Abstract
Shiga toxin-producing E. coli (STEC) are isolated from human patients with bloody diarrhea, hemorrhagic colitis (HC), and hemolytic uremic syndrome (HUS). In the last years, the infections with non-O157 serotypes are increasing their frequency of association with human disease. STEC produce Shiga toxin (Stx) and other virulence factors that could contribute to human pathogenesis. Cattle are the main reservoir and the transmission to humans is through the consumption of undercooked meat, non-pasteurized dairy products, and vegetables or water contaminated with feces. We have previously determined that O130:H11 and O178:H19 serotypes were the most prevalent in dairy cows from Argentina. In the present study, 37 and 25 STEC isolates from dairy cows belonging to O130:H11 and O178:H19 serotypes, respectively, were characterized regarding to their cytotoxicity on Vero cells, stx subtypes, presence of sab and typing by multiple-locus variable-number tandem repeat analysis (MLVA). All strains demonstrated a cytotoxic effect, and in O130:H11 isolates, stx2EDL933 was the predominant subtype. In O178:H19 isolates the main stx2 subtype was stx2vha. The sab gene was detected in 65 and 24% of the isolates belonging to O130:H11 and O178:H19, respectively. Only one MLVA profile was identified among the O130:H11 isolates meanwhile 10 MLVA profiles were detected among the O178:H19 isolates which were grouped in two main clusters. In conclusion, our data show that O130:H11 and O178:H19 STEC isolates encode virulence factors associated with severe human disease and both serotypes should be considered for routinely testing. Our subtyping experiments showed that isolates could be distinguished based on the stx2 subtype and the presence/absence of sab gene, and for isolates belonging to O178:H19, also when the MLVA type was considered. However, MLVA subtyping of O130:H11 isolates will require the development of more specific markers.
Keywords: STEC, dairy cattle, MLVA, Shiga toxin
Introduction
Shiga toxin-producing E. coli (STEC) cause bloody diarrhea, hemorrhagic colitis (HC) and hemolytic uremic syndrome (HUS) in humans (Pearce et al., 2004; Giugno et al., 2007). Most outbreaks have been attributed to O157:H7 serotype (Mora et al., 2004) but infections with non-O157 serotypes are also being frequently associated with HC and HUS (Bettelheim, 2007). In several countries STEC O157:H7 have been frequently isolated from cattle but several studies in Argentina have detected mainly non-O157:H7 serotypes (Meichtri et al., 2004; Padola et al., 2004; Fernández et al., 2010). Cattle are the main reservoir of STEC and the transmission to humans occurs through the consumption of undercooked meat, non-pasteurized dairy products, and vegetables or water contaminated with feces (Hussein and Sakuma, 2005). Direct contact with cattle and dairy farm environment has been reported also as a possible source for STEC human transmission (Oliver et al., 2005).
The main virulence factor of STEC is the production of Shiga toxins (Stx1 and Stx2) (Paton and Paton, 1998; Gyles, 2007). Stx1 group includes few subtypes, while the Stx2 is a more heterogeneous group and comprises an expanding number of subtypes (such as Stx2EDL933, Stx2vha, Stx2vhb, Stx2O118, Stx2dact, Stx2e, Stx2f, and Stx2g). Stx subtypes differ in their degree of association with HC and HUS cases, being Stx2O118 (formerly identified as Stx2d-Ount), Stx2e, Stx2f, and Stx2g not frequently associated with severe human disease (Friedrich et al., 2002; Karch et al., 2005; Prager et al., 2009, 2011). Other virulence factors that could contribute to the pathogenesis are intimin, encoded by the eae gene and responsible for the intimate attachment of STEC to intestinal epithelial cells, an enterohaemolysin (EhxA), an autoagglutinating adhesin (Saa) and a novel STEC autotransporter (Sab) described for first time in a saa-positive O113:H21 strain, which participates in adhesion and biofilm formation (Herold et al., 2009). The ehxA, saa, and sab genes are located in a megaplasmid (Paton and Paton, 1998; Paton et al., 2001; Herold et al., 2009).
In Argentina, O130:H11 and O178:H19 were the most prevalent serotypes isolated from dairy cows (Fernández et al., 2010) and were also identified by Masana et al. (2011) in beef abattoirs and by López et al. (2012) in feedlot cattle. Both serotypes have been isolated from HC and HUS cases in several countries and have been found among human STEC isolates received between 2000–2010 by the CDC National E. coli Reference Laboratory (Blanco et al., 2004; Fremaux et al., 2006; Giugno et al., 2007).
In the present study, we further characterized O130:H11 and O178:H19 STEC isolated by Fernández et al. (2010) from dairy farms regarding their cytotoxicity on Vero cells, stx subtypes, presence of sab gene and typing by multiple-locus variable-number tandem repeat analysis (MLVA), in order to evaluate the genetic diversity of isolates belonging to these serotypes which are prevalent in dairy cattle.
Materials and methods
Bacterial strains
The bacterial strains used in this study were 37 STEC O130:H11 and 25 STEC O178:H19 isolated from dairy cows in five farms (named A, B, C, D, and E) from Argentina (Fernández et al., 2010).
Cytotoxic activity on vero cells
The cytotoxicity of the isolates was evaluated by Vero cells assay. Briefly, each strain was cultured overnight into 25 ml of Microbiological broth (No. 3, Merck) and was centrifuged 120× g (10 min at 4°C) and the supernatant was centrifuged again 17,228× g (10 min at 4°C) and identified as S1. The cell pellet was washed with PBS, resuspended in 3 ml of polymyxin sulfate (0.1 mg/ml) and incubated 30 min. Polymyxin B-treated cultures were centrifuged at 120× g (10 min at 4°C). The supernatant was centrifuged at 17,228× g, 10 min at 4°C, and was identified as S2. Fifty and 25 μl of each one S1 and S2 were inoculated in each one of the 96-well-plates containing 4 × 104 freshly trypsinized Vero cells and were incubated 48 h at 37°C in a 5% CO2 atmosphere. The cell monolayers were fixed with 10% (v/v) formaldehyde and then stained with 0.2% (w/v) crystal violet in phosphate-buffered saline solution. E. coli EDL933 strain was used as positive control and a strain stx positive without cytotoxic effect as negative control (E. coli serotype O15:H21). Wells having 50% or greater cytotoxicity, compared to a standard control well were considered positive.
stx subtyping
The strategy to detect stx2 subtypes was similar to that previously described by Krüger et al. (2011). Briefly, all stx2-positive STEC were subjected to PCR with the primer pair VT2-c/VT2-d, and amplification products were independently digested with restriction endonucleases HaeIII, RsaI, and NciI to detect stx2EDL933, stx2vha, stx2vhb, stx2g, and stx2NV206 (Tyler et al., 1991; Bertin et al., 2001; Krüger et al., 2007). All isolates were also evaluated with the VT2-cm/VT2-f primer set (Pierard et al., 1998) specific for stx2O118 (first termed stx2d by Piérard and renamed stx2O118 as proposed by Scheutz and Strockbine, 2005). The strains used as positive controls for each subtype and the references corresponding to the primers are detailed in Krüger et al. (2011).
sab gene
The detection of the sab gene was performed by PCR using the primers described by Herold et al. (2009) and the following amplification conditions: initial cycle of 94°C for 120 s, 30 cycles with denaturation step (94°C, 30 s), annealing step (54°C, 30 s) and extension step (68°C, 30 s), and a 60 s cycle at 72°C. STEC O20:H19 was used as positive control and Salmonella spp, Staphylococcus aureus, and Pseudomonas aeruginosa as negative controls.
MLVA assay
We performed an MLVA assay that previously showed a high level of discrimination among STEC isolates belonging to different non-O157:H7 serotypes (Schimmer et al., 2008; Bustamante et al., 2010; Franci et al., 2011). The seven VNTR loci studied in this assay were analyzed as described by Bustamante et al. (2010). Representative alleles were sequenced with an ABI PRISM 3730XL genetic analyzer (Macrogen, Korea). The dendrogram was constructed using the UPGMA clustering method implemented by START Vs. 1.0.5 software (Joley et al., 2001). The alleles were indicated in a string order CVN001-CVN002-CVN003-CVN004-CVN007-CVN014-CVN015, named according to the number of tandem repeat sequences. If no amplification product was detected, the allele was designated with an arbitrary number (30).
In all PCR assays, Inbio-Highway (Argentina) DNA polymerase was used.
Results and discussion
Using Vero cell assay, the S1 and S2 supernatants of all isolates from both serotypes demonstrated cytotoxic effect after 48 h post-inoculation on Vero cells.
Among 36 stx2-positive O130:H11 isolates, stx2EDL933 was the predominant subtype (81%), and the other subtype present was stx2vhb (Table 1). Only three isolates harbored both subtypes.
Table 1.
Origin and virulence genotypes of O130:H11 isolates.
| Strain number | Farm | Virulence genotype* | sab | stx2 subtype |
|---|---|---|---|---|
| 1 | A | stx1-ehxA-saa | − | – |
| 2 | A | stx1−stx2-ehxA-saa | − | stx2vhb |
| 3 | A | stx1−stx2-ehxA-saa | − | stx2vhb |
| 4 | A | stx1−stx2-ehxA-saa | − | stx2vhb |
| 5 | A | stx1−stx2-ehxA-saa | − | stx2EDL933 |
| 6 | A | stx1−stx2-ehxA-saa | − | stx2EDL933 |
| 7 | A | stx1−stx2-ehxA-saa | − | stx2EDL933 |
| 8 | B | stx1−stx2-ehxA-saa | − | stx2EDL933 stx2vhb |
| 9 | B | stx1−stx2-ehxA-saa | − | stx2EDL933 stx2vhb |
| 10 | B | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 11 | C | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 12 | C | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 13 | C | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 14 | C | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 15 | C | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 16 | C | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 17 | C | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 18 | C | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 19 | C | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 20 | C | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 21 | C | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 22 | D | stx1−stx2-ehxA-saa | + | stx2EDL933 stx2vhb |
| 23 | D | stx1−stx2-ehxA-saa | − | stx2EDL933 |
| 24 | D | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 25 | D | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 26 | D | stx1−stx2-ehxA-saa | − | stx2vhb |
| 27 | D | stx1−stx2-ehxA-saa | + | stx2vhb |
| 28 | D | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 29 | D | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 30 | D | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 31 | D | stx1−stx2-ehxA-saa | − | stx2vhb |
| 32 | D | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 33 | D | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 34 | D | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 35 | D | stx1−stx2-ehxA-saa | − | stx2EDL933 |
| 36 | D | stx1−stx2-ehxA-saa | + | stx2EDL933 |
| 37 | E | stx1−stx2-ehxA-saa | + | stx2EDL933 |
Previously determined (Fernández et al., 2010).
The most frequent stx2 subtype among O178:H19 isolates was stx2vha (72%), while stx2EDL933 and stx2vhb subtypes were found less frequently (20 and 8%, respectively) and no isolates harboring more than one stx2 subtype were found.
It is interesting to note that the stx2EDL933-positive strains, belonging to either O130:H11 or O178:H19 serotypes, (Tables 1 and 2) corresponded mainly to isolates harboring the profile stx1−stx2-ehxA-saa.
Table 2.
Origin and characterization of O178:H19 isolates.
| Strain number | Farm | Virulence genotype* | sab | stx2 subtype | MLVA profile |
|---|---|---|---|---|---|
| 1 | A | stx2 | − | stx2vha | I1 |
| 2 | A | stx2 | − | stx2vha | I1 |
| 3 | A | stx2 | − | stx2vha | I1 |
| 4 | A | stx2 | − | stx2vha | I1 |
| 5 | A | stx2 | − | stx2vha | I2 |
| 6 | C | stx2 | − | stx2vha | I3 |
| 7 | C | stx2 | − | stx2vha | I5 |
| 8 | D | stx2 | − | stx2vha | I1 |
| 9 | D | stx2 | + | stx2vhb | II2 |
| 10 | E | stx2 | − | stx2vha | I1 |
| 11 | E | stx2 | + | stx2EDL933 | II4 |
| 12 | E | stx2 | − | stx2vhb | I4 |
| 13 | E | stx2 | − | stx2vha | I2 |
| 14 | E | stx2 | − | stx2vha | I1 |
| 15 | B | stx2 | − | stx2vha | I3 |
| 16 | A | stx2 | − | stx2vha | I2 |
| 17 | C | stx1−stx2-ehxA-saa | + | stx2EDL933 | II3 |
| 18 | C | stx1−stx2-ehxA-saa | + | stx2EDL933 | II5 |
| 19 | D | stx2 | − | stx2vha | I2 |
| 20 | D | stx1−stx2-ehxA-saa | + | stx2EDL933 | II2 |
| 21 | D | stx2-ehxA-saa | − | stx2vha | I2 |
| 22 | D | stx2 | − | stx2vha | I1 |
| 23 | E | stx1−stx2-ehxA-saa | + | stx2EDL933 | II1 |
| 24 | C | stx2 | − | stx2vha | I1 |
| 25 | C | stx2 | − | stx2vha | I1 |
Previously determined (Fernández et al., 2010).
The subtypes found in this work have been reported as the predominant sxt2-subtypes in bovine STEC strains in Argentina and other countries (Bertin et al., 2001; Brett et al., 2003; Meichtri et al., 2004; Galli et al., 2010; Krüger et al., 2011) and have been associated with the development of HC and HUS (Friedrich et al., 2002; Persson et al., 2007). In a study performed by Masana et al. (2011) O130:H11 and O178:H19 were also among the most prevalent serotypes found in carcasses and bovine feces sampled at abattoirs in Argentina. In that study, O130:H11 isolates presented the same virulence genotypes (in regard to the presence of stx1, stx2 subtypes, ehxA and saa) as the ones detected in the present report. Regarding to O178:H19, some virulence genotypes (stx2vha; stx1−stx2EDL933-ehxA-saa; stx2vhb) found by Masana et al. (2011) were detected also in the present study, but there were other profiles (stx2NT; stx2EDL933−stx2vha) not shared between these studies.
The gene encoding Sab, a protein which mediates biofilm formation and promotes intestinal adherence, was detected in 65% of the isolates belonging to O130:H11. This study is the first, to our knowledge, to describe O130:H11 as a serotype carrying sab. In O178:H19 isolates sab was detected in 24% of the isolates (Table 2). Buvens et al. (2010) did not detect sab in a STEC O178:H19 strain isolated from HUS. All sab-positive STEC strains identified to date were also positive for ehx as well as saa, all genes located in a megaplasmid, noteworthy, in the present study some of the O178:H19 isolates were sab-positive but negative for ehxA and saa.
Most of the MLVA loci could be amplified, although there were differences between serotypes. To our knowledge this is the first time that STEC O130:H11 is typed by MLVA and notably, only one MLVA profile (5-2-30-9-8-30-6) was detected among these isolates. We have used this MLVA assay to subtype several isolates belonging to different non-O157:H7 serotypes and we found a high level of discrimination (Bustamante et al., 2010; Franci et al., 2011). Other authors have also applied this protocol to successfully resolve outbreaks due to a non-O157 strain (Schimmer et al., 2008). In our experience, this is the first time that all isolates from a same serotype and different origin present a unique MLVA profile. The lack of diversity found in this serotype would indicate that the chosen VNTR loci are not variable enough for typing O130:H11 strains since they did show variability in relation with the presence/absence of sab and also with the stx2 subtype present. Therefore, there is a need to identify VNTR loci that are variable among STEC strains belonging to this serotype.
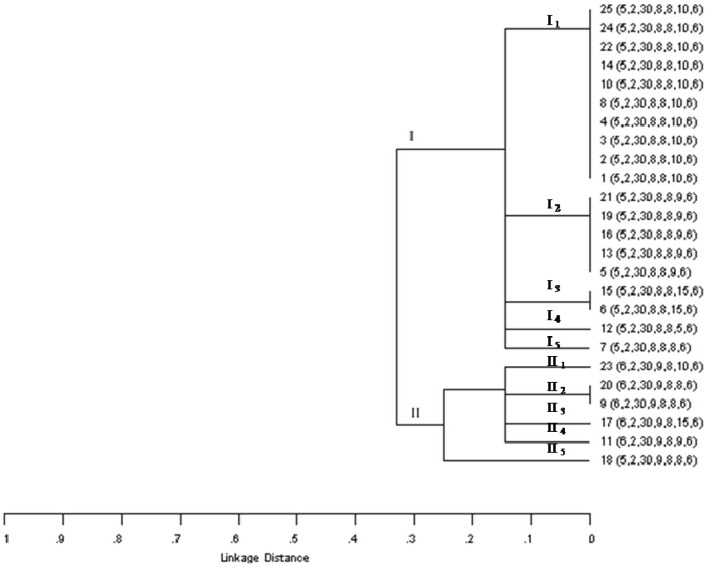
On the other hand, among the 25 O178:H19 isolates, 10 MLVA profiles were detected, which were grouped in two main clusters (Figure 1). A relationship could not be found with regard to MLVA profiles and farm origin (Table 2). Cluster I included isolates from all the farms, and cluster II, isolates from dairy farms C, D, and E. A high variability was found among isolates from farms C and E, detecting in each farm 5 MLVA profiles among 6 isolates (Table 2). All isolates belonging to clade I, were sab-negative and, with the exception of isolate 12, they presented the subtype stx2vha (Table 2). Clade II was the most variable, presenting five different profiles among six isolates. Moreover, isolates 9 and 20 shared the MLVA profile but not their virulence profile. Within this clade, all the isolates were sab-positive and carried stx2EDL933, with the exception of isolate 9 (positive for sab but negative for that stx2 subtype) (Table 2). Although a relationship between the MLVA profile and the stx2 subtype is not expected, with the exception of isolates from a same clone, all stx2vha-positive isolates belonged to cluster I and all stxEDL933-positive isolates, to cluster II. Regarding isolates carrying stx2vhb, one belonged to cluster I and the other to cluster II. Noteworthy, all the MLVA profiles present in these isolates were quite different from the ones detected previously in STEC O178:H19 isolated from minced meat of the same geographic region (Franci et al., 2011). Taking into account all these results, a high genetic variability was evidenced among isolates belonging to this serotype. Our results showed different STEC O178:H19 clonal lineages and determined that some clones may be present in more than one farm.
Figure 1.
Dendrogram based on MLVA profiles of STEC O178:H19 isolated from dairy cows in Argentina. Order of the allele string: CVN001-CVN002-CVN003-CVN004-CVN007-CVN014-CVN015.
Conclusion
The data suggest differences in the genetic variability for the two serotypes. It could be assessed when the stx2 subtype and the presence/absence of sab gene were taken into account, and for isolates belonging to O178:H19, also when the MLVA type was considered. The MLVA typing assay chosen seems not suitable for detecting genetic differences among O130:H11 STEC isolates, and further loci need to be analyzed.
STEC non-O157 serotypes are nowadays frequently associated with outbreaks and sporadic cases of HUS and particularly, O130:H11 and O178:H19 STEC have been isolated from human patients. In our study isolates from dairy cows belonging to these serotypes possess virulence characteristics associated with the development of severe disease in humans and it would be desirable to consider them in the group of serotypes routinely investigated.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Authors thank María R. Ortiz for her technical assistance. This work was supported by FONCYT PICT 2010 PROY 1655, CIC and SECYT-UNCPBA. Nora L. Padola is member of the Scientific Research Commission Prov. Buenos Aires (CIC). Daniel Fernández is a holder of a fellowship from CONICET, Alejandra Krüger, Ana V. Bustamante, A. Mariel Sanso, Analía I. Etcheverría, and Paula M. A. Lucchesi are members of the Research Career of CONICET.
References
- Bertin Y., Boukhors K., Livrelli V., Martin C. (2001). stx2 subtyping of shiga toxin-producing Escherichia coli isolated from cattle in France detection of new stx2 subtype and correlation with additional virulence factors. J. Clin. Microbiol. 39, 3060–3065 10.1128/JCM.39.9.3060-3065.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelheim K. (2007). The non-O157 shiga-toxigenic (verocytotoxigenic) Escherichia coli; under-rated pathogens. Crit. Rev. Microbiol. 33, 67–87 10.1080/10408410601172172 [DOI] [PubMed] [Google Scholar]
- Blanco M., Blanco J. E., Mora A., Dahbi G., Alonso M. P., González E. A., et al. (2004). Serotypes, virulence genes, and intimin types of Shiga toxin producing Escherichia coli isolates from cattle in Spain and identification of a new intimin variant gene (eae). J. Clin. Microbiol. 42, 645–651 10.1128/JCM.42.2.645-651.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett K. N., Hornitzky M. A., Bettelheim K. A., Walker M. J., Djordjevic S. P. (2003). Bovine non-O157 Shiga-toxin 2-containing Escherichia coli isolates commonly possess stx2-EDL933 and/or stx2vhb subtypes. J. Clin. Microbiol. 41, 2716–2722 10.1128/JCM.41.6.2716-2722.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante A. V., Sanso A. M., Lucchesi P. M. A., Parma A. E. (2010). Genetic diversity of O157:H7 and non-O157 verocytotoxigenic Escherichia coli from Argentina inferred by multiple-locus variable-number tandem repeat analysis (MLVA). Int. J. Med. Microbiol. 300, 212–217 10.1016/j.ijmm.2009.08.020 [DOI] [PubMed] [Google Scholar]
- Buvens G., Lauwers S., Piérard D. (2010). Prevalence of subtilase cytotoxin in verocytotoxin-producing Escherichia coli isolated from humans and raw meats in Belgium. Eur. J. Clin. Microbiol. Infect. Dis. 29, 1395–1399 10.1007/s10096-010-1014-z [DOI] [PubMed] [Google Scholar]
- Fernández D., Irino K., Sanz M. E., Padola N. L., Parma A. E. (2010). Characterization of Shiga toxin-producing Escherichia coli isolated from dairy cows in Argentina. Lett. Appl. Microbiol. 51, 377–382 10.1111/j.1472-765X.2010.02904.x [DOI] [PubMed] [Google Scholar]
- Franci T., Sanso A. M., Bustamante A. V., Lucchesi P. M. A., Parma A. E. (2011). Genetic characterization of non-O157 verocytotoxigenic Escherichia coli isolated from raw beef products using multiple-locus variable-number tandem repeat analysis (MLVA). Foodborne Pathog. Dis. 8, 1019–1023 10.1089/fpd.2010.0814 [DOI] [PubMed] [Google Scholar]
- Fremaux B., Raynaud S., Beutin L., Vernozy Rozand C. (2006). Dissemination and persistence of Shiga toxin-producing Escherichia coli (STEC) strains on French dairy farms. Vet. Microbiol. 117, 180–191 10.1016/j.vetmic.2006.04.030 [DOI] [PubMed] [Google Scholar]
- Friedrich A. W., Bielaszewska M., Zhang W. L., Pulz M., Kuczius T., Ammon A., et al. (2002). Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185, 74–84 10.1086/338115 [DOI] [PubMed] [Google Scholar]
- Galli L., Miliwebsky E., Irino K., Leotta G., Rivas M. (2010). Virulence profile comparison between LEE-negative Shiga toxin-producing Escherichia coli (STEC) strains isolated from cattle and humans. Vet. Microbiol. 143, 307–313 10.1016/j.vetmic.2009.11.028 [DOI] [PubMed] [Google Scholar]
- Giugno S. M., Bibiloni N., Rahman R., Miliwebsky E., Chinen I., Rivas M. (2007). Association between uremic hemolytic syndrome and infection by Shiga toxin-producing Escherichia coli. Acta Bioquím. Clín. Latinoam. 41, 27–33 [Google Scholar]
- Gyles C. (2007). Shiga toxin producing Escherichia coli: an overview. J. Animal Sci. 85, E42–E62 10.2527/jas.2006-508 [DOI] [PubMed] [Google Scholar]
- Herold S., Paton J. C., Paton A. W. (2009). Sab, a novel autotransporter of locus of enterocyte effacement-negative shiga-toxigenic Escherichia coli O113:H21, contributes to adherence and biofilm formation. Infect. Immun. 77, 3234–3243 10.1128/IAI.00031-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein H., Sakuma T. (2005). Prevalence of Shiga toxin-producing Escherichia coli in dairy cattle and their products. J. Dairy Sci. 88, 450–465 [DOI] [PubMed] [Google Scholar]
- Joley K. A., Feil E. J., Chan M. S., Maiden M. C. (2001). Sequence type analysis and recombinational tests (START). Bioinformatics 17, 1230–1231 10.1093/bioinformatics/17.12.1230 [DOI] [PubMed] [Google Scholar]
- Karch H., Tarr P. I., Bielaszewska M. (2005). Enterohaemorrhagic Escherichia coli in human medicine. Int. J. Med. Microbiol. 295, 405–418 10.1016/j.ijmm.2005.06.009 [DOI] [PubMed] [Google Scholar]
- Krüger A., Lucchesi P. M. A., Parma A. E. (2007). Evaluation of vt2-subtyping methods for identifying vt2g in verotoxigenic Escherichia coli. J. Med. Microbiol. 56, 1474–1478 10.1099/jmm.0.47307-0 [DOI] [PubMed] [Google Scholar]
- Krüger A., Lucchesi P. M. A., Parma A. E. (2011). Verotoxins in bovine and meat verotoxin-producing Escherichia coli isolates: type, number of variants, and relationship to cytotoxicity. Appl. Environ. Microbiol. 77, 73–79 10.1128/AEM.01445-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López O., Duverne L., Chinen I., Carbonari C., Mazieres J., Deza N., et al. (2012). Shedding and characterization non-O157 of Shiga toxin-producing Escherichia coli strains isolated from beef cattle in one feedlot of Argentina. VTEC (2012). 8th International Symposium on Shiga toxin (verocytotoxin) producing Escherichia coli Infections. Abstract book, P196, 202 [Google Scholar]
- Masana M. O., D'Astek B. A., Palladino P. M., Galli L., Del Castillo L. L., Carbonari C., et al. (2011). Genotypic characterization of non-O157 Shigatoxin-producing Escherichia coli in beef abattoirs of Argentina. J. Food Prot. 12, 2008–2017 10.4315/0362-028X.JFP-11-189 [DOI] [PubMed] [Google Scholar]
- Meichtri L., Miliwebsky E., Gioffré A., Chinen I., Baschkier A., Chillemi G., et al. (2004). Shiga-toxin-producing Escherichia coli in healthy young beef steers from Argentina: prevalence and virulence properties. Int. J. Food Microbiol. 96, 189–198 10.1016/j.ijfoodmicro.2004.03.018 [DOI] [PubMed] [Google Scholar]
- Mora A., Blanco M., Blanco J. E., Alonso M. P., Dhabi G., Thomson-Carter F., et al. (2004). Phage types and genotypes of Shiga toxin-producing Escherichia coli O157:H7 isolates from humans and animals in Spain: identification and characterization of two predominating phage types (PT2 and PT8). J. Clin. Microbiol. 42, 4007–4015 10.1128/JCM.42.9.4007-4015.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. P., Jayarao B. M., Almeida R. A. (2005). Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodborne Pathog. Dis. 2, 115–129 10.1089/fpd.2005.2.115 [DOI] [PubMed] [Google Scholar]
- Padola N. L., Sanz M. E., Blanco J. E., Blanco M., Blanco J., Etcheverria A. I., et al. (2004). Serotypes and virulence genes of Shigatoxigenic Escherichia coli (STEC) isolates from a feedlot in Argentina. Vet. Microbiol. 100, 3–9 10.1016/S0378-1135(03)00127-5 [DOI] [PubMed] [Google Scholar]
- Paton A. W., Paton J. C. (1998). Detection and characterization of Shiga Toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic, E. coli HlyA, rfbO111 and rfbO157. J. Clin. Microbiol. 36, 598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paton A. W., Srimanote P., Woodrow M. C., Paton J. S. (2001). Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga toxigenic Escherichia coli strains that are virulent for humans. Infect. Immun. 69, 6999–7009 10.1128/IAI.69.11.6999-7009.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce M. C., Jenkins C., Vali L., Smith A. W., Knight H. I., Cheasty T., et al. (2004). Temporal shedding patterns and virulence factors of Escherichia coli serogrups O26, O113, O111, O145 and O157in a cohort of beef calves and dairy dams. Appl. Environ. Microbiol. 70, 1708–1716 10.1128/AEM.70.3.1708-1716.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson S., Olsen K. E. P., Scheutz F., Krogfelt K. A., Gerner-Smidt P. (2007). A method for fast and simple detection of major diarrheagenic Escherichia coli in the routine diagnostic laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 13, 516–524 10.1111/j.1469-0691.2007.01692.x [DOI] [PubMed] [Google Scholar]
- Pierard D., Muyldermans G., Moriau L., Stevens D., Lauwers S. (1998). Identification of new verocytotoxin Type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36, 3317–3322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prager R., Fruth A., Busch A., Tietze E. (2011). Comparative analysis of virulence genes, genetic diversity, and phylogeny of Shiga toxin 2g and heat-stable enterotoxin STIa encoding Escherichia coli isolates from humans, animals, and environmental sources. Int. J. Med. Microbiol. 301, 181–191 10.1016/j.ijmm.2010.06.003 [DOI] [PubMed] [Google Scholar]
- Prager R., Fruth A., Siewert U., Strutz U., Tschäpe H. (2009). Escherichia coli encoding Shiga toxin 2f as an emerging human pathogen. Int. J. Med. Microbiol. 299, 343–353 10.1016/j.ijmm.2008.10.008 [DOI] [PubMed] [Google Scholar]
- Scheutz F., Strockbine N. A. (2005). Escherichia, in Bergey's Manual of Systematic Bacteriology, eds Garrity G. M., Brenner D. J., Krieg N. R., Staley J. T. (New York, NY: Springer; ), 607–624 [Google Scholar]
- Schimmer B., Nygard K., Eriksen H. M., Lassen J., Lindstedt B. A., Brandal L. T., et al. (2008). Outbreak of haemolytic uraemic syndrome in Norway caused by stx2-positive Escherichia coli O103:H25 traced to cured mutton sausages. BMC Infect. Dis. 8:41 10.1186/1471-2334-8-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler S. D., Johnson W., Lior H., Wang G., Rozee K. R. (1991). Identification of verotoxin type 2 variant B subunit genes in E. coli by the polymerase chain reaction and restrictionfragment length polymorphism analysis. J. Clin. Microbiol. 29, 1339–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]