes of different length. Vectors were heated to 37, 55, or 65°C and visualized with TEM. Averages are taken from 10 to 20 images with particle counts for each sample ranging from 395 to 5,317 total particles. In all cases the average and standard deviation at each temperature was determined using the percentage of full particles from each image. Smaller capsid fragments (<50% of the shell) were excluded from counting. Genome lengths shown represent single-stranded DNA lengths. All values at 55°C and 65°C were determined to be significantly different from those at 37°C (P < 0.001 by a two-tailed Student t test).
scAAV vectors packaging a similar number of nucleotides are more thermostable than ssAAV vectors.
To explore the phenomenon of AAV genome release further, we developed a more sensitive, fluorescence-based thermal melt assay. Briefly, while impermeable to large fluorophores in the native state, heated AAV capsids would release encapsidated DNA that is free to interact with SYBR gold. This dye is known to exhibit >1,000-fold enhancement in fluorescence upon binding ss/dsDNA, with an excitation maximum of ∼495 nm and emission maximum of ∼537 nm (38). To ensure that genome release is not rate limiting, the virus is held at each temperature for 5 min prior to reading the fluorescent signal. No large differences in fluorescence were observed between 5-min and 30-min heating periods (data not shown). At a critical temperature, we observed a sharp change in the fluorescence emission spectrum of AAV capsids packaging full-length genomes (Fig. 4A). When fluorescence intensities at the emission maxima were plotted as a function of temperature, we observed a sigmoidal profile with a characteristic thermal transition temperature (Fig. 4B). To further explore the applicability of this fluorimetric assay, we carried out similar studies evaluating thermally induced AAV genome release as a function of pH and osmotic pressure. As seen in Fig. 4C and D, AAV capsids appear to resist thermally induced genome release at lower pH and high external osmotic pressure. Next, we explored the phenomenon that self-complementary AAV (scAAV) vectors appeared to be more thermostable than ssAAV in earlier EM studies (Fig. 2). Representative plots of three different AAV vectors packaging ssDNA genomes of different lengths are shown in Fig. 5A. The transition temperature is defined as the temperature required for 50% of the AAV particles to release their preencapsidated genomes. As outlined above, since genome release is irreversible, this measurement can be used to obtain a pseudo-Tm (abbreviated as Tm) rather than a thermodynamically determined melting temperature. It is also important to note that although AAV virions are noted as full or empty in these low-resolution EM studies, it is possible that uncoated capsids and associated genomes exist in multiple states. Such a scenario is supported by previous studies demonstrating the existence of different packaged AAV genome states based on high-resolution cryo-EM studies (39).
Fig 4.
Fluorimetric detection of AAV genome release under different conditions. (A) In the presence of SYBR gold (25 μM), AAV capsids were heated from 37°C to 95°C. At the point of genome release, an increase in fluorescence is observed at 550 nm (excitation, 495 nm), arising from the interaction between SYBR gold and the AAV genome. (B) When plotted and normalized to pretransition and posttransition baselines, a relatively sharp transition is observed in fluorescence at the emission wavelength. (C and D) Encapsidation stability is increased with decreasing pH (C) as well as increasing osmotic pressure (D). All experiments were repeated in triplicate.
Fig 5.

Fluorimetric analysis of AAV capsid uncoating as a function of genome length and self-complementarity. (A) Viral vectors were heated in 2°C increments and held for 5 min at each temperature prior to acquiring fluorescent signal. Three representative curves for different vector genome lengths are shown. Each vector was heated in triplicate along with a premelted control, which did not show a sharp transition. Curves were normalized to pre- and posttransition baselines to yield the percentage of released genomes. (B) Comparison of ssAAV, scAAV, and oversized ssAAV vectors shows an inverse linear correlation of Tm with vector genome size.
Thermal transitions for different ssAAV vectors acquired from the UNC vector core were then plotted as a function of genome length (Fig. 5B). A linear correlation is observed, where Tm is inversely proportional to genome length. These data correlate well with the TEM data shown above. A similar trend is seen with the panel of self-complementary AAV vectors (it should be noted that genome length is shown as total single-strand length in Fig. 5). We also evaluated the thermal profile of AAV vectors packaging oversized ss/scDNA genomes. As mentioned above, the packaging capacity of AAV vectors has been studied extensively. Although numerous reports have demonstrated successful transduction with vectors packaging oversized AAV genomes, the latter have been shown to be fragmented into subgenomic-length DNA (12, 14, 15, 17). Therefore, it is not surprising that ssAAV vectors packaging a 5.8-kb genome exhibit a Tm similar to that of vectors packaging a smaller genome (3.4 kb). Surprisingly, scAAV vectors appear to be more thermally stable than ssAAV vectors in general. In addition, the thermal stability of scAAV vectors appears to be less sensitive to vector genome length than that of ssAAV vectors, as determined by the lower slope in the linear correlation (Fig. 5B).
Heat-induced exposure of VP1 N termini is not dependent on genome size or self-complementarity.
While the current studies are focused primarily on the effects of thermal stimuli on AAV genome release, heat-induced exposure of AAV VP1 N termini has been reported by several groups (21, 22). Using a thermocycler, wild-type AAV, empty AAV capsids, and AAV packaging single-stranded or self-complementary CMV-GFP genomes were heated to temperatures ranging from 40 to 73°C. Samples were then subject to immunoblot analysis using the monoclonal mouse antibody A20, which recognizes a conformational epitope on the intact capsid, and the monoclonal mouse antibody A1, which recognizes a linear epitope in the N terminus of the VP1 capsid protein (30). As observed in Fig. 6, A20 staining is abrogated upon heating to temperatures of >60°C, indicating breakdown of intact capsids. Further, robust A1 staining is observed at 60°C or higher, consistent with earlier studies by other groups. Interestingly, no significant difference was observed between samples regardless of full or empty particles or ssDNA or scDNA genomes. These results suggest that internal capsid pressure exerted by packaged AAV genomes might not play a direct role in externalization of VP1 N termini.
Fig 6.
Exposure of the VP1 N termini is not dependent on genome length or self-complementarity. AAV capsids (5 × 109 vg/well) containing either the wild type (wtAAV), single-stranded CMV-GFP (ssAAV), self-complementary CMV-GFP (scAAV), or no genome at all were heated in a PCR mixture for 30 min. The monoclonal antibodies A20 and A1 were then used to probe the AAV capsid integrity as well as the state of exposure of the buried VP1 N-terminal domain required for infection. No significant difference was observed between genomes with different sizes or self-complementarity.
Tungsten-shadowing EM shows DNA secondary structure for dsDNA vectors.
The intriguing difference in the thermal properties of ssAAV and scAAV vectors suggests that the nature of secondary structures formed within encapsidated viral DNA might affect capsid uncoating. To investigate this phenomenon further, we visualized AAV DNA after DNA release using a combination of transmission electron microscopy and tungsten shadowing (Fig. 7). After heating the ssDNA-packaging AAV vector to 65°C, the released viral genome was observed to collapse upon itself. This is likely due to kinetically driven base pairing within the ssDNA genome. In contrast, the scAAV genome clearly forms a dsDNA structure following release at 65°C. Furthermore, the mutated ITR located in the middle of the AAV genome is clearly observed. The thickness of the DNA in the images was consistent with the width of duplex DNA following coating with tungsten as measured in other studies (31). Consistent with previous reports for other parvoviruses, the majority of viral DNA is observed associated with the capsid. These observations also suggest that a significant level of base pairing may exist in scAAV genomes prior to genome release.
Fig 7.
Tungsten-shadowing EM of released genomes from ssCMV-GFP (A and C) and scCMV-GFP (B and D) vectors. Viral particles were heated to 65°C and then imaged using tungsten-shadowing EM. Grids showed a combination of AAV virions shown as spheres, loose DNA, and virion-associated DNA. Scale bars represent 50 nm. Insets are cartoon representations of the predicted secondary structure.
Molecular dynamics provides insight into genome organization within the capsid.
The persistence length of dsDNA is 50 nm (∼147 bp), which corresponds to a minimum dsDNA ring size of approximately 16 nm in diameter (40). With a capsid shell that has an ∼22-nm inner diameter, one would expect that packaging 2.3-kb dsDNA into the AAV capsid would exert a large amount of internal pressure within the capsid shell (41). It is therefore counterintuitive that scAAV vectors are more thermally stable than ssAAV vectors. Additionally, the observation of dsDNA in electron micrographs after heating suggests that cDNA base pairing could occur within the capsid. We therefore used molecular dynamics simulations to gather insight into the nature of the DNA within the capsid.
In this model, the capsid is represented as a hollow sphere. Packaged DNA is represented as an elastic bead polymer chain of varying flexibility (high for ssDNA and low for dsDNA). Spheres containing ssDNA chains were reduced in diameter from 200 nm to 21.6 nm while performing molecular dynamics to sample conformational space and achieve thermal equilibrium before determining the capsid pressure, measured by the average total force exerted by the genome on the capsid walls divided by the surface area of the capsid. Encapsidated ssDNA chains were modified to have increasing degrees of double-stranded character by increasing the stiffness of the angle constraints between monomers, one at a time, simulating base pairing along an scAAV genome.
Counterintuitively, we find that as the angle stiffness is increased step by step along the confined chain, the pressure exerted by the chain on the capsid walls decreases until about 50% of the genome has been base paired, reaching a capsid pressure of 5.2 ± 0.1 atm (Fig. 8A). Further base pairing causes the capsid internal pressure to rise up to a maximum of 8.3 ± 0.7 atm. Visualizing the MD trajectory of the base pairing simulation provides insight into this phenomenon (Fig. 8B). Before base pairing begins, the ssDNA is largely disordered (blue licorice in Fig. 8B). The small bending energy penalty leaves the beads at the outer surface free to vibrate (due to thermal energy) against the capsid walls, creating pressure. As the chain is base paired by increasing the stiffness sequentially along the length of the chain, the stiffened segment attempts to reduce its bending energy by moving away from the capsid center, forming concentric rings near the surface of the containing sphere (red beads in Fig. 8B, panel ii). Unlike the flexible ssDNA chain, these dsDNA rings are not able to vibrate freely due to their increased stiffness, and thus they exert less pressure on the walls. As the degree of base pairing approaches 50%, the surface of the DNA sphere is almost completely covered by these stiff concentric rings, which stack to form a tight shell around the core of flexible ssDNA, damping their vibrations against the capsid walls (Fig. 8B, panel iii). As base pairing continues along the chain, a second inner layer of concentric rings begins to form, which pushes the outer layer further toward the capsid wall (Fig. 8B, panels iv and v). As the degree of base pairing increases, the force from this inner layer transmitted through the outer shell to the capsid increases, causing the capsid pressure to rise.
Fig 8.
Computational modeling of internal capsid pressure and genome organization. (A) In molecular dynamics simulations, the stiffness of modeled ssDNA chains was increased step by step along the chain to simulate DNA base pairing in scAAV genomes. Pressure exerted on the capsid walls decreases until 50% of the chain has been converted, after which the capsid pressure increases. Error bars represent standard deviations (n = 5). Cartoon representations of predicted secondary structures of AAV genomes at different levels of base-pairing are shown above the curve. (B) Molecular dynamics simulation of progressive change in the organization of flexible ssDNA chains (cyan licorice) while being base paired into stiff dsDNA chains (red beads). Representative snapshots of chain organization within the model capsid at 0% (i), 25% (ii), 50% (iii), 75% (iv), and 100% (v) conversion of ssDNA to dsDNA are shown. dsDNA segments are seen to migrate to the periphery and adjacent to the inner capsid surface, where they may buffer the thermal motion of the flexible ssDNA chain and reduce the pressure exerted on the inner capsid walls during the initial stages of the conversion process. The development of an inner coil of dsDNA after 50% conversion may subsequently contribute to increasing capsid pressures.
We note that several simplifications have been made in constructing the model of the scAAV genome, most notably the omission of electrostatic interaction terms from the force field. Simulations of DNA packaging in bacteriophages in which electrostatics were included showed that DNA-DNA repulsions accounted for up to 51% of the total free energy cost of packaging, suggesting that these forces dominate when the genome is highly confined (42). However, we also note that the genome conformations resulting from these simulations were topologically indistinguishable from those in similar systems where electrostatics was not accounted for (43). The electrostatic interaction energy, although a significant component of the energetics of packaging, depends primarily on the total DNA density and is largely agnostic to conformational details, which are dictated by the elastic bending terms in the force field. Since DNA bending is accurately represented in our model, we believe that, even in the absence of electrostatics, our simplified model correctly predicts the qualitative topology of the scAAV genome and its influence on the measured capsid pressure.
DISCUSSION
Parvoviruses such as minute virus of mice (MVM) have been shown to release their ssDNA genomes in the 3′-to-5′ direction, while leaving the empty virus shell intact (18). Our studies indicate that AAV capsids respond in a similar manner to thermal stimuli. We observed different capsid fragments and morphologies that suggest that empty AAV shells might subsequently unravel from the site of genome release, presumably by loss of the viral proteins comprising the virion shell. Such capsid disassembly intermediates are not observed upon heating purified empty AAV particles (data not shown), which suggests that capsid disassembly might not precede genome release. Interestingly, heat-induced exposure of AAV VP1 N termini has also been reported by several groups (21, 22). In the current study, both VP1 exposure and genome release were observed to occur upon heating capsids to approximately 60°C or higher (Fig. 4 and 6). Paradoxically, we observed that heat-induced exposure of VP1 N termini does not vary significantly between wild-type AAV, empty AAV virion shells, and representative ssAAV or scAAV vectors (Fig. 6). Based on these results, one possible scenario is that parvoviral VP1 exposure and genome release might be structurally distinct events with a certain degree of temporal overlap, as proposed earlier (18). It is important to note that despite our experiments providing insight into biophysical aspects of AAV capsid uncoating, their biological implications remain to be established. Nevertheless, the results underscore the importance of understanding capsid structural transitions that precede VP1 exposure and genome release as well as spatiotemporal aspects of such events in the AAV infectious pathway.
Using a combination of electron microscopy and fluorimetric assays, we observed a trend wherein subgenomic AAV particles require more thermal energy to release their genomes, regardless of whether they are packaging ssDNA or scDNA. In contrast as genome sizes approach wild-type AAV genome length (∼4.7 kb), the capsids appear to acquire an optimal, metastable stable state that can efficiently uncoat. Packaged DNA genomes within bacteriophages can generate pressures of close to 50 atm (41, 44, 45), requiring only minimal stimuli for DNA ejection, but these pressures rapidly drop off as the length of phage DNA is decreased to 78% of wild type (41). One might therefore expect that AAV vector genomes of different subgenomic size would greatly change the internal pressure of the capsid. Thus, it is likely that subgenomic AAV particles are defective in capsid uncoating and genome release within the host cell. Accordingly, incorporation of stuffer DNA and packaging of near-wild-type genomes into AAV capsids might improve infectivity. Further, we also observed that acidic pH and external osmotic pressures exerted by ∼30% PEG 8000 solutions inhibited AAV capsid genome release. One possible explanation for the stabilizing effect of low pH on AAV capsids might be increased interactions between packaged DNA and positively charged histidine side chains within the capsid interior. Additionally, we surmise that the observed increase in capsid stability and genome retention at acidic pH may be beneficial from a physiological standpoint. Enhanced capsid stability within endosomal/lysosomal compartments during intracellular trafficking could help protect viral DNA from degradation by nucleases prior to release into the nucleus. The osmotic pressure effect is corroborated by earlier studies with bacteriophage capsids, wherein genome ejection was observed to be inhibited by pressures comparable to that of the cytoplasm of bacteria (46). Another interesting observation was that AAV vectors packaging genomes larger than wild type exhibit stability equivalent to that of subgenomic particles. This result can be explained by earlier studies demonstrating that packaged genomes for oversized AAV vectors are truncated (12, 14, 15, 17).
Lastly, the observation that scAAV vectors are more thermally stable than ssAAV vectors was unexpected. Since dsDNA is inherently stiffer than ssDNA, one would expect packaging a fully base-paired dsDNA genome into an AAV capsid to be energetically unfavorable. Our results from molecular modeling studies support the notion that internal pressures associated with capsids packaging dsDNA genomes could indeed be higher than those associated with ssDNA-packaging vectors. However, closer analysis of the modeling data revealed that a hybrid AAV genome comprised of ssDNA and dsDNA (50% base pairing) would exert an internal capsid pressure that is lower than that with 100% ssDNA alone. These theoretical predictions are supported indirectly by tungsten-shadowing EM studies, wherein we observed predominantly dsDNA character in released scAAV genomes. In contrast, the ssAAV genome was observed to form a disordered structure consistent with folding of DNA into a local thermodynamic minimum. Therefore, it is tempting to speculate that partially base-paired scAAV genomes might nucleate rapid annealing upon release to form a complete dsDNA structure in the host cell. Although these results suggest that scAAV vectors might be more resistant to uncoating than ssAAV vectors, this apparent defect is likely offset by bypassing second-strand synthesis, which results in rapid onset of transgene expression. In summary, these studies provide a path forward toward characterization of AAV capsid uncoating and genome release in a more relevant physiological setting. When integrated with intracellular trafficking studies and high-resolution structural data, it is possible that the results described here might provide a more complete picture of AAV infection and possibly guide the design of optimal AAV vector genomes for gene therapy applications.
ACKNOWLEDGMENTS
We thank Josh Grieger for providing different AAV vectors for analysis.
This work was supported by NIH grants HL089221 (awarded to A.A.), GM70785 (awarded to S.C.H.), and GM31819 and CA19014 (awarded to J.D.G).
Footnotes
Published ahead of print 26 December 2012
REFERENCES
- 1. Bowles DE, Rabinowitz JE, Samulski RJ. 2006. The genus Dependovirus, p 15–24 In Kerr JR, Cotmore SF, Bloom ME, Linden RM, Parrish CR. (ed), Parvoviruses. Hodder Arnold Publishers, London, United Kingdom [Google Scholar]
- 2. Samulski RJ, Srivastava A, Berns KI, Muzyczka N. 1983. Rescue of adeno-associated virus from recombinant plasmids: gene correction within the terminal repeats of AAV. Cell 33:135–143 [DOI] [PubMed] [Google Scholar]
- 3. Xiao X, Xiao W, Li J, Samulski RJ. 1997. A novel 165-base-pair terminal repeat sequence is the sole cis requirement for the adeno-associated virus life cycle. J. Virol. 71:941–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Agbandje-McKenna M, Kleinschmidt J. 2011. AAV capsid structure and cell interactions. Methods Mol. Biol. 807:47–92 [DOI] [PubMed] [Google Scholar]
- 5. Mingozzi F, High KA. 2011. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 12:341–355 [DOI] [PubMed] [Google Scholar]
- 6. Ding W, Zhang L, Yan Z, Engelhardt JF. 2005. Intracellular trafficking of adeno-associated viral vectors. Gene Ther. 12:873–880 [DOI] [PubMed] [Google Scholar]
- 7. Nonnenmacher M, Weber T. 2012. Intracellular transport of recombinant adeno-associated virus vectors. Gene Ther. 19:649–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McCarty DM. 2008. Self-complementary AAV vectors; advances and applications. Mol. Ther. 16:1648–1656 [DOI] [PubMed] [Google Scholar]
- 9. Hauck B, Zhao W, High K, Xiao W. 2004. Intracellular viral processing, not single-stranded DNA accumulation, is crucial for recombinant adeno-associated virus transduction. J. Virol. 78:13678–13686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ferrari FK, Samulski T, Shenk T, Samulski RJ. 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 70:3227–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fisher KJ, Gao GP, Weitzman MD, DeMatteo R, Burda JF, Wilson JM. 1996. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J. Virol. 70:520–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dong B, Nakai H, Xiao W. 2010. Characterization of genome integrity for oversized recombinant AAV vector. Mol. Ther. 18:87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grieger JC, Samulski RJ. 2005. Packaging capacity of adeno-associated virus serotypes: impact of larger genomes on infectivity and postentry steps. J. Virol. 79:9933–9944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hirsch ML, Agbandje-McKenna M, Samulski RJ. 2010. Little vector, big gene transduction: fragmented genome reassembly of adeno-associated virus. Mol. Ther. 18:6–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lai Y, Yue Y, Duan D. 2010. Evidence for the failure of adeno-associated virus serotype 5 to package a viral genome > or = 8.2 kb. Mol. Ther. 18:75–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu J, Zhao W, Zhong L, Han Z, Li B, Ma W, Weigel-Kelley KA, Warrington KH, Srivastava A. 2007. Self-complementary recombinant adeno-associated viral vectors: packaging capacity and the role of rep proteins in vector purity. Hum. Gene Ther. 18:171–182 [DOI] [PubMed] [Google Scholar]
- 17. Wu Z, Yang H, Colosi P. 2010. Effect of genome size on AAV vector packaging. Mol. Ther. 18:80–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cotmore SF, Hafenstein S, Tattersall P. 2010. Depletion of virion-associated divalent cations induces parvovirus minute virus of mice to eject its genome in a 3′-to-5′ direction from an otherwise intact viral particle. J. Virol. 84:1945–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carrasco C, Carreira A, Schaap IA, Serena PA, Gomez-Herrero J, Mateu MG, de Pablo PJ. 2006. DNA-mediated anisotropic mechanical reinforcement of a virus. Proc. Natl. Acad. Sci. U. S. A. 103:13706–13711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Horowitz ED, Finn MG, Asokan A. 2012. Tyrosine cross-linking reveals interfacial dynamics in adeno-associated viral capsids during infection. ACS Chem. Biol. 7:1059–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Johnson JS, Li C, DiPrimio N, Weinberg MS, McCown TJ, Samulski RJ. 2010. Mutagenesis of adeno-associated virus type 2 capsid protein VP1 uncovers new roles for basic amino acids in trafficking and cell-specific transduction. J. Virol. 84:8888–8902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kronenberg S, Bottcher B, von der Lieth CW, Bleker S, Kleinschmidt JA. 2005. A conformational change in the adeno-associated virus type 2 capsid leads to the exposure of hidden VP1 N termini. J. Virol. 79:5296–5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grieger JC, Choi VW, Samulski RJ. 2006. Production and characterization of adeno-associated viral vectors. Nat. Protoc. 1:1412–1428 [DOI] [PubMed] [Google Scholar]
- 24. Samulski RJ, Chang LS, Shenk T. 1989. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J. Virol. 63:3822–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pulicherla N, Shen S, Yadav S, Debbink K, Govindasamy L, Agbandje-McKenna M, Asokan A. 2011. Engineering liver-detargeted AAV9 vectors for cardiac and musculoskeletal gene transfer. Mol. Ther. 19:1070–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. DiPrimio N, Asokan A, Govindasamy L, Agbandje-McKenna M, Samulski RJ. 2008. Surface loop dynamics in adeno-associated virus capsid assembly. J. Virol. 82:5178–5189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, Diester I, Goshen I, Thompson KR, Deisseroth K. 2010. Molecular and cellular approaches for diversifying and extending optogenetics. Cell 141:154–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gray SJ, Foti SB, Schwartz JW, Bachaboina L, Taylor-Blake B, Coleman J, Ehlers MD, Zylka MJ, McCown TJ, Samulski RJ. 2011. Optimizing promoters for recombinant adeno-associated virus-mediated gene expression in the peripheral and central nervous system using self-complementary vectors. Hum. Gene Ther. 22:1143–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Torikai K, Ito M, Jordan LE, Mayor HD. 1970. Properties of light particles produced during growth of type 4 adeno-associated satellite virus. J. Virol. 6:363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wobus CE, Hugle-Dorr B, Girod A, Petersen G, Hallek M, Kleinschmidt JA. 2000. Monoclonal antibodies against the adeno-associated virus type 2 (AAV-2) capsid: epitope mapping and identification of capsid domains involved in AAV-2-cell interaction and neutralization of AAV-2 infection. J. Virol. 74:9281–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Compton SA, Ozgur S, Griffith JD. 2010. Ring-shaped Rad51 paralog protein complexes bind Holliday junctions and replication forks as visualized by electron microscopy. J. Biol. Chem. 285:13349–13356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Griffith JD, Lee S, Wang YH. 1997. Visualizing nucleic acids and their complexes using electron microscopy. Curr. Opin. Struct. Biol. 7:362–366 [DOI] [PubMed] [Google Scholar]
- 33. Locker CR, Harvey SC. 2006. A model for viral genome packing. Multiscale Modeling Simulation 5:1264–1279 [Google Scholar]
- 34. Cerritelli ME, Cheng N, Rosenberg AH, McPherson CE, Booy FP, Steven AC. 1997. Encapsidated conformation of bacteriophage T7 DNA. Cell 91:271–280 [DOI] [PubMed] [Google Scholar]
- 35. Xie Q, Bu W, Bhatia S, Hare J, Somasundaram T, Azzi A, Chapman MS. 2002. The atomic structure of adeno-associated virus (AAV-2), a vector for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 99:10405–10410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Plimpton S. 1995. Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 117:1–19 [Google Scholar]
- 37. Nosé S. 1984. A molecular dynamics method for simulations in the canonical ensemble. Mol. Phys. 52:255–268 [Google Scholar]
- 38. Tuma RS, Beaudet MP, Jin X, Jones LJ, Cheung CY, Yue S, Singer VL. 1999. Characterization of SYBR gold nucleic acid gel stain: a dye optimized for use with 300-nm ultraviolet transilluminators. Anal. Biochem. 268:278–288 [DOI] [PubMed] [Google Scholar]
- 39. Gerlach B, Kleinschmidt JA, Bottcher B. 2011. Conformational changes in adeno-associated virus type 1 induced by genome packaging. J. Mol. Biol. 409:427–438 [DOI] [PubMed] [Google Scholar]
- 40. Theodorakopoulos N, Peyrard M. 2012. Base pair openings and temperature dependence of DNA flexibility. Phys. Rev. Lett. 108:078104. doi:10.1103/PhysRevLett.108.078104 [DOI] [PubMed] [Google Scholar]
- 41. Ivanovska I, Wuite G, Jonsson B, Evilevitch A. 2007. Internal DNA pressure modifies stability of WT phage. Proc. Natl. Acad. Sci. U. S. A. 104:9603–9608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Petrov AS, Harvey SC. 2008. Packaging double-helical DNA into viral capsids: structures, forces, and energetics. Biophys. J. 95:497–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Locker CR, Fuller SD, Harvey SC. 2007. DNA organization and thermodynamics during viral packing. Biophys. J. 93:2861–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Purohit PK, Inamdar MM, Grayson PD, Squires TM, Kondev J, Phillips R. 2005. Forces during bacteriophage DNA packaging and ejection. Biophys. J. 88:851–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. 2001. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature 413:748–752 [DOI] [PubMed] [Google Scholar]
- 46. Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. 2003. Osmotic pressure inhibition of DNA ejection from phage. Proc. Natl. Acad. Sci. U. S. A. 100:9292–9295 [DOI] [PMC free article] [PubMed] [Google Scholar]