Abstract
The enzyme responsible for carbon dioxide fixation in the Calvin cycle, ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO), is always detected as a phylogenetic marker to analyze the distribution and activity of autotrophic bacteria. However, such an approach provides no indication as to the significance of genomic content and organization. Horizontal transfers of RubisCO genes occurring in eubacteria and plastids may seriously affect the credibility of this approach. Here, we presented a new method to analyze the diversity and genomic content of RubisCO genes in acid mine drainage (AMD). A metagenome microarray containing 7,776 large-insertion fosmids was constructed to quickly screen genome fragments containing RubisCO form I large-subunit genes (cbbL). Forty-six cbbL-containing fosmids were detected, and six fosmids were fully sequenced. To evaluate the reliability of the metagenome microarray and understand the microbial community in AMD, the diversities of cbbL and the 16S rRNA gene were analyzed. Fosmid sequences revealed that the form I RubisCO gene cluster could be subdivided into form IA and IB RubisCO gene clusters in AMD, because of significant divergences in molecular phylogenetics and conservative genomic organization. Interestingly, the form I RubisCO gene cluster coexisted with the form II RubisCO gene cluster in one fosmid genomic fragment. Phylogenetic analyses revealed that horizontal transfers of RubisCO genes may occur widely in AMD, which makes the evolutionary history of RubisCO difficult to reconcile with organismal phylogeny.
INTRODUCTION
The Calvin-Benson-Bassham cycle is the major and most abundant pathway for inorganic carbon fixation (1). In the Calvin cycle, the ribulose-1,5-biphosphate carboxylase/oxygenase (RubisCO), the enzyme responsible for the first rate-limiting step in CO2 fixation (2), is one of the most important enzymes. RubisCO is a bifunctional enzyme that controls the reduction of CO2 and the oxygenolysis of ribulose-1,5-bisphophate (3). The enzyme exists in multiple natural forms in many evolutionarily diverse organisms from all domains of life. RubisCO is a well-studied enzyme because of its essential capacity as the mechanism of primary production in nearly all ecosystems (4–6).
In Bacteria, two forms of RubisCO (forms I and II), sharing 25% to 30% amino acid similarity, are currently recognized (7). Form I RubisCO is a hexadecamer with eight large and eight small subunits (L8S8) and occurs in photo- and chemoautotrophic organisms. Form II protein consists only of large subunits (Ln). It is assumed that the common ancestor of RubisCO is similar to form II RubisCO because it operates successfully under conditions of low O2 and high CO2 concentrations, which are similar to the conditions that existed in the early earth atmosphere (3, 8). In addition to the above-mentioned two forms, form III RubisCO was discovered in some members of Archaea (9, 10). In addition, form IV RubisCO lacks several of the required amino acid residues for the catalytic activity of RubisCO, and is designated a RubisCO-like protein, although it is not involved in the Calvin cycle. It has been discovered in Bacillus subtilis (11), Archaeoglobus fulgidus (10), and Chlorobium tepidum (12).
Because of its functional significance and well-known characteristics, the RubisCO genes have been frequently used as a phylogenetic marker to detect autotrophic populations in situ. However, many previous studies have reported the discrepancy between phylogenies based on the RubisCO gene and those based on other genes (13, 14). Meanwhile, such an approach provides no indication as to what other genetic factors might be provided in phylogenetics. In fact, the knowledge of genetic content and organization is an important basis for analysis of the diversity and evolutionary history of RubisCO genes.
Acid mine drainage (AMD) is the outflow of acidic water from metal or coal mines, which causes worldwide environmental problems that arise largely from microbial activity (15). Since AMD is extremely acidic (pH < 2.0), metal rich, and inhospitable, microorganisms in AMD are usually chemoautotrophic bacteria. Few microorganisms in AMD have been isolated and described (16), since there is no effective isolation method. Here, we focused on an AMD biofilm growing on the surface within a chalcopyrite ore body from Dexing Copper Mine, China. Metagenomic cloning and microarray were introduced to screen and identify the organization and diversity of genes in the region surrounding the RubisCO genes in the AMD community.
MATERIALS AND METHODS
Sample collection, metagenomic library, and microarray construction.
To obtain AMD microbial biofilm, the samples were filtered through a 0.2-μm-pore-size nylon membrane filter. The environmental DNA was extracted as described previously (17). A fosmid library containing a metagenome from AMD was constructed using a CopyControl fosmid library production kit (Epicentre, Madison, WI) according to the manufacturer's protocol. The collection of the library contained a total of 7,776 large-insertion clones. A metagenome microarray was constructed as described previously (18). Each clone was incubated in a shaking incubator at 37°C and 170 rpm in the presence of chloramphenicol (12.5 μg/ml) and an inducer (1 μl/ml) (Epicentre). Cells were harvested the next day, and the fosmid DNA was extracted using a QIAprep Spin Miniprep kit (Qiagen, Germany) according to the manufacturer's protocol. The fosmid DNAs were stored in a final concentration of 40 ng/μl. A 10-μl fosmid DNA sample was transferred to a 384-well microplate, and the DNA samples were diluted 1:1 (vol/vol) with the 40% dimethyl sulfoxide (Sigma). The fosmid DNA samples were arrayed on the glass slide using a Genemachines OmniGrid Accent microarrayer (Genomic Solutions). The glass slide was immersed in boiling water for 5 min, and then immersed in anhydrous ethanol to melt DNA chains (19). In addition, the following controls were spotted to check hybridization, printing, and data analysis: (i) environmental DNA as positive controls, (ii) quantitative and negative controls with Escherichia coli genomic DNA, and (iii) blanks.
RubisCO gene amplification, metagenome microarray hybridization, sequencing, and assembly.
The form I RubisCO large-subunit gene (cbbL) was PCR amplified from environmental DNA extracts with universal primers Rub3_F (5′-GTGCCAGACGTGGATACCG-3′) and Rub3_R (5′-CAACAGCCAGCCCTTCAT-3′). Amplification was performed in 50-μl reaction mixtures containing 25 μl of universal Taq PCR Master Mix (Tiangen Biotech, China), 1 μl of template DNA, 1 μl each of 10 μM forward and reverse primers, and 22 μl of deionized water. The PCR conditions for amplification were as follows: 94°C for 3 min, then 32 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s, followed by a final extension at 72°C for 10 min. PCR products were purified using a QIAquick PCR purification kit (Qiagen, Germany).
PCR products were labeled using a Bioprime DNA labeling kit (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Then, the labeled probes were purified using a QIAquick PCR purification kit (Qiagen, Germany), concentrated into crystallization in a Spendvac, and then resuspended in 20 μl of deionized water. Labeled probes were mixed with hybridization solution. Each microarray hybridization solution contained 20 μl of labeled DNA, 65 μl of formamide (50% [vol/vol]), 19.5 μl of 20× SSC (1× SSC is 150 mM NaCl and 15 mM trisodium citrate), 3.9 μl of 10% sodium dodecyl sulfate (SDS), 9.1 μl of herring sperm DNA (Promega, Madison, WI) (10 mg/ml), and 1.1 μl of DTT (dithiothreitol; 0.1 M) in a total volume of 130 μl. The hybridization solution was incubated at 98°C for 3 min, and then kept at 65°C. The hybridization was performed at 50°C using a HS4800 Pro hybridization station (Tecan, Switzerland). After hybridization, the microarray was scanned using a GenePix 4100A microarray scanner (Axon). The data were analyzed as described previously (20), and each deduced positive clone was tested using PCR amplification and sequenced.
Six of the desired clones were chosen for further analysis. Fosmid DNA was individually isolated using a QIAprep Spin Miniprep kit (Qiagen), and pyrosequenced (Roche 454 GS FLX system; Majorbio, China). For the sequencing process, fosmid DNA fragments were tagged individually with a unique 10-base sequence that is recognized by the analysis software. The average length of reads was 402 bp, and each fosmid had ∼1.2 Mb of DNA data (about 20% of them belonged to the cloning vector). Assembly was performed with the program Newbler (21). All the fosmids were assembled in one single contig (Table 1).
Table 1.
Main features and RubisCO-associated cluster present in the analyzed fosmids
| Fosmid | Length (bp) | %GC content | Coverage (×) | RubisCO-associated genes and characteristicsa |
|---|---|---|---|---|
| DX-8J-22 | 32,275 | 58.37 | 37 | CbbR: A. ferrooxidans ATCC 23270, 99%; CbbL: A. ferrooxidans ATCC 23270, 100%; CbbS: A. ferrooxidans ATCC 23270, 100%; CsoS2: A. ferrooxidans ATCC 53993, 99%; CsoS3: A. ferrooxidans ATCC 23270, 100%; CscA: A. ferrooxidans ATCC 23270, 100%; CscB: A. ferrooxidans ATCC 23270, 100%; CscD: A. ferrooxidans ATCC 23270, 98%; CscE: A. ferrooxidans ATCC 23270, 100%; CbbQ: A. ferrooxidans ATCC 23270, 100%; CbbO: A. ferrooxidans ATCC 23270, 100% |
| DX-4H-17 | 37,399 | 64.53 | 32 | CbbR: Beggiatoa sp. PS, 53%; CbbL: Acidithiomicrobium sp. P2, 90%; CbbS: A. ferrivorans SS3, 69%; CsoS2: H. neapolitanus c2, 45%; CsoS3: H. neapolitanus c2, 58%; CscA: T. denitrificans ATCC 25259, 86%; CscB: A. ferrivorans SS3, 74%; CscE: A. caldus ATCC 51756, 95%; CscD: A. ferrivorans SS3, 88%; CbbO: A. caldus ATCC 51756, 64% |
| DX-4K-26 | 31,477 | 61.01 | 38 | CbbR: A. ferrivorans SS3, 78%; CbbL: A. ferrivorans SS3, 93%; CbbS: A. ferrivorans SS3, 90%; CsoS2: A. ferrivorans SS3, 56%; CsoS3: A. ferrivorans SS3, 57% |
| DX-1A-14 | 32,546 | 64.00 | 36 | CbbR: Beggiatoa sp. PS, 53%; CbbL: Acidithiomicrobium sp. P2, 90%; CbbS: A. ferrivorans SS3, 69%; CsoS2: H. neapolitanus c2, 45%; CsoS3: H. neapolitanus c2, 58% |
| DX-3D-09 | 32,174 | 66.73 | 37 | CbbM: L. cholodnii SP-6, 90%; CbbQ: L. cholodnii SP-6, 82%; CbbO: L. cholodnii SP-6, 69%; CbbL: A. ferrooxidans, 92%; CbbS: T. denitrificans ATCC 25259, 89%; CbbQ: C. metallidurans CH34, 91%; CbbO: T. denitrificans ATCC 25259, 80% |
| DX-7F-24 | 40,978 | 65.85 | 30 | CbbM: L. cholodnii SP-6, 90%; CbbQ: L. cholodnii SP-6, 83%; CbbO: L. cholodnii SP-6, 70% |
Subunit present, closest strain hit, and percent similarity are shown.
Annotation and analysis of genome fragments.
Protein-coding genes were predicted individually using GLIMMER (22) and the RAST server (23), and were further manually curated. Each predicted gene was subsequently searched against the nonredundant NCBI database (http://www.ncbi.nlm.nih.gov/), KEGG database (http://www.genome.jp/kegg/), and COG database (http://www.childrensoncologygroup.org/) using BLAST to ensure that the annotation was consistent. The other unassigned open reading frames (ORFs) were searched using the hmmpfam program of the HMMER package (24). The hidden Markov models for the protein domains were obtained from the Pfam 26.0 database (http://pfam.sanger.ac.uk/). For comparative analysis, BLASTN and BLASTX searches among fosmids and different bacterial genomic islands were carried out, leading to the identification of regions of similarity. To allow the interactive visualization of genomic fragment comparisons, we used the Artemis Comparison Tool (25).
16S rRNA and cbbL gene amplification.
16S rRNA gene sequences were PCR amplified from environmental DNA extracts with the bacterium-specific primer set 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACCTTGTTACGACTT-3′) (15). Amplification was performed in 50-μl reaction mixtures containing 25 μl of universal Taq PCR Master Mix (Tiangen Biotech, China), 1 μl of DNA extracts, 1 μl each of 10 μM forward and reverse primers, and 22 μl of deionized water. The PCR conditions for amplification were as follows: 94°C for 5 min, then 32 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 10 min. Amplification of cbbL sequences from the same environmental DNA extracts was performed as mentioned above.
Clone library construction and sequencing.
PCR products of the 16S rRNA and cbbL genes were visualized on 1.5% agarose gels in 1× Tris-acetate-EDTA (TAE) buffer and purified directly with a QIAquick PCR purification kit (Qiagen, Germany). Purified PCR products of 16S rRNA and cbbL genes were ligated into vector pGM-T (Tiangen Biotech, China) and transformed into competent E. coli DH5α according to the manufacturer's protocol. Plasmids from the 16S rRNA gene and cbbL libraries were subsequently extracted using a Tianprep miniplasmid kit (Tiangen Biotech, China). Clones containing 16S rRNA and putative cbbL genes were screened by restriction fragment length polymorphism (RFLP) with MspI and Hin6 I restriction endonucleases (MBI Fermentas). Restriction fragments were analyzed in 3% (wt/vol) agarose gels in 1× TAE buffer. Selected unique plasmids were sequenced bidirectionally with the vector-specific primers SP6 reverse and T7 promoter. Sequences were edited manually with the DNAstar package (Madison, WI).
Phylogenetic analysis.
Phylogenetic analysis of 16S rRNA and cbbL gene sequences was performed on sequences screened from 16S rRNA gene and cbbL libraries, and representatives from metagenome microarrays (see Fig. 2 and 3). 16S rRNA gene sequences were aligned with sequences from the GenBank database with the CLUSTAL program (26), and phylogenetic trees were constructed by the use of Molecular Evolutionary Genetics Analysis 4.0 software (MEGA, version 4.0) (27). Deduced amino acid sequences for cbbL clones and representatives of 46 cbbL-containing fosmids were aligned with known sequences from the GenBank database with the CLUSTAL program (26), and cbbL gene trees were also constructed by using MEGA 4.0 (27).
Fig 2.
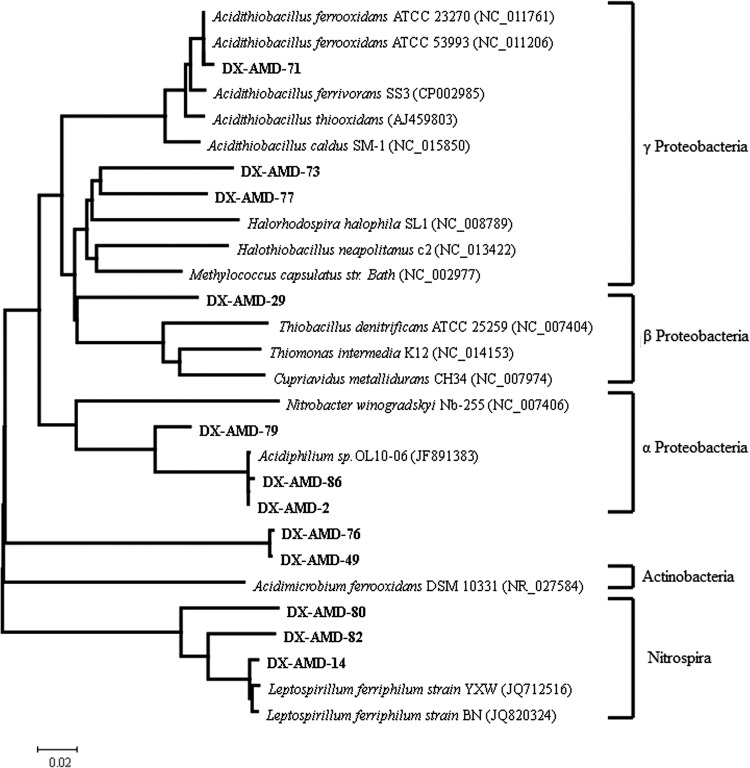
Phylogenetic analysis of 16S rRNA genes. A consensus tree was constructed by the distance (neighbor-joining), maximum-parsimony, and maximum-likelihood methods. Clones obtained from the 16S rRNA clone library were designated DX-AMD-, followed by their number in the clone library. These sequences are shown in bold. Numbers at the nodes indicate the percentage of bootstrap values in 1,000 replications.
Fig 3.
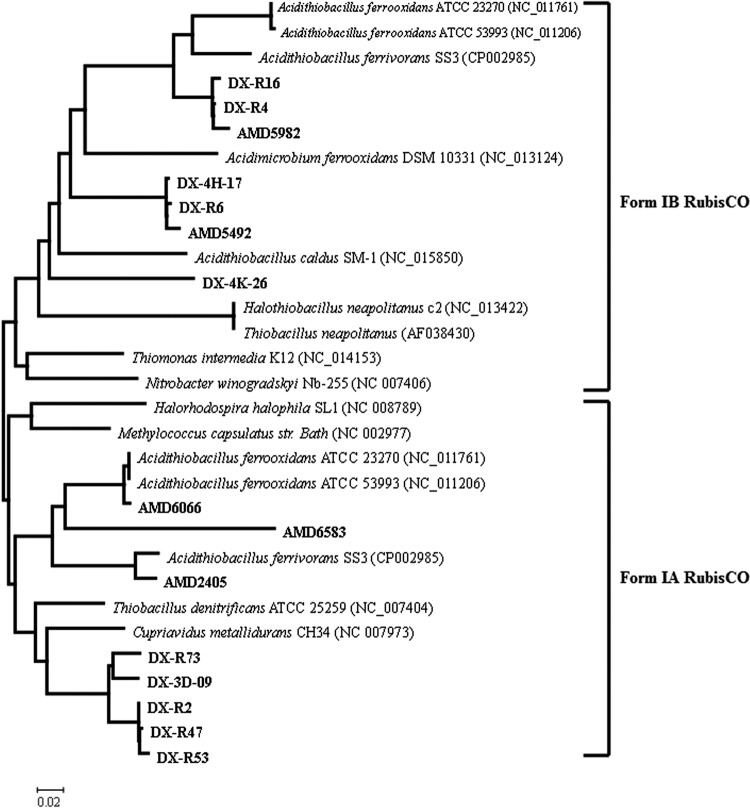
Phylogenetic analysis of cbbL genes. A consensus tree was constructed by distance (neighbor-joining), maximum-parsimony, and maximum-likelihood methods. Clones obtained from microarray hybridization were designated AMD, followed by their number in the fosmid library. Clones obtain from cbbL clone library were designated DX-R, followed by their number in the clone library. DX-3D-09, DX-4H-17, and DX-4K-26 indicate the fully sequenced fosmids. These sequences are shown in bold. Eight representatives of 46 cbbL gene sequences are presented in the phylogenetic tree. Numbers at the nodes indicate the percentage of bootstrap values in 1,000 replications.
Nucleotide sequence accession numbers.
Sequences determined and annotated in this study are available in GenBank under accession numbers JQ815894 to JQ815896 and JX308284 to JX308286. The incomplete cbbL sequences in GenBank have been compiled under accession numbers JX297619 to JX297625 and JQ815897 to JQ815942, and 16S rRNA gene sequences have been compiled under accession numbers JX297607 to JX297618.
RESULTS
Metagenomic library and metagenome microarray hybridization.
Samples were collected from AMD in Dexing Copper Mine, China. The environmental DNA was extracted and used to construct a fosmid library. The library collection contained in total 7,776 large-insertion clones. The genomic fragments cloned in the fosmids had a size of between 30 and 45 kb.
To quickly screen RubisCO genes from the metagenomic library, we constructed a metagenome microarray using the fosmid library. After hybridization and checking by PCR amplification, 46 cbbL-containing fosmids were screened out. And we fully sequenced six fosmids that were selected from the 46 cbbL-containing fosmids. All of the six fosmids showed evidence of coding either a form I or form II RubisCO large subunit. The genomic fragments in the fosmids had a size between 31.4 and 40.9 kb, and all could be assembled in one single contig (Table 1). The total annotations of six genomic fragments are presented in the supplemental material.
RubisCO genes in the fosmids.
The most interesting genomic fragment was found in fosmid DX-3D-09, in which we identified two RubisCO clusters, one belonging to form I and the other belonging to form II (Fig. 1). The subunits CbbL and CbbS are essential for the catalytic activity of the enzyme in form I RubisCO, while CbbM is the sole catalytic subunit in the form II RubisCO enzyme. The catalytic subunit cbbM was complete, as this fosmid contained complete subunits cbbL and cbbS. In addition to this fosmid, complete subunits cbbL and cbbS were also identified in DX-8J-22, DX-4H-17, DX-4K-26, and DX-1A-14, while subunit cbbM was also found in DX-7F-24. In all six of the fosmids, additional subunits in the RubisCO gene cluster were identified, providing more reliable evidence of the presence of a functional RubisCO enzyme.
Fig 1.
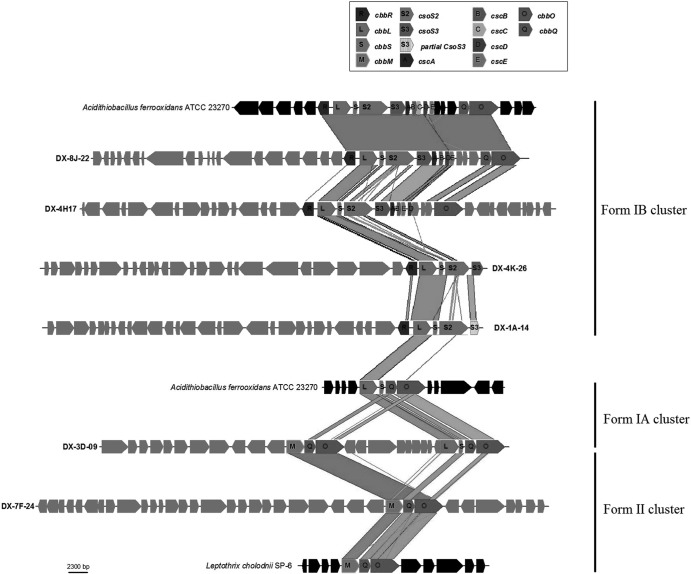
Genes and similarity comparison of environmental fosmids from the AMD metagenomic library containing RubisCO genes. RubisCO cluster genes are highlighted in different shades. Specific additional genes mentioned in the text are also indicated. The two closest genome fragments of cultivated microbes Acidithiobacillus ferrooxidans ATCC 23270 and Leptothrix cholodnii SP-6 are also shown.
Form I RubisCO.
Among the six fosmids, five ORFs were identified as form I large-subunit cbbL from the identification of the conserved catalytic sequence motif KDDE (28) and the small-subunit cbbS located in the immediate downstream region, providing more evidence to distinguish cbbL from cbbM. Comparing RubisCO gene clusters in the six fosmids with different bacterial genomic islands, we could subdivide genes in the form I RubisCO gene cluster into two types: (i) the form IA RubisCO gene cluster and (ii) the form IB RubisCO gene cluster (Fig. 1). Both forms contained catalytic subunits cbbL and cbbS. The form IA RubisCO gene cluster harbored subunits cbbQ and cbbO that are believed to be required in the processes of regulation and posttranslational modification of the RubisCO enzyme (29). The genomic organization of the form IA RubisCO gene cluster is L→S→Q→O. The organization is rather conservative in different organisms. The organization of the form IB RubisCO gene cluster is significantly different from that of the form IA RubisCO gene cluster. Although accessory subunits cbbQ and cbbO may also occur in form IB RubisCO gene clusters, genes encoding the carboxysome are inserted between catalytic subunits cbbLS and accessory subunits cbbQ and cbbO (Fig. 1). This is the most conspicuous difference between form IA and IB RubisCO gene clusters.
Form IA gene cluster.
The only representative of the form IA RubisCO gene cluster was found in fosmid DX-3D-09 (Fig. 1). It shows high overall similarity to other known form IA RubisCO gene clusters. The large-subunit CbbL encoded by fosmid DX-3D-09 has its best hit within Betaproteobacteria, and it is very (93%) similar to the CbbL found in Thiobacillus denitrificans. In addition, the organization of cbbS, cbbQ, and cbbO genes associated with this cbbL gene is L→S→Q→O, typical of the form IA RubisCO gene cluster. The subunits CbbS, CbbQ, and CbbO have their best hits within Betaproteobacteria. These subunits showed highest similarities with Thiobacillus denitrificans ATCC 25259 (89%), Cupriavidus metallidurans CH34 (91%), and Thiobacillus denitrificans ATCC 25259 (80%), respectively (Table 1). The organization of the form I RubisCO gene cluster in fosmid DX-3D-09 is very similar to that of Thiobacillus denitrificans ATCC 25259, but the overall organization of fosmid DX-3D-09 is most like that of Cupriavidus metallidurans, a strain within the class Betaproteobacteria. Interestingly, the same fosmid contains another cluster of RubisCO genes with a form II large-subunit cbbM (see below).
Form IB gene cluster.
Among the other fosmids, four fosmids (DX-1A-14, DX-4H-17, DX-4K-26, and DX-8J-22) contained the form IB RubisCO gene cluster. In the four fosmids, a gene encoding transcriptional regulator CbbR (30) was located in the immediate upstream of cbbL in a divergent orientation. The CbbL protein encoded by fosmid DX-4H-17 (see Table S2 in the supplemental material) has its best hit within Actinobacteria, and it is most (90%) similar to Acidithiomicrobium sp. In the form IB RubisCO gene cluster of fosmid DX-4H-17, the subunit cbbR has its best hit with Beggiatoa sp. PS (53%), cbbS has its best hit with Acidithiobacillus ferrivorans SS3 (69%), and cbbO has its best hit with Acidithiobacillus caldus ATCC 51756 (64%) (Table 1). However, the above-mentioned three bacteria are within the class Gammaproteobacteria. In addition, we could not find the subunit cbbQ in the surrounding region of the form IB RubisCO gene cluster, but a 6-ORF operon encoding carboxysome proteins was identified in the immediate downstream of cbbLS (Fig. 1). In Acidithiobacillus ferrooxidans ATCC 23270 (31) and Synechococcus sp. WH 8102, the RubisCO cluster is located near genes related to the carboxysome (32). In the 6-ORF operon, all of genes are oriented in the direction that is congruent with cbbLS. The gene csoS2 is thought to encode a carboxysome shell protein with a high molecular weight. csoS3, a gene encoding carbonic anhydrase, was immediately downstream of csoS2. Carbonic anhydrase is sequestered with the RubisCO enzyme in the carboxysome to convert bicarbonate to the RubisCO substrate CO2, as it enters the microcompartment from the cytosol (33). The four remaining genes (cscA, cscB, cscD, and cscE) were shown to encode proteins that are components of the carboxysome shell. All 6 of the genes have their best hits within Gammaproteobacteria. The closest strain hits for the 6 genes are summarized in Table 1. The overall organization of fosmid DX-4H-17 has the highest similarity hits to the genomic island of Acidithiobacillus caldus SM-1. As for fosmid DX-8J-22, all genes, including those encoding RubisCO enzymes, are highly similar to those of Acidithiobacillus ferrooxidans (see Table S1 in the supplemental material), and the overall organization of the genes is identical to that of Acidithiobacillus ferrooxidans. So, there is no doubt that the genomic fragment cloned in fosmid DX-8J-22 belongs to Acidithiobacillus ferrooxidans.
The remaining fosmids DX-4K-26 and DX-1A-14 had truncated form IB RubisCO gene clusters. Given the conservative arrangement of carboxysome genes (34), it is possible that a complete operon encoding carboxysome is located in the immediate downstream of cbbLS in the organisms to which these fosmid sequences belong. Fosmid DX-4K-26 contained the subunits cbbR, cbbL, cbbS, and csoS2 (Fig. 1). All of the subunits are most similar to Acidithiobacillus ferrivorans SS3 within the class Gammaproteobacteria. However, the other genes in the further upstream region of these subunits have their best hits within Betaproteobacteria (see Table S3 in the supplemental material). Although none of the individual gene products were most similar to those of Sideroxydans lithotrophicus, the overall organization of genes in fosmid DX-4K-26 is most like that of genes in Sideroxydans lithotrophicus ES-1. The organization of the form IB RubisCO gene cluster in fosmid DX-1A-14 is very similar to that of fosmid DX-4H-17. Also, fosmid DX-1A-14 cbbL is most (90%) similar to Acidithiomicrobium sp. (see Table S4 in the supplemental material).
Form II RubisCO.
Form II RubisCO genes were found in fosmid DX-3D-09 and DX-7F-24 (see Table S6). The form II RubisCO gene clusters in the two fosmids contained subunits cbbM, cbbQ, and cbbO oriented in the same direction. The three subunits in the two fosmids are highly similar (Table 1). The CbbM protein encoded by fosmid DX-3D-09 is 29% similar to CbbL encoded by the same fosmid. CbbM has the conserved catalytic sequence motif GGDFIKNDE, which differentiates form II RubisCO from other members of the RubisCO superfamily (including form I RubisCO) (35). The subunit genes cbbM, cbbQ, and cbbO have their best hits with Leptothrix cholodnii SP-6 (90%, 82%, and 69%, respectively) (see Table S5). Aligning the CbbQ and CbbO amino acid sequences of form I and II RubisCO clusters in fosmid DX-3D-09, respectively, we found that the two CbbQs are 68% similar to each other and the two CbbOs are 40% similar to each other. These results suggest that the cbbQ and cbbO genes in the two clusters may have originated from horizontal gene transfer rather than ancient gene duplication.
Phylogenetic analysis based on 16S rRNA gene and cbbL sequences.
RubisCO genes substitute for 16S rRNA gene sequences in determining phylogenetic relationships in many microbial communities. To test this assertion and determine the phylogenetic affiliation in AMD, 16S rRNA gene sequence recovery was compared with cbbL sequence recovery in the same sample. 16S rRNA gene sequences were designated beginning with “AMD-DX-,” followed by the clone number in the library. The cbbL sequences from cbbL clone library were designated beginning with “DX-R,” followed by the clone number in the library, and the cbbL sequences from metagenome microarray were designated beginning with “AMD,” followed by the clone number in the metagenomic library (Fig. 2 and 3). The phylogenetic tree constructed for cbbL sequences strongly conflicts with the 16S rRNA gene phylogenetic tree. According to the 16S rRNA gene phylogenetic tree, 16S rRNA genes of microorganisms in AMD could be assigned into five bacterial lineages (Alpha-, Beta-, and Gammaproteobacteria, Actinobacteria, and Nitrospira) (Fig. 2). However, the cbbL genes formed two phyletic groups based on the types of RubisCO gene cluster. Some bacteria such as Acidithiobacillus ferrivorans contain both form IA and IB RubisCO gene clusters, but the cbbL genes in the two clusters were assigned into different phyletic groups. Comparing the phylogenetic trees of the 16S rRNA gene and cbbL, more incongruences were found. Nitrobacter winogradskyi Nb-255 belongs to the Alphaproteobacteria group in the 16S rRNA gene phylogenetic tree, and Thiomonas intermedia K12 is a member of the Betaproteobacteria, while they form a phyletic cluster in the cbbL pylogenetic tree. On the other hand, Halorhodospira halophila and Halothiobacillus neapolitanus were assigned into the Gammaproteobacteria group and were closely related to each other in the 16S rRNA gene phylogenetic tree, but they were separately divided into form IB and IA phyletic clusters in the cbbL phylogenetic tree. Similar contradictions occurred between Thiobacillus denitrificans and Thiomonas intermedia. These results revealed that the evolution of the cbbL gene obviously could not reconcile with the organismal phylogeny in AMD.
Focusing on the distribution of cbbL sequences from the library and metagenome microarray, we could find that each monophyletic group formed by experimental sequences contained cbbL sequences from both the cbbL library and the metagenome microarray (Fig. 3). This suggested that the depth of the metagenomic library is sufficient for analysis of RubisCO gene diversity in the AMD environment.
DISCUSSION
The rapid screening of target genes from unculturable bacteria can be achieved by the combination of the metagenomic library and the microarray. Here, we report the first case of RubisCO-associated genomic fragments isolated from AMD using the metagenomic microarray. Six RubisCO genes containing genomic fragments were obtained from microarray screening and sequencing. None of the genomic fragments, with the exception of DX-8J-22, could be assigned into the known bacteria, but they showed the diversity of RubisCO gene clusters in AMD.
Interestingly, two RubisCO gene clusters that separately code for form I and II RubisCO were found in the same fosmid sequence. Previous studies suggested that form I RubisCO can fix carbon dioxide at lower levels of CO2 and that form II RubisCO is activated at higher levels of CO2 (36). The form I RubisCO gene cluster was assigned into form IA and IB RubisCO gene clusters. A primary divergence in the gene organization of the form IB RubisCO gene clusters is the insertion of genes encoding carboxysome, which are lacking in form IA RubisCO gene clusters. Carboxysome is a polyhedral bacterial microcompartment that contains the carbon-concentrating mechanism (CCM) (37). In carboxysome, carbonic anhydrase catalyzes bicarbonate into CO2 in the vicinity of RubisCO to enhance CO2 fixation (34). These results suggested that extremophiles in AMD can control carbon dioxide fixation by various mechanisms, including regulation of the expression of different gene clusters and improvement of the CO2 concentration.
Previous work indicated that discrepancies occurred widely between phylogenies based on RubisCO genes and those based on 16S rRNA genes, but the evidence was much weaker, providing no indication as to the significance of genomic content and organization. The data from the genomic fragments presented here revealed that phylogenetic conflicts occurred widely between the cbbL genes and surrounding genomic fragments. Genes further up- and downstream of the cbbL in fosmid DX-4H-17 showed more congruence to Proteobacteria than to Actinobacteria, within which the cbbL had the best hit. It is possible that these phylogenetic conflicts may result from horizontal gene transfer rather than ancient gene duplication. In fact, extreme environmental conditions such as AMD have a great effect on the occurrence of horizontal gene transfer. The case of fosmid DX-4K-26 is slightly different from those of the above-mentioned two fosmids. All subunits encoded by the RubisCO gene cluster in fosmid DX-4K-26 are most similar to those of Acidithiobacillus ferrivorans SS3. However, it is not enough to determine that the genomic fragment originates from A. ferrivorans SS3. Because all other genes upstream of the RubisCO gene cluster have their best hits within Betaproteobacteria, we deduced that the fragment belongs to Betaproteobacteria. In fact, the RubisCO gene cluster may be acquired by one horizontal gene transfer event. A similar phenomenon occurring in Rhodobactercapsulatus has been reported (38). These data revealed that horizontal transfers of RubisCO genes may occur widely in AMD, which makes the RubisCO gene cluster difficult to reconcile with further up- and downstream genomic fragments. Besides, horizontal transfers of RubisCO genes are significant in ecology, since they may change the microbial carbon dioxide fixation ability, which contributes to niche differentiation in AMD microbial communities.
The cbbL phylogenetic tree revealed the interesting phenomenon that form IA and IB RubisCO gene clusters contain not only the respective genomic organizations but also phylogenetic differences. Previous works showed that some bacteria such as Acidithibacillus caldus ATCC 51756 (39) contain only the form IB RubisCO gene cluster, some bacteria such as Cupriavidus metallidurans CH34 (40) harbor only the form IA RubisCO gene cluster, and some bacteria such as Acidithiobacillus ferrooxidans ATCC 23270 (31) contain RubisCO gene clusters of both forms (41). Nevertheless, cbbL genes preferentially form two phyletic groups based on the forms of the gene cluster rather than on species in the cbbL phylogenetic tree. Thus, we believe that the cbbL genes in form IA and IB RubisCO gene clusters are significantly divergent in molecular evolution and phylogenetics, though whether the assertion is widely applicable in all organisms needs further phylogenetic analysis. Comparing the 16S rRNA gene phylogenetic tree with the cbbL phylogenetic tree, many conflicts were found. Some bacteria that are close in the 16S rRNA gene phylogenetic tree, such as Thiobacillus denitrificans ATCC 25259 and Thiomonas intermedia K12, were divided into form IA and IB phyletic clusters, which suggested that they contain two different form I RubisCO gene clusters, while some bacteria that are distant in the 16S rRNA gene phylogenetic tree form a cluster with a high bootstrap value in the cbbL phylogenetic tree. This is strong evidence of horizontal gene transfer of RubisCO genes. It is unreliable to study the phylogeny based on cbbL sequences without the knowledge of the genetic content and organization of the RubisCO gene cluster in microbes.
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Program (no. 2010CB630901) and the National Natural Science Foundation of China (no. 30770051).
Footnotes
Published ahead of print 18 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03400-12.
REFERENCES
- 1. Shively J, Devore W, Stratford L, Porter L, Medlin L, Stevens S., Jr 1986. Molecular evolution of the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO). FEMS Microbiol. Lett. 37:251–257 [Google Scholar]
- 2. Ellis RJ. 1979. The most abundant protein in the world. Trends Biochem. Sci. 4:241–244 [Google Scholar]
- 3. Tabita F. 1988. Molecular and cellular regulation of autotrophic carbon dioxide fixation in microorganisms. Microbiol. Rev. 52:155–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bainbridge G, Madgwick P, Parmar S, Mitchell R, Paul M, Pitts J, Keys AJ, Parry MAJ. 1995. Engineering Rubisco to change its catalytic properties. J. Exp. Bot. 46:1269 [Google Scholar]
- 5. Kellogg EA, Juliano ND. 1997. The structure and function of RuBisCO and their implications for systematic studies. Am. J. Bot. 84:413 doi:10.2307/2446015 [PubMed] [Google Scholar]
- 6. Selesi D, Schmid M, Hartmann A. 2005. Diversity of green-like and red-like ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes (cbbL) in differently managed agricultural soils. Appl. Environ. Microbiol. 71:175–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tabita FR, Hanson TE, Li H, Satagopan S, Singh J, Chan S. 2007. Function, structure, and evolution of the RubisCO-like proteins and their RubisCO homologs. Microbiol. Mol. Biol. Rev. 71:576–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jordan DB, Ogren WL. 1983. Species variation in kinetic properties of ribulose 1,5-bisphosphate carboxylase/oxygenase. Arch. Biochem. Biophys. 227:425–433 [DOI] [PubMed] [Google Scholar]
- 9. Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058 doi:10.1126/science.273.5278.1058 [DOI] [PubMed] [Google Scholar]
- 10. Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364–370 [DOI] [PubMed] [Google Scholar]
- 11. Kunst F, Ogasawara N, Moszer I, Albertini A, Alloni G, Azevedo V, Bertero M, Bessieres P, Bolotin A, Borchert S. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256 [DOI] [PubMed] [Google Scholar]
- 12. Hanson TE, Tabita FR. 2001. A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 98:4397 doi:10.1073/pnas.081610398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Delwiche CF, Palmer JD. 1996. Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Mol. Biol. Evol. 13:873–882 [DOI] [PubMed] [Google Scholar]
- 14. Uchino Y, Yokota A. 2003. “Green-like” and “red-like” RubisCO cbbL genes in Rhodobacter azotoformans. Mol. Biol. Evol. 20:821–830 [DOI] [PubMed] [Google Scholar]
- 15. Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43 [DOI] [PubMed] [Google Scholar]
- 16. Bond PL, Smriga SP, Banfield JF. 2000. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl. Environ. Microbiol. 66:3842–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou J, Bruns MA, Tiedje JM. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park SJ, Kang CH, Chae JC, Rhee SK. 2008. Metagenome microarray for screening of fosmid clones containing specific genes. FEMS Microbiol. Lett. 284:28–34 [DOI] [PubMed] [Google Scholar]
- 19. Bae JW, Rhee SK, Park JR, Chung WH, Nam YD, Lee I, Kim H, Park YH. 2005. Development and evaluation of genome-probing microarrays for monitoring lactic acid bacteria. Appl. Environ. Microbiol. 71:8825–8835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rhee SK, Liu X, Wu L, Chong SC, Wan X, Zhou J. 2004. Detection of genes involved in biodegradation and biotransformation in microbial communities by using 50-mer oligonucleotide microarrays. Appl. Environ. Microbiol. 70:4303–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chaisson MJ, Pevzner PA. 2008. Short read fragment assembly of bacterial genomes. Genome Res. 18:324–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Aziz R, Bartels D, Best A, DeJongh M, Disz T, Edwards R, Formsma K, Gerdes S, Glass E, Kubal M. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75 doi:10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eddy SR. 2008. A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput. Biol. 4:e1000069 doi:10.1371/journal.pcbi.1000069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 26. Larkin M, Blackshields G, Brown N, Chenna R, McGettigan P, McWilliam H, Valentin F, Wallace I, Wilm A, Lopez R. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 27. Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 28. Imker HJ, Fedorov AA, Fedorov EV, Almo SC, Gerlt JA. 2007. Mechanistic diversity in the RuBisCO superfamily: the “enolase” in the methionine salvage pathway in Geobacillus kaustophilus. Biochemistry 46:4077–4089 [DOI] [PubMed] [Google Scholar]
- 29. Hayashi NR, Arai H, Kodama T, Igarashi Y. 1997. The novel genes, cbbQ and cbbO, located downstream from the RubisCO genes of Pseudomonas hydrogenothermophila, affect the conformational states and activity of RubisCO. Biochem. Biophys. Res. Commun. 241:565–569 [DOI] [PubMed] [Google Scholar]
- 30. Kusian B, Bowien B. 1997. Organization and regulation of cbb CO2 assimilation genes in autotrophic bacteria. FEMS Microbiol. Rev. 21:135–155 [DOI] [PubMed] [Google Scholar]
- 31. Valdés J, Pedroso I, Quatrini R, Dodson RJ, Tettelin H, Blake R, II, Eisen JA, Holmes DS. 2008. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9:597 doi:10.1186/1471-2164-9-597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ. 2007. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J. Mol. Biol. 372:764–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu Y, Feng L, Jeffrey PD, Shi Y, Morel FMM. 2008. Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms. Nature 452:56–61 [DOI] [PubMed] [Google Scholar]
- 34. Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM. 2008. Protein-based organelles in bacteria: carboxysomes and related microcompartments. Nat. Rev. Microbiol. 6:681–691 [DOI] [PubMed] [Google Scholar]
- 35. Elsaied H, Kimura H, Naganuma T. 2002. Molecular characterization and endosymbiotic localization of the gene encoding D-ribulose 1,5-bisphosphate carboxylase-oxygenase (RuBisCO) form II in the deep-sea vestimentiferan trophosome. Microbiology 148(Pt 6):1947–1957 [DOI] [PubMed] [Google Scholar]
- 36. Badger MR, Bek EJ. 2008. Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 59:1525 doi:10.1093/jxb/erm297 [DOI] [PubMed] [Google Scholar]
- 37. Badger MR, Hanson D, Price GD. 2002. Evolution and diversity of CO2 concentrating mechanisms in cyanobacteria. Funct. Plant Biol. 29:161–173 [DOI] [PubMed] [Google Scholar]
- 38. Paoli GC, Soyer F, Shively J, Tabita FR. 1998. Rhodobacter capsulatus genes encoding form I ribulose-1,5-bisphosphate carboxylase/oxygenase (cbbLS) and neighbouring genes were acquired by a horizontal gene transfer. Microbiology 144:219 doi:10.1099/00221287-144-1-219 [DOI] [PubMed] [Google Scholar]
- 39. Valdes J, Quatrini R, Hallberg K, Dopson M, Valenzuela PDT, Holmes DS. 2009. Draft genome sequence of the extremely acidophilic bacterium Acidithiobacillus caldus ATCC 51756 reveals metabolic versatility in the genus Acidithiobacillus. J. Bacteriol. 191:5877–5878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A, Benotmane MA, Leys N, Vallaeys T, Lapidus A. 2010. The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS One 5:e10433 doi:10.1371/journal.pone.0010433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kusano T, Takeshima T, Inoue C, Sugawara K. 1991. Evidence for two sets of structural genes coding for ribulose bisphosphate carboxylase in Thiobacillus ferrooxidans. J. Bacteriol. 173:7313–7323 [DOI] [PMC free article] [PubMed] [Google Scholar]