Abstract
We used the simian immunodeficiency virus mac251 (SIVmac251) macaque model to study the effect of the dose of mucosal exposure on vaccine efficacy. We immunized macaques with a DNA prime followed by SIV gp120 protein immunization with ALVAC-SIV and gp120 in alum, and we challenged them with SIVmac251 at either a single high dose or at two repeated low-dose exposures to a 10-fold-lower dose. Infection was neither prevented nor modified following a single high-dose challenge of the immunized macaques. However, two exposures to a 10-fold-lower dose resulted in protection from SIVmac251 acquisition in 3 out of 12 macaques. The remaining animals that were infected had a modulated pathogenesis, significant downregulation of interferon responsive genes, and upregulation of genes involved in B- and T-cell responses. Thus, the choice of the experimental model greatly influences the vaccine efficacy of vaccines for human immunodeficiency virus (HIV).
INTRODUCTION
The main goal of an effective vaccine for human immunodeficiency virus (HIV) is to protect from acquisition of infection. RV144 is the first human HIV vaccine trial showing that this goal is achievable; the vaccine strategy, a canarypox vector-based ALVAC-HIV given in a prime-boost combination with AIDSVAX B/E, resulted in a modest 31.2% significant efficacy (1).
In the preclinical setting, a vaccine equivalent to that of the Thai trial, tested in the nonhuman primate (NHP) model (2, 3), resulted in different outcomes depending on many experimental choices (4, 5), such as age of the animals, virus used, and dose of the challenge. Vaccination of macaques with ALVAC-HIV-2 vaccine and gp120 protein boost protected adult macaques from acquisition of the minimally pathogenic HIV-2 (4). On the other hand, a vaccine consisting of ALVAC-simian immunodeficiency virus (SIV) Gag-Pol, ALVAC-HIV-1 Env priming, and HIV gp120 protein boosting did not protect macaques from simian-human immunodeficiency virus ku2 (SHIVku2), a CXCR4 tropic challenge virus, but did reduce CD4+ T-cell loss (5). In the pathogenic CCR5-tropic SIVmac251 model, adult macaques vaccinated with ALVAC-SIV expressing Gag-Pol and Env (ALVAC-SIVgpe) and boosted with gp120 envelope protein were not protected from SIVmac251 acquisition after a relatively high-dose mucosal challenge (30 mucosal infectious doses), and the vaccinated animals that became infected were transiently protected from CD4+ T-cell loss (6). In contrast, 10 of the 16 neonatal macaques immunized with ALVAC-SIVgpe alone were protected from SIVmac251 acquisition following exposure to repeated doses of virus (∼104 50% tissue culture infective doses [TCID50] per challenge) (7). Repeated low-dose and high-dose challenges have been compared side by side only in unvaccinated macaques treated or not treated with antiretroviral therapy (8, 9) but not in the same model of macaques vaccinated with the same modality and challenged with an identical stock.
In the present study, we evaluated the effect of SIVmac251 challenge dose on vaccine efficacy. We performed a side-by-side comparison of the level of protection from infection and/or pathogenesis in identically vaccinated animals challenged mucosally with either a single higher dose of 6,100 TCID50 or to repeated lower doses of 470 TCID50 of the same SIVmac251 stock (10). Repeated low-dose and high-dose challenges have been compared side by side only in unvaccinated macaques treated or not treated with antiretroviral therapy (8, 9) but not in macaques vaccinated with the same regimen. We chose a DNA prime and ALVAC-SIVgpe, followed by SIV gp120 protein boost, with the goal of improving upon the limited efficacy observed during the RV144 HIV vaccine trial.
Our results clearly indicate that mucosal exposure to a high dose of SIVmac251 represents an overly stringent challenge that can overwhelm vaccine-induced immune responses that are sufficient to provide a measure of protection against repeated lower-dose challenges. The results demonstrate the importance of properly modeling the viral challenge in assessing relative efficacy of candidate vaccines for HIV.
MATERIALS AND METHODS
Animals and study design.
All of the animals used in this study were colony-bread rhesus macaques (Macaca mulatta) obtained from Covance Research Products (Alice, TX). The animals were housed and maintained in accordance with the standards of the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Advanced BioScience Laboratories, Inc., Institutional Animal Care and Use Committee. All surgery was performed under general anesthesia, and all efforts were made to minimize suffering. All macaques were negative for simian retrovirus, simian T-cell leukemia virus type 1, and herpesvirus B. The major histocompatibility status was determined as previously described (11).
Twenty macaques were immunized at 0 and 8 weeks with intramuscular injection of 2 ml (4 0.5-ml doses) of a DNA-SIVgpe mixture containing a total of 8 mg endotoxin-free SIVmac239-derived optimized plasmid DNAs in phosphate-buffered saline (PBS): 206Sp57gag (1 mg), 209S-MCP3-p39 (1 mg), 99S-SIVmac239 Env (2 mg), 103S-LAMP-pol (2 mg), and 147S-LAMP-NTV (2 mg) (12–14). All animals were boosted with ALVAC-SIVgpe at weeks 21, 40, and either 57 (6,100 TCID50 group) or 61 (470 TCID50 group) (Fig. 1A). Each vaccination with ALVAC-SIVgpe boost consisted of 108 PFU in a volume of 150 μl of PBS delivered intradermally and dispersed over three chest sites on each animal; at weeks 40 and either 57 or 61, all the animals received simultaneously the homologous gp120 protein, adjuvanted in alum, intramuscularly in the opposite thigh of the ALVAC-SIVgpe inoculum. Twenty macaques were used as controls and received alum only at weeks 40 and 57 or 61. All the animals were intrarectally exposed after 4 weeks from infection. A SIVmac251 stock (15) propagated in human cells was utilized at the doses of 6,100 or 470 TCID50.
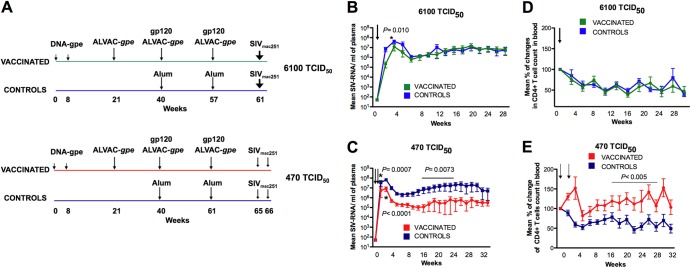
Fig 1.
Effect of vaccination on SIV replication and CD4+ T cells. (A) Immunization regimen. All macaques were immunized with DNA and ALVAC SIVgpe at the time indicated by arrows. The alum-adjuvanted monomeric native SIVmac251 gp120 protein was given at weeks 40 and 57/61. Twenty control animals received alum only. Eight vaccinated and eight control macaques were exposed to a single dose of 6,100 TCID50 of SIVmac251 at week 61. Twelve vaccinated and 12 control macaques were exposed weekly to 470 TCID50 of SIVmac251 until all control animals had detectable plasma virus. Plasma SIV-RNA values are depicted as means ± standard errors for vaccinated (green) and control (blue) animals exposed to one dose of 6,100 TCID50 (B) or two doses of 470 TCID50 (C) of SIVmac251. The arrows indicate the time of exposure, and the P values indicate differences between the vaccinated and controls within groups (Wilcoxon rank sum test). Changes in the CD4+ T-cell count in blood expressed as means ± standard errors of the percentage of change with respect to the prechallenge level in vaccinated and control macaques exposed to 6,100 TCID50 (D) or to 470 TCID50 (E) of SIVmac251. CD4+ T-cell numbers were reconstituted significantly in vaccinated macaques exposed to 470 TCID50 of SIVmac251 (weeks 16 to 30, P < 0.005, by the Wilcoxon rank sum test). The SIV-exposed uninfected animals were excluded from the analysis.
Detection of viral variants by single-genome analysis.
Plasma SIV RNA was quantified by nucleic acid sequence-based amplification (NASBA), as previously described (16). SIV DNA was quantified in the blood and tissue of macaques by quantitative PCR, as previously described (17). Transmitted or founder viruses and their progeny were identified by single-genome amplification of SIV RNA (from plasma at peak viremia and early set point, rectal pinches, or lymph node biopsy specimens), direct amplicon sequencing (ENV PRIMERS), and phylogenetic analysis, as previously described (10). SIV RNA was extracted from specimens, and limiting-dilution PCR of newly synthesized cDNA was performed. Transmitted/founder virus lineages were identified by low-diversity sequences and by single sequences with unique mutations. Phylogenetic trees were generated using ClustalW.
Enumeration of CD4+ T cells in blood and tissues.
CD4+ T-cell counts were periodically determined from whole blood by flow cytometry, as previously described (18).
CD4+ T-cell number, in tissue, was determined in rectal pinch biopsy specimens by immunohistochemistry.
All slides were stained using the Dako autostainer (Dako Inc., Carpinteria, CA). Slides were visualized with epifluorescence illumination using a Zeiss Axioplan 2 microscope (Carl Zeiss) and appropriate filters. Digital images were captured and analyzed by using the Zeiss Axiocam System and Openlab software (Inprovision), as previously described (19). Briefly, the primary antibodies used included monoclonal anti-CD4+ T-cell mouse serum antibody (clone IF6; Vector, Burlingame, CA). Binding of CD4+ T cells was detected using Alexa Fluor 488-labeled polyclonal goat anti-rabbit IgG (Molecular Probes, Eugene, OR). Slides were visualized with epifluorescence illumination using a Zeiss Axioplan 2 microscope (Carl Zeiss Inc., Thornwood, NY) and appropriate filters. Digital images were captured and analyzed by using a Zeiss Axiocam System and Openlab software (Inprovision Inc., Waltham, MA). Only cells with perfect membrane staining, strong fluorescence, and correct cellular structure were considered positive. The number of positive cells is presented as the number of cells per square millimeter.
IFN-γ ELISPOT.
Gamma interferon (IFN-γ) enzyme-linked immunosorbent spot assay (ELISPOT) was performed as described elsewhere (17). Briefly, multiscreen 96-well plates were coated with 50 to 100 μl/well of purified capture antibody mouse human-IFN-γ monoclonal antibodies (MAbs) (B27-MABTECH) at 10 μg/ml in coating buffer (PBS, pH 7.2). Peripheral blood mononuclear cells (PBMCs) were seeded at 50 μl/well (2 × 105 cells/well in triplicate) with the appropriate peptide (1-μg/ml final concentration) or concanavalin A (ConA) (Sigma-Aldrich) as a positive control (2-μg/ml final concentrations). The biotinylated anti-IFN-γ antibody rabbit human-IFN-γ MAbs (MAb7B61; MABTECH) was used for detection. The analyses were performed counting the spots with a microscope and using the software KS ELISPOT (Carl Zeiss Vision GmbH, Germany).
Lymphocyte proliferation assay.
Lymphocyte proliferation assays were performed as described previously (17). Briefly, fresh PBMCs were cultured for 3 days in the absence or the presence of native high-performance liquid chromatography (HPLC)-purified SIVmac251 Gag p27 or Env gp120 proteins (Advanced BioScience Laboratories) or ConA as a positive control. The cells were then pulsed overnight with 1 μCi of [3H]thymidine before harvest. The relative rate of lymphoproliferation was calculated as the fold thymidine incorporation into cellular DNA over medium control (stimulation index).
Intracellular cytokine assay.
The frequency of cytokine-producing memory (CD95+) CD8+ and CD4+ T cells was measured by intracellular cytokine staining. No less than 1 × 106 mononuclear cells were stimulated for 6 h with or without SIV-specific Gag and Env of peptide pools (1 mg/ml) or with phorbol myristate acetate (PMA)-ionomycin (1 mg/ml) as a positive control, in the presence of the costimulatory monoclonal antibodies CD49d and CD28 (0.5 mg/ml; BD Biosciences). ECD-conjugated CD28 was added to the mix to maximize the detection of T cells with high activation thresholds (20). Anti-human CD107a (clone H4A3; BD BioScience) was added to the culture. Following 1 h of stimulation, brefeldin A (Sigma-Aldrich, St. Louis, MO) was added at a final concentration of 10 mg/ml. Cells were stained for 30 min at room temperature with antibodies to CD3 (clone SP34-2), CD4 (clone L200), CD8 (clone RPA-T8), and CD95 (clone DX2), all obtained from BD Biosciences (San Diego, CA). Following surface staining, lymphocytes were permeabilized with Cytofix/Cytoperm solution (BD Biosciences) and stained intracellularly for IFN-γ (clone B27), tumor necrosis factor alpha (TNF-α) (clone MAB11), and interleukin 2 (IL-2) (clone MQ1-17H12), all from BD Biosciences. PBMCs were fixed with 1% paraformaldehyde, and at least 100,000 events were acquired on either a FACSCalibur or LSRII (BD Bioscience). Data analysis was performed with FlowJo (Treestar, CA).
Binding antibody assay.
To detect anti-SIVmac251-binding antibodies, serial dilutions of plasma were incubated with the lysate of SIVmac251 spiked with native purified gp120 Env protein of SIVmac251 bound to microtiter enzyme-linked immunosorbent assay (ELISA) plates, as described elsewhere (21, 21). Endpoint titers were defined as the reciprocal of the highest serum dilution that gave an optical density at 450 nm (OD450) at least 2 standard deviations greater than average values obtained with negative-control serum.
Avidity assay.
A format of previously described capture ELISA for HIV-1 gp120, in which HIV-1 gp120 is captured through its C terminus by a goat antibody, D7324, was adapted for detecting Ab against SIV gp120. Recombinant SIV gp120 protein made from codon-optimized SIVmac239 gp120 fused to the C-terminal tag of HIV-1 gp120 is used as an antigen for the capture ELISA to detect SIV Abs against conformational epitope.
Ab avidity was determined by parallel ELISA. Heat-inactivated plasma samples were serially diluted and applied to a 96-well plate capturing SIVmac239 gp120 as described above in parallel duplicates. After 1 h of incubation, the plate was washed, and half the samples were treated with Tris-buffered saline (TBS), while the paired samples were treated with 1.5 M sodium thiocyanate (NaSCN; Sigma-Aldrich) for 10 min at room temperature. The plate was washed and a goat anti-monkey IgG-detecting Ab (Fitzgerald) was used. The avidity index (%) was calculated by taking the ratio of the NaSCN-treated plasma dilution giving an OD of 0.5 to the TBS-treated plasma dilution giving an OD of 0.5 and multiplying by 100. Plasma of uninfected normal macaques served as negative controls. A high-avidity monkey MAb of 3.11H was included on every plate as the standard.
ADCC assay.
Antibody-dependent cell-mediated cytotoxicity (ADCC) activity mediated by plasma samples was detected by the GranToxiLux (GTL) procedure, as previously described (22). Briefly, CEM.NKRCCR5 target cells (23) were coated with recombinant SIVmac251 gp120 (Advanced Bioscience Laboratories Inc., Kensington, MD) and labeled with a fluorescent target cell marker (TFL4; OncoImmunin, Inc., Gaithersburg, MD) and a viability marker (NFL1; OncoImmunin, Inc.). Labeled target cells were washed and plated at 1 × 104 viable cells/well. Cryopreserved human PBMCs from an HIV-seronegative donor served as effectors and were added to the assay wells at an effector/target ratio of 30:1. Fluorogenic granzyme B (GzB) substrate (OncoImmunin, Inc.) was added to each well to a final concentration of 0.25×. After incubation for 5 min at room temperature, serially diluted serum/plasma samples (4-fold, starting at 1:100) were added to the assay wells. The plates were incubated for 15 min at room temperature, centrifuged for 1 min at 300 × g, and incubated for 1 h at 37°C and 5% CO2. The plates were then washed, and cells were resuspended in 200 μl of 1% fetal bovine serum (FBS)-PBS. A minimum of 2,500 events representing viable target cells was acquired for each well using an LSRII flow cytometer (BD Biosciences, San Jose, CA). Data analysis was performed using FlowJo 8.8.4 software (Tree Star Inc., Ashland, OR). The percentage of GzB activity was defined as the percentage of cells positive for proteolytically active GzB out of the total viable target cell population. The final results are expressed after subtracting the background, represented by the percentage of GzB activity observed in wells containing effector and target T cells in the absence of serum/plasma. The results were considered positive if the percentage of GzB activity after background subtraction was >8%.
Standard neutralization assays.
Neutralization was measured as a reduction in luciferase reporter gene expression after a single round of infection in either TZM-bl or 5.25.EGFP.Luc.M7 (M7-Luc) cells (24). TZM-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program, as contributed by John Kappes and Xiaoyun Wu. M7-Luc cells were kindly provided by Nathaniel R. Landau. For the TZM-bl assay, 200 50% tissue culture infective doses (TCID50) of virus were incubated with serial 3-fold dilutions of test sample for 1 h out of 18 h at 37°C. M7-Luc cells were maintained as previously described (25). For neutralization assays, 500 TCID50 of virus were incubated with serial dilutions of serum samples for 1 h at 37°C. Freshly trypsinized cells were added to each well, in the presence (virus control) or in the absence (background control) of virus. A total of 100 μl of cells was used for measurements of luminescence (Britelite Luminescence Reporter Gene Assay System, PerkinElmer Life Sciences). Neutralization titers are measured as dilution at which relative luminescence units (RLU) are reduced by 50% compared to virus control wells, after subtraction of background RLUs.
Assay stocks of molecularly cloned Env-pseudotyped viruses (SIVmac251CS.41, SIVmac239.23, SIVmac251WY:30) were prepared by transfection in 293T cells and were titrated in TZM-bl cells as described previously (24, 26). Assay stocks of uncloned TCLA-SIVmac251 and SIVmac251CS/2002 were produced in H9 and human PBMCs, respectively, and were titrated in M7-Luc and TZM-bl cells, respectively.
Antibody-dependent cell-mediated viral inhibition.
CEM.NKR-CCR5l (catalog number 4376; AIDS Reagent Program) were infected with SIVmac251 at a multiplicity of infection (MOI) of 0.1 for 48 h with Polybrene (Sigma-Aldrich; 10 μg/ml). Cells were washed in complete medium and plated into 96-well round-bottom microtiter plates at a density of 50,000 cells/well in the presence of heat-inactivated (56 οC) rhesus macaque plasma at a final concentration of 1:100. Fresh human health donor PBM complements were added at a density of 500,000 cells/well to achieve a 10:1 effector-to-target ratio in 200-μl final volumes. All test and control plasma samples were plated in duplicate and cultured for 48 h prior to being washed off (90% volume, 5×), and well volumes were replaced with complete medium. Plates were cultured for an additional 5 days, at which time 50-μl supernatant fluid samples were analyzed for p27 antigen by ELISA (ZeptoMetrix, Buffalo, NY).
Antibody-dependent cell-mediated viral inhibition (ADCVI) activity is expressed as a percent reduction in p27 quantity referenced to negative plasma control wells (mean of 12 nonvaccinated animals). Data reported represent the means from three independent assays.
RNA purification, amplification, and array hybridization.
Cubes (0.5 to 1 cm) of snap-frozen rectal mucosal biopsy specimens were placed into 350 μl of RLT buffer (Qiagen, Valencia, CA), and total RNA was purified using RNeasy minikits (Qiagen) according to the manufacturer's specifications with optional on-column DNase digestion. RNA quality was quantitated by Nanodrop analysis, and Agilent Bioanalyzer capillary electrophoresis was used for quality assessment; all samples had an RNA integrity number (RIN) of >6.9, and the majority were >8.5. RNA was amplified and labeled using the NuGen Ovation Pico system according to kit instructions. Amplified RNA was assessed by Agilent capillary electrophoresis for homogeneity. Amplified RNA was hybridized to Affymetrix GeneChip rhesus macaque genome arrays (Affymetrix, Santa Clara, CA).
Microarray data analysis.
Background adjustment, normalization, and median polish summarization of .CEL files were performed using the robust multichip average (RMA) algorithm. Quality of hybridized chips was assessed after normalization by the distribution of their NUSE and RLE plots using Bioconductor. Downstream analyses were performed using Partek Genomics Suite software version 6.5 (Partek Inc., St. Louis, MO) with the exception of Bayesian analysis, which were performed using limma in Bioconduction. To identify genes potentially regulating vaccine-mediated protection, we contrasted vaccine recipient animals from control animals in the respective challenge groups; vaccine recipients that did not seroconvert were excluded. We initially ran an analysis of variance (ANOVA) between the vaccine and control groups; however, few genes were determined to be significantly different. To increase the sensitivity of detecting novel genes regulating vaccine effects, we utilized Bayesian statistics. To lower the likelihood of false positives, we chose a false discovery rate (FDR) threshold of 0.02 for Bayesian analysis and a fold change cutoff of ±1.5-fold relative to unvaccinated controls. Heat maps were generated using Partek software.
Annotation of the rhesus genome array with human gene symbol identifiers.
Affymetrix annotation of the rhesus GeneChip maps to the rhesus genome, and >11,000 probe sets have provisional gene symbols in the NCBI gene (i.e., “LOC” symbols). To increase the mapping of the array, probe sets lacking definitive annotation were cross-referenced to potential human orthologs identified by the online resources provided by the laboratory of Robert Norgren (version 3, June 2010) (http://www.unmc.edu/rhesusgenechip/) and the InParanoid ortholog database (release 7.0, June 2009) (http://inparanoid.sbc.su.se), using probe set identifiers and Ensembl gene/protein identifiers, respectively. To reduce ambiguity, we have included the latest annotation of the Affymetrix rhesus array (version 32) and gene symbols determined by cross-referencing in Table S1 in the supplemental material.
Statistic analysis.
Statistical analysis involved two-group comparisons of log-transformed frequencies performed using repeated-measure ANOVA and corrected for multiple tests. Correlations were assessed using the Spearman rank correlation method. The statistical software used was SAS version 9.3 (SAS Institute Inc., Cary, NC).
GEO accession numbers.
The microarray data set was submitted to the GEO online repository according to Minimum Information About a Microarray Experiment (MIAME) standards (GEO accession numbers GSE37320, GSE37311, GSE37312).
Nucleotide sequence accession numbers.
All 914 envelope sequences were deposited in GenBank under accession numbers KC116226 to KC117139.
RESULTS
The dose of challenge affects vaccine efficacy.
We randomized 40 macaques in 4 groups based on the expression of the major histocompatibility complex class I (MHC-I) associated with protection from high viral replication (Mamu-A*01, Mamu-B*08, and Mamu-B*17) and based on the expression of protective alleles for TRIM5α (27–29). We vaccinated 20 adult macaques with DNA SIV gag, pol, and env genes followed by (30) three immunizations with recombinant ALVAC-SIVgpe (6) expressing the same genes. At the time of the last two immunizations with ALVAC-SIVgpe, all of the animals were also immunized with native SIVmac251 gp120 formulated in alum (Fig. 1A). Twenty macaques received alum alone as the control (Fig. 1A).
To assess vaccine protection against different doses of SIVmac251, we exposed all animals intrarectally to either a single high dose (6,100 TCID50) or a weekly low dose (470 TCID50) of SIVmac251 (15), 4 weeks after the last immunization, until all controls became infected (two challenges).
Vaccination afforded no protection to the animals exposed to 6,100 TCID50 from either SIVmac251 acquisition or CD4+ T-cell loss. There was only a significant but transient reduction of viral replication compared to that of the controls within the first few weeks after challenge (week 2; P = 0.010; week 3; P = 0.040) (Fig. 1B and Table 1; see also Fig. S1A in the supplemental material). When the three animals that expressed Mamu-A*01 or Mamu-B*08 or Mamu-B*17 alleles were excluded from this analysis, the viral loads of controls and vaccinated animals were no longer significantly different at weeks 2 and 3 (data not shown).
Table 1.
Virus levels in vaccinated and control macaques exposed to 6,100 and 470 TCID50
| Group and infection phase |
P valuea |
||||
|---|---|---|---|---|---|
| SIV RNA in plasma | SIV DNA in rectal mucosa | CD4+ T cells in blood | CD4+ T cells in rectal mucosa | Virus variant | |
| 6100 TCID50 | |||||
| Acute | 0.01/ 0.04 (wk 2/wk 3) | NS | NS | NS | NS |
| Chronic | NS | ND | NS | NS | ND |
| 470 TCID50 | |||||
| Acute | 0.0007 (wk 2) | 0.009 (wk 3) | NS | NS | 0.046 |
| Chronic | 0.0073 (wk 8–34) | ND | <0.005 (wk 16–28) | 0.0021 (wk 11) | ND |
NS, nonsignificant; ND, not determined.
In contrast, in the group of animals exposed to the two lower doses (470 TCID50), vaccination protected three out of 12 immunized macaques from SIVmac251 acquisition, whereas all the control macaques were infected after the last exposure (Fig. 1C; see also Fig. S1B in the supplemental material). The efficacy of protection from acquisition afforded by this vaccine was clearly not significant, due to the low number of animals in each group and, possibly, because 470 TCID50 resulting in 50% risk of infection per exposure can be considered an “intermediate dose” rather than a low dose. The three protected animals, P006, P018, and P010, had undetectable SIV RNA in plasma (<50 copies/ml) (see Fig. S1B) and no detectable T-cell responses in rectal biopsy specimens to the regulatory proteins Tat, Rev, and Nef (not included in the vaccine) at week 2 following virus exposure (data not shown). Of the 3 macaques, only P006 was Mamu-A*01, while P008 and P010 were negative for any protective MHC-I alleles tested, and TRIM5α alleles did not account for the lack of SIVmac251 acquisition in these macaques as we previously demonstrated (27, 28). Indeed, animals P006, P018, and P010, initially protected from SIVmac251 acquisition, were readily infected following reexposure to a single high dose of the same SIVmac251 virus stock (see Fig. S1C). Interestingly, compared with unvaccinated controls, the nine vaccinated animals that became infected after low-dose challenges had significantly less plasma virus during acute infection (P = 0.0007) and thereafter (P = 0.0073) (Fig. 1C and Table 1). When the animals expressing protective MHC-I alleles were excluded from this analysis, the significant difference between vaccinees and controls was retained (data not shown).
To assess whether the vaccine was more effective against the 10-fold-lower dose of virus, we compared plasma virus levels in the vaccinees and controls exposed to the two doses of SIVmac251. A clear trend was observed between the two vaccinated groups when all the animals were analyzed (see Fig. S1D in the supplemental material), and this difference became significant when macaques, with protective alleles, were excluded (Wilcoxon rank test, 7 to 31 weeks postinfection, P = 0.038) (see Fig. S1E). No significant differences were observed between the two control groups, even when animals carrying protective alleles were excluded (data not shown).
The absolute CD4+ T-cell number in blood decreased equivalently during acute infection in the vaccinated and control macaques exposed to 6,100 TCID50 of SIVmac251 (Fig. 1D). Strikingly, however, the vaccinated animals that became infected after challenge with the lower SIVmac251 dose experienced only a modest and transient reduction of CD4+ T cells in the blood (Fig. 1E). CD4+ T cells in the blood differed significantly between vaccinated animals and controls (week 16 to 28, vaccinated versus unvaccinated, P < 0.005; Table 1).
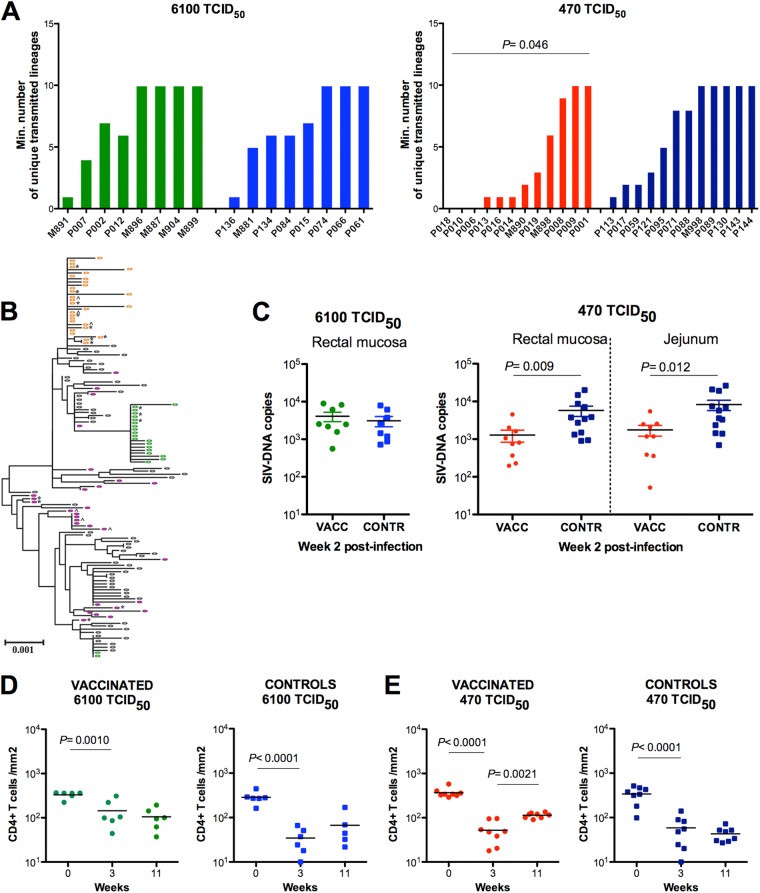
Single-genome amplification (SGA) and direct sequencing of the SIV env gene in the plasma of the infected animals demonstrated unique transmitted viral variants identified by phylogenetic analysis, within the context of the inoculum sequences, as described previously (10). Control and vaccinated animals exposed to 6,100 TCID50 were infected with a median of 6.5 and 8.5 variants, respectively (P = 0.79) (Fig. 2A). Similarly, control animals exposed to the lower doses had a median of 8 virus variants, both in the plasma and at the mucosal site. These results, demonstrating comparable numbers of transmitted variants for control animals exposed to 6,100 or 470 TCID50, likely reflect an underestimation of the total number of variants transmitted. The ability of the SGA method employed to determine the exact number of variants transmitted is challenged as the number of transmitted variants approaches 10, potentially blurring a distinction between the number of variants transmitted with inocula of either 6,100 or 470 TCID50. The results also suggest that even at the lower inoculum of 470 TCID50, more variants were transmitted in controls than is typical of human HIV-1 transmissions or was seen with more extensive titrations of mucosal challenges with SIV. This makes it even more striking that the vaccinated macaques that became infected after challenge with the lower dose had a significant reduction in the number of transmitted variants (median = 1.5; P = 0.046) (Fig. 2A). An example of sequences obtained from the plasma of three vaccinated animals is shown in Fig. 2B, where P013 shows a single transmitted/founder lineage, M896 shows two distinct lineages, and P001 shows at least 10 distinct variants.
Fig 2.
Vaccination affects the number of virus variants transmitted and virus level. (A) Estimated number of transmitted viral lineages at 6,100 TCID50 and 470 TCID50 in vaccinated (green and red bars) and control animals (blue bars). (B) Phylogenetic tree of env gene from the inoculum stock (black ovals), and 3 vaccinated animals challenged with 470 TCID50. Sequences from P013 (orange ovals) and M890 (green ovals) cluster together in 1 or 2 lineages, which demonstrates evidence of 1 and 2 variants establishing systemic infection. Sequences from P001 (purple ovals) are found dispersed throughout the tree, providing evidence of multiple founder viruses (>10). Sequences were obtained from blood plasma with the exception of jejunum (^) or rectal (*) biopsy specimen sequences obtained at week 2 postinfection. (C) Log-transformed viral DNA copies in snap-frozen rectal or jejunal biopsy specimens collected at week 2 after infection in vaccinated and control macaques exposed to either 6,100 TCID50 (left) or 470 TCID50 (right) of SIVmac251. Vaccinated animals are in green or red and the respective controls in blue. P values are calculated using the Wilcoxon rank sum test, excluding the three vaccinated but uninfected animals, P006, P018, and P010. The frequency of CD4+ T cells per mm2 in the rectal mucosa of vaccinated and control macaques was enumerated by immunohistochemistry prior to infection, and at weeks 3 and 11, following the challenge exposure of vaccinated and control macaques to a single dose of 6,100 TCID50 (D) or two doses of 470 TCID50 (E) of SIVmac251. P values are calculated by repeated-measure analysis of variance, and the vaccinated uninfected animals P006, P018, and P010 are excluded from this analysis.
Vaccination also resulted in reduction of SIV DNA levels in the rectum and jejunum of infected vaccinated animals, compared to those of controls exposed to the lower SIVmac251 doses (P = 0.009 and P = 0.012, respectively), but not in those exposed to the high dose of SIVmac251 (Fig. 2C and Table 1). Importantly, while we observed a significant loss of CD4+ T cells in the rectal mucosa during acute infection in both vaccinated and control groups, as expected (Fig. 2D and E), CD4+ T cells were significantly reconstituted over time only in vaccinated macaques exposed to 470 TCID50 (P = 0.0021) (Fig. 2E and Table 1).
The dose of challenge affects immunological parameters associated with infection.
The small number of animals protected from SIVmac251 acquisition in this study did not allow the investigation of correlates of protection from infection. Therefore, we investigated the immunological parameters that may have contributed to the durable control of virus replication and regeneration of CD4+ T cells seen in the vaccinees that became infected. Vaccination induced Gag- and Env-specific CD4+ and CD8+ T-cell responses in the blood of all 20 vaccinated animals, as measured by ELISPOT, intracellular cytokine staining (ICS), and proliferative assays (Fig. 3; see also Fig. S2 in the supplemental material). SIV-specific responses were not detected in unvaccinated animals (data not shown) prior to challenge exposure to SIVmac251, and no significant differences were found between the two vaccinated groups before SIV exposure.
Fig 3.
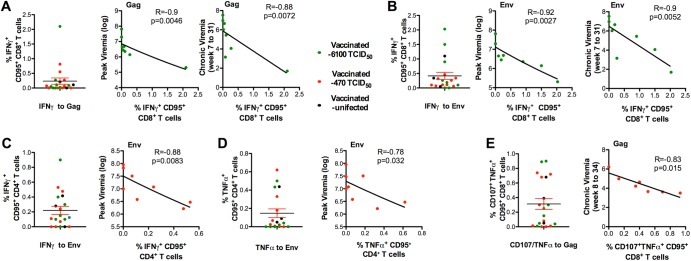
Immune responses that correlate with protection from high plasma virus levels. For panels A to E, single dots represent the frequency in blood of cytokine-producing memory (CD95+) CD4+ or CD8+ T cells from all vaccinated macaques at 3 weeks before SIVmac251 exposure; the background measured from nonstimulated cells has already been subtracted. Vaccinated animals exposed to 470 TCID50 are depicted in red; vaccinated animals exposed to 6,100 TCID50 are depicted in green, controls are in blue, and the protected animals P010, P006, and P018 are in black. The frequency of memory CD8+ T cells producing IFN-γ after stimulation with Gag (A) and Env (B) peptides is shown in the left panel for all vaccinated animals. The correlations between the values of vaccinated macaques exposed to 6,100 TCID50 and the peak viral loads and the median of viral loads measured from weeks 7 to 31 are depicted in the left panels. The frequency of envelope-specific memory CD4+ T cells producing IFN-γ (C) or TNF-α (D) is depicted in the left panel for all vaccinated animals. The correlations between these values and the peak viral loads for vaccinated macaques exposed to 470 TCID50 are depicted in the left panel. The values plotted for the peak virus level correspond to the highest RNA SIV levels measured after viral exposure (week 2 or 3) for each animal. The values plotted for the peak viral load correspond to the highest RNA SIV levels measured in the plasma after viral exposure (week 2) for each animal. (E) Frequency of Gag CD107+ TNF-α+-producing memory (CD95+) CD8+ T cells measured in mononuclear cells from lymph nodes in all macaques (left) and correlation between these values and the median of viral loads measured from weeks 8 to 34 in macaques exposed to the 470 TCID50 dose (right).
No correlation of plasma viremia with cytokine-producing CD4+ T cells was found in the group of vaccinated macaques exposed to 6,100 TCID50 (data not shown and Table 2); there was an inverse correlation of plasma viremia during acute infection with the frequency of Gag and Env (CD95+) CD8+ T cells producing IFN-γ (R = −0.90, P = 0.0046; R = −0.92, P = 0.0027, respectively) (Fig. 3A and B and Table 2), as well as during chronic infection (R = −0.88, P = 0.0072; R = −0.90, P = 0.0052, respectively). In contrast, in vaccinated macaques infected after exposure to 470 TCID50, acute viremia correlated inversely with the frequency of Env-specific, but not Gag-specific, memory (CD95+) CD4+ T cells, producing either IFN-γ or TNF-α measured at 1 week after the last immunization (Fig. 3A and B and Table 2) (Env, R = −0.88, P = 0.0083 for IFN-γ; R = −0.78, P = 0.032 for TNF-α). The vaccinated macaques challenged with 470 TCID50 sustained lower chronic phase plasma viremia correlated with the frequency of Gag-specific CD8+ T cells with an effector function and degranulation ability (CD107+/TNF-α+) (R = −0.83; P = 0.015; median viral load, 8 to 34 weeks) (Fig. 3E and Table 2).
Table 2.
Correlates of protection from a high level of SIV replication in vaccinated macaque
| Group and infection phase | Cytokine; R value; P valuea |
|||
|---|---|---|---|---|
| CD4+ T cells, SIV Env | CD4+ T cells, SIV Gag | CD8+ T cells, SIV Env | CD8+ T cells, SIV Gag | |
| 6,100 TCID50 | ||||
| Acute | NS | NS | IFN-γ; R = −0.92; P = 0.0027 | IFN-γ; R = −0.9; P = 0.0046 |
| Chronic | NS | NS | IFN-γ; R = 0.9; P = 0.0052 | IFN-γ; R = −0.88; P = 0.0072 |
| 470 TCID50 | ||||
| Acute | IFN-γ; R = −0.88; P = 0.0083 | NS | NS | NS |
| TNF-α; R = −0.78; P = 0.032 | NS | NS | NS | |
| Chronic | NS | NS | CD107/TNF-α; R = −0.88; P = 0.0031 | |
P values refer to Spearman correlations between frequencies of T cells producing cytokines measured in blood 1 week after the last immunization and 3 weeks before exposure to SIVmac251. NS, nonsignificant; ND, not done.
Lastly, we did not observe a significant correlation between the levels of SIVmac251 in plasma and the number of Gag-specific IFN-γ-producing cells measured by ELISPOT (see Fig. S2A in the supplemental material) in either vaccinated group or with Gag-specific or Env-specific lymphocyte proliferative responses in blood.
No correlation was found between plasma viremia and the titer of binding and neutralizing antibodies to lab-adapted viruses or titers of antibody-dependent cell-mediated cytotoxicity (ADCC), antibody-dependent cell-mediated viral inhibition (ADCVI), or the avidity to the SIV gp120 measured in plasma at 3 weeks from the last immunization (see Fig. S2C and S2D in the supplemental material; data not shown).
Gene expression profile in the mucosa of vaccinated and control macaques.
Vaccination with recombinant ALVAC induces innate responses that may be able to suppress SIV replication (31–33). We measured mRNA levels of RANTES, MIP-1α, and APOBEC-3G (A3G), by reverse transcription (RT)-PCR in rectal mucosa biopsy specimens collected 1 week after the final boost immunization, 3 weeks prior to SIVmac251 exposure. We found no correlation with the level of expression of these molecules and control of viremia (data not shown).
To gain further insight into the mechanism of protection from viremia induced by vaccination, microarray analysis was performed on RNA from jejunal biopsy specimens collected at 3 weeks postexposure, when no difference in total number of CD4+ T cells was found between the vaccinated groups and controls. In addition, we also collected biopsy specimens at 11 weeks postexposure, when a significant reconstitution of CD4+ T cells was observed in the vaccinated animals exposed to 470 TCID50 but not other vaccinated animals.
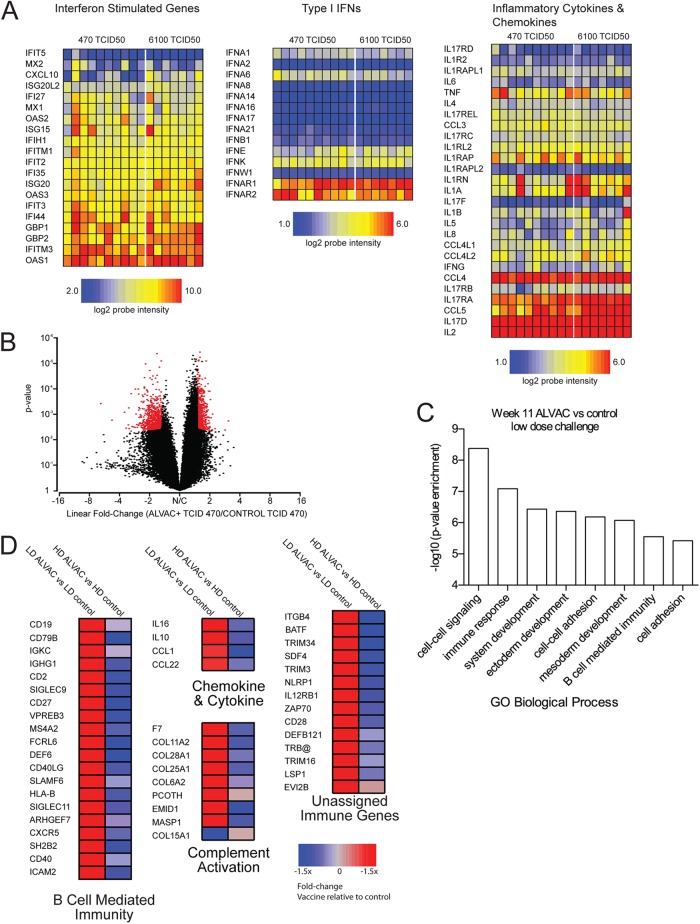
Microarray analysis performed at 3 weeks postexposure on controls revealed no difference in the fold change threshold (FDR) for either the group exposed to 6,100 TCID50 or to 470 TCID50 (data not shown). Indeed, the dose of SIVmac251 exposure per se did not result in differences in the level of immune activation, since expression of the type I IFN, interferon stimulated genes (ISG), or inflammatory cytokines was equally affected at mucosal sites (Fig. 4A), suggesting that differences in the protection afforded by the vaccine was not a direct effect of difference in immune activation.
Fig 4.
Microarray analysis of mucosal gene expression in rhesus macaques vaccinated with ALVAC. cRNA prepared from rectal mucosa punch biopsy specimens was hybridized on Affymetrix rhesus macaque genome arrays. (A) Heat maps of log2 intensities of probe sets representing inflammatory and interferon genes from mucosal biopsy samples taken from control animals at 3 weeks postinfection. Intensity scales for individual gene groups are shown below. (B) Volcano plot of global gene expression in macaques 11 weeks after infection from animals exposed to 470 TCID50 SIVmac251. The x axis indicates the mean ratio of expression expressed in log2 scale; positive values are probe sets overexpressed in vaccinated animals relative to controls. The y axis indicates the significance of differential expression between vaccinated and control animals as determined by Bayesian analysis. Red dots indicate probe sets found to be significantly expressed (FDR ≤ 0.05; expression fold change ≥ ±1.5-fold). (C) Ontology analysis of enriched gene families within significantly expressed probe sets was determined using the PANTHER database (www.panther-db.org) using the human genome as a reference. Significantly, enriched GO pathways are indicated with significance on the y axis. The number of genes within pathways is indicated above individual bars. (D) Heat maps of genes identified by ontology analysis as regulating the immune response at week 11 postinfection; genes were further classified by their GO biological processes or PANTHER protein classes and are displayed as individual heat maps. The left columns of heat maps indicate the average ratio of gene expression of vaccinated to control animals exposed to 470 TCID50; as a comparison, the right columns display the vaccinated-to-control expression in animals challenged with 6,100 TCID50. Gene symbols are indicated at right, and P values for the lower-dose comparison at left. The color scale is indicated at the bottom.
However, at week 11 following infection, a significant difference in gene expression between vaccinated and control animals exposed to 470 TCID50 was readily detectable with 1,386 probe sets (FDR < 0.05; fold change, ±1.5-fold) (Fig. 4B). A complete list of differential probe sets is listed in Table S1 in the supplemental material. In contrast, using the same criteria, no genes were found to be differentially expressed in the vaccinated animals and controls exposed to 6,100 TCID50 (Fig. 4D).
Probe sets with differential expression in the macaques exposed to 470 TCID50 were organized into functional groups using GO ontology (Fig. 4C). A certain number of genes were associated with immune response functions; the top-scoring biological processes are depicted in Fig. 4C. Interestingly, pathways involving tissue development were also significantly enriched, in accordance with the increased number of CD4+ T cells seen in the gut of vaccinated animals compared to the controls (Fig. 2E). Of the individual genes annotated, several were ascribed to regulating B-cell immunity, associated with B-cell memory responses (CD40, CD40LG, CD27 genes), or associated with T cells and NK cells, such as expression of CD2 and two CD2 family members, SLAMF6/NTBA, that regulate both B-lymphocyte and NK cell activation (11). Adding to the evidence that B-cell-related transcripts were overexpressed in mucosal tissue of vaccinated animals exposed to the lower-dose challenge, we noted significantly increased expression of several genes involved in B-cell function (CD19, CD79B, CXCR5, FAIM3, IGKC, IGHG1 genes). Multiple probe sets encoding the B-lymphocyte-related genes for CD20/MS4A1, CXCL13, FAIM3, and SLAMF7/CRACC were among the highest upregulated transcripts in the vaccinated/low-dose-challenge animals, although their expression was slightly above the significance threshold. Of note, the expression of these genes was not changed in the high-dose group relative to their control group. A few chemokines and cytokines with antiviral activity (IL-16, IL-10, CCL1, and CCL22) were also detected with increased expression. Lastly, there was a modest upregulation (1.5- to 1.7-fold) of genes activating complement, F7, MASP1, PCOTH, and a number of collagen proteins (Fig. 4).
Genes that did not segment into GO ontology pathways but had immune function were also upregulated in the low-dose vaccine group as shown in Fig. 4. Several genes were indicative of T cells: TRB@, CD28, ZAP70, LSP1, BATF, IL12RB1, and CD4 genes. The BATF gene has recently been implicated as regulating exhaustion in antigen-specific CD8+ T cells (34). We also noted that tissue from low-dose-vaccinated animals exhibited increased expression of TRIM34, a member of the tripartite motif family, as well as TRIM3. TRIM34 has been demonstrated to have inhibitory capacity against HIV infection in vitro (35); however, this has yet to be determined for TRIM3. Finally, we also noted increased expression of the gene encoding NLRP1/NALP1, which is central to the establishment of the inflammasome (36).
DISCUSSION
Evaluation of candidate vaccine approaches in NHP models is a key aspect of development of HIV vaccines, but it has been controversial as to which models are most relevant and have the greatest predictive value, particularly as the results from clinical vaccine studies have become available. Here, we directly address the relevance of the dose of challenge in detecting vaccine-induced protection from SIVmac251 acquisition or disease, by vaccinating animals with identical vaccines and exposing them to doses of virus that differed by 10-fold.
We observed protection from SIVmac251 acquisition when we used two repeated doses of 470 TCID50 but not when we used a single high dose of virus. Although the number of the protected animals was too low to study correlates, multiple observations support the hypothesis that our vaccine was responsible for the observed protection because only one of the protected animals was Mamu-A*01 positive, and TRIM5 does not restrict the SIVmac251 strain used in this study (27).
Both challenge doses resulted in an equivalent seeding of SIVmac251 at mucosal sites of unvaccinated macaques, and there was 10-fold-less viral DNA at mucosal sites (Fig. 2C) in the vaccinated animals exposed to the lower dose of SIVmac251. Despite this finding, there was no protection from CD4+ T-cell loss in mucosal tissue in both groups of vaccinated macaques, suggesting that the T-cell number may decrease not only by direct SIV infection, but also through indirect mechanisms (37). Importantly, at set point, resolution of CD4+ T cells at a mucosal site was significant only in the group challenged with the 10-fold-less dose of SIVmac251.
Analysis of immune correlates revealed that the frequency of vaccine-induced Env-specific CD4+ T cells and Gag-specific CD8+ T cells correlated with protection from high viral replication in the acute and the chronic phases of infection, respectively, only in the vaccinated animals exposed to the lower dose of virus levels (Table 2). In contrast, both Gag and envelope CD8+ T cells correlated with virus levels in macaques exposed to the single high dose of SIVmac251.
The vaccine regimen used in our study induced measurable titers of binding antibody to Env and neutralization of TCLA SIVmac251 but not to the challenge virus or SIVmac239. ADCC activity was measured in the sera of all vaccinated macaques. ADCC has been correlated with protection from high viral replication in studies where poxvirus- or adenovirus-based strategies were used against low repeated exposures to doses of SIVmac251 that transmitted fewer virus variants than the 470 TCID50 challenge dose used here (38, 39). In the present study, we did not observe a correlation with ADCC regardless of the dose of challenge exposure, suggesting that even the lower dose of SIVmac251 used here may remain too high to discern an antibody correlate of protection. In summary, our results demonstrated that differences in the dose of the inoculum not only influence the evaluation of vaccine efficacy but also result in a different immune correlate of protection.
The significant regeneration of CD4+ T cells levels at both mucosal sites and in blood, observed in vaccinated animals exposed to the low dose of challenge virus, was associated with the downregulation of IFN-responsive genes and pro-inflammatory genes at mucosal sites, suggesting that vaccination was able to modify the host response to the virus, provided that the animals were challenged with an amount of virus close to the threshold of infectivity. Alternatively, the alteration of expression of IFN-responsive and proinflammatory genes at mucosal sites could result from the limitation of infection rather than a direct effect of vaccination modifying the host response to the virus. Chronic immune activation drives CD4+ T-cell depletion in HIV infection (40). Of note, a similar capacity of partial CD4+ T-cell regeneration during chronic infection in spite of unabated viral replication has been observed in the nonpathogenic SIV infection of African monkeys (AGM) or sooty mangabeys (SM) (41–44). Infected AGM show anti-inflammatory profiles and an initial strong, but rapid, control of immune activation through downregulation of type I IFN production, as we have observed in our study (45, 46). Indeed, during acute infection, we found no difference in the expression of genes involved in immune activation at mucosal sites, despite the difference in the levels of SIV DNA found in the two vaccinated groups at mucosal sites. However, vaccinated macaques exposed to 470 TCID50, but not those exposed to the high dose, had attenuated type II and I IFN responses in the gut at week 11. The downregulation of proapoptotic signals, such as TRAIL or TNFSF10, and upregulation of IL-10 are similar in infected AGM and infected vaccinated macaques exposed to the lower dose of SIVmac251 (45). The decrease of local inflammation may have contributed to the regeneration of CD4+ T cells (46). Accordingly, many of the significant differences found in the low-dose group suggested better maintenance of total B, T, and NK function. We also observed a significant enrichment of pathways involving tissue development observed in this group, suggesting that potential homeostatic mechanisms may be acting to protect the mucosal sites. The upregulation of genes for BATF may indicate differentiation of CD4+ T cells into Th17, as this type of gut-homed, inflammatory immune cells are dependent of this transcription factor, and they are absent in BATF-knockout mice (47, 48). BATF expression has not been analyzed in AGM, but SIVagm infection results in no change in the level of IL-17-expressing cells in lymphoid organs (41). Taken together, these data indicate that the DNA/ALVAC-gp120 vaccine regimen, by decreasing viral replication, alters the local immune environment in a manner that promotes reduction of immune activation, maintenance of immune function, and prolonged protection from high viral replication and CD4+ T-cell loss.
Supplementary Material
ACKNOWLEDGMENTS
We thank N. Letvin, J. Mascola, and M. Roederer for providing the SIVmac251 virus stock, J. Treece, D. Weiss, and P. Markham for animal husbandry and care, V. Kalyanaraman and L. Ajayi for antibody binding assays, H. K. Chung and E. Lee for the quantitative analysis of viral RNA and DNA, and Teresa Habina for manuscript editing.
This work was supported with federal funds from the National Cancer Institute, National Institutes of Health, and in part with contract no. HHSN261200800001E.
Footnotes
Published ahead of print 16 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02863-12.
REFERENCES
- 1. Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, SMde Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 2. Franchini G, Gurunathan S, Baglyos L, Plotkin S, Tartaglia J. 2004. Poxvirus-based vaccine candidates for HIV: two decades of experience with special emphasis on canarypox vectors. Expert. Rev. Vaccines 3(Suppl 1):S75–S88 [DOI] [PubMed] [Google Scholar]
- 3. Vaccari M, Poonam P, Franchini G. 2010. Phase III HIV vaccine trial in Thailand: a step toward a protective vaccine for HIV. Expert. Rev. Vaccines 9:997–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Franchini G, Robert-Guroff M, Tartaglia J, Aggarwal A, Abimiku AG, Benson J, Markham PD, Limbach K, Hurteau G, Fullen J, Aldrich K, Miller N, Sadoff J, Paoletti E, Gallo RC. 1995. Highly attenuated HIV type 2 recombinant poxviruses, but not HIV-2 recombinant Salmonella vaccines, induce long-lasting protection in rhesus macaques. AIDS Res. Hum. Retroviruses 11:909–920 [DOI] [PubMed] [Google Scholar]
- 5. Pal R, Venzon D, Santra S, Kalyanaraman VS, Montefiori DC, Hocker L, Hudacik L, Rose N, Nacsa J, Edghill-Smith Y, Moniuszko M, Hel Z, Belyakov IM, Berzofsky JA, Parks RW, Markham PD, Letvin NL, Tartaglia J, Franchini G. 2006. Systemic immunization with an ALVAC-HIV-1/protein boost vaccine strategy protects rhesus macaques from CD4+ T-cell loss and reduces both systemic and mucosal simian-human immunodeficiency virus SHIVKU2 RNA levels. J. Virol. 80:3732–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pal R, Venzon D, Letvin NL, Santra S, Montefiori DC, Miller NR, Tryniszewska E, Lewis MG, Vancott TC, Hirsch V, Woodward R, Gibson A, Grace M, Dobratz E, Markham PD, Hel Z, Nacsa J, Klein M, Tartaglia J, Franchini G. 2002. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J. Virol. 76:292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Van Rompay KK, Abel K, Lawson JR, Singh RP, Schmidt KA, Evans T, Earl P, Harvey D, Franchini G, Tartaglia J, Montefiori D, Hattangadi S, Moss B, Marthas ML. 2005. Attenuated poxvirus-based simian immunodeficiency virus (SIV) vaccines given in infancy partially protect infant and juvenile macaques against repeated oral challenge with virulent SIV. J. Acquir. Immune Defic. Syndr. 38:124–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu J, Keele BF, Li H, Keating S, Norris PJ, Carville A, Mansfield KG, Tomaras GD, Haynes BF, Kolodkin-Gal D, Letvin NL, Hahn BH, Shaw GM, Barouch DH. 2010. Low-dose mucosal simian immunodeficiency virus infection restricts early replication kinetics and transmitted virus variants in rhesus monkeys. J. Virol. 84:10406–10412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McDermott AB, Mitchen J, Piaskowski S, De SI, Yant LJ, Stephany J, Furlott J, Watkins DI. 2004. Repeated low-dose mucosal simian immunodeficiency virus SIVmac239 challenge results in the same viral and immunological kinetics as high-dose challenge: a model for the evaluation of vaccine efficacy in nonhuman primates. J. Virol. 78:3140–3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Keele BF, Li H, Learn GH, Hraber P, Giorgi EE, Grayson T, Sun C, Chen Y, Yeh WW, Letvin NL, Mascola JR, Nabel GJ, Haynes BF, Bhattacharya T, Perelson AS, Korber BT, Hahn BH, Shaw GM. 2009. Low-dose rectal inoculation of rhesus macaques by SIVsmE660 or SIVmac251 recapitulates human mucosal infection by HIV-1. J. Exp. Med. 206:1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Knapp LA, Lehmann E, Piekarczyk MS, Urvater JA, Watkins DI. 1997. A high frequency of Mamu-A*01 in the rhesus macaque detected by PCR-SSP and direct sequencing. Tissue Antigens 50:657–661 [DOI] [PubMed] [Google Scholar]
- 12. Kulkarni V, Jalah R, Ganneru B, Bergamaschi C, Alicea C, Avon G, Patel V, Zhang GM, Chowdhury B, Broderick KE, Sardesai NY, Valentin A, Rosati M, Felber BK, Pavlakis GN. 2011. Comparison of immune responses generated by optimized DNA vaccination against SIV antigens in mice and macaques. Vaccine 29:6742–6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rosati M, Bergamaschi C, Valentin A, Kulkarni V, Jalah R, Alicea C, Patel V, von Gegerfelt AS, Montefiori DC, Venzon DJ, Khan AS, Draghia-Akli R, Van Rompay KK, Felber BK, Pavlakis GN. 2009. DNA vaccination in rhesus macaques induces potent immune responses and decreases acute and chronic viremia after SIVmac251 challenge. Proc. Natl. Acad. Sci. U. S. A. 106:15831–15836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rosati M, Valentin A, Jalah R, Patel V, Avon G, Bergamaschi C, Alicea C, Weiss D, Treece J, Pal R, Markham PD, Marques ET, August JT, Khan A, Draghia-Akli R, Felber BK, Pavlakis GN. 2008. Increased immune responses in rhesus macaques by DNA vaccination combined with electroporation. Vaccine 26:5223–5229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Letvin NL, Rao SS, Dang V, Buzby AP, Korioth-Schmitz B, Dombagoda D, Parvani JG, Clarke RH, Bar L, Carlson KR, Kozlowski PA, Hirsch VM, Mascola JR, Nabel GJ. 2007. No evidence for consistent virus-specific immunity in simian immunodeficiency virus-exposed, uninfected rhesus monkeys. J. Virol. 81:12368–12374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Romano JW, Williams KG, Shurtliff RN, Ginocchio C, Kaplan M. 1997. NASBA technology: isothermal RNA amplification in qualitative and quantitative diagnostics. Immunol. Invest. 26:15–28 [DOI] [PubMed] [Google Scholar]
- 17. Vaccari M, Mattapallil J, Song K, Tsai WP, Hryniewicz A, Venzon D, Zanetti M, Reimann KA, Roederer M, Franchini G. 2008. Reduced protection from simian immunodeficiency virus SIVmac251 infection afforded by memory CD8+ T cells induced by vaccination during CD4+ T-cell deficiency. J. Virol. 82:9629–9638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vaccari M, Trindade CJ, Venzon D, Zanetti M, Franchini G. 2005. Vaccine-induced CD8+ central memory T cells in protection from simian AIDS. J. Immunol. 175:3502–3507 [DOI] [PubMed] [Google Scholar]
- 19. Vaccari M, Boasso A, Ma ZM, Cecchinato V, Venzon D, Doster MN, Tsai WP, Shearer GM, Fuchs D, Felber BK, Pavlakis GN, Miller CJ, Franchini G. 2008. CD4+ T-cell loss and delayed expression of modulators of immune responses at mucosal sites of vaccinated macaques following SIV(mac251) infection. Mucosal Immunol. 1:497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Waldrop SL, Davis KA, Maino VC, Picker LJ. 1998. Normal human CD4+ memory T cells display broad heterogeneity in their activation threshold for cytokine synthesis. J. Immunol. 161:5284–5295 [PubMed] [Google Scholar]
- 21. Hel Z, Tsai WP, Tryniszewska E, Nacsa J, Markham PD, Lewis MG, Pavlakis GN, Felber BK, Tartaglia J, Franchini G. 2006. Improved vaccine protection from simian AIDS by the addition of nonstructural simian immunodeficiency virus genes. J. Immunol. 176:85–96 [DOI] [PubMed] [Google Scholar]
- 22. Pollara J, Hart L, Brewer F, Pickeral J, Packard BZ, Hoxie JA, Komoriya A, Ochsenbauer C, Kappes JC, Roederer M, Huang Y, Weinhold KJ, Tomaras GD, Haynes BF, Montefiori DC, Ferrari G. 2011. High-throughput quantitative analysis of HIV-1 and SIV-specific ADCC-mediating antibody responses. Cytometry A 79:603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Trkola A, Matthews J, Gordon C, Ketas T, Moore JP. 1999. A cell line-based neutralization assay for primary human immunodeficiency virus type 1 isolates that use either the CCR5 or the CXCR4 coreceptor. J. Virol. 73:8966–8974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Montefiori DC. 2005. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. Chapter 12:Unit 12.11 [DOI] [PubMed] [Google Scholar]
- 25. Brandt SM, Mariani R, Holland AU, Hope TJ, Landau NR. 2002. Association of chemokine-mediated block to HIV entry with coreceptor internalization. J. Biol. Chem. 277:17291–17299 [DOI] [PubMed] [Google Scholar]
- 26. Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fenizia C, Keele BF, Nichols D, Cornara S, Binello N, Vaccari M, Pegu P, Robert-Guroff M, Ma ZM, Miller CJ, Venzon D, Hirsch V, Franchini G. 2011. TRIM5alpha does not affect simian immunodeficiency virus SIV(mac251) replication in vaccinated or unvaccinated Indian rhesus macaques following intrarectal challenge exposure. J. Virol. 85:12399–12409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Letvin NL, Rao SS, Montefiori DC, Seaman MS, Sun Y, Lim SY, Yeh WW, Asmal M, Gelman RS, Shen L, Whitney JB, Seoighe C, Lacerda M, Keating S, Norris PJ, Hudgens MG, Gilbert PB, Buzby AP, Mach LV, Zhang J, Balachandran H, Shaw GM, Schmidt SD, Todd JP, Dodson A, Mascola JR, Nabel GJ. 2011. Immune and genetic correlates of vaccine protection against mucosal infection by SIV in monkeys. Sci. Transl. Med. 3:81ra36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yeh WW, Rao SS, Lim SY, Zhang J, Hraber PT, Brassard LM, Luedemann C, Todd JP, Dodson A, Shen L, Buzby AP, Whitney JB, Korber BT, Nabel GJ, Mascola JR, Letvin NL. 2011. The TRIM5 gene modulates penile mucosal acquisition of simian immunodeficiency virus in rhesus monkeys. J. Virol. 85:10389–10398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rosati M, von Gegerfelt A, Roth P, Alicea C, Valentin A, Robert-Guroff M, Venzon D, Montefiori DC, Markham P, Felber BK, Pavlakis GN. 2005. DNA vaccines expressing different forms of simian immunodeficiency virus antigens decrease viremia upon SIVmac251 challenge. J. Virol. 79:8480–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cocchi F, DeVico AL, Garzino-Demo A, Arya SK, Gallo RC, Lusso P. 1995. Identification of RANTES, MIP-1 alpha, and MIP-1 beta as the major HIV-suppressive factors produced by CD8+ T cells. Science 270:1811–1815 [DOI] [PubMed] [Google Scholar]
- 32. Harenberg A, Guillaume F, Ryan EJ, Burdin N, Spada F. 2008. Gene profiling analysis of ALVAC infected human monocyte derived dendritic cells. Vaccine 26:5004–5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ryan EJ, Harenberg A, Burdin N. 2007. The canarypox-virus vaccine vector ALVAC triggers the release of IFN-gamma by natural killer (NK) cells enhancing Th1 polarization. Vaccine 25:3380–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quigley M, Pereyra F, Nilsson B, Porichis F, Fonseca C, Eichbaum Q, Julg B, Jesneck JL, Brosnahan K, Imam S, Russell K, Toth I, Piechocka-Trocha A, Dolfi D, Angelosanto J, Crawford A, Shin H, Kwon DS, Zupkosky J, Francisco L, Freeman GJ, Wherry EJ, Kaufmann DE, Walker BD, Ebert B, Haining WN. 2010. Transcriptional analysis of HIV-specific CD8+ T cells shows that PD-1 inhibits T cell function by upregulating BATF. Nat. Med. 16:1147–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang F, Hatziioannou T, Perez-Caballero D, Derse D, Bieniasz PD. 2006. Antiretroviral potential of human tripartite motif-5 and related proteins. Virology 353:396–409 [DOI] [PubMed] [Google Scholar]
- 36. Martinon F, Burns K, Tschopp J. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10:417–426 [DOI] [PubMed] [Google Scholar]
- 37. Doitsh G, Cavrois M, Lassen KG, Zepeda O, Yang Z, Santiago ML, Hebbeler AM, Greene WC. 2010. Abortive HIV infection mediates CD4 T cell depletion and inflammation in human lymphoid tissue. Cell 143:789–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barouch DH, Liu J, Li H, Maxfield LF, Abbink P, Lynch DM, Iampietro MJ, San Miguel A, Seaman MS, Ferrari G, Forthal DN, Ourmanov I, Hirsch VM, Carville A, Mansfield KG, Stablein D, Pau MG, Schuitemaker H, Sadoff JC, Billings EA, Rao M, Robb ML, Kim JH, Marovich MA, Goudsmit J, Michael NL. 2012. Vaccine protection against acquisition of neutralization-resistant SIV challenges in rhesus monkeys. Nature 482:89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gomez-Roman VR, Patterson LJ, Venzon D, Liewehr D, Aldrich K, Florese R, Robert-Guroff M. 2005. Vaccine-elicited antibodies mediate antibody-dependent cellular cytotoxicity correlated with significantly reduced acute viremia in rhesus macaques challenged with SIVmac251. J. Immunol. 174:2185–2189 [DOI] [PubMed] [Google Scholar]
- 40. Giorgi JV, Hultin LE, McKeating JA, Johnson TD, Owens B, Jacobson LP, Shih R, Lewis J, Wiley DJ, Phair JP, Wolinsky SM, Detels R. 1999. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J. Infect. Dis. 179:859–870 [DOI] [PubMed] [Google Scholar]
- 41. Beaumier CM, Harris LD, Goldstein S, Klatt NR, Whitted S, McGinty J, Apetrei C, Pandrea I, Hirsch VM, Brenchley JM. 2009. CD4 downregulation by memory CD4+ T cells in vivo renders African green monkeys resistant to progressive SIVagm infection. Nat. Med. 15:879–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Diop OM, Gueye A, Dias-Tavares M, Kornfeld C, Faye A, Ave P, Huerre M, Corbet S, Barre-Sinoussi F, Muller-Trutwin MC. 2000. High levels of viral replication during primary simian immunodeficiency virus SIVagm infection are rapidly and strongly controlled in African green monkeys. J. Virol. 74:7538–7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gordon SN, Klatt NR, Bosinger SE, Brenchley JM, Milush JM, Engram JC, Dunham RM, Paiardini M, Klucking S, Danesh A, Strobert EA, Apetrei C, Pandrea IV, Kelvin D, Douek DC, Staprans SI, Sodora DL, Silvestri G. 2007. Severe depletion of mucosal CD4+ T cells in AIDS-free simian immunodeficiency virus-infected sooty mangabeys. J. Immunol. 179:3026–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pandrea I, Apetrei C, Dufour J, Dillon N, Barbercheck J, Metzger M, Jacquelin B, Bohm R, Marx PA, Barre-Sinoussi F, Hirsch VM, Muller-Trutwin MC, Lackner AA, Veazey RS. 2006. Simian immunodeficiency virus SIVagm.sab infection of Caribbean African green monkeys: a new model for the study of SIV pathogenesis in natural hosts. J. Virol. 80:4858–4867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Jacquelin B, Mayau V, Targat B, Liovat AS, Kunkel D, Petitjean G, Dillies MA, Roques P, Butor C, Silvestri G, Giavedoni LD, Lebon P, Barre-Sinoussi F, Benecke A, Muller-Trutwin MC. 2009. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J. Clin. Invest. 119:3544–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kornfeld C, Ploquin MJ, Pandrea I, Faye A, Onanga R, Apetrei C, Poaty-Mavoungou V, Rouquet P, Estaquier J, Mortara L, Desoutter JF, Butor C, Le Grand R, Roques P, Simon F, Barre-Sinoussi F, Diop OM, Muller-Trutwin MC. 2005. Antiinflammatory profiles during primary SIV infection in African green monkeys are associated with protection against AIDS. J. Clin. Invest. 115:1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Betz BC, Jordan-Williams KL, Wang C, Kang SG, Liao J, Logan MR, Kim CH, Taparowsky EJ. 2010. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med. 207:933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schraml BU, Hildner K, Ise W, Lee WL, Smith WA, Solomon B, Sahota G, Sim J, Mukasa R, Cemerski S, Hatton RD, Stormo GD, Weaver CT, Russell JH, Murphy TL, Murphy KM. 2009. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature 460:405–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.