Abstract
Foamy viruses (FVs) are the least known retroviruses commonly found in primates, cats, horses, and cattle. Although FVs are considered apathogenic, simian and feline FVs have been shown to be associated with some transient health abnormalities in animal models. Currently, data regarding the course of infection with bovine FV (BFV) are not available. In this study, we conducted experimental infections of natural (cattle) and heterologous (sheep) hosts with the BFV100 isolate and monitored infection patterns in both hosts during the early phase postinoculation as well as after long-term infection. Four calves and six sheep inoculated with BFV100 showed no signs of pathology but developed persistent infection, as confirmed by virus rescue, consistent detection of BFV-specific antibodies, and presence of viral DNA. In both hosts, antibodies against BFV Gag and Bet appeared early after infection and persisted at high and stable levels while seroreactivity toward Env was consistently detectable only in BFV-infected sheep. Interestingly, the BFV proviral DNA load was highest in lung, spleen, and liver and moderate in leukocytes, while salivary glands contained either low or undetectable DNA loads in calves or sheep, respectively. Additionally, comparison of partial BFV sequences from inoculum and infected animals demonstrated very limited changes after long-term infection in the heterologous host, clearly less than those found in BFV field isolates. The persistence of BFV infection in both hosts suggests full replication competence of the BFV100 isolate with no requirement of genetic adaptation for productive replication in the authentic and even in a heterologous host.
INTRODUCTION
Due to unique features in their replication strategy and molecular biology, foamy viruses (FVs) comprise the subfamily of Spumaretrovirinae within the retroviruses. FVs are widespread in different nonhuman primates (NHPs), collectively termed simian FVs (SFVs), and in cats (feline FV, or FFV), cattle (bovine FV, or BFV), and horses (equine FV, or EFV) in which they establish a lifelong, persistent infection (1, 2). However, target organs or target cells for either productive replication and/or persistent infection are ill defined for any of the known FVs. In addition, other parameters of FV replication for instance, concerning the kinetics of virus replication or host-mediated immune response, are far from being fully understood.
FVs are considered to be the most ancient RNA viruses of vertebrates, with an extremely low evolution rate and a high degree of cospeciation with their hosts (3). However, even in the face of this pronounced cospeciation, SFVs have been shown to repeatedly cross species barriers to other NHPs or even humans (4, 5, 6, 7, 8, 9). It is currently unknown whether genetic adaptation to the new host occurs, for instance, in genes that are at the forefront of host-pathogen interactions (10, 11). Here, host-encoded antiviral restriction factors like APOBEC3 cytidine deaminases and the viral proteins counteracting this restriction, like the lentiviral Vif and the FV Bet proteins, are prominent examples for rapid coevolution and cospeciation (12, 13, 14, 15).
In all authentic host species as well as in zoonotically SFV-infected humans, FV infections have not been associated with a defined disease although modulation of host immunity has been described (16). In addition, a cofactorial role of FVs in the genesis and expression of a multifactorial disease is still possible and has never been ruled out for any FV. In line with this, the tissue range of SFV replication has been shown to be expanded in simian immunodeficiency virus (SIV)-positive monkeys (17). Issues related to the pathogenic potential of FVs are of significant importance for the field since (i) FV-based vectors for gene therapy and vaccination are currently being developed in several labs, (ii) different SFVs have been shown to have a high capacity of interspecies transmissions to either other NHPs or humans, and (iii) BFV is present in the human food chain via meat or dairy products from BFV-positive cattle (18, 19, 20, 21, 22, 23).
BFV is present in a high percentage of livestock cattle in different parts of the world (21, 24, 25, 26, 27, 28). Transmission of BFV may occur through close contact pre- or perinatally, for instance, via colostrum or milk (29, 30). In fact, we have recently shown that BFV can be reproducibly isolated from the cellular fraction of raw milk (21, 22). Whether BFV can be transmitted via infected foodstuff to other species, including humans, is under investigation. It is also unknown whether BFV-related FVs are also present in other ruminants. Previous work, in fact, reported the presence of an FV-like virus in sheep (31).
In order to determine the replication and immunogenicity of BFV in its homologous host and in a related but different host, infection studies of calves and sheep were performed. Surprisingly, BFV spread and replicated to similar degrees in both the homologous and heterologous hosts. In addition, we did not identify consistent genetic changes of BFV upon replication for 3 years in sheep although signs of immune escape were detectable.
MATERIALS AND METHODS
Virus and cell cultures.
Canine fetal thymus cells infected with the Polish BFV100 isolate (Cf2Th/BFV100) (28) were used in this study. BFV100 was propagated by cocultivation of Cf2Th/BFV100 with uninfected Cf2Th cells maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 10% fetal calf serum (Sigma) and passaged at a ratio of 1:5 every week. Cf2Th/BFV100 cells were used for the inoculation of calves and sheep at the time of maximum syncytium formation. Four additional BFV isolates (isolates C1 to C4) used for sequence analysis were obtained from four Holstein-Friesian cows (6 to 10 years old) from two areas of Poland more than 250 km apart from each other (28).
Animals and experimental inoculations.
All animals used in this study were purchased from local suppliers and adapted to the housing and feeding conditions in the animal facilities of the National Veterinary Research Institute in Pulawy before the beginning of the study. The studies were approved by the local ethical commission and in accordance with national regulations for animal experimentation.
Six male calves of Holstein-Friesian breed (6 weeks old) seronegative for BFV, bovine leukemia virus (BLV), and bovine virus diarrhea virus (BVDV) were used. Four calves (calves 1 to 4) were inoculated intravenously with 5 × 106 Cf2Th/BFV100 cells while two calves (calves 5 and 6) were inoculated with the same amount of uninfected Cf2Th cells as controls. Additionally, eight Polish sheep of a long-wool breed (6 weeks old) that were seronegative for Maedi-Visna virus (MVV) were used. Prior to experimental inoculation, two sheep (sheep A and B) were intraperitoneally treated with 180 ml of sterile 4% aqueous sodium thioglycolate solution, as described previously, to stimulate macrophage formation, a procedure used to facilitate bovine immunodeficiency virus (BIV) infection of sheep (32). After 3 days, these sheep were inoculated intraperitoneally with 5 × 106 Cf2Th/BFV100 cells, and 4 weeks later BFV infection was confirmed by nested PCR and Gag, Env, and Bet enzyme-linked immunosorbent assays (ELISAs). At this time, the remaining six sheep were treated with sodium thioglycolate as described above. Three days later, 160 ml of whole blood from each of the two BFV-inoculated sheep was drawn into anticoagulant (EDTA-Na) and pooled. Eighty milliliters of the pooled blood was inoculated into each of four uninfected sheep (sheep 1 to 4) while the remaining two sheep (sheep 5 and 6) were used as controls and intraperitoneally (i.p.) inoculated with 80 ml of their own pooled blood. Throughout the experiment the control calves and sheep were kept isolated from the BFV-infected animals. Blood samples of experimentally inoculated animals, except two sheep which served as blood donors, were taken every 2 weeks up to 3 months postinoculation (p.i.). Hematology was carried out throughout this period. Calves and sheep were euthanized at 5 and 36 months p.i., respectively, and samples of blood, liver, spleen, lymph nodes, brain cortex, cerebellum, salivary glands, muscles, bone marrow, and lungs were collected and frozen at −70°C until DNA extraction.
Naturally infected cows.
Four Holstein-Friesian cows at the age of 5 to 7 years (cows 1 to 4), previously identified as seropositive for BFV, were slaughtered. Samples of blood, liver, spleen, lymph nodes, salivary glands, muscles, and lungs were collected from them. Since sample collection was performed prior to official sampling for bovine spongiform encephalopathy (BSE) examination, brain tissue was not available. All tissue samples were frozen at −70°C until DNA extraction.
Virus isolation.
For virus isolation, a total of 5 × 106 peripheral blood leukocytes (PBLs) from inoculated calves and sheep were cocultivated with 5 × 105 BFV-free Cf2Th cells (as determined by nested integrase [IN] PCR) which were maintained as described above in 25-cm2 flasks (Nunc). The nonadherent cells were removed after 24 h, and the cells were cultivated until development of multinucleated syncytia typical for infections with FVs. In some cases, indirect immunofluorescence (IIF) was performed to confirm the presence of BFV. For IIF, cells grown on glass coverslips were fixed in ice-cold 100% methanol and 0.02% EGTA and used directly for immunostaining. Polyclonal rabbit immune serum specific for recombinant BFV Gag (1:5,000 in phosphate-buffered saline [PBS] containing 3% bovine serum albumin [BSA]) was used as the primary antibody. Following 1 h of incubation, cells were washed three times for 10 min in 0.1% Tween 20 in PBS and incubated with Alexa-488-coupled anti-rabbit IgG diluted 1:2,000 in PBS with 3% BSA (Invitrogen). For nuclear staining, Hoechst 33342 (Sigma, Warsaw, Poland) was added at a 1:1,000 dilution. Cells were incubated for 1 h, washed as described above, and examined for BFV Gag-specific immunofluorescence.
Antibody detection.
A glutathione S-transferase (GST) capture ELISA was used to examine antibody responses to BFV proteins in sera of experimentally infected calves and sheep and naturally infected cows as described previously (21). In brief, 96-well microtiter plates (Thermo Labsystems, Dreieich, Germany) were coated with glutathione casein, blocked with 0.2% (wt/vol) casein and 0.05% (vol/vol) Tween 20 in PBS (blocking buffer), and then incubated with cleared Escherichia coli lysates at a concentration of 0.25 μg/μl (total lysate in blocking buffer) containing the GST tag or GST-X fusion proteins (where X is BFV-Gag, BFV-Bet, or BFV-Env). For preabsorption of GST-binding antibodies, all sera were incubated at a dilution of 1:100 in blocking buffer containing 2 μg/μl total lysate of a GST tag-expressing E. coli culture prior to application on the coated plates. Preabsorbed serum samples were incubated for 1 h at room temperature (RT) in the coated ELISA plate wells, washed, and incubated for 1 h at RT with protein G peroxidase conjugate as the secondary antibody (1:5,000; Sigma, Munich, Germany). Tetramethylbenzidine (TMB; Sigma, Warsaw, Poland) (0.1 mg per 1 ml of acetate buffer) was added as a substrate. The reaction was stopped with 1 M H2SO4 and read at 450 nm in a Multiskan ELISA reader (Thermo Labsystems). For each serum sample, absorbance of the GST tag was determined and subtracted from the absorbance with the GST-X protein to calculate the specific reactivity against the BFV antigens. Antibody levels were expressed as net absorbance values and optical density measurements at a wavelength (λ) of 450 nm (OD450) were done in duplicates. The threshold for each antigen was estimated as the mean plus 2 standard deviations (SD) calculated for the OD values obtained from sera of mock-infected animals.
DNA preparation.
Total DNA was extracted from 5 × 106 PBLs or 25 mg of the respective organs of experimentally inoculated calves and sheep as well as naturally infected cows (10 mg of spleen samples) and from cell cultures of BFV isolates (C1, C2, C3, C4, and BFV100) using a DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer's instructions. DNA concentration was measured spectrophotometrically using GeneQuant (GE Healthcare), and samples were stored at −20°C until used.
BFV DNA detection and quantification.
Quantitative PCR (qPCR) was used as described previously (22). The primers Int3 (TCCCGCCTAAAGCTGATAGA) and Int4 (CAAACCTGAAATGGCTTGGT) (Oligo IBB, Warsaw, Poland) were designed to match 100% of the American and Chinese BFV isolates (GenBank accession numbers U94514 and AY134750.1) and flank 241 bp of the pol gene region encoding the viral integrase. A QuantiTect SYBR green kit (Qiagen) was used for PCR amplification as recommended with the addition of 0.2 μM each primer and 0.5 μg of genomic DNA. Reactions were carried out in a 7500 Real-Time PCR System (Applied Biosystems) under the following conditions: initial incubation and polymerase (Pol) activation at 95°C for 15 min, denaturation at 94°C for 30 s, annealing at 54°C for 45 s, and elongation at 72°C for 1 min for 40 cycles. The reactions were done in duplicates. When the calculated SD exceeded 25%, the PCRs were obligatorily repeated. qPCR was standardized to a dilution series of a BFV Pol plasmid (100 to 104 copies of plasmid DNA per reaction mixture) which was run in parallel to each assay. The specificity of the PCR was confirmed by melting temperature (Tm) analysis of amplification products during the reaction.
Nested PCR was applied as a confirmatory test for BFV infection in blood leukocytes of experimentally inoculated calves and sheep, as well as in in vitro cell cultures. Primers Int 1 (TGGACAGGAGAGGAGAGGAA) and Int 2 (GCAGTGCCAGTGAGATGTGT) were used in the primary PCR, while in the nested PCR, primers Int 3 and Int 4 (the same as in qPCR) were used to amplify a 430-bp fragment and 241-bp fragment, respectively. The reaction mixture for both PCRs consisted of 2.5 U of DyNazyme II DNA polymerase (Thermo Scientific), 1× PCR buffer with 1.5 mM MgCl2, 0.1 mM each deoxynucleoside triphosphate (dNTP), and 0.2 μM each primer. One microgram of genomic DNA was used as the template in the primary PCR, while in the nested PCR, 1/10 of the primary mixture was used. Both PCRs were carried out in a TGradient thermocycler (Biometra). Standard PCR conditions were used for both rounds of amplification with the exception of an annealing temperature of 57°C in the primary PCR and 54°C in the nested reaction. Nested PCR products were separated in a 1% agarose gel and visualized by ethidium bromide staining.
PCR amplification, molecular cloning, and sequencing.
Nested PCR was performed using genomic DNA from PBLs of infected sheep at 36 months p.i. and DNA extracted from the Cf2Th/BFV100 cells (28) originally used as the inoculum. The following primers (MWG Biotech) within the bet, env surface (SU), and long terminal repeat (LTR) regions were used in this study: bet-1BS, CGGTTGGATCTCCACGAAGG; bet-1AS, TAGCTTGATGAGCCATCCTC; bet-2S, GCAAGAGTAGCCTGCAGACG; bet-2AS, GTCCAATTCTGGTGTTCCTC; BFV-LTR-1S, TTACTTGCCCGGAGGATTGG; BFV-LTR-1AS, TAGTGATCTGGAAGGTAAGC; BFV-LTR-2S, CTTATGGATGGAGCCTTATGG; BFV-LTR-2AS, CTTACCACAGCCTGGAAGTC; nEnv-S1, TGGACTCTAGTAGTCTCACC; nEnv-AS1, CTTAGAAAGCGTGGTAATGGC; nEnv-S2, TGTCATTAGAGGACTTCAGG; nEnv-AS2, TTGATTGTCCTGCTATCTGG. All primers were designed to match 100% of known BFV sequences (GenBank accession numbers U94514 and AY134750.1). The first reaction mixture for bet, env, and LTR amplification contained 2.5 U of Taq DNA polymerase (New England BioLabs), 1× PCR buffer with 1.5 mM MgCl2, 0.2 μM each primer, 0.1 mM dNTP mix (New England BioLabs), and 1 μg of genomic DNA. The temperature profile of the bet PCR was as follows: initial denaturation at 94°C for 3 min, denaturation at 94°C for 45 s, annealing at 54°C for 30 s, elongation at 72°C for 1 min, and final elongation at 72°C for 10 min. Nested amplification was performed under similar conditions using a 1/10 volume of the first PCR as the template. Initial PCRs for BFV env and LTR were performed using the same DNA as in the bet PCR and a similar temperature profile: initial denaturation at 94°C for 3 min, denaturation at 94°C for 45 s, annealing at 52°C for 45 s, elongation at 72°C for 2 min, and final elongation at 72°C for 10 min. Nested amplification was done under similar conditions, with a modified annealing temperature of 54°C for 45 s, using a 1/10 volume of the first PCR as the template. The resulting amplicons were analyzed on 1% agarose gels, cloned into the TOPO-TA vector (Invitrogen), and sequenced by GATC (Konstanz, Germany). The primary nucleotide sequences of available independent PCR clones of each sheep, the original Cf2Th/BFV100 cells, and four other Polish isolates of BFV were determined for env, bet, and the LTR. Multiple alignments of these clones were done with Geneious software (33).
Nucleotide sequence accession numbers.
Consensus nucleotide sequences of BFV isolates were deposited in the GenBank under the following accession numbers: for the LTR, JX308297, JX308296, JX308298, and JX308299; for bet, JX308300, JX308301, JX308302, and JX308303; for env, JX308292, JX308293, JX308294, and JX308295.
RESULTS
BFV infection of calves and sheep and clinical observations.
Four Holstein-Friesian calves (calves 1 to 4; 6 weeks old) were inoculated intravenously with 5 × 106 Cf2Th cells productively infected with BFV100 in order to study whether the cell culture-adapted Polish BFV100 isolate is replication competent in cattle and to determine the molecular, virological, and immunological parameters of BFV replication in its authentic host. Infection via BFV-infected cells was chosen since BFV is known to be highly cell associated, thus preventing classical infection with cell-free virus preparations (34). In parallel, two calves (calves 5 and 6) were inoculated with the same amount of uninfected Cf2Th cells as controls.
In a corresponding study, eight Polish sheep of a long-wool breed (6 weeks old) were used for experimental BFV infection in order to study BFV replication in a heterologous but closely related host species. For initial amplification of BFV in sheep, 5 × 106 Cf2Th/BFV100 cells were intraperitoneally inoculated into two sheep (donor sheep A and B). Four weeks later, when BFV infection was confirmed by PCR and ELISA, blood from BFV-infected sheep A and B was pooled and used for inoculation and long-term studies of BFV infection of sheep. For this purpose, four sheep (6 weeks old; sheep 1 to 4) were each inoculated with 80 ml of pooled infectious blood, while two other sheep were inoculated with 80 ml of their own pooled blood as negative controls (sheep 5 and 6).
All BFV-infected and uninfected animals (calves and sheep) were routinely checked for health status or signs of disease. No infection-related symptoms could be attributed to experimental BFV infection. Blood samples were taken every 2 weeks up to 3 months postinoculation (p.i.) from all animals except the two donor sheep. All animals remained healthy throughout the study period. There were no significant fluctuations in any of the hematological parameters. Leukocyte counts also remained within reference ranges in all animals. All calves and sheep were euthanized at 5 and 36 months p.i., respectively, and samples of blood, liver, spleen, lymph nodes, brain cortex, cerebellum, salivary glands, muscles, bone marrow, and lungs were collected.
Virus isolation.
BFV reisolation from infected calves and sheep was done by cocultivation of peripheral blood leukocytes (PBLs) with permissive Cf2Th cells (35). Samples were scored positive when FV-specific syncytia containing at least 8 nuclei were clearly detectable. Using this method, BFV was periodically rescued from calves and sheep in the interval between initiation of BFV infection and euthanasia of animals (Table 1). No restrictions due to the use of fetal calf serum as a cell culture medium supplement were observed. In Cf2Th cocultures derived from BFV-infected calves, typical syncytia indicative of BFV infection were clearly visible after the first subculture (days 4 and 5), while in cultures from sheep, this effect was not apparent until the second subculture (days 7 and 8). However, since formation of syncytia in cocultures with sheep PBLs was weak and could be misread as spontaneous syncytium-like structures of uninfected Cf2Th cells, selected cultures were assayed by indirect immunofluorescence using a BFV Gag-specific rabbit antiserum. In all cocultures from infected calves and sheep, BFV-specific signals indicative of BFV infection were observed (Fig. 1 demonstrates this in sheep 3). Uninfected control animals always scored negative in all assays.
Table 1.
Results of virus isolation from blood of BFV infected calves and sheep
| Virus isolation sample no.a | Relative CPE by animal and treatment:b |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calf |
Sheep |
|||||||||||
| Infected |
Mock infected |
Infected |
Mock infected |
|||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 1 | 2 | 3 | 4 | 5 | 6 | |
| 1 | +++ | ++ | ++ | +++ | − | − | + | + | + | + | − | − |
| 2 | ++ | ++ | ++ | ++ | − | − | + | + | + | + | − | − |
| 3 | + | + | + | + | − | − | + | + | + | + | − | − |
Virus isolation was performed at months 1, 2, and 3 p.i. for calves and at years 1, 2, and 3 p.i. for sheep. For both groups, samples 1, 2, and 3 correspond to the respective sampling period.
CPE, cytopathic effect, scored as follows: +++, strong; ++, medium; +, weak; −, none.
Fig 1.
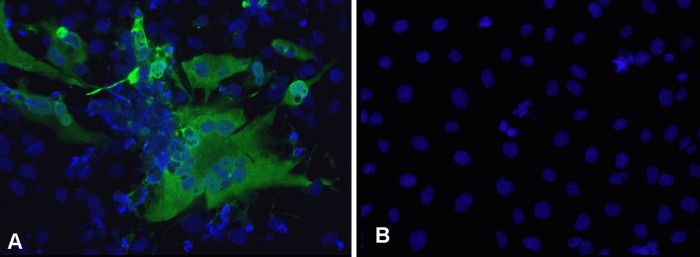
Detection of BFV Gag protein in cocultures by indirect immunofluorescence. Cf2Th cells cocultivated with PBLs of sheep 3 (A) and uninfected Cf2Th cells (B). Indirect immunofluorescence was done using rabbit BFV Gag-specific antiserum and anti-rabbit-Alexa-488. Nuclei were stained with Hoechst 33342.
BFV-specific seroreactivity of infected calves and sheep.
Sera from all BFV- and mock-inoculated calves and sheep as well as naturally infected cows were tested with a generic BFV GST ELISA (21) for antibodies against BFV Gag, Bet, and part of the Env leader protein (Elp) SU ectodomain.
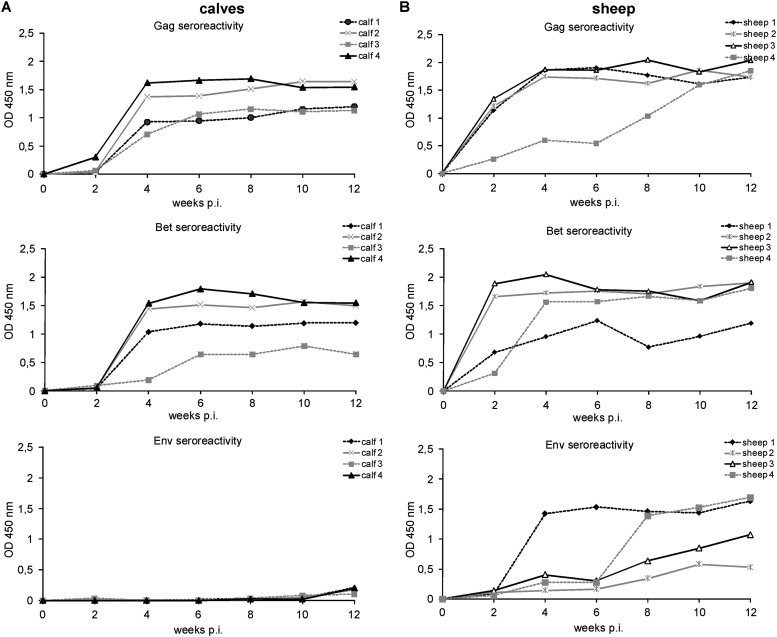
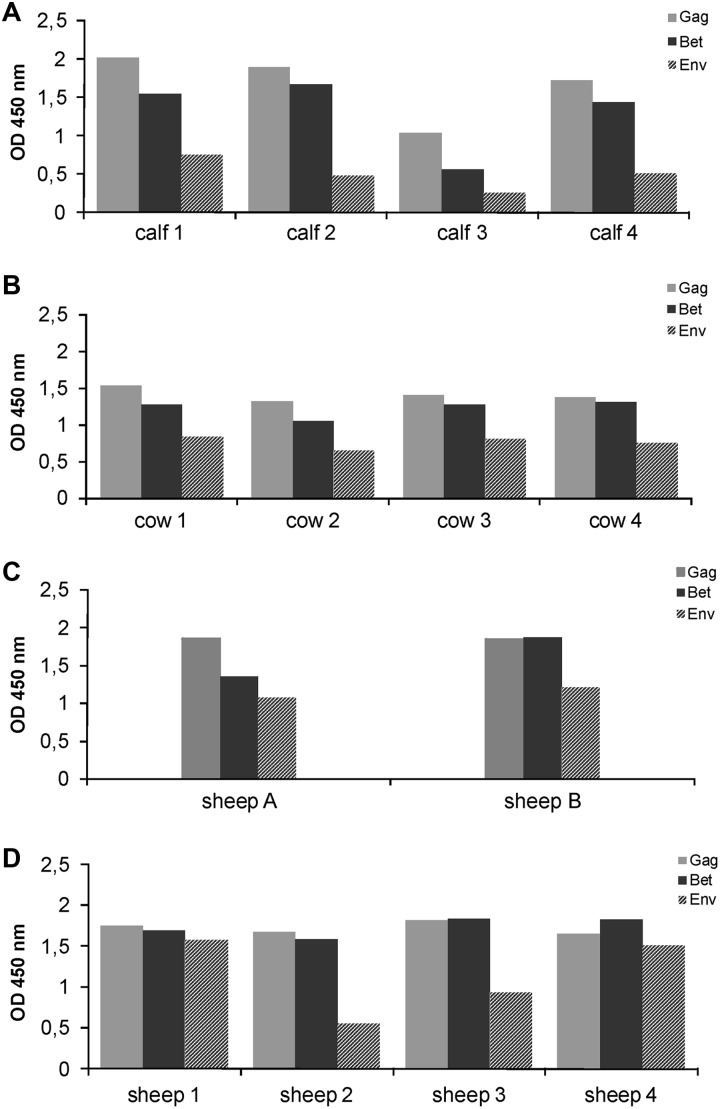
In all BFV-infected calves, Gag-specific antibodies were first detected at 4 weeks postinoculation (p.i.) (Fig. 2A), while Bet-specific antibodies appeared at 4 weeks p.i. in three animals and at 6 weeks p.i. in the remaining calf. Seroreactivity against Gag and Bet increased very rapidly in the first 4 to 6 weeks p.i. and was maintained at high levels until 12 weeks p.i., as usually seen in full-blown BFV infections of cattle (21). Env SU-specific antibodies were not observed in any animal until week 12. Additionally, serum samples of all inoculated calves tested again at the time of euthanasia showed high reactivity against Gag and Bet antigens but still only low Env SU-specific reactivity (Fig. 3A). To compare reactivity levels against BFV proteins between experimentally infected calves and naturally infected adult cattle, sera from four naturally infected dairy cows were tested by ELISA. Reactivity levels of these sera for Gag and Bet antigens were similar to those seen in the calves. Surprisingly, reactivity against Env was present in all four cows (Fig. 3B) but at clearly lower levels than Gag- and Bet-specific reactivity.
Fig 2.
Seroreactivity to BFV proteins in calves and sheep at the early stage of infection. The humoral response to BFV antigens was tested by GST ELISA during the first 12 weeks p.i. in four calves experimentally inoculated with Cf2Th/BFV100 (A) and in four sheep inoculated with blood of two donor sheep (B). Background-subtracted OD450 values are given.
Fig 3.
Seroreactivity to BFV proteins in calves, sheep, and cows upon euthanasia/slaughtering. The humoral response to BFV antigens was tested by GST ELISA at 5 months p.i. in four calves inoculated with Cf2Th/BFV100 (A) and in four cows naturally infected with BFV (B) and at 36 months p.i. in two donor sheep inoculated with Cf2Th/BFV100 (C) and four sheep inoculated with the blood of two donor sheep (D). Background-subtracted OD450 values are given.
In sheep inoculated with blood of BFV-infected animals A and B, antibodies specific for Gag were detected at 2 weeks p.i. in three sheep and at 4 weeks p.i. in one sheep, while antibodies specific to Bet protein appeared at 2 weeks p.i. in two animals and at 4 weeks p.i. in the other two (Fig. 2B). The reactivity of the serum increased very rapidly and was maintained at high levels until the end of experiment at 36 months p.i. In contrast to the experimentally infected calves, Env seroconversion occurred in all infected sheep early after infection. One sheep displayed BFV Env-specific antibodies already at 4 weeks p.i., while the remaining three animals seroconverted at 8 weeks p.i. The reactivity of sheep serum against BFV Env protein was lower than that against BFV Gag and Bet. Since Env seroreactivity differences observed in calves and sheep in the early phase of infection may be due to different inocula, we tested late-phase (at 36 months p.i.) serum samples collected from two sheep inoculated with Cf2Th/BFV100 (Fig. 3C) and four sheep inoculated with blood of BFV-infected animals (Fig. 3D). The reactivity of sera from all experimentally infected sheep at 36 months p.i. was mainly against Gag and Bet antigens. Env reactivity was also observed but, similar to results in naturally infected cows, varied between animals. Clearly, the reactivity levels of BFV-specific antibodies at the late phase of infection were similar in sheep infected with different inocula and were compatible with levels observed in sheep early after infection as well as in naturally infected cows. Mock-infected calves and sheep did not show reactivity against any of the BFV antigens.
Detection of BFV DNA genomes and quantification of proviral loads.
Detection and quantification of integrated and free BFV DNA in samples collected from BFV- and mock-inoculated calves and sheep were done by qPCR and nested PCR specific for sequences within the BFV integrase (IN) domain (22). Genomic DNA extracted from calf PBLs collected at biweekly intervals during the first 12 weeks of the experiment as well as DNA from different tissues sampled at 5 months p.i. (liver, spleen, lymph nodes, brain cortex, cerebellum, salivary gland, muscle, thymus, bone marrow, and lung) was analyzed by both PCR tests. Sheep genomic DNA extracted from PBLs early during infection, from different tissues collected 3 years p.i., and from cows naturally infected with BFV was analyzed by qPCR only.
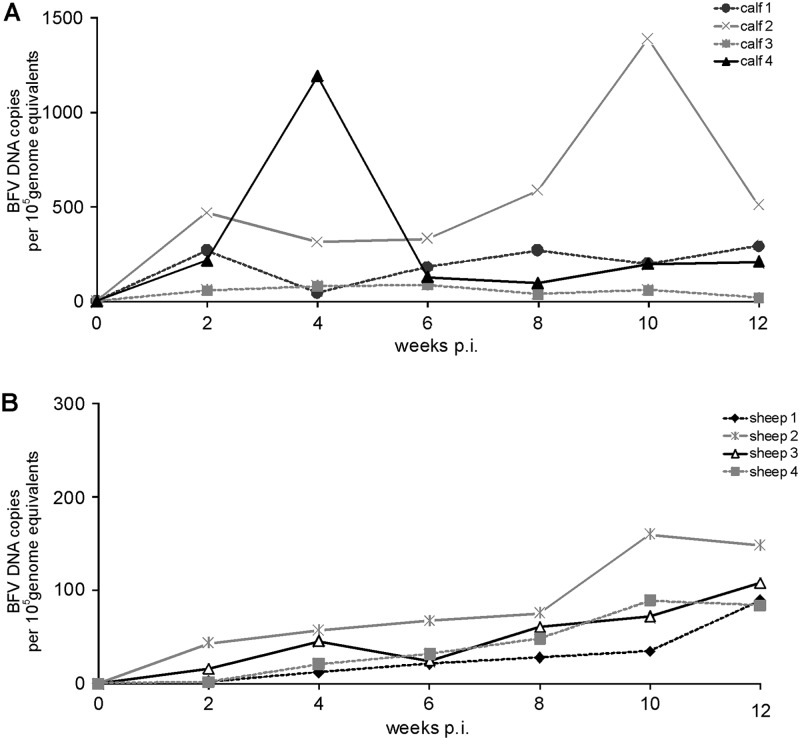
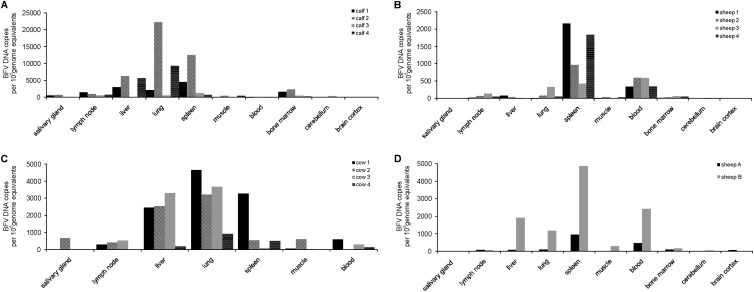
In PBLs of all experimentally infected calves, BFV DNA was detected by qPCR starting at 2 weeks p.i. (Fig. 4A). During the following weeks, viral DNA load in calf PBLs ranged from 20 to almost 1,400 copies of BFV DNA per 100,000 genome equivalents and varied between individual animals. No amplification of BFV DNA was observed in control samples of mock-infected animals. Furthermore, qPCR with different tissues demonstrated the presence of BFV DNA in all tissues except brain cortex (Fig. 5A; note that different scales were used to present the BFV DNA copy numbers in the graphs). High copy numbers of BFV DNA were detected in lung, spleen, PBLs, liver, lymph nodes, and bone marrow; however, the variability among the four calves was significant. The highest copy numbers of BFV DNA were observed in lung, at 22,250 copies per 100,000 genome equivalents in calf 2 and over 9,000 in calf 4, and spleen, at about 12,500 copies per 100,000 genome equivalents in calf 2. No viral DNA was detectable in PBLs from mock-inoculated animals (data not shown). BFV DNA detection by qPCR was also performed with DNA extracted from PBLs of the four BFV-inoculated sheep collected in biweekly intervals during the first 12 weeks and from different tissues taken at 36 months p.i. (liver, spleen, lymph nodes, brain cortex, cerebellum, salivary gland, muscles, thymus, bone marrow, and lungs). Similar to the BFV-infected calves, the variation of BFV copy numbers in sheep tissues late after experimental infection was significant. The results of these assays showed initially (2 weeks p.i.) the presence of very low copy numbers of viral DNA in leukocytes of all four BFV-inoculated sheep, with an average of 15 copies per 100,000 genome equivalents (Fig. 4B). The BFV viral load increased slightly starting at 4 weeks p.i. in all four sheep until 12 weeks p.i. Viral load of the BFV-infected sheep determined at 36 months p.i. by qPCR with different tissues demonstrates the presence of BFV DNA in almost all samples tested. BFV was poorly detectable in cerebellum and brain cortex and undetectable in salivary glands (Fig. 5B). The highest copy number of BFV DNA was observed in spleen, where it reached about 2,200 copies per 100,000 genome equivalents in sheep 1 and over 1,800 copies per 100,000 genome equivalents in sheep 4. High copy numbers of BFV DNA were also observed in PBLs, liver, and lung. Interestingly, similar patterns of BFV DNA load were observed in organs of the two donor sheep inoculated with Cf2Th/BFV100. The highest viral load was observed in spleen (almost 4,000 copies per 100,000 genome equivalents), blood, liver, and lung (Fig. 5D). No amplification of BFV DNA was observed in control samples of mock-inoculated animals (data not shown).
Fig 4.
BFV DNA load in PBLs of calves and sheep at the early phase of infection. The load of BFV DNA was determined by qPCR in PBLs of four calves experimentally inoculated with Cf2Th/BFV100 (A) and four sheep experimentally inoculated with blood of donor sheep (B) during the first 12 weeks p.i. Note that different scales to present the BFV DNA copy numbers have been used in the two panels.
Fig 5.
BFV DNA load in tissues of claves, sheep, and cows. The load of BFV DNA was determined by qPCR at the end of experiment in organs of four calves experimentally inoculated with Cf2Th/BFV100 (A), four sheep experimentally inoculated with the blood of donor sheep (B), four cows naturally infected with BFV (C), and two donor sheep experimentally inoculated with Cf2Th/BFV100 (D). Note that different scales to present the BFV DNA copy numbers have been used in the graphs.
To study the situation in presumably long-term-BFV-infected cattle, viral copy numbers in four outgrown, naturally BFV-infected dairy cattle (cows 1 to 4) were analyzed (Fig. 5C). BFV copy numbers detected in PBLs of all four cows were similar to those obtained from long-term experimentally infected sheep. However, the viral load observed in all other cattle tissues was clearly higher than in sheep. Interestingly, the distribution of viral DNA observed in naturally infected cows reflects the situation seen in experimentally inoculated calves, especially in the lung, spleen, and liver. The highest copy number of BFV DNA was observed in lung, where it reached almost 4,700 copies per 100,000 genome equivalents in cow 1 and almost 3,700 copies per 100,000 genome equivalents in cow 3. Furthermore, the detection of BFV DNA in salivary glands was possible only in two cows.
Analysis of BFV sequences in experimentally infected sheep and naturally infected cattle.
To molecularly characterize the genetic variability of BFV within cattle and after experimental transmission to a heterologous host, sequences from three regions of the BFV genome were analyzed. The bet gene was chosen as FV Bet is the viral antagonist of cellular antiviral APOBEC3 cytidine deaminase restriction factors and likely subjected to host-specific adaptations (36). The SU region of env was analyzed since Env is the main viral protein exposed to the host's immune system and thus prone to escape mutations, as known for other retroviruses. Although, in contrast to orthoretroviruses, FV Env is well conserved between different primate and nonprimate FVs, SU antigenic regions tend to show a considerable degree of variability, even including serotype specificity in FFV (37, 38, 39). Finally, we analyzed the long terminal repeat (LTR) region since it is known to modulate retroviral gene expression and determine cell tropism, as was shown for BLV and Maedi-Visna virus (40, 41). The initial PCR yielded fragments of 1.2 to 1.8 kb, either encompassing the complete bet gene, the 3′ part of env SU, or almost the complete LTR. In the respective nested PCRs, DNA fragments of 569 bp for bet, 874 for the LTR, and 915 for env were amplified for DNA fragment purification, cloning, sequencing, and bioinformatics. As the template, total genomic DNA extracted 3 years p.i. from inoculated sheep was used. To compare genomic changes after experimental transmission into sheep with the intrinsic genetic variability of BFV in its natural host, cell culture-adapted BFV isolates obtained from cattle in the late 1990s from different parts of Poland were analyzed. Isolates C3 (65206) and C4 (42357) are from the same farm, and isolate C1 (80195) also comes from northeast Poland. Isolate C2 (99907) and the Polish reference isolate BFV100 (28) are from central Poland, more than 250 km away from the other sampling site. In most cases, several independent clones of each sheep or cattle BFV isolate per PCR were analyzed.
In most of the sheep-derived clones, only very few mutations per clone were noted in all analyzed genomic regions (Table 2). Even the lowest mutation rate values (0.06 to 0.38% for bet and 0.08 to 0.55% for env) were higher than those due to the Taq DNA polymerase errors (0.028%). The highest rates of mutations were observed in the LTR region (0.49 to 0.71%) but did not affect any known regulatory sequence/motif (42). Point mutations observed in two out of four bet clones of sheep 2 resulted in frameshift mutations. Similarly, in all env clones derived from sheep 2, single-nucleotide deletions (2/3 clones) and an insertion (1/3 clones) led to frameshift mutations and truncation of the Env protein. Although about 70% and 40% of the single-nucleotide mutations in the sheep-derived BFV bet and env sequences, respectively, led to changes in the amino acid sequences, there was no consistent pattern of adaptive changes detectable.
Table 2.
Frequency of nucleotide and amino acid mutations in BFV isolates from infected sheep
| Genomic region | Sheep no. | No. of clones tested | Avg no. of nucleotide mutations per clone | Avg no. of amino acid changes per clone | Mutation rate (% of nucleotide sequence) |
|---|---|---|---|---|---|
| bet | 1 | 3 | 1.25 | 0.8 | 0.13 |
| 2 | 4 | 3.6 | 2.2a | 0.38 | |
| 3 | 5 | 1.25 | 1.0 | 0.13 | |
| 4 | 5 | 0.6 | 0.6b | 0.06 | |
| LTR | 1 | 1 | 5.0 | NCc | 0.58 |
| 2 | 3 | 4.3 | NC | 0.49 | |
| 3 | 4 | 6.25 | NC | 0.71 | |
| 4 | 1 | 6.0 | NC | 0.69 | |
| env | 1 | 3 | 1.7 | 0.3 | 0.18 |
| 2 | 3 | 5 | 2.0a | 0.55 | |
| 3 | 3 | 0.7 | 0.7 | 0.08 | |
| 4 | 2 | 4 | 1.5 | 0.44 |
Frameshift mutation.
Nonsense mutation.
NC, noncoding.
The analysis of the four different BFV isolates from Polish cattle compared to BFV100 showed rather random polymorphic changes in env clones of all isolates. In the LTR, however, similar patterns of mutation among the C1, C3, and C4 isolates were observed. Furthermore, the LTR region was much more variable in all clones than the two other analyzed genomic regions (Table 3). There were at least five polymorphic sites present in this region in almost all clones of isolates C1, C3, and C4. Additionally, a single pattern of polymorphism (A to G) in a short fragment of 569 nucleotides (nt) from bet was also observed at position 92 only in isolates C1, C3, and C4. This result showed a clear correlation between geographic origin of the isolates and the pattern of genomic changes and may be a result of genetic alterations in local BFV strains as mutations were not detected in isolate C2 or the reference Polish isolate BFV100 (GenBank accession number JX307861) (43).
Table 3.
Frequency of nucleotide and amino acid mutations in BFV isolates from infected cows
| Genomic region | Isolate no. | No. of clones tested | Avg no. of nucleotide mutations per clone | Avg no. of amino acid changes per clone | Mutation rate (% of nucleotide sequence) |
|---|---|---|---|---|---|
| bet | C1 | 5 | 1.0 | 1.0 | 0.17 |
| C2 | 4 | 2 | 1.25 | 0.35 | |
| C3 | 4 | 3 | 2.6 | 0.53 | |
| C4 | 5 | 8.8 | 6.2 | 1.55 | |
| LTR | C1 | 4 | 17.25 | NCc | 1.97 |
| C2 | 3 | 4 | NC | 0.46 | |
| C3 | 2 | 17.5 | NC | 2.01 | |
| C4 | 3 | 16.75 | NC | 1.92 | |
| env | C1 | 5 | 2.2 | 2.2b | 0.25 |
| C2 | 5 | 3 | 1.8a | 0.33 | |
| C3 | 5 | 3.6 | 2.8a,b | 0.40 | |
| C4 | 3 | 4 | 2.3 | 0.45 |
Frameshift mutation.
Nonsense mutation.
NC, noncoding.
DISCUSSION
To determine the molecular and virological parameters of BFV replication in natural and heterologous hosts, infection of calves and sheep with BFV-infected Cf2Th cells or blood from BFV-infected sheep was performed. Both modes of infection resulted in persistent infections of calves and sheep, showing that cell culture-adapted BFV100 has maintained the capacity of replicating in its authentic and heterologous hosts. Establishment of productive BFV infections in calves and sheep was confirmed by virus isolation from leukocytes of all infected animals. While full BFV replication and reisolation of replication-competent virus from experimentally infected calves was anticipated, reisolation of BFV from infected sheep at 3 years p.i. clearly confirms for the first time that BFV can also replicate in a heterologous host species, as previously observed for other FVs (44, 45, 46, 47). In addition, BFV was rescued from both infected animal hosts even in the presence of BFV-specific antibodies, confirming that BFV infection is not cleared by the host immune system. Additional parameters of BFV infection, such as the humoral immune response to BFV proteins and presence of BFV DNA in animal blood and organs, were determined and confirmed the persistence of BFV in both hosts. Experimental BFV infection of neither calves nor sheep resulted in pathology. Surprisingly, not even changes in hematological parameters, typical for viral infections, were observed. However, we cannot exclude the possibility that, due to the setup of sample collection, we missed transient fluctuations, for instance, of lymphocyte counts. Similarly, SFV-infected primates do not develop overt signs of pathology (48, 49). Although some studies of pathogenicity in heterologous hosts have shown transient immunodeficiency following SFV and prototypic foamy virus (PFV) infections of rabbits (45) and mice (16), it was not confirmed by others (16, 44). However, central nervous system malfunction and muscle degeneration leading to death were reported in transgenic mice expressing the PFV accessory proteins under the control of the viral LTR promoter (50).
Although infection patterns measured by virus isolation, BFV DNA load, and humoral immune response to BFV proteins were similar in both experimental models, distinguishing features of homologous and heterologous host infection were clearly observed. They may be the result of an individual condition of the infected animal or the effect of genetic divergence between both host species; however, other factors cannot be excluded, and the different modes of inoculation may also have played a role.
Antibodies against BFV proteins were observed in all experimentally infected animals. Seroconversion occurred at 4 weeks p.i. in calves and sheep inoculated with culture-grown virus while it happened at 2 weeks p.i. in most sheep inoculated with infectious blood. Surprisingly, sheep, as a heterologous model genetically distant from the natural host, developed stronger serological responses to BFV. This discordance could be the result of different inoculation modes (51), and perhaps a higher dose of the virus was injected in sheep with BFV-infected blood. At the early phase of infection (until week 12 p.i.), the humoral response in calves was directed only against Gag and Bet, while seroreactivity to Env was observed at low levels at 5 months p.i. In contrast, in sheep, seroreactivity against Env was observed also early after infection but clearly later than that against Gag and Bet. Moreover, reduced antibody reactivity against Env was observed in sheep 2 which may be due to the appearance of frameshift mutations in env sequence of sheep 2, resulting in the production of truncated and thus unstable proteins. Remarkably, previous experimental studies also involving heterologous hosts of FV infections, such as rabbits, also reported the induction of Env-specific antibodies (44). Furthermore, in four naturally BFV-infected cows involved in our study, seroreactivity against Env was observed but was clearly weaker than that against the other viral antigens used. This is in line with previous findings that reactivity against Env is detectable only in a fraction of BFV-infected cows (21). Some differences may also be explained by using a bacterially expressed GST-Elp-SU fusion protein with the altered conformation lacking important protein modifications. On the other hand, these differences in seroreactivities may be caused by host-specific differences to control BFV replication. Presumably, BFV is better suppressed in calves than in sheep, resulting in the observed pattern in Env-specific seroreactivity. In addition, our current findings confirm the diagnostic value of Gag and Bet as antigens for BFV serodiagnostics, as previously shown also for FFV (21, 52, 53).
BFV DNA was found in almost all tissues of infected animals, concurring with reports of primate FVs in natural and experimental hosts (44, 47, 48, 49). Although BFV DNA was detectable in blood leukocytes of all infected calves and sheep starting at 2 weeks p.i., a gradual increase in the viral load was subsequently detectable in calves, while in sheep viral load increased much more slowly. The highest BFV DNA load was observed in lungs, liver, and spleen of experimentally inoculated calves at the early phase p.i., while in long-term-infected sheep and naturally infected cows, much lower overall DNA loads were detected. Detection of FV DNA in a particular tissue does not necessarily indicate active virus replication since alveolar macrophages and areas of lymphocyte aggregation or lymphoid proliferation are known to act as latent reservoirs of other retroviruses like Maedi-Visna virus or human immunodeficiency virus (HIV) (54, 55, 56, 57). Furthermore, since lymphocytes have been previously identified as targets of FV infection (58), the presence of viral DNA in lungs can be explained by BFV-infected lymphocyte and monocyte-derived macrophage infiltration or by high vascularization and an enrichment of infected leukocytes. This phenomenon was previously reported by histological examination of lung tissues from cats experimentally infected with FFV (59). Similarly, the high BFV load in spleen, liver, and lymph nodes may be due to the high number of lymphocytes in these organs (59). Surprisingly, in contrast to the results from cats infected with FFV, our study shows that BFV load can be detected in bone marrow of all calves and sheep. The low level of BFV DNA in the other tissues may suggest BFV latency without virus replication in non-lymphocyte-associated organs. Since we tested only for the presence of BFV DNA, it is thus not possible to distinguish sites of active BFV replication from those of proviral latency without viral gene expression.
Strikingly, we noted variations of BFV DNA loads between the same organs of homologous and heterologous hosts. In calves and cows, the highest BFV DNA load was noted in lung, spleen, and liver, while the most loaded organs in sheep were the spleen and blood. This may be related to the host but not to the inoculation mode since similar patterns were observed in sheep inoculated by infected blood and infected Cf2Th cells. Furthermore, quite high interindividual variability, especially concerning the BFV load, was clearly observable in all animal groups. Such individual variations between the animals cannot be resolved in our small-scale study, and it is thus impossible to draw more specific conclusions.
Most interesting, there seems to be a link between BFV DNA load and BFV seroreactivity in calves. Interestingly, the calves with the initial transient increase in BFV viral loads (calves 2 and 4) were those with the highest postmortem viral loads (in lung and liver) and the strongest Gag and Bet reactivity levels. Furthermore, it seems that in calf 4, an initial burst of BFV load was quickly controlled by the immune system since the appearance of Gag-specific antibodies 4 weeks p.i. clearly correlated with a decrease in the BFV DNA load. However, immune control of BFV may not be stable since calf 2 failed to control BFV DNA load in the blood at 10 weeks p.i.
To test whether infection of sheep as a heterologous host leads to adaptative changes of the BFV genome, BFV sequences obtained from sheep at 36 months p.i. were compared to the original virus used as the inoculum. The characterization of the genetic variability of bet, env, and the LTR showed only limited sequence variation, which was, however, clearly higher than that artificially generated by Taq DNA polymerase. Analysis of the LTR sequences showed that this was the most variable region in sheep-derived clones, but similar to bet and env variability, no conserved pattern of mutations was observed in the regions analyzed. However, the enrichment of nonsynonymous changes in coding sequences may be indicative of BFV adaptation. Changes in Bet, which counteracts host antiviral proteins, might help the virus to survive or increase replication in a new environment (12, 60).
The majority of mutations resulting in amino acid changes in bet were observed in sheep 2, which also carried loss-of-function frameshift mutations in env. Sheep 2, in fact, showed the lowest Env reactivity and increased BFV DNA load in blood at week 10 p.i., suggesting that these changes may be responsible for immune escape or that they led to an optimal adaptation to the new host. Since BFV was reisolated from this sheep after 3 years, replication-competent and replication-deficient BFV coexisted in this animal. Further studies including experimental infection for longer periods of time and deep-sequencing analysis of the virus quasi-species within the animals will be required to unravel the mechanisms of BFV persistence and escape from innate/intrinsic and adaptive immune control.
The low-level genetic changes of BFV seen in long-term-infected sheep are in contrast to the detection of geographic variants of BFV in Poland. The field isolates were from aged cows naturally infected with BFV, presumably for an extended period of time. Similar to findings in sheep-derived clones, the LTR sequences analyzed were also the most variable regions of BFV field isolates. Furthermore, there were clear polymorphic sites observed in both the LTR and bet. Almost 80% of the mutations in Bet resulted in amino acid changes indicative of positive, adaptive evolution of this protein. Additionally, conserved polymorphism in the LTR and bet in BFV isolates from the same geographic region may suggest the presence of defined local geographic variants. However, env sequences did not reveal geographic clustering, and, thus, it is highly unlikely that these variants represent distinct serotypes which have been described and identified for SFVs and FFV (37, 38, 48, 61, 62, 63).
Our findings, which confirm the ability of BFV to establish persistent infection not only in its natural host but also in heterologous animals, are in line with previous studies on SFV or FFV and show the utility of sheep as an animal model to study the biology of spumavirus infection (47). Furthermore, the results clearly demonstrate the capacity of BFV to productively replicate in heterologous hosts. In cases of cross-species transmissions, such events can dramatically increase virulence and pathogenicity, as demonstrated for HIV in the human population. However, studies on FV infection as a result of interspecies transmission (5, 6, 7, 19) have not yet confirmed changes in virus pathogenicity. Nonetheless, the risk of disease development due to adaptation to the new host cannot be formally excluded for any FV. Since sheep are livestock animals, the new animal model established in this study might also be an attractive model for evaluating and optimizing novel FV vector-based gene transfer strategies.
ACKNOWLEDGMENTS
This study was funded by grant 2PO6K04327 from the Polish Ministry of Science and Computerization to J.K. and by grant P23/08//A81/07 from the Else Kröner-Fresenius Stiftung to M.L.
We greatly appreciate Bozena Krawczak (NVRI) for excellent technical assistance, Lutz Gissmann (DKFZ) for the continuous support of this study, and Janet Lei (DKFZ) for constructive proofreading.
Footnotes
Published ahead of print 16 January 2013
REFERENCES
- 1. Meiering CD, Linial ML. 2001. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 14:165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Saïb A. 2003. Non-primate foamy viruses. Curr. Top. Microbiol. Immunol. 277:197–211 [DOI] [PubMed] [Google Scholar]
- 3. Switzer WM, Salemi M, Shanmugam V, Gao F, Cong ME, Kuiken C, Bhullar V, Beer BE, Vallet D, Gautier-Hion A, Tooze Z, Villinger F, Holmes EC, Heneine W. 2005. Ancient co-speciation of simian foamy viruses and primates. Nature 434:376–380 [DOI] [PubMed] [Google Scholar]
- 4. Schweizer M, Falcone V, Gange J, Turek R, Neumann-Haefelin D. 1997. Simian foamy virus isolated from an accidentally infected human individual. J. Virol. 71:4821–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wolfe ND, Switzer WM, Carr JK, Bhullar VB, Shanmugam V, Tamoufe U, Prosser AT, Torimiro JN, Wright A, Mpoudi-Ngole E, McCutchan FE, Birx DL, Folks TM, Burke DS, Heneine W. 2004. Naturally acquired simian retrovirus infections in central African hunters. Lancet 363:932–937 [DOI] [PubMed] [Google Scholar]
- 6. Jones-Engel L, Engel GA, Schillaci MA, Rompis A, Putra A, Suaryana KG, Fuentes A, Beer B, Hicks S, White R, Wilson B, Allan JS. 2005. Primate-to-human retroviral transmission in Asia. Emerg. Infect. Dis. 11:1028–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calattini S, Betsem EB, Froment A, Mauclere P, Tortevoye P, Schmitt C, Njouom R, Saïb A, Gessain A. 2007. Simian foamy virus transmission from apes to humans, rural Cameroon. Emerg. Infect. Dis. 13:1314–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Betsem E, Rua R, Tortevoye P, Froment A, Gessain A. 2011. Frequent and recent human acquisition of simian foamy viruses through apes' bites in central Africa. PLoS Pathog. 7:e1002306 doi:10.1371/journal.ppat.1002306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mouinga-Ondémé A, Caron M, Nkoghé D, Telfer P, Marx P, Saïb A, Leroy E, Gonzalez J-P, Gessain A, Kazanji M. 2012. Genetic diversity and evolution cross-species transmission of simian foamy virus to humans in rural Gabon, Central Africa. J. Virol. 86:1255–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Callahan ME, Switzer WM, Matthews AL, Roberts BD, Heneine W, Folks TM, Sandstrom PA. 1999. Persistent zoonotic infection of a human with simian foamy virus in the absence of an intact orf-2 accessory gene. J. Virol. 73:9619–9624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rua R, Betsem E, Calattini S, Saib A, Gessain A. 2012. Genetic characterization of simian foamy viruses infecting humans. J. Virol. 86:13350–13359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Löchelt M, Romen F, Bastone P, Muckenfuss H, Kirchner N, Kim YB, Truyen U, Rösler U, Battenberg M, Saib A, Flory E, Cichutek K, Münk C. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. U. S. A. 102:7982–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neil S, Bieniasz P. 2009. Human immunodeficiency virus, restriction factors, and interferon. J. Interferon Cytokine Res. 29:569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Perković M, Schmidt S, Marino D, Russell RA, Stauch B, Hofmann H, Kopietz F, Kloke BP, Zielonka J, Ströver H, Hermle J, Lindemann D, Pathak VK, Schneider G, Löchelt M, Cichutek K, Münk C. 2009. Species-specific inhibition of APOBEC3C by the prototype foamy virus protein Bet. J. Biol. Chem. 284:5819–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zielonka J, Marino D, Hofmann H, Yuhki N, Löchelt M, Münk C. 2010. Vif of feline immunodeficiency virus from domestic cats protects against APOBEC3 restriction factors from many felids. J. Virol. 84:7312–7324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Santillana-Hayat M, Rozain F, Bittoun P, Chopin-Robert C, Lasneret J, Peries J, Canivet M. 1993. Transient immunosuppressive effect induced in rabbits and mice by the human spumaretrovirus prototype HFV (human foamy virus). Res. Virol. 144:389–396 [DOI] [PubMed] [Google Scholar]
- 17. Murray SM, Picker LJ, Axthelm MK, Linial ML. 2006. Expanded tissue targets for foamy virus replication with simian immunodeficiency virus-induced immunosuppression. J. Virol. 80:663–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bastone P, Truyen U, Löchelt M. 2003. Potential of zoonotic transmission of non-primate foamy viruses to humans. J. Vet. Med. B Infect. Dis. Vet. Public Health 50:417–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Switzer WM, Bhullar V, Shanmugam V, Cong ME, Parekh B, Lerche NW, Yee JL, Ely JJ, Boneva R, Chapman LE, Folks TM, Heneine W. 2004. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J. Virol. 78:2780–2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rethwilm A. 2007. Foamy virus vectors: an awaited alternative to gammaretro- and lentiviral vectors. Curr. Gene Ther. 7:261–271 [DOI] [PubMed] [Google Scholar]
- 21. Romen F, Backes P, Materniak M, Sting R, Vahlenkamp TW, Riebe R, Pawlita M, Kuzmak J, Löchelt M. 2007. Serological detection systems for identification of cows shedding bovine foamy virus via milk. Virology 364:123–131 [DOI] [PubMed] [Google Scholar]
- 22. Materniak M, Sieradzki Z, Kuźmak J. 2010. Detection of bovine foamy virus in milk and saliva of BFV seropositive cattle. Bull. Vet. Inst. Pulawy 54:461–465 [Google Scholar]
- 23. Lindemann D, Rethwilm A. 2011. Foamy virus biology and its application for vector development. Viruses 3:561–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Woode GN. 1972. Isolation of bovine syncytial virus in Britain. Vet. Rec. 91:363. [DOI] [PubMed] [Google Scholar]
- 25. Clarke JK, McFerran JB, Nelson RT. 1973. The isolation of a strain of bovine syncytial virus in Northern Ireland. Res. Vet. Sci. 14:117–119 [PubMed] [Google Scholar]
- 26. Greig AS. 1979. A syncytium regression test to detect antibodies to bovine syncytial virus. Can. J. Comp. Med. 43:112–114 [PMC free article] [PubMed] [Google Scholar]
- 27. Jacobs RM, Pollari FL, McNab WB, Jefferson B. 1995. A serological survey of bovine syncytial virus in Ontario: associations with bovine leukemia and immunodeficiency-like viruses, production records, and management practices. Can. J. Vet. Res. 59:271–278 [PMC free article] [PubMed] [Google Scholar]
- 28. Materniak M, Bicka L, Kuźmak J. 2006. Isolation and partial characterization of bovine foamy virus from Polish cattle. Pol. J. Vet. Sci. 9:207–211 [PubMed] [Google Scholar]
- 29. Malmquist WA, van der Maaten MJ, Boothe AD. 1969. Isolation, immunodiffusion, immunofluorescence and electron microscopy of a syncytial virus of lymphosarcomatous and apparently normal cattle. Cancer Res. 29:188–200 [PubMed] [Google Scholar]
- 30. Gould EA, Allan GM, Logan EF, McFerran JB. 1978. Detection of antibody to bovine syncytial virus and respiratory syncytial virus in bovine fetal serum. J. Clin. Microbiol. 8:233–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Flanagan M. 1992. Isolation of a spumavirus from a sheep. Aust. Vet. J. 69:112–113 [DOI] [PubMed] [Google Scholar]
- 32. Jacobs RM, Smith HE, Whetstone CA, Suarez DL, Jefferson B, Valli VE. 1994. Haematological and lymphocyte subset analyses in sheep inoculated with bovine immunodeficiency-like virus. Vet. Res. Commun. 18:471–482 [DOI] [PubMed] [Google Scholar]
- 33. Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson RH, Oginnusi AA, Ladds PW. 1983. Isolations and serology of bovine spumavirus. Aust. Vet. J. 60:147. [DOI] [PubMed] [Google Scholar]
- 35. Renshaw RW, Gonda MA, Casey JW. 1991. Structure and transcriptional status of bovine syncytial virus in cytopathic infections. Gene 105:179–184 [DOI] [PubMed] [Google Scholar]
- 36. Münk C, Hechler T, Chareza S, Löchelt M. 2010. Restriction of feline retroviruses: lessons from cat APOBEC3 cytidine deaminases and TRIM5alpha proteins. Vet. Immunol. Immunopathol. 134:14–24 [DOI] [PubMed] [Google Scholar]
- 37. Winkler IG, Flügel RM, Löchelt M, Flower RL. 1998. Detection and molecular characterisation of feline foamy virus serotypes in naturally infected cats. Virology 247:144–151 [DOI] [PubMed] [Google Scholar]
- 38. Zemba M, Alke A, Bodem J, Winkler IG, Flower RL, Pfrepper K, Delius H, Flügel RM, Löchelt M. 2000. Construction of infectious feline foamy virus genomes: cat antisera do not cross-neutralize feline foamy virus chimera with serotype-specific Env sequences. Virology 266:150–156 [DOI] [PubMed] [Google Scholar]
- 39. Lindemann D, Goepfert PA. 2003. The foamy virus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 277:111–129 [DOI] [PubMed] [Google Scholar]
- 40. Agnarsdóttir G, Thorsteinsdóttir H, Oskarsson T, Matthíasdóttir S, Haflidadóttir BS, Andrésson OS, Andrésdóttir V. 2000. The long terminal repeat is a determinant of cell tropism of maedi-visna virus. J. Gen. Virol. 81:1901–1905 [DOI] [PubMed] [Google Scholar]
- 41. Zhao X, Jimenez C, Sentsui H, Buehring GC. 2007. Sequence polymorphisms in the long terminal repeat of bovine leukemia virus: evidence for selection pressures in regulatory sequences. Virus Res. 124:113–124 [DOI] [PubMed] [Google Scholar]
- 42. Renshaw RW, Casey JW. 1994. Analysis of the 5′ long terminal repeat of bovine syncytial virus. Gene 141:221–224 [DOI] [PubMed] [Google Scholar]
- 43. Hechler T, Materniak M, Kehl T, Kuźmak J, Löchelt M. 2012. Complete genome sequences of two novel European clade bovine foamy viruses from Germany and Poland. J. Virol. 86:10905–10906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saïb A, Neves M, Giron ML, Guillemin MC, Valla J, Peries J, Canivet M. 1997. Long-term persistent infection of domestic rabbits by the human foamy virus. Virology 228:263–268 [DOI] [PubMed] [Google Scholar]
- 45. Hooks JJ, Detrick-Hooks B. 1979. Simian foamy virus-induced immunosuppression in rabbits. J. Gen. Virol. 44:383–390 [DOI] [PubMed] [Google Scholar]
- 46. Brown P, Moreau-Dubois MC, Gajdusek DC. 1982. Persistent asymptomatic infection of the laboratory mouse by simian foamy virus type 6: a new model of retrovirus latency. Arch. Virol. 71:229–234 [DOI] [PubMed] [Google Scholar]
- 47. Falcone V, Schweizer M, Neumann-Haefelin D. 2003. Replication of primate foamy viruses in natural and experimental hosts. Curr. Top. Microbiol. Immunol. 277:161–180 [DOI] [PubMed] [Google Scholar]
- 48. Hooks JJ, Gibbs CJ., Jr 1975. The foamy viruses. Bacteriol. Rev. 39:169–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Falcone V, Leupold J, Clotten J, Urbanyi E, Herchenröder O, Spatz W, Volk B, Böhm N, Toniolo A, Neumann-Haefelin D, Schweizer M. 1999. Sites of simian foamy virus persistence in naturally infected African green monkeys: latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology 257:7–14 [DOI] [PubMed] [Google Scholar]
- 50. Bothe K, Aguzzi A, Lassmann H, Rethwilm A, Horak I. 1991. Progressive encephalopathy and myopathy in transgenic mice expressing human foamy virus genes. Science 253:555–557 [DOI] [PubMed] [Google Scholar]
- 51. Eschbaumer M, Wäckerlin R, Rudolf M, Keller M, König P, Zemke J, Hoffmann B, Beer M. 2010. Infectious blood or culture-grown virus: a comparison of bluetongue virus challenge models. Vet. Microbiol. 146:150–154 [DOI] [PubMed] [Google Scholar]
- 52. Hahn H, Baunach G, Brautigam S, Mergia A, Neumann-Haefelin D, Daniel MD, McClure MO, Rethwilm A. 1994. Reactivity of primate sera to foamy virus Gag and Bet proteins. J. Gen. Virol. 75:2635–2644 [DOI] [PubMed] [Google Scholar]
- 53. Bleiholder A, Mühle M, Hechler T, Bevins S, vande Woude S, Denner J, Löchelt M. 2011. Pattern of seroreactivity against feline foamy virus proteins in domestic cats from Germany. Vet. Immunol. Immunopathol. 143:292–300 [DOI] [PubMed] [Google Scholar]
- 54. Brodie SJ, Pearson LD, Zink MC, Bickle HM, Anderson BC, Marcom KA, DeMartini JC. 1995. Ovine lentivirus expression and disease. Virus replication, but not entry, is restricted to macrophages of specific tissues. Am. J. Pathol. 146:250–263 [PMC free article] [PubMed] [Google Scholar]
- 55. Franchini M, Walker C, Henrard DR, Suter-Gut D, Braun P, Villiger B, Suter M. 1995. Accumulation of activated CD4+ lymphocytes in the lung of individuals infected with HIV accompanied by increased virus production in patients with secondary infections. Clin. Exp. Immunol. 102:231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Carrozza ML, Mazzei M, Bandecchi P, Arispici M, Tolari F. 2003. In situ PCR-associated immunohistochemistry identifies cell types harbouring the Maedi-Visna virus genome in tissue sections of sheep infected naturally. J. Virol. Methods 107:121–127 [DOI] [PubMed] [Google Scholar]
- 57. McNeilly TN, Baker A, Brown JK, Collie D, Maclachlan G, Rhind SM, Harkiss GD. 2008. Role of alveolar macrophages in respiratory transmission of Visna/Maedi virus. J. Virol. 82:1526–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. von Laer D, Neumann-Haefelin D, Heeney JL, Schweizer M. 1996. Lymphocytes are the major reservoir for foamy viruses in peripheral blood. Virology 221:240–244 [DOI] [PubMed] [Google Scholar]
- 59. German AC, Harbour DA, Helps CR, Gruffydd-Jones TJ. 2008. Is feline foamy virus really apathogenic? Vet. Immunol. Immunopathol. 123:114–118 [DOI] [PubMed] [Google Scholar]
- 60. Russell RA, Wiegand HL, Moore MD, Schäfer A, McClure MO, Cullen BR. 2005. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 79:8724–8731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. McClure MO, Bieniasz PD, Schulz TF, Chrystie IL, Simpson G, Aguzzi A, Hoad JG, Cunningham A, Kirkwood J, Weiss RA. 1994. Isolation of a new foamy retrovirus from orangutans. J. Virol. 68:7124–7130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bieniasz PD, Rethwilm A, Pitman R, Daniel MD, Chrystie I, McClure MO. 1995. A comparative study of higher primate foamy viruses, including a new virus from a gorilla. Virology 207:217–228 [DOI] [PubMed] [Google Scholar]
- 63. Schweizer M, Neumann-Haefelin D. 1995. Phylogenetic analysis of primate foamy viruses by comparison of pol sequences. Virology 207:577–582 [DOI] [PubMed] [Google Scholar]