Abstract
The renewed interest in controlling Staphylococcus aureus infections using their natural enemies, bacteriophages, has led to the isolation of a limited number of virulent phages so far. These phages are all members of the Twortlikevirus, displaying little variance. We present two novel closely related (95.9% DNA homology) lytic myoviruses, Romulus and Remus, with double-stranded DNA (dsDNA) genomes of 131,333 bp and 134,643 bp, respectively. Despite their relatedness to Staphylococcus phages K, G1, ISP, and Twort and Listeria phages A511 and P100, Romulus and Remus can be proposed as isolates of a new species within the Twortlikevirus genus. A distinguishing feature for these phage genomes is the unique distribution of group I introns compared to that in other staphylococcal myoviruses. In addition, a hedgehog/intein domain was found within their DNA polymerase genes, and an insertion sequence-encoded transposase exhibits splicing behavior and produces a functional portal protein. From a phage therapy application perspective, Romulus and Remus infected approximately 70% of the tested S. aureus isolates and displayed promising lytic activity against these isolates. Furthermore, both phages showed a rapid initial adsorption and demonstrated biofilm-degrading capacity in a proof-of-concept experiment.
INTRODUCTION
In addition to asymptomatic colonization of the skin and mucosal surfaces, Staphylococcus aureus is responsible for various infections of the skin, systemic infections, and sepsis (1–3). Like many other bacteria, S. aureus produces biofilms which are difficult to eradicate (4). In combatting this versatile opportunistic pathogen, bacteriophages have become an interesting alternative to antibiotics. Staphylococcus phages belong to the viral families Myoviridae, Podoviridae, and Siphoviridae, with the last being temperate so far and therefore not suitable for phage therapy.
The myoviruses infecting S. aureus appear to be genotypically and proteomically related and have been classified into the subfamily Spounavirinae, genus Twortlikevirus (5, 6). This genus includes staphylococcal phages K (7), G1, Twort (8), Sb-1 (9), ISP (10, 11), A5-W, Staph1N, Fi200W, P4W, 676Z, A3R, MSA6 (12, 13), SA11 (14), and GH15 (15). Their double-stranded genomes contain 127,188 bp to 140,194 bp, display a G+C content of 30.04 to 30.60%, and contain 183 to 217 open reading frames (ORFs). Staphylococcus phage 812 likely is a twortlikevirus as well, but only fragments of its genome sequence were deposited in the National Center for Biotechnology Information database (16).
Phages ISP, Sb-1, 812, and K were able to infect antibiotic-resistant S. aureus isolates (9, 11, 17, 18) and therefore show promising therapeutic potential. For phage K, its ability to remove and prevent S. aureus biofilms was reported as well (19), while the administration of phage Sb-1 was proved to be effective in a cystic fibrosis patient (9).
A characteristic feature of twortlikeviruses infecting S. aureus is the interruption of genes with self-splicing mobile elements. This is illustrated by introns encoding endonucleases within the endolysin genes and the DNA polymerase genes of K and G1 (7, 8). For phage Twort, multiple self-splicing group I introns were experimentally demonstrated within a gene with unknown function and within the ribonucleotide reductase large-subunit gene (20, 21). Further, two introns were predicted within the large terminase gene as well as an intein-encoding region in the helicase gene (8). Phages Sb-1, ISP, Staph1N, Fi200W, P4W, 676Z, A3R, MSA6, and A5-W display an intron distribution similar to those of K and G1 (13), whereas phage GH15 lacks all previously described introns (15). Although splicing in phages was first believed to be a regulatory mechanism of DNA metabolism (22, 23), the discovery of introns in late genes (8, 21, 24) challenges the relevance of this hypothesis. So far, no other solid prediction of the meaning of splicing in phages can be made. Moreover, the appearance of split genes is more plentiful in phages infecting Gram-positive bacteria (25).
In this report, we present a microbiological and molecular examination of two newly isolated phages, Romulus and Remus, as well as a proof-of-concept experiment which illustrates their biofilm-degrading potential. Furthermore, both phages carry introns, a hedgehog/intein domain, and an insertion sequence in their genome-interrupting multiple open reading frames. Notably, Romulus and Remus represent a new species within the genus Twortlikevirus, which calls for expanding the existing taxonomy within this genus.
MATERIALS AND METHODS
Bacterial isolates and culture conditions.
For phage enrichment purposes, a collection of S. aureus isolates was obtained from J. Verhaegen (Laboratory of Clinical Bacteriology and Mycology, Campus Gasthuisberg, UZ Leuven, Belgium) and the Institut Pasteur of Brussels (Belgium). Additional Staphylococcus isolates were provided by M. Vaneechoutte (Department of Clinical Chemistry, Microbiology and Immunology, Ghent University, Belgium), T. Ito (Department of Bacteriology, Juntendo University, Tokyo, Japan), the Centre for Food and Microbial Technology (University of Leuven, Belgium), the Department of Pathology, Bacteriology and Poultry Diseases (Ghent University, Belgium), A. Toledo-Arana (Instituto de Agrobiotecnología and Departamento de Producción Agraria, Universidad Pública de Navarra, Spain), and C. Loc-Carrillo (VA SLC Health Care System, Department of Orthopaedics, Salt Lake City, UT) for host range screening. Isolates were grown in liquid Mueller-Hinton (MH) medium at 37°C, on solid MH medium (1.5% agar), or in MH soft agar overlays (0.35% agar).
Sampling and bacteriophage enrichment.
Raw inlet sewage water was collected in sterile Falcon tubes and homogenized by shaking at 4°C, after which bacteria and large waste particles were removed by centrifugation (30 min at 3,000 × g and 4°C, Sorvall Legend RT+; Thermo Fisher Scientific Inc., Waltham, MA) and filtration (Durapore polyvinylidene difluoride [PVDF] membrane, 0.45 μm; Millipore Corporation, Billerica, MA). After enrichment (26) in liquid MH medium (overnight at 37°C) against a selected S. aureus host strain, chloroform was added and the bacterial debris was removed by centrifugation (30 min at 3,000 × g and 4°C). Finally, potential phages were collected by centrifugation (90 min at 28,000 × g and 4°C), and the resulting pellet was dissolved in phage buffer (10 mM Tris-HCl, 10 mM MgSO4, 150 mM NaCl; pH 7.5) and spotted on a soft agar lawn containing the selected host. The presence of phages was evaluated after incubation (overnight at 37°C). In case of lysis spots, multiple single-plaque isolations were performed to obtain a pure phage isolate.
Bacteriophage amplification and purification.
High-titer phage stocks were obtained through amplification in liquid MH medium. Following visible lysis of the liquid culture, the lysate was incubated with chloroform for 10 min to kill any residual bacteria. Phages were collected through centrifugation (30 min at 3,000 × g and 4°C) Phage particles were purified and concentrated by polyethylene glycol (PEG) 8000 precipitation (27) and separated from contaminants by sedimentation in a CsCl (MP Biomedicals, Solon, OH) gradient {1.33, 1.45, 1.50, and 1.70 g/cm3 in SM buffer (50 mM Tris-HCl, 100 mM NaCl, 8 mM MgSO4, 0.01% gelatin [Sigma-Aldrich]; pH 7.5)} in Ultra-Clear tubes (Beckman Coulter, Inc., Fullerton, CA). Finally, the phage preparation was dialyzed against phage buffer using Slide-A-Lyzer G2 dialysis cassettes (2K MWCO; Pierce, Rockford, IL) and stored at 4°C.
Electron microscopic imaging.
Phage particles were collected by centrifugation at 25,000 × g for 1 h and washed twice in 0.1 M ammonium acetate (pH 7.0) using a Beckman high-speed centrifuge and a JA-18.1 fixed-angle rotor. Following deposition on carbon-coated copper grids and staining with 2% (wt/vol) potassium phosphotungstate (pH 7.0), phages were visualized in a Philips EM 300 transmission electron microscope (28).
Host range screening.
The host range was analyzed by spotting serial dilutions of phage on a soft agar lawn of S. aureus, Staphylococcus epidermidis, and Staphylococcus haemolyticus isolates. The results were confirmed with a plaque assay which permitted assessment of the efficiency of plating, i.e., the relative phage titer on a bacterial strain compared to the maximum titer observed.
Killing curve and adsorption curve.
For adsorption experiments, 100-μl phage samples were taken at fixed intervals (20 s and 1, 2, 5, 7, 10, 15, 20, and 25 min) and transferred to 850 μl of liquid MH medium supplemented with 50 μl of chloroform to lyse the remaining bacteria (26, 29). The number of unadsorbed or reversibly adsorbed phages was determined, and for each time point during the adsorption experiment, the adsorption rate constant was calculated. This value, k, is calculated by the equation where P represents the number of unadsorbed or reversibly adsorbed phages, P0 the initial number of phages, B the bacterial titer, and t the time after infection.
To generate a killing curve at different multiplicities of infection (MOI; the ratio of phage particles to host cells), a bacterial culture was infected at an optical density at 600 nm (OD600) of 0.3. The OD600 of the infected cultures was monitored every 20 min for 6 h and compared with that of an uninfected bacterial culture.
Biofilm degradation.
Biofilm degradation was examined using the MBEC assay (30, 31) in which 96 polystyrene pegs (Nunc-Immuno TSP; Thermo Fischer Scientific, Tournai, Belgium) are used to grow 96 identical biofilms. Overnight cultures grown in liquid MH medium were diluted (1:200) in 200 μl of MH medium supplemented with 1% (wt/vol) glucose. The peg lid was placed in the microtiter plate, sealed with Parafilm, and incubated at 37°C without shaking. Phage treatment of the biofilms occurred by placing the peg lid in 200 μl of phage buffer containing a specific amount of phages. To quantify the attached biofilms, the pegs were first washed briefly with phage buffer. Next, the biofilm mass was stained with 200 μl of a 0.1% (wt/vol) crystal violet (Merck) solution in 5% (vol/vol) isopropanol, 5% (vol/vol) methanol, and 90% (vol/vol) phosphate-buffered saline (PBS; 137 mM NaCl, 10 mM phosphate, 2.7 mM KCl; pH 7.4) for 30 min, washed again, and air dried for 30 min. The bound crystal violet was removed with 200 μl of 33% glacial acetic acid. The absorbance of crystal violet at 600 nm was measured using a Multiskan RC (Thermo Labsystems, Vantaa, Finland).
Results of treatment with different phages and titers were compared to treatment with phage buffer using a two-tailed Student t test (P < 0.05).
Biophysical stability.
The pH stability of the phage particles was tested by incubation in pH buffer (150 mM potassium chloride, 10 mM potassium dihydrogen phosphate, 10 mM sodium citrate, 10 mM boric acid; adjusted to pH 1 to pH 13) for 24 h at room temperature. Titration of the phage samples and comparison with a control sample incubated in phage buffer permitted the calculation of the relative survival of the phages. Similarly, the temperature stability was examined by incubation in phage buffer at different temperatures (−20°C, 4°C, 16°C, 37°C, 42°C, and 50°C), with 4°C as a control.
DNA isolation and genome sequencing.
Isolation of phage DNA was performed as described earlier (27). Sequencing of phage Remus was performed by Sanger shotgun sequencing and 454 sequencing (Plate-forme d'Analyses Génomiques at Laval University, Quebec, Quebec, Canada). Raw sequences arising from Sanger sequencing were filtered for vector contamination, and poor-quality reads were eliminated using the Pregap4 program (32). These were combined with the raw 454 sequence reads and assembled using the MIRA sequence assembly package, version 3.2.1.15 (33). The resulting assembly was imported into the Gap5 program (32), in which it was edited and proofread. Additional shotgun sequencing was done to elucidate ambiguities in A and T stretches. The phage Romulus genome was sequenced by 454 sequencing and upon comparison with that of phage Remus was verified with shotgun sequencing.
In silico analysis.
Phage genomes were autoannotated using MyRAST (http://blog.theseed.org/servers/presentations/t1/running-a-job-with-the-desktop-rast.html) and then proofread in Kodon (Applied Maths, Austin, TX). Shine-Dalgarno sequences of these predicted open reading frames and potential alternative start codons were checked manually. Using BLASTP (34) and HHpred (35) with databases available in August 2011 and in June 2012, putative protein functions were assigned for Remus and Romulus, respectively. Promoters were predicted using MEME (36), followed by manual verification. TransTerm (37) and ARNold (38, 39) were used to detect potential terminators, and the free energy of their secondary structures was calculated using Mfold (40). tRNAs were predicted using tRNAscan-SE (41) and ARAGORN (42). A comparative genome figure was generated using CGView (43), and DNA homology between phages was examined with EMBOSS stretcher (44, 45).
Proteome analysis.
Phage proteins were dried by extraction with methanol-chloroform (1:0.75, vol/vol). The protein pellet was resuspended in loading buffer (1% [wt/vol] sodium dodecyl sulfate [SDS], 6% [wt/vol] sucrose, 100 mM 1,4-dithiothreitol, 10 mM Tris-HCl [pH 6.8], 0.0625% [wt/vol] bromophenol blue) (46) and loaded onto a 12% polyacrylamide gel. Following electrophoresis (SDS-PAGE) (1 h at 200 V), the gel was stained with Simply Blue SafeStain (Invitrogen) and fragmented and trypsinized according to the protocol of Shevchenko et al. (47). The eluted peptides were subjected to electrospray ionization tandem mass spectrometry (ESI-MS-MS) on a LCQ Classic (ThermoFinnigan, San Jose, CA) equipped with a nano-LC column switching system as described earlier (48). The resulting MS-MS data were analyzed using Mascot, version 2.3.01, and Sequest, version 1.2.0.208, against a local database of all possible phage proteins.
Nucleotide sequence accession numbers.
The nucleotide sequences of Romulus and Remus have been deposited in the NCBI database under accession number JX846613.
RESULTS
Isolation and morphology.
Three phage propagation strains (Staphylococcus aureus PS47, PS92, and PSPB) and two clinical isolates (S. aureus KS8 and KS13) were selected for phage enrichment. Both clinical isolates carry genes conferring resistance to methicillin, penicillin, tetracycline, and aminoglycosides (11). Analysis of 23 sewage samples of the UZ Leuven Campus Gasthuisberg (Belgium), collected in January 2009, resulted in successful phage enrichment against PS47 and PSPB. In each case, plaques were about 1 mm in diameter and clear. Restriction analysis of both phage isolates (data not shown) revealed two similar but slightly different patterns, implying the isolation of two closely related lytic phages. The phages were named after Romulus and Remus, twin brothers and founders of Rome.
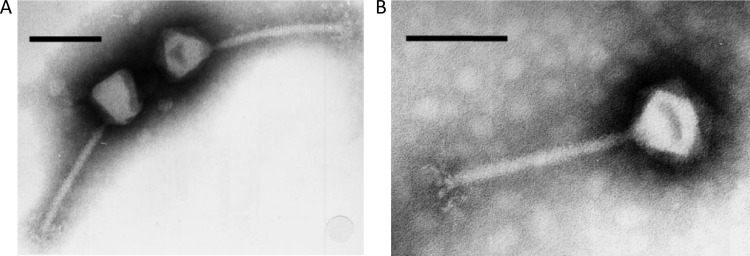
Based on their morphology examined by transmission electron microscopy (Fig. 1), Romulus and Remus were classified as members of the family Myoviridae. Both phages possess an isometric head with a diameter of 90 nm and a contractile tail with a length of 204 nm and a width of 17 nm. These dimensions correspond to those of other twortlikeviruses infecting S. aureus (5). Consequently, phage SA11, closely related to Romulus and Remus, was presumably erroneously described as a siphovirus (14).
Fig 1.
Morphology of Romulus and Remus. Shown are transmission electron microscopic images of myoviruses Romulus (A) and Remus (B) negatively stained with 2% (wt/vol) potassium phosphotungstate (pH 7.0). Scale bars represent 100 nm.
Host range screening.
A collection of 90 S. aureus isolates, 9 S. haemolyticus isolates, and 1 S. epidermidis isolate was used to assess the host range and corresponding efficiency of plating (see Table S1 in the supplemental material). For completeness, the host range of phage ISP was added as additional isolates were examined compared to earlier results (11). Romulus and Remus were able to infect 69% and 68% of all tested S. aureus isolates, respectively, which is considerably lower than the 87% infected by ISP. Further, while all human S. aureus isolates were sensitive to ISP infection, Romulus and Remus infected 28 and 30 out of 36 S. aureus isolates from patients, respectively. In contrast to the 21 phage propagation strains infected by ISP, only 19 and 18 of the 31 phage propagation strains were sensitive to Romulus and Remus, respectively. However, all the pig isolates which were insensitive to ISP infection were killed by Romulus and Remus. Similar to the case with ISP (11), none of the S. haemolyticus or S. epidermidis isolates showed sensitivity to Romulus and Remus infection. In general, Romulus and Remus displayed a moderate to low efficiency of plating, while a notable number of human isolates was infected with a high efficiency of plating by Remus. S. aureus isolates from rabbits are infected with a high efficiency of plating by all three phages.
Infection parameters.
To examine the infection parameters, the adsorption to and subsequent infection of Staphylococcus aureus subsp. aureus Rosenbach ATCC 6538 at different MOIs in liquid culture were studied. Adsorption experiments were performed in the presence of approximately 3 ×108 bacterial cells/ml. The adsorption assay (data not shown) indicated that approximately 60% of the Romulus and Remus particles were irreversibly adsorbed within 1 min, with adsorption rate constants of 2.1 × 10−9 ml/min and 2.2 × 10−9 ml/min, respectively. Furthermore, for both phages 80% of the viral particles were bound after 25 min, with adsorption rate constants of 2.6 × 10−11 ml/min and 3.7 × 10−11 ml/min, respectively. These adsorption rate constants illustrate an initial rapid adsorption followed by a reduced rate leading to maximum adsorption, as was already observed for Escherichia phage T2 in 1961 (49).
Romulus and Remus showed a similar infection pattern on S. aureus subsp. aureus Rosenbach ATCC 6538 (data not shown) and were both able to kill the culture at MOIs of 1 and 10. In case of both phages, new phage particles were liberated after a lytic cycle of approximately 40 min. At an MOI of 0.1 the optical density at 600 nm of the bacterial culture decreased, but not to a steady minimum within the time period of the experiment. In contrast, S. aureus subsp. aureus Rosenbach ATCC 6538 was only attenuated by ISP (11). Considering therapeutic implementation, killing curves were generated as well for S. aureus KS23 isolated from a patient at the UZ Leuven Campus Gasthuisberg (11). Again, the optical densities upon infection with Romulus and Remus progressed in parallel (data not shown). Further, the bacterial culture was killed for all three MOIs tested, while ISP could only cause a decrease in the optical density of which the minimum was not reached within the time period of the experiment (data not shown).
Biofilm degradation: proof-of-concept experiment.
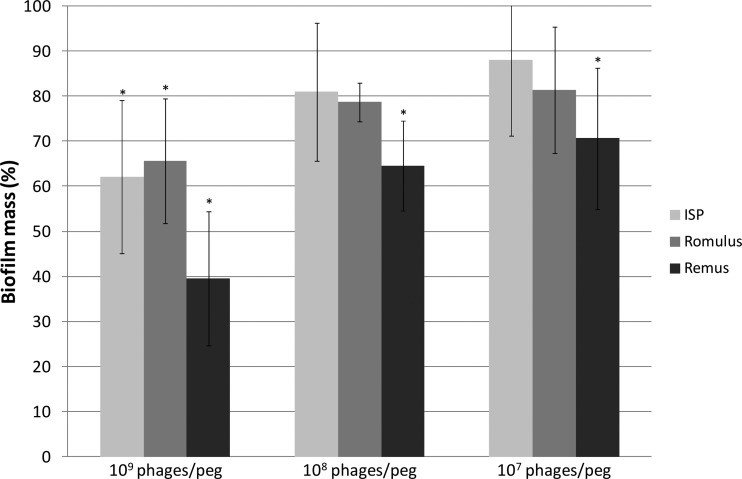
Before a proof-of-concept experiment was carried out to illustrate the biofilm-degrading potential of phages ISP, Romulus, and Remus, S. aureus PS47—a phage propagation strain characterized by a high relative biofilm-forming capacity (data not shown) and sensitivity to all three phages (see Table S1 in the supplemental material)—was selected. Using the MBEC assay (30, 31), we evaluated the effects of ISP, Romulus, and Remus on a 24-h pregrown biofilm of S. aureus PS47 (Fig. 2). Phage treatment for 2 to 6 h did not result in any substantial biofilm degradation, in contrast to when phages were applied for 24 h. The effects of ISP and Romulus treatments were significant only when 109 phages per peg were applied, while 107 to 109 Remus particles per peg resulted in a significant decrease of biofilm mass. When applied at 109 phages per peg, ISP, Romulus, and Remus were able to degrade the S. aureus PS47 biofilm by approximately 37.8%, 34.4%, and 60.4%, respectively. S. aureus Xen29, used to examine the biofilm-degrading activity of phage K (19), is considered to display an intermediate biofilm-degrading capacity using the MBEC assay in this study (data not shown). Hence, the biofilm-degrading potentials of phages ISP, Romulus, Remus, and K cannot be compared due to the variation in biofilm-forming capacities between S. aureus PS47 and S. aureus Xen29.
Fig 2.
Phage-mediated biofilm degradation. S. aureus PS47 biofilms grown on pegs for 24 h were treated with 109, 108, or 107 PFU of ISP, Romulus, or Remus for 24 h. Remaining biofilm mass was represented relative to the negative control, treatment with phage buffer. Equality of variances was analyzed using an F test; an asterisk marks significant values examined with a two-tailed Student t test (P < 0.05). Mean values and corresponding standard deviations were based on six independent experiments.
Biophysical stability.
To determine optimal storage and application conditions, the survival of Romulus and Remus at different temperatures and pH values was tested. Both phages appeared to be stable at 4°C and 16°C. At 37°C, the phage titers dropped by approximately 1 logarithmic unit, and at 42°C and 50°C, the logarithmic decrease was considerable. This should be taken into account in case of medical use. The pH range 5 to 9 caused no loss of phage infectivity, while a pH of 10 led to a logarithmic reduction of 1.5 and 4.5 units for Romulus and Remus, respectively. Like phage ISP (11), Romulus and Remus are therefore not suitable for oral administration without manipulation.
Genome analysis. (i) General features.
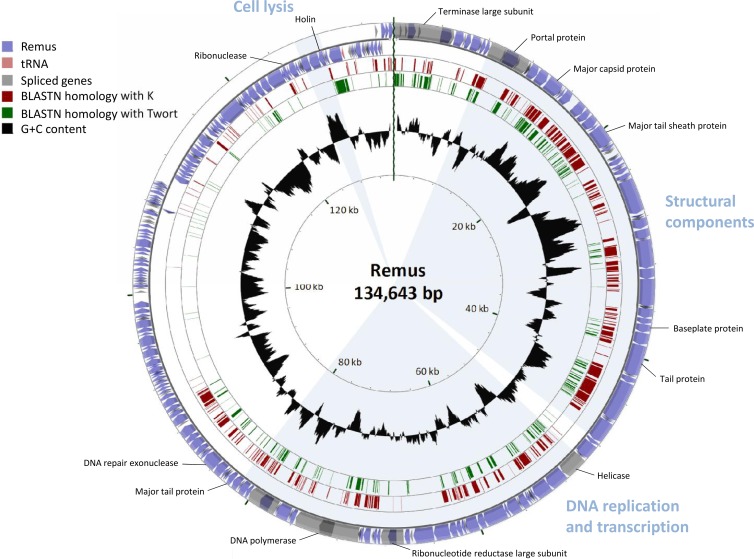
The linear double-stranded DNA of Remus with terminally redundant ends comprises 134,643 bp (JX846612) encoding 189 putative ORFs (Fig. 3; see alsoTable S2 in the supplemental material). The 131,333-bp genome of Romulus (JX846613), on the other hand, is highly similar to that of Remus, except for a 4.4-kb deletion resulting in the absence of 12 putative ORFs (ORF123 to ORF134), a 1.1-kb insertion leading to three additional putative genes (ORF140A to ORF140C), and two point mutations (see Table S3 in the supplemental material). Forty-three of these predicted genes are transcribed from the minus strand, including the lysis module. A combination of prediction programs followed by manual verification predicted 61 and 60 promoters and 26 and 25 terminators for Remus and Romulus, respectively. Furthermore, one tRNA encoding tRNASer was detected in an intergenic region. In addition, Romulus and Remus do not encode virulence-associated or toxic proteins and are therefore potentially safe to use in phage therapy.
Fig 3.
BLASTN genome comparison of the phages representing the three species identified within Twortlikevirus. The outer ring presents the ORFs of the circularized phage Remus genome (blue) and the numbering of every 10 ORFs. The two other rings display the BLASTN homology between Remus on one hand and K (red) and Twort (green) on the other hand. The inner ring shows the G+C content of the Remus genome (black). The three functional modules in the genome and some major predicted gene functions are indicated.
(ii) Comparative genome analysis.
Bioinformatic analysis of the Romulus and Remus genomes revealed 95.9% identity at the nucleotide level. A DNA comparison of both phage genomes with the genomes of K (AY176327), G1 (AY954969), Twort (NC_007021), Sb-1 (HQ163896), and ISP (FR852584) and the recently published phages Staph1N (JX080300), Fi200W (JX080303), P4W (JX080305), 676Z (JX080302), A3R (JX080301), MSA6 (JX080304), A5-W (EU418428), SA11 (JX194239), and GH15 (JQ686190) is represented in Table 1. The homology with the closely related phages (>90% nucleotide homology) K, G1, Sb-1, and ISP is as low as the homology between K, G1, Sb-1, and ISP on one hand and Twort on the other hand (approximately 55%). Furthermore, SA11 and the eight phages described by Łobocka et al. (13) and Gu and coworkers (15) (nucleotide identity between 88.6% and 99.9%) display a nucleotide identity between 53.2% and 58.7% to Romulus and Remus. In addition, Romulus, Remus, and SA11 share only 53.2%, 53.7%, and 55.8% DNA identity, respectively, with phage Twort. Figure 3 shows a BLASTN comparison of the genomes of phages Remus, K, and Twort.
Table 1.
Comparative nucleotide analysis between genomes of S. aureus phages present in the NCBI databasea
| Phage | % Nucleotide identity |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K | G1 | Twort | ISP | A5-W | Staph1N | Fi200W | P4W | 676Z | A3R | MSA6 | Sb-1 | GH15 | SA11 | Romulus | Remus | |
| K | 100 | 90.3 | 56.1 | 90.6 | 91.4 | 91.3 | 89.2 | 89.8 | 89.4 | 93.5 | 88.6 | 96.6 | 80.0 | 58.4 | 57.6 | 57.9 |
| G1 | 90.3 | 100 | 55.2 | 99.5 | 98.4 | 98.3 | 97.5 | 97.0 | 97.4 | 92.0 | 97.0 | 91.0 | 84.5 | 57.8 | 55.1 | 55.8 |
| Twort | 56.1 | 55.2 | 100 | 55.3 | 55.4 | 55.4 | 55.8 | 55.9 | 55.8 | 56.6 | 55.4 | 55.9 | 55.2 | 55.8 | 54.5 | 54.9 |
| ISP | 90.6 | 99.5 | 55.3 | 100 | 98.7 | 98.8 | 97.9 | 97.4 | 97.8 | 92.4 | 97.3 | 90.6 | 84.9 | 57.8 | 55.2 | 55.9 |
| A5-W | 91.4 | 98.4 | 55.4 | 98.7 | 100 | 99.9 | 97.3 | 98.1 | 97.3 | 93.2 | 96.1 | 90.6 | 84.5 | 58.0 | 55.5 | 56.2 |
| Staph1N | 91.3 | 98.3 | 55.4 | 98.8 | 99.9 | 100 | 97.4 | 98.2 | 97.4 | 93.3 | 96.2 | 90.5 | 84.6 | 58.0 | 55.5 | 56.1 |
| Fi200W | 89.2 | 97.5 | 55.8 | 97.9 | 97.3 | 97.4 | 100 | 99.1 | 99.6 | 94.1 | 95.4 | 88.9 | 83.9 | 57.5 | 54.9 | 55.6 |
| P4W | 89.8 | 97.0 | 55.9 | 97.4 | 98.1 | 98.2 | 99.1 | 100 | 99.0 | 94.7 | 94.9 | 89.2 | 83.6 | 57.6 | 55.1 | 55.8 |
| 676Z | 89.4 | 97.4 | 55.8 | 97.8 | 97.3 | 97.4 | 99.6 | 99.0 | 100 | 94.3 | 95.3 | 88.8 | 83.9 | 57.5 | 54.9 | 55.6 |
| A3R | 93.5 | 92.0 | 56.6 | 92.4 | 93.2 | 93.3 | 94.1 | 94.7 | 94.3 | 100 | 90.0 | 92.5 | 81.5 | 58.0 | 56.4 | 57.1 |
| MSA6 | 88.6 | 97.0 | 55.4 | 97.3 | 96.1 | 96.2 | 95.4 | 94.9 | 95.3 | 90.0 | 100 | 88.8 | 82.7 | 58.7 | 56.3 | 56.9 |
| Sb-1 | 96.6 | 91.0 | 55.9 | 90.6 | 90.6 | 90.5 | 88.9 | 89.2 | 88.8 | 92.5 | 88.8 | 100 | 79.7 | 58.4 | 57.7 | 57.9 |
| GH15 | 80.0 | 84.5 | 55.2 | 84.9 | 84.5 | 84.6 | 83.9 | 83.6 | 83.9 | 81.5 | 82.7 | 79.7 | 100 | 57.3 | 55.4 | 56.0 |
| SA11 | 58.4 | 57.8 | 55.8 | 57.8 | 58.0 | 58.0 | 57.5 | 57.6 | 57.5 | 58.0 | 58.7 | 58.4 | 57.3 | 100 | 82.1 | 84.6 |
| Romulus | 57.6 | 55.1 | 53.2 | 55.2 | 55.5 | 55.5 | 54.9 | 55.1 | 54.9 | 56.4 | 56.3 | 57.7 | 55.4 | 82.1 | 100 | 95.9 |
| Remus | 57.9 | 55.8 | 54.9 | 55.9 | 56.2 | 56.1 | 55.6 | 55.8 | 55.6 | 57.1 | 56.9 | 57.9 | 56.0 | 84.6 | 95.9 | 100 |
The nucleotide identity between the genomes of each set of two phages was calculated using EMBOSS stretcher. Homologies lower than 60% are shaded. Complete (100%) homology is indicated by bold type.
Despite this relatively low DNA similarity, Romulus and Remus display the same modular genome organization as K, G1, Twort, and ISP. Three major modules encoding virion proteins, DNA metabolism functions, and proteins necessary for cell lysis can be distinguished. Predicted structural proteins of Romulus, Remus, and SA11 show approximately 70 to 80% sequence identity to proteins of the other twortlikeviruses infecting S. aureus and approximately 60% identity to proteins of the Listeria phages A511 and P100 (5, 50, 51). In general, other proteins encoded by the Romulus and Remus genomes displayed an average 50% homology with proteins of K, G1, ISP, and Twort. For 60 and 58% of the proteins predicted for Remus and Romulus, no function could be assigned using BLASTP or HHpred, and for 109 and 118 proteins, no database counterpart was found at all. The lysis module, for example, appeared to be significantly different from the module observed in phages K, G1, ISP, and Twort. While the holins of Romulus and Remus are homologous to the Twort holin, the endolysin, interrupted by a group I intron in K, G1, and ISP, is replaced by two presumed endolysins homologous to siphoviral endolysins in Romulus and Remus.
(iii) Genes interrupted by self-splicing elements.
The presence of mobile elements in essential genes of Romulus and Remus is a characteristic feature among most known staphylococcal myoviruses (7, 8, 11, 13). In the case of Romulus and Remus, six protein-encoding genes were found to be interrupted by six coding and two noncoding introns, an insertion sequence, and a hedghoge/intein domain. The distribution of the coding introns, however, is dissimilar from those observed before in S. aureus phages (7, 8, 11, 13).
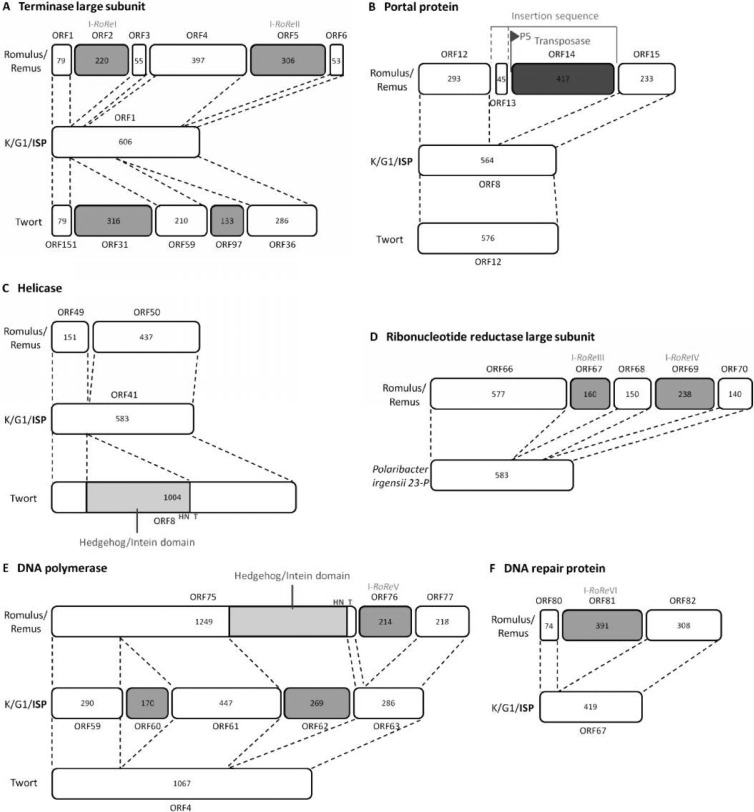
First, the terminase large-subunit gene is fragmented by three group I introns of which the first and the third encode a homing endonuclease (Fig. 4A). The three introns were shown to be unrelated, whereas the embedded homing endonucleases, named I-RoReI and I-RoReII, appeared to be mutually homologous, implying a common evolutionary ancestor. In addition, HHpred analysis revealed relatedness of these homing endonucleases with Bacillus thuringiensis phage 03058-36 I-Bth0305I, the first and until now only member of the EDxHD family of homing endonucleases discovered in a phage genome (52) (Table 2). Furthermore, I-RoReI and I-RoReII share homology with the homing endonucleases residing within the group I intron in the terminase large-subunit genes of Staphylococcus phages MSA6 (I-MsaI) and Twort (8, 13).
Fig 4.
Split genes in Romulus and Remus. Shown is a graphical representation of the splicing events in the terminase large-subunit gene (A), the gene encoding the portal protein (B), the helicase gene (C), the ribonucleotide reductase large-subunit gene (D), the DNA polymerase gene (E), and the gene encoding a DNA repair protein (F) in phages Romulus and Remus compared to those in phages K, G1, ISP, and Twort or Polaribacter irgensii 23-P. The open reading frames are presented as boxes, and the size of the corresponding amino acid sequence is indicated within each box. Group I intron-encoded homing endonucleases are depicted in blue. The insertion sequence-encoded transposase is depicted in dark blue. P5 represents the predicted promoter preceding ORF14. In the cases of K, G1, and ISP, the ORF numbering corresponds to that of ISP. H, histidine; N, asparagine; T, threonine.
Table 2.
Overview of the predicted homing endonucleases embedded in group I introns in the Romulus and Remus genomesa
| Homing endonuclease | ORF | Target gene | Homolog | Homing endonuclease family | Prediction program | E value |
|---|---|---|---|---|---|---|
| I-RoReI | ORF2 | Terminase large subunit | I-Bth0305I (B. thuringiensis phage 0305Φ8–36) | EDxHD | HHpred | 6.00E−17 |
| I-RoReII | ORF5 | Terminase large subunit | I-Bth0305I (B. thuringiensis phage 0305Φ8–36) | EDxHD | HHpred | 6.00E−17 |
| I-RoReIII | ORF67 | Ribonucleotide reductase large subunit | I-TevI (E. coli phage T4) | GIY-YIG | BLASTP/HHpred | 6.00E−10/1.10E−24 |
| I-RoReIV | ORF69 | Ribonucleotide reductase large subunit | I-HmuI (B. subtilis phage SPO1) | HNH | HHpred | 7.20E−40 |
| I-RoReV | ORF76 | DNA polymerase | I-HmuI (B. subtilis phage SPO1) | HNH | HHpred | 1.70E−21 |
| I-RoReVI | ORF81 | DNA repair protein | I-DmoI (Desulfurococcus mobilis) | LAGLIDADG | HHpred | 2.50E−24 |
For each predicted homing endonuclease, the encoding ORF and the target gene are given. Further, the homologous homing endonuclease, its family, the prediction program, and corresponding E value are indicated.
An insertion sequence comprising a transposase gene (ORF14) and an additional gene (ORF13) was detected between ORF12 and ORF15, both homologous to parts of the gene encoding the portal protein in K, G1, ISP, and Twort (Fig. 4B). The transposase gene is preceded by the predicted promoter P5. According to BLAST analysis, the transposase in Romulus and Remus is homologous to the IS605 family transposase OrfB. The insertion sequences of this family typically comprise two genes encoding OrfA and OrfB, serving together as the functional transposase (53). ORF13 and ORF14 are transcribed from the same strand, whereas the IS605 family gene encoding OrfA is transcribed from the reverse strand (54). This raises the question of whether ORF13 is part of the insertion sequence. This question is strengthened by the imperfect palindromic sequences (29 to 30 nucleotides [nt]), which replace the inverted terminal repeats in the IS605 family of insertion sequences (55), and the direct target repeats (7 nt) flanking ORF14.
Third, the helicase gene of Romulus and Remus is split into two open reading frames, ORF49 and ORF50 (Fig. 4C). The insert is approximately 245 nucleotides in length and probably constitutes a group I intron lacking an open reading frame. The interruption occurs at the same location as where a hedgehog/intein (Hint) domain is found in the helicase gene of phage Twort (8).
Next, the ribonucleotide (or ribonucleoside disphosphate) reductase large-subunit gene is interrupted by two group I introns (Fig. 4D). Again, the introns are not related. The embedded homing endonucleases, I-RoReIII and I-RoReIV, are homologous to I-TevI and I-HmuI, members of the GIY-YIG and HNH families, respectively (56, 57) (Table 2). The ribonucleotide reductase large-subunit gene itself is related to the Polaribacter irgensii 23-P ribonucleotide reductase large-subunit gene and does not show any protein homology with the ribonucleotide reductase large-subunit gene of phages K, G1, Twort, ISP, Sb-1, A5-W, Staph1N, Fi200W, P4W, 676Z, A3R, or MSA6.
The DNA polymerase gene not only is interrupted by a group I intron but also contains a Hint domain, an autoprocessing domain which undergoes a protein splicing reaction (58) (Fig. 4E). This Hint domain is located at the end of ORF75, with predicted N- and C-terminal boundaries at amino acid positions 732 and 1140, and typically contains a histidine and an asparagine residue at the C terminus, while a threonine residue follows the downstream splice site. The group I intron codes for a homing endonuclease, I-RoReV, which is also related to I-HmuI (Table 2).
The sixth group I intron-invaded gene is the gene coding for a DNA repair protein (Fig. 4F). This intron codes for I-RoReVI, an endonuclease similar to I-DmoI, an archaeal intron-encoded homing endonuclease belonging to the LAGLIDADG family (Table 2); BLASTP analysis showed homology with other members of this family as well (data not shown). Remarkably, an LAGLIDADG homing endonuclease has never been reported for a phage genome before. The homolog of this DNA repair protein in Staphylococcus phages MSA6 and 812 is interrupted by a group I intron as well (13, 16). In addition, the embedded homing endonuclease in Romulus and Remus, I-RoReVI, resembles the corresponding homing endonuclease in MSA6 and 812 (13, 16).
It should be mentioned that the above-mentioned group I introns were predicted with bioinformatic tools and therefore require experimental evidence.
Structural proteome analysis of Remus.
Mass spectrometric analysis of 20 gel slices of the Remus structural proteome resulted in the identification of 19 proteins (Table 3). Homologs of 18 of these proteins were already identified in earlier proteome studies (11, 59), while Gp42 of Remus can now be considered a component of the virus particle as well. Various Remus proteins were detected throughout the SDS-PAGE gel, illustrated by the major capsid (Gp18) and the major tail sheath (Gp25) protein, suggesting their abundant presence in the Remus virion. Similar observations were made for phage ISP (11).
Table 3.
Characteristics of the ESI-MS-MS-identified proteins of bacteriophage Remusa
| Gp | Band no. | Molecular mass (kDa) | No. of identified peptides | Sequence coverage (%) | HHpred search | E value | ISP homolog | Molecular mass (kDa) |
|---|---|---|---|---|---|---|---|---|
| Gp12 | 7 | 33.8 | 4 | 15.0 | Portal protein (HK97) | 5.8E−32 | Gp8 | 64.1 |
| Gp15 | 7 | 26.1 | 4 | 19.7 | Portal protein (HK97) | 2.2E−30 | Gp8 | 64.1 |
| Gp16 | 19 | 28.8 | 2 | 9.8 | Prohead protease (HK97) | 9.5E−34 | Gp9 | 28.6 |
| Gp18 | 1, 2, 4–12 | 51.0 | 16 | 52.9 | Major capsid protein (HK97) | 5.2E−05 | Gp11 | 51.2 |
| Gp21 | 11 | 33.6 | 2 | 9.3 | Gp14 | 33.7 | ||
| Gp25 | 5–9, 11, 14–16, 18–20 | 64.6 | 24 | 51.9 | Tail sheath protein (Mu) | 3.1E−61 | Gp18 | 64.5 |
| Gp26 | 18, 19 | 12.4 | 4 | 55.4 | Tail tube protein (T4) | 0.74 | Gp19 | 15.9 |
| Gp36 | 14 | 34.9 | 1 | 5.7 | Cell wall peptidase (NlpC) | 2.7E−24 | Gp28 | 34.6 |
| Gp38 | 13 | 30.9 | 9 | 36.0 | Baseplate structural protein (T4) | 96 | Gp30 | 29.3 |
| Gp39 | 16 | 20.1 | 2 | 13.3 | Gp31 | 20.0 | ||
| Gp41 | 9 | 39.2 | 3 | 9.2 | Baseplate J-like protein (P2) | 8.5E−43 | Gp33 | 39.2 |
| Gp42 | 3, 5 | 102.7 | 6 | 9.0 | Tail protein (P2) | 1.6E−4 | Gp34 | 116.3 |
| Gp43 | 17 | 19.2 | 3 | 20.8 | Baseplate structural protein (T4) | 3.7E−35 | Gp35 | 19.2 |
| Gp44 | 1, 2, 4–6 | 129.4 | 20 | 24.9 | Gp36 | 129.1 | ||
| Gp46 | 6, 14 | 72.8 | 5 | 9.7 | Gp38 | 72.6 | ||
| Gp85 | 15, 16, 18, 19 | 23.4 | 8 | 61.4 | Gp70 | 23.2 | ||
| Gp86 | 16, 18–20 | 18.2 | 6 | 54.7 | Major tail protein (lambda) | 9.5E−21 | Gp71 | 17.9 |
| Gp87 | 20 | 7.6 | 2 | 50.0 | Gp72 | 7.8 | ||
| Gp97 | 19 | 17.7 | 3 | 19.7 | Gp83 | 17.8 |
For each gene product (Gp), the band number(s) (according to Fig. 5), the molecular mass, the number of identified peptides (>99% protein identification probability with manual validation), and the protein sequence coverage are listed. Furthermore, the putative protein function according to HHpred and the corresponding E value are shown, as well as homologous ISP proteins and their molecular masses.
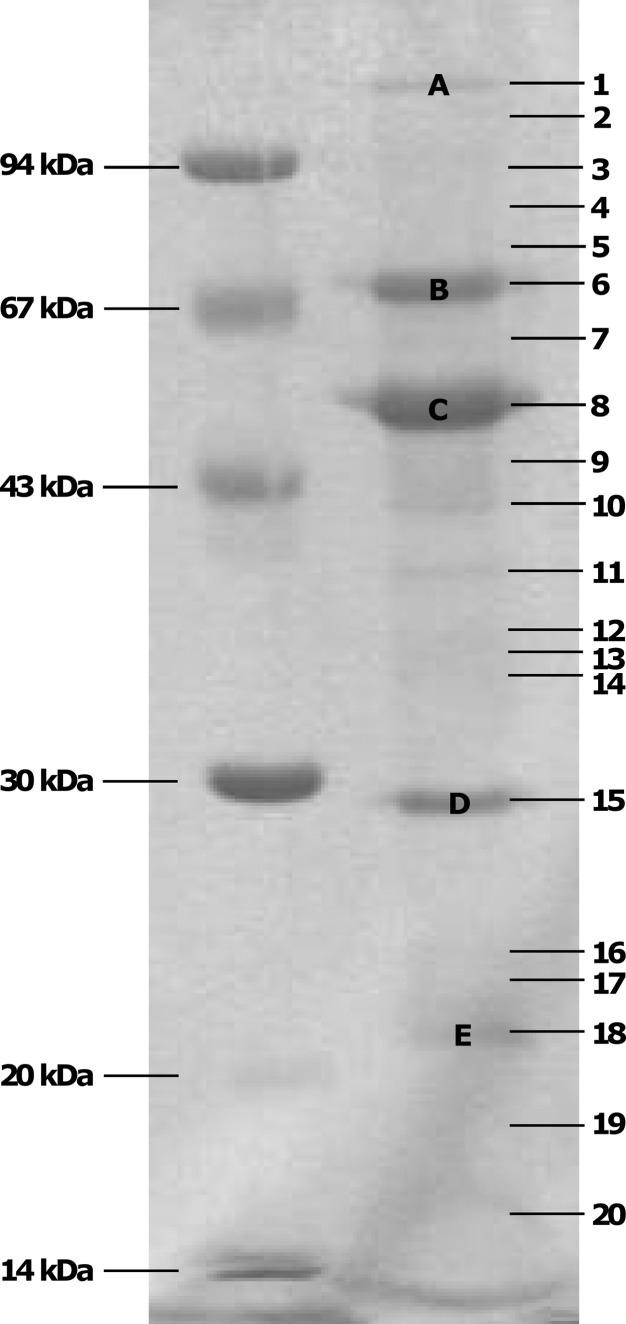
The five major protein bands (Fig. 5) of the Remus structural proteome are very similar to the bands visualized for phage ISP (11). However, band D of Remus is localized several kilodaltons lower. Because for ISP no corresponding molecular mass was found, this band was thought to be a proteolytically processed form of the tail sheath protein (59, 60). If this hypothesis is correct, the major tail sheath protein of Remus (Gp25), of which the molecular mass corresponds to that of the major tail sheath protein of ISP, is modified in a different way. For Gp12 as well as Gp15 of Remus, both corresponding to the portal proteins of K, G1, and ISP encoded by only one open reading frame, peptides were detected in band number 7 (Table 3).
Fig 5.
Coomassie G-250-stained SDS-PAGE gel of the structural proteome of bacteriophage Remus. A low-molecular-mass marker is indicated on the left. The numbers on the right correspond to the analyzed gel pieces of which the identified proteins are listed in Table 2. Prominent protein bands are indicated with capital letters.
A thorough HHpred analysis in which a higher E value was also taken into account (Table 3) permitted us to confirm and even specify the function of several identified structural proteins. For several Remus proteins, homology with Escherichia phages T4, HK97, and P2 was detected. Moreover, the tail protein Gp26 is homologous to the tail tube protein of T4, and Gp38, Gp41, and Gp43 are proposed to be components of the Remus baseplate. The molecular masses of Gp26 (12.4 kDa) and Gp42 (102.7 kDa) are markedly lower than the 15.9 kDa and 116.3 kDa of the putative tail tube protein and corresponding tail protein of phage ISP, probably resulting in different dimensions of the contractile myoviral tail.
DISCUSSION
The presence of the pathogenic bacterium S. aureus is often associated with hospital environments (61, 62), while bacteriophages are expected to be widespread in locations populated by their bacterial host. Therefore, this research focused on hospital sewage for phage isolation, resulting in the isolation of the novel phages Romulus and Remus. Phage isolation attempts in this research, however, have revealed that the isolation of new virulent Staphylococcus phages is not a straightforward approach, illustrated by the limited number of virulent Staphylococcus phages described in the scientific literature (7–9, 11–15, 63, 64).
Like the majority of the known virulent Staphylococcus phages, Romulus and Remus were assigned to the genus Twortlikevirus based on their morphological features, including a characteristic baseplate at the end of the tail (5, 13). Furthermore, both phages were shown to infect a considerable number of S. aureus isolates, which was also observed for twortlikeviruses K, ISP, Sb-1, and MSA6 (9, 11, 12, 17). From a genome perspective, Romulus and Remus are very closely related to each other and to phage SA11 but are substantially different from the other twortlikeviruses infecting S. aureus. At the DNA level, Romulus, Remus, and SA11 display no more than 60% homology with these staphylococcal myoviruses, suggesting that they represent a new species within Twortlikevirus (Fig. 3). This places Romulus, Remus, and SA11 in a new clade within Twortlikevirus (representative phage, Remus), next to and clearly divergent from phages K, G1, ISP, A5-W, Staph1N, Fi200W, P4W, 676Z, A3R, MSA6, and Sb-1 on one hand (representative phage, K) and phage Twort on the other hand (representative phage, Twort).
Although Staphylococcus phages MSA6, Twort, and 812 are quite distantly related to Romulus and Remus at the DNA level, these phages contain similar group I intron-embedded homing endonucleases. Interestingly, the related homing endonucleases target functional homologs. In addition to the ribonucleotide reductase large-subunit gene of Bacillus subtilis bacteriophage SPβ (24), the DNA polymerase gene of Romulus and Remus provides a second example of a group I intron and an intein coding sequence within the same gene. Furthermore, an insertion sequence in Romulus and Remus is unusually located between ORF12 and ORF15, genes encoding parts of the portal protein. For Gp12 and Gp15, peptides were detected in the same band. This implies that both genes are transcribed and translated into one mature, functional protein, suitable to be incorporated in the phage particle despite the presence of an insertion sequence. For the first time, evidence was provided for intron-like splicing behavior of an insertion sequence-encoded transposase.
Despite their close relatedness, the differences in host range and efficiency of plating between Romulus and Remus are remarkable. Their host range is, however, relatively narrow compared to that of phage ISP, known for its broad host range, including human clinical isolates (10, 11). Nonetheless, the in vitro infection patterns of Romulus and Remus appeared more promising than those of ISP, implying their suitability for efficient control of sensitive S. aureus isolates. Furthermore, the biofilm-degrading potential of Romulus and Remus makes their therapeutic application even more attractive. In consideration of their therapeutic implementation, the temperature and pH range in which Romulus and Remus are stable were determined.
To conclude, Romulus and Remus are considered to be members of a new species within Twortlikevirus, containing a unique scale of group I introns, an intein, and an insertion sequence in their genomes. In addition, the characterization of both phage isolates, comprising microbiological as well as molecular aspects, demonstrated their appropriateness for therapeutic application.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Federal Public Service of Health, Food Chain Safety and Environment, Belgium (contract RT 08/6 PHAGE).
We thank H.-W. Ackermann for performing the electron microscopic imaging.
Footnotes
Published ahead of print 9 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02763-12.
REFERENCES
- 1. Jarraud S, Mougel C, Thioulouse J, Lina G, Meugnier H, Forey F, Nesme X, Etienne J, Vandenesch F. 2002. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lowy FD. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520–532 [DOI] [PubMed] [Google Scholar]
- 3. Veien NK. 1998. The clinician's choice of antibiotics in the treatment of bacterial skin infection. Br. J. Dermatol. 139:30–36 [DOI] [PubMed] [Google Scholar]
- 4. Patel R. 2005. Biofilms and antimicrobial resistance. Clin. Orthop. Relat. Res. 437:41–47 [DOI] [PubMed] [Google Scholar]
- 5. Klumpp J, Lavigne R, Loessner MJ, Ackermann HW. 2010. The SPO1-related bacteriophages. Arch. Virol. 155:1547–1561 [DOI] [PubMed] [Google Scholar]
- 6. Lavigne R, Darius P, Summer E, Seto D, Mahadevan P, Nilsson A, Ackermann H, Kropinski A. 2009. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 9:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O'Flaherty S, Coffey A, Edwards R, Meaney W, Fitzgerald GF, Ross RP. 2004. Genome of staphylococcal phage K: a new lineage of Myoviridae infecting gram-positive bacteria with a low G+C content. J. Bacteriol. 186:2862–2871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kwan T, Liu J, DuBow M, Gros P, Pelletier J. 2005. The complete genomes and proteomes of 27 Staphylococcus aureus bacteriophages. Proc. Natl. Acad. Sci. U. S. A. 102:5174–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kvachadze L, Balarjishvili N, Meskhi T, Tevdoradze E, Skhirtladze N, Pataridze T, Adamia R, Topuria T, Kutter E, Rohde C, Kutateladze M. 2011. Evaluation of lytic activity of staphylococcal bacteriophage Sb-1 against freshly isolated clinical pathogens. Microb. Biotechnol. 4:643–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Merabishvili M, Pirnay J-P, Verbeken G, Chanishvili N, Tediashvili M, Lashkhi N, Glonti T, Krylov V, Mast J, Van Parys L, Lavigne R, Volckaert G, Mattheus W, Verween G, De Corte P, Rose T, Jennes S, Zizi M, De Vos D, Vaneechoutte M. 2009. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS One 4:e4944 doi:10.1371/journal.pone.0004944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vandersteegen K, Mattheus W, Ceyssens P-J, Bilocq F, De Vos D, Pirnay J-P, Noben J-P, Merabishvili M, Lipinska U, Hermans K, Lavigne R. 2011. Microbiological and molecular assessment of bacteriophage ISP for the control of Staphylococcus aureus. PLoS One 6:e24418 doi:10.1371/journal.pone.0024418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kwiatek M, Parasion S, Mizak L, Gryko R, Bartoszcze M, Kocik J. 2012. Characterization of a bacteriophage, isolated from a cow with mastitis, that is lytic against Staphylococcus aureus strains. Arch. Virol. 157:225–234 [DOI] [PubMed] [Google Scholar]
- 13. Łobocka M, Hejnowicz MS, Dąbrowski K, Gozdek A, Kosakowski J, Witkowska M, Ulatowska MI, Weber-Dąbrowska B, Kwiatek M, Parasion S, Gawor J, Kosowska H, Głowacka A. 2012. Genomics of staphylococcal Twort-like phages—potential therapeutics of the post-antibiotic era. Adv. Virus Res. 83:143–216 [DOI] [PubMed] [Google Scholar]
- 14. Kim MS, Myung H. 2012. Complete genome of Staphylococcus aureus phage SA11. J. Virol. 86:10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gu J, Liu X, Lu R, Li Y, Song J, Lei L, Sun C, Feng X, Du C, Yu H, Yang Y, Han W. 2012. Complete genome sequence of Staphylococcus aureus bacteriophage GH15. J. Virol. 86:8914–8915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaspárek P, Pantùcek R, Kahánková J, Rùzicková V, Doskar J. 2007. Genome rearrangements in host-range mutants of the polyvalent staphylococcal bacteriophage 812. Folia Microbiol. (Praha) 52:331–338 [DOI] [PubMed] [Google Scholar]
- 17. O'Flaherty S, Ross RP, Meaney W, Fitzgerald GF, Elbreki MF, Coffey A. 2005. Potential of the polyvalent anti-Staphylococcus bacteriophage K for control of antibiotic-resistant staphylococci from hospitals. Appl. Environ. Microbiol. 71:1836–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pantùček R, Rosypalová A, Doškaø J, Kailerová J, Rùžičková V, Borecká P, Š. Snopková Horváth R, Götz F, Rosypal S. 1998. The polyvalent staphylococcal phage φ812: its host-range mutants and related phages. Virology 246:241–252 [DOI] [PubMed] [Google Scholar]
- 19. Kelly D, McAuliffe O, Ross RP, Coffey A. 2012. Prevention of Staphylococcus aureus biofilm formation and reduction in established biofilm density using a combination of phage K and modified derivatives. Lett. Appl. Microbiol. 54:286–291 [DOI] [PubMed] [Google Scholar]
- 20. Landthaler M, Begley U, Lau NC, Shub DA. 2002. Two self-splicing group I introns in the ribonucleotide reductase large subunit gene of Staphylococcus aureus phage Twort. Nucleic Acids Res. 30:1935–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Landthaler M, Shub DA. 1999. Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: introns in multiple genes, a single gene with three introns, and exon skipping by group I ribozymes. Proc. Natl. Acad. Sci. U. S. A. 96:7005–7010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goodrich-Blair H, Scarlato V, Gott JM, Xu M-Q, Shub DA. 1990. A self-splicing group I intron in the DNA polymerase gene of Bacillus subtilis bacteriophage SPO1. Cell 63:417–424 [DOI] [PubMed] [Google Scholar]
- 23. Gott JM, Shub DA, Belfort M. 1986. Multiple self-splicing introns in bacteriophage T4: evidence from autocatalytic GTP labeling of RNA in vitro. Cell 47:81–87 [DOI] [PubMed] [Google Scholar]
- 24. Lazarevic V, Soldo B, Düsterhöft A, Hilbert H, Mauël C, Karamata D. 1998. Introns and intein coding sequence in the ribonucleotide reductase genes of Bacillus subtilis temperate bacteriophage SPβ. Proc. Natl. Acad. Sci. U. S. A. 95:1692–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Edgell DR, Belfort M, Shub DA. 2000. Barriers to intron promiscuity in bacteria. J. Bacteriol. 182:5281–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Adams MH. 1959. Bacteriophages. Interscience Publishers Inc., New York, NY [Google Scholar]
- 27. Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. Ackermann HW. 2009. Basic phage electron microscopy, p 113–126 In Clokie MRJ, Kropinski AM. (ed), Bacteriophages: methods and protocols, vol 1. Isolation, characterization, and interactions. Humana Press, Clifton, NJ: [DOI] [PubMed] [Google Scholar]
- 29. Ceyssens PJ, Lavigne R, Mattheus W, Chibeu A, Hertveldt K, Mast J, Robben J, Volckaert G. 2006. Genomic analysis of Pseudomonas aeruginosa phages LKD16 and LKA1: establishment of the phiKMV subgroup within the T7 supergroup. J. Bacteriol. 188:6924–6931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ceri H, Olson M, Morck D, Storey D, Read R, Buret A, Olson B. 2001. The MBEC assay system: multiple equivalent biofilms for antibiotic and biocide susceptibility testing. Methods Enzymol. 337:377–385 [DOI] [PubMed] [Google Scholar]
- 31. Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 37:1771–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bonfield JK, Whitwham A. 2010. Gap5—editing the billion fragment sequence assembly. Bioinformatics 26:1699–1703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chevreux B, Wetter T, Suhai S. 1999. Genome sequence assembly using trace signals and additional sequence information. Comput. Sci. Biol. Proc. German Conf. Bioinformatics 99:45–56 [Google Scholar]
- 34. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 35. Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248 doi:10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p 28–36 In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology AAAI Press, Menlo Park, CA: [PubMed] [Google Scholar]
- 37. Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL. 2000. Prediction of transcription terminators in bacterial genomes. J. Mol. Biol. 301:27–33 [DOI] [PubMed] [Google Scholar]
- 38. Gautheret D, Lambert A. 2001. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J. Mol. Biol. 313:1003–1011 [DOI] [PubMed] [Google Scholar]
- 39. Macke T, Ecker D, Gutell R, Gautheret D, Case DA, Sampath R. 2001. RNAMotif—a new RNA secondary structure definition and discovery algorithm. Nucleic Acids Res. 29:4724–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32:11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stothard P, Wishart DS. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21:537–539 [DOI] [PubMed] [Google Scholar]
- 44. Myers EW, Miller W. 1988. Optimal alignments in linear space. Comput. Appl. Biosci. 4:11–17 [DOI] [PubMed] [Google Scholar]
- 45. Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277 [DOI] [PubMed] [Google Scholar]
- 46. Moak M, Molineux IJ. 2004. Peptidoglycan hydrolytic activities associated with bacteriophage virions. Mol. Microbiol. 51:1169–1183 [DOI] [PubMed] [Google Scholar]
- 47. Shevchenko A, Wilm M, Vorm O, Mann M. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850–858 [DOI] [PubMed] [Google Scholar]
- 48. Lavigne R, Noben JP, Hertveldt K, Ceyssens PJ, Briers Y, Dumont D, Roucourt B, Krylov VN, Mesyanzhinov VV, Robben J, Volckaert G. 2006. The structural proteome of Pseudomonas aeruginosa bacteriophage phiKMV. Microbiology 152:529–534 [DOI] [PubMed] [Google Scholar]
- 49. Baylor MB, Silver SD. 1961. Studies on the additivity of action of genes affecting host range in coliphage T2. Virology 14:167–176 [DOI] [PubMed] [Google Scholar]
- 50. Carlton RM, Noordman WH, Biswas B, de Meester ED, Loessner MJ. 2005. Bacteriophage P100 for control of Listeria monocytogenes in foods: genome sequence, bioinformatic analyses, oral toxicity study, and application. Regul. Toxicol. Pharmacol. 43:301–312 [DOI] [PubMed] [Google Scholar]
- 51. Klumpp J, Dorscht J, Lurz R, Bielmann R, Wieland M, Zimmer M, Calendar R, Loessner MJ. 2008. The terminally redundant, nonpermuted genome of Listeria bacteriophage A511: a model for the SPO1-like myoviruses of gram-positive bacteria. J. Bacteriol. 190:5753–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Taylor GK, Heiter DF, Pietrokovski S, Stoddard BL. 2011. Activity, specificity and structure of I-Bth0305I: a representative of a new homing endonuclease family. Nucleic Acids Res. 39:9705–9719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kersulyte D, Akopyants NS, Clifton SW, Roe BA, Berg DE. 1998. Novel sequence organization and insertion specificity of IS605 and IS606: chimaeric transposable elements of Helicobacter pylori. Gene 223:175–186 [DOI] [PubMed] [Google Scholar]
- 54. Kersulyte D, Mukhopadhyay AK, Shirai M, Nakazawa T, Berg DE. 2000. Functional organization and insertion specificity of IS607, a chimeric element of Helicobacter pylori. J. Bacteriol. 182:5300–5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Barabas O, Ronning DR, Guynet C, Hickman AB, Ton-Hoang B, Chandler M, Dyda F. 2008. Mechanism of IS200/IS605 family DNA transposases: activation and transposon-directed target site selection. Cell 132:208–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Goodrich-Blair H, Shub DA. 1996. Beyond homing: competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell 84:211–221 [DOI] [PubMed] [Google Scholar]
- 57. Quirk SM, Bell-Pedersen D, Belfort M. 1989. Intron mobility in the T-even phages: high frequency inheritance of group I introns promoted by intron open reading frames. Cell 56:455–465 [DOI] [PubMed] [Google Scholar]
- 58. Hall TMT, Porter JA, Young KE, Koonin EV, Beachy PA, Leahy DJ. 1997. Crystal structure of a hedgehog autoprocessing domain: homology between hedgehog and self-splicing proteins. Cell 91:85–97 [DOI] [PubMed] [Google Scholar]
- 59. Eyer L, Pantùček R, Zdráhal Z, Konečná H, Kašpárek P, Rùžičková V, Hernychová L, Preisler J, Doškaø J. 2007. Structural protein analysis of the polyvalent staphylococcal bacteriophage 812. Proteomics 7:64–72 [DOI] [PubMed] [Google Scholar]
- 60. Chibani-Chennoufi S, Dillmann M-L, Marvin-Guy L, Rami-Shojaei S, Brüssow H. 2004. Lactobacillus plantarum bacteriophage LP65: a new member of the SPO1-like genus of the family Myoviridae. J. Bacteriol. 186:7069–7083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Boyce JM, Havill NL, Otter JA, Adams NM. 2007. Widespread environmental contamination associated with patients with diarrhea and methicillin-resistant Staphylococcus aureus colonization of the gastrointestinal tract. Infect. Control Hosp. Epidemiol. 28:1142–1147 [DOI] [PubMed] [Google Scholar]
- 62. Boyce JM, Potter-Bynoe G, Chenevert C, King T. 1997. Environmental contamination due to methicillin-resistant Staphylococcus aureus: possible infection control implications. Infect. Control Hosp. Epidemiol. 18:622–627 [PubMed] [Google Scholar]
- 63. Son J-S, Lee S-J, Jun S, Yoon S, Kang S, Paik H, Kang J, Choi Y-J. 2010. Antibacterial and biofilm removal activity of a podoviridae Staphylococcus aureus bacteriophage SAP-2 and a derived recombinant cell-wall-degrading enzyme. Appl. Microbiol. Biotechnol. 86:1439–1449 [DOI] [PubMed] [Google Scholar]
- 64. Vybiral D, Takáč M, Loessner M, Witte A, von Ahsen U, Bläsi U. 2003. Complete nucleotide sequence and molecular characterization of two lytic Staphylococcus aureus phages: 44AHJD and P68. FEMS Microbiol. Lett. 219:275–283 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.