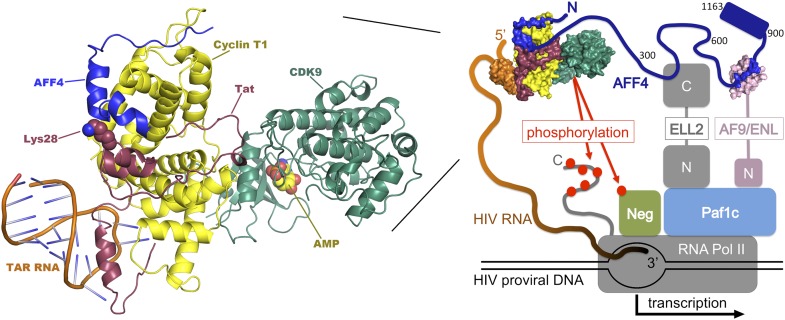
Figure 1.
Transcription of DNA in a cell infected by HIV. The HIV DNA that has been integrated into the host cell's genome is transcribed by the cellular enzyme RNA polymerase II (RNA Pol II), but this process usually pauses after fewer than 50 nucleotides have been transcribed. In order to synthesize long transcripts, the HIV genome encodes a protein called Tat (shown here in maroon), which binds newly formed viral RNA transcripts (orange) at a hairpin-shaped structure called TAR. The Tat:TAR complex then binds a protein complex called P-TEFb (which is composed of cyclin T1 [yellow] and CDK9 [green]), plus a scaffolding protein, AFF4 (blue; numbers correspond to amino-acid positions within the protein). Note that Tat and AFF4 may interact directly on the cyclin T1 surface and that acetylation of a lysine residue at position 28 in Tat (which has been shown to enhance Tat activity; Ott et al., 2011) may help regulate this interaction. The AFF4 tethers additional elongation factors (ELL-2 and either ENL or AF9; only shown in the right-hand figure, ‘N' and ‘C' represent the N and C terminal domains) to produce a ‘super elongation complex' that regulates the transition to efficient transcriptional elongation. Left, expanded model of the P-TEFb:AFF4:Tat:TAR complex. Right, overview of the interactions within the super elongation complex, including interactions with RNA Pol II and the Paf1 complex (Paf1c), which is a positive regulator of transcription. Phosphorylation (red dots) by P-TEFb of negative factors that inhibit transcription (NELF and DSIF, abbreviated ‘Neg') and of the C-terminal tail of RNA Pol II, releases the stalled polymerase and increases transcription of the viral genome. The composite model shown here was assembled from structures of human Cdk9:cyclin T1:AFF4 (pdb 4imy), Tat:p-TEFb (pdb 3mi9), and EIAV Tat:TAR (pdb 2w2h).