Abstract
Incorporation of the human immunodeficiency virus type 1 (HIV-1) envelope glycoproteins into assembling particles is crucial for virion infectivity. Genetic and biochemical data indicate that the matrix (MA) domain of Gag and the cytoplasmic tail of the transmembrane glycoprotein gp41 play an important role in coordinating Env incorporation; however, the molecular mechanism and possible role of host factors in this process remain to be defined. Recent studies suggested that Env incorporation is mediated by interactions between matrix and tail-interacting protein of 47 kDa (TIP47; also known as perilipin-3 and mannose-6-phosphate receptor-binding protein 1), a member of the perilipin, adipophilin, TIP47 (PAT) family of proteins implicated in protein sorting and lipid droplet biogenesis. We have confirmed by nuclear magnetic resonance spectroscopy titration experiments and surface plasmon resonance that MA binds TIP47. We also reevaluated the role of TIP47 in HIV-1 Env incorporation in HeLa cells and in the Jurkat T-cell line. In HeLa cells, TIP47 overexpression or RNA interference (RNAi)-mediated depletion had no significant effect on HIV-1 Env incorporation, virus release, or particle infectivity. Similarly, depletion of TIP47 in Jurkat cells did not impair HIV-1 Env incorporation, virus release, infectivity, or replication. Our results thus do not support a role for TIP47 in HIV-1 Env incorporation or virion infectivity.
INTRODUCTION
Incorporation of the Env glycoprotein complex into assembling HIV-1 particles is critical for viral infectivity and replication. Env is synthesized as an ∼160-kDa precursor polyprotein (gp160) in the endoplasmic reticulum (ER) and is processed into the mature surface glycoprotein gp120 and transmembrane glycoprotein gp41 during transport to virus assembly sites on the plasma membrane (1). Although many aspects of the HIV-1 assembly pathway have been deciphered over the past 2 decades (2, 3), the mechanism of Env recruitment and incorporation remains poorly understood. Several non-mutually exclusive models for Env incorporation have been suggested (1, 4): (i) an active model, in which Env is recruited through a direct interaction with the viral structural polyprotein Gag; (ii) a passive model, in which Env present at assembly sites is randomly incorporated; (iii) a Gag-Env cotargeting model, in which both Gag and Env localize to a common membrane microdomain to increase the local concentration of viral proteins at assembly sites; and (iv) an indirect Gag-Env interaction model, in which Env incorporation requires the formation of a ternary complex composed of Env, Gag, and a host cellular factor. The active model is supported by studies showing that mutations in the matrix protein (MA) or the gp41 cytoplasmic tail (CT) reduce levels of incorporation and that second-site compensatory mutations in MA can rescue other MA substitutions or a small gp41 CT deletion that blocks Env incorporation (5–11). The passive model for Env incorporation derives support from pseudotyping experiments, wherein foreign (non-HIV-1) or HIV-1 CT-deleted viral Env glycoproteins are incorporated into HIV-1 particles in the presumed absence of a direct Gag-Env interaction (12, 13). The colocalization of Gag and Env in cholesterol-enriched plasma membrane microdomains (lipid rafts) that become part of the virion lipid bilayer is consistent with the Gag-Env cotargeting model (14).
The indirect Gag-Env interaction model posits that a host factor interacts with MA and/or the gp41 CT to promote Env incorporation. The observation that truncation of the gp41 CT blocks Env incorporation in most T-cell lines and in primary cell types (T cells and monocyte-derived macrophages) but has only a modest effect in several laboratory cell lines (15, 16) supports the notion that a host factor(s) bridges MA and the gp41 CT to recruit Env into particles. Recent studies suggested that tail-interacting protein of 47 kDa (TIP47) may serve such a bridging role (17–19): HIV-1 Gag and TIP47 coimmunoprecipitated in an in vivo pulldown assay; TIP47-MA binding was detectable in vitro in a quantitative yeast two-hybrid assay and in glutathione S-transferase (GST) pulldowns; TIP47 overexpression in HeLa cells increased Env incorporation into virions; depletion of TIP47 in cell-based assays decreased Env packaging; and MA and gp41 CT mutations that impair TIP47 binding reduced Env incorporation (17–19).
TIP47 is a soluble protein that forms oligomers in solution, a process promoted by 11-mer helix repeat elements within its ∼150 N-terminal residues. These N-terminal residues do not appear to be required for TIP47 binding to cellular partners (e.g., mannose 6-phosphate receptors [M6PRs]), but they are required for function (20, 21). TIP47 was originally proposed to mediate the trafficking of M6PRs from late endosomes to the trans-Golgi network by selectively binding the cytoplasmic domains of these cargo receptors (21–24). More-recent phylogenetic, structural, and protein function studies indicated that TIP47 is a member of the perilipin/ADRP/TIP47 (PAT) protein family (20, 25). PAT proteins bind intracellular lipids either constitutively or in response to metabolic stimuli and are important in lipid droplet biogenesis (20, 25–27). X-ray crystallographic studies revealed that the C-terminal domain of TIP47 has a structure similar to that of the N-terminal domain of apolipoprotein E, suggesting a role for TIP47 in protein recruitment to lipid droplets and lipid biogenesis (20). TIP47 adopts an extended conformation in solution, suggesting that N- and C-terminal domains are well separated, perhaps facilitating independent functions of the two terminal regions (28).
To understand further the role of TIP47 in HIV-1 MA binding and Env incorporation, we have cloned, expressed, and purified the intact TIP47 protein and deletion mutants lacking the N terminus. Nuclear magnetic resonance (NMR) spectroscopy-detected titration experiments were performed with myristylated (myr) and unmyristylated [myr(−)] forms of HIV-1 MA. Surface plasmon resonance (SPR) assays were also performed to evaluate MA-TIP47 binding. Our results confirm that TIP47 binds MA and, contrary to expectations based on TIP47-M6PR binding (21), suggest that the N terminus of TIP47 is required for this interaction. However, we were unable to verify a role for TIP47 in HIV-1 Env glycoprotein incorporation into virions.
MATERIALS AND METHODS
Sample preparation for NMR.
The DNAs encoding a C-terminally His-tagged TIP47 (residues 1 to 434) and a C-terminally His-tagged TIP47 fragment from which the N-terminal 112 residues were deleted (Δ1–112 TIP47), were subcloned and transformed into Rosetta 2 (DE3) pLysS expression cells (Novagen). Cells were grown in flasks of Luria broth medium, and unlabeled TIP47 and Δ1–112 TIP47 were induced with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at an A600 reading of 0.6. The cells were harvested and lysed with a microfluidizer (Microfluidics) and clarified by centrifugation, and the target protein was applied to a cobalt affinity resin (BD Biosciences). The resin was washed extensively (50 mM sodium phosphate, pH 7.0, 500 mM sodium chloride, 5 mM dithiothreitol [DTT], 10 mM imidazole), and the proteins were eluted with wash buffer containing 250 mM imidazole. Gel filtration chromatography was used to purify the proteins to homogeneity (GE Healthcare). Electrospray mass spectrometry results indicated calculated molecular masses of 36,088.6 Da for Δ1-112 TIP47 and 47855.7 Da for intact TIP47. 15N isotopically labeled Δ1-112 TIP47 was overexpressed in M9 minimal medium supplemented with 99.9% enriched 15N-ammonium chloride as the sole nitrogen source (Isotec) and purified as above. 15N-labeled myristylated (myr) and unmyristylated (myr−) HIV-1 MA samples were prepared as previously described (29, 30). The purified TIP47 and MA samples were simultaneously exchanged into an NMR buffer consisting of 50 mM sodium phosphate [pH 7.0], 5 mM DTT, and 10% D2O. Titrations with 15N-labeled Δ1-112 TIP47 were collected at pH 5.5.
NMR spectroscopy.
Two-dimensional (2D) 1H, 15N heteronuclear single quantum coherence (1H-15N HSQC) NMR titration data were obtained with a Bruker Avance 600 MHz NMR spectrometer equipped with a cryoprobe, using protein concentrations of 25 to 200 μM (35°C). Data were processed using NMRPIPE (31) and analyzed using NMRView (32).
Recombinant protein production and purification.
The MA region of the HIV-1 gag gene was amplified from plasmid pLAI (a generous gift from E. Kilareski and B. Wigdahl, Drexel University College of Medicine), using primers designed to facilitate ligation-independent cloning into the vector pETHSUL.1 (LabLife) (33). This vector is designed for the insertion of genes of interest in frame with an N-terminal SUMO tag (33). The recombinant pETHSUL plasmid was verified for the presence of MA insert by restriction digestion and sequence analysis (Genewiz, Inc., South Plainfield, NJ). The resultant vector was designated pSUMO-MA. The purification of H6SUMO-Gag was achieved via immobilized metal affinity chromatography (IMAC) using a Talon cobalt resin affinity column (CloneTech Laboratories, Inc.). The Escherichia coli strain BL21(DE3) Codon+-RIL (Stratagene) was used for expression of H6SUMO-MA from pSUMO-MA. Two milliliters of LB, containing 100 μg/ml ampicillin and 50 μg/ml chloramphenicol, was inoculated with a single transformed colony, and the culture was allowed to grow at 37°C for 9 h. Twenty-five milliliters of the preculture was used to inoculate 50 ml of the autoinducing medium ZYP-5052 (34) containing 100 μg/ml ampicillin and 50 μg/ml chloramphenicol. The culture was grown at 30°C for 16 h. Cells were harvested by centrifugation at 3,000 rpm for 20 min at 4°C, and the pellet was resuspended in 10 ml phosphate-buffered saline (PBS) (Roche) containing 2.5 mM imidazole. Cells were lysed by sonication, and the supernatant was clarified by centrifugation at 10,000 rpm (SS-34, Sorvall RC 5C Plus) for 20 min at 4°C. The supernatant was removed and applied to a Talon cobalt resin affinity column (CloneTech Laboratories), previously equilibrated with PBS (Roche). Loosely bound proteins were removed via 7 column volumes of PBS containing 7.5 mM imidazole. Tightly associated proteins were eluted in 3 column volumes of PBS containing 250 mM imidazole. The eluates were then pooled and made 1 mM with respect to EDTA. To this pooled sample was added 10 μg of a recombinant His6-tagged form of the catalytic domain (dtUD1) of the Saccharomyces cerevisiae SUMO hydrolase (33). Cleavage was allowed to proceed for 4 h at 18°C. Following cleavage, the sample was dialyzed at 4°C overnight against 2 liters of PBS to remove any imidazole. After dialysis, the dtUD1-catalyzed cleavage reaction was subjected to a second cobalt affinity purification using the Talon cobalt resin affinity column (CloneTech Laboratories). In this purification step, however, the cleaved MA protein passes straight through the column due to removal of the 6-histidine tag. Subsequently, the subtractively purified MA was dialyzed against 25 mM Tris-HCl [pH 8.0]–10% glycerol, at 4°C overnight. This dialyzed sampled was then filtered and loaded onto a 5-ml Hi-Trap Q HP (GE Healthcare). The flowthrough, containing the MA, was concentrated, flash frozen in liquid nitrogen, and stored at −80°C.
The human TIP47 gene was amplified from total cDNA generated from the CEM human T lymphoblastoid cell line using primers designed to facilitate ligation-independent cloning into the vector pETHSUL (33). TIP47 was expressed in E. coli BL21(DE3) Codon+-RIL (Stratagene) and purified by the subtractive purification methodology outlined by Hynson et al. (28). In addition to the full-length TIP47, an N-terminally truncated mutant of TIP47 was created, based on the work of Hickenbottom et al. (20). All vectors were verified by DNA sequencing. Expression and purification were carried out as for full-length TIP47.
The cytoplasmic tail of the cation-dependent (CD) mannose 6-phosphate receptor (M6PR) was produced from its expression vector (generously provided by Susanne Pfeffer, Stanford University) and purified as previously described (22, 35, 36). A C-terminally truncated mutant Rab9A GTPase (37) was overproduced from plasmid pET28b-Rab9(1–177), generously provided by E. J. Meehan (University of Alabama, Huntsville, AL). E. coli strain BL21(DE3) (Stratagene) harboring pET28b-Rab9(1–177) was grown in superbroth at 37°C. Overproduction of the His tag fusion protein was induced at an A600 of ∼2.0 with 1 mM IPTG for 2 h. Rab9(1–177) was purified using the same protocol as that used for HIV-1 MA purification, except that all buffers contained 10 μM GTP (Sigma). The eluates from the purification were pooled and then dialyzed at 4°C overnight against 2 liters of PBS containing 10 μM GTP to remove excess imidazole. Protein was stored at 4°C until used.
SPR binding assays.
Binding studies were performed at 25°C using a Biacore 3000 optical biosensor equipped with a CM4 research grade sensor chip. Immobilization of proteins to sensor surfaces was achieved using standard amine coupling methods. Analysis of the direct binding of the CD-M6PR cytoplasmic tail to full-length and mutant TIP47 was achieved by passage over surfaces to which the TIP47 proteins had been immobilized directly using standard amine coupling. Different concentrations of CD-M6PR cytoplasmic tail diluted in PBS running buffer were passed over these surfaces at a flow rate of 50 μl/min for 3 min of association and 10 min of dissociation. Specific regeneration of the surfaces was not needed between injections due to the rapid dissociation rate. Analysis of the direct binding of the Rab9(1–177) protein was achieved by passage over surfaces to which the human TIP47 proteins had been immobilized directly. Different concentrations of Rab9(1–177) diluted in running buffer (PBS containing 10 μM GTP) were passed over these surfaces at a flow rate of 50 μl/min for 3 min of association and 10 min of dissociation. Specific regeneration of the surfaces between injections was achieved by pulses of 1.3 M NaCl–35 mM NaOH until the baseline returned to the preexposure value. For MA binding assays, different concentrations of MA, diluted in PBS running buffer, were passed over TIP47-containing surfaces at a flow rate of 50 μl/min for 3 min of association and 10 min of dissociation. Specific regeneration of the surfaces between injections was achieved by pulses of 10 mM glycine, pH 1.5, until the baseline returned to the preexposure value.
SPR data analysis was performed using BIAEvaluation 4.0 software (Biacore Inc., NJ). The responses of a buffer injection and responses from a reference flow cell were subtracted to account for nonspecific binding. Experimental data were fitted to a simple 1:1 binding model with a parameter included for mass transport. The average kinetic parameters (association [ka[rsqb] and dissociation [kd] rates) generated from a minimum of four data sets were used to define equilibrium association (KA) and dissociation constants (KD).
Cell culture, plasmids, transfections, and RNA interference (RNAi).
Cell lines 293T, TZM-bl (obtained from J. Kappes through the NIH AIDS Research and Reference Reagent Program), and HeLa were maintained in Dulbecco's modified Eagle's medium (DMEM). Jurkat T cells were maintained in RPMI-1640 medium. All media were supplemented with 5 or 10% fetal bovine serum (FBS; HyClone), penicillin, and streptomycin unless otherwise indicated.
The full-length molecular clone pNL4-3 (38) was used in this study. Previous TIP47 studies (17, 19) used pHXB2R and pNL(AD8) clones. pNL(AD8) (39, 40) consists of pNL4-3 sequences encoding gp120 and the N-terminal portion (ectodomain) of gp41 from the CCR5-tropic clone pAD8. pNL(AD8) thus contains MA and gp41 cytoplasmic tail coding regions identical to (derived from) those of pNL4-3. pHXB2R and pNL4-3 are closely related molecular clones, and, importantly, are identical at residues implicated in TIP47 binding (residues 12, 15, 16, and 30 of MA and the YW motif in the gp41 CT) (19).
To deplete TIP47 in HeLa cells, 1 × 105 HeLa cells were transfected in suspension with 30 pmol of a pool of four TIP47 small interfering RNAs (siRNAs) (SMARTpool, Dharmacon) using RNAiMAX reagent (Invitrogen) and seeded in a 12-well plate in medium supplemented with 5% FBS. The next day, cells were transferred to a 6-well plate. Three days posttransfection with siRNA, cells were transfected with 3 μg of the pNL4-3 HIV-1 molecular clone (38) using Lipofectamine LTX (Invitrogen). Approximately 24 h posttransfection with pNL4-3, cells were metabolically radiolabeled with [35S]Cys for 4 h. Virus-containing supernatants were filtered and subjected to ultracentrifugation (41, 42). Cell and virus lysates were immunoprecipitated with HIV immunoglobulin (HIV-Ig; obtained from the NIH AIDS Research and Reference Reagent Program) and analyzed by SDS-PAGE and fluorography. Bands were quantified by phosphorimager analysis using QuantityOne software (Bio-Rad).
To overexpress TIP47 in HeLa cells, 0.6 × 106 HeLa cells were seeded on a 6-well plate. Cells were cotransfected with 2 μg of pNL4-3 and 0.5 or 1 μg of a plasmid containing the TIP47 open reading frame (ORF) driven by the cytomegalovirus (CMV) promoter (EX-U0354-MO2 from GeneCopoeia) or a green fluorescent protein (GFP)-expressing control plasmid (pmaxGFP from Lonza). Approximately 24 h postinfection, supernatants containing released virions were collected and cell and virus lysates were assayed for Env incorporation, virus release, and cellular levels of TIP47 as described above.
For short hairpin RNA (shRNA) transductions, five TIP47 MISSION shRNAs and the SCH002 nontargeting (NT) shRNA were obtained from Sigma: TRCN0000029679 NM_005817.2-904s1c1, TRCN0000029680 NM_005817.2-329s1c1, TRCN0000029681 NM_005817.2-1044s1c1, TRCN0000029682 NM_005817.2-1267s1c1, and TRCN0000029683 NM_005817.2-550s1c1, here referred to as TIP47 shRNAs 1, b, 2, c, and d, respectively. As a negative control, the SCH002 NT shRNA (Sigma) was used. C. Berlioz-Torrent (Institut Cochin, Paris, France) (19) kindly provided an anti-TIP47 shRNA pLKO.1-TIP1 (TIP1sensTRC sequence, CCGGAAGACTGTCTGCGACGCAGCACTCGAGTGCTGCGTCGCAGACAGTCTTTTTTTG, and TIP1 antisenseTRC sequence, AATTCAAAAAAAGACTGTCTGCGACGCAGCACTCGAGTGCTGCGTCGCAGACAGTCTT), cloned into pLKO.1-TRC and here referred to as TIP47 shRNA 3. shRNAs were packaged in 293T cells by cotransfecting with pCMV delta R8.2 (43) and pHCMV-G (44) at a 1:1:0.1 ratio using Lipofectamine LTX reagent (Invitrogen) and a total of 2 μg DNA. Twenty-four hours posttransfection, media were changed, and virus-containing supernatant was collected 48 h posttransfection. To deplete TIP47 in the Jurkat T-cell line, 3 × 105 Jurkat cells were infected with 50 to 75 μl of shRNA vector virus. Once cells had reached a density of 1 × 106 cells/ml, shRNA-transduced cells were selected with 1 μg/ml of puromycin. From the five tested shRNAs against TIP47, only two were found to consistently deplete TIP47. These two shRNAs were used independently to deplete TIP47 in all the experiments with Jurkat cells and were compared to the TIP1 (TIP47 3) shRNA (19). TIP47-depleted and NT-transduced Jurkat cells were infected with vesicular stomatitis virus (VSV)-G-pseudotyped HIV-1 prepared in 293T cells. Twenty-four hours postinfection, cell and virus lysates were assayed for Env incorporation, virus release, and cellular levels of TIP47 as described above.
Western blot analysis.
Proteins were separated by SDS-PAGE (10% acrylamide) and transferred to polyvinylidene difluoride membranes using the iBlot Dry blotting system (Invitrogen). TIP47 protein was detected with goat anti-TIP47 IgG (sc-14723; Santa Cruz) at a 1:600 dilution, followed by horseradish peroxidase-conjugated mouse anti-goat IgG (Thermo Scientific). Alpha tubulin was detected with mouse anti-tubulin IgG (T6074; Sigma) at a 1:10,000 dilution, followed by horseradish peroxidase-conjugated goat anti-mouse IgG. Proteins were visualized by Western Lightning (Perkin-Elmer) and scanned using the FluorChem SP Imaging system (Alpha Innotech), followed by exposure to X-ray film. Levels of TIP47 and tubulin from cell lysates were quantified using Quantity One software (Bio-Rad). Standard deviations (SD) and standard errors of the mean (SEM) were calculated from at least three independent experiments.
RIPA analysis.
Radioimmunoprecipitation assays (RIPA) were performed as previously described (41, 42, 45, 46). Approximately 18 h following transfection with pNL4-3 or infection with VSV-G-pseudotyped HIV-1, cells were washed and metabolically labeled in labeling medium (RPMI 1640 without l-Cys [MP Biomedicals no. 091646454] containing 5% FBS and 500 μCi [35S]l-Cys /well) for 4 to 6 h at 37°C. Released virions were collected from culture supernatants by 0.45-μm filtration and ultracentrifugation at 125,000 × g for 45 min. Cell and virion samples were solubilized in lysis buffer (0.5% Triton X-100, 300 mM NaCl, 50 mM Tris [pH 7.5], and Complete protease inhibitor cocktail [Roche]). Cell lysates were precleared by adsorption with protein A agarose in RIPA buffer (0.1% Triton X-100, 300 mM NaCl, 50 mM Tris [pH 7.5]) and 0.1% bovine serum albumin (BSA). Virion and precleared cell lysates were immunoprecipitated at 4°C with HIV-Ig bound to protein A agarose beads. Immunoprecipitated cell lysates were washed three times with RIPA buffer and once with SDS-deoxycholic acid wash buffer (0.1% SDS, 300 mM NaCl, 50 mM Tris [pH 7.5], and 2.5 mM deoxycholic acid). Immunoprecipitated virus lysates were washed once with RIPA buffer. Immunoprecipitated proteins were eluted in Laemmli sample buffer at 99°C for 5 min and resolved by SDS-PAGE in 12% acrylamide with AcrylAide cross-linker (FMC Corp.). After fixation and dehydration of the gel, labeled proteins were detected by exposure to phosphor screens and quantified using Quantity One software (Bio-Rad). The percentage of Env incorporation was calculated as the amount of virion gp120 relative to the amount of virion capsid protein (CA). Virus release efficiency was calculated as the amount of virion CA over the total sum of cell and virion Gag (Pr55Gag, p41, p25, and p24). Standard deviations and standard errors of the mean were calculated from at least three independent experiments.
Infectivity assays.
Virus stocks used for infections were collected by 0.45-μm filtration of supernatant from transfected HeLa cells or infected Jurkat cells and then normalized for reverse transcriptase (RT) activity as previously described (42). For single-cycle infectivity assays, 5 × 104 TZM-bl cells/well were infected in the presence of 20 μg DEAE-dextran (GE Healthcare) per ml with four different virus inputs. Infected cells were lysed 48 h postinfection using Glo Lysis buffer (Promega) and assayed for luciferase activity using a luciferase assay kit (Promega) as previously described (47). Standard deviations and standard errors of the mean were calculated from at least three independent experiments.
Analysis of virus replication kinetics.
To evaluate HIV-1 replication kinetics, 2 × 106 Jurkat cells were transfected with 2 μg of pNL4-3 in 200 μl of 0.7 mg/ml of DEAE-dextran and incubated for 15 min at 37°C. Cells were then washed with transfection buffer (25 mM Tris-HCl [pH 7.4], 0.6 mM Na2HPO4, 5 mM KCl, 140 mM NaCl, 0.7 mM CaCl2, 0.5 mM MgCl2) and resuspended in 1 ml of RPMI 1640 supplemented with 10% FBS. Cultures were split 1:3 every 2 to 3 days, at which time culture supernatants were collected to monitor RT activity (41).
RESULTS AND DISCUSSION
Recombinantly produced TIP47 is competent to bind its natural ligands.
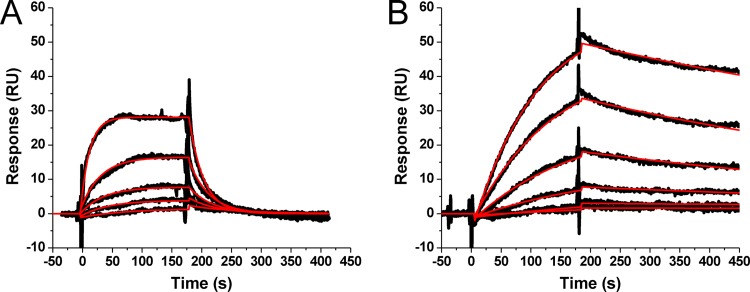
It has been demonstrated that preparations of TIP47, either recombinantly produced or endogenously purified, have low binding activity (21, 35, 36, 48). We therefore employed a surface plasmon resonance (SPR)-based interaction assay using two native ligands of TIP47, CD-M6PR and Rab9, to verify and quantify the activity of our full-length recombinant protein. Representative sensorgrams for these interactions are shown in Fig. 1. The apparent equilibrium dissociation constants (KD) derived from fitting of the sensorgrams are in good agreement with values previously reported (35). These results demonstrate that our recombinant TIP47 protein is correctly folded and active (35, 36, 49–51).
Fig 1.
Interaction of purified TIP47 with its natural ligands in SPR assays. (A) Sensorgrams depicting the interaction of the cytoplasmic tail of CD-M6PR with sensor chip-immobilized human TIP47. Results for GST-tagged CD-M6PR cytoplasmic tail protein at concentrations of 0.134, 0.269, 0.538, 1.075, and 2.15 μM are shown. (B) Sensorgrams depicting the interaction of the Rab9 GTPase with sensor chip-immobilized human TIP47. Results for His-tagged Rab9(1–177) proteins at concentrations of 8.6, 17.3, 34.6, 134.3, and 276.6 nM are shown. Black lines indicate experimental data, whereas red lines indicate fitting to a 1:1 Langmuir binding model with a parameter included for mass transport. Kinetic values are as follows: for CD-M6PR-TIP47, ka = (1.3 ± 0.2) × 104 M−1 · s−1; kd = 4.5 × 10−2 · s−1; KD = 3.6 μM; for Rab9-TIP47, ka = (4.2 ± 2.0) × 104 M−1 · s−1; kd = (1.0 ± 0.1) × 10−3 · s−1; KD = 24.4 nM.
The N terminus of TIP47 is required for its interaction with HIV-1 MA.
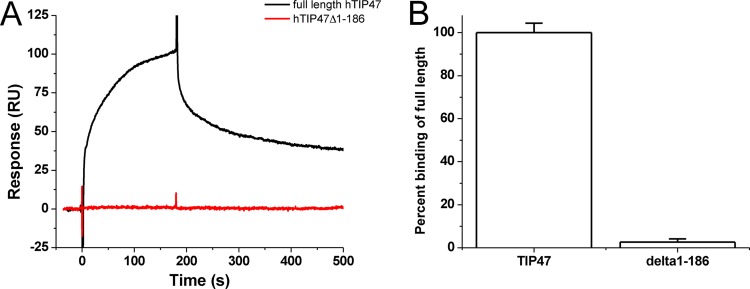
To confirm the reported MA-TIP47 interaction (19) and define the domain of TIP47 required for binding, we performed SPR-based interaction analyses with purified MA protein and full-length or N-terminally truncated TIP47 protein (Fig. 2). We observed that HIV-1 MA binds well to the full-length TIP47 protein but does not bind the Δ1–186 TIP47 mutant. This implies that the N terminus of TIP47 is required for its interaction with HIV-1 MA. To place these results in the context of previous TIP47 binding studies, Hanna et al. showed that TIP47 residues 152 to 186 are required for Rab9 interaction (50) and Krise et al. demonstrated that the M6PR cytoplasmic domain interacts with the Δ1–186 TIP47 mutant (35).
Fig 2.
The N terminus of TIP47 is required for its interaction with HIV-1 MA in SPR assays. HIV-1 MA at a concentration of 250 nM was passed over full-length TIP47 and Δ1–186 TIP47, and the responses were recorded. (A) Representative sensorgrams for these interactions; (B) mean responses of three experiments in graphical form. Error bars represent 1 standard deviation.
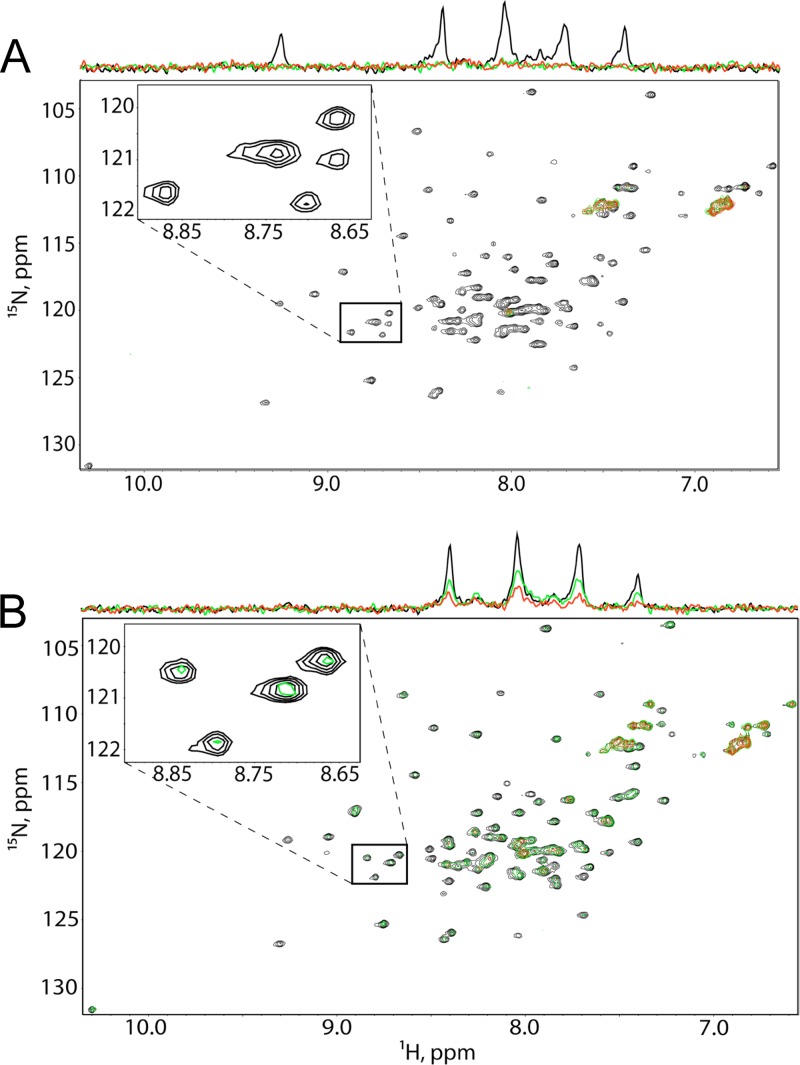
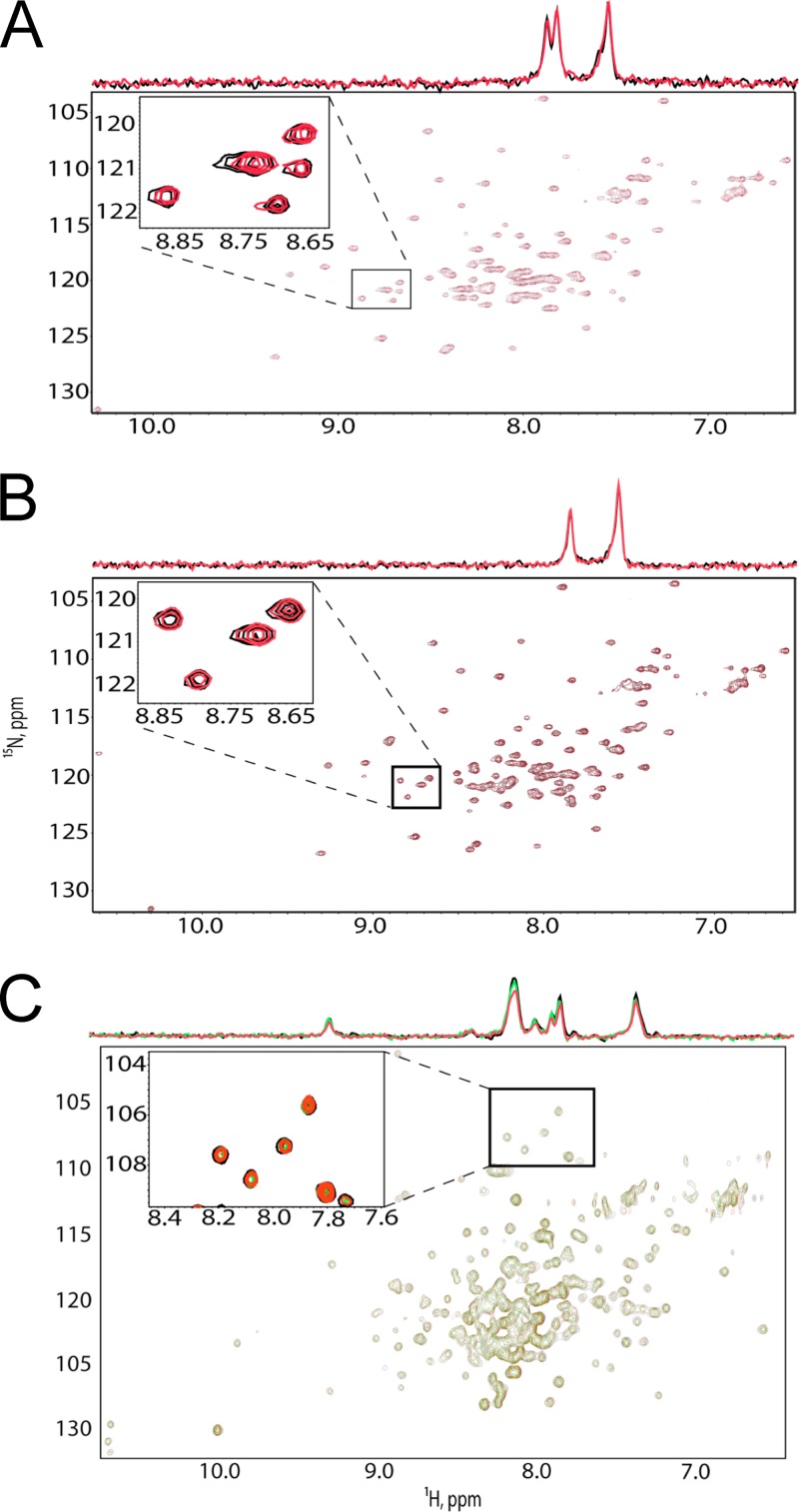
To confirm these binding results by an independent approach, we performed 1H-15N HSQC experiments to probe for TIP47-MA binding. 1H-15N HSQC signals for the backbone amides of 15N-labeled myristylated HIV-1 MA (myrMA) exhibited significant signal broadening upon titration with full-length TIP47 (Fig. 3A), which again indicates a direct interaction. The signals were nearly undetectable at TIP47/MA ratios above 1.0, consistent with tight binding and a dissociation equilibrium constant of less than ∼100 μM. Similar results were obtained for the unmyristylated MA [myr(−)MA] protein, indicating that the myristyl group is not required for TIP47-MA binding (Fig. 3B). Consistent with the SPR data, titrations with TIP47 lacking the N-terminal oligomerization domain (Δ1–112 TIP47) did not affect the 1H-15N HSQC spectra obtained for 15N-labeled myrMA and myr(−)MA (Fig. 4A and B, respectively), which suggests no interaction. Similarly, titration of 15N-labeled Δ1–112 TIP47 with myr(−)MA did not affect the 1H-15N HSQC spectrum of Δ1–112 TIP47 (Fig. 4C). These findings again indicate that the N-terminal domain of TIP47 is required for MA binding. However, neither the SPR nor the NMR approach excludes the possibility that oligomerization-dependent interactions between MA and the C-terminal domain of TIP47 may also exist.
Fig 3.
MA interacts with TIP47 in NMR-based assays. 1H-15N HSQC NMR spectra obtained upon titration of 15N-labeled HIV-1 myrMA (50 μM) (A) and myr(−)MA (50 μM) (B) with intact TIP47; [TIP47]:[MA] = 0:1 (black), 0.5:1 (green), and 1:1 (red).
Fig 4.
The N terminus of TIP47 is required for its interaction with TIP47 in NMR-based assays. (A, B) 1H-15N HSQC NMR spectra obtained for 15N-labeled HIV-1 myrMA (50 μM) (A) and myr(−)MA (50 μM) (B) upon titration with Δ1–112 TIP47; [Δ1–112 TIP47]:[MA] = 0:1 (black), 1:1 (red). (C) 1H-15N HSQC NMR spectra obtained for 15N-labeled Δ1–112 TIP47 upon titration with myrMA; [MA]:[Δ1–112 TIP47] = 0:1 (black), 2:1 (green), and 4:1 (red).
Depletion of TIP47 in HeLa cells does not impair HIV-1 Env incorporation, virus release, or infectivity.
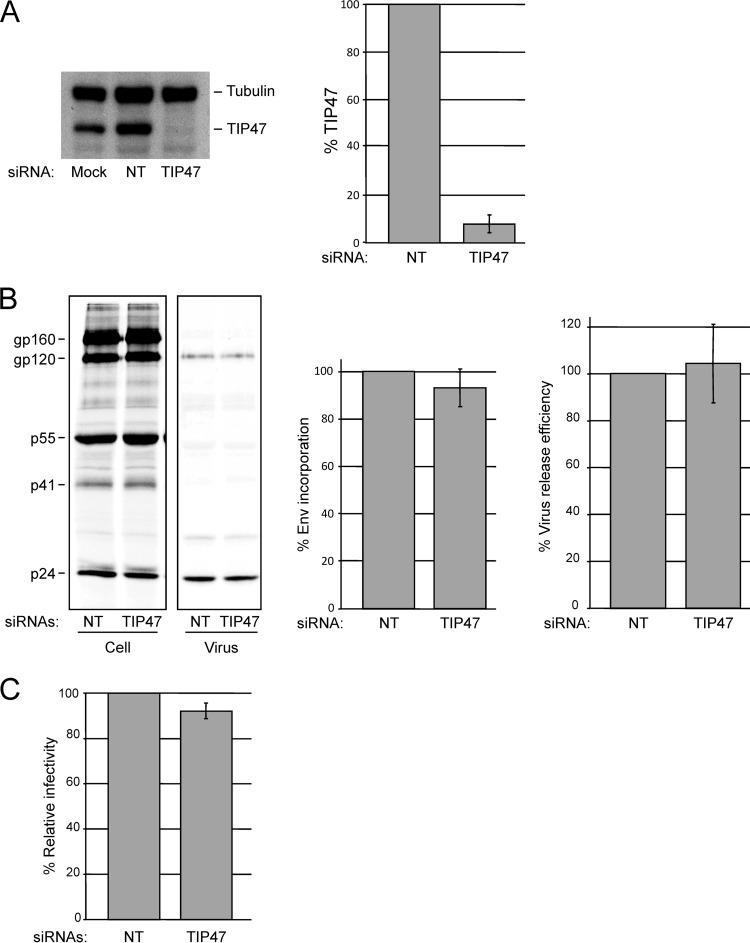
To assess the putative role of TIP47 in Env incorporation, we depleted TIP47 in HeLa cells with a pool of four siRNAs. Nontargeting (NT) siRNA was used as a negative control. Cells were transfected with the pNL4-3 HIV-1 molecular clone and metabolically labeled with [35S]Cys. Cell and virus lysates were immunoprecipitated with HIV-Ig. Levels of TIP47 were analyzed in parallel by quantitative Western blotting. Although TIP47 protein levels were significantly reduced (∼90 to 95% depletion) (Fig. 5A), no significant decrease in Env (gp120) incorporation or virus release efficiency was observed (Fig. 5B). To evaluate the effect of TIP47 depletion on specific particle infectivity, viral supernatants were harvested from control and TIP47-depleted cells, normalized for RT activity, and used to infect the TZM-bl indicator cell line. No effect of TIP47 depletion on single-round infectivity was observed (Fig. 5C). These results indicate that depletion of 90 to 95% of TIP47 in HeLa cells does not affect Env incorporation, virus release efficiency, or HIV-1 infectivity.
Fig 5.
Depletion of TIP47 in HeLa cells does not impair HIV-1 Env incorporation, virus release, or infectivity in HeLa cells. Cells were transfected with 30 pmol nontargeting siRNA (NT) or a pool of four TIP47-specific siRNAs. (A) Transfected cell lysates were probed with antibodies specific for tubulin and TIP47 (left panel); the ratio of TIP47 to tubulin was quantified by using a FluorChem SP Imaging System (Alpha Innotech) (right panel) (means ± SD; n = 3). (B) Control and TIP47-depleted cells were transfected with pNL4-3 and metabolically radiolabeled with [35S]Cys; cell and virus lysates were immunoprecipitated with HIV-Ig. The relative ratios of virus-associated gp120 to p24(CA) and virus release efficiencies were quantified by phosphorimager analysis (means ± SD; n = 3). (C) Virus-containing supernatants were harvested from control and TIP47-depleted cells, normalized for RT activity, and used to infect the TZM-bl indicator cell line. Graphs show average levels of infectivity with a virus input of 50,000 RT cpm. Luciferase activity was measured 2 days postinfection (means ± SD, n = 3).
Overexpression of TIP47 in HeLa cells does not enhance HIV-1 Env incorporation or virion infectivity.
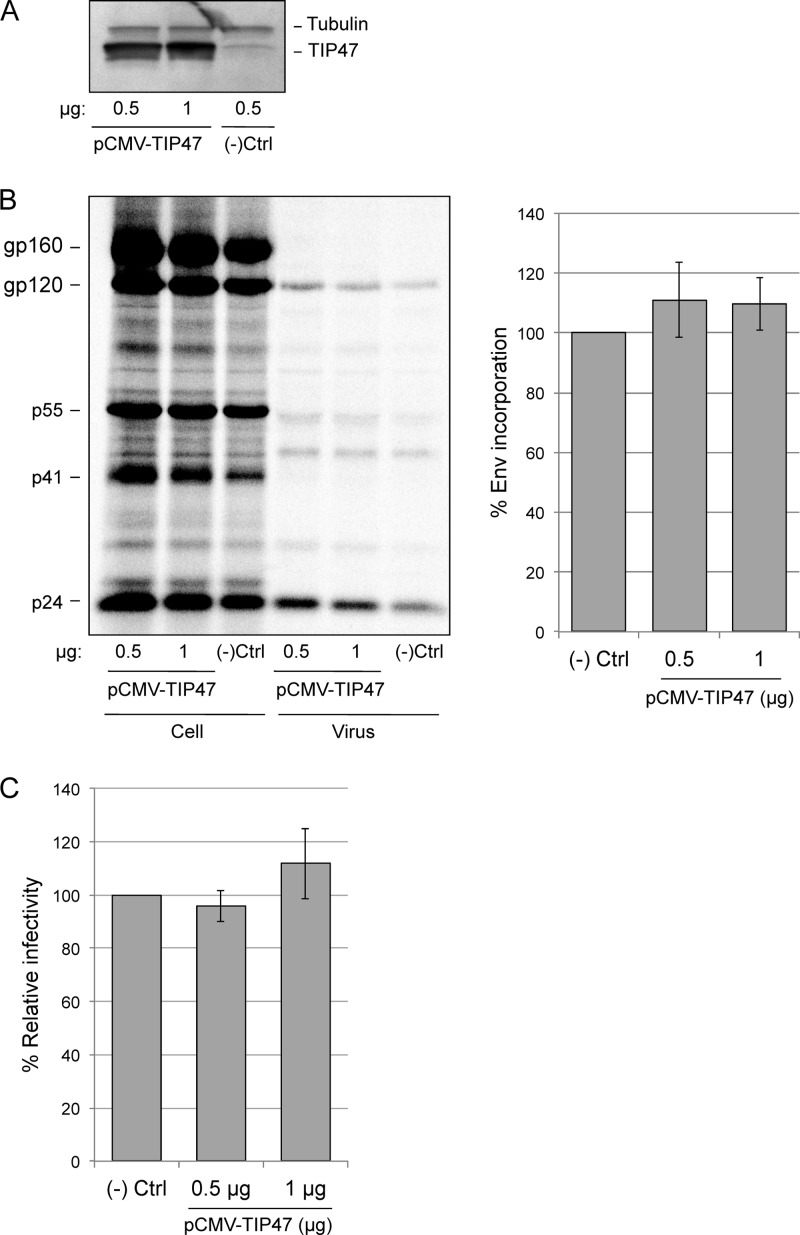
Lopez-Verges and coworkers (19) reported that overexpression of TIP47 in HeLa cells increases Env incorporation. To confirm this finding, HeLa cells were transfected with TIP47 or GFP (negative control) expression vectors and metabolically labeled with [35S]Cys. Cell and virus lysates were immunoprecipitated with anti-HIV Ig. Despite markedly increased levels of TIP47 expression (Fig. 6A), no increase in HIV-1 Env incorporation was observed, as determined by the ratio of gp120/p24(CA) (Fig. 6B). We also measured the specific infectivity of virus released from TIP47-overexpressing cells in the TZM-bl single-cycle assay. No significant effect on particle infectivity was observed (Fig. 6C). These results demonstrate that TIP47 overexpression in HeLa cells does not enhance either Env incorporation or infectivity.
Fig 6.
Overexpression of TIP47 in HeLa cells does not enhance HIV-1 Env incorporation or virion infectivity. HeLa cells were cotransfected with pNL4-3 and either GFP [(−)Ctrl] or TIP47 expression vectors (pCMV-TIP47) at the indicated DNA concentrations. (A) Transfected cell lysates were probed with antibodies specific for tubulin and TIP47. (B) Cells were metabolically radiolabeled with [35S]Cys; cell and virus lysates were immunoprecipitated with HIV-Ig. The relative ratios of virus-associated gp120 to p24(CA) were quantified by phosphorimager analysis (± SEM; n = 3). Using a paired t test, both samples overexpressing TIP47 were considered not significantly different from the negative control (two-tailed P values > 0.3) (C) Virus-containing supernatants were harvested from control and TIP47-overexpressing cells, normalized for RT activity, and used to infect the TZM-bl indicator cell line with four different virus inputs. Graphs show average levels of infectivity with a virus input of 50,000 RT cpm. Luciferase activity was measured 2 days postinfection (± SEM; n = 3).
Depletion of TIP47 in Jurkat cells does not disrupt HIV-1 Env incorporation, virus release, infectivity, or replication.
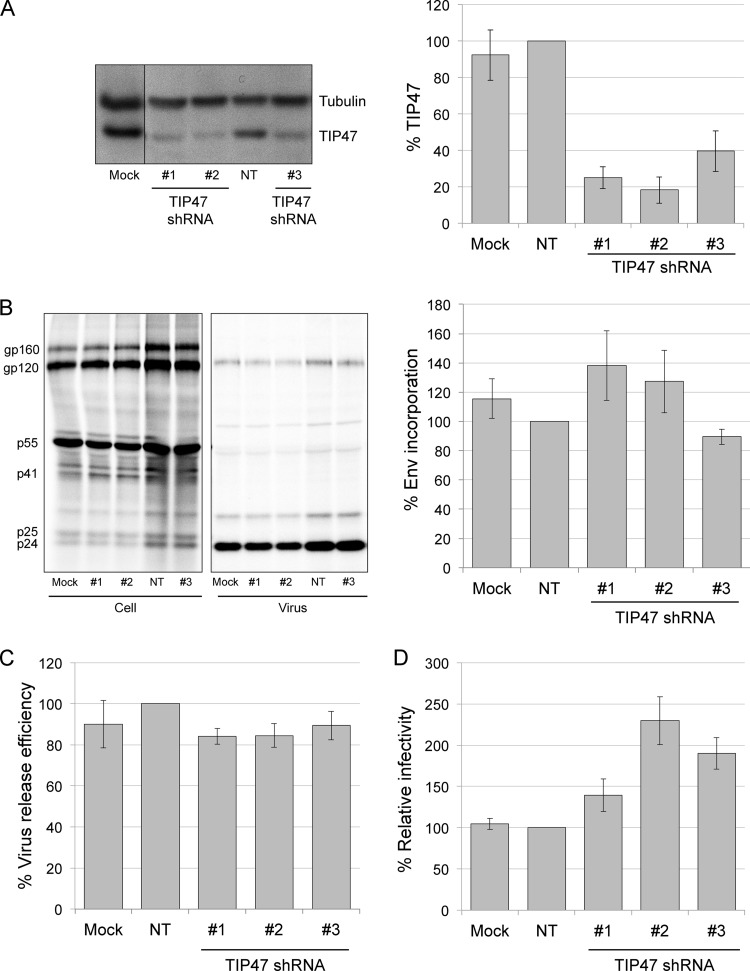
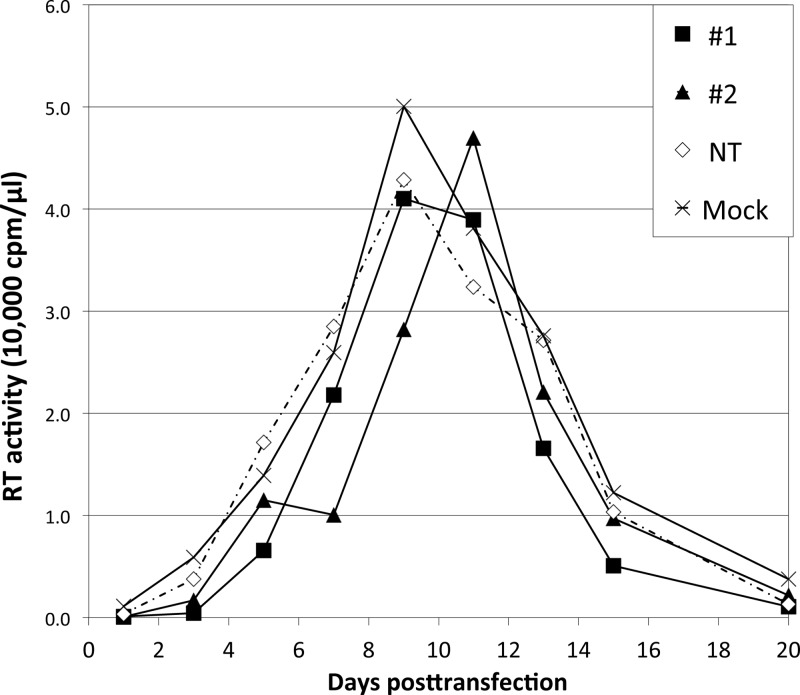
We previously demonstrated that the requirement for the gp41 CT in Env incorporation is cell type dependent; in HeLa cells, gp41 CT truncation had only a small effect on Env incorporation, whereas in most T-cell lines (e.g., Jurkat) the gp41 CT is required for Env incorporation and virus replication (16). These observations raised the possibility that TIP47 could play a cell type-dependent role in Env incorporation. We therefore depleted TIP47 in the Jurkat T-cell line by transducing with TIP47-specific shRNAs. Of five shRNAs tested, three yielded consistent knockdowns in the range of 60% for shRNA 3 and ∼80% for shRNAs 1 and 2 (Fig. 7A); the others failed to produce reliable depletion (data not shown). To determine whether TIP47 depletion in Jurkat cells affects Env incorporation or virus release, TIP47-depleted cells were infected with VSV-G-pseudotyped HIV-1. Infected cells were then washed and labeled with [35S]Cys for 4 h. Cell and virus lysates were immunoprecipitated, and Env incorporation and virus particle production were analyzed. We did not observe a significant decrease in Env (gp120) incorporation (Fig. 7B) or virus release efficiency (Fig. 7C). As a control, we compared levels of Env incorporation between wild-type (WT) (NL4-3) and a gp41 CT truncation mutant (NL4-3/CTdel-144) in Jurkat cells by applying the same VSV-G pseudotyping approach as the one used for TIP47-depleted Jurkat cells. As we reported previously (16), the CTdel-144 mutant displayed a 10-fold decrease in Env incorporation in Jurkat cells relative to the WT (data not shown). To determine the effect of TIP47 knockdown on HIV-1 infectivity, TZM-bl cells were infected with supernatants from TIP47-depleted, HIV-1-infected Jurkat cells. We did not observe a decrease in HIV-1 infectivity (Fig. 7D) but saw a small increase that was of marginal statistical significance (see legend to Fig. 7D). Finally, we tested whether TIP47 is required for the establishment of a spreading HIV-1 infection. TIP47-depleted Jurkat cells were transfected with pNL4-3, and virus replication was monitored by RT activity. We observed that HIV-1 replication in TIP47-depleted Jurkat cells peaked either on the same day as, or 2 days later than, replication in cells transduced with the nontargeting shRNA (Fig. 8). We observed a small amount of cytotoxicity in TIP47-depleted cells, likely accounting for the very minor delays in peak RT production in some cultures. TIP47 protein levels were analyzed close to the end of each replication assay; TIP47 knockdown efficiencies were calculated to be ∼80% (data not shown). Because this level of TIP47 knockdown diminished after approximately 1 month, shRNA transductions were performed de novo for each experiment.
Fig 7.
Depletion of TIP47 in Jurkat cells does not disrupt HIV-1 Env incorporation, infectivity, or virus release. (A) TIP47 depletion in Jurkat cells using shRNAs. TIP47 and tubulin were detected by Western blotting of puromycin-resistant Jurkat cells after transduction with TIP47-targeted shRNAs or nontargeting (NT) shRNA. Average protein levels relative to tubulin are shown. Error bars indicate SEM (n = 3). (B) HIV-1 Env incorporation efficiency in TIP47-depleted Jurkat cells. Cells were metabolically radiolabeled with [35S]Cys; cell and virus lysates were immunoprecipitated with HIV-Ig (left panel). Average levels of Env incorporation are shown (right panel). Error bars indicate SEM (n = 4). Using a paired t test, none of the results were considered significantly different from the NT control results (P > 0.05). (C) HIV-1 release efficiency in TIP47-depleted Jurkat cells. Average virus release efficiency normalized to NT shRNA-transduced cells is shown. Error bars indicate SEM (n = 4). Using a paired t test, none of the results were considered significantly different from the NT control results (P > 0.01). (D) Infectivity of HIV-1 produced from TIP47-depleted Jurkat cells. Infectivity was detected by luciferase activity in infected TZM-bl cells, and results were averaged. Infectivity assays were performed with four different virus inputs, normalized for RT activity. Graphs show average levels of infectivity with a virus input of 25,000 RT cpm. Error bars indicate SEM (n = 4). Using a paired t test, the increases in infectivity were marginally significant relative to the controls (P = 0.1 for shRNA 2 compared to mock sample, and P = 0.02 for shRNA 2 compared to NT sample).
Fig 8.
TIP47 depletion does not significantly affect HIV-1 replication in the Jurkat T-cell line. Jurkat cells transduced with shRNA 1 or 2 targeting TIP47 (#1 or #2) or nontargeting shRNA (NT) or Jurkat cells with no shRNA (mock) were transfected with the HIV-1 molecular clone pNL4-3 and passaged by dilution with fresh medium every 2 to 3 days. Culture supernatants were collected at each passage, and virus replication was monitored by RT activity.
We note that Manrique and coworkers were able to detect an interaction between TIP47 and an HIV-1 gp41 CT-GST fusion but observed that TIP47 did not increase the binding of HIV-1 MA to the gp41 CT (52).
In summary, the results of this study confirm the findings of Lopez-Verges et al. (19) indicating that HIV-1 MA binds to TIP47. However, our findings do not confirm a role for TIP47 in HIV-1 Env incorporation. It is possible that low levels of TIP47 could be sufficient to promote Env incorporation and that our depletions were not complete enough to induce an Env incorporation defect. However, the TIP47 depletion efficiencies observed in this study are similar to those achieved in studies that reported a requirement for TIP47 in Env incorporation, which was not quantified by Lopez-Verges et al. (19) but found to be in the 60 to 85% range by Bauby et al. (17). Further work will be required to delineate the physiological relevance to HIV-1 replication, if any, of the interaction between MA and TIP47.
ACKNOWLEDGMENTS
We thank members of the Freed laboratory for helpful discussion and critical review of the manuscript. The HIV-Ig and TZM-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program. We thank C. Berlioz-Torrent for sharing reagents and for discussion of our results.
This research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH, by the Intramural AIDS Targeted Antiviral Program, and by NIH/NIAID grants 1R03AI078790-01A1 (S.C.) and AI30917 (M.F.S.).
Footnotes
Published ahead of print 16 January 2013
REFERENCES
- 1. Checkley MA, Luttge BG, Freed EO. 2011. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J. Mol. Biol. 410:582–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balasubramaniam M, Freed EO. 2011. New insights into HIV assembly and trafficking. Physiology 26:236–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2:a006924 doi:10.1101/cshperspect.a006924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Johnson MC. 2011. Mechanisms for Env glycoprotein acquisition by retroviruses. AIDS Res. Hum. Retroviruses 27:239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dorfman T, Mammano F, Haseltine WA, Gottlinger HG. 1994. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 68:1689–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Freed EO, Martin MA. 1996. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J. Virol. 70:341–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Freed EO, Martin MA. 1995. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J. Virol. 69:1984–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mammano F, Kondo E, Sodroski J, Bukovsky A, Gottlinger HG. 1995. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J. Virol. 69:3824–3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murakami T, Freed EO. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J. Virol. 74:3548–3554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reil H, Bukovsky AA, Gelderblom HR, Gottlinger HG. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yu X, Yuan X, Matsuda Z, Lee TH, Essex M. 1992. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J. Virol. 66:4966–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cronin J, Zhang XY, Reiser J. 2005. Altering the tropism of lentiviral vectors through pseudotyping. Curr. Gene Ther. 5:387–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Page KA, Landau NR, Littman DR. 1990. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J. Virol. 64:5270–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Waheed AA, Freed EO. 2010. The role of lipids in retrovirus replication. Viruses 2:1146–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Akari H, Fukumori T, Adachi A. 2000. Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J. Virol. 74:4891–4893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Murakami T, Freed EO. 2000. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc. Natl. Acad. Sci. U. S. A. 97:343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bauby H, Lopez-Verges S, Hoeffel G, Delcroix-Genete D, Janvier K, Mammano F, Hosmalin A, Berlioz-Torrent C. 2010. TIP47 is required for the production of infectious HIV-1 particles from primary macrophages. Traffic 11:455–467 [DOI] [PubMed] [Google Scholar]
- 18. Blot G, Janvier K, Le Panse S, Benarous R, Berlioz-Torrent C. 2003. Targeting of the human immunodeficiency virus type 1 envelope to the trans-Golgi network through binding to TIP47 is required for env incorporation into virions and infectivity. J. Virol. 77:6931–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lopez-Verges S, Camus G, Blot G, Beauvoir R, Benarous R, Berlioz-Torrent C. 2006. Tail-interacting protein TIP47 is a connector between Gag and Env and is required for Env incorporation into HIV-1 virions. Proc. Natl. Acad. Sci. U. S. A. 103:14947–14952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hickenbottom SJ, Kimmel AR, Londos C, Hurley JH. 2004. Structure of a lipid droplet protein; the PAT family member TIP47. Structure 12:1199–1207 [DOI] [PubMed] [Google Scholar]
- 21. Sincock PM, Ganley IG, Krise JP, Diederichs S, Sivars U, O'Connor B, Ding L, Pfeffer SR. 2003. Self-assembly is important for TIP47 function in mannose 6-phosphate receptor transport. Traffic 4:18–25 [DOI] [PubMed] [Google Scholar]
- 22. Diaz E, Pfeffer SR. 1998. TIP47: a cargo selection device for mannose 6-phosphate receptor trafficking. Cell 93:433–443 [DOI] [PubMed] [Google Scholar]
- 23. Lombardi D, Soldati T, Riederer MA, Goda Y, Zerial M, Pfeffer SR. 1993. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 12:677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Riederer MA, Soldati T, Shapiro AD, Lin J, Pfeffer SR. 1994. Lysosome biogenesis requires Rab9 function and receptor recycling from endosomes to the trans-Golgi network. J. Cell Biol. 125:573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bulankina AV, Deggerich A, Wenzel D, Mutenda K, Wittmann JG, Rudolph MG, Burger KN, Honing S. 2009. TIP47 functions in the biogenesis of lipid droplets. J. Cell Biol. 185:641–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bickel PE, Tansey JT, Welte MA. 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta 1791:419–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wolins NE, Rubin B, Brasaemle DL. 2001. TIP47 associates with lipid droplets. J. Biol. Chem. 276:5101–5108 [DOI] [PubMed] [Google Scholar]
- 28. Hynson RM, Jeffries CM, Trewhella J, Cocklin S. 2012. Solution structure studies of monomeric human TIP47/perilipin-3 reveal a highly extended conformation. Proteins 80:2046–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Massiah MA, Starich MR, Paschall C, Summers MF, Christensen AM, Sundquist WI. 1994. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J. Mol. Biol. 244:198–223 [DOI] [PubMed] [Google Scholar]
- 30. Tang C, Loeliger E, Luncsford P, Kinde I, Beckett D, Summers MF. 2004. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. U. S. A. 101:517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6:277–293 [DOI] [PubMed] [Google Scholar]
- 32. Johnson BA. 2004. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278:313–352 [DOI] [PubMed] [Google Scholar]
- 33. Weeks SD, Drinker M, Loll PJ. 2007. Ligation independent cloning vectors for expression of SUMO fusions. Protein Expr. Purif. 53:40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Studier FW. 2005. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41:207–234 [DOI] [PubMed] [Google Scholar]
- 35. Krise JP, Sincock PM, Orsel JG, Pfeffer SR. 2000. Quantitative analysis of TIP47-receptor cytoplasmic domain interactions: implications for endosome-to-trans Golgi network trafficking. J. Biol. Chem. 275:25188–25193 [DOI] [PubMed] [Google Scholar]
- 36. Orsel JG, Sincock PM, Krise JP, Pfeffer SR. 2000. Recognition of the 300-kDa mannose 6-phosphate receptor cytoplasmic domain by 47-kDa tail-interacting protein. Proc. Natl. Acad. Sci. U. S. A. 97:9047–9051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen L, DiGiammarino E, Zhou XE, Wang Y, Toh D, Hodge TW, Meehan EJ. 2004. High resolution crystal structure of human Rab9 GTPase: a novel antiviral drug target. J. Biol. Chem. 279:40204–40208 [DOI] [PubMed] [Google Scholar]
- 38. Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Freed EO, Englund G, Martin MA. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 69:3949–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Freed EO, Martin MA. 1994. HIV-1 infection of non-dividing cells. Nature 369:107–108 [DOI] [PubMed] [Google Scholar]
- 41. Freed EO, Martin MA. 1994. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J. Virol. 68:2503–2512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Willey RL, Bonifacino JS, Potts BJ, Martin MA, Klausner RD. 1988. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc. Natl. Acad. Sci. U. S. A. 85:9580–9584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263–267 [DOI] [PubMed] [Google Scholar]
- 44. Yee JK, Miyanohara A, LaPorte P, Bouic K, Burns JC, Friedmann T. 1994. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc. Natl. Acad. Sci. U. S. A. 91:9564–9568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Checkley MA, Luttge BG, Soheilian F, Nagashima K, Freed EO. 2010. The capsid-spacer peptide 1 Gag processing intermediate is a dominant-negative inhibitor of HIV-1 maturation. Virology 400:137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luttge BG, Shehu-Xhilaga M, Demirov DG, Adamson CS, Soheilian F, Nagashima K, Stephen AG, Fisher RJ, Freed EO. 2008. Molecular characterization of feline immunodeficiency virus budding. J. Virol. 82:2106–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kiernan RE, Ono A, Englund G, Freed EO. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Burguete AS, Sivars U, Pfeffer S. 2005. Purification and analysis of TIP47 function in Rab9-dependent mannose 6-phosphate receptor trafficking. Methods Enzymol. 403:357–366 [DOI] [PubMed] [Google Scholar]
- 49. Ganley IG, Carroll K, Bittova L, Pfeffer S. 2004. Rab9 GTPase regulates late endosome size and requires effector interaction for its stability. Mol. Biol. Cell 15:5420–5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hanna J, Carroll K, Pfeffer SR. 2002. Identification of residues in TIP47 essential for Rab9 binding. Proc. Natl. Acad. Sci. U. S. A. 99:7450–7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zentner IJ, Cocklin S. Analysis of protein-protein interactions using surface plasmon resonance biosensing, in press. Research Signpost Publishing, Kerala, India [Google Scholar]
- 52. Manrique JM, Affranchino JL, Gonzalez SA. 2008. In vitro binding of simian immunodeficiency virus matrix protein to the cytoplasmic domain of the envelope glycoprotein. Virology 374:273–279 [DOI] [PubMed] [Google Scholar]