Abstract
Open reading frame 2 (ORF2) of the feline calicivirus (FCV) genome encodes a capsid precursor that is posttranslationally processed to release the mature capsid protein (VP1) and a small protein of 124 amino acids, designated the leader of the capsid (LC). To investigate the role of the LC protein in the virus life cycle, mutations and deletions were introduced into the LC coding region of an infectious FCV cDNA clone. Three cysteine residues that are conserved among all vesivirus LC sequences were found to be critical for the recovery of FCV with a characteristic cytopathic effect in feline kidney cells. A cell-rounding phenotype associated with the transient expression of wild-type and mutagenized forms of the LC correlated with the cytopathic and growth properties of the corresponding engineered viruses. The host cellular protein annexin A2 was identified as a binding partner of the LC protein, consistent with a role for the LC in mediating host cell interactions that alter the integrity of the cell and enable virus spread.
INTRODUCTION
Members of the family Caliciviridae are small nonenveloped viruses that contain a positive-sense single-stranded RNA genome. Feline calicivirus (FCV) is in the genus Vesivirus of the family and has been an important model for studying calicivirus replication because it grows efficiently in cell culture and has a reverse genetics system (1–5). The RNA genomes of caliciviruses range in size from ∼6.7 to 8.5 kb and typically encode 8 or 9 viral proteins from two (Sapovirus, Nebovirus, and Lagovirus) or three (Norovirus and Vesivirus) open reading frames (ORFs). A unique ORF4 in the murine norovirus genome that encodes a protein reported to downregulate the innate immune response has been identified (6). The 5′ end of the viral RNA genome is covalently linked to a VPg protein; the 3′ end of the genome is polyadenylated (2, 7–9). The infectious virion is composed of 180 copies of the major capsid protein VP1 and 1 to 10 copies of the minor structural protein VP2, which together form a capsid around the VPg-linked genome (10–15). The genomic RNA serves as a template for translation of the ORF1 polyprotein that is proteolytically cleaved by the viral protease to release the nonstructural proteins (16, 17), and an ∼2.2- to 2.6-kb subgenomic bicistronic RNA template is used for the translation of VP1 and VP2 (7, 18). A genetic feature unique to members of the genus Vesivirus is expression of the major capsid protein from ORF2 as a precursor protein (5, 18–20). This precursor is processed in trans by the viral protease to release two proteins: the leader of the capsid (LC) and the mature capsid protein (VP1) (5, 19, 21, 22). The function of the LC protein is not clear, but cleavage of the precursor to release LC and VP1 is essential for the recovery of infectious virions (5). Transient expression of the LC was reported to enhance replication of a human norovirus RNA replicon (23), and an increase in the level of mRNA for the low-density lipoprotein receptor (LDLR) was observed (24). We showed previously that the FCV LC can tolerate the insertion of foreign proteins such as green fluorescent protein and DsRed between amino acids 88 and 89, and recombinant viruses expressing fluorescent markers were used to visualize a calicivirus infection in real time (1).
In this study, we used sequence comparisons, transient-expression experiments, and reverse genetics to investigate the role of the LC protein in the vesivirus life cycle. The FCV LC was shown to be critical in the production of virus with characteristic cytopathic effect (CPE) and in the spread in feline kidney cell monolayers, and key amino acid residues involved in this activity were mapped. Evidence was found for an interaction of the LC protein with cellular annexin A2, a protein reported to be involved in the life cycle of other positive-sense single-stranded RNA viruses.
MATERIALS AND METHODS
Viruses and cells.
Feline calicivirus strain vR6, derived from the infectious cDNA clone of the Urbana strain designated pR6, was described previously (4) and is designated the wild-type (wt) virus in this study. Crandell-Rees feline kidney (CRFK) cells were grown in maintenance medium that contained Dulbecco's modified Eagle's medium (Lonza Inc., Allendale, NJ) with added penicillin (250 U/ml; Mediatec Inc., Manassas, VA), streptomycin (250 μg/ml; Mediatec Inc.), and l-glutamine (2 mM; Mediatec Inc.) and was supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen Inc., Carlsbad, CA).
Bioinformatic analysis of LC sequences.
Eighty-eight LC sequences of viruses in the genus Vesivirus from the GenBank database were used for alignment in the program ClustalX 2.1 (25). To address the diversity in nucleotide sequences and gene lengths, the program GeneDoc was used to optimize the alignment (26).
A Bayesian phylogenetic tree was inferred using the software program MrBayes 3.2 (27). The parameters employed included the general time-reversible (GTR) model with a gamma distribution of substitution rates. Convergence was achieved after 12 million generations. The first 25% of the trees were excluded as burn-in, and tree topologies were calculated from the consensus of the remaining tree samples. The tree was displayed using FigTree software (28).
The amino acid identities between the sequences included in the analysis were calculated using the pairwise distances algorithm of the MEGA5 program. The amino acid pairwise distances (p distances) were plotted on the x axis, and the frequencies were represented on the y axis. The cutoff value for differentiating between lineages was defined as the percentage value that best discriminated between the intralineage and interlineage distances.
Construction of recombinant full-length FCV cDNA clones.
Standard recombinant DNA methods were employed to generate recombinant FCV full-length (FL) clones, as described previously (1, 4). To introduce a unique KpnI cleavage site into the 5′ end of the FCV VP1 sequence (downstream of the LC and VP1 border), the FL clone pR6 (4) was modified with a QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA), using the primer pair 5′-CTGCCCCAGAGCAAGGtACcGTGGTTGGAGGAG (designated Urb-VP1-KpnI_F) and 5′-CTCCTCCAACCACgGTaCCTTGCTCTGGGGCAG (designated Urb-VP1-KpnI_R). The sequences of the primers corresponded to nucleotides (nt) 5705 to 5735 of the FCV genome and included two synonymous nucleotide changes (nucleotide changes are indicated by lowercase letters) to introduce a unique KpnI site (underlined). A clone with the engineered KpnI site was identified by sequence analysis and the resulting plasmid, pR6*, was used in the construction of FL FCV clones with truncated or chimeric LC protein coding sequences.
To construct FL clones with consecutive in-frame deletions extending from the 5′ and 3′ ends of the LC region, sense and antisense primers were used to amplify DNA fragments from the plasmid pR6*. Purified DNA fragments were digested with BstBI and KpnI and ligated into the BstBI-KpnI-linearized pR6* vector, replacing the LC coding sequence with truncated LC sequences. Clones with the desired truncations were screened by PCR and confirmed by sequence analysis, and the resulting plasmids were designated pR6*-LC-ΔA–B, where A–B corresponds to the span of LC amino acids deleted in a particular construct.
To generate a recombinant FL clone that expressed the LC as a fusion protein with unique tags, PCR amplification was performed with a pCI-NV-VP1 clone (29) as the template to produce a DNA fragment encoding a FLAG tag and a 22-amino-acid sequence of the Norwalk virus VP1 that is recognized by the monoclonal antibody NV10 (our unpublished data). The fragment was bordered by an amino acid linker sequence (GGS) and the PmeI and AflII restriction enzyme sites. The DNA fragment was purified, digested with PmeI and AflII, and ligated into the PmeI-AflII-linearized pR6-LC vector. Clones with the desired mutation were screened by sequence analysis, and the resulting plasmid was designated pR6-LC-NV10 (see Fig. 8A).
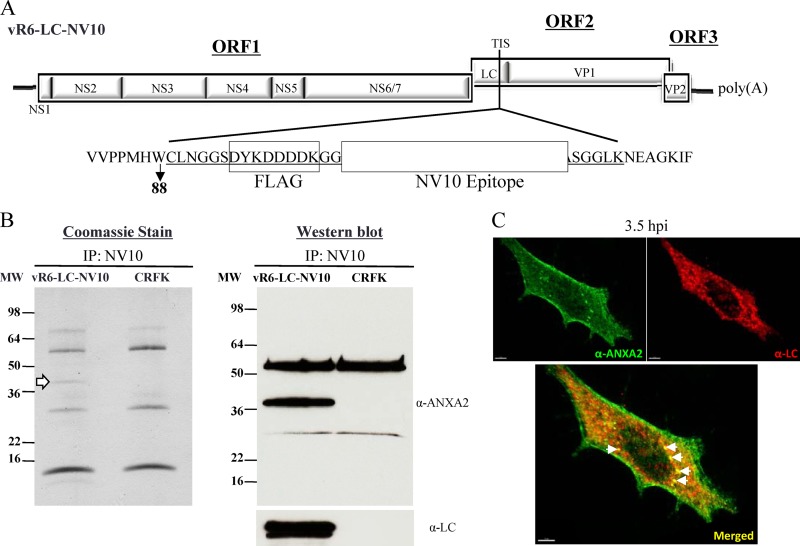
Fig 8.
Coimmunoprecipitation of recombinant LC from infected CRFK cells. (A) A recombinant FL FCV clone in which two unique tags were introduced into the LC coding sequence at the TIS (between amino acids 88 and 89). The entire heterologous sequence inserted into the LC is underlined, and the two unique tags are boxed and labeled. FLAG refers to the commercially available FLAG tag, and the white box corresponds to the NV10 epitope of the Norwalk virus VP1 capsid protein, as described in Materials and Methods. (B) Coomassie blue stain and Western blot analyses of protein samples immunoprecipitated (IP) with the NV10 monoclonal antibody from vR6-LC-NV10- or mock-infected CRFK lysates separated by SDS-PAGE. The Coomassie blue stain revealed a band that was consistently observed at ∼36 kDa, as indicated by the white arrow. The Western blot was first probed with an anti-ANXA2 monoclonal antibody, stripped, and then reprobed with anti-LC hyperimmune sera. (C) Confocal images of CRFK cells infected with vR6 (MOI, 10). Cells were fixed 3.5 hpi. Immunofluorescence analysis was performed to detect ANXA2 (green) and LC (red). The bottom panel shows a merged image in which a yellow signal indicates colocalization of ANXA2 and LC. White arrows indicate sites of colocalization of ANXA2 and LC.
To generate recombinant clones that contained an entire or partial heterologous LC, San Miguel sea lion virus type 5 (SMSV-5) Hom-1 or mink calicivirus 9 LC sequences were PCR amplified and cloned into the pR6* vector using the unique BstBI and KpnI restriction sites (see Fig. 9A). A cDNA consensus clone of SMSV-5 Hom-1 was used as the starting template for the SMSV LC clone (Sosnovtsev et al., unpublished data), and cDNA generated from mink calicivirus 9 RNA (30) was the starting template for the mink calicivirus LC clone.
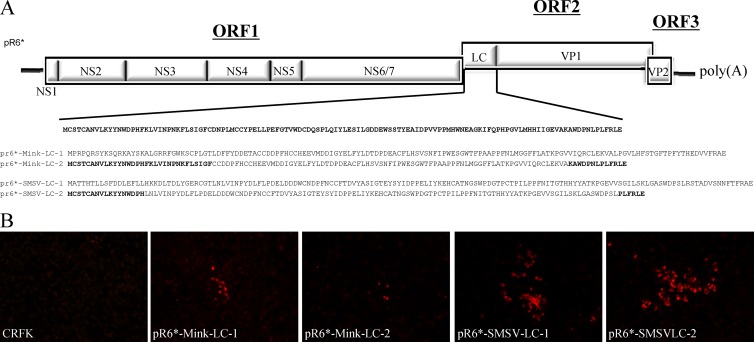
Fig 9.
FL LC-chimeric FCV clones. (A) Recombinant FL FCV clones in which the FCV LC was replaced by either the entire or partial SMSV-5 Hom-1 or mink calicivirus 9 LC sequence. (B) Fluorescence microscopy analysis of capsid expression in CRFK cells at P1 from the various recombinant FCV mutants.
To introduce alanine substitutions into the LC sequence of the FL clone, a QuikChange XL site-directed mutagenesis kit (Stratagene) was used with sense and antisense primers and pR6 as a template. The clones were screened by sequence analysis, and those that were positive for the substitution were selected as templates for PCR amplification of a region that encompassed the unique BstBI and SpeI restrictions sites (upstream and downstream, respectively, of the LC coding sequence). The purified DNA fragments were then digested with BstBI and SpeI and ligated into the BstBI-SpeI-linearized pR6 vector. The reconstructed clones were screened by sequence analysis, and plasmids containing the desired substitutions were selected and designated pR6-LC-X#, where X corresponds to the amino acid residue present in the wt Urbana strain, # indicates the amino acid position within the LC coding sequence, and Z corresponds to the acquired or engineered amino acid substitution at that position. FL Urbana clones with the compensatory mutations were generated using the same strategy of primer mutagenesis with the QuikChange mutagenesis kit (Stratagene). The sequences of the oligonucleotides used in this study are available upon request.
Recovery of virus.
Virus was recovered from plasmid DNA with the modified vaccinia virus Ankara (MVA)-T7 expression system, as described previously (16). Briefly, confluent CRFK cell monolayers in 6-well plates (approximately 2 × 106 cells) were infected with MVA-T7 (a gift from Bernard Moss) at a multiplicity of infection (MOI) of 10 and incubated for 1 h at 37°C. The supernatant was removed, and 2 ml of maintenance medium was added to the cells. Transfections were carried out using PolyJet DNA in vitro transfection reagent (SignaGen, Rockville, MD) and prepared according to instructions from the manufacturer. Following incubation for 24 to 48 h at 37°C, medium from the transfected cell monolayer was transferred to a fresh cell monolayer, and cells were monitored for the development of viral CPE, which appeared as cell rounding and detachment of cells from the surface.
To confirm that recovered virus originated from the engineered constructs, reverse transcriptase (RT)-PCR products derived from viral RNA were analyzed by direct sequencing. RNA was purified using an RNeasy minikit (Qiagen, Valencia, CA). To control for the presence of DNA from the original plasmid used to synthesize the RNA for transfection, PCR was performed on the isolated viral RNA in the absence of RT. For sequencing analyses of the full-length genomes of passage 9 (P9) of vR6-LC-C40A and P3 of pR6-LC-C40A,S29P,Y41C, the 5′ ends were amplified with an Invitrogen 5′ RACE system for rapid amplification of cDNA ends and then sequenced.
Plaque assays.
CRFK cells were seeded onto 6-well plates, and upon confluence, the monolayers were infected with serial dilutions of virus prepared in Dulbecco's modified Eagle's medium. Plates were incubated for 60 min (at 37°C in 5% CO2) with gentle agitation every 15 min. The inocula were removed, and monolayers were overlaid with 2 ml maintenance medium containing 1% agarose (Invitrogen). The plates were then incubated for approximately 48 h at 37°C in a humidified 5% CO2 incubator. Cells were fixed with 10% formaldehyde, and the agarose overlay was removed. The fixed cells were stained with 1% (wt/vol) crystal violet solution and then washed.
Plaque size was calculated using the software program GraphClick (Arizona Software). Statistical analysis of the differences in plaque sizes was performed using Prism software from GraphPad (La Jolla, CA).
pCI plasmid construction.
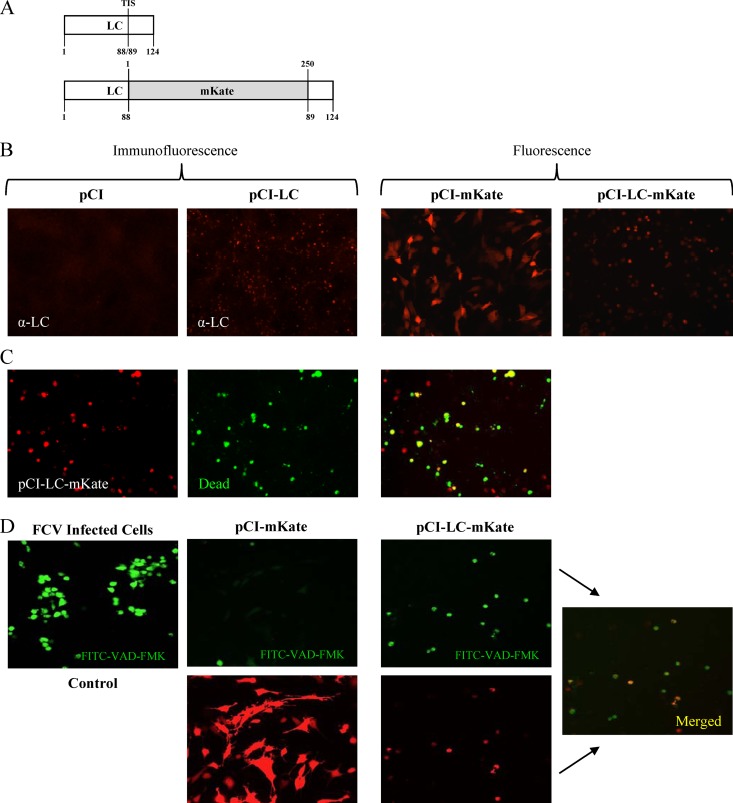
To construct eukaryotic expression clones for transient-expression experiments, the LC sequence was cloned into the pCI expression vector (Promega, Madison, WI). The LC region was PCR amplified using pR6 as a template, with primers that introduced a SalI restriction site at the 5′ end and a stop codon followed by a NotI restriction site at the 3′ end. DNA fragments were purified, digested with SalI and NotI, and ligated into a SalI-NotI-linearized pCI vector. The clones were screened by sequence analysis, and a plasmid containing the correct sequence was selected and designated pCI-LC (see Fig. 5).
Fig 5.
Analysis of transient expression of the LC in CRFK cells. (A) Schematic representations of the wild-type LC protein and the recombinant LC-mKate fusion protein. The transposon insertion site (TIS) is indicated for the wild-type LC, and the location of the insertion of the red fluorescent protein mKate is shown. (B) Immunofluorescence and fluorescence analyses of CRFK cells transfected with the indicated plasmids. The LC was detected using hyperimmune sera, and expression of mKate and LC-mKate was visualized in live cells. (C) Representative images of CRFK cells transfected with pCI-LC-mKate and treated with the Live/Dead reagent (Invitrogen) that stains dead cells green. (D) Fluorescence microscopy of CRFK cells transfected with either pCI-mKate or pCI-LC-mKate and treated with FITC-VAD-FMK. FCV-infected cells were used as a positive control for apoptosis induction.
pCI-LC clones that contained the red fluorescent protein mKate2 fused to the LC were generated in two steps. First, the mKate2 coding sequence was PCR amplified using the pmKate2-N plasmid as a template (Evrogen, Moscow, Russia) to include the bordering KpnI and AflII restriction sites and a linker sequence (GGSGGS). Then, the PCR-amplified mKate2 was digested, purified, and ligated into the LC transposon insertion site (TIS) of the pR6-LC FL clone between amino acids (aa) 88 and 89, previously identified to tolerate insertions without abrogating the recovery of virus (1). Individual colonies were screened by PCR to identify plasmids that contained the insert. Colonies positive for the insert were grown overnight and confirmed by sequence analysis. A plasmid containing the correct sequence was selected and designated pR6-LC-mK. The LC-mKate fusion protein was amplified using pR6-LC-mK as a template, with primers that introduced a SalI restriction site at the 5′ end and a stop codon followed by a NotI restriction site at the 3′ end. DNA fragments were purified, digested with SalI and NotI, and ligated into a SalI-NotI-linearized pCI vector. The clones were screened by sequence analysis, and a plasmid containing the correct sequence was selected and designated pCI-LC-mK.
Transient-expression assays.
Transfections were performed with the PolyJet DNA in vitro transfection reagent (SignaGen) according to the instructions from the manufacturer. The viability of transfected cells was assessed with the Invitrogen Live/Dead fixable dead cell stain kit.
Detection of activated caspases in cells expressing LC.
Activated caspases were detected as described previously (31). Briefly, CRFK cells transfected with pCI-mKate or pCI-LC-mKate were incubated with a 10 μM fluorescein isothiocyanate-Val-Ala-Asp-fluoromethylketone (FITC-VAD-FMK) in situ marker (Promega) for 20 min at 37°C. The cells were then washed with phosphate-buffered saline and directly analyzed by fluorescence microscopy.
Microscopy analysis.
Fluorescence and immunofluorescence images were visualized using a Leica DMI4000 B microscope (Leica Microsystems, Buffalo Grove, IL), and images were captured with a QImaging Retiga-2000R camera (Surrey, BC, Canada). Images were processed using iVision 4.0.14 software (BioVision, Exton, PA). Confocal microscopy images were obtained with a Leica TCS-SP5 confocal microscope (Leica Microsystems) equipped with a white laser and using a 63× 1.4-numerical-aperture oil immersion objective.
Immunoprecipitation assays.
Confluent CRFK cells (∼1 × 107) were infected with vR6-LC-NV10 at an MOI of 5. Four hours postinfection (hpi), infected and mock-infected cells were disrupted with lysis buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris HCl [pH 8.0]). Samples were mixed and incubated on ice for 30 min. The cell lysates were clarified by centrifugation at 10,000 × g for 10 min at 4°C. Clarified lysates were incubated with 1 μg of biotinylated NV10 antibody (our unpublished data) for 30 min at 4°C. Prewashed magnetic MyOne streptavidin T1 Dynabeads (250 μg) (Invitrogen) were then added to the samples and incubated at 4°C for 30 min. Recombinant LC was purified using a magnetic rack to isolate the magnetic beads, and the bead-protein complexes were washed four times with lysis buffer. SDS-PAGE sample buffer (Invitrogen) was added to release the protein complexes from the beads, which were then analyzed by SDS-PAGE. Bands of interest were excised from the gel and submitted for mass spectrometry analysis. Data processing and databank searching were performed with PD 1.2 and Mascot software (Matrix Science, Beachwood, OH). The data were searched against protein sequences deposited in the National Center for Biotechnology Information nonredundant protein database (NCBInr, April 2011) and a reverse-sequence decoy database. Proteins were identified using a false-discovery rate cutoff of 1% and a minimum of two peptides per protein.
Western blot analysis.
Cell lysates were heated at 95°C for 5 min in 1× Tris-glycine SDS sample buffer (Invitrogen) containing 5% β-mercaptoethanol. Western blot analysis was performed using 4 to 20% Tris-glycine SDS-PAGE gels (Lonza). Proteins were transferred to nitrocellulose membranes by dry blotting using an iBlot apparatus (Invitrogen), and membranes were incubated with guinea pig anti-FCV virion serum, rabbit anti-LC serum (1), or a murine monoclonal antibody specific for annexin A2 (BD Biosciences, San Jose, CA). The binding of the primary antibodies was detected with goat anti-guinea pig immunoglobulins (Kirkegaard & Perry Laboratories, Gaithersburg, MD), anti-rabbit immunoglobulins (Thermo Scientific, Waltham, MA), or anti-mouse immunoglobulins (Thermo Scientific) conjugated with horseradish peroxidase (HRP), followed by development with SuperSignal chemiluminescent substrate (Thermo Scientific).
To reprobe, Western blot membranes were treated with Restore Plus Western blot stripping buffer (Thermo Scientific), followed by incubation with HRP-conjugated anti-β-actin monoclonal antibody. A SeeBlue Plus2 prestained standard (Invitrogen) was used to estimate protein molecular weights.
RESULTS
Phylogenetic analysis of the leader of the capsid.
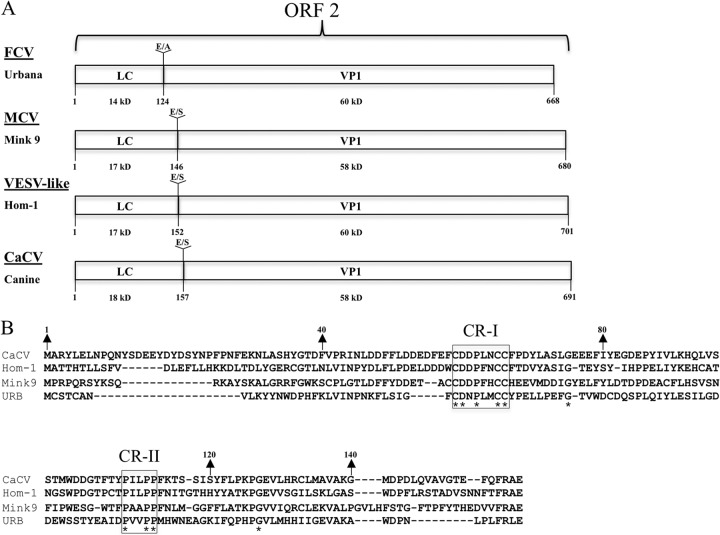
Vesivirus LC proteins are heterogeneous in size (14 to 18 kDa) and have marked sequence diversity across the genus (Fig. 1). Linear sequence analysis using bioinformatic programs did not identify functional domains or motifs in representative sequences from the four different clusters. LC sequences do not share significant similarity with any protein that has been submitted to GenBank. Despite the sequence diversity of LC sequences, two conserved regions were identified and designated conserved region I (CR-I) and CR-II (Fig. 1B). CR-I is a cysteine-rich region highlighted by three conserved cysteines, and CR-II is a proline-rich region that contains three conserved prolines.
Fig 1.
Vesivirus LC sequences. (A) The organization of ORF2 from representative vesiviruses: FCV (Urbana), mink calicivirus 9 (MCV), VESV (Hom-1), and CaCV. Sequences of the dipeptide cleavage sites and their positions between the LC and VP1 are indicated. The calculated molecular mass of each protein is shown. (B) Amino acid alignment of the LC sequences of the four representative vesiviruses. Asterisks indicate residues conserved across all of the LC proteins that were analyzed. CR-I and CR-II refer to conserved regions.
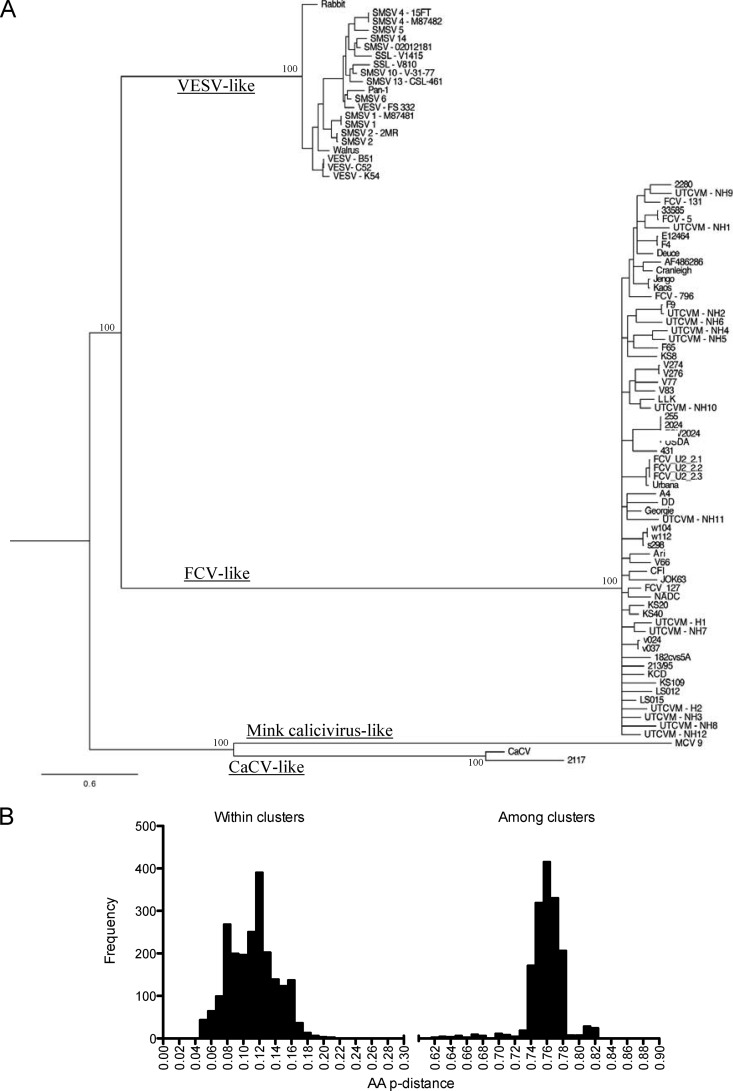
A Bayesian phylogenetic analysis of 88 LC nucleotide sequences showed four main evolutionary clusters or lineages, designated here as FCV-like, vesicular exanthema of swine (VESV)-like, canine calicivirus (CaCV)-like, and mink calicivirus-like (Fig. 2A). Consistent with a previous report (32), isolate 2117, which was identified as a contaminating agent in fetal bovine serum, clustered closely with CaCV (Fig. 2A).
Fig 2.
Phylogenetic analysis of LC sequences. (A) Bayesian phylogenetic analysis of 88 LC nucleotide sequences showing the presence of four major clusters. (B) Amino acid p distances within and between the four clusters.
A graphic representation comparing the amino acid p-distance values within all four clusters showed high levels of similarity within clusters and a marked lack of similarity between clusters (Fig. 2B). The amino acid p distances ranged from 0.02 to 0.23 within clusters and from 0.62 to 0.82 between clusters (Fig. 2B).
N- and C-terminal deletions of the LC and virus viability.
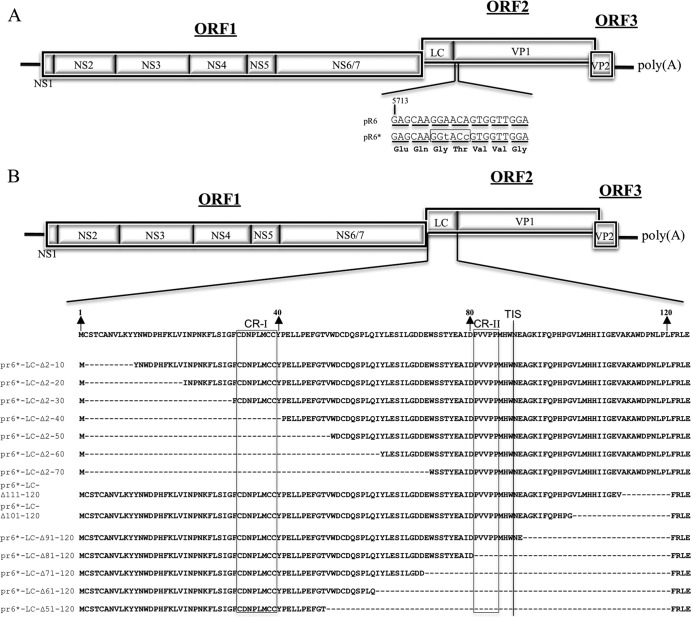
In order to facilitate the mutagenesis of LC within the FL clone pR6 cDNA backbone, two silent mutations were introduced at positions 5721 and 5724 of the virus genome (GGA5721→GGT5721 and ACA5724→ACC5724) to generate a unique KpnI restriction site at the beginning of the VP1 coding sequence. The newly generated vector was designated pR6* (Fig. 3A) and contained BstBI and KpnI restriction sites bordering the LC coding region that allowed the replacement of wt LC with genetically modified LC. The presence of the new restriction enzyme sites did not affect the recovery of virus from pR6*, and the silent mutations remained stable when examined by sequence analysis of the recovered virus at passage 3 (P3).
Fig 3.
(A) Recombinant FL FCV clones with truncated LC sequences. The FCV genome is organized into three open reading frames (ORFs 1 to 3), with the ORF1 encoding nonstructural proteins 1 (NS1) through NS6/7. ORF2 encodes a capsid protein precursor that is cleaved by the viral proteinase (NS6/7) to yield LC and VP1. ORF3 encodes minor structural protein VP2. Two silent nucleotide changes (lowercase letters) were engineered into the FL FCV plasmid pR6 near the beginning of the VP1 coding sequence in ORF2 to create unique restriction enzyme site KpnI (boxed) in plasmid pR6* in order to facilitate cloning. Nucleotide triplets corresponding to VP1 codons are underlined, and the encoded amino acids are indicated below. (B) Plasmid constructions containing engineered deletions of the LC in the context of the FL genome. Amino acid numbers corresponding to the LC are indicated with arrows, regions conserved across all vesiviruses are boxed (CR-I and CR-II), and the transposon insertion site (TIS) is indicated with a line. The names of the FL clones refer to the LC amino acids that have been deleted.
To determine whether all or part of the LC is essential for the recovery of infectious virus, a series of 14 FL clones that contained sequential 10-aa deletions in the LC protein, beginning from either the N or C terminus, was generated (Fig. 3B). Cleavage of the capsid precursor is essential for the recovery of infectious virus (5), and the highly conserved C-terminal residues (FRLE) were present to maintain the cleavage site (E/A). The MVA-T7-based recovery system for FCV (33) was used to test the mutant FL clones for the recovery of infectious virus. Following FL plasmid transfection/MVA-T7 infection, cell culture medium was transferred to a fresh monolayer of CRFK cells (P1) at 24 to 48 h posttransfection. The cells were monitored for CPE for up to three passages. Table 1 summarizes whether overt CPE can be observed at P3 for each construct. In addition, RNA was extracted from the cell culture medium and RT-PCR was performed using FCV-specific primers as a more-sensitive detection method to detect low levels of viral replication. No clear CPE was observed at P3 for any of the deletion constructs, although two C-terminal deletion constructs (vR6*-LC-Δ111–120 and vR6*-LC-Δ91–120) were positive by RT-PCR. Sequence analysis of the amplicon generated from the RNA of vR6*-LC-Δ111–120 confirmed that the replicating virus lacked aa 111 to 120. RNA extracted from vR6*-LC-Δ91–120 yielded a weak RT-PCR amplicon that was not sufficient for sequence analysis.
Table 1.
Effects of LC deletions and alanine substitutions of conserved LC residues on virus recovery
| Construct | Passage 3 results |
|
|---|---|---|
| CPE | RT-PCR | |
| pR6*-LC-Δ2–10 | − | − |
| pR6*-LC-Δ2–20 | − | − |
| pR6*-LC-Δ2–30 | − | − |
| pR6*-LC-Δ2–40 | − | − |
| pR6*-LC-Δ2–50 | − | − |
| pR6*-LC-Δ2–60 | − | − |
| pR6*-LC-Δ2–70 | − | − |
| pR6*-LC-Δ111–120 | − | + |
| pR6*-LC-Δ101–120 | − | − |
| pR6*-LC-Δ91–120 | − | +/−a |
| pR6*-LC-Δ81–120 | − | − |
| pR6*-LC-Δ71–120 | − | − |
| pR6*-LC-Δ61–120 | − | − |
| pR6*-LC-Δ51–120 | − | − |
| pR6-LC-C33A | − | + |
| pR6-LC-D34A | + | + |
| pR6-LC-P36A | + | + |
| pR6-LC-C39A | − | + |
| pR6-LC-C40A | − | + |
| pR6-LC-P81A | + | + |
| pR6-LC-P84A | + | + |
| pR6-LC-P85A | + | + |
| pR6-LC-P81A,P84A,P85A | − | + |
+/−, a faint PCR amplicon was detected by gel electrophoresis.
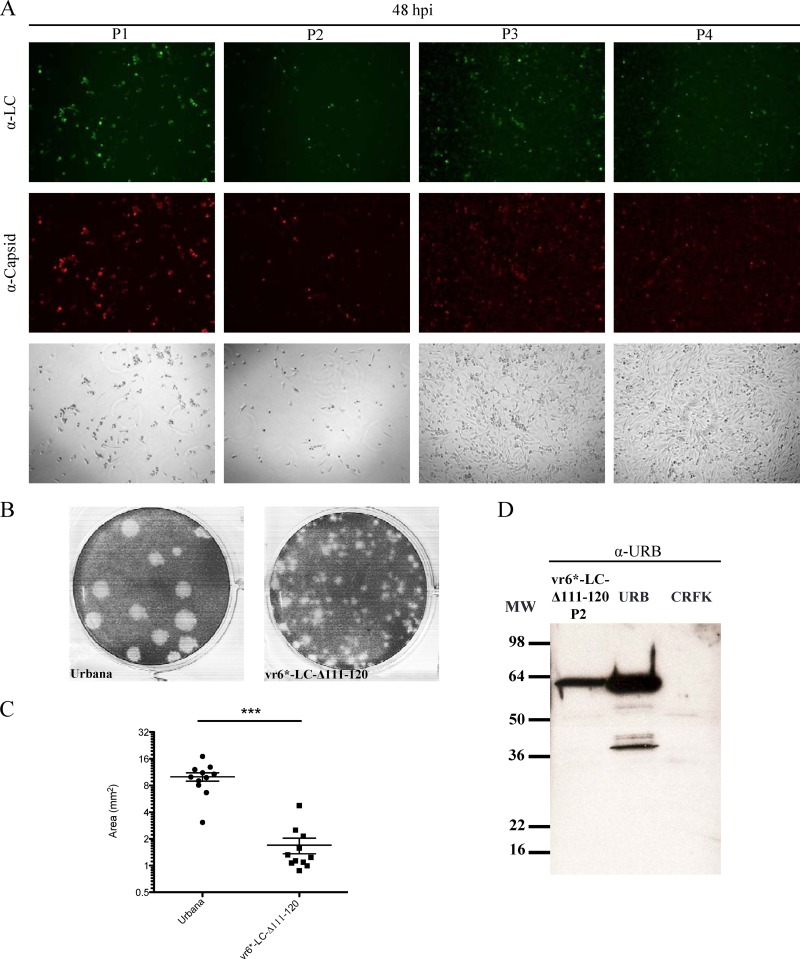
The virus recovered from the vR6*-LC-Δ111–120 construct was investigated further. While complete CPE was observed at P1 and P2 during the initial recovery experiment, no CPE was detected by P3. However, evidence for continuing replication was supported by positive immunofluorescence assay results at P3 and P4 (Fig. 4A) and the detection of viral RNA by RT-PCR, as noted above. Figure 4A depicts CRFK monolayers at 48 hpi for each corresponding passage, highlighting the clearly visible CPE observed at P1 and P2, in contrast to the intact monolayers observed at P3 and P4. The plaque phenotype of the mutant virus recovered from vR6*-LC-Δ111–120 at P1 was compared to that of the wild-type Urbana FCV strain (Fig. 4B). The plaques generated from vR6*-LC-Δ111–120 were approximately five times smaller than those produced by wild-type virus (P < 0.0001) (Fig. 4C).
Fig 4.
Analysis of replication of vR6*-LC-Δ111–120. (A) Fluorescence microscopy analysis of LC and capsid expression in CRFK cells from a series of passages of vR6*-LC-Δ111-120 at the same time point. Bright-field images are shown to demonstrate the differences between the CPE observed at P1 and P2 versus P3 and P4. CRFK cells were stained 48 hpi with hyperimmune sera raised against either FCV virions or recombinant LC. (B) Representative image of plaque assays performed simultaneously, comparing wild-type FCV (Urbana) and P1 of vR6*-LC-Δ111-120. (C) Plaque size of viruses in panel B, measured using the software program GraphClick. The statistical significance of differences in size was calculated using an unpaired t test (P < 0.0001). (D) Western blot analysis of a cell lysate from P2 of vR6*-LC-Δ111-120. A cell lysate of Urbana-infected cells (URB) was included as a positive control, and an uninfected cell lysate (CRFK) was included as a negative control. MW, molecular weight.
One possible explanation for the growth attenuation of vR6*-LC-Δ111–120 in cell culture was reduced efficiency in cleavage of the capsid precursor, which has been reported to severely affect virus growth and limit viral replication to one virus life cycle (5). A Western blot analysis using cell lysates from P2 of vR6*-LC-Δ111–120 was performed to determine whether uncleaved capsid precursor protein can be detected. Using hyperimmune sera specific to the Urbana virion, only mature cleaved capsid protein (60 kDa) was detected, indicating that the C-terminal deletions did not affect cleavage of the capsid precursor (Fig. 4D).
Transient expression of LC and LC-mKate in CRFK cells.
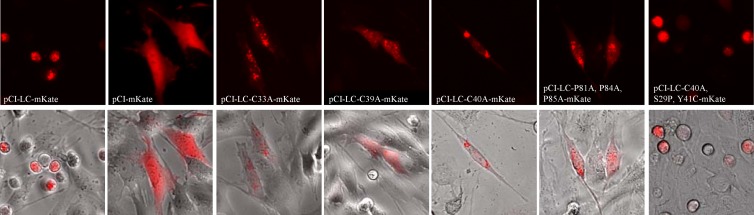
The deletional mutagenesis analysis suggested that expression of the entire LC protein sequence was required to recover virus with characteristic CPE. The complete LC coding sequence of the Urbana strain was cloned into a pCI eukaryotic expression vector to investigate the effect of LC expression alone on cells (Fig. 5A). In order to monitor LC expression in live cells, a recombinant LC protein was generated by cloning the red fluorescent protein mKate into the TIS (between aa 88 and 89 of the coding sequence of the LC), which was designated pCI-LC-mKate. We showed previously that the LC can tolerate both small (hemagglutinin [HA] or FLAG) and large (green or red fluorescent protein) insertions in this TIS without affecting the recovery of infectious virus (1). It should be noted that the insertion of foreign sequences into the TIS preserved expression of the entire LC sequence, albeit with the N terminus (aa 1 to 88) and C terminus (aa 89 to 124) separated by an in-frame insertion.
Expression of wt LC and LC-mKate in CRFK cells resulted in the appearance of rounded cells similar to those observed during viral infection (Fig. 5B). To test whether the cell-rounding phenotype was associated with cell death, a commercially available reagent that reacts with free amines in the cytoplasm of dead cells and emits a green signal when exposed to UV light was added. The majority of cells positive for LC-mKate expression were positive also for the reactive dye (Fig. 5C), indicating that overexpression of the LC was toxic to CRFK cells and caused cell death. To determine if the cell death caused by transient LC expression was associated with apoptosis, which is induced in FCV-infected cells (31, 34, 35), a cell-permeable fluorogenic peptide marker (FITC-VAD-FMK) was employed to detect activated caspases in LC-mKate-expressing cells. The majority of cells positive for LC-mKate were positive also for activated caspases (Fig. 5D), suggesting that apoptosis was induced in cells expressing LC.
Scanning alanine mutagenesis of conserved residues in the LC.
The sequence analysis identified two conserved regions (CRs) in the analyzed LC sequences (Fig. 1B), which we designated CR-I and CR-II. To evaluate the significance of these conserved regions within the FCV LC N-terminal region, scanning alanine mutagenesis was performed. Eight new FL constructs, in which a single conserved cysteine or proline residue was replaced by alanine (Fig. 6A), were generated and transfected into MVA-T7-infected cells. Recovery of virus was monitored by CPE and RT-PCR for three passages, as described above.
Fig 6.
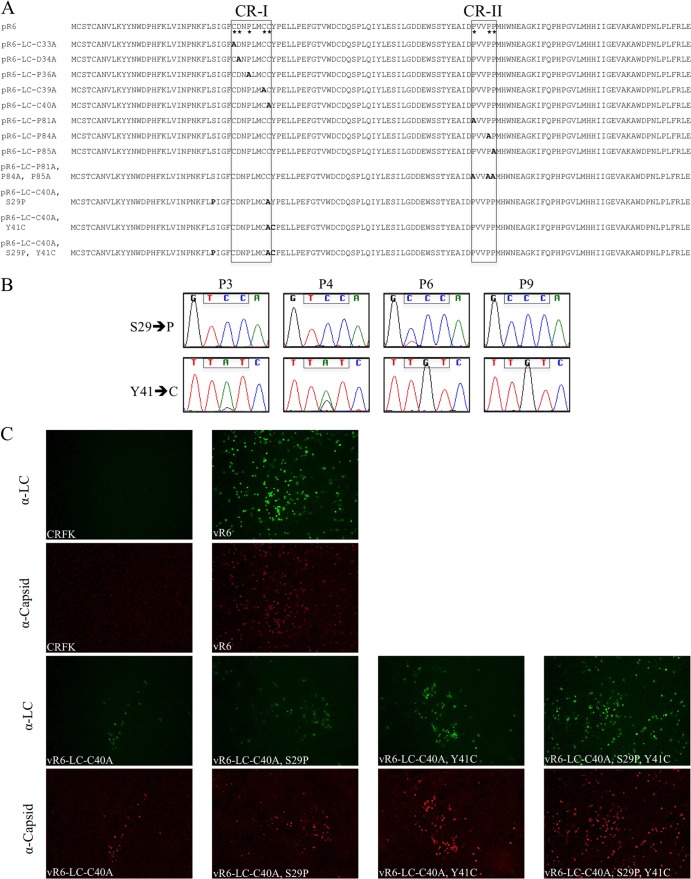
Alanine replacement of conserved residues in the LC in the FL FCV cDNA clone and analysis of virus capsid and LC expression. (A) FL plasmid constructions containing engineered alanine replacements of the conserved residues (*) from CR-I and CR-II. Additionally, three constructs that contain the C40A substitution and either one or both compensatory mutations (S29P and Y41C) are shown. The names of the FL clones refer to the LC amino acids that are replaced, their position according to the coding sequence of the LC, and the amino acid substitutions. (B) Chromatograms of LC codons (boxed nucleotides) at positions 29 and 41 corresponding to several different passages of vR6-LC-C40A. (C) Fluorescence microscopy analysis of LC and capsid expression in CRFK cells at P1 from the various FCV mutants. vR6-infected and mock-infected CRFK cells were included as positive and negative controls, respectively.
Replacement of any of the three prolines (at positions 81, 84, and 85) in CR-II with alanine had no effect on the recovery of virus (Table 1), and the CPE was similar to that of the wt. The genetic stability of the recovered viruses was examined at P3 and no reversions were detected, indicating that these residues were not essential for the recovery of infectious virus. Similarly, when D34 or P36 of CR-I was replaced by alanine, virus was readily recovered and the substitutions were stable through P3. Replacement of all three proline residues in CR-II with alanine residues did not result in overt CPE, but low levels of replication were detected (Table 1).
Alanine substitutions introduced at position C33, C39, or C40 of CR-I resulted in failure to recover cytopathic viruses by P3, although all three passages were positive by RT-PCR, indicating low levels of replication (Table 1). Cell culture media from each of the three viruses were passaged in an attempt to isolate a revertant virus that would cause characteristic CPE. Viruses containing the C33A and C39A substitutions were negative by RT-PCR at P6, indicating that these mutations were deleterious for virus growth. In contrast, the C40A mutant remained RT-PCR positive through P9, at which point an overt CPE phenotype was restored (Table 2). Sequence analysis of viral RNA from the C40A mutant at P9 revealed that the alanine substitution remained stable but that two new nonsynonymous mutations had been acquired in the LC coding sequence. A point mutation at position 5398 (codon 29 of the LC) resulted in the replacement of a UCC (Ser) codon by a CCC (Pro) codon, and a point mutation at position 5435 (codon 41 of the LC) resulted in the replacement of a UAU (Tyr) codon by a UGU (Cys) codon (Fig. 6B). Sequence analysis of the full-length genome revealed that there were two synonymous mutations and one nonsynonymous mutation in ORF1 (Table 3).
Table 2.
Serial passage of recombinant viruses and analysis of virus replication
| Virus construct | Passage 5 results |
Passage 6 results |
Passage 9 results |
|||
|---|---|---|---|---|---|---|
| CPE | RT PCR | CPE | RT PCR | CPE | RT PCR | |
| pR6-LC-C33A | − | +/−a | − | − | NTb | NT |
| pR6-LC-C39A | − | +/− | − | − | NT | NT |
| pR6-LC-C40A | − | + | − | + | + | + |
+/−, a faint PCR amplicon was detected by gel electrophoresis.
NT, not tested.
Table 3.
Sequence analysis of vR6-LC-C40A at P9 compared with that of vR6 (wild type)
| Nucleotide sequence (wt→vR6-LC-C40A) | Encoded amino acid (wt→vR6-LC-C40A) | aa position in ORF1 (corresponding viral protein[s]) |
|---|---|---|
| GAT→GAC | D→D | 674 (NS3) |
| AGA→AGG | R→R | 789 (NS4) |
| GAG→AAG | E→K | 1702 (NS6-7) |
Sequence analysis was performed with several passages of the revertant virus to examine if the substitutions in the LC arose simultaneously or separately. A mixed population was observed at codon 41 of the LC as early as P3, and the point mutation resulting in the Y41C substitution was clearly dominant by P6 (Fig. 6B). The substitution at codon 29 appeared following the acquisition of the Y41C substitution around P6 and was fixed in the population by P9 (Fig. 6B), when the characteristic CPE phenotype was restored.
In order to examine whether one or both mutations within the LC coding sequence were sufficient to suppress the C40A mutation, three new FL clones that introduced individual substitutions (pR6-LC-C40A,S29P or pR6-LC-C40A,Y41C) or both (pR6-LC-C40A,S29P,Y41C) in the context of the C40A background were generated (Fig. 6A). Rapidly spreading virus was not recovered from either of the individual constructs at P1 as judged by the lack of visible CPE, although the presence of infected cells was more readily detected when either the S29P or Y41C substitution was present by immunofluorescence analysis (Fig. 6C). However, when the two substitutions were present together, virus with characteristic CPE was recovered at P1 (Fig. 6C). Full-length genomic sequence analysis of P3 of vR6-LC-C40A,S29P,Y41C revealed that no additional mutations were present in the genome (including those listed in Table 3), indicating that these two substitutions in the LC were sufficient to suppress the C40A mutation.
Effects of alanine substitutions on the cell-rounding phenotype.
The importance of the conserved cysteines of CR-I and prolines of CR-II of the LC in the cell-rounding phenotype that was observed following transient expression of LC and LC-mKate was examined. Images of the transient expression of LC-mKate in CRFK cells were captured using modified pCI-LC-mKate clones in which the conserved residues were replaced by alanine. As shown above (Fig. 5B), a cell-rounding phenotype was consistently observed when the wt sequence of the LC was expressed. In contrast, expression of the mutagenized LC proteins resulted in a heterogeneous phenotype in which cells positive for LC expression did not cause cell rounding (Fig. 7). These data established a correlation between the ability of the LC to induce a cell-rounding phenotype and virus spread, as viruses bearing these substitutions did not grow well (Table 1). We next investigated whether the compensatory mutations that allowed viruses with the C40A mutation to become cytopathic were associated with cell rounding. A pCI-LC-mKate clone that contained the C40A substitution in addition to the two compensatory mutations (S29P and Y41C) was generated. Expression of this modified LC restored a homogeneous cell-rounding phenotype, further supporting a link between the cell-rounding phenotype induced by the LC and the ability of FCV to spread efficiently.
Fig 7.
Alanine replacement of conserved residues in the LC in the pCI-LC-mKate clone and analysis of cell morphology in transient-expression experiments. Fluorescence microscopy analysis of CRFK cells transfected with LC-mKate, LC-mKate clones containing alanine replacements of conserved cysteines in the CR-I, and an LC-mKate-C40A clone that contained the two compensatory mutations required to suppress the defect in virus spread caused by the C40A substitution was performed. Fluorescent images are shown in the top row, and corresponding merged images (bright field and fluorescent) are shown below.
Identification of host cellular partners of the LC.
To identify cellular proteins that interact with the LC during FCV infection, a FL clone that fused two unique epitopes (FLAG and NV10, a 22-amino-acid peptide in Norwalk virus VP1 that is recognized by the monoclonal antibody NV10 [our unpublished data]) to the LC coding region was generated (Fig. 8A). Two epitope tags were included in order to allow for tandem affinity immunoprecipitation if it was required. Infectious virus was readily recovered from the recombinant FL clone, and the stability of the epitope insertions was confirmed after P3.
Coimmunoprecipitation assays were performed using biotinylated monoclonal antibody NV10 in combination with streptavidin-conjugated beads to purify the recombinant LC. A 36-kDa protein coprecipitating with the recombinant LC was consistently observed when the pulled-down protein complex was analyzed by SDS-PAGE followed by Coomassie blue staining (Fig. 8B). The protein was submitted for mass spectrometry analysis and identified as annexin A2 (ANXA2), a host cellular protein associated with the cytoskeleton and cell motility (36, 37). Western blot analysis using a monoclonal antibody against ANXA2 confirmed the specificity of the interaction (Fig. 8B). In an immunofluorescence assay, ANXA2 was detected on the plasma membrane and also had a punctate-like distribution in the cytoplasm (Fig. 8C), consistent with previous reports that ANXA2 colocalizes with endosomes (38). Colocalization of ANXA2 and LC in the cytoplasm was observed in vR6 (wild type)-infected CRFK cells (Fig. 8C).
FCV LC chimeras.
Phylogenetic analysis of the LC protein showed marked diversity among vesiviruses in different genetic clusters. We explored whether the LC proteins representing the VESV-like or mink calicivirus-like clusters can replace the LC protein of FCV. Two chimeric recombinant FL FCV clones were constructed in which the entire LC sequence of the FCV LC was replaced with the LC sequence from mink calicivirus 9 (19% identity when compared to the Urbana LC protein) or SMSV-5 Hom-1 (VESV-like) (22% identity when compared to the Urbana LC protein) (Fig. 9A). Recovery experiments did not yield rapidly spreading virus as judged by the lack of visible CPE, but evidence for a low level of viral replication was found when cells from P1 were analyzed by an immunofluorescence assay for VP1 expression. To test whether partial replacement of the FCV LC was better tolerated, two more constructs were generated in which only an internal portion of the FCV LC (aa 33 to 111 for pR6-Mink-LC-2 and aa 18 to 118 for pR6-SMSV-LC-2) was replaced by one corresponding to mink calicivirus or SMSV while the FCV ends were preserved (Fig. 9A). The SMSV LC (corresponding to the VESV-like cluster) was better tolerated than the mink calicivirus LC, in that larger foci of positive cells were observed at P1 (Fig. 9B), and there was no visible difference between the full-length substitutions and the internal portion substitutions. The virus medium from each construct was passaged two additional times up to P3. Viral RNA was detected at P3 for both pR6-SMSV-LC constructs, providing further evidence that these viruses can replicate at low levels. Sequence analysis of the pR6-SMSV-LC viruses collected at P3 revealed that the chimeric LC was stable and that there were no acquired mutations in the LC region. RT-PCR of the recombinant viruses expressing mink calicivirus LC protein (entire or partial) from P3 was negative, indicating that even though there was evidence of some replication at P1 (Fig. 9B), there was no recovery of chimeric viruses that were passaged successfully.
DISCUSSION
Vesiviruses of the family Caliciviridae encode a capsid precursor in ORF2 which is cleaved by the viral protease to release the major capsid protein VP1 and the leader of the capsid (LC). The role of the LC in the viral replication cycle is not known, although the LC does enhance the human norovirus replicon when provided in trans (23). Furthermore, the LC does not share significant homology with any known proteins. The goal of this study was to employ the FCV reverse genetics system, in addition to transient-expression experiments, to study the functional role of the LC in the FCV life cycle. We determined that the LC is essential for the production of virus with characteristic CPE, and we identified residues critical for the recovery of infectious particles. Furthermore, a correlation was determined between the ability of the LC to cause cell rounding in CRFK cells and the generation of rapidly spreading virus.
Our phylogenetic analysis of LC sequences determined that there are four different LC clusters: FCV-like, VESV-like, CaCV-like, and mink calicivirus-like. The LC sequences are strikingly divergent across clusters but share a high level of amino acid identity within clusters. A noteworthy observation is that the predicted secondary structure of all LC sequences appears to be conserved (data not shown). Attempts to express soluble recombinant LC protein for structural analysis were unsuccessful, but it is possible that LC proteins have similar tertiary structures. When different vesivirus LC sequences replaced FCV LC in the FL clone, low levels of viral replication were detected, but cytopathic virus was not recovered. Although vesivirus LC proteins likely share similar functions, the striking species clustering of the LC might be related to specific interactions between host and viral proteins.
The deletional mutagenesis analysis provided insight into functional domains of the LC, and essential domains for viral replication were mapped to the N-terminal region (aa 1 to 88) of the LC. Virus was not recovered from any of the N-terminal deletion constructs, although a possible effect on the RNA secondary-structure signals involved in translation of the subgenomic RNA cannot be ruled out. The 5′ end of the FCV subgenomic RNA has been mapped to nt 5296/5297 (39), which is located 17/18 nt upstream of the LC AUG, and RNA elements important for FCV subgenomic RNA production can be inferred from work with other caliciviruses. For example, an RNA element upstream of the subgenomic RNA has been shown to be essential for murine norovirus replication and proposed to be important for FCV (40). These data are consistent with a report that mapped the subgenomic promoter of another calicivirus, rabbit hemorrhagic disease virus, to a 50-nt region upstream of the beginning of the subgenomic RNA (41). It is not known whether downstream signals that drive translation of the calicivirus subgenomic RNA are present in the subgenomic region. In the alphaviruses, Frolov and Schlesinger characterized an RNA secondary structure downstream of the initial AUG that is important for translation of the subgenomic RNA in Sindbis virus (42, 43). Preliminary analysis of the FCV LC sequences employing the Vienna RNA Web suite (44), which detects conserved secondary structures based on aligned sequences using the RNAalifold server, identified a conserved stem-loop downstream of the LC AUG (nt 5327 to 5338). Mutagenesis studies of the stem-loop will be needed to determine its significance, if any, in translation of the subgenomic RNA in FCV.
The small-plaque phenotype of the vR6*-LC-Δ111–120 virus at P1 suggested involvement of the C-terminal region in virus spread. This virus grew relatively well for the first two passages but lost the appearance of CPE by P3. It should be noted that the virus recovery system is plasmid based and involves transfection of the FL plasmid DNA into CRFK cells infected at a high MOI with a modified vaccinia virus to provide T7 polymerase in trans. The recovery system is robust, and the first pool of virus generated from the transfection was likely sufficient in titer to infect the entire monolayer at P1. However, as the virus was serially passaged, the inability to spread efficiently in the monolayer might have reduced the number of infected cells. No visible CPE was observed by P3, although infected cells were detected (Fig. 4A) and viral RNA was purified and sequenced. The mechanism by which virus spread was inhibited will require further investigation.
The diversity of LC sequences limited the number of conserved amino acid residues for scanning alanine mutagenesis. Mutagenesis studies focused on two conserved regions, CR-I (cysteine rich) and CR-II (proline rich), to fine map residues critical for the recovery of infectious virus. It is possible that the conserved proline motif of CR-II is involved in important protein-protein interactions with cellular proteins that contain an SH3 domain (45, 46). However, replacement of individual prolines had no visible effect on the recovery of virus through three passages, and the substitutions were stable. In contrast, certain cysteines in CR-I proved important for the recovery of viruses with characteristic CPE. Furthermore, serial passage of the virus containing the C40A substitution resulted in compensatory mutations that restored the cytopathic phenotype, suggesting a strong selective pressure to restore the function of the LC in CPE. The additional mutations observed in the adapted C40A virus will require further study. Proline is present at LC position 29 in a few FCV strains from the 1970s and 1980s (JOK63, F4, V83, and 182cvs5A), as well as a more recent strain from 1998 (UTCVM_NH2) and a virulent systemic FCV strain from 2002 (UTCVM_H2). There is no distinct phenotype that can be associated with the presence of proline at position 29 in wt isolates reported in the literature, so at this point it is difficult to interpret the significance of this substitution. The tyrosine-to-cysteine substitution at position 41 (Y41C) was not expected, since the tyrosine at position 41 is conserved in nearly all reported FCV LC sequences. It will be interesting to explore whether the loss of this highly conserved tyrosine, which lies within a predicted Janus kinase 2 tyrosine phosphorylation motif, YXX[L/I/V] (47), affects viral fitness. However, the presence of a cysteine in this region might be critical in restoring a structurally important disulfide bond. The subcellular localization of the mutant LC proteins might suggest that there is misfolding leading to the formation of aggregates (Fig. 7). Further investigation will be required to determine if the C40A mutation affects the native conformation of LC.
One limitation of transient-expression experiments is that the behavior of a single viral protein out of the context of a viral infection and overexpressed might not reveal meaningful insight into its normal function. In our studies, we established a correlation between the cell-rounding phenotype observed in transient-expression experiments and the recovery of virus with characteristic CPE. The C40A substitution in the LC (located within the N-terminal region) resulted in loss of the homogeneous cell-rounding phenotype in transfected cells and resulted in a growth-attenuated virus in the context of the FL clone. More importantly, when the substitutions acquired during viral replication to compensate for the C40A mutation (S29P and Y41C) were introduced into the pCI-LC-mKate vector, a homogeneous cell-rounding phenotype in transfected cells was restored.
Our coimmunoprecipitation assays identified annexin A2 (ANXA2) as a cellular protein interacting with the LC during infection. Annexin A2 is a member of the annexin family of proteins, which are characterized as proteins that bind negatively charged phospholipids in a Ca2+-dependent manner and contain several conserved structural domains (annexin repeats) (36). ANXA2 is a multifunctional protein and is involved in many cellular processes, such as fibrinolysis, exocytosis, endocytosis, cell-cell adhesion, and cell motility (36, 37, 48). ANXA2 has been linked to the life cycle of other positive-sense single-stranded RNA viruses, such as hepatitis C virus (49–51) and HIV (52–54). In both cases, a viral protein has been reported to interact with ANXA2 in infected cells, and silencing of ANXA2 affected the infectivity of the viruses. Experiments are in progress to clone and sequence the feline ANXA2 from CRFK cells in order to perform small interfering RNA experiments and determine the significance of ANXA2 in the life cycle of FCV.
These findings, along with our previous discovery that the LC can tolerate the insertion of foreign sequences twice its own length between amino acids 88 and 89 (1), lead us to propose that the LC contains at least two domains to mediate its function in viral CPE: an N-terminal domain (within aa 1 to 88) that is associated with cell rounding and death and a C-terminal domain (within aa 89 to 124) that is involved in virus spread. Although the precise role of this unique vesivirus protein and its mechanism of action require further investigation, additional study might give insight into how caliciviruses interact with host cells to produce progeny in one of the fastest replication cycles among viruses.
ACKNOWLEDGMENTS
We thank Albert Z. Kapikian for helpful suggestions and Rachel Dexter for technical support. We also thank Lily Koo, Stephen Becker, and Owen M. Schwartz of the NIAID Biological Imaging Section for assistance with confocal imaging.
This research was supported by the Intramural Research Program of the NIH, NIAID.
Footnotes
Published ahead of print 26 December 2012
REFERENCES
- 1. Abente EJ, Sosnovtsev SV, Bok K, Green KY. 2010. Visualization of feline calicivirus replication in real-time with recombinant viruses engineered to express fluorescent reporter proteins. Virology 400:18–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mitra T, Sosnovtsev SV, Green KY. 2004. Mutagenesis of tyrosine 24 in the VPg protein is lethal for feline calicivirus. J. Virol. 78:4931–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sosnovtsev S, Green KY. 1995. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology 210:383–390 [DOI] [PubMed] [Google Scholar]
- 4. Sosnovtsev SV, Belliot G, Chang KO, Onwudiwe O, Green KY. 2005. Feline calicivirus VP2 is essential for the production of infectious virions. J. Virol. 79:4012–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sosnovtsev SV, Sosnovtseva SA, Green KY. 1998. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 72:3051–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McFadden N, Bailey D, Carrara G, Benson A, Chaudhry Y, Shortland A, Heeney J, Yarovinsky F, Simmonds P, Macdonald A, Goodfellow I. 2011. Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4. PLoS Pathog. 7:e1002413 doi:10.1371/journal.ppat.1002413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Herbert TP, Brierley I, Brown TD. 1997. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J. Gen. Virol. 78:1033–1040 [DOI] [PubMed] [Google Scholar]
- 8. Neill JD, Mengeling WL. 1988. Further characterization of the virus-specific RNAs in feline calicivirus infected cells. Virus Res. 11:59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schaffer FL, Ehresmann DW, Fretz MK, Soergel MI. 1980. A protein, VPg, covalently linked to 36S calicivirus RNA. J. Gen. Virol. 47:215–220 [DOI] [PubMed] [Google Scholar]
- 10. Chen R, Neill JD, Estes MK, Prasad BV. 2006. X-ray structure of a native calicivirus: structural insights into antigenic diversity and host specificity. Proc. Natl. Acad. Sci. U. S. A. 103:8048–8053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen R, Neill JD, Prasad BV. 2003. Crystallization and preliminary crystallographic analysis of San Miguel sea lion virus: an animal calicivirus. J. Struct. Biol. 141:143–148 [DOI] [PubMed] [Google Scholar]
- 12. Peterson JE, Studdert MJ. 1970. Feline picornavirus: structure of the virus and electron microscopic observations on infected cell cultures. Arch. Gesamte Virusforsch. 32:249–260 [PubMed] [Google Scholar]
- 13. Prasad BV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287–290 [DOI] [PubMed] [Google Scholar]
- 14. Prasad BV, Matson DO, Smith AW. 1994. Three-dimensional structure of calicivirus. J. Mol. Biol. 240:256–264 [DOI] [PubMed] [Google Scholar]
- 15. Sosnovtsev SV, Green KY. 2000. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology 277:193–203 [DOI] [PubMed] [Google Scholar]
- 16. Sosnovtsev SV, Garfield M, Green KY. 2002. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J. Virol. 76:7060–7072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sosnovtseva SA, Sosnovtsev SV, Green KY. 1999. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J. Virol. 73:6626–6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neill JD, Reardon IM, Heinrikson RL. 1991. Nucleotide sequence and expression of the capsid protein gene of feline calicivirus. J. Virol. 65:5440–5447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carter MJ, Milton ID, Turner PC, Meanger J, Bennett M, Gaskell RM. 1992. Identification and sequence determination of the capsid protein gene of feline calicivirus. Arch. Virol. 122:223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fretz M, Schaffer FL. 1978. Calicivirus proteins in infected cells: evidence for a capsid polypeptide precursor. Virology 89:318–321 [DOI] [PubMed] [Google Scholar]
- 21. Carter MJ. 1989. Feline calicivirus protein synthesis investigated by Western blotting. Arch. Virol. 108:69–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tohya Y, Shinchi H, Matsuura Y, Maeda K, Ishiguro S, Mochizuki M, Sugimura T. 1999. Analysis of the N-terminal polypeptide of the capsid precursor protein and the ORF3 product of feline calicivirus. J. Vet. Med. Sci. 61:1043–1047 [DOI] [PubMed] [Google Scholar]
- 23. Chang KO, George DW, Patton JB, Green KY, Sosnovtsev SV. 2008. Leader of the capsid protein in feline calicivirus promotes replication of Norwalk virus in cell culture. J. Virol. 82:9306–9317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chang KO. 2009. Role of cholesterol pathways in norovirus replication. J. Virol. 83:8587–8595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 26. McCarthy M, Estes MK, Hyams KC. 2000. Norwalk-like virus infection in military forces: epidemic potential, sporadic disease, and the future direction of prevention and control efforts. J. Infect. Dis. 181(Suppl 2):S387–S391 [DOI] [PubMed] [Google Scholar]
- 27. Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61:539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fankhauser RL, Noel JS, Monroe SS, Ando T, Glass RI. 1998. Molecular epidemiology of “Norwalk-like viruses” in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571–1578 [DOI] [PubMed] [Google Scholar]
- 29. Parra GI, Abente EJ, Sandoval-Jaime C, Sosnovtsev SV, Bok K, Green KY. 2012. Multiple antigenic sites are involved in blocking the interaction of GII.4 norovirus capsid with ABH histo-blood group antigens. J. Virol. 86:7414–7426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Evermann JF, Smith AW, Skilling DE, McKeirnan AJ. 1983. Ultrastructure of newly recognized caliciviruses of the dog and mink. Arch. Virol. 76:257–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sosnovtsev SV, Prikhod'ko EA, Belliot G, Cohen JI, Green KY. 2003. Feline calicivirus replication induces apoptosis in cultured cells. Virus Res. 94:1–10 [DOI] [PubMed] [Google Scholar]
- 32. Oehmig A, Buttner M, Weiland F, Werz W, Bergemann K, Pfaff E. 2003. Identification of a calicivirus isolate of unknown origin. J. Gen. Virol. 84:2837–2845 [DOI] [PubMed] [Google Scholar]
- 33. Sosnovtsev SV, Sosnovtseva S, Green KY. 1996. Recovery of feline calicivirus from plasmid DNA containing a full-length copy of the genome, p 125–130 In Chasey D, Gaskell RM, Clarke IN. (ed), The 1st international symposium on caliciviruses. European Society for Veterinary Virology and Central Veterinary Laboratory, Reading, United Kingdom [Google Scholar]
- 34. Natoni A, Kass GE, Carter MJ, Roberts LO. 2006. The mitochondrial pathway of apoptosis is triggered during feline calicivirus infection. J. Gen. Virol. 87:357–361 [DOI] [PubMed] [Google Scholar]
- 35. Roberts LO, Al-Molawi N, Carter MJ, Kass GE. 2003. Apoptosis in cultured cells infected with feline calicivirus. Ann. N. Y. Acad. Sci. 1010:587–590 [DOI] [PubMed] [Google Scholar]
- 36. Gerke V, Moss SE. 2002. Annexins: from structure to function. Physiol. Rev. 82:331–371 [DOI] [PubMed] [Google Scholar]
- 37. Rescher U, Ludwig C, Konietzko V, Kharitonenkov A, Gerke V. 2008. Tyrosine phosphorylation of annexin A2 regulates Rho-mediated actin rearrangement and cell adhesion. J. Cell Sci. 121:2177–2185 [DOI] [PubMed] [Google Scholar]
- 38. Morel E, Gruenberg J. 2009. Annexin A2 binding to endosomes and functions in endosomal transport are regulated by tyrosine 23 phosphorylation. J. Biol. Chem. 284:1604–1611 [DOI] [PubMed] [Google Scholar]
- 39. Neill JD. 2002. The subgenomic RNA of feline calicivirus is packaged into viral particles during infection. Virus Res. 87:89–93 [DOI] [PubMed] [Google Scholar]
- 40. Simmonds P, Karakasiliotis I, Bailey D, Chaudhry Y, Evans DJ, Goodfellow IG. 2008. Bioinformatic and functional analysis of RNA secondary structure elements among different genera of human and animal caliciviruses. Nucleic Acids Res. 36:2530–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morales M, Barcena J, Ramirez MA, Boga JA, Parra F, Torres JM. 2004. Synthesis in vitro of rabbit hemorrhagic disease virus subgenomic RNA by internal initiation on (−)sense genomic RNA: mapping of a subgenomic promoter. J. Biol. Chem. 279:17013–17018 [DOI] [PubMed] [Google Scholar]
- 42. Frolov I, Schlesinger S. 1996. Translation of Sindbis virus mRNA: analysis of sequences downstream of the initiating AUG codon that enhance translation. J. Virol. 70:1182–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Frolov I, Schlesinger S. 1994. Translation of Sindbis virus mRNA: effects of sequences downstream of the initiating codon. J. Virol. 68:8111–8117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gruber AR, Lorenz R, Bernhart SH, Neubock R, Hofacker IL. 2008. The Vienna RNA websuite. Nucleic Acids Res. 36:W70–W74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kay BK, Williamson MP, Sudol M. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 14:231–241 [PubMed] [Google Scholar]
- 46. Reebye V, Frilling A, Hajitou A, Nicholls JP, Habib NA, Mintz PJ. 2012. A perspective on non-catalytic Src homology (SH) adaptor signalling proteins. Cell. Signal. 24:388–392 [DOI] [PubMed] [Google Scholar]
- 47. Argetsinger LS, Kouadio JL, Steen H, Stensballe A, Jensen ON, Carter-Su C. 2004. Autophosphorylation of JAK2 on tyrosines 221 and 570 regulates its activity. Mol. Cell. Biol. 24:4955–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grieve AG, Moss SE, Hayes MJ. 2012. Annexin A2 at the interface of actin and membrane dynamics: a focus on its roles in endocytosis and cell polarization. Int. J. Cell Biol. 2012:852430 doi:10.1155/2012/852430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Backes P, Quinkert D, Reiss S, Binder M, Zayas M, Rescher U, Gerke V, Bartenschlager R, Lohmann V. 2010. Role of annexin A2 in the production of infectious hepatitis C virus particles. J. Virol. 84:5775–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lai CK, Jeng KS, Machida K, Lai MM. 2008. Association of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J. Virol. 82:8838–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saxena V, Lai CK, Chao TC, Jeng KS, Lai MM. 2012. Annexin A2 is involved in the formation of hepatitis C virus replication complex on the lipid raft. J. Virol. 86:4139–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr, Sowder RC, II, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J. Virol. 80:9039–9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rai T, Mosoian A, Resh MD. 2010. Annexin 2 is not required for human immunodeficiency virus type 1 particle production but plays a cell type-dependent role in regulating infectivity. J. Virol. 84:9783–9792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ryzhova EV, Vos RM, Albright AV, Harrist AV, Harvey T, Gonzalez-Scarano F. 2006. Annexin 2: a novel human immunodeficiency virus type 1 Gag binding protein involved in replication in monocyte-derived macrophages. J. Virol. 80:2694–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]