Abstract
The phosphoprotein (P) of vesicular stomatitis virus (VSV) plays essential roles in viral RNA synthesis. It associates with nascent nucleoprotein (N) to form N0-P (free of RNAs), thereby preventing the N from binding to cellular RNAs and maintaining the N in a viral genomic RNA encapsidation-competent form for transcription and replication. The contributions of phosphorylation of P to transcription and replication have been studied intensively, but a concrete mechanism of action still remains unclear. In this study, using a VSV minigenome system, we demonstrated that a mutant of P lacking N-terminal phosphorylation (P3A), in which the N-terminal phosphate acceptor sites are replaced with alanines (S60/A, T62/A, and S64/A), does not support transcription and replication. However, results from protein interaction assays showed that P3A self-associates and interacts with N and the large protein (L) as efficiently as P does. Furthermore, purified recombinant P3A from Sf21 cells supported transcription in an in vitro transcription reconstitution assay. We also proved that P3A is not distributed intranuclearly in vivo. CsCl gradient centrifugation showed that P3A is incapable of preventing N from binding to cellular RNAs and therefore prevents functional template formation. Taken together, our results demonstrate that N-terminal phosphorylation is indispensable for P to prevent N from binding to nonviral RNAs and to maintain the N-specific encapsidation of viral genomic RNA for functional template formation.
INTRODUCTION
Vesicular stomatitis virus (VSV) is a nonsegmented negative-strand RNA virus that serves as the prototype of the rhabdovirus group. The active RNA polymerase complex of VSV is composed of the large protein (L) and its cofactor, the phosphoprotein (P) (1), and both are required for the transcription and replication of the viral negative-sense single-stranded RNA genome, which is tightly encapsidated by nucleoprotein (N) (2, 3). P is a multifunctional protein: it associates with newly synthesized N during infection to prevent N from binding to cellular RNAs and maintains the replication-competent form of N by specifically encapsidating the nascent viral genomic/antigenomic RNA (4–6). On the other hand, it may also help to protect the L protein from proteolytic degradation (7). In addition, P also forms a tripartite replicase complex with L and N (L-N-P) to convert the N-RNA template to positive-sense genomic RNA in vivo. Via mutational and biochemical studies, P of the VSV Indiana serotype has been divided arbitrarily into three domains, i.e., N-terminal domain I (residues 1 to 137), domain II (residues 211 to 244), and domain III (residues 245 to 265), and a hypervariable hinge region (residues 138 to 210).
Like the P's of many other negative-strand RNA viruses, the P of VSV is phosphorylated in infected cells and virions (8–10). The P of the VSV Indiana serotype has been shown to be phosphorylated in two different domains: N-terminal domain I, in which phosphorylation sites were mapped to S60, T62, and S64, and C-terminal domain II, in which phosphorylation sites were mapped to S226 and S227. Since P was shown to be phosphorylated in specific domains involved in regulating transcription in vitro, phosphorylation of P and its role in transcription have been investigated intensively (11–18). The conclusions derived from these investigations have demonstrated that the phosphorylation of P is important for its functionality in various ways. Several studies employing in vitro and in vivo experiments have suggested that N-terminal phosphorylation of P of the VSV Indiana serotype by casein kinase II (CKII) is essential for its transcription function (11, 12, 14–16, 18). In contrast, Spadafora et al. (17) showed that the N-terminal phosphorylation of P may not be essential for VSV RNA synthesis but may play a role in the self-association of P and interaction with L. In addition, an infectious VSV in which N-terminal phosphorylation of P was absent at all three sites could not be recovered, which confirms that N-terminal phosphorylation of P is absolutely critical for viral growth and multiplication (19). Since a defect at any step of the viral life cycle definitely disrupts viral growth, it is unclear how N-terminal phosphorylation of P plays a critical role in transcription or growth and multiplication.
In the present study, we sought to examine the effect of a lack of N-terminal P phosphorylation in VSV serotype Indiana on (i) the transcription activity of P in vitro and in vivo, (ii) P self-association or interaction with N or L, and (iii) N binding of cellular RNAs and viral genomic RNA for functional template formation. Our results show that N-terminal phosphorylation of P is critical for viral genomic RNA encapsidation and template function in VSV.
MATERIALS AND METHODS
Abs.
A polyclonal anti-Myc antibody (Ab) (sc789), a monoclonal Ab (sc-40), and an anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) monoclonal Ab (sc-32233) were purchased from Santa Cruz Biotechnology, Inc. A glutathione S-transferase (GST) polyclonal Ab and protein G-Sepharose 4 Fast Flow were obtained from Amersham Biosciences. The anti-hemagglutinin (anti-HA; clone HA-7) and anti-Flag monoclonal Abs used were from Sigma, and the anti-six-His monoclonal Ab (ab18184) used was from Abcam Inc. Anti-VSV L polyclonal Abs were raised in rabbits and supplied by Invitrogen, as described by Chen et al. (20).
Cells and viruses.
HeLa and BHK-21 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. Recombinant vaccinia virus (vTF7-3) carrying the bacteriophage T7 RNA polymerase gene was propagated as described by Chen et al. (20).
Plasmid constructs.
Plasmids encoding HA-tagged P (AD-P) and the P3A mutant (S60A, T62A, S64A) lacking N-terminal phosphorylation (AD-P3A), Myc-tagged N (BD-N) and P (BD-P) or Flag-tagged L (plasmids pBS-N and pBS-L), and a VSV minigenome (pVSV-CAT2) have been described previously (20, 21). cDNA encoding P3A was amplified from AD-P3A and cloned in frame with the open reading frame encoding the GAL4 DNA binding domain (GAL4-BD) and a Myc tag into pGBKT7 (Clontech) (BD-P3A). cDNAs encoding P and P3A were amplified from AD-P and AD-P3A and cloned in frame with GST into pGEX-6P3. A replication-defective minigenome (ΔTr) was constructed by deleting 47 nucleotides from the trailer region of plasmid pVSV-CAT2. Plasmids encoding P-RFP and P3A-RFP were constructed by cloning the relevant open reading frames into the vector pDsRed-Express-N1 via BglII and Acc65I restriction sites, respectively. A plasmid encoding the ΔLe-Tr minigenome was constructed by deleting 19 nucleotides from both the 3′ terminus and the 5′ terminus of plasmid pVSV-CAT2. All constructs were verified by DNA sequencing.
VSV minigenome assay.
BHK-21 cells in six-well plates were infected with vTF7-3 and transfected with plasmids pBS-N, pBS-L, AD-P or AD-P3A, and pVSV-CAT2 or the replication-defective minigenome (ΔTr). After 40 h, cell lysates were subjected to a chloramphenicol acetyltransferase (CAT) enzyme-linked immunosorbent assay (ELISA) for detection of the CAT expression level as described by Chen et al. (20). The expression levels of P and P3A were detected by Western blotting with a monoclonal anti-HA antibody. All assays were repeated at least three times for accuracy.
RNA extraction and relative quantitative real-time RT-PCR.
RNA extraction was performed using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. The RNA was eluted in 40 μl RNase-free water. One microgram of RNA of each sample was treated with DNase I (Invitrogen) and used for reverse transcription (RT) using a reverse transcriptase (Roche) following the manufacturer's instructions. For RT, the primer 5′-GCTGAACGGTCTGGTTATAG-3′ was used for cDNA/antigenome detection, and oligo(dT) was used for cDNA/mRNA detection. Real-time PCR was performed on a real-time PCR system (Bio-Rad), using Eva Green Super mix (Bio-Rad) with a gene-specific primer for antigenomic cDNA, CAT mRNA, or β-actin mRNA. β-Actin was used as a reference.
Yeast mating.
Plasmids encoding binding domain (BD)-baits and activation domain (AD)-preys were transformed into Saccharomyces cerevisiae AH109 (type a) and Y187 (type α) (Clontech), respectively, and yeast mating was performed as described by Chen et al. (20).
In vivo coimmunoprecipitation.
Recombinant proteins were expressed by transient transfection. HeLa cells in six-well plates were infected with vTF7-3 for 1 h at a multiplicity of infection of 10, washed, and then transfected with the appropriate plasmids in the presence of Lipofectamine 2000 (Invitrogen). After 24 h of transfection, cell lysates were prepared as described by Chen et al. (20). In experiments to examine self-association of P3A, Myc-tagged P or P3A was coexpressed with HA-tagged P or P3A in various combinations. To examine interactions between P3A and N, Myc-tagged N was coexpressed with HA-tagged P or P3A. Precleared supernatants of lysates were incubated with a polyclonal anti-Myc Ab for 2 h at 4°C with rotation. After centrifugation, supernatants were mixed with protein G Sepharose 4 Fast Flow medium and incubated overnight. To detect interactions between P3A and the L protein, Flag-tagged L protein was coexpressed with Myc-tagged P or P3A. Precleared supernatants of lysates were incubated with a polyclonal anti-L Ab and mixed with protein G Sepharose 4 Fast Flow medium as described above. Beads were collected and washed five times with washing buffer as described elsewhere (20). The beads were boiled in Laemmli buffer, and bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and detected by Western blotting using an anti-Myc, anti-HA, or anti-Flag monoclonal Ab.
In vitro transcription of VSV genomic RNA supported by P and P3A.
Recombinant hexahistidine (His)-tagged P and P3A and Flag-tagged L were expressed in Sf21 insect cells by use of the Bac-to-Bac baculovirus expression system and used for in vitro transcription reconstitution reactions with purified N-RNA (75 ng) as described by Chen et al. (21). [α-32P]GTP-labeled transcripts were analyzed by electrophoresis on a 5% polyacrylamide gel containing 7 M urea, followed by autoradiography.
GST pulldown assays.
GST, GST-P, or GST-P3A was expressed in BL21 cells, treated with the lysis buffer provided by a ProFound GST pulldown protein-protein interaction kit (Pierce), and incubated for 30 min. After centrifugation at 13,000 rpm for 30 min, equal amounts of supernatants were mixed with 200 ng purified His-tagged P protein as described by Chen et al. (20). GST pulldown assays were performed with a ProFound GST pulldown protein-protein interaction kit according to the manufacturer's protocol. Proteins in supernatants and eluted proteins from GST pulldown beads were detected by Western blotting with anti-HA, anti-GST, or anti-N Ab.
Nuclear and cytoplasmic extraction.
BHK-21 cells were first infected with vTF7-3 and then transfected with AD-P or AD-P3A. At 40 h posttransfection, the nuclear and cytoplasmic extracts were prepared following the manufacturer's instructions for nuclear and cytoplasmic extraction reagents (Thermo). Protein was analyzed by Western blotting using anti-HA Ab.
RFP-labeled P and P3A localization analysis.
HeLa cells in six-well plates were transfected with expression plasmids for P-red fluorescent protein (P-RFP) or P3A-RFP. After 40 h of transfection, the medium was discarded. Cells were washed three times with ice-cold phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde for 20 min at room temperature. The cells were then washed three times with PBS, and nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). Cells were imaged on a fluorescence microscope.
N-RNA purification by CsCl gradient centrifugation and RT-PCR.
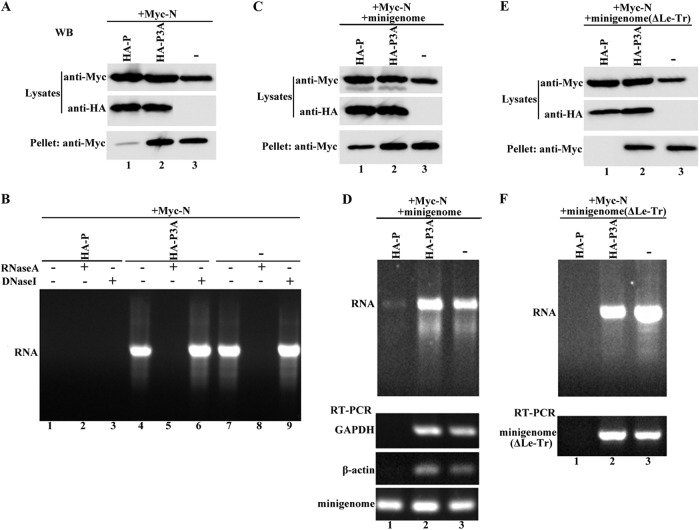
BHK-21 cells (cultured in 10-cm dishes) were infected with vTF7-3 for 1 h and then transfected with plasmid combinations encoding the following: (i) Myc-N alone or Myc-N plus HA-P or HA-P3A and (ii) the former plasmids plus a plasmid minigenome or the ΔLe-Tr minigenome. After 40 h, the cells were lysed in 600 μl of lysis buffer (10 mM Tris-HCl [pH 7.8], 0.15 M NaCl, 1 mM EDTA, 1% NP-40, and protease inhibitor cocktail [Roche]), incubated at 4°C for 30 min, and centrifuged at 13,000 × g at 4°C for 20 min to remove insoluble components. The clear supernatant was loaded onto a discontinuous 25 to 40% (wt/wt) CsCl gradient (CsCl in 10 mM Tris-HCl [pH 7.8], 0.15 M NaCl, 1 mM EDTA) (25%, 1.5 ml; 30% 1.5 ml; and 40%, 1.5 ml) and centrifuged at 250,000 × g at 20°C for 1 h (Hitachi P55ST2 rotor). The contents of a visible band were collected and subjected to centrifugation at 200,000 × g at 4°C for 1 h, and the resultant pellet was resuspended in 100 μl of PBS. Fifty microliters of the sample was analyzed by Western blotting. The other 50 μl of the sample was used for purification of RNA by phenol-chloroform extraction. The RNA was analyzed in a 1% agarose gel and used for the following RT-PCR. In order to detect binding of the plasmid minigenome and the ΔLe-Tr minigenome by N, RT was carried out using primer 1 (5′-ACGAAGACAAACAAACCATT-3′) for plasmid minigenome RNA and primer 2 (5′-TATTATCATTAAAAGGCTCAGGAG-3′) for ΔLe-Tr minigenome RNA, followed by PCR with Taq DNA polymerase (Fermentas). In order to detect binding of cellular RNAs by the N protein, RT-PCR was carried out with specific primers for cellular GAPDH RNA and β-actin RNA.
RESULTS
P3A does not support VSV minigenome transcription and replication in vivo.
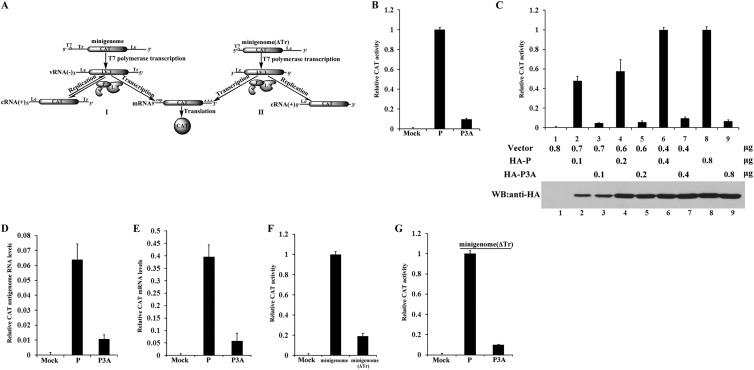
Previous studies showed that the transcription activity of P3A was inactive in a minigenome system (16). To confirm this finding, we used a different VSV minigenome system that contained trailer (Tr) and leader (Le) sequences of the genome and a reporter gene (the CAT gene) flanked by these sequences and which expressed the CAT protein when coexpressed with three support proteins of VSV, i.e., N, P, and L (Fig. 1A, panel I). As shown in Fig. 1B, the relative CAT expression supported by P3A was only about 8% of that supported by P, although the expression levels of P3A and P were similar (data not shown), thereby indicating that P3A was barely active in the minigenome assay. Next, we gradually increased the expression of P and P3A, and relative CAT expression increased gradually as the expression of P increased (from 52% to 100%) (Fig. 1C, upper panel, lanes 2, 4, 6, and 8). However, CAT expression remained below 10% despite increases in P3A expression (Fig. 1C, bottom panel, lanes 3, 5, 7, and 9). Note that we retained the same quantities of total plasmid DNA for all assays. Clearly, these findings confirmed that N-terminal phosphorylation of P is required for minigenome transcription.
Fig 1.
N-terminal phosphorylation of P is essential for VSV minigenome transcription in vivo. (A) Schematic diagram of minigenome and replication-defective minigenome (ΔTr) systems. The negative-strand minigenome or ΔTr minigenome could be generated from transcription driven by T7 RNA polymerase in cells. Of the two, the former was encapsidated for transcription into the reporter gene mRNA, resulting in expression of CAT and its use as a template for replication by a complex of N, P, and L, but the latter was encapsidated and transcribed for reporter gene expression only. (B) BHK cells expressing T7 polymerase were transfected with plasmids encoding N, L, minigenome RNA, and P or P3A. Relative CAT expression levels in lysates were measured via CAT ELISA. The relative CAT expression level of cells transfected with a plasmid encoding P, as a positive control, was defined as 100%. (C) BHK cells expressing T7 polymerase were transfected with increasing quantities of plasmids encoding P or P3A together with the indicated plasmids in the minigenome system. Relative CAT expression levels were measured via CAT ELISA, and expression of P and P3A was detected via Western blot analysis (WB) with anti-HA antibody. (D and E) Transfection was performed as described for panel B. Relative CAT antigenomic RNA levels and mRNA levels were analyzed via RT-PCR, using β-actin as a reference gene. (F) BHK cells expressing T7 polymerase were transfected with plasmids encoding N, P, L, and the ΔTr minigenome, and relative CAT expression levels were measured as described for panel C. The CAT expression level of cells transfected with the minigenome, as a positive control, was defined as 100%. (G) The ΔTr minigenome is supported by P3A. The assay described for panel B was performed, but with the minigenome replaced with the ΔTr minigenome.
To directly examine the activity of P3A in transcription and replication at the RNA level, we also performed real-time RT-PCR to identify the transcription and replication products of the minigenome that were supported by P and P3A. Relative CAT antigenomic RNA levels and mRNA levels severely declined, to around 15% and 12%, respectively, when supported by P3A compared to those supported by P (Fig. 1D and E), thereby indicating that P3A supports neither transcription nor replication in vivo. This also raised the possibility that defective replication may interfere with transcription. Although viral genome RNA [vRNA(−)] generated by T7 RNA polymerase could be utilized directly for CAT transcription (Fig. 1A, panel I), N has been reported to be more efficient for encapsidating the vRNA(−) generated from P-L RNA polymerase than that generated from T7 RNA polymerase for transcription (22). Thus, we deleted the Tr sequences that were required as promoters of P-L polymerase for replication to generate vRNA(−) from cRNA(+) and constructed a replication-defective minigenome (ΔTr) that could transcribe only vRNA(−) generated by T7 RNA polymerase (Fig. 1A, panel II). Clearly, compared to the relative CAT activity of the minigenome (Fig. 1A, panel I), the relative CAT activity of the replication-defective minigenome (ΔTr) declined 5-fold in the presence of P (Fig. 1F), thereby suggesting that replication significantly enhanced transcription activity in the minigenome system. Using this replication-defective minigenome (ΔTr), we again compared relative CAT expression levels supported by P and P3A. The results showed that P3A could not support CAT expression, thus suggesting that transcription was also inactive when supported by P3A, independent of replication. Taken together, these data suggest that N-terminal phosphorylation of P is absolutely required for its transcription and replication functions in vivo.
P3A self-associates in vivo and in vitro.
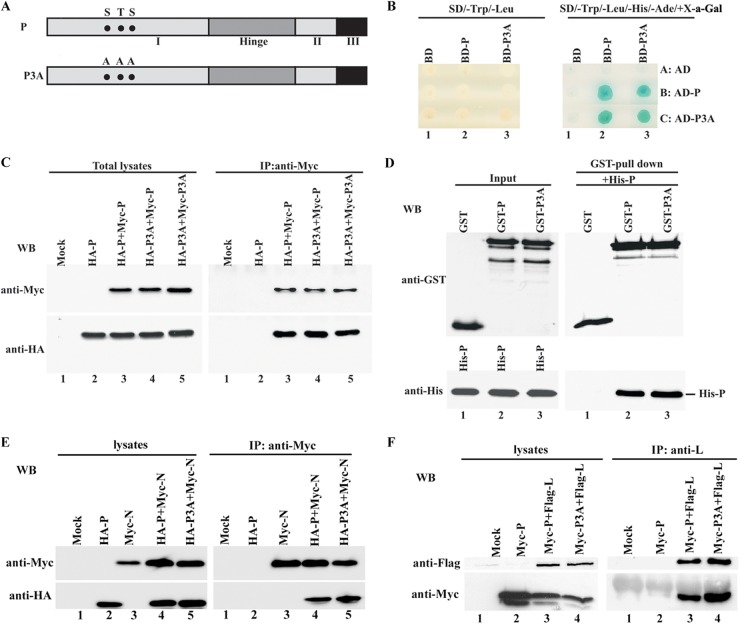
Figure 2A depicts the boundaries of the proposed domains of P and the three mutation sites leading to a lack of N-terminal phosphorylation. We previously showed that P self-association is indispensable for VSV transcription (20). Thus, we sought to determine whether a lack of N-terminal phosphorylation disrupts the self-association of P. To do so, we first used a yeast two-hybrid system to test P-P3A and P3A-P3A interactions. The cDNAs expressing P and P3A were fused in frame, individually, with the cDNAs encoding GAL4-BD and GAL4-AD. The recombinant plasmids encoding BD-P and BD-P3A were transformed into Saccharomyces cerevisiae AH109, and the recombinant plasmids encoding AD-P and AD-P3A were transformed into Saccharomyces cerevisiae Y187. Blue yeast growth on SD/−Trp/−Leu/−His/−Ade/+X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) plates was observed after the BD-P strain was mated with the AD-P or AD-P3A strain and after the BD-P3A strain was mated with the AD-P or AD-P3A strain (Fig. 2B, right panel, lanes 2B, 2C, 3B, and 3C). No yeast appeared in the plates with combinations of the BD strain plus the AD-P or AD-P3A strain or the BD-P or BD-P3A strain plus the AD strain (Fig. 2B, right panel, lanes 1B, 1C, 2A, and 3A). However, diploid cells were all able to grow on the SD/−Trp/−Leu plates, thereby confirming that yeast mating occurred and that fusion proteins were expressed well in all mating combinations (Fig. 2B, left panel). This finding suggests that N-terminal phosphorylation is not required for the self-association of P in the yeast two-hybrid system.
Fig 2.
Self-association of P3A and interaction of P3A with N or L. (A) Domain structures of P and P3A. P structures with three functionally defined domains (I, II, and III) and a hinge region are shown. The phosphate acceptor sites S60, T62, and S64 and their respective mutations to alanine (A60, A62, and A64) are indicated with solid circles. (B) Interaction of P3A with P and itself in a yeast two-hybrid assay. A yeast mating assay was performed between yeast AH109 strains (type a) expressing BD and BD-baits and Y187 strains (type α) expressing AD and AD-preys, as indicated. Experiments were repeated at least three times for accuracy. (C) Myc-tagged P or P3A and HA-tagged P or P3A were coexpressed in HeLa cells as indicated. (Left) Protein expression was detected via Western blot analysis using anti-Myc and anti-HA monoclonal Abs. (Right) Immunoprecipitation (IP) was performed using anti-Myc polyclonal Ab, and immune complexes were detected with anti-Myc and anti-HA monoclonal Abs. (D) Cell lysates of E. coli BL21 expressing GST, GST-P, or GST-P3A were mixed with His-P purified from Sf21 cells to allow their direct interaction. GST pulldown assays were performed. Input (left) and bound (right) proteins were assayed via Western blotting using anti-GST and anti-His Abs. (E) Myc-tagged N and HA-tagged P or P3A were coexpressed in HeLa cells as indicated. Protein expression (left) and immunoprecipitation (right) were detected as described for panel C. (F) Flag-tagged L and Myc-tagged P or P3A were coexpressed in HeLa cells as indicated. (Left) Protein expression was detected via Western blot analysis using anti-Flag and anti-Myc monoclonal Abs. (Right) Immunoprecipitation was performed using anti-L polyclonal Ab, and immune complexes were detected using anti-Flag and anti-Myc monoclonal Abs.
Next, we performed coimmunoprecipitation analyses in vivo by coexpressing Myc-P and HA-P3A or Myc-P3A and HA-P3A in HeLa cells. Myc- and HA-tagged fusion proteins were well expressed at a high rate in HeLa cells (Fig. 2C, left panel, lanes 2, 3, 4, and 5). When total cell lysates were subjected to immunoprecipitation using anti-Myc polyclonal antibody, HA-P3A was coimmunoprecipitated with Myc-P as efficiently as HA-P was (Fig. 2C, right panel, lanes 3 and 4). Furthermore, HA-P3A was also efficiently coimmunoprecipitated with Myc-P3A (Fig. 2C, right panel, lane 5). As a negative control, HA-P alone did not coimmunoprecipitate with anti-Myc antibody in the absence of Myc-P (Fig. 2C, right panel, lane 2). These results confirm that N-terminal phosphorylation is indeed not involved in the self-association of P in vivo, and they further support the results obtained from the yeast two-hybrid assay.
To further prove that the interaction observed in vivo was direct, we also performed an in vitro GST pulldown experiment with GST-fused P or P3A expressed in bacteria. When the proteins were mixed with equal amounts of highly pure His-tagged P (His-P) from Sf21 cells, both GST-P and GST-P3A, but not GST alone, were able to pull down His-P (Fig. 2D, right panel). Thus, taken together, these results confirm that P3A is able to self-associate in vivo and in vitro.
P3A interacts with N and L.
Previous studies showed that P acts as a bridge to connect the N-RNA template and polymerase L. Thus, we sought to determine whether P3A could associate with N and L. The results from our coimmunoprecipitation experiments clearly showed that both HA-P and HA-P3A could be coimmunoprecipitated similarly with Myc-N when they were coexpressed with Myc-N (Fig. 2E, right panel, lanes 4 and 5). These results strongly suggest that N-terminal phosphorylation is not required for binding of P to N. Similarly, when Myc-P or Myc-P3A was coexpressed with Flag-L, both Myc-P and Myc-P3A were also coimmunoprecipitated at the same intensity by anti-L antibody (Fig. 2F, right panel, lanes 3 and 4), thereby indicating that N-terminal phosphorylation is also dispensable for interaction of P with L.
P3A supports transcription in vitro.
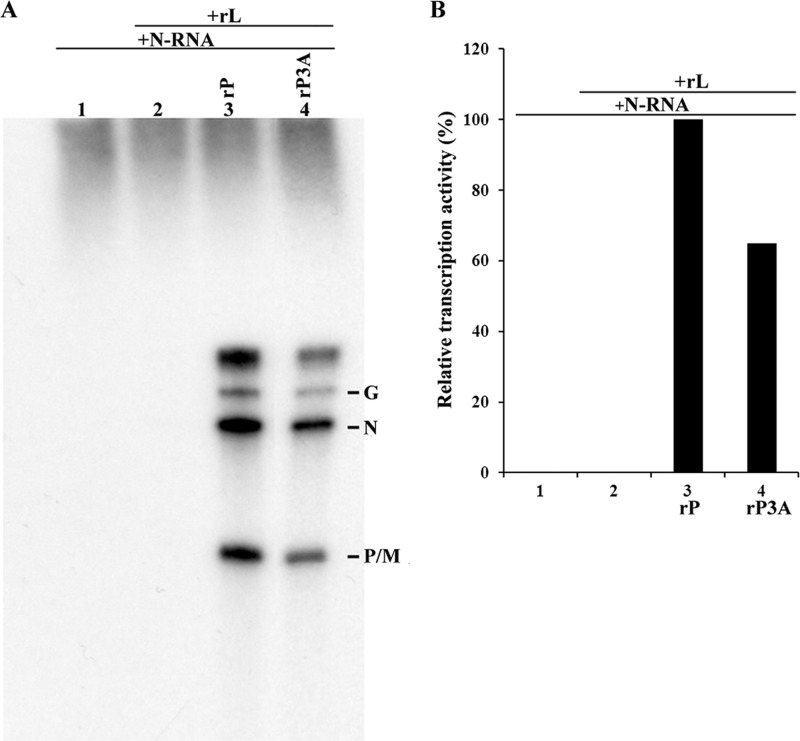
To determine whether N-terminal phosphorylation of P is required for transcription in vitro, we performed an in vitro transcription reconstitution experiment. Purified His-P or P3A and Flag-tagged L expressed in Sf21 cells by use of recombinant baculovirus were incubated with the N-RNA template purified from VSV virions in the presence of [α-32P]GTP at 30°C, and the resulting mRNA products were analyzed via urea-polyacrylamide gel electrophoresis. The added N-RNA template, alone or in the presence of Flag-L only, had no VSV polymerase activity (Fig. 3A, lanes 1 and 2), and the accumulated mRNAs (G, N, and P/M) were clearly discerned in the presence of Flag-L, His-P, and His-P3A (Fig. 3A, lanes 3 and 4). Transcription activity supported by P3A was at least 60% of that supported by P (Fig. 3B, lanes 3 and 4). This result clearly demonstrates that P3A supports VSV genome transcription in vitro.
Fig 3.
Transcription activity of P3A in vitro. (A) Recombinant P or P3A and L were subjected to in vitro transcription with the N-RNA template as described in Materials and Methods. [α-32P]GTP-labeled transcripts were analyzed via urea–5% PAGE followed by autoradiography. The positions of the viral mRNAs are indicated on the right. (B) Relative transcription activities (percentages) of P and P3A in transcription reactions. The relative transcription activity of P was defined as 100%. All assays were repeated at least three times for accuracy.
Localization of P3A within cells to the same level as that of P.
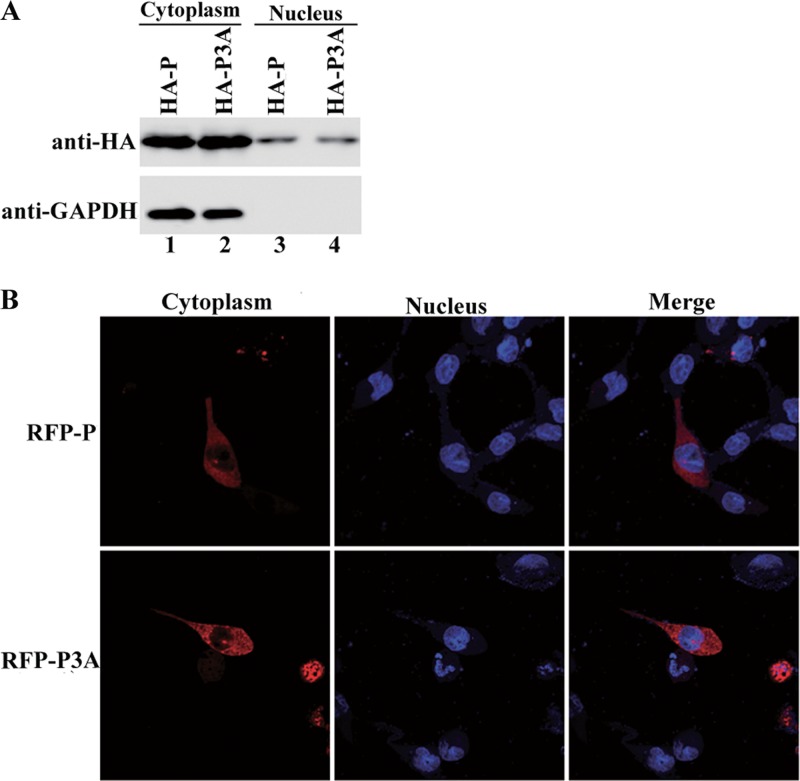
Like most other nonsegmented negative-strand RNA viruses, VSV replicates in the cytoplasm. Having confirmed that P3A cannot support minigenome transcription in vivo but can support in vitro viral genome transcription, we hypothesized that the absence of N-terminal phosphorylation results in a relocalization of P within cells that is unsuitable for transcription and replication. Thus, HA-tagged P and P3A were expressed in HeLa cells, and proteins expressed in the cytoplasm and nucleus were separated to assess the localization of P and P3A. GAPDH, a protein that localizes exclusively to the cytoplasm, was detected in the cytoplasmic fraction only, not the nuclear fraction (Fig. 4A, bottom panel, lanes 1 and 2), thereby indicating that the nucleus-cytoplasm separation assay was accurate. When the cytoplasmic and nuclear fractions were probed for P and P3A, high levels of the proteins remained in the cytoplasm (Fig. 4A, upper panel, lanes 1 and 2), thereby suggesting that the absence of N-terminal phosphorylation of P did not shift P from the cytoplasm to the nucleus. Furthermore, RFP-fused P and P3A were also expressed separately in HeLa cells. The results showed that P and P3A were evenly distributed throughout the cytoplasm (Fig. 4B). Thus, the localization levels of P3A within cells were the same as those of P.
Fig 4.
Localization of P3A within cells. (A) HA-tagged P and P3A were expressed in BHK cells. Nuclear and cytoplasmic extracts were analyzed via Western blotting using anti-HA antibody. GAPDH was used as a reference for cytoplasmic extracts. (B) RFP-fused P or P3A was expressed in HeLa cells as described in Materials and Methods. Localization of P and P3A within the cells was detected via immunofluorescence microscopy. The nucleus was stained with DAPI.
P3A is unable to prevent N from binding to cellular RNAs and does not support functional template formation.
Having excluded the aforementioned possibilities for rendering P3A inactive in transcription and replication in vivo, we still sought to determine what disrupts the function of P3A in vivo. Note that in contrast to in vitro transcription reconstitution experiments, in which the N-RNA template is directly available, in the in vivo minigenome assay, vRNA(−) must be encapsidated specifically by N for the formation of the template, and P is indispensable for this process. Thus, we hypothesized that P3A is incapable of driving the formation of a functional template. To test this hypothesis, we first sought to determine whether P3A was still able to prevent N from binding to cellular RNAs. Myc-N was expressed alone or coexpressed with P or P3A, cell lysates were centrifuged, and clear supernatants were collected and loaded onto a discontinuous CsCl gradient for further centrifugation as described in Materials and Methods. A visible band indicating the N-RNA complex was collected, pelleted, and resuspended in PBS for Western blot analysis and nucleic acid extraction. As shown in Fig. 5B, the nucleic acids extracted from the pellet ran as a smear with a bright main band in a 1% agarose gel when N was expressed alone (Fig. 5B, lane 7), and they were unaffected by DNase I treatment (Fig. 5B, lane 9) but digested by RNase A treatment (Fig. 5B, lane 8), thereby demonstrating that N binds to cellular RNAs. No RNAs were detected when N was coexpressed with P (Fig. 5B, lane 1), thereby indicating that P prevents N from binding to nonviral RNAs. However, when it was coexpressed with P3A, N bound to cellular RNAs as efficiently as when it was expressed alone (Fig. 5B, lanes 4, 5, and 6), even though P3A and P were expressed at similar levels (Fig. 5A, middle panel, lanes 1 and 2). Correspondingly, when it was coexpressed with P, the N in the pellet was also significantly decreased compared to that when N was expressed alone or coexpressed with P3A (Fig. 5A, bottom panel), even though N was expressed at similar levels in the presence of P3A and P and at lower levels alone in lysates (Fig. 5A, top panel), indicating that the formation of N0-P (free of cellular RNAs) substituted for N-RNAs in the presence of P. These data strongly demonstrate that N-terminal phosphorylation of P is absolutely required to prevent N from binding to nonviral RNAs.
Fig 5.
Ability of N binding of cellular RNAs and viral genomic RNA in the presence of P and P3A. (A and B) P3A is incapable of preventing N from binding to cellular RNAs. BHK cells were transfected with plasmids encoding N alone or N plus P or P3A. (A) Lysates from transfected cells were centrifuged, and an aliquot of 10 μl of supernatant was analyzed via Western blotting to detect N, P, and P3A expression. The remaining supernatant was analyzed via CsCl gradient centrifugation as described in Materials and Methods. After centrifugation, the pellet was used for N detection (A) and further purification of RNAs (B). The RNAs or RNAs treated with RNase A or DNase I were analyzed in a 1% agarose gel. (C and D) Assay performed as described for panels A and B, but with the addition of minigenome expression for all transfection combinations. CAT antigenomic RNA and GAPDH and β-actin RNAs were amplified via RT-PCR, using RNAs in the pellet as a template. (E and F) The assay described for panels C and D was performed, but the minigenome was replaced with the ΔLe-Tr minigenome.
Next, we sought to assess how N encapsidated the viral genomic RNA in the presence of P and P3A when the minigenome was added in the aforementioned experiment. After coexpressing plasmids encoding the minigenome with those encoding N or N plus P or P3A, the same assay was performed, and the results were similar to those shown in Fig. 5A and B for both proteins (Fig. 5C) and RNAs (Fig. 5D, top panel). RT-PCR was further performed using RNAs from different transfection combinations as templates to detect vRNA(−) of the minigenome and mRNAs of the cellular GAPDH or β-actin gene. As expected, the CAT reporter gene representing the vRNA(−) of the minigenome could be detected clearly after RT-PCR (Fig. 5D, bottom panel, lane 1), although the RNA template was almost invisible in the agarose gel (Fig. 5D, top panel, lane 1), but mRNAs for GAPDH and β-actin, representing cellular RNAs, could not be observed at all when the minigenome was coexpressed with N plus P, thereby suggesting that N specifically bound to viral genomic RNA in the presence of P. In contrast, after RT-PCR, mRNAs for GAPDH and β-actin could be detected when the minigenome was coexpressed with N or N plus P3A (Fig. 5D, bottom panel, lanes 2 and 3), indicating that N bound nonspecifically to nonviral RNAs, despite the presence of viral genomic RNAs. Interestingly, we also detected products of the CAT reporter gene representing vRNA(−) of the minigenome after performing RT-PCRs using the same RNAs as templates, indicating that N bound not only to nonviral RNAs but also to viral RNAs when N was expressed alone or coexpressed with P3A. We predict that when N is coexpressed with P3A or expressed alone, its binding to viral genomic RNA may be unspecific or it can bind to any available RNA species, including viral genomic RNA. Previous studies have suggested that the specific binding of viral RNA by N occurs via the recognition of specific RNA sequences within the leader region that act as encapsidation nucleation signals (23, 24). This means that the specific binding would be abolished, but nonspecific binding would be unaffected, if the leader region was deleted. Therefore, we constructed a minigenome (ΔLe-Tr) in which the leader and trailer regions were deleted; the trailer region was deleted because complementary sequences in the trailer region may also act as encapsidation nucleation signals. The aforementioned experiment was performed again, using the ΔLe-Tr minigenome to replace the original minigenome. Our results agreed with previous studies (23, 24): N failed to bind to ΔLe-Tr minigenome RNA in spite of the presence of P (Fig. 5E and F, lanes 1), indicating that encapsidation nucleation signals in the leader region were indispensable for specific binding. However, the binding of ΔLe-Tr minigenome RNA by N was unaffected when N was coexpressed with P3A or expressed alone (Fig. 5E and F, lanes 2 and 3), indicating that the binding of viral RNA under such conditions is nonspecific. Taken together, our results show that the lack of N-terminal phosphorylation of P not only abolishes the ability of P to prevent N from binding to cellular RNAs but also abolishes the activity of P in viral genome-specific encapsidation and functional template formation.
DISCUSSION
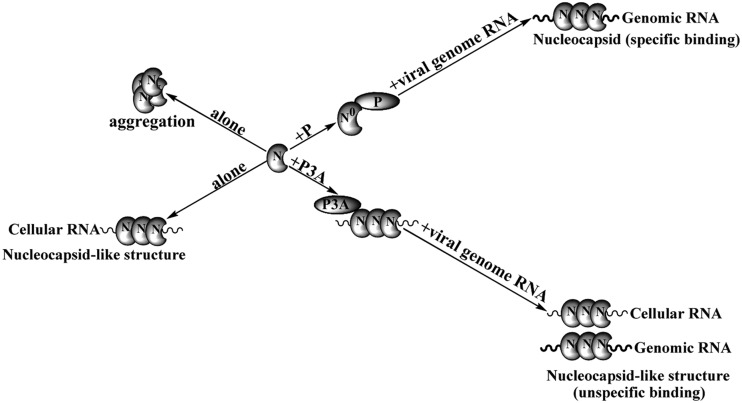
In this study, using a mutant lacking N-terminal phosphorylation (P3A) to examine the effect of P on different steps of viral gene expression in detail, we showed that N-terminal phosphorylation of P is indispensable for preventing N from binding to cellular RNAs and for functional template formation. From these data, we propose a model in which P and P3A regulate template formation differently (Fig. 6). On one hand, when N is expressed alone, it aggregates and can be pelleted by low-speed centrifugation (21); on the other hand, it binds to cellular RNAs to form nucleocapsid-like structures. However, P prevents N from aggregating and from binding to cellular RNAs by forming N0-P complexes; in the presence of viral genomic RNA, P acts as a chaperone and conducts N0 specifically to encapsidate viral genomic RNA for the formation of a nucleocapsid that is a functional template for transcription and replication. Although P3A was also able to prevent N from aggregating (data not shown), its ability to prevent N from binding to cellular RNAs was completely abolished, and N still bound to cellular RNAs to form nucleocapsid-like structures. In such situations, even in the presence of viral genomic RNA, N bound only nonspecifically to viral genomic RNA to form a nonfunctional template incapable of transcription and replication.
Fig 6.
Proposed model of the role of N-terminal phosphorylation of P in facilitating viral RNA-specific encapsidation by N and functional template formation. P prevents N not only from aggregating but also from binding to cellular RNAs. Instead, the N0-P complex forms and maintains N-specific encapsidation of viral genomic RNA. P3A prevents N from aggregating but is unable to prevent it from binding to cellular RNAs. Alternatively, N binds to any RNAs in the presence of P3A, irrespective of sequence specificity (see the text for details).
The P's of nonsegmented negative-strand RNA viruses are multifunctional, playing critical roles in viral genome-specific encapsidation for functional template formation and in subsequent transcription and replication. Previous studies have indicated that the P's of respiratory syncytial virus (RSV) (25) and Sendai virus (SeV) (26) play roles in the transcription and replication of these viruses. A recent study also showed that a phosphorylation site within the P of parainfluenza virus 5 plays a positive role in viral mRNA synthesis and virus growth (27).
Pattnaik et al. (16) previously concluded that P3A is nearly inactive (2 to 3% of the transcription level seen with P) by using a VSV minigenome system in which an antigenomic positive-sense minigenome first transcribed via T7 RNA polymerase must be replicated for subsequent transcription and reporter gene expression. To test their conclusion, we first performed an in vivo transcription assay using a different VSV minigenome system in which genomic-sense vRNA(−), first transcribed via T7 RNA polymerase, in principle, was subsequently transcribed and replicated separately (20). Our results confirmed their conclusion (Fig. 1B); even when P3A expression gradually increased, P3A was still barely (3 to 10%) active in supporting transcription (Fig. 1C). We did not observe an increase in the transcription activity of P3A in a concentration-dependent fashion in vivo, as previously described (17). Furthermore, the results from real-time RT-PCR to detect products of transcription and replication suggested that not only transcription but also replication was severely impaired (Fig. 1F and G). This finding was somewhat surprising, because a previous study of the replication of defective interfering (DI) particles showed that N-terminal phosphorylation was not required for the replicative function of P (16). In this assay, the RNA of the DI contains at its 5′ terminus 2,163 nucleotides from the 5′ terminus of the VSV genome and at its 3′ terminus an inverted complement of the first 45 nucleotides of the 5′ terminus which is absent at the 3′ terminus of standard VSV and our minigenome (28). These terminal signals determine the dominant replication ability of DI and interfere specifically with the replication of standard VSV (29). Thus, we also replaced the 3′-terminal sequences of the minigenome with an inverted complement of the first 45 nucleotides of the 5′ terminus and constructed a DI minigenome which completely simulates the real DI. The replication efficiency of the DI minigenome increased 3-fold compared with the minigenome in the presence of P3A (data not shown), indicating that P3A effectively supports the replication of the DI minigenome, as described by Pattnaik et al. (16). It is probable that specific signals within the 3′ terminus of DI regulate its encapsidation by N and its replication, which does not require N-terminal phosphorylation of P.
In our minigenome system, in principle, transcription was independent of replication; however, using a similar minigenome system to study the role of matrix proteins of Ebola virus in regulating viral genome replication and transcription, Hoenen et al. (22) demonstrated that replication significantly facilitated transcription, as the transcription activity dramatically decreased when replication was defective. This was because N encapsidated vRNA(−) transcribed via T7 RNA polymerase much less efficiently than that replicated via P-L RNA polymerase. The results from our replication-defective minigenome (ΔTr) confirmed these findings (Fig. 1F) and raised the possibility that transcription inactivity of P3A may derive from lower replication levels. However, using a replication-defective minigenome (ΔTr), we confirmed that the transcription inactivity of P3A was independent of replication, which excluded the aforementioned possibility (Fig. 1G). Thus, P3A is separately inactive in supporting transcription and replication.
Previous studies indicated that N-terminal phosphorylation is required for the self-association of P, which facilitates its interaction with L as well as the N-RNA template and its subsequent transcription activity (14, 15, 30). Here we provided direct evidence that N-terminal phosphorylation of P was not required for its self-association via three panels of experiments: a yeast two-hybrid assay, in vivo coimmunoprecipitation, and an in vitro GST pulldown test. This finding confirms those of several studies demonstrating that phosphorylation of the P's of rabies virus and Sendai virus is not required for their oligomerization (31–33), indicating that P of VSV behaves like the P's of rabies virus and Sendai virus. Furthermore, our in vivo coimmunoprecipitation assay also confirmed that N-terminal phosphorylation of P is dispensable for the interaction of P with N and L, although Gao and Lenard (14, 15) proposed that N-terminal phosphorylation may be involved in interaction of P with L. Moreover, our results also support a previous study revealing that phosphorylation of VSV P is not required for N-P complex formation (34). Thus, the transcriptional inactivity of P3A in vivo may not be caused by its self-association, as it interacted with L and N.
Previous in vitro transcription reconstitution studies showed that purified P expressed in Escherichia coli was neither phosphorylated nor active in transcription unless it was provided with cellular casein kinase II, which is known to phosphorylate residues in the N terminus of P (12, 35). Our in vitro transcription reconstitution experiment using highly purified P or P3A and L proteins expressed in Sf21 cells, which have similar posttranslational modifications to those expressed in mammalian cells, showed that P3A still maintained a relatively high activity compared with wild-type P (>60% of the P level), which could reflect the concentration dependence of P3A for full activity, as suggested by Spadafora et al. (17).
Note, however, that COS cell lysates expressing L and P3A either separately or together were barely (5 to 10%) active in supporting transcription in the in vitro transcription reconstitution assay. It is possible that a cellular factor(s) disrupted the transcription function of P3A via associating with P3A and was disassociated from P3A after purification, or a different secondary structure or conformation may have been present when P3A was synthesized in two different cellular milieus (36). This question will not be answered until the structure of P3A is resolved.
In general, our results indicate that N-terminal phosphorylation of P is not essential for the aforementioned protein-protein interactions and is also not required for the transcription process per se. But these findings are somewhat discordant with results showing that N-terminal phosphorylation of P of the Indiana serotype by CKII is essential for transcription in vitro (14, 15). A reasonable explanation is that unidentified sites of phosphorylation by CKII that are also crucial for transcription activity exist in P, or CKII not only phosphorylates P but also plays an additional role in modifying P that is lacking in E. coli. In addition, despite extensive studies of phosphorylation of P, the localization of P3A within cells has not been explored and might be involved in the disruption of its function. Our nucleus-cytoplasm separation and localization experiments were based on this prediction.
The most important finding in this study was that P3A was incapable of preventing N from binding to cellular RNAs, resulting in the inability of N to specifically encapsidate viral genomic RNA for functional template formation. To our knowledge, this is the first report to demonstrate that phosphorylation of P is involved in viral genome encapsidation and template function. Our results accord well with previous studies that reported that P can prevent the binding of N to nonspecific RNAs in vitro (5). P has been shown to interact with the terminal sequences of the viral genome (37, 38). Thus, we are interested in determining whether N and P utilize overlapping sequences to encapsidate nucleation signals and interact or, alternatively, whether only N0, P, or both contribute to viral genomic RNA-specific recognition by N0-P. We are currently working to identify specific nucleotides within the leader or trailer region that are required for specific binding by N0-P.
ACKNOWLEDGMENTS
We thank Markeda Wade for professionally editing the manuscript.
This work was supported by grants from the National Natural Sciences Foundation of China (grant 81271816) and the National Basic Research Program (973) of China (grant 2012CB518906).
Footnotes
Published ahead of print 2 January 2013
REFERENCES
- 1. Emerson SU, Wagner RR. 1972. Dissociation and reconstitution of the transcriptase and template activities of vesicular stomatitis B and T virions. J. Virol. 10:297–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Banerjee AK. 1987. Transcription and replication of rhabdoviruses. Microbiol. Rev. 51:66–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Banerjee AK, Barik S. 1992. Gene expression of vesicular stomatitis virus genome RNA. Virology 188:417–428 [DOI] [PubMed] [Google Scholar]
- 4. Davis NL, Arnheiter H, Wertz GW. 1986. Vesicular stomatitis virus N and NS proteins form multiple complexes. J. Virol. 59:751–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Masters PS, Banerjee AK. 1988. Complex formation with vesicular stomatitis virus phosphoprotein NS prevents binding of nucleocapsid protein N to nonspecific RNA. J. Virol. 62:2658–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Peluso RW, Moyer SA. 1988. Viral proteins required for the in vitro replication of vesicular stomatitis virus defective interfering particle genome RNA. Virology 162:369–376 [DOI] [PubMed] [Google Scholar]
- 7. Canter DM, Perrault J. 1996. Stabilization of vesicular stomatitis virus L polymerase protein by P protein binding: a small deletion in the C-terminal domain of L abrogates binding. Virology 219:376–386 [DOI] [PubMed] [Google Scholar]
- 8. Bell JC, Prevec L. 1985. Phosphorylation sites on phosphoprotein NS of vesicular stomatitis virus. J. Virol. 54:697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clinton GM, Huang AS. 1981. Distribution of phosphoserine, phosphothreonine and phosphotyrosine in proteins of vesicular stomatitis virus. Virology 108:510–514 [DOI] [PubMed] [Google Scholar]
- 10. Hsu CH, Morgan EM, Kingsbury DW. 1982. Site-specific phosphorylation regulates the transcriptive activity of vesicular stomatitis virus NS protein. J. Virol. 43:104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barik S, Banerjee AK. 1991. Cloning and expression of the vesicular stomatitis virus phosphoprotein gene in Escherichia coli: analysis of phosphorylation status versus transcriptional activity. J. Virol. 65:1719–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barik S, Banerjee AK. 1992. Phosphorylation by cellular casein kinase II is essential for transcriptional activity of vesicular stomatitis virus phosphoprotein P. Proc. Natl. Acad. Sci. U. S. A. 89:6570–6574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen JL, Das T, Banerjee AK. 1997. Phosphorylated states of vesicular stomatitis virus P protein in vitro and in vivo. Virology 228:200–212 [DOI] [PubMed] [Google Scholar]
- 14. Gao Y, Lenard J. 1995. Cooperative binding of multimeric phosphoprotein (P) of vesicular stomatitis virus to polymerase (L) and template: pathways of assembly. J. Virol. 69:7718–7723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gao Y, Lenard J. 1995. Multimerization and transcriptional activation of the phosphoprotein (P) of vesicular stomatitis virus by casein kinase-II. EMBO J. 14:1240–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pattnaik AK, Hwang L, Li T, Englund N, Mathur M, Das T, Banerjee AK. 1997. Phosphorylation within the amino-terminal acidic domain I of the phosphoprotein of vesicular stomatitis virus is required for transcription but not for replication. J. Virol. 71:8167–8175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spadafora D, Canter DM, Jackson RL, Perrault J. 1996. Constitutive phosphorylation of the vesicular stomatitis virus P protein modulates polymerase complex formation but is not essential for transcription or replication. J. Virol. 70:4538–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takacs AM, Barik S, Das T, Banerjee AK. 1992. Phosphorylation of specific serine residues within the acidic domain of the phosphoprotein of vesicular stomatitis virus regulates transcription in vitro. J. Virol. 66:5842–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Das SC, Pattnaik AK. 2004. Phosphorylation of vesicular stomatitis virus phosphoprotein P is indispensable for virus growth. J. Virol. 78:6420–6430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen M, Ogino T, Banerjee AK. 2006. Mapping and functional role of the self-association domain of vesicular stomatitis virus phosphoprotein. J. Virol. 80:9511–9518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen M, Ogino T, Banerjee AK. 2007. Interaction of vesicular stomatitis virus P and N proteins: identification of two overlapping domains at the N terminus of P that are involved in N0-P complex formation and encapsidation of viral genome RNA. J. Virol. 81:13478–13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoenen T, Jung S, Herwig A, Groseth A, Becker S. 2010. Both matrix proteins of Ebola virus contribute to the regulation of viral genome replication and transcription. Virology 403:56–66 [DOI] [PubMed] [Google Scholar]
- 23. Blumberg BM, Giorgi C, Kolakofsky D. 1983. N protein of vesicular stomatitis virus selectively encapsidates leader RNA in vitro. Cell 32:559–567 [DOI] [PubMed] [Google Scholar]
- 24. Blumberg BM, Leppert M, Kolakofsky D. 1981. Interaction of VSV leader RNA and nucleocapsid protein may control VSV genome replication. Cell 23:837–845 [DOI] [PubMed] [Google Scholar]
- 25. Lu B, Ma C-H, Brazas R, Jin H. 2002. The major phosphorylation sites of the respiratory syncytial virus phosphoprotein are dispensable for virus replication in vitro. J. Virol. 76:10776–10784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hu CJ, Kato A, Bowman MC, Kiyotani K, Yoshida T, Moyer SA, Nagai Y, Gupta KC. 1999. Role of primary constitutive phosphorylation of Sendai virus P and V proteins in viral replication and pathogenesis. Virology 263:195–208 [DOI] [PubMed] [Google Scholar]
- 27. Sun D, Luthra P, Xu P, Yoon H, He B. 2011. Identification of a phosphorylation site within the P protein important for mRNA transcription and growth of parainfluenza virus 5. J. Virol. 85:8376–8385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meier E, Harmison G, Keene JD, Schubert M. 1984. Sites of copy choice replication involved in generation of vesicular stomatitis virus defective-interfering particle RNAs. J. Virol. 51:515–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pattnaik AK, Ball LA, LeGrone A, Wertz GW. 1995. The termini of VSV DI particle RNAs are sufficient to signal RNA encapsidation, replication, and budding to generate infectious particles. Virology 206:760–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Das T, Gupta AK, Sims PW, Gelfand CA, Jentoft JE, Banerjee AK. 1995. Role of cellular casein kinase II in the function of the phosphoprotein (P) subunit of RNA polymerase of vesicular stomatitis virus. J. Biol. Chem. 270:24100–24107 [DOI] [PubMed] [Google Scholar]
- 31. Curran J, Boeck R, Lin-Marq N, Lupas A, Kolakofsky D. 1995. Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology 214:139–149 [DOI] [PubMed] [Google Scholar]
- 32. Gigant B, Iseni F, Gaudin Y, Knossow M, Blondel D. 2000. Neither phosphorylation nor the amino-terminal part of rabies virus phosphoprotein is required for its oligomerization. J. Gen. Virol. 81:1757–1761 [DOI] [PubMed] [Google Scholar]
- 33. Tarbouriech N, Curran J, Ebel C, Ruigrok RW, Burmeister WP. 2000. On the domain structure and the polymerization state of the Sendai virus P protein. Virology 266:99–109 [DOI] [PubMed] [Google Scholar]
- 34. Takacs AM, Das T, Banerjee AK. 1993. Mapping of interacting domains between the nucleocapsid protein and the phosphoprotein of vesicular stomatitis virus by using a two-hybrid system. Proc. Natl. Acad. Sci. U. S. A. 90:10375–10379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gupta AK, Das T, Banerjee AK. 1995. Casein kinase II is the P protein phosphorylating cellular kinase associated with the ribonucleoprotein complex of purified vesicular stomatitis virus. J. Gen. Virol. 76:365–372 [DOI] [PubMed] [Google Scholar]
- 36. Mathur M, Das T, Chen JL, Chattopadhyay D, Banerjee AK. 1997. Display of disparate transcription phenotype by the phosphorylation negative P protein mutants of vesicular stomatitis virus, Indiana serotype, expressed in E. coli and eucaryotic cells. Gene Expr. 6:275–286 [PMC free article] [PubMed] [Google Scholar]
- 37. Isaac CL, Keene JD. 1982. RNA polymerase-associated interactions near template promoter sequences of defective interfering particles of vesicular stomatitis virus. J. Virol. 43:241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Keene JD, Thornton BJ, Emerson SU. 1981. Sequence-specific contacts between the RNA polymerase of vesicular stomatitis virus and the leader RNA gene. Proc. Natl. Acad. Sci. U. S. A. 78:6191–6195 [DOI] [PMC free article] [PubMed] [Google Scholar]