Abstract
AlgE is a monomeric 18-stranded β-barrel protein required for secretion of the extracellular polysaccharide alginate in Pseudomonas aeruginosa. To assess the molecular mechanism of alginate secretion, AlgE was subjected to site-specific and FLAG epitope insertion mutagenesis. Except for β-strands 6 and 10, epitope insertions into the transmembrane β-strands abolished localization of AlgE to the outer membrane. Interestingly, an epitope insertion into β-strand 10 produced alginate and was only detectable in outer membranes isolated from cells grown on solid media. The deletion of nine C-terminal amino acid residues destabilized AlgE. Replacement of amino acids that constitute the highly electropositive pore constriction showed that individual amino acid residues have a specific function in alginate secretion. Two of the triple mutants (K47E+R353A+R459E and R74E+R362A+R459E) severely reduced alginate production. Mutual stability analysis using the algE deletion mutant PDO300ΔalgE(miniCTX) showed the periplasmic alginate biosynthesis proteins AlgK and AlgX were completely destabilized, while the copy number of the inner membrane c-di-GMP receptor Alg44 was reduced. Chromosomal integration of algE restored AlgK, AlgX, and Alg44, providing evidence for a multiprotein complex that spans the cell envelope. Periplasmic turn 4 of AlgE was identified as an important region for maintaining the stability of the putative multiprotein complex.
INTRODUCTION
Pseudomonas aeruginosa is an opportunistic human pathogen of particular relevance to cystic fibrosis (CF) patients, whose lungs are susceptible to severe and chronic infections by the bacterium. In the CF lung, P. aeruginosa converts to a highly mucoid phenotype, which is characterized by the overproduction of alginate (1). Alginate serves as an extracellular matrix component that enables the formation of differentiated biofilms, which confer resistance to antibiotics and prevent phagocytosis by the immune system of the host (2, 3). Alginate is an unbranched random polymer consisting of β-d-mannuronic acid and its C-5 epimer, α-l-guluronic acid. Most of the genes required for alginate biosynthesis are located on a single operon under the control of the AlgD promoter (4). Products of these genes are involved in precursor biosynthesis, polymerization, modification, and secretion (5, 6). Alginate precursor biosynthesis also requires AlgC, the gene for which is located outside this operon. AlgC is also involved in rhamnolipid and lipopolysaccharide biosynthesis (7). Recently, it was proposed that AlgC also has a role in controlling the amount of polysaccharides Pel, Psl, and alginate produced by P. aeruginosa (8). The precursor for alginate biosynthesis, GDP mannuronic acid, is synthesized from fructose-6-phosphate by the concerted actions of AlgA, AlgC, and AlgD (1). Polymerization of alginate requires two inner membrane proteins, Alg8 and Alg44 (9, 10). Alg8 is predicted to have multiple membrane-spanning regions and a large cytoplasmic glycosyltransferase domain, while Alg44 has one transmembrane domain, a cytoplasmic PilZ c-di-GMP binding domain, and a periplasmic domain which is predicted to be similar to the membrane fusion protein MexA, from the MexAB-OprM multidrug efflux pump (9, 11, 12). The nascent polymannuronate chain is believed to enter the periplasmic space into a scaffold formed by AlgK, -G, -X, -L, -44 E, and the proteins involved in alginate acetylation, AlgJ, -I, and -F (5). AlgK is an outer membrane lipoprotein with multiple copies of the tetratricopeptide repeat protein-protein interaction motif (13). AlgK has been demonstrated to interact with AlgX, which in turn interacts with MucD, a serine protease involved in regulation of alginate biosynthesis (14). Deletion of algK, algE, or algX results in secretion of free uronic acids, which are degradation products of high-molecular-weight alginate (15–19). The alginate lyase AlgL is thought to play a dual role in alginate biosynthesis, both as a component of the putative periplasmic scaffold and degrading mislocalized periplasmic alginate (20, 21). Free uronic acids are believed to be secreted when the integrity of the scaffold/complex is compromised and alginate escapes into the periplasm and is exposed to the alginate lyase AlgL. AlgG is an acetylation-sensitive epimerase that converts nonacetyled d-mannuronate to its C-5 epimer, l-guluronate, at the polymer level (22). Alginate secreted by P. aeruginosa can also be selectively O-acetylated at the O-2′ and/or O-3′ positions of mannuronate residues by the actions of AlgI, AlgJ, and AlgF (23). The recently solved structure of AlgE revealed that it is a monomeric outer membrane 18-stranded β-barrel porin (24). An isogenic deletion mutant of algE has shown that AlgE is essential for alginate production (16), but the presence of free uronic acids indicates that not only is AlgE required for the secretion of full-length alginate but also that the protein may play a role in the formation of the periplasmic scaffold/complex. The periplasmic turns of AlgE have been suggested as potential sites for mediating its interaction with the other periplasmic components required for alginate biosynthesis (24). The AlgE pore is lined with highly conserved, charged amino acid residues, which have been suggested to confer selectivity toward alginate and/or facilitate its efficient secretion across the outer membrane (24).
In the present study, a site-specific mutagenesis approach, guided by the structure of AlgE, was applied to assess the role the membrane segments, periplasmic turns, and pore-constricting residues play in alginate secretion and the stability, subcellular localization, and assembly of the proposed multiprotein periplasmic complex. Chromosomal AlgE variants that contain FLAG epitope insertions in the periplasmic turns have also been used to gain insight into the proposed interaction(s) between AlgE and other components of the periplasmic scaffold.
MATERIALS AND METHODS
Construction of FLAG epitope insertion variants.
The FLAG epitope was inserted into seven transmembrane segments and six periplasmic turns by site-directed, ligase-independent mutagenesis (25). In brief, two PCR products were made for each FLAG epitope insertion by using pGEM-TEasy:algE (16) as the template. One primer pair was FFLAG and the corresponding reverse primer, Rs (e.g., algEM2FFLAG and algEM2Rs), and the second with primer pair was Fs and corresponding reverse primer RFLAG (e.g., algEM2Fs and algEM2RFLAG) (see Table S1 in the supplemental material). An AlgE variant, AlgEtrC9, with 9 amino acids removed from the C terminus, was created using primer algEN-HiSDNd and reverse primer algECtr9. Plasmid template was removed at the end of the PCR by addition of 10 μl of D-buffer (20 mM MgCl2, 20 mM Tris [pH 8.0], 5 mM dithiothreitol) containing 10 units of DpnI and incubating the mixture at 37°C for 60 min. The DpnI-treated products were mixed for the hybridization reaction with 10 μl of 5× H-buffer (750 mM NaCl in 125 mM Tris [pH 9.0] and 100 mM EDTA pH 8.0 with a final pH of 8.5), 15 μl of each PCR product, and autoclaved water to bring the volume to 50 μl. The hybridization mixture was incubated at 99°C for 3 min, followed by two cycles of 65°C for 5 min and 30°C for 15 min. Twenty microliters of hybridized product was used to transform Escherichia coli competent TOP 10 cells. Selection of cells containing the new plasmid was achieved on LB plates containing ampicillin at a concentration of 75 μg/ml of medium. The insertion of the 24-bp FLAG epitope was confirmed by sequencing the open reading frame. The resulting pGEM-TEasy plasmids containing the different algE-(FLAG) insertions were hydrolyzed with BamHI and HindIII, and the resulting 1,497-bp fragments were ligated into pBBR1MCS-5 to produce the following plasmids: pBBR1MCS-5:algEM2FLAG, pBBR1MCS-5:algEM4FLAG, pBBR1MCS-5:algEM6FLAG, pBBR1MCS-5:algEM8FLAG, pBBR1MCS-5:algEM10FLAG, pBBR1MCS-5:algEM12FLAG, pBBR1MCS-5:algEM14FLAG, pBBR1MCS-5:algET2FLAG, pBBR1MCS-5:algET4FLAG, pBBR1MCS-5:algET5FLAG, pBBR1MCS-5:algET6FLAG, pBBR1MCS-5:algET8-1FLAG, pBBR1MCS-5:algET8-2FLAG, pBBR1MCS-5:algET8-3FLAG, pBBR1MCS-5:algECtr9. The pBBR1MCS-5-carrying variants of algE were transferred into PDO300ΔalgE (16) by electroporation and selection of the transformants on pseudomonas isolation agar (PIA) plates containing 300 μg/ml of gentamicin (see Table S2 in the supplemental material).
Generation of site-specific variants.
Site-directed mutants of AlgE were created as described above and resulted in the construction of the following plasmids: pBBR1MCS-5:algEK47E, pBBR1MCS-5:algER74E, pBBR1MCS-5:algER129E, pBBR1MCS-5:algER154E, pBBR1MCS-5:algER353A, pBBR1MCS-5:algEH364A, pBBR1MCS-5:algER362A, pBBR1MCS-5:algER365A, pBBR1MCS-5:algER459E, pBBR1MCS-5:algER461E, pBBR1MCS-5:algER152A, pBBR1MCS-5:algEN164A, pBBR1MCS-5:algER481E, pBBR1MCS-5:algEE130A, pBBR1MCS-5:algED162A, pBBR1MCS-5:algEE189A, pBBR1MCS-5:algED193A, pBBR1MCS-5:algEE368A, pBBR1MCS-5:algED485A. Using pGEMTeasy:algER459E as a template, double amino acid substitution variants pGEMT-easy:algER353A+R459E, pGEM-Teasy:algER362A+R459E, and pGEM-Teasy:algER368A+R459E were made. The pGEMTeasy plasmids containing different site-directed mutants of algE were hydrolyzed with BamHI and HindIII, and the resulting 1,473-bp fragments were ligated into pBBR1MCS-5, resulting in the following plasmids: pBBR1MCS-5:algER353A+R459E, pBBR1MCS-5:algER362A+R459E, and pBBR1MCS-5:algER368A+R459E. To make triple mutants, plasmids pBBR1MCS-5:algEK47E and pBBR1MCS-5:algER74E were hydrolyzed with HindIII and EcoRI, and the resulting 407-bp fragments were ligated into HindIII- and EcoRI-digested pBBR1MCS-5:algER353A+R459E, pBBR1MCS-5:algER362A+R459E, and pBBR1MCS-5:algER368A+R459E, resulting in plasmids pBBR1MCS-5:algEK47E+R353A+R459E, pBBR1MCS-5:algER74E+R353A+R459E, pBBR1MCS-5:algEK47E+R362A+R459E, pBBR1MCS-5:algER74E+R362A+R459E, pBBR1MCS-5:algEK47E+R368A+R459E, and pBBR1MCS-5:algER74E+R368A+R459E. The pBBR1MCS-5 plasmids carrying different site-directed variants of algE were transferred into PDO300ΔalgE (16) through electroporation (see Table S2). All of the restriction enzymes were purchased from Roche.
Alginate quantification.
Bacterial cultures were grown overnight, and cells from 2 ml of bacterial culture were harvested and washed twice with sterile saline buffer. Aliquots 200 μl of cells were spread on a PIA plate and incubated at 37°C for 72 h. Cells were scraped off the plates and washed twice with sterile saline solution while keeping the supernatant with dissolved alginate for subsequent precipitation. Cell pellets were freeze-dried, and the final weight was determined. Supernatant with dissolved alginate was precipitated with 1 volume of ice-cold isopropanol, and alginate was harvested and freeze-dried. For further purification, alginate was dissolved in buffer A (50 mM Tris-HCl [pH 7.4] and 10 mM MgCl2) to a final concentration of 0.5% (wt/vol). After alginate was solubilized in buffer A, 15 μg/ml of DNase and RNase was added, and the solution was incubated at 37°C for 6 h with shaking. Pronase E was added to a final concentration of 20 μg/ml, and the solution was incubated again for 18 h at 37°C in a shaking incubator. The final solution was dialyzed against 5 liters of Milli-Q water for 48 h at 4°C in tubing with a molecular mass cutoff 12 kDa (ZelluTrans, Roth). After dialysis, the alginate was precipitated against 1 volume of isopropanol and freeze-dried for subsequent uronic acid quantification.
The amount of free uronic acid in 2-ml aliquots from overnight-grown cultures was measured. Cells were pelleted by centrifugation, the supernatant was filtered through vivaspin-500 (GE Healthcare) centrifugal filter devices with a molecular mass cutoff of 10 kDa, and the flowthrough was collected. The uronic acid content of the flowthrough, which contained the free uronic acids and short-chain alginate degradation products, was determined as described below.
Uronic acid assay.
Alginate quantification was performed using the uronic acid assay as described previously (26). Alginic acid from brown seaweed was used as a standard. Briefly, alginate samples were dissolved in Milli-Q water at concentrations of between 0.25 and 0.05 mg/ml. Aliquots of 200 μl of these samples were mixed with 1.2 ml of tetraborate solution (12.5 mM disodium tetraborate in concentrated sulfuric acid) and incubated on ice for 10 min. This mixture was incubated at 100°C for 5 min and then cooled on ice for 5 min. A volume of 20 μl of m-hydroxybiphenyl reagent [0.15%, wt/vol] hydroxybiphenyl in 125 mM NaOH) was added to the reaction mixture, and the mixture was vortexed for 1 min. For each sample or dilution, a negative control was assayed using 125 mM NaOH instead of using hydroxybiphenyl reagent. The uronic acid concentrations were determined spectrophotometrically at a wavelength of 520 nm.
Isolation of whole envelopes and OMs.
Strains of P. aeruginosa were grown overnight in LB medium with appropriate antibiotics. Cells were harvested by centrifugation at 6,000 × g for 30 min at 4°C and washed twice with an equal volume of 10 mM HEPES (pH 7.4) buffer. Cells were suspended in 10 ml of 10 mM HEPES buffer with 1 Complete mini-EDTA-free protease inhibitor cocktail tablet (Roche) and sonicated on ice for 12 cycles with 15 s of sonication followed by 15 s of cool down on ice. Cellular debris and unbroken cells were removed by centrifugation at 8,000 × g for 45 min at 4°C. The whole-envelope fraction was isolated by centrifugation at 100,000 × g for 1 h at 4°C and washed. To isolate the outer membranes (OMs), the envelope fraction was resuspended in 1 volume of 10 mM HEPES buffer containing 0.7% (wt/vol) N-laurosyl-sarcosine, and the suspension was incubated at room temperature for 20 min with shaking to solubilize the inner membrane. This mixture was centrifuged at 100,000 × g for 1 h. The pellet was resuspended in 10 ml of 10 mM HEPES buffer and centrifuged again at 100,000 × g to remove residual detergent. The resulting sediments represented the OM fraction. The total protein concentration of each respective fraction, the envelope and OM, was determined using a Quant-iT protein assay kit (Invitrogen).
Analysis of outer membrane proteins.
Total protein (25 μg) was loaded and separated by SDS-PAGE using 8% polyacrylamide gels. The resulting gels were either stained with Coomassie blue stain or Western blotted using the iBlot dry blotting system (Invitrogen). After blotting, the nitrocellulose membrane was blocked with 5% (wt/vol) skim milk in Tris-buffered saline containing 0.05% (vol/vol) Tween 20 for 1 h at room temperature. Anti-Alg44 (1:10,000), anti-AlgK (1:10,000), anti-AlgG (1:1,000), anti-AlgX (1:7,000), and anti-AlgE (1:5,000) polyclonal antibodies, raised in rabbits against the respective purified proteins, were used as primary antibodies, and anti-IgG anti-rabbit antibodies, labeled with horseradish peroxidase were used as secondary antibodies. The membrane was washed three times, and bound antibodies were resolved with SuperSignal West Pico chemiluminescent substrate (Thermoscientific, Rockford, IL) and developed on X-ray film (Kodak, Rochester, NY).
Chromosomal integration.
The promoter region at bp −879 relative to the algD open reading frame was amplified by forward (PalgPstIF) and reverse (PalgHindIIIR) primers. After A tailing, the fragment was ligated into pGEM-TEasy vector (Promega, Madison, WI), and the fidelity of the sequence was verified. The promoter region of algD, 879 bp, was hydrolyzed from pGEM-TEasy by using PstI and HindIII. Various variants of algE (T2F, T4F, T5F, T6F, M6F, and T8F-1) and wild-type algE were hydrolyzed using HindIII and BamHI from the respective pGEM-TEasy backbones. Purified algD promoter region and individual algE fragments were ligated together into the integration proficient mini-CTX::lacZ-based plasmid, hydrolyzed with PstI and BamHI, to generate the mini-CTX::PalgET2F, mini-CTX::PalgET4F, mini-CTX::PalgET5F, mini-CTX::PalgET6F, and mini-CTX::PalgE plasmids. These plasmids were then transferred into PDO300ΔalgE strains by electroporation and selected on medium containing tetracycline at a concentration of 150 μg/ml. Integration into the chromosome was confirmed through PCR using PserUP and PserDOWN primers (27). The backbone of the mini-CTX plasmid was removed by transferring the pFLP2 plasmid through electroporation, and subsequently pFLP2 was cured by cultivating for 24 h on mineral salt medium containing 5% (wt/vol) sucrose (28). Tetracycline- and carbenicillin-sensitive cells were analyzed by PCR to confirm the removal of the mini-CTX-lacZ backbone. Similarly, the mini-CTX::lacZ plasmid was transferred into PDO300 and PDO300ΔalgE and confirmed by PCR.
RESULTS
Mutational analysis of residues involved in the secretion of alginate.
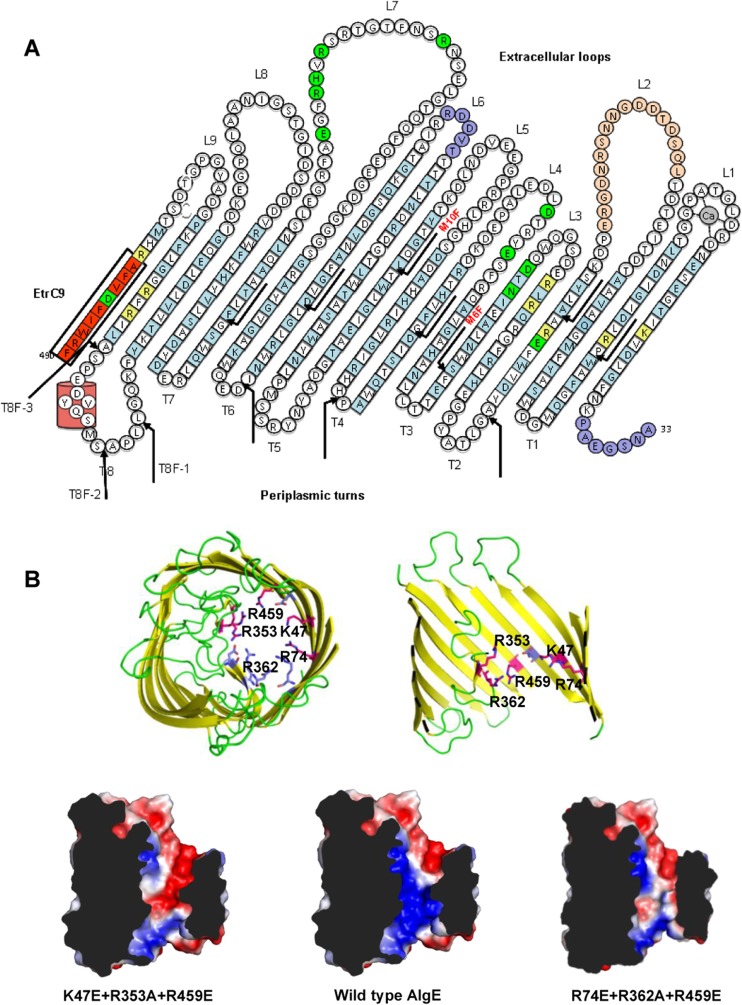
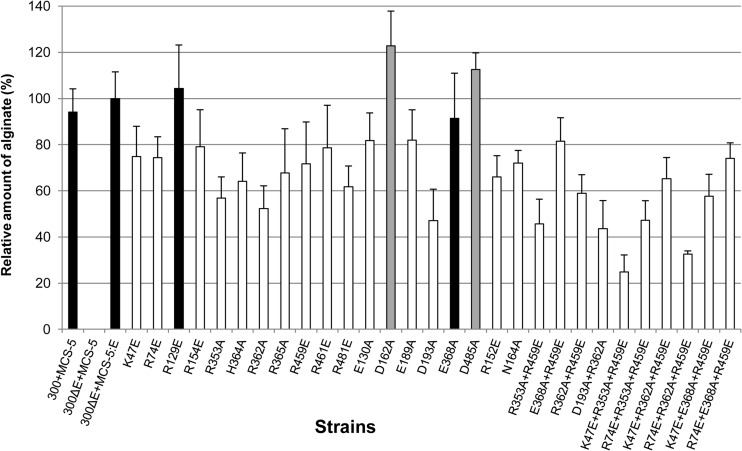
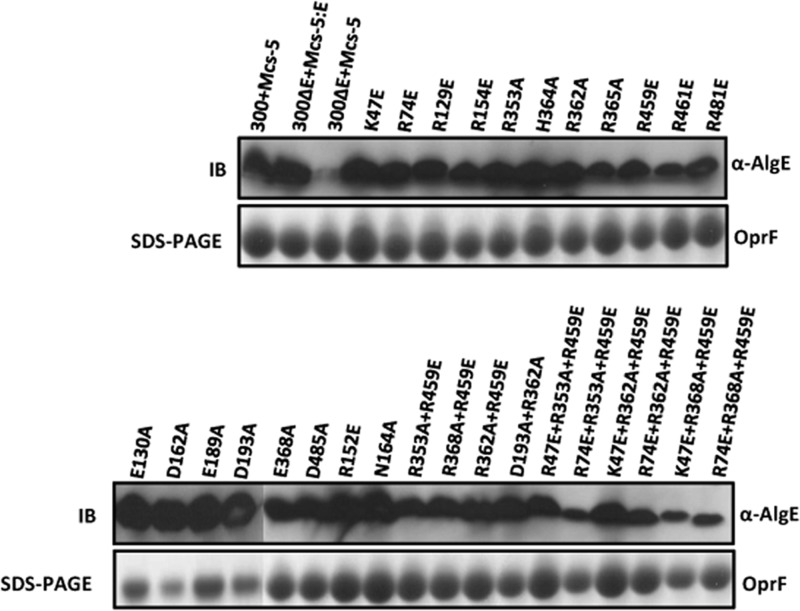
Site-directed mutagenesis of AlgE was performed to identify amino acid residues that are critical for alginate secretion. Highly conserved amino acid residues that line the pore of the β-barrel, constrict the channel opening, and contribute to its electropositive surface were selected, as these residues may confer specific binding and/or selectivity toward its polyanionic substrate, alginate (24). To assess the role of these residues single, double, and triple amino acids substitution variants of AlgE were created. Residues K47, R74, R129, R152, D162, N164, R353, R362, R459, and D485 were selected, as they are highly conserved and constrict the channel. Most of these residues were found conserved in P. aeruginosa, P. putida, P. entomophilia, P. mendocina, P. fluorescence, P. syringae, and Azotobacter vinelandii (24). To reduce the positive electrostatic field inside the AlgE lumen, positively charged arginine and lysine residues were substituted with glutamate and alanine, while asparagine and negatively charged aspartate residues were replaced with alanine (Fig. 1). The amount of secreted alginate mediated by these AlgE variants was quantified. Except for R129E, D162A, and D485A, mutation of each of the selected amino acid residues resulted in reduced production of alginate compared to the wild type (Fig. 2). OMs derived from PDO300ΔalgE expressing the various mutated algE genes were subjected to SDS-PAGE and Western blot analysis, using primary anti-AlgE polyclonal rabbit antibodies. All site-specific mutations were structurally tolerated, and the respective AlgE variants were able to localize to the OM (Fig. 2).
Fig 1.
Membrane topology and structural models of AlgE and its variants. (A) Topology model of AlgE, based on the X-ray crystal structure (modified from J. C. Whitney et al. [24]). Arrows indicate the positions at which FLAG epitopes were inserted. “EtrC9” indicates the location of residues 482 to 490, which were deleted at the C terminus of the protein. Amino acid residues forming the transmembrane β-sheet are indicated in squares. Amino acid residues are represented by single-letter codes. Amino acid residues with side chains pointing into the barrel lumen are shown in blue. Alanine- and glutamic acid-substituted amino acids residues are shown in green and yellow, respectively. The amino acid residues which could not be modeled in the crystal structure are shown in blue-purple. Ca in L1 represents a calcium ion. (B) Cartoon representation of AlgE, as shown from the extracellular surface (upper left) and in the plane of the outer membrane (upper right), with the top exposed to the cell surface and the bottom exposed to the periplasm. In each model, the pore-forming residues are indicated using a sticks representation. Residues shown in pink, K47E, R74E, R353A, R459E, and R362A, constitute residues in the two triple mutants with significantly reduced alginate production. Electrostatic surface representation of wild-type AlgE and the AlgE K47E+R353A+R459E and R74E+R362A+R459E triple mutants (lower panel). Note that the positive electrostatic field (blue) inside the putative alginate translocation pathway is replaced by a negative electrostatic field (red) in both triple mutants. Mutated residues were modeled as their lowest-energy rotamers in COOT (29). Electrostatics were generated using Pymol.
Fig 2.
Relative amounts of alginate produced by P. aeruginosa PDO300ΔalgE harboring various plasmids. The amounts of alginate (normalized to the amount of AlgE in OM) produced by PDO300ΔalgE complemented with the algE site-specific mutants are presented relative to the amounts produced by P. aeruginosa PDO300ΔalgE complemented with wild-type algE. The amount of alginate produced by wild-type algE-complemented PDO300ΔalgE, given as 100%, corresponds to 0.729 g/g of cell dry weight. Quantification of bands was performed by using the gel analysis software UN-SCAN-IT gel 6.1 (Silkscientific). Experiments were conducted in triplicates, and the error bars represent the standard deviations of the means. An unpaired Student t test was applied, and P values of ≤0.05 were considered significant. Variants with no significant change in alginate production are shown in black, those with an increase in alginate production are shown in gray, and those with a reduction in alginate production are shown in white. 300+MCS-5 indicates PDO300 carrying the pBBR1MCS-5 plasmid; 300ΔE+MCS-5 shows PDO300ΔalgE carrying the pBBR1MCS-5 plasmid; 300ΔE+MCS-5:E depicts PDO300ΔalgE carrying the pBBR1MCS-5::algE plasmid. Variants of AlgE are designated by the single-letter code and number for each amino acid in wild-type AlgE that was targeted, followed by the single-letter code of the amino acid to which the residue was mutated.
It was previously shown that deletions or insertions in loop L7, located between β-strands 13 and 14, are not tolerated and completely ablate alginate production (16). The structure of AlgE revealed that this loop is folded inside the lumen of the β-barrel, where it restricts the size of the pore opening and helps stabilize the structure (24). We therefore replaced amino acids R353, R362, H364, R365, and E368 with alanine to further explore the role of loop L7 in defining the specificity of the pore and AlgE function. Mutation of R353, R362, H364, and R365 resulted in at least a 32% reduction in alginate production, while the E368A variant did not show any significant difference in alginate production (Fig. 3). Substitutions were also made to other residues which line the inside the AlgE pore (E130A, R154E, E189A, D193A, R461E, and R481E), and various double (R353A+R459E, R368A+R459E, R362A+R459E, and D193A+R362A) and triple (K47E+R353A+R459E, R74E+R353A+R459E, K47E+R362A+R459E, R74E+R362A+R459E, K47E+E368A+R459E, and R74E+E368A+R459E) mutants were created. All of these variants showed a decrease in the amount of alginate produced compared to wild-type AlgE. The largest reductions in alginate production were observed for the K47E+R353A+R459E and R74E+R362A+R459E triple mutants, which showed 75% and 67% decreases in alginate production, respectively, compared to wild-type AlgE (Fig. 2).
Fig 3.
Outer membrane localization of AlgE and its variants. Outer membranes from planktonic cultures of P. aeruginosa PDO300ΔalgE and P. aeruginosa PDO300ΔalgE harboring various plasmids carrying the AlgE gene were isolated and analyzed by immunoblotting (IB). Constitutively expressed oprF was used as a loading control (bottom panel). Only the relevant parts of the gels and the immunoblot are shown.
Structure-function analysis using FLAG epitope insertion mutagenesis.
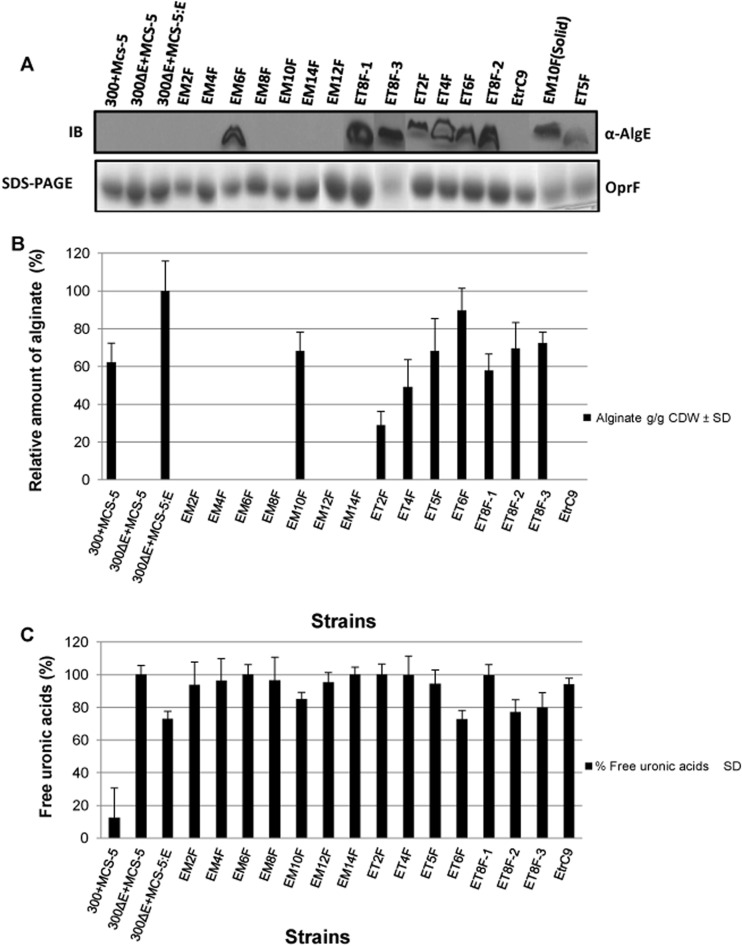
To assess the structural and functional importance of different regions of AlgE, FLAG epitopes were inserted in transmembrane regions and periplasmic turns by using a site-directed mutagenesis approach (25). FLAG epitopes were inserted in alternating β-strands, starting from β-strand 2 through β-strand 14, and also in selected periplasmic turns to generate the AlgE variants. The variants with the FLAG insertion in transmembrane regions were designated EM2F, EM4F, EM6F, EM8F, EM10F, EM12F and EM14F, and variants with insertion in periplasmic turns were referred to as ET2F, ET4F, ET5F, ET6F, ET8F-1, ET8F-2, and ET8F-3 (Fig. 1A). These variants were used to investigate the role of these regions in protein folding and stability and the ability of the periplasmic turns to interact with other periplasmic components of the alginate synthesis machinery. The selection of the periplasmic turns was based on the AlgE structure. Plasmids carrying these variants of AlgE were then introduced into PDO300ΔalgE, and their subcellular localization and ability to restore alginate production were assessed. Variants of AlgE with FLAG epitope insertions in β-strands 2, 4, 8, 12, and 14 (EM2F, EM4F, EM8F, EM12F, and EM14F) were unable to localize to the OM when cells were grown in either planktonic mode or on solid medium, as shown by their absence in the OM fractions analyzed by immunoblotting (Fig. 4A). These variants were unable to restore alginate production to PDO300ΔalgE (Fig. 4B) and produced 100% free uronic acids (Fig. 4C). Insertion in β-strand 6 (EM6F) was tolerated with respect to subcellular localization to the OM but was not functional, as no high-molecular-weight alginate was detected (Fig. 4A and B). This variant produced 100% free uronic acids (Fig. 4C). The EM10F variant of AlgE with the FLAG epitope inserted in β-strand 10 was the only β-strand insertion tested that was capable of restoring alginate production (Fig. 4B). While the EM10F variant could not be detected in the OM when cells were grown in liquid medium, it was detected in OMs isolated from cells grown on solid medium (Fig. 4A), and these results were therefore consistent with our alginate quantification assay, which measured the amount of alginate produced after growth of the cells on solid medium (Fig. 4B).
Fig 4.
Localization of alginate and free uronic acid production by FLAG epitope insertion variants of AlgE. (A) The OM fraction isolated from planktonic cultures (unless mentioned from solid medium) of P. aeruginosa PDO300ΔalgE harboring various plasmids, showing the absence or presence of AlgE and its variants. The presence of AlgE and its variants in the OM can be seen in the immunoblots (IB) probed with anti-AlgE antibodies (upper panel). EM10F(Solid) indicates that the OM was isolated from PDO300ΔalgE (MCS5::algEM10F) cells grown on solid medium. Constitutively expressed oprF was used as a loading control (bottom panel). Only the relevant parts of the different gels and the immunoblots are shown. The bands shown here are not from a single blot. Some bands were pasted in after cutting from a different blot. (B) The amount of alginate produced was assessed by growing cells on solid medium, and results are presented relative to the amount of alginate produced by P. aeruginosa PDO300ΔalgE (pBBRMCS-5::algE). (C) The amount of free uronic acid was assessed after overnight growth in liquid culture and is given as the percent ratio between the filtrate and the supernatant. Experiments were conducted in triplicate. Error bars show the standard deviations of the mean values. 300+MCS-5 indicates PDO300 carrying the pBBR1MCS-5 plasmid; 300ΔE+MCS-5 shows PDO300ΔalgE carrying the pBBR1MCS-5 plasmid; 300ΔE+MCS-5:E depicts PDO300ΔalgE carrying the pBBR1MCS-5::algE plasmid. Variants of AlgE are indicated with the first letter E, indicating wild-type AlgE, followed by M for the membrane segment or T for the periplasmic turn, followed by a number that indicates the position in AlgE as defined by the structure shown schematically in Fig. 1. The letter F is used as an abbreviation for the FLAG epitope.
Insertions of FLAG epitopes in the periplasmic turns were tolerated as judged by their insertion into the OM and the restoration of alginate production, as quantified on solid medium (Fig. 4A and B). AlgE variants with insertions in periplasmic turns T2, T4, T5, and T8-1 produced 100% free uronic acids in liquid culture (Fig. 4C). Variants of AlgE with insertions in periplasmic turns T6, T8-2, and T8-3 produced 70 to 80% free uronic acids, compared to the algE deletion mutant (Fig. 4C). The ET6F variant produced the most alginate. The levels of both alginate and uronic acids produced by this variant were comparable to the amounts produced by the algE deletion mutant complemented with the wild-type algE gene (Fig. 4B and C). Deletion of 9 amino acids at the C terminus of AlgE abolished localization to the OM and alginate production (Fig. 4A and B) and produced 100% free uronic acids (Fig. 4C).
Mutual stability effect of AlgE and its variants on components of the proposed periplasmic scaffold.
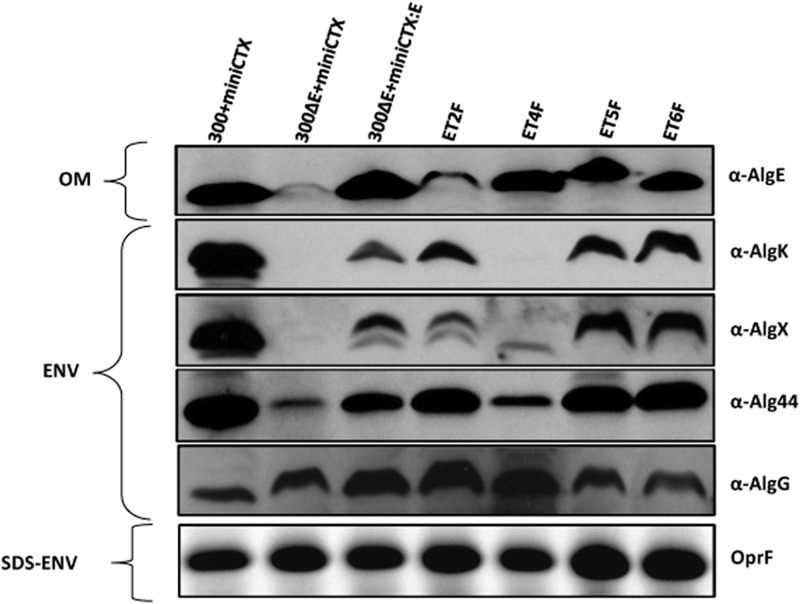
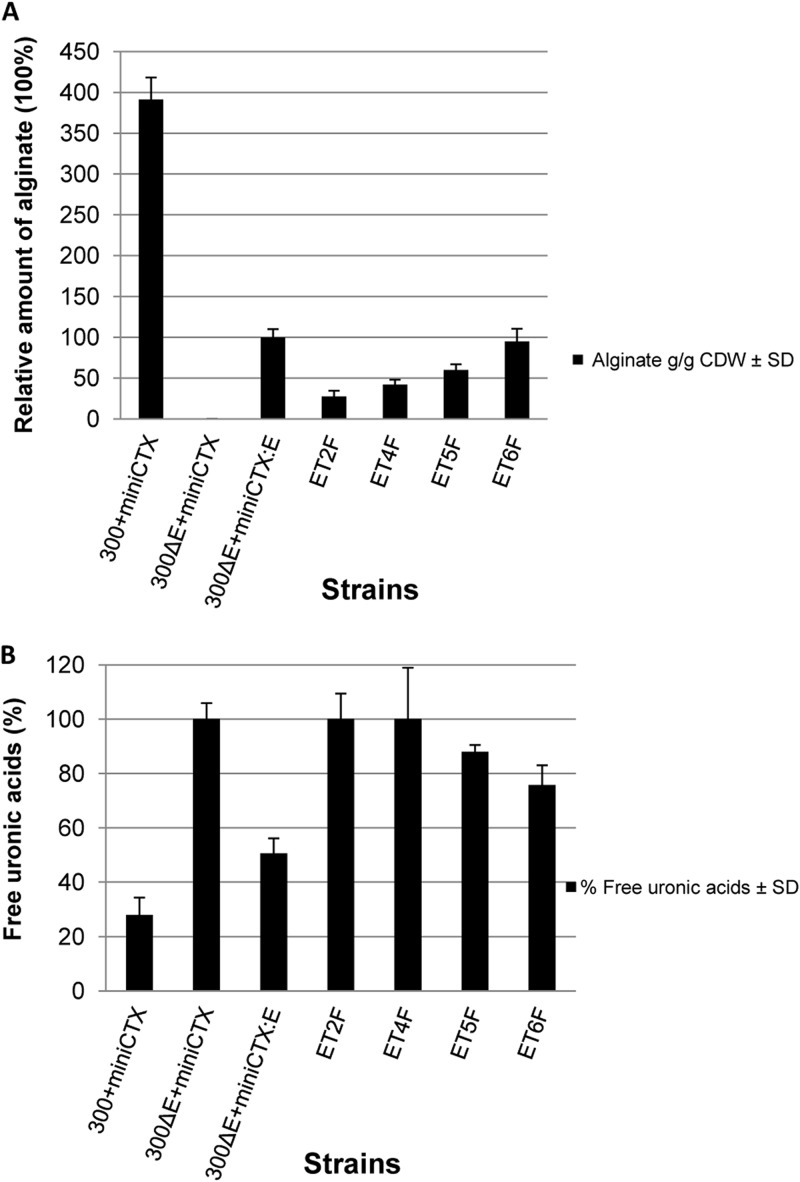
It has been proposed that the alginate biosynthesis machinery forms a supramolecular complex (10) and that AlgE interacts with AlgK (13, 24). As deletion of AlgE might affect the stability of other components of the alginate biosynthesis machinery, we integrated wild-type algE and its variants encoding FLAG insertions in periplasmic turns T2, T4, T5, and T6 into the chromosome under the control of the native algD promoter by using the mini-CTX-lacZ vector that integrates at the attB site. ET2F, ET4F, and ET5F were selected, as they produce 100% free uronic acids during planktonic growth, suggesting a role in stabilizing alginate biosynthesis complex. The ET6F variant was used as the FLAG epitope control. OMs and envelope fractions were isolated, and the OMs were subjected to immunoblotting analysis using an anti-AlgE antibody (Fig. 5). Wild-type AlgE and all the variants tested were detected in the OM, in agreement with the results we observed in our in trans complementation studies (Fig. 4A). The AlgE variant (ET2F) was present in lower copy number. As expected AlgE was absent from the OM of PDO300ΔalgE(miniCTX) (negative control) (Fig. 5). The ET2F variant was detected at wild-type levels in OMs isolated from cells grown on solid media (data not shown). Envelope fractions isolated from all the strains were probed with primary anti-AlgK, anti-AlgX, anti-AlgG, and anti-Alg44 (rabbit) antibodies. In envelope fractions isolated from PDO300ΔalgE(miniCTX), AlgK and AlgX were not detected, and the copy number of Alg44 was reduced relative to PDO300ΔalgE(miniCTX:algE) (Fig. 5). Expression of algE in trans in PDO300ΔalgE(pBBR1MCS-5:algE) restored the presence of AlgK or AlgX only when cells were isolated from solid media (data not shown). However, chromosomally integrated algE and variants (ET2F, ET5F, and ET6F) restored AlgK, AlgX, and Alg44 even in the planktonic mode of growth (Fig. 5). Interestingly, the chromosomally integrated variant of AlgE, ET4F, destabilized AlgK and AlgX, to the point that AlgK could not be detected. In this variant, the copy number of Alg44 appeared to be reduced, while AlgX was present as a lower-molecular-weight band, suggesting potentially proteolytic degradation (Fig. 5). A small reduction in the level of AlgG was observed in PDO300ΔalgE(miniCTX) and AlgE variants ET5F and ET6F compared to PDO300ΔalgE(miniCTX:algE) (Fig. 5). Chromosomal integration of algE and the T2F, T4F, T5F, and T6F variants restored alginate production to various extents and produced various amounts of free uronic acids (Fig. 6A and B).
Fig 5.
Effects of AlgE and its variants on the stability of other components of the alginate biosynthesis machinery. Genes encoding AlgE and its variants were integrated into the chromosome of P. aeruginosa PDO300ΔalgE by using the mini-CTX integration plasmid. Outer membranes and envelope fractions were isolated, and immunoblotting was performed using specific primary antibodies as indicated. OM, outer membrane; ENV, envelope fraction; SDS-ENV, protein from SDS-PAGE of envelope fraction. 300+mini-CTX indicates PDO300 carrying the integration-proficient mini-CTX plasmid; 300ΔE+miniCTX shows PDO300ΔalgE carrying the mini-CTX plasmid; 300ΔE+miniCTX:E depicts PDO300ΔalgE carrying the mini-CTX::algE plasmid. Variants of AlgE are indicated with the first letter E, indicating wild-type AlgE, followed by M for the membrane segment or T for the periplasmic turn, followed by a number to indicate the position in AlgE, as defined by the structure shown schematically in Fig. 1. The letter F is used as an abbreviation for the FLAG epitope. Constitutively expressed outer membrane protein OprF was used as a loading control.
Fig 6.
Alginate and free uronic acids produced by P. aeruginosa with chromosomal integration of genes for AlgE and its variants. (A) The amount of alginate, relative to the amount of alginate produced by PDO300ΔalgE (mini-CTX::algE). (B) The amount of free uronic acids, given as a percentage ratio between the filtrate and supernatant. Experiments were conducted in triplicate, and the error bars represent the standard deviations of the mean values. 300+miniCTX indicates PDO300 carrying the integration-proficient mini-CTX plasmid; 300ΔE+miniCTX shows PDO300ΔalgE carrying the mini-CTX plasmid; 300ΔE+miniCTX:E depicts PDO300ΔalgE carrying the mini-CTX::algE plasmid. Variants of AlgE are indicated as first letter E indicating wild-type AlgE, followed by M for the membrane segment or T for the periplasmic turn, followed by a number indicating their respective position from the N terminus of AlgE; F stands for the FLAG epitope insertion.
DISCUSSION
OM proteins are synthesized in the cytoplasm and secreted into the periplasm through Sec translocons (29). After cleavage of the N-terminal signal sequence in the periplasm, the mature proteins are sequestered by periplasmic chaperones, which transport them to the β-barrel assembly machinery (BAM), with its core component, BamA, located in the OM. The BAM, with the aid of the periplasmic chaperones, fold and insert β-barrel proteins into the OM (30). This machinery is responsible for the correct folding and insertion of AlgE in the outer membrane. The structure of AlgE revealed that the inside of its β-barrel is highly positively charged (Fig. 1B), as might be expected for the selection and/or efficient secretion of negatively charged alginate. The proposed conduit for alginate secretion is lined with the highly conserved charged residues K47, R74, R129, R152, D162, N164, R353, R362, R459, and D485, as well as the less-well-conserved residues E130, R154, E189, D193, H364, R365, E368, R461, and R481 (24). Except for R129, mutation of each of the positively charged residues resulted in a decrease in the amount of alginate produced, while the replacement of the negatively charged D485 and D162 with alanine showed an increase in alginate production (Fig. 2). Substitutions to alanine of R353, R362, H364, and R365 located on loop 7 led to reduced alginate production, suggesting an important role of this loop not only in protein stability, as previously described (16), but also in alginate secretion (24). The identification of a role for loop 7 in substrate recognition is not unique to AlgE, as other substrate-specific porins, e.g., OccD1, OccK1 (31), and LamB, have residues located on comparable channel constriction loops that define substrate specificity in these systems. To further investigate the impact of site-directed mutagenesis on the function of AlgE, double and triple variants were created. Assessment of the K47E+R353A+R459E and R74E+R362A+R459E triple mutants suggested a decrease in the net positive charge and an increase in the net negative charge inside the lumen (Fig. 1B). The increased electronegative potential might interfere with selection and/or efficient secretion of the negatively charged substrate, alginate. This was further supported by the observation that substitution of negatively charged D162 or E485 with alanine increased alginate production (Fig. 2). All of the site-specific variants of AlgE were detected in the OM (Fig. 3).
AlgE was originally described as a general diffusion protein, but recently it has been proposed to form a substrate-specific pore (24, 32). The substrate specificity is supported by the observation that the structure of AlgE superimposes with other substrate-specific porins of P. aeruginosa, such as OccD1 and OccK1, with root mean square deviations of 2.7 Å and 2.9 Å over 302 and 306 equivalent Cα positions, respectively (24). The OccD family of proteins is involved in the uptake of low-molecular-weight substrates, a highly divergent function compared to the proposed function of AlgE. The electropositive conduit (8 Å in diameter) of AlgE also suggests its specificity for alginate (24). Our data showed that changing the conduit to a more electronegative potential impacted alginate production and hence suggested that AlgE exhibits properties as a substrate-specific porin. As perhaps expected, none of the AlgE site-specific or multiple point variants showed a complete loss of functionality. This was similar to results obtained previously for the substrate-specific maltoporin LamB, where replacement of all six residues found to be involved in carbohydrate transport was required to abolish substrate binding (33). The secretion of alginate by site-specific AlgE variants is further explained by the fact that sugar-protein interactions are generally weak, and thus even in the triple mutants some alginate would still be secreted by the force of alginate polymerization.
To study the structural and functional importance of different regions of AlgE in AlgE stability, protein-protein interactions, and secretion of alginate, we inserted FLAG epitopes into transmembrane regions and periplasmic turns. Transmembrane β-strands are amphipathic in nature, and disruption of this amphipathicity by inserting a foreign epitope can affect the protein's ability to fold and insert into the OM. Unexpectedly, AlgE variant M6F was detected in the OM; however, it could not restore alginate production (Fig. 4A and B). These results suggest that the FLAG insertion at this site, 3 amino acids upstream of periplasmic turn 3 (Fig. 1A), does not affect the ability of the 9 residues upstream to form β-strand 6. In this case, the FLAG epitope and 3 residues downstream of it would be pushed into the periplasm and thus could result in a large and strongly charged periplasmic turn 3, which in turn could interfere with the assembly of the alginate polymer and the secretion complex and/or the ability of the AlgE variant to secrete alginate properly. Of the FLAG epitope insertions into the transmembrane regions, the EM10F variant is unique in that it was only detected in OMs isolated from solid medium. (Fig. 5A). This is consistent with the alginate quantification results, which were done from solid medium (Fig. 5B). It has been shown previously that alginate is overproduced by P. aeruginosa when grown in the biofilm mode (34). Growth conditions which favor alginate production might require upregulation of algE gene expression and/or more efficient assembly of the alginate biosynthesis/secretion machinery. This could have led to the presence of detectable copy numbers of the AlgE variant M10F.
The production of 100% free uronic acids by ET2F, ET4F, ET5F, and ET8F-1 during planktonic growth of cells harboring the respective plasmids, i.e., multiple copies of each gene (Fig. 4C), suggested that FLAG epitope insertions in these periplasmic turns might interfere with the ability of AlgE to interact with other proteins of the alginate biosynthesis machinery. However, previously it was shown that deletion of the large periplasmic turn, T8, resulted in the production of 50% free uronic acids when the respective AlgE variant was carried by a plasmid (24). This suggested that insertion of the FLAG epitope in turn 8 (ET8F-1) could interfere with AlgE's function by locking T8 into an open conformation or by destabilizing the neighboring turns and interfering with the ability of AlgE to interact with periplasmic scaffold proteins. This would cause destabilization of the alginate polymerization/secretion multiprotein complex, which resulted in production of 100% free uronic acid. The T8 deletion variant was not considered for integration into the chromosome, because the deletion of T8 produced 50% free uronic acids and the possibility that ET8F-1 could have destabilized a neighboring turn. Future studies should attempt to try to explain the exact function of T8 in alginate secretion and biosynthesis.
It has been proposed that hydrophobic residues at the C terminus of β-barrel porins, especially the terminal residue, play a role in targeting these proteins to the OM. For example, the C-terminal Phe of PhoE was shown to be required for efficient localization of the protein to the OM (35). AlgE has a comparable signature sequence of W-R-F at its C terminus, and thus it was perhaps not surprising that truncation of the last 9 residues, EtrC9, resulted in AlgE not successfully translocated to the OM; hence, it was not detected in the OM (Fig. 4A, B, and C).
The alginate biosynthesis machinery is proposed to span the entire envelope fraction and to form a multiprotein complex. This multiprotein complex is proposed to guide the alginate through the periplasm to AlgE. This assumes that components of this machinery are interacting with each other at least when alginate is produced. Indeed, it has been proposed that AlgK, which contains at least 9.5 tetratricopeptide-like protein-protein interaction motifs, interacts with AlgE (13, 24). Previous experimental data have suggested an interaction between AlgK with AlgX, and of AlgX and the periplasmic serine protease, MucD, a negative regulator of alginate biosynthesis (14, 15). Our assessment of the impact of AlgE and its FLAG insertion variants on the stability of other components of the alginate biosynthesis machinery suggests that AlgE interacts with AlgK and/or AlgX (Fig. 5). As an OM lipoprotein, AlgK is likely to be in close proximity to AlgE, and since deletion of AlgK caused mislocalization of AlgE, the data presented here propose that AlgK and AlgE directly interact with each other (13). This is further supported by the recent finding that the functional homologues of AlgE and AlgK, HmsH and HmsF of Yersinia pestis, which are required for the secretion of poly-β-1,6-N-acetyl-d-glucosamine (PGA) were found to interact (36). Destabilization of the periplasmic component AlgX could be an indirect consequence of destabilization of AlgK, which interacts with AlgX. It is unlikely that AlgE interacts directly with Alg44, the proposed copolymerase, because for Alg44 to directly interact with AlgE it would require Alg44 to have a large periplasmic domain that is capable of spanning the entire periplasmic region (∼200 Å) (37). Such a large domain cannot be deduced from bioinformatics analysis of Alg44. Based on the homology models of Alg44, it thus seems more likely that Alg44 interacts with AlgX and/or AlgK. These results support the hypothesis that a multiprotein complex that spans the entire envelope region is required for alginate secretion (10). Polymerization of alginate has been previously shown to require components present in both the inner and outer membranes (10).
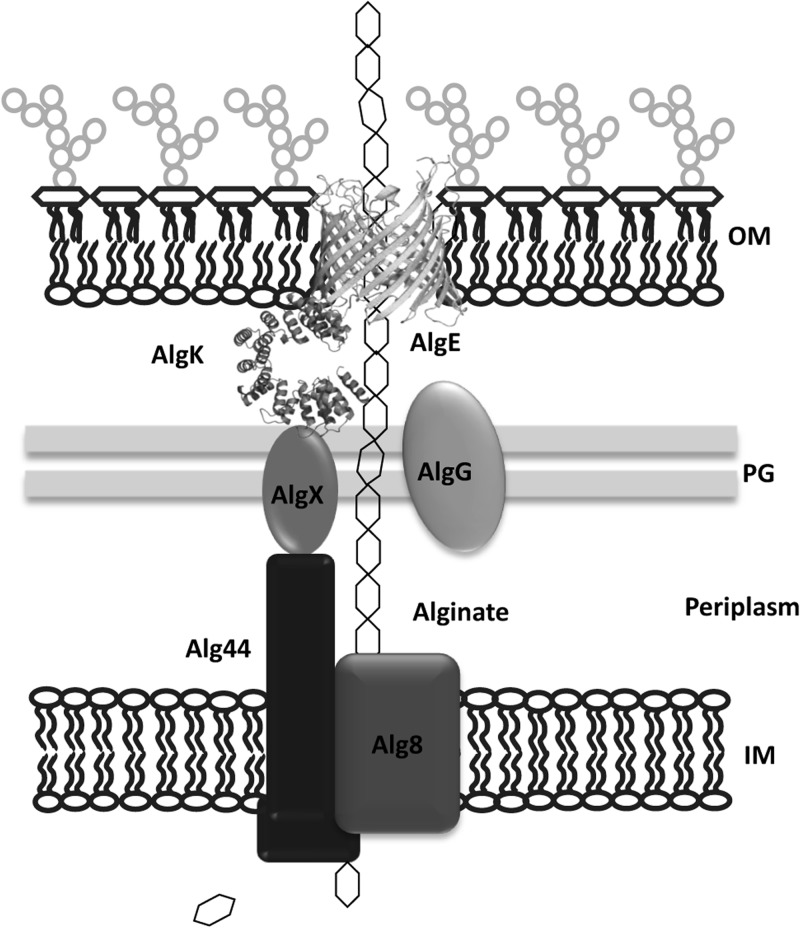
Mutual stability analysis further showed that periplasmic turns T5 and T6 can play a role in stabilizing AlgG through either a direct or indirect interaction (Fig. 5). The results for AlgE variant ET4F, like that observed for the EM10F variant, suggest a difference in expression of the alginate operon or stability of respective alginate biosynthesis protein when bacteria are grown in liquid culture and solid media, as this variant produced ∼45% full-length alginate relative to the wild-type protein when alginate production was analyzed in solid medium (Fig. 6A). This level of alginate production would not be expected given the complete destabilization of AlgK and the production of 100% uronic acids in an algK deletion mutant (17). Interestingly, AlgE variant ET4F did not affect the stability of AlgK, AlgX, and Alg44 when cells were harvested from solid medium (unpublished data). However, our results presented here are consistent with free uronic acids analysis performed from liquid culture. The T4 turn is the smallest of all the periplasmic turns (with only 2 amino acids protruding into the periplasm) tested in this study (Fig. 1A), and while there is a discrepancy between the solid and liquid media results, they do suggest that any protein possibly interacting with this turn should localize close to the OM, a criterion that could be fulfilled by AlgK. Based on the findings in this study, a model has been generated that shows the potential involvement of AlgE in stabilizing members of the periplasmic scaffold via specific protein-protein interactions (Fig. 7). Future and ongoing studies are aimed at providing experimental evidence for the existence of a multiprotein complex by identifying direct protein-protein interactions.
Fig 7.
Model of proposed AlgE interactions based on our mutual stability results. The protein-protein interactions depicted above are based on mutual stability analysis using AlgE and its variants, along with results from I. D. Hay, et al. (14). The analysis of the impacts of FLAG tag variants of AlgE on the mutual stability of other components of the alginate biosynthesis machinery suggested that AlgK may interact with periplasmic turns 4 and 8 of AlgE, although we cannot rule out the involvement of other periplasmic turns. Alginate is represented as a chain of hexagons. OM, outer membrane; IM, inner membrane; PG, peptidoglycan.
ACKNOWLEDGMENTS
This work is supported by grants from Massey University Research Fund to B.H.A.R. Z.U.R. is supported by a doctoral scholarship and research grant from Higher Education Commission Pakistan.
We thank John C. Whitney and P. Lynne Howell (Toronto University Canada) for assistance in designing a site-specific mutagenesis approach and for providing antibodies. The provision of specific antibodies against alginate biosynthesis proteins by Dennis E. Ohman (Virginia Commonwealth University) is gratefully acknowledged.
Footnotes
Published ahead of print 18 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03960-12.
REFERENCES
- 1. Hay ID, Rehman ZU, Ghafoor A, Rehm BHA. 2010. Bacterial biosynthesis of alginates. J. Chem. Technol. Biotechnol. 85:752–759 [Google Scholar]
- 2. Ghafoor A, Hay ID, Rehm BH. 2011. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 77:5238–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pier GB, Coleman F, Grout M, Franklin M, Ohman DE. 2001. Role of alginate O acetylation in resistance of mucoid Pseudomonas aeruginosa to opsonic phagocytosis. Infect. Immun. 69:1895–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chitnis CE, Ohman DE. 1993. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol. Microbiol. 8:583–593 [DOI] [PubMed] [Google Scholar]
- 5. Franklin MJ, Nivens DE, Weadge JT, Howell PL. 2011. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel, and Psl. Front. Microbiol. 2:167 doi:10.3389/fmicb.2011.00167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rehm BH. 2010. Bacterial polymers: biosynthesis, modifications and applications. Nat. Rev. Microbiol. 8:578–592 [DOI] [PubMed] [Google Scholar]
- 7. Olvera C, Goldberg JB, Sanchez R, Soberon-Chavez G. 1999. The Pseudomonas aeruginosa algC gene product participates in rhamnolipid biosynthesis. FEMS Microbiol. Lett. 179:85–90 [DOI] [PubMed] [Google Scholar]
- 8. Ma L, Wang J, Wang S, Anderson EM, Lam JS, Parsek MR, Wozniak DJ. 2012. Synthesis of multiple Pseudomonas aeruginosa biofilm matrix exopolysaccharides is post-transcriptionally regulated. Environ. Microbiol. 14:1995–2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Remminghorst U, Rehm BH. 2006. Alg44, a unique protein required for alginate biosynthesis in Pseudomonas aeruginosa. FEBS Lett. 580:3883–3888 [DOI] [PubMed] [Google Scholar]
- 10. Remminghorst U, Rehm BH. 2006. In vitro alginate polymerization and the functional role of Alg8 in alginate production by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 72:298–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65:876–895 [DOI] [PubMed] [Google Scholar]
- 12. Oglesby LL, Jain S, Ohman DE. 2008. Membrane topology and roles of Pseudomonas aeruginosa Alg8 and Alg44 in alginate polymerization. Microbiology 154:1605–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Keiski CL, Harwich M, Jain S, Neculai AM, Yip P, Robinson H, Whitney JC, Riley L, Burrows LL, Ohman DE, Howell PL. 2010. AlgK is a TPR-containing protein and the periplasmic component of a novel exopolysaccharide secretin. Structure 18:265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hay ID, Schmidt O, Filitcheva J, Rehm BH. 2012. Identification of a periplasmic AlgK-AlgX-MucD multiprotein complex in Pseudomonas aeruginosa involved in biosynthesis and regulation of alginate. Appl. Microbiol. Biotechnol. 93:215–227 [DOI] [PubMed] [Google Scholar]
- 15. Gutsche J, Remminghorst U, Rehm BH. 2006. Biochemical analysis of alginate biosynthesis protein AlgX from Pseudomonas aeruginosa: purification of an AlgX-MucD (AlgY) protein complex. Biochimie 88:245–251 [DOI] [PubMed] [Google Scholar]
- 16. Hay ID, Rehman ZU, Rehm BH. 2010. Membrane topology of outer membrane protein AlgE, which is required for alginate production in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 76:1806–1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jain S, Ohman DE. 1998. Deletion of algK in mucoid Pseudomonas aeruginosa blocks alginate polymer formation and results in uronic acid secretion. J. Bacteriol. 180:634–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Monday SR, Schiller NL. 1996. Alginate synthesis in Pseudomonas aeruginosa: the role of AlgL (alginate lyase) and AlgX. J. Bacteriol. 178:625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Robles-Price A, Wong TY, Sletta H, Valla S, Schiller NL. 2004. AlgX is a periplasmic protein required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 186:7369–7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Albrecht MT, Schiller NL. 2005. Alginate lyase (AlgL) activity is required for alginate biosynthesis in Pseudomonas aeruginosa. J. Bacteriol. 187:3869–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jain S, Ohman DE. 2005. Role of an alginate lyase for alginate transport in mucoid Pseudomonas aeruginosa. Infect. Immun. 73:6429–6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Franklin MJ, Chitnis CE, Gacesa P, Sonesson A, White DC, Ohman DE. 1994. Pseudomonas aeruginosa AlgG is a polymer level alginate C5-mannuronan epimerase. J. Bacteriol. 176:1821–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Franklin MJ, Ohman DE. 2002. Mutant analysis and cellular localization of the AlgI, AlgJ, and AlgF proteins required for O acetylation of alginate in Pseudomonas aeruginosa. J. Bacteriol. 184:3000–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Whitney JC, Hay ID, Li C, Eckford PD, Robinson H, Amaya MF, Wood LF, Ohman DE, Bear CE, Rehm BH, Howell PL. 2011. Structural basis for alginate secretion across the bacterial outer membrane. Proc. Natl. Acad. Sci. U. S. A. 108:13083–13088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chiu J, Tillett D, Dawes IW, March PE. 2008. Site-directed, ligase-independent mutagenesis (SLIM) for highly efficient mutagenesis of plasmids greater than 8kb. J. Microbiol. Methods 73:195–198 [DOI] [PubMed] [Google Scholar]
- 26. Blumenkrantz N, Asboe-Hansen G. 1973. New method for quantitative determination of uronic acids. Anal. Biochem. 54:484–489 [DOI] [PubMed] [Google Scholar]
- 27. Hoang TT, Kutchma AJ, Becher A, Schweizer HP. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59–72 [DOI] [PubMed] [Google Scholar]
- 28. Schlegel HG, Kaltwasser H, Gottschalk G. 1961. A submersion method for culture of hydrogen-oxidizing bacteria: growth physiological studies. Arch. Mikrobiol. 38:209–222 [PubMed] [Google Scholar]
- 29. du Plessis DJ, Nouwen N, Driessen AJ. 2011. The Sec translocase. Biochim. Biophys. Acta 1808:851–865 [DOI] [PubMed] [Google Scholar]
- 30. Knowles TJ, Scott-Tucker A, Overduin M, Henderson IR. 2009. Membrane protein architects: the role of the BAM complex in outer membrane protein assembly. Nat. Rev. Microbiol. 7:206–214 [DOI] [PubMed] [Google Scholar]
- 31. Eren E, Vijayaraghavan J, Liu J, Cheneke BR, Touw DS, Lepore BW, Indic M, Movileanu L, van den Berg B. 2012. Substrate specificity within a family of outer membrane carboxylate channels. PLoS Biol. 10:e1001242 doi:10.1371/journal.pbio.1001242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hancock RE, Brinkman FS. 2002. Function of Pseudomonas porins in uptake and efflux. Annu. Rev. Microbiol. 56:17–38 [DOI] [PubMed] [Google Scholar]
- 33. Denker K, Orlik F, Schiffler B, Benz R. 2005. Site-directed mutagenesis of the greasy slide aromatic residues within the LamB (maltoporin) channel of Escherichia coli: effect on ion and maltopentaose transport. J. Mol. Biol. 352:534–550 [DOI] [PubMed] [Google Scholar]
- 34. Hentzer M, Teitzel GM, Balzer GJ, Heydorn A, Molin S, Givskov M, Parsek MR. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Struyve M, Moons M, Tommassen J. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141–148 [DOI] [PubMed] [Google Scholar]
- 36. Abu Khweek A, Fetherston JD, Perry RD. 2010. Analysis of HmsH and its role in plague biofilm formation. Microbiology 156:1424–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cuthbertson L, Mainprize IL, Naismith JH, Whitfield C. 2009. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in Gram-negative bacteria. Microbiol. Mol. Biol. Rev. 73:155–177 [DOI] [PMC free article] [PubMed] [Google Scholar]