Abstract
Foamy viruses (FVs) are unique among retroviruses in performing genome reverse transcription (RTr) late in replication, resulting in an infectious DNA genome, and also in their unusual Pol biosynthesis and encapsidation strategy. In addition, FVs display only very limited Gag and Pol processing by the viral protease (PR) during particle morphogenesis and disassembly, both thought to be crucial for viral infectivity. Here, we report the generation of functional prototype FV (PFV) particles from mature or partially processed viral capsid and enzymatic proteins with infectivity levels of up to 20% of the wild type. Analysis of protein and nucleic acid composition, as well as infectivity, of virions generated from different Gag and Pol combinations (including both expression-optimized and authentic PFV open reading frames [ORFs]) revealed that precursor processing of Gag, but not Pol, during particle assembly is essential for production of infectious virions. Surprisingly, when processed Gag (instead of Gag precursor) was provided together with PR-deficient Pol precursor during virus production, infectious, viral DNA-containing particles were obtained, even when different vector or proviral expression systems were used. Although virion infectivity was reduced to 0.5 to 2% relative to that of the respective parental constructs, this finding overturns the current dogma in the FV literature that viral PR activity is absolutely essential at some point during target cell entry. Furthermore, it demonstrates that viral PR-mediated Gag precursor processing during particle assembly initiates intraparticle RTr. Finally, it shows that reverse transcriptase (RT) and integrase are enzymatically active in the Pol precursor within the viral capsid, thus enabling productive host cell infection.
INTRODUCTION
Retrovirus-encoded enzymatic activities, especially those with protease (PR), reverse transcriptase (RT), and integrase (IN) functions, play essential roles in the viral replication cycle. For example, orthoretroviral Gag processing by the viral PR, initiated upon incorporation of all viral components in the budding virion, is a highly ordered process essential for generation of infectious particles (reviewed in references 1 and 2). In the case of human immunodeficiency virus type 1 (HIV-1), viral PR-mediated Gag precursor processing involves a sequential cascade of events (3, 4). Generation of mature matrix (MA), capsid (CA), and nucleocapsid (NC) subunits, as well as other peptides, is kinetically controlled by different cleavage rates at individual sites (4). Alterations of the retroviral Gag cleavage pattern have detrimental effects on particle morphology and infectivity. The controls and triggers behind retroviral PR activation during assembly remain largely unknown.
Spumaviruses or foamy viruses (FVs) comprise the only genus of the subfamily of Spumaretrovirinae in the family Retroviridae. FV replication involves several unique characteristics that distinguish these viruses from the orthoretroviruses (5). An important difference is the expression of FV Pol as a separate precursor protein from a singly spliced mRNA and not as a Gag-Pol fusion protein from unspliced viral genomic RNA (6, 7). Thus, the FV Pol precursor cannot be packaged into newly forming capsids in an orthoretroviral-like fashion as part of a Gag-Pol fusion protein; instead, FVs directly incorporate the Pol protein into assembling virions (5). Although the mechanism of FV Pol encapsidation is only partially understood, it seems clear that viral RNA (vRNA) is necessary for efficient prototype FV (PFV) Pol particle incorporation (8–10). Furthermore, it is assumed that the vRNA serves as a bridging molecule between Gag and Pol during assembly. There are conflicting results related to the requirement of additional protein-protein interactions between both proteins for PFV Pol incorporation (11–13). Interestingly, only the unprocessed Pol precursor protein appears to be efficiently packaged into assembling PFV particles (9, 14).
The sequential events of FV structural protein maturation have not been determined in much detail, but processing of Gag and Pol precursors by the viral PR shows some striking differences in comparison to orthoretroviruses. The 127-kDa PFV Pol precursor (p127Pol) is autocatalytically processed into only two subunits, an 85-kDa PR-RT fusion protein (p85PR-RT) and a 40-kDa IN protein (p40IN) (reviewed in reference 15). The PR and RT domains appear to be enzymatically active in the p127Pol precursor, whereas efficient viral DNA (vDNA) genome integration activity was reported to require Pol precursor processing (14). Like orthoretroviral proteases, PFV PR is active only upon formation of a symmetric homodimer (16). However, mature PFV p85PR-RT was shown to be monomeric in solution and to lack proteolytic activity under physiologically relevant conditions in vitro (17). The mechanism of PR activation is a controversial subject (18, 19). Whereas Lee et al. (19) reported a role of the IN domain within the precursor protein for inducing PR dimerization and enzymatic activity, Hartl and colleagues (18) described the stimulation of PFV PR activity by a PFV vRNA element, the protease-activating RNA motif (PARM). PFV PR dimerization by PARM involves two purine-rich sequences and seems to be mediated by RNA binding to the RT domain with no need for the IN domain (18, 20).
PFV Pol proteolytic activity is reported to be essential for productive infection, not only for Gag and Pol precursor processing in the course of viral assembly but also for further Gag maturation during entry steps in the target cell (reviewed in reference 21). PFV Gag lacks classical orthoretroviral MA, CA, and NC domains. During the course of viral assembly only a limited processing of the p71Gag precursor into an N-terminal p68Gag and a C-terminal p3Gag subunit is observed (reviewed in reference 15). Capsids of released infectious PFV particles contain p71Gag precursor and the p68Gag subunit at ratios of 1:1 to 1:4 (22). The smaller mature p3Gag subunit has so far eluded detection in cell lysates and viral particle preparations, and its function for replication remains unclear (23). Gag precursor processing during capsid assembly is known to be essential for correct capsid morphogenesis, intraparticle reverse transcription (RTr) of the packaged viral RNA genome, and viral infectivity (23–25). Whereas failure of Gag processing during assembly completely abolishes infectivity, particles composed of p68Gag alone show wild-type (wt) morphology but are up to 100-fold less infectious (23, 25). Unlike other retroviruses, further processing of FV Gag at secondary Gag cleavage sites by the viral PR during virus entry was reported to be essential for productive infection (26, 27). This additional Gag maturation is thought to be required for complete disassembly and subsequent transport of the preintegration complex into the nucleus of the target cell.
As with the regulation of retroviral protease activation, little is known about what controls and triggers the time point of RTr initiation during retroviral replication (reviewed in reference 28). In orthoretroviruses, RTr is connected to uncoating upon target cell entry. The details of these processes, including the timing, location, and mechanism, are poorly understood and subject to considerable debate. The orthoretroviral NC subunit of Gag is known to be a potent nucleic acid chaperone. NC not only is involved in selective packaging of the genomic vRNA but also facilitates annealing of complementary sequences and the strand transfer and exchange reactions during RTr. Only recently, a role of NC in the timing of RTr was discovered. NC domains with mutations in the zinc fingers reduced vRNA packaging and also led to a strong enhancement of intraparticle RTr in released HIV-1 virions (29, 30).
Unlike orthoretroviruses but similar to hepatitis B virus, FV genome RTr is initiated to a large extent during or shortly after capsid assembly (7, 31). Samples of released FV virions contain large amounts of double-stranded vDNA (ds-vDNA) in addition to vRNA, with vDNA constituting up to 20% of all particle-associated viral nucleic acid molecules. This particle-associated vDNA appears to be the predominant form of the infectious genome of FVs. Further RTr is observed during early steps of FV target cell entry and is thought to be important for infectivity at low multiplicities of infection (32). The mechanism controlling the timing and extent of FV RTr at different steps in the replication cycle is poorly characterized.
In this study, we report for the first time the identification of conditions that allow the assembly of infectious particles of a complex retrovirus from mature or partially processed capsid and polymerase protein subunits, thereby obviating the need for prior precursor protein processing by the viral protease. Using different PFV production systems, we evaluated the requirement of FV protease function during different steps of the viral replication cycle. Furthermore, we identified the initiation event resulting in intraparticle RTr of packaged vRNA in newly assembled FV capsids during virus egress.
MATERIALS AND METHODS
Cell culture.
The human kidney cell line 293T (33), the human fibrosarcoma cell line HT1080 (34), and the PFV indicator cell line HT1080 PLNE were cultivated in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum and antibiotics. HT1080 PLNE cells are a clonal variant of the parental HT1080 cells that were stably transfected with a pcDNA3.1 zeo expression vector having the cytomegalovirus (CMV) promoter replaced by the HSRV2 PFV U3-R region (−777 to +6) and driving expression of a simian virus 40 (SV40) nuclear localization signal (NLS)-tagged enhanced green fluorescent protein (EGFP).
Recombinant DNA.
The CMV-driven proviral expression vector (prov) pczHSRV2 (wt) and its variant pczHSRV2 iPR (iPR), expressing a Pol protein with enzymatically inactive PR (iPR) domain (D24A mutation), were described previously (Fig. 1A) (31, 35). For this study, variant pczHSRV2 p68 (p68), containing a gag open reading frame (ORF) with a translation stop codon at the p68Gag/p3Gag cleavage site, was generated, as well as a variant, pczHSRV2 p68 iPR (p68 iPR), which combines both mutations.
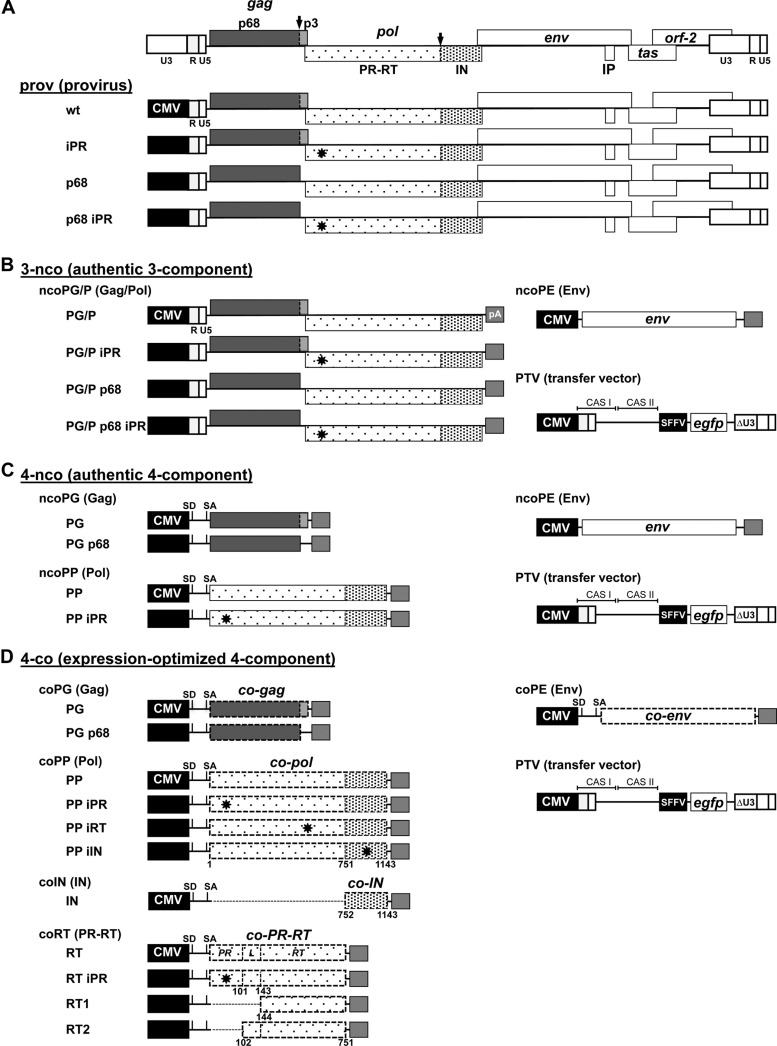
Fig 1.
PFV production systems. Schematic illustration of the different PFV production systems used in the study. Three systems based on authentic PFV ORFs were utilized and include systems for production of the following: prov, replication-competent virus based on CMV (cytomegalovirus)-driven proviral expression constructs (A); 3-nco, replication-deficient vectors based on CMV-driven Gag/Pol and Env packaging constructs and CMV-driven PFV transfer vector (PTV) containing an egfp marker gene expression cassette (B); and 4-nco, replication-deficient vectors based on separate CMV-driven packaging vectors for Gag, Pol, and Env and the previously mentioned transfer vector (C). (D) A fourth replication-deficient vector system (4-co) is based on separate packaging constructs with expression-optimized ORFs encoding PFV Gag, Pol, and Env and the same transfer vector mentioned above. R, long terminal repeat region (LTR); U5, LTR unique 5′ region; U3, LTR unique 3′ region; ΔU3: enhancer-promoter deleted U3 region; IP, internal promoter; CAS, cis-acting sequence; SFFV, spleen focus-forming virus U3 promoter; SD, splice donor; SA, splice acceptor; PR, protease domain; RT, reverse transcriptase domain; L, linker domain between PR and RT; IN: integrase domain; pA: bovine growth hormone polyadenylation site. Major PFV PR cleavage sites in PFV Gag and Pol are indicated by black arrows; iPR (D24A), iRT (DD312–315GAAA), and iIN (D936A) mutations are indicated with stars throughout the pol ORF; authentic ORFs are indicated by boxes with continuous lines; expression-optimized ORFs are indicated by boxes with dashed lines. Numbers indicate amino acid positions in Pol.
The packaging vectors of the original four-component PFV vector system (4-nco) consisting of the pcziPG4 (ncoPG), pcziPol (ncoPP), and pczHFVenvEM002 (ncoPE) vectors with authentic PFV ORFs were described previously (Fig. 1C) (36). For this study, the variant Gag packaging construct pcziPG4 p68 (ncoPG p68), encoding a C-terminally truncated PFV Gag (amino acids [aa] 1 to 621), was generated, having the p3 domain replaced by a stop codon. Furthermore, a PR-inactive variant (D24A mutation), pcziPol iPR (ncoPP iPR), based on the pcziPol packaging construct was generated. Plasmid puc2MD9 (PTV), encoding a packageable PFV vector RNA containing a spleen focus-forming virus U3 region (SFFV U3)-driven egfp marker gene expression cassette, was used as a transfer vector (37).
In the course of this study a three-component PFV vector system (3-nco) was generated (Fig. 1B) that is based on the Env packaging construct pczHFVenv EM002 (ncoPE) of the 4-nco vector system and the puc2MD9 (PTV) transfer vector. In this system PFV Gag and Pol are expressed from a single newly generated Gag/Pol packaging vector, pczPG/P (ncoPG/P) harboring the CMV-driven 5′ part of the human spumaretrovirus-2 (HSRV2) proviral genome starting at the 5′ long terminal repeat (LTR) R region and ending after the Pol translation stop. Furthermore, variants of this Gag/Pol packaging vector were generated containing a gag ORF with a translation stop codon at the p68Gag/p3Gag cleavage site (pczPG/P p68) or a pol ORF with an enzymatically inactive PR domain (pczPG/P iPR, with a D24A mutation) or a combination of both mutations (pczPG/P p68 iPR).
The expression-optimized packaging constructs for PFV Gag (pcoPG4 [coPG]), PFV Env (pcoPE [coPE]), and Pol (pcoPP [coPP]), as well as the EGFP-expressing transfer vector puc2MD9 (PFV transfer vector [PTV]) (Fig. 1D), have been previously described (12, 37). For some experiments, the variant expression-optimized PFV Pol expression constructs pcoPP1 (coPP iPR, with an inactive PR domain as a result of a D24A mutation [35, 38]), pcoPP2 (coPP iRT, with an inactive RT domain as a result of a DD312–315GAAA mutation, [31]), or pcoPP3 (coPP iIN, with an inactive IN domain as a result of a D936A mutation[39]) were used. The expression-optimized PR-RT and IN subunit constructs pcoRT (coRT) and pcoIN (coIN) used in this study are schematically shown in Fig. 1D. Both are based on the parental full-length Pol packaging construct pcoPP (coPP) having either the IN subunit coding region replaced by a stop codon (coRT) or the PR-RT subunit replaced by a translation start codon (coIN). pcoRT iPR (coRT iPR) is a variant of pcoRT containing the D24A mutation (35, 38) for enzymatic inactivation of the PR domain. The expression plasmid pcoRT1 (coRT1) encodes a minimal RT protein (aa 144 to 596), which has the N-terminal PR core domain (PR) and the intervening flexible linker sequence deleted (40) (Fig. 1D). In contrast, pcoRT2 (coRT2) encodes a protein comprising the flexible linker and minimal RT domain (aa 102 to 596), having only the N-terminal PR core domain deleted (Fig. 1D). Furthermore, a C-terminally truncated Gag packaging construct coding for PFV Gag aa 1 to 621 (pcoPG p68 [coPG p68]) was generated by replacing the p3 domain with a stop codon.
All constructs were generated by recombinant PCR techniques and verified by sequencing analysis. Primer sequences and additional details are available upon request.
Viral supernatant production and analysis of transduction efficiency.
Recombinant PFV particles were harvested from transiently transfected 293T cells as previously described (12, 36, 37). Briefly, PFV supernatants derived from the different PFV production systems were generated by polyethyleneimine (PEI)-mediated cotransfection of 293T cells with the individual components of the respective systems using 16 μg of total DNA per 10-cm dish. For the proviral system (prov) pczHSRV2 and puc2MD9 vectors were mixed at a ratio of 1:1 for titration on H1080 cells, whereas puc2MD9 was omitted for titrations on HT1080 PLNE indicator cells. For the authentic three-component vector system (3-nco), packaging vectors for Gag/Pol and Env and the transfer vector were mixed at a ratio of 2:1:1. The packaging components of the authentic four-component vector system (4-nco) were cotransfected at a ratio of 1:1:1:1 as described by Heinkelein et al. (36). The packaging plasmids and transfer vector of the expression-optimized four-component vector system (4-co) were cotransfected at a ratio of 4:1:2:28 unless indicated otherwise in the legends. At this experimentally determined relative ratio of the individual components, the highest infectious titers of the 4-co vector system were obtained (data not shown). In some experiments involving the Pol subunit packaging constructs pcoRT and pcoIN, the amount of transfer vector was reduced to 21 or 14 parts in all samples while the total DNA amount was kept constant by the addition of pUC19 DNA if necessary. In experiments involving cotransfection of different Gag packaging constructs at various relative ratios, the total amount of Gag expression construct and its ratio to the other vector components were kept constant. For supernatant production in 12-well plates, Polyfect transfection using 3 μg of total DNA per well and identical plasmid ratios was employed. Viral supernatants were harvested 48 h after transfection.
For transduction efficiency analysis of viral particles containing EGFP-expressing PFV transfer vectors, 2 × 104 HT1080 cells were plated in 12-well plates 24 h before infection. For particle preparations derived by transfection of proviral expression constructs lacking an EGFP marker gene, HT1080 PLNE cells were plated instead. The target cells were incubated with 1 ml of plain cell-free viral supernatant or serial dilutions thereof for 4 to 6 h. Determination of the percentage of EGFP-expressing cells by flow cytometry analysis was performed at 72 h postinfection (p.i.) for the replication-deficient PFV production systems and at 24 to 72 h p.i. for the replication-competent system. The values of the percentage of EGFP-positive cells were used for titer determination as previously described (41). All transduction experiments were repeated at least three times. To compare the infectivity in repetitive experiments, the titer obtained for wild-type particles (derived from full-length wild-type Gag and Pol proteins) in individual experiments was set to an arbitrary value of 100%. The other values were then normalized as a percentage of the wild-type value.
Purification of viral particles.
The cell-free supernatant of transiently transfected 293T cells was harvested by sterile filtration through a 0.45-μm-pore-size filter, and viral particles were concentrated by ultracentrifugation through a 20% sucrose cushion at 4°C and 25,000 rpm for 3 h in an SW32 rotor. The viral pellet was resuspended in phosphate-buffered saline (PBS).
Subtilisin digest.
Subtilisin treatment of concentrated particles was performed as previously described (42, 43). Briefly, half of each purified particle pellet resuspended in PBS was incubated in a digestion mix containing final concentrations of 1 mM CaCl2, 50 mM Tris-HCl, pH 8.0, and 25 μg/ml subtilisin. The mock-treated other half was incubated with the digestion mix including PBS instead of subtilisin. The digest was stopped by the addition of phenylmethylsulfonyl fluoride (PMSF) at a final concentration of 100 μg/ml to each reaction mixture prior to the addition of 2× sodium dodecyl sulfate (SDS) protein sample buffer Coomassie (PPPC; 100 mM Tris-HCl [pH 6.8], 24% glycerol, 8% SDS, 2% dithiothreitol, 0.02% Coomassie blue G-250).
Biochemical analysis of cell lysates, viral particles, and antisera.
Preparation of cell lysates from one transfected 10-cm cell culture dish was performed by incubation with 0.6 ml of lysis buffer for 20 min at 4°C followed by centrifugation through a QIAshredder (Qiagen). All protein samples were mixed with 2×PPPC prior to separation by SDS-PAGE using 7.5% polyacrylamide gels. Immunoblotting using polyclonal antisera specific for PFV Gag (44), PFV IN (942054), simian foamy virus type 1 (SFV-1) PR-RT (67C6), and PFV Env leader peptide (Env-LP) (35), as well as hybridoma supernatants specific for PFV Gag (clone SGG-1) (36), PFV PR-RT (clone 15E10) or PFV IN (clone 3E11) (45), PFV Env surface unit (Env-SU) (clone P3E10) (46, 47), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (G8795; Sigma) was performed as previously described (35). The chemiluminescence signal was digitally recorded using an LAS-3000 imager and quantified using ImageGauge in the linear range of the sample signal intensities as described previously (44).
qPCR analysis.
Preparation of quantitative PCR (qPCR) samples was performed as previously described (12, 42). For qPCR analysis, 4 μl of each reverse transcriptase reaction mixture in a total volume of 25 μl was added in duplicates into microtiter plates and analyzed using a Brilliant II QPCR Kit (Stratagene) and an Mx4000 Multiplex Quantitative PCR System (Stratagene) or Applied Biosystems 7300 Real Time PCR system. Primers and PCR conditions have been previously described (12). All obtained values were compared to a standard curve consisting of serial dilutions of the puc2MD9 transfer vector or pczHSRV2 proviral expression construct. All sample values included were in the linear range of the standard curve with a span from 10 to more than 108 copies. The values for the DNA or RNA content of the investigated viral particles were normalized to amounts of released Gag protein determined by quantitative Western blot analysis of particle lysates, and values are expressed as a percentage of the wild type (generated by transfection of pcoPG4, pcoPP, pcoPE, and puc2MD9).
RESULTS
Enhanced virus production by an expression-optimized PFV vector system.
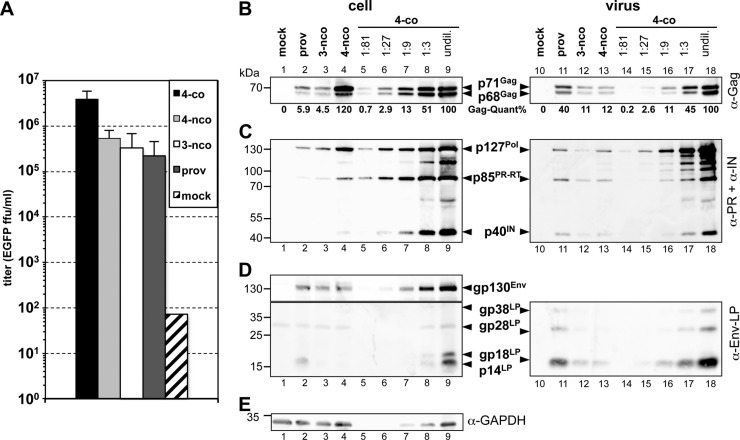
We have recently developed a new replication-deficient four-component PFV vector system (4-co) employing expression-optimized packaging constructs (Fig. 1D) (12, 36, 37). Titers of viral supernatants generated by transient transfection of 293T cells with this vector system (4-co) were improved up to 10-fold in comparison to 4-component (4-nco) or 3-component (3-nco) vector systems based on authentic PFV sequences and up to 20-fold in comparison to proviral (Prov) expression constructs (Fig. 2A). Increased viral titers of the 4-co vector system were largely the result of a higher physical particle release (Fig. 2B to D, lanes 10 to 18). This was a consequence of higher cellular expression levels of all viral components achieved by expression optimization of the packaging components of the 4-co vector system in comparison to the other virus production systems employing authentic PFV ORFs (Fig. 2B to E, lanes 1 to 9) as well as optimization of the relative ratios of the individual vector components (data not shown).
Fig 2.
Comparison of the features of the different PFV production systems. 293T cells were transiently transfected with the components of the different PFV production systems as indicated. The proviral constructs (prov) were cotransfected with the EGFP-expressing PFV transfer vector puc2MD9 at a ratio of 1:1. (A) Cell-free viral supernatants of the wild-type controls of the individual PFV production systems were titrated on HT1080 target cells using a flow cytometric marker gene assay, and titers were determined at 24 to 72 postransduction. For the proviral constructs, similar titers were obtained when they were not cotransfected with puc2MD9 and titrated on HT1080 PLNE cells, containing a Tas-inducible nuclear EGFP expression cassette under the control of the PFV LTR (data not shown). ffu, focus-forming units. (B to E) Cell lysates (cell) as well as viral particle preparations (virus), concentrated by ultracentrifugation through 20% sucrose, were analyzed by Western blotting. Serial dilutions of the samples of the expression-optimized four-component vector system (4-co, lanes 5 to 9 and 14 to 18) were loaded to determine their relative expression levels in comparison to the other PFV production systems. undil, undiluted. (B) Rabbit polyclonal antibodies specific for PFV Gag (α-Gag). (C) Mouse monoclonal antibody mixture specific for PFV PR and PFV IN (α-PR+α-IN). (D) Rabbit polyclonal antibodies specific for PFV Env-LP (α-Env-LP). (E) Mouse monoclonal antibodies specific for GAPDH (α-GAPDH).
Encapsidation of mature Pol subunits into PFV virions.
Previous investigations using expression systems employing the authentic PFV pol ORF demonstrated that PFV Pol is packaged only in its precursor state (9, 14). Coexpression of individual mature PR-RT or IN subunits in the context of such packaging systems in 293T cells did not result in any detectable encapsidation of the mature Pol subunits or in any measurable infectivity of viral supernatants. Expression optimization of the PFV pol ORF reduced the amount of Pol packaging vector (pcoPP) required to obtain a comparable cellular expression level achieved with a corresponding Pol packaging construct harboring an authentic PFV pol ORF (pcziPol) by about 128-fold (data not shown). This enabled us to investigate whether a particle incorporation of mature Pol subunits might be possible under conditions of higher intracellular Pol levels and, if so, whether it would be sufficient to generate infectious vector particles.
We addressed this by establishing separate expression-optimized packaging constructs for mature PFV PR-RT (pcoRT) and IN (pcoIN) subunits (Fig. 1D). 293T cells were cotransfected with identical DNA amounts of packaging constructs for these mature subunits (coRT+coIN) or for the Pol precursor (coPP), together with the remaining components (Gag, Env, and vRNA) of the expression-optimized replication-deficient PFV vector system. Analysis of the cell lysates revealed increased intracellular amounts of the mature Pol subunits p85PR-RT and p40IN in samples cotransfected with the mature Pol subunit expression constructs (coRT+coIN) in comparison to parental coPP precursor expression construct-transfected samples (Fig. 3A, upper panel, lanes 1 and 2). Interestingly, upon simultaneous cotransfection of packaging plasmids for both mature Pol subunits, p85PR-RT and p40IN (coRT+coIN) proteins were detected in particle preparations, apparently in amounts exceeding those achieved by cotransfection of the expression-optimized Pol precursor protein packaging vector (Fig. 3A, lower panel, lanes 1 and 2). Both mature Pol subunits (coRT and coIN) were also incorporated individually into particles when coexpressed together with the other vector components (Fig. 3A, lower panel, lanes 5 and 6). This suggested that the process of apparent particle association of mature Pol subunits is not dependent on the simultaneous presence of both proteins.
Fig 3.
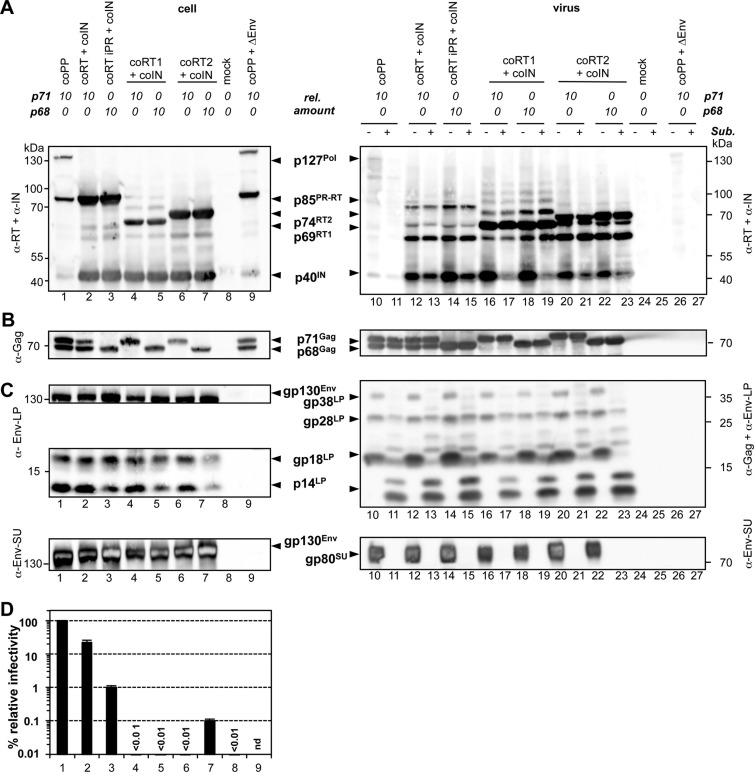
Infectious PFV vector particles generated by separate PFV Pol subunit packaging constructs. 293T cells were cotransfected with identical amounts of PFV transfer vector puc2MD9 (for lanes 4, 13, and 14, the amount was reduced to 2/3), identical amounts of Gag and Env packaging vectors pcoPG4 and pcoPE, and different amounts (starting with 0.91 μg of DNA per 10-cm dish as relative amount 1; lanes 1, 11, and 12) of the various Pol packaging constructs. Pol packaging constructs used contained expression-optimized ORFs for the full-length wild-type PFV Pol protein (coPP; lanes 1, 11, and 12) or variants thereof with enzymatically inactive reverse transcriptase (coPP iRT; lane 7) or integrase (coPP iIN; lane 8), as indicated. Alternatively, packaging constructs harboring expression-optimized ORFs for the mature PR-RT (coRT) or IN (coIN) subunit were cotransfected together (coRT+coIN, lanes 2 to 4, 13, and 14) or alone (coRT, lanes 5, 17, and 18; coIN, lanes 6, 15, and 16), as indicated. As a control, the Env packaging construct was omitted in a sample containing the wild-type full-length Pol packaging construct (coPP+ΔEnv, lanes 9, 19, and 20), or cells were transfected only with pUC19 DNA (mock, lanes 10, 21, and 22). The relative amount (rel. amount) of the individual Pol packaging construct used in comparison to the full-length wild-type PFV Pol protein (coPP)-encoding construct is indicated on top. The total amount of transfected DNA (16 μg) was kept constant in all samples by addition of pUC19 DNA. (A to E) Proteins of viral particle samples (virus) purified by ultracentrifugation through 20% sucrose and cell lysates (cell) harvested 48 h posttransfection. For the experiments shown in panels C to E, samples were either digested with subtilisin (+) or mock incubated (−) prior to lysis. Subsequently, samples were separated by SDS-PAGE and analyzed by Western blotting using mouse monoclonal antibody mixture specific for PFV PR and PFV IN (α-PR+α-IN) (A and D), rabbit polyclonal antibodies specific for PFV Gag (α-Gag) (B and E), or rabbit polyclonal antibodies specific for PFV Env-LP (α-Env-LP) or mouse monoclonal antibodies specific for PFV Env-SU (α-Env-SU) (C). The identity of the individual proteins is indicated on the right. (F) Three days postinfection, relative infectivities of 293T cell culture supernatants were determined with an egfp marker gene transfer assay. The values obtained using full-length expression-optimized pol ORF expression plasmid (coPP, column 1) were arbitrarily set to 100%. Sample values were normalized for capsid protein release. Absolute titers of these plain supernatants were (4.31 ± 0.75) × 106 EGFP-positive focus-forming units (FFU)/ml. Means and standard deviations of four independent experiments are shown. (G) Relative nucleic acid composition of mutant particles as determined with PFV Pol-specific primers and TaqMan probe. Following DNase I digestion of intact, purified particles, nucleic acids were isolated, and the relative amount of vector RNA and DNA copies was determined in comparison to the wild type by qPCR. The mean values and standard deviations of the relative RNA and DNA contents of at least three independent experiments are shown. Sample values were normalized for capsid protein release. Differences between means of the wild type and the individual mutants were analyzed by Welch's t test (*, P < 0.01).
Since it was previously reported (43) that subtilisin-sensitive, non-particle-associated Pol precursor aggregates are found in PFV particle preparations, we also examined particle association of the different Pol proteins by subtilisin digestion (Fig. 3C to E). Efficient digestion of proteins on the lumenal side was monitored by disappearance of the Env-SU subunit and reduction of the molecular weight of the Env-LP subunit due to removal of its extracellular domains (Fig. 3C). Whereas Pol proteins were no longer detectable in the ΔEnv control samples (Fig. 3D, lanes 19 and 20) after subtilisin digestion, mature p85PR-RT and p40IN were only partially subtilisin sensitive in samples generated by wild-type (coPP) or mature (coRT and coIN) packaging constructs (Fig. 3D, lanes 11 to 18). Particles generated from mature Pol packaging constructs contained larger amounts of particle-associated p85PR-RT and p40IN than the wild type, even after the proteolytic digest removing non-particle-associated proteins. Taken together, the data indicate that overexpression of mature PFV Pol subunits in cells leads to their efficient capsid incorporation.
Infectious PFV particles derived from individual, packaged Pol subunits.
Next, we asked whether encapsidation of the mature Pol subunits is compatible with their natural functions in the replication cycle that result in productive infection of host cells. Several features demonstrated the functionality of these particle-associated mature subunits. First, packaging of the p85PR-RT subunit resulted in a concentration-dependent processing of particle-associated Gag, which was not dependent on IN copackaging (Fig. 3B, lower panel, lanes 2 to 5). However, despite an apparent increase in particle-associated amounts of p85PR-RT in virion samples generated by cotransfection of identical amounts of pcoRT and pcoIN as in the wild-type control, Gag processing in particles was greatly diminished (Fig. 3B, lower panel, lanes 1 and 2). In contrast, cell-associated Gag processing was nearly identical under these conditions (Fig. 3B, upper panel, lanes 1 and 2). A wild-type-like particle-associated Gag processing was observed only upon further increase of the amounts of pcoRT and pcoIN transfected (Fig. 3B, lower panel, lanes 1 and 4).
Second, virions derived from cotransfection of packaging constructs for both mature Pol subunits together with the other vector components were infectious and resulted in a dose-dependent, stable transgene expression when infectivity was analyzed by a marker gene transfer assay (Fig. 3F, columns 2 to 4). When the same amount of mature subunit packaging constructs as in the wild-type control was used, vector infectivity was reduced to 3% of the wild-type level (Fig. 3F, columns 1 and 2). However, the infectivity could be elevated to about 20% relative to the wild type by increasing the amounts of the mature Pol subunit packaging constructs in the transfection mixture (Fig. 3F, columns 1 to 4). Interestingly, even cotransfection of the pcoRT expression construct alone was sufficient for Gag processing and transgene expression in target cells although infectivity was reduced a further 200-fold (0.06% of the wild-type level) in comparison to the simultaneous cotransfection of expression constructs for both mature subunits (20% of the wild-type level) (Fig. 3B and F, lanes 4 and 5). A similar relative reduction in infectivity (100-fold, or 0.8% of the wild-type level) was observed for samples generated by cotransfection of a full-length Pol packaging construct harboring an enzymatically inactive IN domain (coPP iIN) compared to the Pol wild-type control (Fig. 3F, columns 1 and 8). In both cases, marker gene expression was only transient and declined over time (data not shown), similar to previously described integrase-inactive retroviral vector systems (48, 49). In all other samples, marker gene expression was stable and constant over time (data not shown).
Taken together, these data demonstrate that mature PFV Pol subunits can be functionally incorporated into secreted virions if present at high intracellular concentrations, achieved in this study by use of expression-optimized packaging constructs. Furthermore, if both Pol subunits are provided in parallel, the produced PFV vector particles are infectious, resulting in stable transduction of target cells.
Intraparticle genome reverse transcription by individually packaged mature PFV Pol subunits.
Efficient intraparticle RTr of the viral genome is a hallmark of FVs and contributes to the majority of infectivity of FV particle preparations (7, 31).
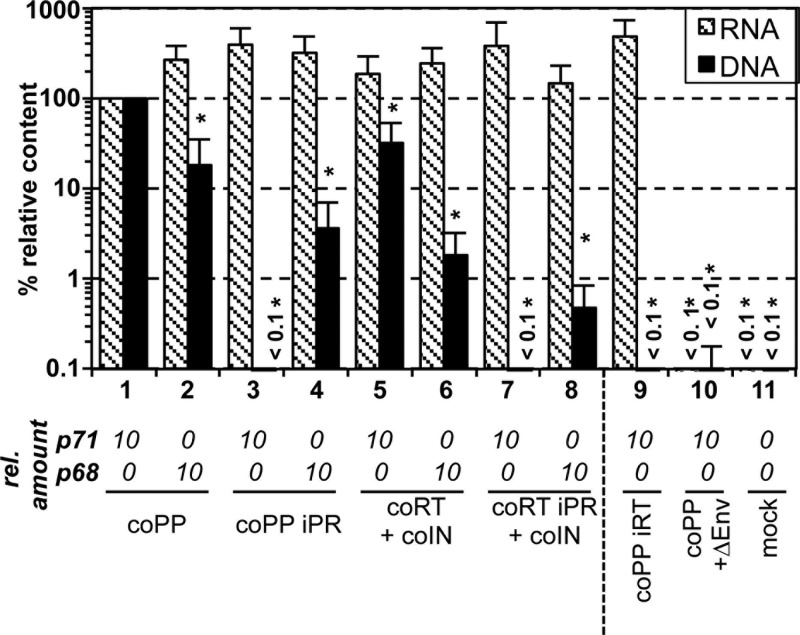
When we examined the nucleic acid composition of particles generated by cotransfecting the Gag, Env, and vRNA components of the expression-optimized vector system together with packaging constructs for both mature Pol subunits (coRT+coIN) or the different subunits individually (coRT and coIN), similar amounts of packaged vRNA were detected (Fig. 3G, striped bars). In contrast, significant differences were observed in the levels of particle-associated reverse transcript (vDNA) (Fig. 3G, black bars). Transfection of the coRT subunit expression construct alone or in combination with the coIN construct was sufficient for efficient intraparticle genome RTr (Fig. 3G, columns 2 to 5). The amount of particle-associated vDNA of samples generated by cotransfection of the individual mature Pol subunit expression constructs correlated well with the respective particle infectivity (Fig. 3F and G, columns 2 to 4). In virion samples generated by cotransfection of the Gag, Env, and vRNA vector components and the coRT or coIN packaging vector alone, only the coRT-derived particles contained detectable amounts of vDNA (Fig. 3G, columns 5 and 6). However, the level of reverse transcription was reduced 2.3-fold in comparison to analogous samples generated with the same amount of packaging vectors for both subunits (Fig. 3G, columns 4 and 5). This suggests that the presence of IN might enhance the reverse transcription efficiency of the packaged vRNA.
Thus, encapsidation of the mature PR-RT subunit, achieved by high intracellular protein expression, is sufficient for intraparticle reverse transcription of the packaged viral RNA genome. Furthermore, the time point of reverse transcription within the replication cycle is not altered. This further highlights the functionality of the particle-associated mature Pol subunits.
Conditions of PFV Pol PR activity dispensability for detectable viral particle infectivity.
PFV Gag precursor processing by the viral PR is thought to be essential for correct capsid morphogenesis, which is a prerequisite for intraparticle genome RTr and viral infectivity (23–25, 42). In addition, PFV IN activity, essential for stable transduction of target cells, is believed to require autocatalytic PFV Pol precursor processing (14). Furthermore, additional PFV PR-mediated Gag processing events during viral entry were reported to be instrumental for capsid disassembly and productive infection of target cells (26).
The possibility of assembling infectious PFV viral particles from mature Pol subunits presented an opportunity to examine in more detail the dependence of individual steps of the viral replication cycle on viral PR activity. The results shown above already indicated that autocatalytic processing of the PFV Pol precursor after its encapsidation into newly formed virions is not absolutely essential for viral replication so long as the mature subunits are incorporated into released particles instead.
For further examinations, we generated variants of the expression-optimized mature PR-RT subunit packaging construct (coRT iPR) as well as the full-length Pol precursor packaging construct (coPP iPR) with enzymatically inactivate PR domains (Fig. 1D). Furthermore, a C-terminal truncation mutant of the expression-optimized Gag packaging construct (pcoPG p68) lacking the p3 domain was generated (Fig. 1D). Subsequently, vRNA and Env expression vectors were cotransfected with different types and combinations of Pol and Gag packaging constructs into 293T cells. Cotransfecting the p68Gag- and the p71Gag-expressing packaging constructs pcoPG (p71) and pcoPG p68 (p68) at different ratios was employed to reconstitute the p71Gag/p68Gag protein capsid composition in the absence of an active viral protease during particle assembly.
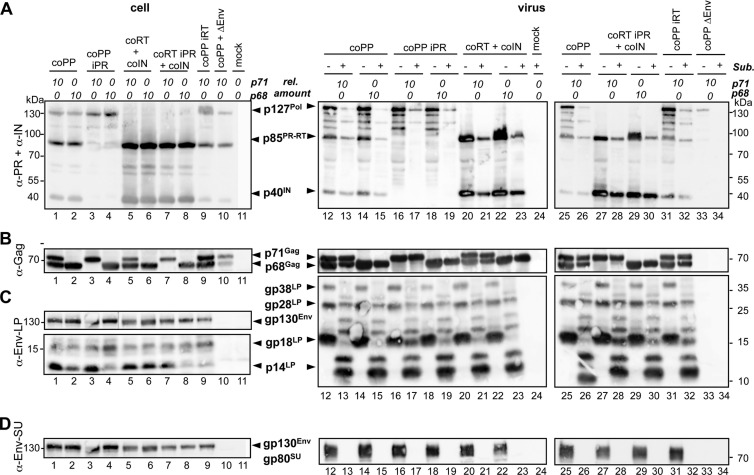
No major differences in the cellular expression levels and particle association of different wild-type Pol constructs relative to their respective Pol iPR mutants were observed (Fig. 4A). Subtilisin digestion of particles prior to lysis and immunodetection confirmed the previous observation that the amounts of p85PR-RT and p40IN protein were higher in samples generated with packaging constructs for separate mature Pol subunits than in full-length Pol precursor packaging construct-derived samples (Fig. 4A, lanes 12 to 23 and 25 to 32). The enzymatic activity status of the Pol domain in the different Pol packaging constructs did not alter this result. Furthermore, the type of Gag packaging construct (p71Gag versus p68Gag) cotransfected did not influence Pol expression, processing, or particle association (Fig. 4A). As expected, Pol and Gag precursor processing was dependent on an enzymatically active PR domain (Fig. 4A and B).
Fig 4.
Biochemical analysis of PFV particles derived from Pol packaging constructs with enzymatically inactive PR domains. 293T cells were cotransfected with identical amounts of PFV transfer vector puc2MD9, Env packaging vector pcoPE, different ratios of expression-optimized PFV Gag packaging constructs coding for p71Gag and p68Gag as indicated, and various Pol packaging constructs harboring expression-optimized ORFs for the following: the full-length wild-type PFV Pol protein (coPP, lanes 1, 2, 12 to 15, 25, and 26); a variant thereof with enzymatically inactive PR (coPP iPR, lanes 3, 4, and 16 to 19) or enzymatically inactive RT (coPP iRT, lanes 9, 31, and 32), the mature PR-RT and IN subunits (coRT+coIN, lanes 5, 6, and 20 to 23); the mature PR-RT subunit with enzymatically inactive PR and IN subunit (coRT iPR+coIN, lanes 7, 8, and 27 to 30). As a control, the Env packaging construct was omitted in a sample containing the wild-type full-length Pol packaging construct (coPP+ΔEnv, lanes 10, 33, and 34), or cells were transfected only with pUC19 DNA (mock, lanes 11 and 24). The total amount of transfected PFV Gag packaging construct was kept constant in all samples. The ratios of p71Gag (p71) to p68Gag (p68) packaging constructs are indicated on top. Western blot analysis of cell lysates (cell) and pelleted viral supernatants (virus) digested with subtilisin (+) or mock (−) incubated using a mouse monoclonal antibody mixture specific for PFV PR and PFV IN (α-PR+α-IN) (A), rabbit polyclonal antibodies specific for PFV Gag (α-Gag) (B), rabbit polyclonal antibodies specific for PFV Env-LP (α-Env-LP) (C), or mouse monoclonal antibodies specific for PFV Env-SU (α-Env-SU) (D). The identities of the individual proteins are indicated on the right.
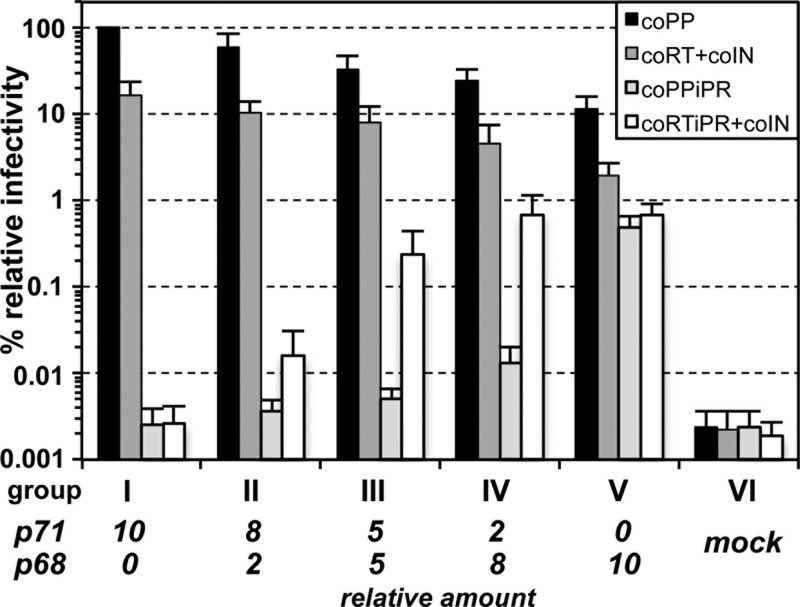
For the infectivity analysis, samples of viruses with different Pol proteins were grouped according to the ratio of p71Gag/p68Gag packaging constructs used during particle production (Fig. 5, groups I to V). Particles derived by cotransfection of the p71Gag expression construct pcoPG (p71) in combination with either a wild-type full-length Pol packaging construct (coPP) or a mixture of the wild-type mature Pol subunit packaging constructs (coRT+coIN) were highly infectious (15% of wt) (Fig. 5, group I). In contrast, combinations of pcoPG (p71) with either a PR-inactive, full-length Pol packaging construct (coPP iPR) or a mixture of the mature Pol subunit packaging constructs (coRT iPR+coIN) with enzymatically inactive PR yielded only noninfectious particles (<0.003% of wt) (Fig. 5, group I). Increasing the relative amounts of pcoPG p68 (p68) in the Gag packaging plasmid mixture in combination with a wild-type full-length Pol packaging construct (coPP) or a combination of the wild-type mature subunit packaging constructs (coRT+coIN) resulted in a dose-dependent decline in viral infectivity down to 10% and 2% of wt, respectively (Fig. 5, groups II to IV).
Fig 5.
Infectious PFV particles derived from Pol packaging constructs with enzymatically inactive PR domains. 293T cells were cotransfected with identical amounts of PFV transfer vector puc2MD9, Env packaging vector pcoPE, and different ratios of expression-optimized PFV Gag packaging constructs coding for p71Gag and p68Gag (indicated below the x axis) as well as various Pol packaging constructs harboring expression-optimized ORFs for the full-length wild-type PFV Pol protein (coPP), a variant thereof with enzymatically inactive PR (coPPiPR), the mature PR-RT and IN subunits (coRT+coIN), or the mature PR-RT subunit with enzymatically inactive PR and IN subunit (coRTiPR+coIN) as indicated. At 3 days postinfection, relative infectivities of extracellular 293T cell culture supernatants were determined with an egfp marker gene transfer assay. The values obtained using a full-length expression-optimized pol ORF expression plasmid (coPP) were arbitrarily set to 100%. Absolute titers of these plain supernatants were (4.99 ± 2.74) × 106 EGFP-positive FFU/ml. Means and standard deviations of at least four independent experiments are shown.
To our great surprise, the opposite was true in combination with the mixture of the PR-inactive mature subunit packaging constructs (coRT iPR+coIN) and even for the PR-inactive full-length Pol packaging construct (coPP iPR) (Fig. 5, groups II to IV). Here, a pcoPG p68 (p68) dose-dependent increase in particle infectivity (up to 0.7% and 0.5% of wt, respectively) was observed (Fig. 5) that resulted in stable marker gene expression in target cells (data not shown). Although both types of PR-inactive Pol packaging constructs yielded similar maximal vector infectivity, differences in the p68Gag dose dependency were observed (Fig. 5, groups II to V). The highest titers (0.5% of wt) for the full-length iPR Pol mutant (Fig. 5, coPP iPR) were obtained when only p68Gag was provided (Fig. 5, group V). In contrast, for vector particles generated with PR-inactive mature subunit packaging constructs (coRT iPR+coIN) already small amounts of p68Gag were sufficient to produce detectable amounts of infectious particles (Fig. 5, group II). Furthermore, maximal infectivity (0.7% of wt) was obtained at lower relative levels of p68Gag than required for particles containing full-length iPR Pol (coPP iPR) (Fig. 5).
Comparing the relative infectivity of different virus types within individual virus groups, in particular those generated by providing only p71Gag (Fig. 5, group I) or only p68Gag (Fig. 5, group V), yielded more interesting results. In group I, PR inactivation of both types of Pol packaging components completely abolished particle infectivity (at least 10,000- to 50,000-fold reduced), whereas in group V PR-deficient particles showed only a 3- to 15-fold reduced relative infectivity in comparison to their respective PR-active counterparts (Fig. 5, groups I and V).
Taken together these results confirm previous observations that PFV Gag precursor processing by the viral protease during assembly is indispensable for viral infectivity (23, 25, 38). More importantly, however, they indicate that, in contrast to current belief, PR enzymatic activity of PFV Pol is not absolutely essential during virus entry in target cells although infectivity of PR-inactive virions was always significantly lower than that of respective virions with active PR domains.
Structural requirements of the PR domain for viral infectivity.
Our data indicated that the enzymatic activity of the viral PR is not necessary for FV infectivity if Gag precursor processing is mimicked by supplying the mature cleavage product during particle assembly. Recent structural analysis data of Hartl et al. (40) suggest that the PFV PR-RT subunit is subdivided into N-terminal PR (aa 1 to 101) and C-terminal RT (aa 144 to 571) core domains, which are connected by a flexible linker peptide (aa 102 to 143) (Fig. 1D). Therefore, we examined next which PR-RT subdomains are structurally relevant for viral infectivity in context of the mature PR-RT subunit. Two N-terminal truncation mutants of the coRT subunit packaging construct were generated (Fig. 1D). The first, coRT1, lacks sequences encoding the core PR domain and flexible linker peptide (aa 1 to 143), whereas the second, coRT2, has only the N-terminal core PR domain (aa 1 to 101)-encoding sequences removed (Fig. 1D).
Biochemical characterization of viral particles generated with these different coRT-derived packaging constructs in combination with the other vector components (vRNA, Gag, Env, and coIN) revealed that the RT1 and RT2 proteins, independently of subtilisin treatment, were found in higher levels in particle lysates than the parental PR-RT protein (Fig. 6A, lanes 12 to 23). As expected, no Gag precursor protein processing was observed when the RT1 or RT2 protein was coexpressed with p71Gag (Fig. 6B, lanes 4 to 6, 16, 17, 20, and 21).
Fig 6.
Dependence of viral infectivity on PR structure. 293T cells were cotransfected with identical amounts of PFV transfer vector puc2MD9, Env packaging vector pcoPE, and various Pol packaging constructs harboring expression-optimized ORFs for the full-length wild-type PFV Pol protein (coPP, lanes 1, 10, and 11), the mature PR-RT and IN subunits (coRT+coIN, lanes 2, 12, and 13), the mature PR-RT subunit with enzymatically inactive PR and IN subunit (coRT iPR+coIN, lanes 3, 14, and 15), the full PR domain-deleted PR-RT subunit and IN subunit (coRT1+coIN, lanes 4, 5, and 16 to 19), and the core PR-deleted PR-RT subunit and IN subunit (coRT2+coIN, lanes 6, 7, and 20 to 23) as well as different ratios of expression-optimized PFV Gag packaging constructs coding for p71Gag and p68Gag as indicated. As a control, the Env packaging construct was omitted in a sample containing the wild-type full-length Pol packaging construct (coPP +ΔEnv, lanes 9, 26, and 27), or cells were transfected only with pUC19 DNA (mock, lanes 8, 24, and 25). The total amount of transfected PFV Gag packaging was kept constant in all samples. The ratios of p71Gag (p71) to p68Gag (p68) packaging constructs are indicated on top. The total amount of transfected DNA (16 μg) was kept constant in all samples by addition of pUC19 DNA. (A to C) Western blot analysis of Pol and Gag expression in cell lysates (cell) and concentrated virus (virus) either digested with subtilisin (+) or mock (−) incubated using a polyclonal antibody mixture specific for SFV-1 PR-RT and PFV IN (α-RT+α-IN) (A), a rabbit polyclonal antibody specific for PFV Gag (α-Gag) (B), and a rabbit polyclonal antibody specific for PFV Env-LP (α-Env-LP) or mouse monoclonal antibodies specific for PFV Env-SU (α-Env-SU) (C). (D) At 3 days postinfection, transduction rates of HT1080 cells were measured using an egfp marker gene transfer assay. The values obtained using full-length expression-optimized pol ORF expression plasmid (coPP) were arbitrarily set to 100%. Absolute titers of these plain supernatants were (4.38 ± 0.96) × 106 EGFP-positive FFU/ml. Means and standard deviations of at least four independent experiments are shown. In this assay the sample coPP+ΔEnv was not determined (nd).
Analysis of viral infectivity showed that removal of the PR core domain and the flexible linker (coRT1) reduced the infectivity in p68Gag-derived particles to below the detection limit of the assay (0.01% of the wild-type level) (Fig. 6D, bar 5). In contrast, significant residual viral infectivity (0.1% of the wild-type level) was detectable for the coRT2 packaging construct, which had only the PR core domain deleted (Fig. 6D, bar 7). However, infectivity of the coRT2-derived particles (0.1% of the wild-type level) was reduced an additional 10-fold in comparison to the coRT iPR-derived particles containing an enzymatically inactive PR domain (1% of the wild-type level) (Fig. 6D, bars 1, 3, and 7).
These results suggest that the PR core domain of PFV PR-RT is not absolutely required for, but seems to enhance, viral infectivity. In contrast, the flexible linker appears to be absolutely essential for viral infectivity.
Gag precursor maturation is an initiation step for intraparticle reverse transcription.
Previous mutagenesis analysis of the PFV Gag p68/p3 cleavage site revealed that Gag precursor processing is absolutely essential for viral infectivity (23, 25), which was confirmed by the data presented above. Furthermore, Enssle and colleagues (23) reported that in cells transfected with proviral expression constructs producing a noncleavable p71Gag protein, no intracellular viral cDNA synthesis was detectable by Southern blotting.
To determine how Gag and Pol precursor processing influences FV intraparticle RTr of encapsidated vRNA, we examined the nucleic acid composition of particles differing in their Gag and Pol composition. The variation in Gag and Pol composition had no statistically significant influence on vRNA packaging (Fig. 7, striped bars). In contrast, large differences were observed for the level of intraparticle RTr of the packaged vRNA genome, which correlated well with the corresponding particle infectivity (compare Fig. 7, black bars, and 5, groups I and V). Using a p68Gag expression construct in combination with PR-active Pol packaging constructs yielded particles with 5- to 10-fold reduced vDNA content in comparison to respective p71Gag-derived wild-type particles (Fig. 7, black bars 1, 2, 5, and 6). In particles composed of p71Gag with PR-inactive Pol proteins, no intraparticle vDNA was detectable (Fig. 7, black bars 3 and 7), whereas in corresponding p68Gag-derived particles, vDNA was readily measurable (Fig. 7, black bars 4 and 8). The vDNA levels of the latter were only 3- to 5-fold reduced compared to respective p68Gag virions containing PR-active Pol (Fig. 7, compare black bars 2 to 4 and 6 to 8). Unlike Gag, Pol processing was not required for intraparticle RTr since vDNA was present in p68Gag-derived virions generated by coexpression of the PR-inactive Pol precursor coPP iPR (Fig. 7, black bar 4). Interestingly, the level of intraparticle vDNA in corresponding p68Gag-derived virions generated by coexpression of the mature coRT iPR and coIN Pol subunits was about 10-fold lower than in p68Gag-derived virions derived from coPP iPR (Fig. 7, black bars 4 and 8), whereas the infectivity levels of both types of particles were nearly identical (Fig. 5, group V). This result suggests that the IN domain embedded in the precursor protein might have a lower enzymatic activity than the mature IN subunit. This would explain the higher relative infectivity of the coRT iPR+coIN-derived particles than of the coPP iPR-derived particles when values were normalized for the amount of particle-associated vDNA.
Fig 7.
Correlation of released particle-associated nucleic acid composition with Gag processing and Pol enzymatic activities. 293T cells were cotransfected with identical amounts of PFV transfer vector puc2MD9, Env packaging vector pcoPE, and various Pol packaging constructs harboring expression-optimized ORFs for the full-length wild-type PFV Pol protein (coPP), a variant thereof with enzymatically inactive PR (coPP iPR), or enzymatically inactive RT (coPP iRT), the mature PR-RT and IN subunits (coRT+coIN), the mature PR-RT subunit with enzymatically inactive PR and IN subunit (coRT iPR+coIN), and different ratios of expression-optimized PFV Gag packaging constructs coding for p71Gag or p68Gag as indicated. As a control the Env packaging construct was omitted in a sample containing the wild-type full-length Pol packaging construct (coPP+ΔEnv), or cells were transfected only with pUC19 DNA (mock). The total amount of transfected PFV Gag packaging was kept constant in all samples. The ratios of p71Gag (p71) to p68Gag (p68) packaging constructs are indicated below the x axis. The relative nucleic acid composition of mutant particles was determined with PFV Pol-specific primers and TaqMan probe. Following DNase I digestion of intact, purified particles, nucleic acids were isolated, and the relative amount of vector RNA and DNA copies was determined in comparison to the wild type by qPCR. The mean values and standard deviations of the relative RNA and DNA contents of at least three independent experiments are shown. Sample values were normalized for capsid protein release. Differences between means of the wild type and the individual mutants were analyzed by Welch's t test (* P < 0.01).
These data strongly imply that the major event initiating FV intraparticle RTr is Gag precursor cleavage by the viral PR. Both the unprocessed Pol precursor and the mature PR-RT subunit are capable of intraparticle RTr, an activity that seems to be inhibited by p71Gag.
Generation of infectious PR-deficient PFV particles is not an artifact of PFV ORF expression optimization.
The major novel and unexpected observation reported in this study is that infectious PFV vector particles were obtained by combining p68Gag and PR-deficient p127Pol precursor-encoding packaging constructs for virus production. It is possible that this phenotype is mainly the result of the increased viral protein expression and virus production achieved by the expression-optimized four-component PFV vector system (4-co) employed in most experiments of this study. To ensure that this was not the case, we also examined analogous combinations of Gag and Pol mutants in the context of several other PFV production systems based on authentic PFV ORFs (Fig. 1A to C). We used the original authentic-ORF-containing four-component vector system (4-nco) developed by Heinkelein et al. (36). Furthermore, we generated and used an authentic-ORF-containing three-component vector system (3-nco), which includes a Gag/Pol packaging construct containing PFV genomic sequences starting at the 5′ LTR R region and terminating after the translation stop of the Pol ORF harboring natural splice sites. Finally, we generated and characterized mutant proviral expressions constructs (prov) expressing only p68Gag, a p127Pol precursor protein with an enzymatically inactive PR domain, or a combination of both. The relative expression levels of PFV proteins and virus titers achieved for the wild-type forms of the different PFV productions systems are shown in Fig. 2 and were discussed in detail above. Figure 8 summarizes the infectivity analysis of viral supernatants generated by the different PFV production systems containing various combinations of the mutant Gag and Pol proteins. The results demonstrated that only the absolute titers of samples derived from different production systems varied, as shown in Fig. 2A for the wild-type samples. This affected the sensitivity and dynamic range of the flow cytometric infectivity assay for the individual PFV production system. However, the infectious phenotype of p68Gag/iPR particles was completely reproducible in all systems examined (Fig. 8). Similar relative differences in infectivities between wild-type particles and p68Gag and p68Gag/iPR particles were observed with all PFV production systems.
Fig 8.
PR-deficient, infectious PFV particles generated by production systems with authentic-codon usage. 293T cells were transiently transfected with the components of the different PFV production systems as indicated. The proviral constructs (prov) were cotransfected with the EGFP-expressing PFV transfer vector puc2MD9 (PTV) at a ratio of 1:1. Cell-free viral supernatants of various combinations of Gag and Pol mutants in the context of the individual PFV production systems were titrated on HT1080 target cells using a flow cytometric marker gene assay, and titers were determined at 72 posttransduction. For the proviral constructs, similar titers were obtained when they were not cotransfected with puc2MD9 and titrated on HT1080 PLNE cells, containing a Tas-inducible nuclear EGFP expression cassette under the control of the PFV LTR (data not shown). The values obtained using full-length gag (p71) and pol ORFs with an enzymatically active PR domain (Pol) were arbitrarily set to 100%. Absolute titers of these plain supernatants are shown in Fig. 2A. Means and standard deviations of at least three independent experiments are shown.
DISCUSSION
Previously it was reported that virion encapsidation of mature PFV Pol subunits, when expressed as separate proteins from individual packaging constructs, could not be detected (9, 14). We demonstrate in this study that mature PFV Pol subunits can be functionally incorporated into PFV capsids, either individually or in combination, when intracellular protein levels are highly elevated, achieved here by using corresponding expression-optimized subunit packaging constructs. Whether encapsidation of the mature Pol subunits is an active process involving binding of the subunit to specific viral structures or, rather, a passive incorporation during budding due to their high overall concentration in the cytoplasm cannot be deduced from our experiments. However, the requirement of a high intracellular protein concentration for mature subunit packaging strongly indicates that at low cellular Pol levels, as probably found in natural infections or achieved by vector systems using authentic PFV ORFs, Pol is preferentially packaged as precursor protein. This is presumably due to specific interaction of the Pol precursor with cis-acting sequences of the viral genome and potential Gag-Pol protein interactions.
Although Pol precursor-independent incorporation of mature PFV Pol subunits by cellular overexpression represents an artificial system, its analysis might allow some insights into the function of the mature subunits found in authentic FV particles, which can only be generated by proteolytic processing of the encapsidated Pol precursor. First, encapsidation of the mature PR-RT subunit alone was sufficient for Gag processing and intraparticle genome RTr in a dose-dependent manner. Gag processing appeared not to be affected by copackaging of the mature IN subunit, but the amount of intraparticle vDNA was increased about 2-fold. Thus, the mature IN subunit seems not to influence the PR activity of the mature PR-RT subunit. This is in line with a recent report on PFV PR-RT subunit encapsidation as a Gag-PR-RT fusion protein demonstrating the dispensability of the IN subunit for particle-associated Gag processing (20). However, the increased vDNA content of IN subunit-containing particles observed in our study indicates that the IN subunit might have a yet undiscovered function as a potential cofactor for intraparticle RTr.
Second, although the mature Pol subunits were present in larger amounts in particles derived from mature subunit packaging constructs than in respective particles derived from Pol precursor packaging constructs, the level of intraparticle vDNA, which correlated well with particle infectivity, was much lower. Furthermore, a much higher particle-associated level of PR-RT subunit, packaged as mature subunit, was required to achieve a wild-type-like Gag processing pattern. These observations might suggest that in authentic PFV particles the encapsidated Pol precursor, rather than its autocatalytic processing products, preferentially or more efficiently processes the Gag precursor and reverse transcribes the encapsidated vRNA. This can also mean that Gag precursor processing occurs prior to maturation of the Pol precursor. Comparison of the enzymatic activities of recombinant mature PR-RT to that of processing-resistant Pol precursor in vitro may provide further information on this aspect. However, such in vitro enzyme assays also represent simplified artificial systems, which might not completely reflect the natural situation found in wild-type particles. In line with this, a couple of noninfectious FV Gag mutants were described that failed to undergo natural intraparticle RTr, but Pol proteins extracted from such mutant capsid displayed no apparent defects in in vitro reverse transcription assays using nonviral substrates (12, 42).
The identification of conditions for production of infectious FV particles lacking any viral protease activity was the most striking and unexpected discovery of this study. Although the infectivity levels of PR-deficient particles were only 0.5 to 2% of the wild-type level, to our knowledge this is the first report of an infectious retrovirus particle which can be assembled from its mature capsid and either precursor or mature enzymatic subunits without need for further viral protein processing by the viral protease. Importantly, this phenotype is fully reproducible with three different virus production systems based on authentic PFV sequences, including proviral constructs, and is therefore neither solely the result of nor dependent on expression optimization of packaging constructs. The characterization of various combinations of Gag and Pol mutants in the context of the different PFV production systems demonstrates that Gag precursor and not Pol processing is the initiating event for intraparticle RTr and viral infectivity. In line with these results, Enssle et al. (23) previously observed that inactivation of the Gag p68/3 cleavage site alone prevents intracellular accumulation of the viral cDNA. Furthermore, restoration of intraparticle RTr of proviral constructs with PR-inactive Pol by p68Gag provision in trans was also previously noticed by Lehmann-Che et al. (26). What prevents intraparticle RTr and infectivity of p71Gag-derived particles is currently unclear. We can envision different possibilities. First, p71Gag capsids, which show morphological abnormalities (i.e., many incompletely closed horseshoe-like capsid structures), might not provide the correct “microenvironment” for RTr. This incorrect microenvironment could result from a lack of essential Gag-Pol interactions required for RTr initiation, perhaps due to structural differences between p71Gag and p68Gag. Alternatively, the p3 domain as part of the p71Gag precursor might exert a direct or indirect inhibitory effect on RT enzymatic activity, similar to the function reported for NC in HIV-1 particles, thereby preventing RTr in p71Gag particles (30, 50). Separation of the PFV Gag p3 domain from the precursor by viral protease-mediated Gag precursor processing may then relieve this inhibitory function. It is possible that released mature p3Gag might subsequently even act as a stimulatory factor for RT enzymatic activity in the FV capsid. The latter hypothesis could also explain the lower infectivity of FV particles composed of only p68Gag, which we show here for the first time is the consequence of reduced intraparticle RTr. The reduced infectivity and vDNA content of p68Gag particles might be the direct consequence of a missing potential stimulatory activity for RTr by mature p3Gag, which is not available in this type of virions. However, p3Gag has not been detected in infected cells or viral particles so far, nor does it contain any known nucleic acid binding activities as reported for HIV NC (28).
Finally, our results overturn the current dogma in FV literature that viral PR activity is absolutely essential at some point during target cell entry (26, 27). Although the results do not formally exclude the possibility that further Gag processing by PR occurs and enhances disassembly, the infectivity of p68Gag-derived virions containing PR-inactive Pol proteins clearly refutes the claim that further Gag processing at secondary cleavage sites upon target cell entry is absolutely required for productive FV infection (26, 27). All p68Gag/iPR PFV particles were only 3- to 20-fold less infectious than their respective p68Gag/PR-active counterparts. But in all cases, a clearly detectable and stable transgene expression was obtained upon target cell transduction. Therefore, our results suggest that the apparent noninfectious phenotype of PFV particles with mutations at putative secondary Gag cleavage sites described by Pfrepper et al and Lehmann-Che et al. (26, 27) is not a consequence of a processing defect of capsids by the viral PR during viral entry but is, rather, the result of a yet uncharacterized disassembly defect of this Gag mutant. The different requirements of PR activity for FV entry in the study of Lehmann-Che et al. (26) and our study might be explained by the different combinations of Gag and Pol mutants examined. Our results for the expression-optimized vector system demonstrate that virions containing a PR-deficient Pol precursor protein obtain their maximal infectivity only in the absence of p71Gag. Furthermore, we show that PR-deficient proviral constructs expressing only p68Gag instead of Gag precursor show low but detectable infectivity. These conditions were not examined by Lehmann-Che et al. (26) since the PR-deficient provirus-derived particles, reconstituted with p68Gag in trans, had a ratio of p71Gag to p68Gag of about 1:1, and the very low-level infectivity of such particles might thus have been below the detection limit of the infectivity assays employed.
In summary, this is the first report of the generation of infectious retroviral particles exclusively from mature capsid and polymerase subunits. Furthermore, we demonstrate for different PFV production systems, based on authentic or expression-optimized ORFs, that Gag precursor processing by the viral PR during assembly appears to be absolutely required for generation of infectious particles as it is essential for intraparticle reverse transcription initiation. In contrast to previous assumptions, our data show that further viral PR-mediated Gag processing during virus entry, although not formally excluded, is not absolutely required for productive infection of target cells.
ACKNOWLEDGMENTS
We thank Welkin Johnson, Andrea Kirmaier, and Michael Thompson for critically reading the manuscript.
E.M. was partially supported by a DIGS-BB fellowship. This work was supported by grants from the DFG (Li621/3-3, Li621/4-1, Li621/4-2, and Li621/6-1) to D.L.
The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 2 January 2013
REFERENCES
- 1. Briggs JA, Krausslich HG. 2011. The molecular architecture of HIV. J. Mol. Biol. 410:491–500 [DOI] [PubMed] [Google Scholar]
- 2. Ganser-Pornillos BK, Yeager M, Sundquist WI. 2008. The structural biology of HIV assembly. Curr. Opin. Struct. Biol. 18:203–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pettit SC, Moody MD, Wehbie RS, Kaplan AH, Nantermet PV, Klein CA, Swanstrom R. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68:8017–8027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Krausslich HG. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rethwilm A. 2010. Molecular biology of foamy viruses. Med. Microbiol. Immunol. 199:197–207 [DOI] [PubMed] [Google Scholar]
- 6. Enssle J, Jordan I, Mauer B, Rethwilm A. 1996. Foamy virus reverse transcriptase is expressed independently from the Gag protein. Proc. Natl. Acad. Sci. U. S. A. 93:4137–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yu SF, Baldwin DN, Gwynn SR, Yendapalli S, Linial ML. 1996. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science 271:1579–1582 [DOI] [PubMed] [Google Scholar]
- 8. Heinkelein M, Leurs C, Rammling M, Peters K, Hanenberg H, Rethwilm A. 2002. Pregenomic RNA is required for efficient incorporation of pol polyprotein into foamy virus capsids. J. Virol. 76:10069–10073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Peters K, Wiktorowicz T, Heinkelein M, Rethwilm A. 2005. RNA and protein requirements for incorporation of the pol protein into foamy virus particles. J. Virol. 79:7005–7013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wiktorowicz T, Peters K, Armbruster N, Steinert AF, Rethwilm A. 2009. Generation of an improved foamy virus vector by dissection of cis-acting sequences. J. Gen. Virol. 90:481–487 [DOI] [PubMed] [Google Scholar]
- 11. Lee EG, Linial ML. 2008. The C terminus of foamy retrovirus Gag contains determinants for encapsidation of Pol protein into virions. J. Virol. 82:10803–10810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Müllers E, Uhlig T, Stirnnagel K, Fiebig U, Zentgraf H, Lindemann D. 2011. Novel functions of prototype foamy virus Gag glycine- arginine-rich boxes in reverse transcription and particle morphogenesis. J. Virol. 85:1452–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stenbak CR, Linial ML. 2004. Role of the C terminus of foamy virus Gag in RNA packaging and Pol expression. J. Virol. 78:9423–9430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roy J, Linial ML. 2007. Role of the foamy virus Pol cleavage site in viral replication. J. Virol. 81:4956–4962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Flügel RM, Pfrepper KI. 2003. Proteolytic processing of foamy virus Gag and Pol proteins. Curr. Top. Microbiol. Immunol. 277:63–88 [DOI] [PubMed] [Google Scholar]
- 16. Dunn BM, Goodenow MM, Gustchina A, Wlodawer A. 2002. Retroviral proteases. Genome Biol. 3:REVIEWS3006 doi:10.1186/gb-2002-3-4-reviews3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hartl MJ, Mayr F, Rethwilm A, Wohrl BM. 2010. Biophysical and enzymatic properties of the simian and prototype foamy virus reverse transcriptases. Retrovirology 7:5 doi:10.1186/1742-4690-7-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hartl MJ, Bodem J, Jochheim F, Rethwilm A, Rosch P, Wohrl BM. 2011. Regulation of foamy virus protease activity by viral RNA: a novel and unique mechanism among retroviruses. J. Virol. 85:4462–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee EG, Roy J, Jackson D, Clark P, Boyer PL, Hughes SH, Linial ML. 2011. Foamy retrovirus integrase contains a Pol dimerization domain required for protease activation. J. Virol. 85:1655–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Spannaus R, Hartl MJ, Wohrl BM, Rethwilm A, Bodem J. 2012. The prototype foamy virus protease is active independently of the integrase domain. Retrovirology 9:41 doi:10.1186/1742-4690-9-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindemann D, Rethwilm A. 2011. Foamy virus biology and its application for vector development. Viruses 3:561–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cartellieri M, Rudolph W, Herchenröder O, Lindemann D, Rethwilm A. 2005. Determination of the relative amounts of Gag and Pol proteins in foamy virus particles. Retrovirology 2:44 doi:10.1186/1742-4690-2-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Enssle J, Fischer N, Moebes A, Mauer B, Smola U, Rethwilm A. 1997. Carboxy-terminal cleavage of the human foamy virus Gag precursor molecule is an essential step in the viral life cycle. J. Virol. 71:7312–7317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fischer N, Heinkelein M, Lindemann D, Enssle J, Baum C, Werder E, Zentgraf H, Müller JG, Rethwilm A. 1998. Foamy virus particle formation. J. Virol. 72:1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zemba M, Wilk T, Rutten T, Wagner A, Flügel RM, Löchelt M. 1998. The carboxy-terminal p3Gag domain of the human foamy virus Gag precursor is required for efficient virus infectivity. Virology 247:7–13 [DOI] [PubMed] [Google Scholar]
- 26. Lehmann-Che J, Giron ML, Delelis O, Lochelt M, Bittoun P, Tobaly-Tapiero J, de The H, Saib A. 2005. Protease-dependent uncoating of a complex retrovirus. J. Virol. 79:9244–9253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pfrepper KI, Löchelt M, Rackwitz HR, Schnolzer M, Heid H, Flügel RM. 1999. Molecular characterization of proteolytic processing of the Gag proteins of human spumavirus. J. Virol. 73:7907–7911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mougel M, Houzet L, Darlix JL. 2009. When is it time for reverse transcription to start and go? Retrovirology 6:24 doi:10.1186/1742-4690-6-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Houzet L, Morichaud Z, Didierlaurent L, Muriaux D, Darlix JL, Mougel M. 2008. Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic Acids Res. 36:2311–2319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thomas JA, Bosche WJ, Shatzer TL, Johnson DG, Gorelick RJ. 2008. Mutations in human immunodeficiency virus type 1 nucleocapsid protein zinc fingers cause premature reverse transcription. J. Virol. 82:9318–9328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moebes A, Enssle J, Bieniasz PD, Heinkelein M, Lindemann D, Bock M, McClure MO, Rethwilm A. 1997. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 71:7305–7311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zamborlini A, Renault N, Saib A, Delelis O. 2010. Early reverse transcription is essential for productive foamy virus infection. PLoS One 5:e11023 doi:10.1371/journal.pone.0011023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DuBridge RB, Tang P, Hsia HC, Leong PM, Miller JH, Calos MP. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rasheed S, Nelson-Rees WA, Toth EM, Arnstein P, Gardner MB. 1974. Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer 33:1027–1033 [DOI] [PubMed] [Google Scholar]
- 35. Lindemann D, Pietschmann T, Picard-Maureau M, Berg A, Heinkelein M, Thurow J, Knaus P, Zentgraf H, Rethwilm A. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762–5771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heinkelein M, Dressler M, Jarmy G, Rammling M, Imrich H, Thurow J, Lindemann D, Rethwilm A. 2002. Improved primate foamy virus vectors and packaging constructs. J. Virol. 76:3774–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stirnnagel K, Lüftenegger D, Stange A, Swiersy A, Müllers E, Reh J, Stanke N, Grosse A, Chiantia S, Keller H, Schwille P, Hanenberg H, Zentgraf H, Lindemann D. 2010. Analysis of prototype foamy virus particle-host cell interaction with autofluorescent retroviral particles. Retrovirology 7:45 doi:10.1186/1742-4690-7-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Konvalinka J, Löchelt M, Zentgraf H, Flügel RM, Kräusslich HG. 1995. Active foamy virus proteinase is essential for virus infectivity but not for formation of a Pol polyprotein. J. Virol. 69:7264–7268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Enssle J, Moebes A, Heinkelein M, Panhuysen M, Mauer B, Schweizer M, Neumann-Haefelin D, Rethwilm A. 1999. An active foamy virus integrase is required for virus replication. J. Gen. Virol. 80:1445–1452 [DOI] [PubMed] [Google Scholar]
- 40. Hartl MJ, Wohrl BM, Rosch P, Schweimer K. 2008. The solution structure of the simian foamy virus protease reveals a monomeric protein. J. Mol. Biol. 381:141–149 [DOI] [PubMed] [Google Scholar]
- 41. Ho Y-P, Schnabel V, Swiersy A, Stirnnagel K, Lindemann D. 2012. A small-molecule-controlled system for efficient pseudotyping of prototype foamy virus vectors. Mol. Ther. 20:1167–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mannigel I, Stange A, Zentgraf H, Lindemann D. 2007. Correct capsid assembly mediated by a conserved YXXLGL motif in prototype foamy virus Gag is essential for infectivity and reverse transcription of the viral genome. J. Virol. 81:3317–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swiersy A, Wiek C, Reh J, Zentgraf H, Lindemann D. 2011. Orthoretroviral-like prototype foamy virus Gag-Pol expression is compatible with viral replication. Retrovirology 8:66 doi:10.1186/1742-4690-8-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stange A, Luftenegger D, Reh J, Weissenhorn W, Lindemann D. 2008. Subviral particle release determinants of prototype foamy virus. J. Virol. 82:9858–9869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Imrich H, Heinkelein M, Herchenroder O, Rethwilm A. 2000. Primate foamy virus Pol proteins are imported into the nucleus. J. Gen. Virol. 81:2941–2947 [DOI] [PubMed] [Google Scholar]
- 46. Duda A, Stange A, Luftenegger D, Stanke N, Westphal D, Pietschmann T, Eastman SW, Linial ML, Rethwilm A, Lindemann D. 2004. Prototype foamy virus envelope glycoprotein leader peptide processing is mediated by a furin-like cellular protease, but cleavage is not essential for viral infectivity. J. Virol. 78:13865–13870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu SF, Eastman SW, Linial ML. 2006. Foamy virus capsid assembly occurs at a pericentriolar region through a cytoplasmic targeting/retention signal in Gag. Traffic 7:966–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Banasik MB, McCray PB., Jr 2010. Integrase-defective lentiviral vectors: progress and applications. Gene Ther. 17:150–157 [DOI] [PubMed] [Google Scholar]
- 49. Deyle DR, Li Y, Olson EM, Russell DW. 2010. Nonintegrating foamy virus vectors. J. Virol. 84:9341–9349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Didierlaurent L, Houzet L, Morichaud Z, Darlix JL, Mougel M. 2008. The conserved N-terminal basic residues and zinc-finger motifs of HIV-1 nucleocapsid restrict the viral cDNA synthesis during virus formation and maturation. Nucleic Acids Res. 36:4745–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]