Abstract
Most betacoronaviruses possess an hemagglutinin-esterase (HE) protein, which appears to play a role in binding to or release from the target cell. Since this HE protein possesses an acetyl-esterase activity that removes acetyl groups from O-acetylated sialic acid, a role as a receptor-destroying enzyme has been postulated. However, the precise function of HE and of its enzymatic activity remains poorly understood. Making use of neutralizing antibody and of molecular clones of recombinant human coronavirus OC43 (HCoV-OC43), our results suggest that the HE protein of this HCoV could be associated with infection of target cells and, most notably, is important in the production of infectious viral particles. Indeed, after transfecting BHK-21 cells with various cDNA infectious clones of HCoV-OC43, either lacking the HE protein or bearing an HE protein with a nonfunctional acetyl-esterase enzymatic activity, we were reproducibly unable to detect recombinant infectious viruses compared to the reference infectious HCoV-OC43 clone pBAC-OC43FL. Complementation experiments, using BHK-21 cells expressing wild-type HE, either transiently or in a stable ectopic expression, demonstrate that this protein plays a very significant role in the production of infectious recombinant coronaviral particles that can subsequently more efficiently infect susceptible epithelial and neuronal cells. Even though the S protein is the main viral factor influencing coronavirus infection of susceptible cells, our results taken together indicate that a functionally active HE protein enhances the infectious properties of HCoV-OC43 and contributes to efficient virus dissemination in cell culture.
INTRODUCTION
Coronaviruses are widespread in nature and can infect several different species (1), in which they cause mainly respiratory and enteric pathologies, with neurotropic and neuroinvasive properties in various hosts including humans, cats, pigs, and rodents (reviewed in references 2 and 3). They are part of the family Coronaviridae, within the order Nidovirales, and they are classified into three different genera, namely, Alphacoronavirus, Betacoronavirus, and Gammacoronavirus (4). Coronaviruses form a group of enveloped viruses that have the largest genome among RNA viruses. This nonsegmented 30-kb positive single-stranded polyadenylated RNA of ∼30 kb possesses four or five genes encoding structural proteins (S, E, M, and N; hemagglutinin-esterase [HE] protein for the genus Betacoronavirus) and several genes encoding nonstructural proteins. Moreover, the genus Betacoronavirus is divided into four different lineages: A, B, C, and D. Members of lineage A include the species murine coronavirus (often still referred to as mouse hepatitis virus [MHV]), human coronavirus HKU1 (HCoV-HKU1), and betacoronavirus-1, which comprises the porcine hemagglutinin encephalomyelitis virus (PHEV), bovine coronavirus (BCoV), and HCoV-OC43 (4), which all possess an hemagglutinin-esterase (HE) protein in the viral envelope (5, 6). This HE protein displays 30% identity to the subunit 1 of the HEF protein of the influenza C virus (7). Like the spike protein (S), the large type 1 transmembrane glycosylated viral protein responsible for the recognition of the cellular receptor used by the coronaviruses to infect susceptible cells (8), the HE protein, present in species of the Betacoronavirus genus, is also a type 1 transmembrane protein (9), and it interacts with different types of sialic acid, associated with an apparent role in hemagglutination. Furthermore, since the HE protein possesses an acetyl-esterase activity that removes acetyl groups from O-acetylated sialic acid, a role as a receptor-destroying enzyme has been postulated, a function that may be important early during infection, possibly in virus binding to or later during the release of viral particles from infected cells at the end of the replication cycle of betacoronaviruses (10, 11). Even though the precise role and function of the HE protein remains incompletely understood, the structure of the BCoV HE protein in complex with its receptor is now unraveled and this will certainly help our understanding of its biological role (12). The S protein of HCoV-OC43 was shown to interact with 9-O-acetyl sialic acid and is believed to be the major hemagglutinin of this virus (13), as well as being the protein responsible for binding to the cellular receptor (14). However, the HCoV-OC43 HE protein may, like the HE protein of BCoV (12, 15, 16), also interact with this particular sialic acid. We present here results that indicate that the HE protein plays, in conjunction with the S protein, an important and significant role in HCoV-OC43 infectious properties and therefore in the infection of cells and the spread of infection in cell cultures.
MATERIALS AND METHODS
Cell lines, pBAC-OC43 cDNA clones, and corresponding recombinant viruses.
The HRT-18 cell line (kindly provided by David Brian, University of Tennessee) was cultured in minimal essential medium alpha (MEM-α; Life Technologies) supplemented with 10% (vol/vol) fetal bovine serum (FBS; PAA) and used to propagate all of the different viruses, as previously described (17), and in infection and neutralization experiments. The BHK-21 cell line (ATCC-CCL10) was also cultured in MEM-α supplemented with 10% (vol/vol) FBS and used for transfection. The LA-N-5 cell line (kindly provided by Stephan Ladisch, George Washington University School of Medicine) was cultured in RPMI 1640 supplemented with 15% (vol/vol) FBS, 10 mM HEPES, 1 mM sodium pyruvate, and 100 μM nonessential amino acids (Gibco-Invitrogen). For use in experiments, LA-N-5 cells were differentiated into human neurons, as previously described (18). Briefly, cells were seeded in Cell+ 24-well (1.25 × 104 cells) plates (Sarstedt) in RPMI medium supplemented with 10% (vol/vol) FBS, 10 mM HEPES, 1 mM sodium pyruvate, and 100 μM nonessential amino acids. The next day, and every 2 days for 6 days, medium was replaced with the same medium containing 10 μM all-trans retinoic-acid (Sigma-Aldrich).
The recombinant wild-type reference HCoV-OC43 virus, designated here HCoV-OC43 rOC/ATCC, was obtained from the full-length cDNA clone pBAC-OC43FL (19). Using a QuikChange multisite-directed mutagenesis kit (Agilent-Stratagene), this full-length cDNA clone was used to generate different pBAC-OC43-HE mutants. The cDNA clones containing a point mutation in the HE gene either at nucleotide position 8 of the coding sequence to replace the amino acid residue leucine at position 3 by a STOP codon (pBAC-OC43-HEstop) or at nucleotide position 347 of the HE coding sequence to mutate the amino acid residue at position 116 from a serine to a phenylalanine (S116F), designated pBAC-OC43-HES116F, were generated by reverse genetics.
Results obtained by others (12) and sequence alignment of the HCoV-OC43 HE protein with other members of the Betacoronavirus genus using the PFAM software program (http://www.sanger.ac.uk/Software/Pfam/) revealed that the putative active site for O-acetyl-esterase activity, FGDS, is located between amino acid residues 37 to 40. To abolish the O-acetyl-esterase activity of the HE protein (HE0), the serine at position 40 was mutated to a threonine to modify the active site to FGDT. To generate the corresponding pBAC-OC43 HE mutant, we mutated the nucleotide position 119 in the HE coding sequence and this cDNA molecular clone was designated pBAC-OC43-HES40T.
Overlapping PCR was used to obtain a cDNA clone with complete deletion of the HE gene (pBAC-OC43-delHE). Sequencing was performed to confirm that there were no other mutations in the genomes of the different pBAC-OC43 clones.
Primary cultures of mouse central nervous system (CNS) cells.
Embryos at 12 to 14 days of gestation were removed from pregnant anesthetized CD1 mice. Cortical cell cultures were obtained by a modification of a previously described protocol (20). Briefly, cortical cell cultures were harvested in Hanks balanced salt solution (HBSS; without Ca2+ and Mg2+), supplemented with 1 mM sodium pyruvate and 10 mM HEPES buffer. Cortical cell cultures were incubated in 1 ml of trypsin-EDTA supplemented with 20 μg of DNase I/ml for 5 min at 37°C. The trypsin solution was then slowly discarded with a sterile pipette and replaced by HBSS (without Ca2+ and Mg2+) supplemented with 10% (vol/vol) FBS. The samples were then briefly centrifuged at 250 × g, and the supernatant was removed and replaced by 5 ml of HBSS. Cortical cell cultures were then triturated, and the cells were dispersed in the same medium. Samples were centrifuged for 1 min at 1,000 × g, and the pellet was resuspended in 1 ml of neurobasal medium, supplemented with 0.5 mM l-glutamine and B27 supplement (Life Technologies) per brain. Cells were then seeded at ∼106/cm2 and grown on poly-d-lysine-treated glass coverslips in the same medium that was replaced after 4 days in culture. The cultures were ready for infection after 7 days in culture.
Infection of human cell lines and of primary mouse CNS cultures.
LA-N-5 or HRT-18 cells were infected at a multiplicity of infection (MOI) of 0.5 or mock infected and then incubated at 37 or 33°C, respectively, for 2 h, washed with phosphate-buffered saline (PBS), and incubated at 37°C with fresh RPMI 1640 medium supplemented with 2.5% (vol/vol) FBS or MEM-α supplemented with 1% (vol/vol) FBS, respectively, for different periods of time before being analyzed by immunofluorescence. For regular infection and neutralization experiments, the MOI was 0.5, and for the complementation experiments on HRT-18 cells and primary mouse CNS cultures, infection was made at an approximate MOI of 0.0001 without PBS washing and incubated at 33°C for 48 h (HRT-18) or 37°C for 24 and 48 h for primary cultures before being analyzed by immunofluorescence assay (IFA).
Neutralization assay.
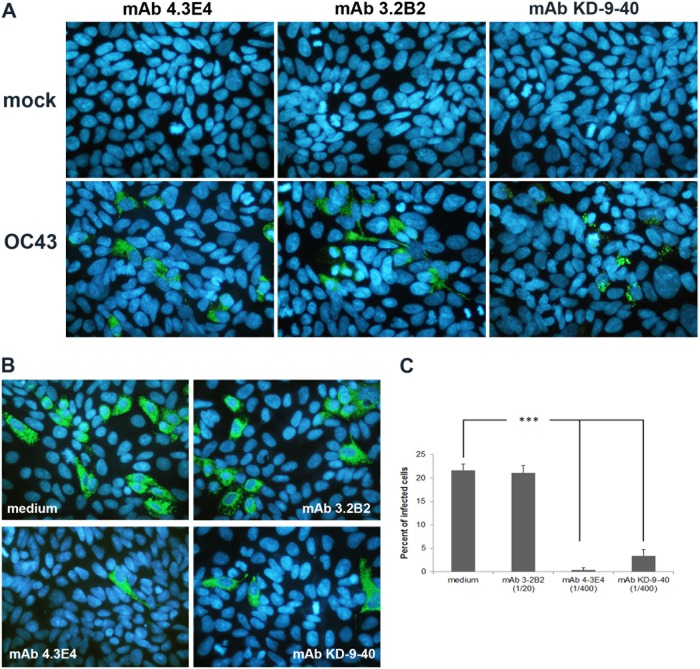
A series of monoclonal antibodies (MAbs) against the HE and S proteins of either HCoV-OC43 or BCoV were used to evaluate the relative importance of the S and HE proteins of HCoV-OC43 during infection of susceptible cells. MAbs 4.3E4 and 3.2B2, which both recognize the HCoV-OC43 S protein, served as positive and negative controls of neutralization, respectively. MAb KD-9-40 against the HE protein of BCoV (kindly provided by Sylvia van Drunen Littel-van den Hurk, University of Saskatchewan, Saskatchewan, Canada) also recognizes the HE protein of HCoV-OC43 (see Fig. 6). Therefore, it was used to evaluate the importance of the HCoV-OC43 HE protein during infection. Wild-type recombinant HCoV-OC43 (rOC/ATCC; 105 50% tissue culture infective doses [TCID50]) was preincubated with the different antibodies (1/20 of ascites fluid for MAb 3.2B2 and 1/400 of ascites fluid for MAbs 4.3E4 and KD-9-40) for 1 h at 37°C before being inoculated into cells for 2 h at 33°C (HRT-18) or 37°C (LA-N-5). The cells were washed with PBS and incubated at 33°C with fresh MEM-α supplemented with 1% (vol/vol) FBS (HRT-18) or at 37°C with fresh RPMI medium supplemented with 2.5% (vol/vol) FBS (LA-N-5) for 24 h before being analyzed by IFA. Statistical significance was estimated by analysis of variance (ANOVA), followed by post hoc Dunnett's (two-sided) analysis.
Fig 6.
Detection of the HE protein in infected cells and partial neutralization of HCoV-OC43 infection by HE-specific MAb. (A) Detection of S protein (MAbs 4.3E4 and 3.2B2) or HE protein (MAb KD-9-40) in HRT-18 cells at 24 h postinfection. The green cells in the bottom panels are infected cells. (B) Before infection, wild-type recombinant HCoV-OC43 was preincubated with medium or with MAb 3.2B2 or MAb 4.3E4 or KD-9-40. At 24 h postinfection, the cells were analyzed by immunofluorescence to detect the S protein. (C) Semiquantitative IFA analysis determined that the neutralization of infection by the KD-9-40 MAb was to a level of almost 90% compared to infection in the presence of medium alone or in the presence of non-neutralizing antibody 3.2B2. The results are representative of three independent experiments. Statistical significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Transfection of BHK-21 cells and production of recombinant viruses.
BHK-21 cells were seeded in six-well plates (Corning) at 106 cells/well and transfected the next day using Lipofectamine LTX (Life Technologies) according to the manufacturer's instructions, as modified previously (19). Briefly, 5 μg of either pBAC-OC43FL, pBAC-OC43-delHE (devoid of the HE gene), pBAC-OC43-HEstop (mutated at nucleotide 8, which replaces the amino acid residue leucine by a STOP codon), pBAC-OC43-HES40T (point mutation at nucleotide 119, introducing a change of amino acid at position 40 from a serine to a threonine [S40T]), or pBAC-OC43-HES116F, which harbors a single point mutation at nucleotide 347 that introduces a change of amino acid at position 116 from a serine to a phenylalanine (S116F), was evaluated. The medium was changed 8 h after transfection and replaced by MEM-α supplemented with 10% (vol/vol) FBS, and the cells were incubated 3 days at 37°C. The transfected BHK-21 cells were lysed by three cycles of freeze-thaw and clarified by centrifugation at 2,500 × g for 10 min. The supernatant (P0) served to inoculate HRT-18 cells in order to amplify the viral stocks. The medium from this first round of amplification (P1) was recovered and served for a second round of viral amplification on HRT-18 cells from which we recovered the supernatant (P2). The production of infectious viral particles corresponding to the different pBAC-OC43 cDNA clones was titrated by an immunoperoxidase assay (IPA).
Titration of infectious virus using an IPA.
An IPA was performed on HRT-18 cells, as previously described (21). Briefly, the primary antibody used was MAb 1-10C3 directed against the S protein of HCoV-OC43. The secondary antibody was horseradish peroxidase-conjugated goat anti-mouse immunoglobulin (KPL). Immune complexes were detected by incubation with 0.025% (wt/vol) 3,3′-diaminobenzidine tetrahydrochloride (Bio-Rad) and 0.01% (vol/vol) hydrogen peroxide in PBS, and infectious virus titers were calculated by the Karber method, as previously described (21).
IFA and semiquantitative analysis to determine percentage of cells positive for viral proteins.
Cells were fixed with 4% (wt/vol) paraformaldehyde for 20 min at room temperature and permeabilized with methanol at −20°C for 5 min. The presence of the HCoV-OC43 proteins in HRT-18, BHK-21, and LA-N-5 cells was investigated using either primary mouse MAb 4.3E4 (1/500 from ascites fluid) directed against the S protein of HCoV-OC43, MAb KD-9-40 (1/500 from ascites fluid) directed against the HE protein of the serologically related BCoV, or MAb 4E11.3 (1/1,000 from ascites fluid) directed against the N protein of the serologically related porcine coronavirus PHEV (kindly provided by the late Serge Dea, INRS-Institut Armand-Frappier, Laval, Québec, Canada). The secondary antibody was an anti-mouse Alexa Fluor 488-conjugated antibody (Life Technologies/Molecular Probes).
For primary cultures of the mouse CNS, the primary antibodies were a rabbit antiserum (1/1,000) against the structural proteins of BCoV (kindly provided by Serge Dea) and a mouse MAb against the neuron-specific MAP2 protein (1/500; BD Pharmingen, catalog no. 556320) for 1 h at room temperature and washed three times with PBS. The cells were then incubated for 1 h at room temperature with anti-rabbit Alexa Fluor 488- and anti-mouse Alexa Fluor 568-conjugated secondary antibodies (1/1,500; Life Technologies/Molecular Probes). After three washes with PBS, fixed cells were incubated for 5 min with DAPI (4′,6′-diamidino-2-phenylindole; Sigma-Aldrich) at 1 μg/ml to stain the DNA in the nucleus.
To determine the percentages of cells positive for viral proteins, 20 fields containing a total of either 100 to 125 cells (HRT-18) or between 50 and 70 cells (LA-N-5) at a magnification of ×400 using a Nikon Eclipse E800 microscope were counted for each condition tested in three independent experiments. Green cells were scored as HCoV-OC43 protein positive over the total amount of cells (DAPI stained in blue for HRT-18 and LA-N-5). The statistical significance was estimated by ANOVA, followed by post hoc Dunnett's (two-sided) analysis. Precise quantification of infected neurons in primary cultures of mouse CNS was not determined.
Acetyl-esterase activity assay in cells.
Transfected BHK-21 cells and infected HRT-18 cells grown on coverslips were fixed 30 s at room temperature with a solution of citrate-acetone-formaldehyde (25, 65, and 8% [vol/vol]), and their acetyl-esterase activity was evaluated using an alpha-naphtyl acetate esterase kit (91-A; Sigma-Aldrich) according to the manufacturer's instructions, as previously reported for the MHV-S HE protein (22).
RNA isolation, RT-PCR, and real-time PCR.
Total RNA was extracted from transfected BHK-21 cells using the Qiagen RNA extraction kit according to the manufacturer's instructions and quantified using a ND1000 spectrophotometer (NanoDrop). Portions (5 μg) or total RNA were reversed transcribed for reverse transcription-PCR (RT-PCR) and quantitative real-time PCR using the Superscript III first-strand kit with oligo(dT) primer (Life Technologies) according to the manufacturer's instructions. RT-PCR was performed using Accuprime Pfx Supermix (Life Technologies) with 1 cycle at 95°C for 5 min, followed by 35 cycles at 95°C for 30 s, 55°C for 45 s, and 68°C for 2 min with the primers A-forward (5′-GGA TCC ATG TTT TTG CTT CCT AGA TT-3′) and B-reverse (5′-ACC GGT AAC TGG TAG CGG ATC ATA CA-3′), which amplify a fragment of 1,175 bp, corresponding to the complete coding sequence of the HE gene without the transmembrane domain. Real-time quantitative PCR was performed using the SsoFastEvagreen supermix qPCR kit (Bio-Rad) with the Rotor-Gene 3000 (Corbett Life Science) with 1 cycle at 95°C for 1 min, followed by 40 cycles at 95°C for 10 s and 60°C for 30 s, with the primers C-forward (5′-TAT TTG TCA GGT TGT GAC GA-3′) and D-reverse (5′-CCA TAA ATA ACA CCA GTG TCT T-3′), which amplify a fragment of 124 bp between nucleotides 577 and 701 of the coding sequence of the HE gene. Modulation of the HE gene expression from quantitative real-time PCR was analyzed using the 2−ΔΔCT method (23) and normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) expression.
Flow cytometry.
Guinea pig antiserum, which was made in-house by immunization of a guinea pig with inactivated HCoV-OC43 and which recognizes the structural proteins of HCoV-OC43, was diluted in PBS containing 2% (wt/vol) bovine serum albumin at 1/1,000 and was used to detect the HE protein by flow cytometry on PFA-fixed BHK-21 cells.
RESULTS
The absence of wild-type enzymatically active HE protein strongly abrogates the production of infectious HCoV-OC43.
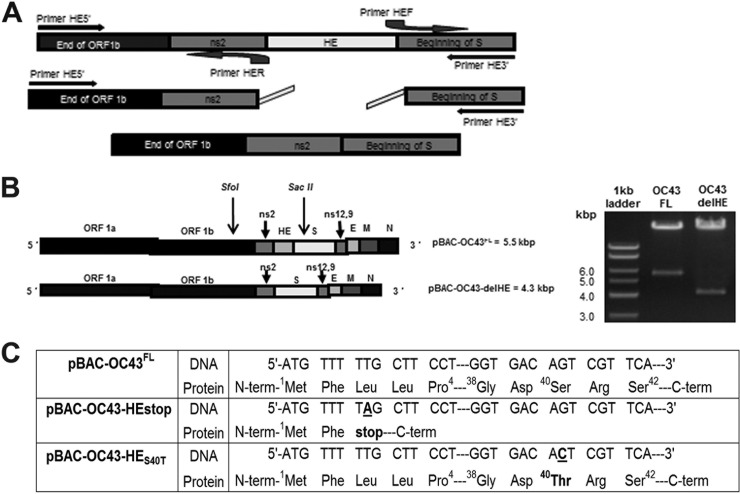
Making use of our cDNA molecular clone pBAC-OC43FL (19), we were able to obtain cDNA molecular clones with either a deleted or a mutated HE gene. These different mutants either lack the HE gene (pBAC-OC43-delHE) or possess a mutated HE gene (pBAC-OC43-HEstop and pBAC-OC43-HES40T) (Fig. 1).
Fig 1.
The cDNA molecular clone pBAC-OC43FL was used to design strategies of deletion/inactivation of the gene encoding the HCoV-OC43 HE protein. (A) Three-step overlapping PCR was used to delete the gene encoding the HE protein. (B) The deletion of 1.2 kbp (the complete HE gene) was detected by the digestion of the reference and mutagenized pBAC-OC43 cDNA infectious clones by the SfoI and SacII restriction enzymes. (C) Site-directed mutagenesis allowed recovery of cDNA infectious clones with nonsense mutation pBAC-OC43HEstop, by the introduction of a STOP codon at amino acid Leu3 (nucleotide position 8), or with a single point mutation to change the serine at position 40 to a threonine, mutation S40T (pBAC-OC43-HES40T), which inactivates the putative active site of the O-acetyl-esterase.
The cDNA molecular pBAC-OC43 clones without the HE gene or with the HE gene encoding an enzymatically inactive HE protein (HE0) produce viral S protein but no HE protein bearing acetyl-esterase activity and no detectable infectious recombinant HCoV-OC43.
Making use of the different molecular clones described in Fig. 1, we wanted to evaluate the importance of the HE protein and of its acetyl-esterase enzymatic activity in the production of recombinant infectious HCoV-OC43. As shown in Fig. 2A, and as previously reported (19), transfection of BHK-21 cells by the pBAC-OC43FL led to the detection of wild-type HCoV-OC43 recombinant infectious virus (rOC/ATCC). However, by transfecting BHK-21 cells with the mutants pBAC-OC43-delHE, pBAC-OC43-HEstop or pBAC-OC43-HES40T, the level of production of infectious virus was reproducibly under the limit of detection, with the one exception of the molecular clone of pBAC-OC43-HEstop-A1 (Fig. 2A). In fact, even though the sequence of the HE gene of this particular molecular cDNA clone possessed the introduced STOP codon prior to transfection of BHK-21 cells, sequencing of the viral RNA after the second round of amplification on HRT-18 cells (P2) showed that a reversion had occurred at nucleotide position 8, to replace the STOP codon by a tryptophan at amino acid residue 3, allowing the corresponding recombinant virus to grow in culture.
Fig 2.
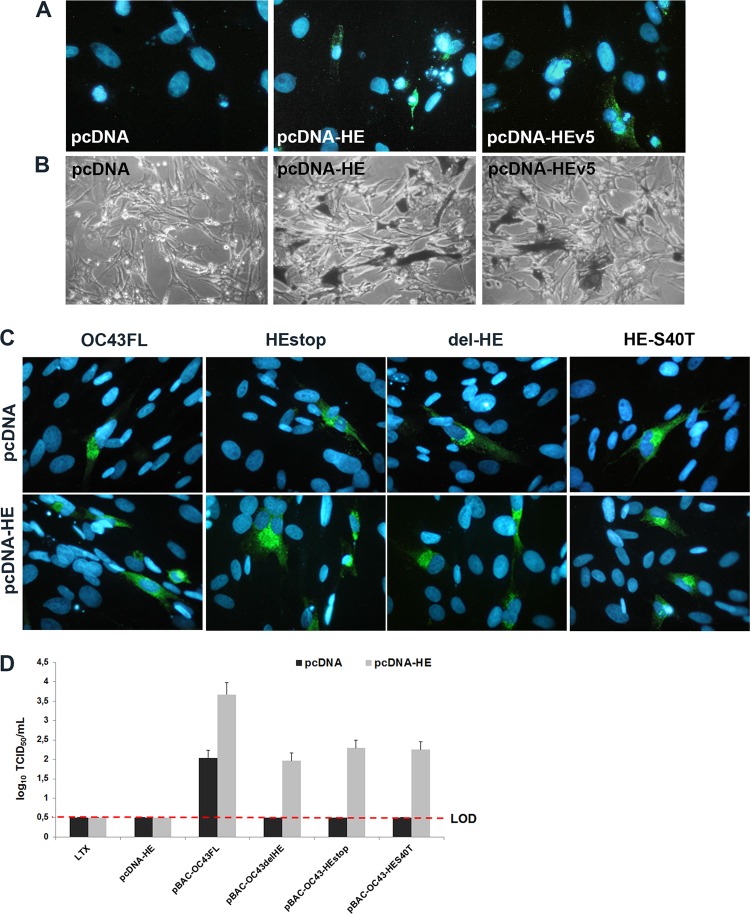
The acetyl-esterase enzymatic activity of the HE protein of the HCoV-OC43 plays an important role in production of detectable infectious viral particles. (A) Titration of recombinant infectious HCoV-OC43 particles after transfection of BHK-21 cells at passage 0 (P0), harvested directly from transfected BHK-21 cells, and amplified between P1 and P2 on HRT-18 cells. nd, not detected. (B) IFA confirmed that the S protein was produced in transfected BHK-21 cells at equivalent levels for all of the pBAC-OC43 molecular clones and that the HE protein was produced only in BHK-21 cells transfected with pBAC-OC43FL or pBAC-HES40T. (C) Acetyl-esterase activity was only detected in BHK-21 cells transfected with the pBAC-OC43FL, compared to all pBAC-OC43 HE mutants and to mock-transfected BHK-21 cells (Lipofectamine LTX).
Considering these observations, we wanted to evaluate whether the production of viral proteins after transfection of BHK-21 cells was normal compared to wild-type pBAC-OC43FL. Making use of an MAb against the S protein of HCoV-OC43 (4.3E4), immunofluorescence analysis confirmed that the viral S protein was indeed produced in transfected BHK-21 cells, at levels (same amount of positive green cells) equivalent for all of the pBAC-OC43 molecular clones (Fig. 2B). On the other hand, this same approach revealed that the HE protein was also produced at equivalent levels after transfection of BHK-21 cells with either pBAC-OC43FL or pBAC-HES40T molecular clones but that no HE protein was produced after transfection by the pBAC-OC43-HEstop and pBAC-OC43-delHE clones (Fig. 2B). Furthermore, an acetyl-esterase assay revealed that only pBAC-OC43FL produced an active form of HE protein, compared to all of the pBAC-OC43 HE mutants and to mock-transfected BHK-21 cells (Fig. 2C). Transfection of cells with a pBAC-OC43 molecular clone pBAC-OC43-HES116F (mutation in the HE protein at a potential site of N-glycosylation not located in the domain responsible for the acetyl-esterase activity) did produce detectable infectious recombinant HCoV-OC43 virus at a level comparable to pBAC-OC43FL. This pBAC-OC43-HES116F molecular clone confirmed that it was possible to mutate the HE protein in other domains and produce easily detectable and significant amounts of infectious HCoV-OC43 particles.
Stable BHK-21 clones ectopically expressing the HCoV-OC43 HE protein complement the recombinant pBAC HE mutants.
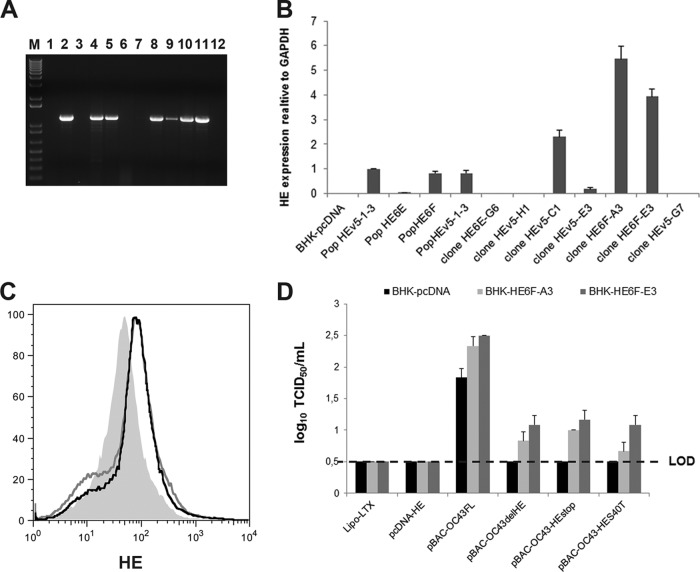
Considering that the S protein was detected after transfection of BHK-21 cells with all of the different pBAC-OC43-HE mutant molecular clones and the appearance of a revertant virus after two rounds of amplification (Fig. 2A), we sought to evaluate whether exogenous wild-type HE protein could help generate detectable amounts of infectious HCoV-OC43 by complementation. Therefore, BHK-21 subpopulations and clones ectopically expressing the HE gene were selected and analyzed (Fig. 3A). Quantitative real-time PCR on a series of BHK-21 subpopulations and clones resistant to G418 (1 mg/ml) revealed different levels of HE expression, and two clones (HE6F-A3 and HE6F-E3) that express the HE mRNA at the highest levels (Fig. 3B) were analyzed by flow cytometry to quantitate HE protein expression. The two clones indeed express the HE protein at a low, but significant, level compared to BHK-21 cells transfected by the control vector (Fig. 3C). Using these BHK-21 clones, which ectopically express the HE protein, we were able to detect infectious HCoV-OC43 particles after transfection by the pBAC-OC43-HE mutants compared to BHK-21 cells transfected by the control vector pcDNA, from which no infectious virus was detected (Fig. 3D).
Fig 3.
BHK-21 cells ectopically expressing the HCoV-OC43 wild-type HE protein complement recombinant pBAC HE mutants. (A) BHK-21 subpopulations and clones analyzed by RT-PCR revealed ectopic expression of the HE. Samples were run on a 1% (wt/vol) agarose gel, and amplicons were revealed with ethidium bromide. Lane M, molecular weight markers; lanes 1 to 5, subpopulation pcDNA, HEv5-1-3, HE6E, HE6F, and HEv5-2-13, respectively; lanes 6 to 12, individual clones HE6E-G6, HEv5-H1, HEv5-C1, HEv5-E3, HE6F-A3, HE6F-E3, and HEv5-G7, respectively. (B) Quantitative real-time PCR revealed different levels of HE expression, with the two clones (HE6F-A3 and HE6F-E3) expressing the highest levels of the HE mRNA. (C) Flow cytometry analysis revealed that these two clones expressed the HE protein compared to BHK-21 cells transfected by the control vector pcDNA alone. (D) Transfection of the BHK-21 HE6F-A3 and HE6F-E3 clones by the pBAC-OC43-HE mutants produced detectable amounts of infectious HCoV-OC43 particles compared to BHK-21 cells transfected by the control vector pcDNA, from which no infectious virus was detected. LOD, limit of detection.
Transient expression in BHK-21 cells of HCoV-OC43 HE protein with acetyl-esterase activity is more efficient than stable BHK-21 clones in complementing pBAC HE mutants, leading to production of higher titers of infectious recombinant HCoV-OC43.
Even though infectious HCoV-OC43 was detected by complementing the pBAC-OC43-HE mutants with the BHK-21 clones stably expressing the HCoV-OC43 HE protein (Fig. 3D), we sought to increase this production by using an alternative experimental approach. The pcDNA-HE or the pcDNA-HEv5, which are vectors expressing the wild-type HE protein, either alone or fused with a v5 epitope, respectively, were compared to the pcDNA vector alone to evaluate the production of the HE protein (Fig. 4A) and the expression of its acetyl-esterase activity (Fig. 4B). These vectors were used in cotransfection of BHK-21 cells with the different pBAC-OC43 molecular clones. This experimental approach led to a significant increase in the number of cells producing viral antigens after transfection (Fig. 4C) and led to the production of detectable levels of infectious virus from pBAC-OC43-HE mutants compared to cotransfection of cells with the pcDNA vector without HE (Fig. 4D). Furthermore, this alternative experimental approach successfully led to an important increase in infectious virus production, which was indeed 10 times more efficient compared to the use of BHK-21 clones that constitutively express the HE protein (Fig. 4D compared to Fig. 3C).
Fig 4.
Transient expression of HCoV-OC43 wild-type HE protein is more efficient than stably expressing BHK-21 clones to complement pBAC-HE mutants. (A) Expression of the wild-type HE protein after transfection of BHK-21 cells with either pcDNA-HE or pcDNA-HEv5, compared to pcDNA vector alone. (B) Detection of O-acetyl-esterase activity in BHK-21 cells transfected with either pcDNA-HE or pcDNA-HEv5 compared to pcDNA alone. (C) Immunofluorescence analysis (detection of the S protein) of BHK-21 cells cotransfected by either pcDNA vector alone (top panels) or by pcDNA-HE (bottom panels), together with all of the pBAC-OC43 molecular clones. (D) Production of infectious virus after transfection of BHK-21 cells by the pBAC-OC43FL and the different pBAC-OC43-HE mutant infectious clones cotransfected with either pcDNA vector alone or pcDNA-HE. LOD, limit of detection.
The HE-complemented recombinant HCoV-OC43 can make a first round of infection of human epithelial cells and primary cultures from murine CNS but are defective in producing infectious progeny virions that can spread in cell culture.
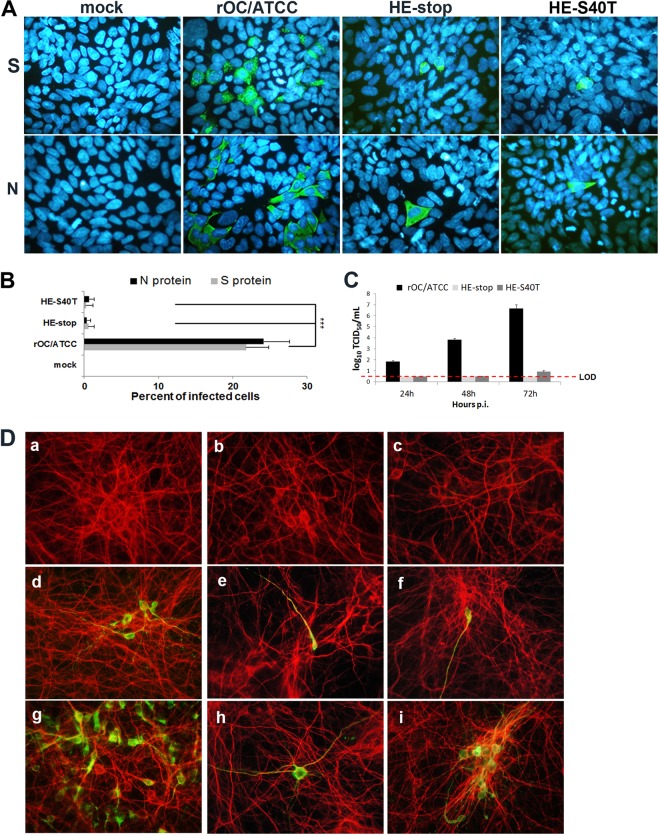
Since exogenous HE protein allowed the production of detectable infectious virus after the transfection of BHK-21 cells with all of the different pBAC-OC43-HE mutants, we sought to determine whether these new infectious particles were able to efficiently infect susceptible cells and to evaluate whether the incapacity of HCoV-OC43 to produce completely active HE protein would impact the capacity of the virus to produce new infectious particles that could disseminate in cell culture. Considering the low infectious virus titers produced for the complemented HE mutants (Fig. 4D; 102 TCID50/ml), susceptible HRT-18 cells were infected at an MOI of 0.0001 with infectious virus that was produced after cotransfection of BHK-21 cells with the pcDNA-HE vector in conjunction with either pBAC-OC43-HEstop or pBAC-OC43-HES40T and compared to infection by wild-type HCoV-OC43 (rOC/ATCC). At 48 h postinfection, wild-type virus had spread within the culture, and viral S and N proteins were detected (Fig. 5A). Semiquantitative analysis revealed that these viral antigens were present in ca. 25% of the cells (Fig. 5B). Even though the two types of HE-complemented infectious virus produced by cotransfecting the pcDNA-HE vector and pBAC-OC43-HEstop or pBAC-OC43-HES40T were able to infect HRT-18 cells (Fig. 5A), their incapacity to de novo synthesize new HE protein appeared to affect their efficient spread within the culture as monitored by the fact that <1% of the cells produced viral S or N proteins (Fig. 5B). Furthermore, since we previously reported that HCoV-OC43 is neuroinvasive (24) and neurotropic, the neuron being its main cell target in the CNS (20, 25), we wanted to evaluate whether HE-complemented infectious virus was able to infect mixed primary cultures from murine CNS, replicate, and eventually disseminate within this culture. Starting with an approximate MOI of 0.0001, ca. 5 to 10% of neurons were already positive for viral antigens after 24 h postinfection by the wild-type virus (Fig. 5D, panel d), whereas <1% of neurons produced viral antigens 24 h postinfection by either HE mutant virus (Fig. 5D, panels e and f). At 48 h postinfection, wild-type virus had clearly disseminated to about half of the available neurons (Fig. 5D, panel g), whereas the percentage of neurons infected by the complemented virus produced by the cotransfection of pcDNA-HE plus the pBAC-OC43-HEstop molecular clone was still very low and was estimated to represent <1% (Fig. 5D, panel h). The percentage of infected neurons was the same for the HE-complemented virus produced by the cotransfection of pcDNA-HE plus the pBAC-OC43-delHE molecular clone (data not shown). At the same time postinfection, the percentage of neurons infected by the HE-complemented virus produced by the cotransfection of pcDNA-HE and the pBAC-OC43-HES40T molecular clone had increased significantly (between 3 and 5%; Fig. 5D, panel i) as a small but significant amount of new infectious viruses were produced (Fig. 5C).
Fig 5.
HE-complemented recombinant HCoV-OC43 infects human epithelial cells and primary cultures of murine CNS but is defective in producing new infectious virions that can spread to surrounding noninfected cells. (A) IFA at 48 h postinfection of HRT-18 cells. (Upper panels) Green indicates cells producing the viral S protein. (Lower panels) Green indicates cells producing the viral N protein. Blue is DAPI staining of the nucleus. (B) Semiquantitative analysis revealed that the S protein (green) was present in ca. 25% of the cells for wild-type virus compared to <1% for the two HE-complemented infectious virus. The results are representative of three independent experiments. Statistical significance: *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Production of infectious particles after infection of primary murine CNS cells. (D) IFA at 24 and 48 h postinfection in mixed primary cultures from murine CNS. Subpanels: d and g, wild-type virus (rOC/ATCC); e and h, HE-complemented HEstop mutant virus; f and i, HE-complemented HES40T mutant virus. Red represents the microtubule-associated protein 2 (MAP-2) staining in neurons. Green indicates viral antigens and yellow indicates colocalization (merge) of red (MAP-2) and green (viral antigens). Panel a, mock-infected cells; panels b and c, cells infected with control noncomplemented virus, which are lysates of BHK-21 cells cotransfected with pBAC-OC43-HEstop or pBAC-OC43-HES40T with empty pcDNA vector, respectively. The results for primary cultures are representative of two independent experiments.
The HE protein may enhance HCoV-OC43 infection of susceptible human cells.
Since HE-complemented mutant viruses were able to infect epithelial cells (HRT-18) and primary cultures from murine CNS, we sought to evaluate whether the HE protein could play a role in this infection. In order to study this potential role of the HE protein of HCoV-OC43, we performed a neutralization assay using a specific MAb that was raised against the closely related bovine coronavirus (BCoV) HE protein (26). Our results clearly indicate that this MAb also recognizes the HE protein of HCoV-OC43 in an infected human epithelial cell line (HRT-18; Fig. 6A), as well as the human neuronal LA-N-5 cells (data not shown), and suggest that it decreased infection of HRT-18 (Fig. 6B) and LA-N-5 (data not shown) cells by almost 90% (Fig. 6C), suggesting a potential role for the HE protein in enhancing the S-protein-mediated infection of susceptible cells by HCoV-OC43
DISCUSSION
Even though the coronavirus S protein is the main viral protein involved in receptor recognition on the cell surface, the HE protein, present in several betacoronaviruses, may represent a second receptor-binding protein (27) and play an important role in infection (26, 28). In the present study we made use of various experimental tools, most notably of HE mutants produced with our pBAC-OC43 cDNA clone (19), to demonstrate that a fully functional HE protein plays an important role in the production of detectable levels of infectious HCoV-OC43 particles, suggesting that this protein may also influence viral infectivity and eventual spread in cell culture.
The precise role of the HE protein of betacoronaviruses in general, and of HCoV-OC43 in particular remains elusive. Our data showing that transfection of BHK-21 cells led to the production of equal amounts of viral S protein with either pBAC-OC43FL, pBAC-OC43-delHE, pBAC-OC43-HEstop, or pBAC-OC43-HES40T (Fig. 2) indicate that these cDNA clones can replicate as efficiently as the pBAC-OC43FL. This observation suggests, as expected, that the HE protein does not participate in the transcription of the subgenomic RNA that will be translated into proteins by the infected cells. On the other hand, the observation that the level of production of infectious virus was reproducibly under the limit of detection for all of the cDNA encoding HE mutant viruses, with the one exception of the molecular clone of pBAC-OC43-HEstop-A1 (Fig. 2A), indicate that the production of a fully enzymatically active HE protein is important at some point during the replicative cycle of HCoV-OC43 in order to give rise to viruses that are able to infect susceptible cells. Inhibiting the acetyl-esterase activity of the BCoV HE protein was also previously shown to strongly abrogate BCoV replication, by up to a 1,000-fold (11).
Our pBAC-OC43FL system previously used to identify specific S protein amino acid residues as important molecular determinants of virus-induced neuropathogenesis (29, 30) and neuronal cell death (31, 32) has proven very stable since no mutations appeared in the S gene of recombinant HCoV-OC43 produced after transfection of BHK-21 cells and two rounds of amplification on HRT-18 cells (29, 30, 32, 33). Therefore, results presented in Fig. 2A, concerning the unique molecular clone pBAC-OC43-HEstop-A-1, from which the STOP codon has reverted to allow for the production of infectious virus at a low titer (102.5 TCID50/ml) after the same two rounds of amplification on HRT-18 cells, suggest that there is some sort of selective pressure for recombinant viruses with a functional HE protein. RNA mutation may appear without production of viral particles, and we cannot completely rule out the possibility that only mutated RNA was formed without virion production before the second round of amplification on HRT-18 cells. However, based on our experimental design (detailed in Materials and Methods), our data rather suggest that this appear highly unlikely and indicate that infectious virus had to be produced from the beginning. Indeed, if a revertant with a different HE sequence compared to wild-type HCoV-OC43 (Fig. 2) is able to arise after two rounds of amplification on HRT-18 cells (P2), this indicates that viral particles must have been produced already after transfection of BHK-21 cells (P0) but at a level that was under the limit of detection. However, after several attempts at showing viral particles by electron microscopy, which included the use of colloidal gold-conjugated antibodies, we were never able to observe viral particles in transfected BHK-21 cells. This probably could be due to the very low production of recombinant virus particles, even for the wild-type HCoV-OC43 produced after transfection (P0) with the pBAC-OC43FL (Fig. 2A).
Our data concerning the complementation of HE mutant viruses using BHK-21 cells indicate that there is a dose-dependent effect of the HE protein on the production of infectious virus. Moreover, the relatively low levels of expression of the HE mRNA and protein in HE-expressing BHK-21 clones, combined to the fact that we were never able to detect its expression in HRT-18 subpopulations or isolated clones (data not shown), strongly suggest that the long-term ectopic expression of the HE protein is deleterious to the cell in contrast to the ectopic expression of the HE protein of BCoV (34). These results suggest that, in order to remain viable, the cells had to restrain the level of the HCoV-OC43 HE protein expression and, in the end, this limited the production of infectious HCoV-OC43 particles. Therefore, it appears logical that an approach where the HCoV-OC43 HE protein is expressed only transiently would be more efficient for complementation of the HE-defective mutant viruses (Fig. 4), producing up to 10 times more infectious virus compared to the BHK-21 clones HE6F-A3 and HE6F-E3 presented in Fig. 3. This approach of transient HE expression was also efficient to significantly increase the number of cells that produced the S viral protein after transfection (Fig. 4C). A hypothesis that could explain this observation is that recombinant viruses produced in the process of transfection would be able to infect surrounding BHK-21 cells, which are susceptible to HCoV-OC43 infection. Indeed, since a strong acetyl-esterase activity, related to the production of the HE protein, was already detectable at 24 h posttransfection, cells that produce the HE protein may also generate more newly produced infectious virus particles (Fig. 4D), which in turn would be able to infect surrounding cells Moreover, one can imagine that HE-deficient HCoV-OC43 particles were produced and that the functional HE protein independently produced in the transfected BHK-21 cells (Fig. 4) helped prevent a possible aggregation of viral particles, and to regain their capacity to infect a susceptible cell, as previously suggested (6, 35).
Results presented in Fig. 5 emphasize that the HCoV-OC43 HE protein plays an important role in the production of infectious virions that are able to infect susceptible cells and to eventually spread in cell culture. The HE protein of MHV-JHM was previously shown to affect the spread of the virus in the CNS without any influence on the production of infectious virus. However, in contrast to the latter report (27), it appears that the acetyl-esterase function of the HCoV-OC43 HE protein is important for optimal production of new infectious virions and eventual virus spreading. Indeed, even though the HE-complemented virus produced by cotransfection of pcDNA-HE and pBAC-OC43-HES40T partially disseminated within primary cultures of murine CNS, associated with a low production of new infectious virus, it remained far less effective than wild-type HCoV-OC43 (rOC/ATCC). Nevertheless, the fact that this HE-complemented mutant virus was able to produce this low amount of infectious virions that could partially spread in primary cell cultures (Fig. 5) suggests that HCoV-OC43 does not completely rely on the acetyl-esterase function of the HE protein for spread within the CNS.
The HE protein possesses both sialic acid binding properties and an acetyl-esterase enzymatic activity, the latter being postulated to be associated with a receptor-destroying enzymatic function (reviewed in reference 6). Over the years, a debate has been on-going about the fact that these two different activities of the HE protein could contribute to viral entry and/or release from the cell surface (reviewed in references 6, 10, and 36). Further studies are currently being pursued to determine the relative contribution of the HCoV-OC43 HE protein at early steps of infection, during viral entry, or at later stages, when the virus is assembling or has assembled and is ready to be released from the cell where it is produced. Our current results point toward an important role of the HE protein in helping to produce viral particles that are infectious and that will somehow more efficiently infect susceptible cells. Even though the precise mechanism remains to be elucidated, one can hypothesize that a fully enzymatically active HE could release the S protein from the 9-O-acetyl sialic acid on the cell surface. Indeed, HCoV-OC43 was previously shown to interact with this type of sialic acid (14) that could even serve as a receptor determinant for infection (37). In such a model, in which the S protein is responsible for primary attachment of HCoV-OC43 to the cell surface (6), deletion of the HE protein or inactivation of its acetyl-esterase activity could prevent fusion of cellular and viral membranes induced by the S protein, which would inhibit an efficient entry of HCoV-OC43 into susceptible cells.
The HCoV-OC43 and BCoV are closely related, as they share a 92% degree of nucleotide identity over the genome (38, 39), and this similarity increases to ca. 95% for the HE gene and protein alone (40). The BCoV HE protein was previously suggested to play a role in infection of susceptible cells, as suggested by inhibition of infection by neutralizing MAbs (26). Therefore, as was suggested for BCoV (26), our data suggest that the HE protein of HCoV-OC43 can affect viral infectivity. Moreover, MAb KD-9-40 directed against BCoV, used here, was previously shown to inhibit the acetyl-esterase activity of BCoV (41) and, as previously cited, this activity was suggested to be important for BCoV replication (11).
Even though, for most coronaviruses, the S protein is incorporated into the envelope in intracellular vesicles near the ERGIC area, coronaviruses go through the secretory pathway to be released from the cells after going through the Golgi apparatus. Considering that 9-O-acetylation of sialic acid linked to glycoproteins takes place in the Golgi apparatus in various cells, including the BHK-21 cell line (33), the S protein (incorporated or not in the virion) could interact with 9-O-acetylated glycoproteins present in this subcellular compartment. In the absence of a fully enzymatically active HE, this potential interaction of the S protein with 9-O-acetyl-sialic acid would not be cleaved off, which may affect the release of newly produced viral particles or the infectivity of these particles. Furthermore, even though lentiviruses assemble very differently than coronaviruses, data obtained using a pseudotyped lentivirus system suggest that the absence of a fully enzymatically active HE protein reduces the amount of S protein incorporated in the viral membrane and therefore decreases viral infectivity (C. Zhang et al., data not shown).
Taken together, our results show that infectious viruses (found either extracellularly in the cell supernatant or intracellularly) were never detected after transfection of BHK-21 cells with pBAC-OC43 cDNA clones without HE protein or bearing an acetyl-esterase deficient HE protein (HE0). The present study clearly indicates for the first time that the O-acetyl-esterase activity of HE protein of an HCoV plays a significant and important role in producing infectious virions and therefore in subsequent infection of cells by these newly produced viral particles to disseminate in cell culture. Furthermore, since HCoV-OC43 is neuroinvasive in both mice and humans, our results strongly suggest that HE represents an important viral factor that may influence the outcome of a CNS infection by this HCoV.
ACKNOWLEDGMENTS
This study was supported by discovery grant 42619-2009 from the National Sciences and Engineering Research Council of Canada and by operating grant MT-9203 from the Institute of Infection and Immunity of the Canadian Institutes of Health Research (CIHR) to P.J.T., who is the holder of the Tier-1 (Senior) Canada Research Chair in Neuroimmunovirology award. J.D. gratefully acknowledges studentship support from a CIHR Neuroinflammation Training Grant and the National Sciences and Engineering Research Council of Canada.
We thank Marcel Desrosiers, INRS-Institut Armand-Frappier, for technical assistance with the flow cytometry data and Hélène Jacomy and Élodie Brison for the establishment of primary cultures from the mouse CNS.
Footnotes
Published ahead of print 2 January 2013
REFERENCES
- 1. Vabret A, Dina J, Brison E, Brouard J, Freymuth F. 2009. Human coronaviruses. Pathol. Biol. 57:149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Buchmeier MJ, Lane TE. 1999. Viral-induced neurodegenerative disease. Curr. Opin. Microbiol. 2:398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Talbot PJ, Desforges M, Brison E, Jacomy H. 2011. Coronaviruses as encephalitis-inducing infectious agents. p 185–202 In Tkachev S. (ed), Non-flavivirus encephalitis. In-Tech, Rijeka, Croatia [Google Scholar]
- 4. de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya A, Holmes KV, Perlman S, Poon L, Rottier PJM, Talbot PJ, Woo PCY, Ziebuhr J. 2011. Coronaviridae, p 806–828 In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Academic Press, Inc, New York, NY [Google Scholar]
- 5. Brian DA, Hogue BG, Kienzle TE. 1995. The coronavirus hemagglutinin esterase glycoprotein, p 165–179 In Siddell SG. (ed), The Coronaviridae. Plenum Press, Inc, New York, NY [Google Scholar]
- 6. de Groot RJ. 2006. Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses. Glycoconj. J. 23:59–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luytjes W, Bredenbeek PJ, Noten AF, Horzinek MC, Spaan WJ. 1988. Sequence of mouse hepatitis virus A59 mRNA 2: indications for RNA recombination between coronaviruses and influenza C virus. Virology 166:415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cavanagh D. 1995. The coronavirus surface glycoprotein, p 73–113 In Siddell SG. (ed), The Coronaviridae. Plenum Press, Inc, New York, NY [Google Scholar]
- 9. Hogue BG, Machamer CE. 2008. Coronavirus structural proteins and virus assembly, p 179–200 In Perlman S, Gallagher T, Snijder EJ. (ed), Nidoviruses. ASM Press, Washington, DC [Google Scholar]
- 10. Rottier PJ. 1990. Background paper. Coronavirus M and HE: two peculiar glycoproteins. Adv. Exp. Med. Biol. 276:91–94 [DOI] [PubMed] [Google Scholar]
- 11. Vlasak R, Luytjes W, Leider J, Spaan W, Palese P. 1988. The E3 protein of bovine coronavirus is a receptor-destroying enzyme with acetylesterase activity. J. Virol. 62:4686–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zeng Q, Langereis MA, van Vliet AL, Huizinga EJ, de Groot RJ. 2008. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc. Natl. Acad. Sci. U. S. A. 105:9065–9069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kunkel F, Herrler G. 1993. Structural and functional analysis of the surface protein of human coronavirus OC43. Virology 195:195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vlasak R, Luytjes W, Spaan W, Palese P. 1988. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc. Natl. Acad. Sci. U. S. A. 85:4526–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schultze B, Gross HJ, Brossmer R, Herrler G. 1991. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J. Virol. 65:6232–6237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schultze B, Wahn K, Klenk HD, Herrler G. 1991. Isolated HE-protein from hemagglutinating encephalomyelitis virus and bovine coronavirus has receptor-destroying and receptor-binding activity. Virology 180:221–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mounir S, Talbot PJ. 1992. Sequence analysis of the membrane protein gene of human coronavirus OC43 and evidence for O-glycosylation. J. Gen. Virol. 73:2731–2736 [DOI] [PubMed] [Google Scholar]
- 18. Hill DP, Robertson KA. 1998. Differentiation of LA-N-5 neuroblastoma cells into cholinergic neurons: methods for differentiation, immunohistochemistry and reporter gene introduction. Brain Res. Brain Res. Protoc. 2:183–190 [DOI] [PubMed] [Google Scholar]
- 19. St-Jean JR, Desforges M, Almazan F, Jacomy H, Enjuanes L, Talbot PJ. 2006. Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone. J. Virol. 80:3670–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jacomy H, Fragoso G, Almazan G, Mushynski WE, Talbot PJ. 2006. Human coronavirus OC43 infection induces chronic encephalitis leading to disabilities in BALB/c mice. Virology 349:335–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lambert F, Jacomy H, Marceau G, Talbot PJ. 2008. Titration of human coronaviruses, HCoV-229E and HCoV-OC43, by an indirect immunoperoxidase assay. Methods Mol. Biol. 454:93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Regl G, Kaser A, Iwersen M, Schmid H, Kohla G, Strobl B, Vilas U, Schauer R, Vlasak R. 1999. The hemagglutinin-esterase of mouse hepatitis virus strain S is a sialate-4-O-acetylesterase. J. Virol. 73:4721–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 24. Arbour N, Day R, Newcombe J, Talbot PJ. 2000. Neuroinvasion by human respiratory coronaviruses. J. Virol. 74:8913–8921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jacomy H, Talbot PJ. 2003. Vacuolating encephalitis in mice infected by human coronavirus OC43. Virology 315:20–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Deregt D, Babiuk LA. 1987. Monoclonal antibodies to bovine coronavirus: characteristics and topographical mapping of neutralizing epitopes on the E2 and E3 glycoproteins. Virology 161:410–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kazi L, Lissenberg A, Watson R, de Groot RJ, Weiss SR. 2005. Expression of hemagglutinin esterase protein from recombinant mouse hepatitis virus enhances neurovirulence. J. Virol. 79:15064–15073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Deregt D, Gifford GA, Ijaz MK, Watts TC, Gilchrist JE, Haines DM, Babiuk LA. 1989. Monoclonal antibodies to bovine coronavirus glycoproteins E2 and E3: demonstration of in vivo virus-neutralizing activity. J. Gen. Virol. 70:993–998 [DOI] [PubMed] [Google Scholar]
- 29. Brison E, Jacomy H, Desforges M, Talbot PJ. 2011. Glutamate excitotoxicity is involved in the induction of paralysis in mice after infection by a human coronavirus with a single point mutation in its spike protein. J. Virol. 85:12464–12473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jacomy H, St-Jean JR, Brison E, Marceau G, Desforges M, Talbot PJ. 2010. Mutations in the spike glycoprotein of human coronavirus OC43 modulate disease in BALB/c mice from encephalitis to flaccid paralysis and demyelination. J. Neurovirol. 16:279–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Favreau DJ, Desforges M, St-Jean JR, Talbot PJ. 2009. A human coronavirus OC43 variant harboring persistence-associated mutations in the S glycoprotein differentially induces the unfolded protein response in human neurons as compared to wild-type virus. Virology 395:255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Favreau DJ, Meessen-Pinard M, Desforges M, Talbot PJ. 2012. Human coronavirus-induced neuronal programmed cell death is cyclophilin D dependent and potentially caspase dispensable. J. Virol. 86:81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dumermuth E, Beuret N, Spiess M, Crottet P. 2002. Ubiquitous 9-O-acetylation of sialoglycoproteins restricted to the Golgi complex. J. Biol. Chem. 277:18687–18693 [DOI] [PubMed] [Google Scholar]
- 34. Popova R, Zhang X. 2002. The spike but not the hemagglutinin/esterase protein of bovine coronavirus is necessary and sufficient for viral infection. Virology 294:222–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smits SL, Gerwig GJ, van Vliet AL, Lissenberg A, Briza P, Jkamerling JP, Vlasak R, de Groot RJ. 2005. Nidovirus sialate-O-acetylesterases: evolution and substrate specificity of coronaviral and toroviral receptor-destroying enzymes. J. Biol. Chem. 280:6933–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Weiss SR, Leibowitz JL. 2011. Coronavirus pathogenesis, Adv. Virus Res. 81:85–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krempl C, Schultze B, Herrler G. 1995. Analysis of cellular receptors for human coronavirus OC43. Adv. Exp. Med. Biol. 380:371–374 [DOI] [PubMed] [Google Scholar]
- 38. St-Jean JR, Jacomy H, Desforges M, Vabret A, Freymuth F, Talbot PJ. 2004. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J. Virol. 78:8824–8834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vijgen L, Keyaerts E, Moes E, Thoelen I, Wollants E, Lemey P, Vandamme AM, Van Ranst M. 2005. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J. Virol. 79:1595–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang XM, Kousoulas KG, Storz J. 1992. The hemagglutinin/esterase gene of human coronavirus strain OC43: phylogenetic relationships to bovine and murine coronaviruses and influenza C virus. Virology 186:318–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Parker MD, Yoo D, Babiuk LA. 1990. Expression and secretion of the bovine coronavirus hemagglutinin-esterase glycoprotein by insect cells infected with recombinant baculoviruses. J. Virol. 64:1625–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]