Abstract
The rapid spread of highly pathogenic avian influenza (HPAI) H5N1 virus underscores the importance of effective antiviral treatment. Previously, we developed human monoclonal antibodies 65C6 and 100F4 that neutralize almost all (sub)clades of HPAI H5N1. The conserved 65C6 epitope was mapped to the globular head of HA. However, neither the 100F4 epitope nor the neutralization mechanism by these antibodies was known. In this study, we determined the 100F4 epitope and unraveled a neutralization mechanism by antibodies 65C6 and 100F4.
TEXT
Influenza A virus infection is a persistent threat to public health worldwide. High-affinity neutralizing antibodies against conserved epitopes could provide immunity to diverse influenza virus strains and protection against future pandemic viruses. Previously, we developed human monoclonal antibodies (MAb) 65C6 and 100F4 that potently neutralize all H5 clades and subclades of highly pathogenic avian influenza (HPAI) H5N1 viruses except subclade 7.2 (Fig. 1A) and defined a conformational 65C6 epitope (1). In this study, we determined the 100F4 epitope and dissected the neutralization mechanism by these antibodies.
Fig 1.
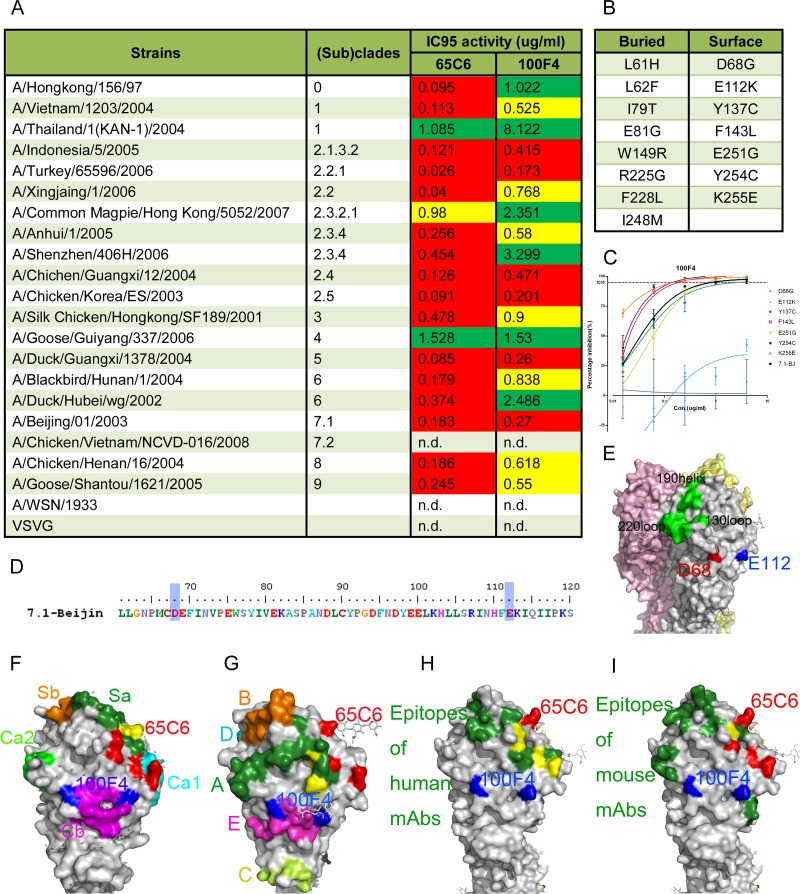
Amino acid residues involved in the neutralization epitope of 100F4. (A) Broad neutralization activity (95% inhibitory concentration [IC95]) of antibodies 65C6 and 100F4 against a panel of H5N1 pseudotypes reproduced from reference 1. Green, >1 μg/ml required to reach IC95; yellow, between 0.5 and 1 μg/ml required to reach IC95; red, <0.5 μg/ml required to reach IC95. (B) List of 15 single amino acid mutants that antibody 100F4 can no longer bind to, obtained by using yeast Saccharomyces cerevisiae that displays a random mutagenesis library of HA fragment comprising amino acid residues 51 to 260 in fine epitope mapping. Among them, 8 amino acid mutants are underneath the HA surface and the other 7 are on the surface of HA. (C) Titration of antibody 100F4 against H5N1 pseudotypes expressing 7 single surface amino acid mutants compared to the results against H5N1 pseudotype expressing the parental HA. (D) Amino acid residues D68 and E112 (D72 and E116 in H3 numbering) involved in 100F4 epitope are highlighted by blue shading. (E) Amino acid residues D68 and E112 are highlighted in red and blue, respectively, in space-filling model of HA. Yellow, pink, and grey each indicate one of three monomers that make up an HA trimer. (F) 100F4 and 65C6 epitopes in the context of known neutralization epitopes in the H1 HA structure (Protein Data Bank [PDB] structure accession number IRU7), as follows: Ca1, cyan; Ca2, light green; Cb, magenta; Sa, forest green; and Sb, orange. Yellow, overlap amino acid residues between the 65C6 epitope and the Sa site. (G) 100F4 and 65C6 epitopes in the context of known neutralization epitopes in H3 HA structure (PBD structure accession number 2VIU), as follows: site A, forest green; site B, orange; site C, yellow-green; site D, cyan; and site E, magenta. Yellow, overlap amino acid residues between the 65C6 epitope and site A. (H) 100F4 and 65C6 epitopes in the context of known neutralization epitopes detected by human MAb in H5 HA structure (PBD structure accession number 2ibx). Neutralization epitopes detected by human MAb are highlighted in forest green, and amino acid residues that overlap amino acid residues of the 65C6 epitope are highlighted in yellow. (I) 100F4 and 65C6 epitopes in the context of known neutralization epitopes detected by mouse MAb in H5 HA structure (PBD structure accession number 2ibx). Neutralization epitopes detected by mouse MAb are highlighted in forest green, and amino acid residues that overlap amino acid residues of 65C6 epitope are highlighted in yellow.
To map the 100F4 epitope, a yeast display analysis was carried out similarly to the way we mapped the 65C6 epitope (1, 2). Figure 1B shows the 15 single amino acid mutations in H5 hemagglutinin (HA) that abolish the binding of antibody 100F4. Among these, the 7 residues at positions 68,112, 137, 143, 251, 254, and 255 were on the HA surface, while the rest were underneath the surface.
To test whether these 7 surface mutations would affect neutralization by antibody 100F4, genes encoding 7 full-length H5 HA single mutants derived from H5N1 strain A/Beijing/01/03 subclade 7.1 were constructed and used to generate H5N1 pseudotypes. The resistance of H5N1 pseudotypes to antibody 100F4 was measured with the pseudotype-based neutralization assay (3). Compared to the wild-type subclade 7.1 H5N1 pseudotype, only H5N1 pseudotypes expressing H5 HA mutants with mutations at position 68 or 112 (72 or 116 according to H3 numbering) were dramatically resistant to antibody 100F4 (Fig. 1C and D). On the HA surface, these two resistant residues are adjacent to each other (Fig. 1E), but they are next to the Cb in H1 HA and site E in H3 HA (4–7) (Fig. 1F and G). The 100F4 epitope does not overlap any known epitopes in the head region detected by human and mouse MAb (Fig. 1H and I). Thus, the 100F4 epitope is a new conserved conformational epitope on the globular head and away from the receptor binding site (RBS). In contrast, the 65C6 epitope partially overlaps with Sa in H1 HA at residue 161 (K165 according to H3 numbering) and with site A in H3 HA at residues 118 and 121 (T122 and F125 according to H3 numbering) (4–7). In addition, the 65C6 epitope also partially overlaps epitopes detected by some human MAb, i.e., FLA5.10 at P118 (P122 according to H3 numbering), FLD21.140 at S121, Y164, and T167 (S125, Y168, and T171 according to H3 numbering) (8), AVFLuigG01 at P118, Y164, and T167 (P122, Y168, and T171 according to H3 numbering) (9) (Fig. 1H), and mouse MAb NR2728 at S121 (S125 according to H3 numbering) (10) (Fig. 1I). In addition, the binding of antibodies 100F4 and 65C6 to their epitopes is also different. Single mutations at position 68 or 112 almost or completely abolish the neutralization by antibody 100F4 (Fig. 1C), whereas single mutations at positions 118, 121, 161, 164, and 167 only show 3- to 5-fold reductions in neutralization by antibody 65C6 (1).
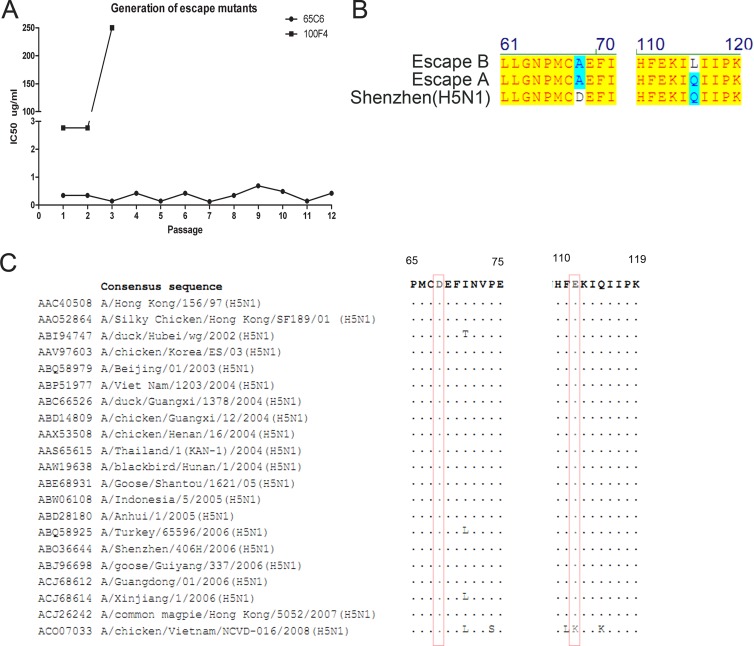
This could explain why after 2 rounds of antibody-driven mutagenesis (11), escape mutants from antibody 100F4 were readily detected, whereas even after 12 rounds of antibody-driven mutagenesis, no escape mutants from antibody 65C6 were detected (Fig. 2A). Sequencing of escape mutants purified by plaque assay revealed the same Asp-to-Ala mutation at position 68 in both escape mutants (Fig. 2B). Sequence alignment shows that at position 68, all strains tested have an identical Asp residue, whereas at position 112, only subclade 7.2 has a Lys residue, while other clades and subclades have a Glu residue, which explains why 100F4 neutralizes all H5 clades and subclades except subclade 7.2 (1) (Fig. 1A and 2C).
Fig 2.
Determination of antibody-driven escape mutants. (A) Antibody-driven escape mutants were generated through a series of passages. (B) Point mutations found in two 100F4 antibody-driven escape clones compared to the sequence of parental H5 HA. (C) Sequence alignment of amino acid residues 65 to 75 and 110 to 119 among 21 HA covering all H5 clades and subclades. Residues 68 and 112 are highlighted in boxes.
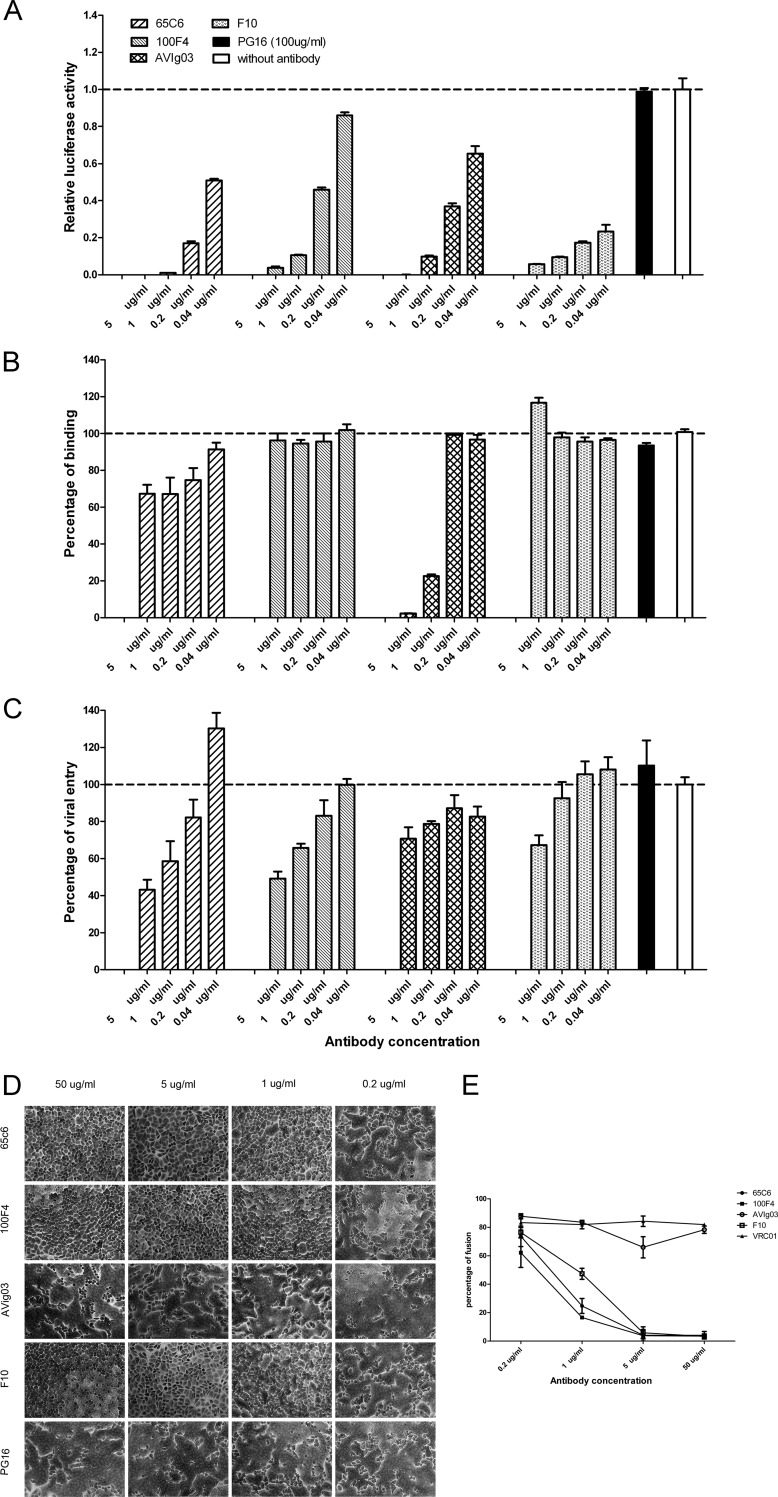
What could be their neutralization mechanism, since both of the conserved epitopes 65C6 and 100F4 are present in the head region outside the RBS? To determine the mechanism, we first carried out pseudotype-based neutralization, attachment, postattachment, and fusion assays (12, 13) using five antibodies, PG16, 65C6, 100F4, F10, and AVFluIg03 (AVIg03). Antibody PG16 recognizes HIV-1 gp120 and was used as a negative control (14). Antibody AVFluIg03, an anti-H5 HA human antibody, recognizes the RBS of the HA head and blocks virus attachment to cells (15). Antibody F10 recognizes a conserved epitope in the stalk region that blocks virus-cell fusion but not virus attachment to cells (16). As shown by the data in Figure 3A, all four anti-HA antibodies but not antibody PG16 exhibited dose-dependent neutralization against H5N1 viruses. In the attachment assay, antibody AVIg03 but not antibodies F10 and PG16 dramatically inhibited virus attachment to the cell targets, whereas antibody 65C6 exhibited low but measurable inhibition of virus attachment and antibody 100F4 did not have any inhibition at all (Fig. 3B). In the postattachment assay, antibodies AVIg03 and PG16 had minimal effects on postattachment events of virus infection, whereas both antibody 65C6 and 100F4, like antibody F10, dramatically inhibited postattachment events of virus infection (Fig. 3C).
Fig 3.
Determination of effects of antibodies 65C6 and 100F4 on virus neutralization, attachment, postattachment, and fusion. (A) Neutralization activity against H5N1 pseudotype A/Shenzhen/406H/06 by indicated concentrations of antibodies 65C6, 100F4, AVIg03, F10, and PG16. The neutralization activity was calculated as follows: relative luciferase activity (RLA) of pseudotype with a given concentration of an antibody/RLA of pseudotype alone without antibodies. (B) Effects of indicated concentrations of antibodies 65C6, 100F4, AVIg03, F10, and PG16 on binding of H5N1 pseudotype A/Shenzhen/406H/06 to MDCK cells compared to binding of pseudotype alone (without antibodies). (C) Effects of indicated concentrations of antibodies 65C6, 100F4, AVIg03, F10, and PG16 on postattachment entry of H5N1 pseudotype A/Shenzhen/406H/06 into MDCK cells compared to entry of pseudotype alone (without antibodies). (D) Effects of antibodies 65C6, 100F4, AVIg03, F10, and PG16 on pH 5.0-triggered, HA-mediated cell-cell fusion among influenza H5 HA-transduced HeLa cells. (E) Mean and standard deviation of fusion percentages in each image in panel D. The fusion percentages were calculated as described in the text.
To determine whether antibodies 65C6 and 100F4 block low-pH-triggered, HA-mediated fusion, we transfected a plasmid expressing H5 HA into HeLa cells using a lentiviral vector (17). A fusion assay was performed as described previously (16, 18). The results in Figure 3D show that both antibody 65C6 and 100F4, like antibody F10, dramatically inhibited low-pH-triggered, HA-mediated fusion in a dose-dependent fashion. To quantify the fusion data shown in Figure 3D, each image was divided into four areas. The numbers of cells and nuclei in each area were counted manually. The fusion percentage was calculated with the following formula: (number of nuclei − number of cells)/number of nuclei. For each dose of a given antibody, the mean and standard deviation of the fusion percentages were then calculated. Clearly, like antibody F10, antibodies 65C6 and 100F4 also block low-pH-triggered, HA-mediated fusion in a dose-dependent manner. At and above 1 μg/ml, they significantly block fusion compared to the results for control antibody PG16 (all P < 0.001) (Fig. 3E). These four experiments have been repeated three times with similar results. Thus, consistent with the epitope-mapping study, antibody 100F4 recognizes an epitope that is totally away from the RBS, and its binding to the epitope does not interfere with virus attachment to the receptor but blocks low-pH-triggered, HA-mediated fusion, while antibody 65C6 recognizes an epitope that is outside but nearby the RBS (1), and its binding to the epitope has low but measurable spatial hindrance of virus attachment to the receptor but blocks postattachment and low-pH-triggered, HA-mediated fusion much more dramatically.
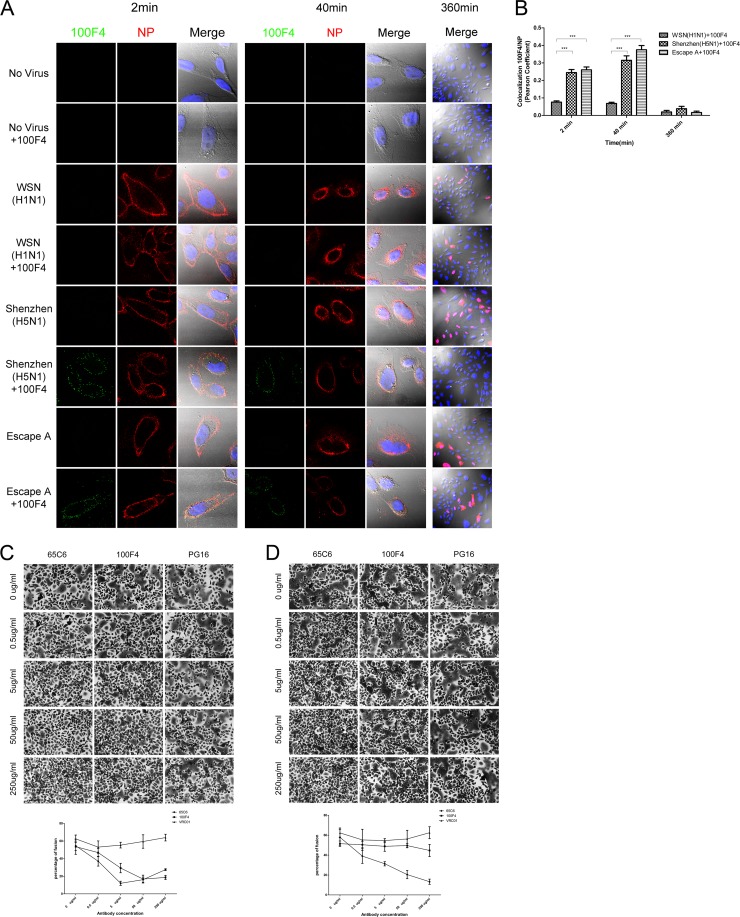
To determine whether the antibodies could block the internalization of viruses, we incubated antibody 100F4 at 37°C for 1 h with three different viruses, A/WSN/33 (H1N1), A/Shenzhen/406H/06 (H5N1), and the 100F4 escape mutant A (Fig. 2B, escape A), and then added the mixture onto CHO-K1 cells and incubated them at 4°C for 1 h. After two washes with PBS to remove unbound viruses and antibodies, the temperature was rapidly elevated to 37°C to initiate infection. At 2, 40, and 360 min postinfection, the cells were rinsed extensively, fixed, and stained with mouse anti-influenza NP MAb (Millipore) followed by Alexa 555-conjugated goat anti-mouse IgG antibody (Molecular Probes) and CF488-conjugated goat anti-human IgG antibody (Sigma). Fluorescence images of the viruses and antibodies at 2, 40, and 360 min postinfection are shown in Figure 4A. These images show that adding antibody 100F4 alone onto CHO-K1 cells resulted in no antibody detected either on the cell surface or inside the cells, as for the no-virus and no-antibody controls, whereas adding antibody 100F4 with A/Shenzhen/406H/06 and escape A resulted in antibody mainly detected on the cell surface at 2 min postinfection and in the perinuclear region at 40 min postinfection (Fig. 4A, first and fourth columns), indicating that after antibody 100F4 binding to A/Shenzhen/406H/06 and escape A, the virus and antibody complex still could bind to the HA receptor and be internalized, like the unbounded viruses. Moreover, at 2 min postinfection, all three viruses were mainly detected on the cell surface and very few viruses were internalized, regardless of whether viruses were bound or not bound with antibody 100F4 (Fig. 4A, second column). In contrast, at 40 min postinfection, significant viral transport toward the perinuclear region was observed, similar to what was reported by Lakadamyali et al. (19), regardless of whether viruses were bound or not bound with antibody 100F4 (Fig. 4A, fifth column). Furthermore, at 360 min postinfection, significant amounts of viral NP were observed in the cytosol and nuclei in cells infected with A/WSN/33 (H1N1), A/Shenzhen/406H/06 (H5N1), and escape A without antibody 100F4 or in cells infected with A/WSN/33 (H1N1) and escape A with antibody 100F4 but not in cells infected with A/Shenzhen/406H/06 (H5N1) with antibody 100F4 (Fig. 4A, seventh column). To quantify the colocalization between NP and antibody 100F4, the Pearson's correlation coefficient was analyzed using the JACoP plugin in FIJI software (20) and calculated to obtain an unbiased evaluation of the extent of colocalization between them. The coefficients were significantly higher in A/Shenzhen/406H/06 with antibody 100F4 (P < 0.001) and in escape A with antibody 100F4 (P < 0.001) than in the A/WSN/33 control with antibody 100F4 at 2 and 40 min postinfection (Fig. 4B). Similar patterns of internalization and colocalization were also observed with antibody 65C6 (M. R. Qian, H. X. Hu, and P. Zhou, data not shown).
Fig 4.
Bound antibodies 65C6 and 100F4 are internalized and block low-pH-induced fusion. (A) Confocal microscopy of virus internalization into CHO-K1 cells with or without antibody 100F4. Cell samples were collected at 2, 40, and 360 min postinfection as indicated. Green, antibody 100F4 staining; red, viral NP staining; blue, DAPI staining for nucleus. (B) Colocalization of antibody 100F4 and viral NP, as determined by Pearson's correlation coefficients, was quantified at 2, 40, and 360 min postinfection. ***, P < 0.001. (C) Blockage of low-pH-triggered, wild-type H5 HA-mediated fusion by antibodies 65C6 and 100F4 but not PG16 control. Mean and standard deviation of fusion percentage in each image is shown below. (D) Blockage of low-pH-triggered, escape A of H5 HA-mediated fusion by antibody 65C6 but not by antibody 100F4 and PG16 control. Mean and standard deviation of fusion percentage in each image is shown below.
Finally, to determine whether antibodies 65C6 and 100F4 could block low-pH-triggered, HA-mediated fusion mediated by escape A of H5 HA, we transiently transfected plasmids expressing the wild-type and escape A of H5 HA into HeLa cells using Lipofectamine plus (Invitrogen) (17). The fusion assay was performed as described above. Both antibody 65C6 and 100F4 but not the PG16 control inhibited low-pH-triggered fusion mediated by wild-type H5 HA in a dose-dependent fashion (Fig. 4C), whereas only antibody 65C6 but not antibodies 100F4 and PG16 inhibited low-pH-triggered fusion mediated by escape A of H5 HA in a dose-dependent fashion (Fig. 4D). Thus, taking these results together with the confocal image data shown in Figure 4A, we conclude that although antibody 100F4 can still bind escape A of H5 HA and internalize with it into the perinuclear region, antibody 100F4 no longer blocks low-pH-triggered fusion mediated by escape A of H5 HA.
In summary, we unraveled a neutralization mechanism by antibodies 65C6 and 100F4 that differs from two other well-known mechanisms (21), i.e., antibodies that recognize the RBS and block binding of virus to receptors (12, 22–28) and antibodies that recognize the fusion domain of HA2 and block HA-mediated virus-cell membrane fusion (16, 18, 22, 29–37). Both antibody 65C6 and 100F4 recognize epitopes outside the RBS and bind virus before and after virus attaches to target cells, similar to the mouse MAbs described by Edwards and Dimmock (13). Antibody-bound viruses are internalized into the perinuclear region. However, antibody-bound wild-type H5 HA on the virion surface fails to mediate low-pH-triggered membrane fusion, whereas antibody-bound escape A of H5 HA still does.
ACKNOWLEDGMENTS
We thank L. Naldini at the University of Turin Medical School, Turin, Italy, for the lentiviral transfer vector.
This work was supported by research grants from the Chinese National Science Foundation (number 30671922), National Science and Technology Major Projects (numbers 2009ZX10004-105 and 2009ZX10004-016), and the Li Kai-Shing Foundation of Hong Kong.
Footnotes
Published ahead of print 26 December 2012
REFERENCES
- 1. Hu H, Voss J, Zhang G, Buchy P, Zuo T, Wang L, Wang F, Zhou F, Wang G, Tsai C, Calder L, Gamblin SJ, Zhang L, Deubel V, Zhou B, Skehel JJ, Zhou P. 2012. A human antibody recognizing a conserved epitope of H5 hemagglutinin broadly neutralizes highly pathogenic avian influenza H5N1 viruses. J. Virol. 86:2978–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zuo T, Shi X, Liu Z, Guo L, Zhao Q, Guan T, Pan X, Jia N, Cao W, Zhou B, Goldin M, Zhang L. 2011. Comprehensive analysis of pathogen-specific antibody response in vivo based on an antigen library displayed on surface of yeast. J. Biol. Chem. 286:33511–33519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsai C, Caillet C, Hu H, Zhou F, Ding H, Zhang G, Zhou B, Wang S, Lu S, Buchy P, Deubel V, Vogel FR, Zhou P. 2009. Measurement of neutralizing antibody responses against H5N1 clades in immunized mice and ferrets using pseudotypes expressing influenza hemagglutinin and neuraminidase. Vaccine 27:6777–6790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31:417–427 [DOI] [PubMed] [Google Scholar]
- 5. Stray SJ, Pittman LB. 2012. Subtype- and antigenic site-specific differences in biophysical influences on evolution of influenza virus hemagglutinin. Virol. J. 9:91 doi:10.1186/1743-422X-9-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Skehel JJ, Stevens DJ, Daniels RS, Douglas AR, Knossow M, Wilson IA, Wiley DC. 1984. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc. Natl. Acad. Sci. U. S. A. 81:1779–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Webster RG, Laver WG. 1980. Determination of the number of nonoverlapping antigenic areas on Hong Kong (H3N2) influenza virus hemagglutinin with monoclonal antibodies and the selection of variants with potential epidemiological significance. Virology 104:139–148 [DOI] [PubMed] [Google Scholar]
- 8. Khurana S, Suguitan AL, Jr, Rivera Y, Simmons CP, Lanzavecchia A, Sallusto F, Manischewitz J, King LR, Subbarao K, Golding H. 2009. Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med. 6:e1000049 doi:10.1371/journal.pmed.1000049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cao Z, Meng J, Li X, Wu R, Huang Y, He Y. 2012. The epitope and neutralization mechanism of AVFluIgG01, a broad-reactive human monoclonal antibody against H5N1 influenza virus. PLoS One 7:e38126 doi:10.1371/journal.pone.0038126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han T, Sui J, Bennett AS, Liddington RC, Donis RO, Zhu Q, Marasco WA. 2011. Fine epitope mapping of monoclonal antibodies against hemagglutinin of a highly pathogenic H5N1 influenza virus using yeast surface display. Biochem. Biophys. Res. Commun. 409:253–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, Cinatl J, ter Meulen J, Lasters I, Carsetti R, Peiris M, de Kruif J, Goudsmit J. 2008. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 3:e3942 doi:10.1371/journal.pone.0003942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oh HL, Akerstrom S, Shen S, Bereczky S, Karlberg H, Klingstrom J, Lal SK, Mirazimi A, Tan YJ. 2010. An antibody against a novel and conserved epitope in the hemagglutinin 1 subunit neutralizes numerous H5N1 influenza viruses. J. Virol. 84:8275–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Edwards MJ, Dimmock NJ. 2001. Hemagglutinin 1-specific immunoglobulin G and Fab molecules mediate postattachment neutralization of influenza A virus by inhibition of an early fusion event. J. Virol. 75:10208–10218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun L, Lu X, Li C, Wang M, Liu Q, Li Z, Hu X, Li J, Liu F, Li Q, Belser JA, Hancock K, Shu Y, Katz JM, Liang M, Li D. 2009. Generation, characterization and epitope mapping of two neutralizing and protective human recombinant antibodies against influenza A H5N1 viruses. PLoS One 4:e5476 doi:10.1371/journal.pone.0005476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, Wan H, Murakami A, Yammanuru A, Han T, Cox NJ, Bankston LA, Donis RO, Liddington RC, Marasco WA. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Follenzi A, Ailles LE, Bakovic S, Geuna M, Naldini L. 2000. Gene transfer by lentiviral vectors is limited by nuclear translocation and rescued by HIV-1 pol sequences. Nat. Genet. 25:217–222 [DOI] [PubMed] [Google Scholar]
- 18. Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, Silacci C, Fernandez-Rodriguez BM, Agatic G, Bianchi S, Giacchetto-Sasselli I, Calder L, Sallusto F, Collins P, Haire LF, Temperton N, Langedijk JP, Skehel JJ, Lanzavecchia A. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333:850–856 [DOI] [PubMed] [Google Scholar]
- 19. Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. 2003. Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 100:9280–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bolte S, Cordelieres FP. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232 [DOI] [PubMed] [Google Scholar]
- 21. Han T, Marasco WA. 2011. Structural basis of influenza virus neutralization. Ann. N. Y. Acad. Sci. 1217:178–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Prabhu N, Prabakaran M, Ho HT, Velumani S, Qiang J, Goutama M, Kwang J. 2009. Monoclonal antibodies against the fusion peptide of hemagglutinin protect mice from lethal influenza A virus H5N1 infection. J. Virol. 83:2553–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu X, Tsibane T, McGraw PA, House FS, Keefer CJ, Hicar MD, Tumpey TM, Pappas C, Perrone LA, Martinez O, Stevens J, Wilson IA, Aguilar PV, Altschuler EL, Basler CF, Crowe JE., Jr 2008. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 455:532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lim AP, Wong SK, Chan AH, Chan CE, Ooi EE, Hanson BJ. 2008. Epitope characterization of the protective monoclonal antibody VN04-2 shows broadly neutralizing activity against highly pathogenic H5N1. Virol. J. 5:80 doi:10.1186/1743-422X-5-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang J, Yoshida R, Kariya Y, Zhang X, Hashiguchi S, Nakashima T, Suda Y, Takada A, Ito Y, Sugimura K. 2010. Characterization of human single-chain antibodies against highly pathogenic avian influenza H5N1 viruses: mimotope and neutralizing activity. J. Biochem. 148:507–515 [DOI] [PubMed] [Google Scholar]
- 26. Chen Y, Qin K, Wu WL, Li G, Zhang J, Du H, Ng MH, Shih JW, Peiris JS, Guan Y, Chen H, Xia N. 2009. Broad cross-protection against H5N1 avian influenza virus infection by means of monoclonal antibodies that map to conserved viral epitopes. J. Infect. Dis. 199:49–58 [DOI] [PubMed] [Google Scholar]
- 27. Simmons CP, Bernasconi NL, Suguitan AL, Mills K, Ward JM, Chau NV, Hien TT, Sallusto F, Ha do, Farrar QJ, de Jong MD, Lanzavecchia A, Subbarao K. 2007. Prophylactic and therapeutic efficacy of human monoclonal antibodies against H5N1 influenza. PLoS Med. 4:e178 doi:10.1371/journal.pmed.0040178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, Kepler TB, Liao HX, Harrison SC. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc. Natl. Acad. Sci. U. S. A. 108:14216–14221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324:246–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Corti D, Suguitan AL, Jr, Pinna D, Silacci C, Fernandez-Rodriguez BM, Vanzetta F, Santos C, Luke CJ, Torres-Velez FJ, Temperton NJ, Weiss RA, Sallusto F, Subbarao K, Lanzavecchia A. 2010. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J. Clin. Invest. 120:1663–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burioni R, Canducci F, Mancini N, Clementi N, Sassi M, De Marco D, Diotti RA, Saita D, Sampaolo M, Sautto G, Pianezze M, Clementi M. 2010. Monoclonal antibodies isolated from human B cells neutralize a broad range of H1 subtype influenza A viruses including swine-origin influenza virus (S-OIV). Virology 399:144–152 [DOI] [PubMed] [Google Scholar]
- 33. Kashyap AK, Steel J, Oner AF, Dillon MA, Swale RE, Wall KM, Perry KJ, Faynboym A, Ilhan M, Horowitz M, Horowitz L, Palese P, Bhatt RR, Lerner RA. 2008. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc. Natl. Acad. Sci. U. S. A. 105:5986–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kashyap AK, Steel J, Rubrum A, Estelles A, Briante R, Ilyushina NA, Xu L, Swale RE, Faynboym AM, Foreman PK, Horowitz M, Horowitz L, Webby R, Palese P, Lerner RA, Bhatt RR. 2010. Protection from the 2009 H1N1 pandemic influenza by an antibody from combinatorial survivor-based libraries. PLoS Pathog. 6:e1000990 doi:10.1371/journal.ppat.1000990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yoshida R, Igarashi M, Ozaki H, Kishida N, Tomabechi D, Kida H, Ito K, Takada A. 2009. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog. 5:e1000350 doi:10.1371/journal.ppat.1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Okuno Y, Isegawa Y, Sasao F, Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 67:2552–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang TT, Tan GS, Hai R, Pica N, Petersen E, Moran TM, Palese P. 2010. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 6:e1000796 doi:10.1371/journal.ppat.1000796 [DOI] [PMC free article] [PubMed] [Google Scholar]