Abstract
Bacteria often use pheromones to coordinate group behaviors in specific environments. While high cell density is required for pheromones to achieve stimulatory levels, environmental cues can also influence pheromone accumulation and signaling. For the squid symbiont Vibrio fischeri ES114, bioluminescence requires pheromone-mediated regulation, and this signaling is induced in the host to a greater extent than in culture, even at an equivalent cell density. Our goal is to better understand this environment-specific control over pheromone signaling and bioluminescence. Previous work with V. fischeri MJ1 showed that iron limitation induces luminescence, and we recently found that ES114 encounters a low-iron environment in its host. Here we show that ES114 induces luminescence at lower cell density and achieves brighter luminescence in low-iron media. This iron-dependent effect on luminescence required ferric uptake regulator (Fur), which we propose influences two pheromone signaling master regulators, LitR and LuxR. Genetic and bioinformatic analyses suggested that under low-iron conditions, Fur-mediated repression of litR is relieved, enabling more LitR to perform its established role as an activator of luxR. Interestingly, Fur may similarly control the LitR homolog SmcR of Vibrio vulnificus. These results reveal an intriguing regulatory link between low-iron conditions, which are often encountered in host tissues, and pheromone-dependent master regulators.
INTRODUCTION
Many bacteria transmit diffusible pheromone signals within and between species to coordinate group functions such as biofilm formation, antibiotic production, and infection. Such signaling is widespread among diverse bacteria (1–3), and it is especially common and well studied among the Proteobacteria, which use various signals, including acyl-homoserine lactones (4, 5). The accumulation of pheromones to stimulatory levels often depends on high cell densities, giving rise to the term “quorum sensing” to describe such behavior (6); however, environmentally responsive regulators control both the synthesis of pheromones and responsiveness to these signals, rendering such signaling dependent on environmental context as well as cell density (7–9). We have sought to elucidate this interconnection of environment-specific regulation and pheromone signaling in the model symbiont Vibrio fischeri.
V. fischeri is a bioluminescent gammaproteobacterium that monospecifically colonizes the light organ of the Hawaiian bobtail squid, Euprymna scolopes (10, 11). Bioluminescence is a colonization factor for V. fischeri (12, 13), and it is regulated in part by LuxR-LuxI pheromone-mediated regulation, as described below (14). This highly tractable symbiosis serves as a model system for studying host-microbe interactions and how bacterial pheromone-mediated gene regulation functions during a natural infection (15).
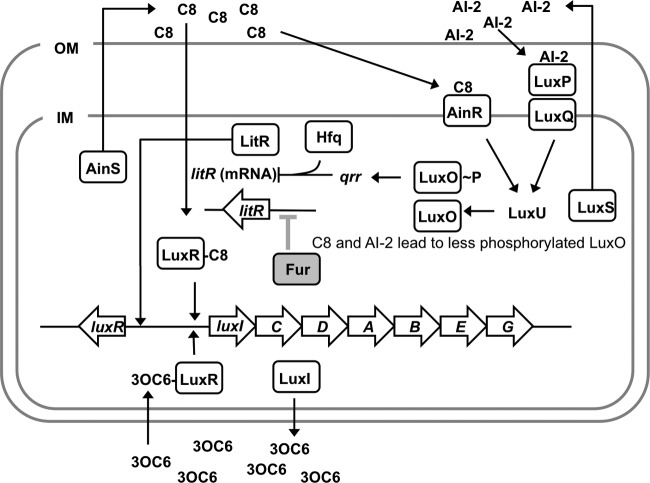
In V. fischeri, the luxCDABEG genes underlie bioluminescence and are downstream of luxI in the lux operon (Fig. 1). LuxI produces the pheromone autoinducer N-(3-oxo-hexanoyl)-l-homoserine lactone (3OC6) (16), which can combine with LuxR to activate expression of the lux operon (17–19). Bioluminescence in V. fischeri is influenced by two additional autoinducers; octanoyl-homoserine lactone (C8) (20, 21) and the product of LuxS (22), which is called autoinducer-2 (AI-2). Figure 1 illustrates a current model of the interconnected signaling cascades of 3OC6, C8, and AI-2, based on homology to other systems and studies of V. fischeri (20, 22–28).
Fig 1.
Model of pheromone-mediated regulation of the lux operon in V. fischeri ES114. Large block arrows correspond to genes including luxR (encoding a pheromone-dependent transcriptional regulator [VF_A0925]), luxI (encoding an acyl-homoserine lactone synthase [VF_A0924]), genes for bioluminescence (luxCDABEG [VF_A0918 to VF_A0923]), and litR (encoding another pheromone-controlled regulator [VF_2177]). 3OC6 and, to a lesser extent, C8 bind LuxR and enable it to stimulate transcription of the lux operon (among other genes). C8 and AI-2 are thought to be detected by AinR and LuxP/LuxQ, respectively. When C8 and AI-2 levels are elevated, AinR and LuxQ initiate a regulatory cascade via LuxU, resulting in less phosphorylation of LuxO. Phosphorylated LuxO (LuxO-P) increases transcription levels of the regulatory RNA Qrr, which, together with Hfq, represses expression of LitR. LitR activates transcription of LuxR, among other genes. Thus, C8 and AI-2 lead to increased levels of LitR in a pheromone signaling circuit conserved in many Vibrio species. This model is derived from experimental data, genomic predictions, and work with related bacterial species (see the text) (reviewed in reference 44). The putative role of Fur in the regulatory circuit, as described in this paper, is highlighted in gray. OM, outer membrane; IM, inner membrane. (Reprinted from reference 37.)
In V. fischeri, LuxR and LitR are considered pheromone-dependent master regulators. Based on the current model (Fig. 1), LuxR activates transcription of the lux operon and other genes in response to 3OC6 and, to a lesser extent, in response to C8. LitR levels are enhanced by elevated levels of C8 or AI-2, and LitR activates transcription of luxR and other genes (29). V. fischeri LuxR-type regulators are absent from most Vibrio species, but LitR is a homolog of the quorum-sensing master regulators in Vibrio cholerae (30), Vibrio parahaemolyticus (31), Vibrio vulnificus (32), and Vibrio harveyi (33).
The influence of environmental context on pheromone-dependent regulation is dramatically evident in V. fischeri ES114, a strain typical of isolates from the E. scolopes light organ. Even at similar high cell densities, ES114 cells produce less 3OC6 and are ∼1,000 times dimmer in culture than in the host (34). Moreover, lux expression appears heterogeneous in different light organ microenvironments (35). Recent work has identified several regulatory inputs controlling ES114's pheromone signal systems (36–38). Others demonstrated a link between low iron levels and increased luminescence in strain MJ1 (39). In transgenic Escherichia coli, the ferric uptake regulator (Fur) did not directly control the luxR-luxICDABEG locus (40); however, this experimental setup would not have accounted for regulation through LitR or other regulators that are absent from E. coli.
Low iron levels are often faced by symbiotic bacteria in host tissues and have been implicated in the V. fischeri-squid symbiosis (41, 42). For example, we recently demonstrated that the heme uptake system in V. fischeri ES114 is repressed by Fur but is induced under low-iron conditions and during symbiotic colonization (42). We therefore investigated the possible connection between iron and luminescence regulation in V. fischeri ES114. Strain ES114 is significantly different from MJ1 (43, 44), which was isolated from a fish, and the response of ES114 to iron is not well understood. Here we describe how changes in iron levels influence luminescence through Fur-mediated regulation of LitR.
MATERIALS AND METHODS
Media and growth conditions.
V. fischeri strains were grown in either LBS medium (45), ASWT medium (42), or SWTO medium (36) at 28°C or 24°C. E. coli strains were grown in either LB medium (46) or brain heart infusion broth (Difco) at 37°C. Antibiotic selection for V. fischeri and E. coli strains was performed as described previously (47). Plasmids were maintained in E. coli strain DH5α (48), except for plasmids with the R6Kγ origin of replication, which were maintained in strain DH5αλpir (47) or in strain CC118λpir (49), in the case of plasmid pEVS104 (50). As a chelator, ethylenediamine-N,N′-diacetic acid (EDDA) or 2,2′-bipyridyl was added to V. fischeri cultures at a final concentration of 1 μM or 100 μM, respectively, with the latter added from a stock solution prepared at 100 mM in dimethyl sulfoxide (DMSO).
Strain and plasmid construction.
Bacterial strains, plasmids, and oligonucleotides used in this study are presented in Table 1. For constructing V. fischeri mutants, plasmids bearing mutant alleles were mobilized into V. fischeri by triparental mating using CC118λpir pEVS104 as a conjugative helper. Transconjugants were selected with appropriate antibiotics and screened for allelic exchange using PCR and antibiotic resistance markers. To construct the ΔryhB mutant, the sequence upstream of ryhB was PCR amplified by using primers prNL66 and prNL67.2 and cloned into pCR-Blunt II-TOPO, resulting in plasmid pNL35. The sequence downstream of ryhB was PCR amplified by using primers prNL68 and prNL69 and cloned into SmaI-digested pEVS122, resulting in plasmid pNL36. AvrII-digested pNL35 was ligated into AvrII-digested pNL36, resulting in plasmid pNL49. The ΔryhB allele on plasmid pNL49 was exchanged into wild-type V. fischeri ES114, resulting in strain NL58. To construct the Δfur litR double mutant, the litR::erm allele on plasmid pJLB96 was exchanged into V. fischeri Δfur strain YLM111, resulting in strain ANS63. To construct the V. fischeri ES114 litR promoter-reporter plasmid, a region containing 374 bp upstream of the ATG start codon and 71 bp into the coding region of litR in V. fischeri ES114 was PCR amplified with primers ASlitRP2 and ASlitRP3. This product was digested with SphI and NheI and then cloned into the same restriction sites of pAKD701 to generate the PlitR-lacZ transcriptional reporter plasmid pAS120. To construct the V. vulnificus C7184 smcR promoter reporter plasmid, a region containing 391 bp upstream of the ATG start codon and 19 bp into the coding region of smcR in V. vulnificus C7184 was PCR amplified with primers ASvv1634P1 and ASvv1634P2. This product was digested with SphI and NheI and then cloned into the same restriction sites of pAKD701 to generate the PsmcR-lacZ transcriptional reporter plasmid pAS123. To construct the V. cholerae hapR promoter reporter plasmid, a region containing 295 bp upstream of the ATG start codon and 24 bp into the coding region of hapR in V. cholerae CB98-41 was PCR amplified with primers ASvcA0115P1 and ASvcA0115P2. This product was digested with SphI and NheI and then cloned into the same restriction sites of pAKD701 to generate the PhapR-lacZ transcriptional reporter plasmid pAS128.
Table 1.
Strains, plasmids, and oligonucleotides used in this work
| Strain, plasmid, or oligonucleotide | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F′ endA1 hsdR17 glnV44 thi-1 recA1 gyrA96 (Nxr) relA1 Δ(lacIZYA-argF)U169 deoR[ϕ80dlacIΔ(lacZ)M15] | 48 |
| DH5αλpir | λpir derivative of DH5α | 47 |
| CC118λpir | Δ(ara-leu) araD Δlac74 galE galK phoA20 thi-1 rpsE rpsB argE(Am) recA λpir | 49 |
| Vibrio vulnificus C7184 | Wild-type strain | Brett Macey |
| Vibrio cholerae CB98-41 | Wild-type strain | Christopher J. Grim |
| Vibrio fischeri | ||
| ANS63 | ES114 litR::erm Δfur | This study |
| ES114 | Wild-type isolate from E. scolopes light organ | 51 |
| JB13 | ES114 luxO::erm | 36 |
| JB19 | ES114 litR::erm | 36 |
| JB22 | ES114 lacIq PA1/34-luxCDABEG | 12 |
| NL58 | ES114 ΔryhB | this study |
| YLM111 | ES114 Δfur | 42 |
| Plasmids | ||
| pAKD701 | Promoterless lacZ; oriV R6Kγ oriVpES213 oriT Knr | 53 |
| pAKD702 | Promoterless lacZ; oriVR6Kγ oriVpES213 oriT Cmr | 43 |
| pAKD912 | pAKD701 containing the ES114 VF_1225 promoter region; oriVR6Kγ oriVpES213 oriT Knr | 42 |
| pAS120 | pAKD701 containing the ES114 litR promoter region; oriVR6Kγ oriVpES213 oriT Knr | This study |
| pAS123 | pAKD701 containing the V. vulnificus C7184 smcR promoter region; oriVR6Kγ oriVpES213 oriT Knr | This study |
| pAS128 | pAKD701 containing the V. cholerae CB98-41 hapR promoter region; oriVR6Kγ oriVpES213 oriT Knr | This study |
| pEVS104 | Conjugative helper; oriVR6Kγ oriT Knr | 50 |
| pEVS122 | oriVR6Kγ oriT Ermr | 47 |
| pJLB96 | litR::erm allele; oriVColE1 oriT Ermr Cmr | 36 |
| pJLB170 | pAKD702 containing the ES114 luxR promoter region; oriVR6Kγ oriVpES213 oriT Cmr | 43 |
| pNL49 | ΔryhB allele; oriVR6Kγ oriVColE1 oriT Ermr Knr | This study |
| Oligonucleotidesb | ||
| prNL66 | GGCGGTAATGCTGCCTGTTGCCCAAGGCATAAA | This study |
| prNL67.2 | GGCCCCTAGGAAATAGTGCGGATAACTCCGTGTGCGTATTCCCT | This study |
| prNL68 | GGCCCCTAGGAGCAGTGGTGGACGTACAAACGTATTACCA | This study |
| prNL69 | CCAATAAGGTTCGCCACCATGTAATCTAAACTATCGGTTTC | This study |
| ASlitRP2 | TAGCTAGCATATCAAGTAATTGTTCTTTGC | This study |
| ASlitRP3 | TAGCATGCACTATCTCACTTATTCGTTG | This study |
| ASvv1634P1 | TAGCATGCACTGTACTCAATGTTTTATAGTTGC | This study |
| ASvv1634P2 | TAGCTAGCTCTTTGCGATTGAGTCCATAG | This study |
| ASvcA0115P1 | TAGCATGCACCATTCTCGTTGTGTTGG | This study |
| ASvcA0115P2 | TAGCTAGCGCGTTTTTCGATTGATGCG | This study |
Knr, kanamycin resistance; Cmr and cat, chloramphenicol resistance; Ermr and erm, erythromycin resistance; Nxr, nalidixic acid resistance. Plasmid replication origins are designated oriV with a subscript, indicating the source, and oriT indicates the RP4 origin of transfer.
Oligonucleotides are in the 5′-to-3′ orientation, with introduced restriction sites underlined.
Luminescence assays.
To assay luminescence, V. fischeri cultures were grown overnight in LBS medium and diluted 1:1,000 into either 25 ml SWTO medium in 125-ml flasks or 50 ml SWTO medium in 250-ml flasks. Media were supplemented with 43 μM or 2 mM FeSO4 or with 20 mM trisodium citrate, as indicated. Cultures were incubated at 24°C with shaking at 200 rpm. At the indicated time points, 0.5-ml samples were removed, and the cell density was measured at a 595-nm wavelength (optical density at 595 nm [OD595]), using a BioPhotometer (Brinkman Instruments, Westbury, NY). The cuvette was then shaken to aerate the sample, and luminescence was measured by using a Glomax 20/20 luminometer (Promega, Madison, WI) with a 10-s integration setting. Luminescence values were normalized to the OD595.
β-Galactosidase assays.
V. fischeri strains harboring lacZ-based transcriptional reporter plasmids were grown as described above for luminescence assays. For strains containing reporter plasmids pAKD912 and pJLB170, cells were harvested at an OD595 of ∼1.0, while strains harboring reporter plasmid pAS120 (PlitR), pAS123 (PsmcR), or pAS128 (PhapR) were harvested at an OD595 of ∼0.5. Cells were collected by centrifugation, and the supernatant was discarded. Cell pellets were frozen at −20°C overnight, and β-galactosidase assays were performed to determine Miller units, as described previously (12). All β-galactosidase assays were performed with V. fischeri.
Nucleotide sequence accession numbers.
Sequences for the fragments upstream of smcR and hapR were deposited in GenBank under accession numbers JX519291 and JX519292, respectively.
RESULTS
Iron limitation affects luminescence in V. fischeri ES114.
To manipulate the iron available to V. fischeri ES114, we supplemented the medium with a chelator and/or ferrous sulfate. In medium supplemented with 20 mM trisodium citrate as an iron chelator, ES114 induced luminescence at a lower OD595 and displayed an approximately 3- to 4-fold increase in peak luminescence (Fig. 2A). To test whether this effect on luminescence was the result of sodium ions or their influence on osmolarity (54), 60 mM NaCl was added to cultures, which had no effect on growth or luminescence under these conditions (data not shown).
Fig 2.

Effect of citrate on luminescence. Shown is luminescence as a function of cell density (A) or peak luminescence per OD595 (B) for wild-type (WT) ES114 cultures grown in aerobic shake flasks in SWTO medium supplemented with 43 μM FeSO4 without further additions or supplemented with 20 mM citrate or with 20 mM citrate and 2 mM additional FeSO4. In panel B, lowercase letters shared between bars indicate no statistically significant difference (P > 0.9), whereas different letters indicate a significant difference (P < 0.001), based on a one-way analysis of variance and Tukey's honestly significant difference test. Data are representative of at least three independent experiments. Error bars (some too small to visualize) indicate standard deviations (n = 3) for the one experiment shown in each panel.
Supplementing the medium with an alternative iron chelator, either EDDA or 2,2′-bipyridyl, also resulted in earlier luminescence by ES114 (data not shown). However, addition of 2,2′-bipyridyl or EDDA inhibited ES114 growth, possibly owing to these chelators' reported cell permeability (55, 56), and we found it difficult to reproducibly limit iron without restricting growth severely. In addition to acting as a chelator, citrate can also be used as a carbon source by ES114; however, we found that citrate had similar effects on luminescence in a citrate synthase and aconitase double mutant that cannot metabolize citrate (data not shown), suggesting that the citrate addition was a useful nontoxic approach to manipulate availability of extracellular iron for luminescence assays.
To test further whether the effect of citrate on luminescence was due to iron limitation, we added iron to the medium along with citrate. The brighter luminescence of wild-type cultures supplemented with citrate as a chelator was reversed by additional supplementation with 2 mM FeSO4 (Fig. 2B). These data suggest that 20 mM citrate leads to an increase in luminescence in ES114 as a result of citrate's chelating effect lowering iron availability.
Citrate supplementation causes derepression of the Fur-regulated heme uptake system.
Previous studies of members of the Vibrionaceae found that Fur mediates many responses to iron limitation (57–59). Typically, under iron-replete conditions, coordination of one Fe2+ to each Fur monomer allows dimerized Fur to bind DNA at a “Fur box” and repress transcription, while low-iron conditions result in derepression of Fur-regulated genes (60). To test whether the addition of 20 mM exogenous citrate causes derepression of the Fur regulon, we assayed expression of the Fur-repressed heme uptake gene cluster promoter using the lacZ transcriptional reporter on plasmid pAKD912. This transcriptional reporter was previously shown to have elevated activity under low-iron conditions in a Fur-dependent manner (42). This reporter showed greater activity in the Δfur mutant than in the wild type, and, as we predicted, neither citrate nor iron supplementation affected the reporter in the Δfur background (Fig. 3). In contrast, in the wild-type background, the reporter was derepressed in medium containing citrate, and this elevated expression level was reversed by supplementation with 2 mM iron (Fig. 3). These data indicate that supplementing the medium with citrate results in derepression of Fur-regulated transcripts, such as those encoding the heme uptake system.
Fig 3.
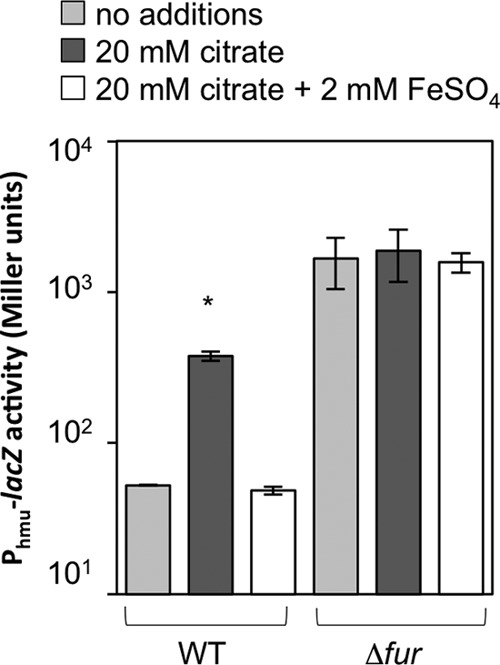
Citrate addition affects expression of a Fur-regulated reporter. Shown is β-galactosidase reporter activity for wild-type and Δfur mutant V. fischeri strains with the Fur-dependent PVF_1225-lacZ reporter plasmid pAKD912 grown in SWTO medium supplemented with 43 μM FeSO4 without further additions or supplemented with 20 mM citrate or with 20 mM citrate and 2 mM additional FeSO4. Cells were harvested at an OD595 of ∼1.0. The asterisk indicates a significant difference from other medium conditions within a strain (P < 0.001), based on an analysis of variance and Tukey's honestly significant difference test. Data are representative of at least three independent experiments. Error bars indicate standard deviations (n = 2).
The effect of iron limitation on luminescence is largely Fur dependent.
Given the results described above and the prominent role of Fur in other members of the Vibrionaceae, we hypothesized that Fur may modulate luminescence in response to iron levels, repressing luminescence when cells are iron replete. Consistent with our hypothesis, Δfur mutant cultures showed enhanced induction of luminescence at a low OD595 (less than 1.0), similar to that observed for wild-type cultures supplemented with citrate (Fig. 4A). The luminescence of Δfur mutant cultures did not attain the same maximal luminescence level as that of the wild-type cultures at a higher OD595; however, citrate had little effect on luminescence in the Δfur mutant at any cell density (Fig. 4A). Citrate did not alter the timing of luminescence induction in the Δfur mutant (Fig. 4A), and addition of 2 mM FeSO4 to citrate-supplemented Δfur mutant cultures did not affect luminescence (Fig. 4B).
Fig 4.
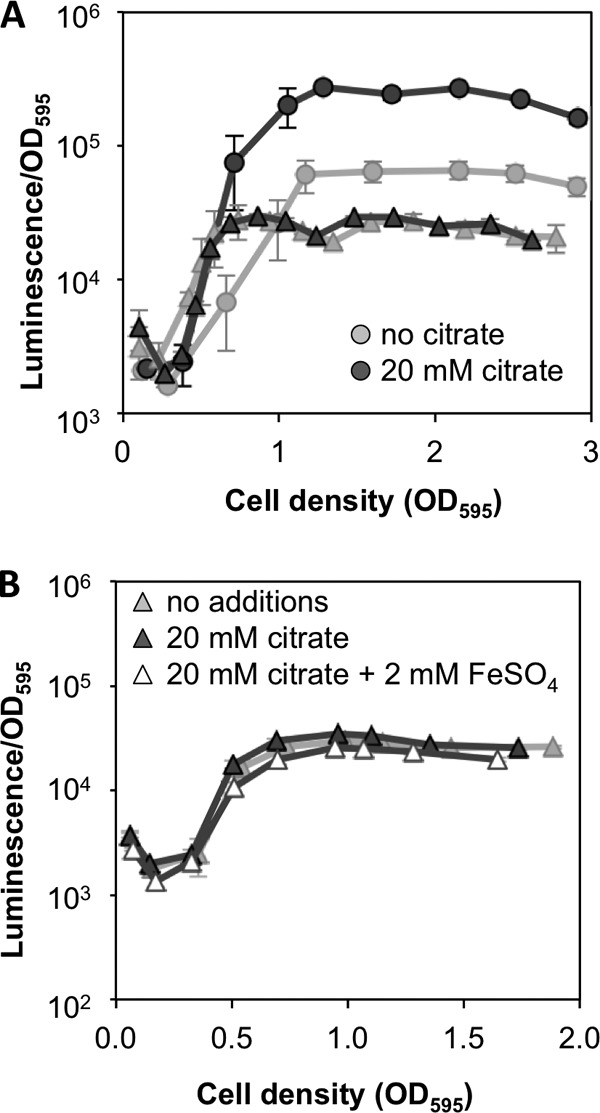
Role of Fur in the response of luminescence to low iron. (A) Cultures of wild-type ES114 (circles) or the Δfur mutant YLM111 (triangles) were grown in aerobic shake flasks in SWTO medium supplemented with 43 μM FeSO4 and either 20 mM citrate or no citrate. Error bars indicate standard deviations (n = 2). (B) Cultures of Δfur mutant strain YLM111 were grown in SWTO medium supplemented with 43 μM FeSO4 without further additions or supplemented with 20 mM citrate or with 20 mM citrate and 2 mM additional FeSO4. Error bars (some too small to visualize) indicate standard deviations (n = 3). The data in each panel are from one experiment representative of at least three independent experiments.
In 8 out of 11 experiments, we observed a small (8 to 29%) but statistically significant (P < 0.05) increase in peak luminescence for the Δfur mutant in the presence of citrate. The magnitude of this difference is so small that it may not be apparent on the log-scale y axes of Fig. 4 and 5, despite statistical significance. Thus, taken together, our data suggest the likely possibility of a fur-independent effect of citrate on luminescence. Importantly, however, such a <30% fur-independent effect of citrate on luminescence would appear too small to account for the >300% effect in the wild type. Moreover, the results of adding iron suggest that any small increase in luminescence observed for citrate-supplemented Δfur mutant cultures is not an iron-mediated effect. Taken together, the data described above indicate that decreased iron availability induces brighter luminescence in ES114 when iron chelators are added to cultures and that this response requires Fur-mediated regulation.
Fig 5.

Regulatory determinants of luminescence induction in response to low iron. V. fischeri cultures were grown in aerobic shake flasks in SWTO medium supplemented with 43 μM FeSO4 either without or with 20 mM citrate. Error bars indicate standard deviations (n = 3). Asterisks indicate a significant effect of citrate addition on luminescence of the strain (P < 0.03), as determined by using Student's t test, while the dagger indicates variable statistical significance (P value ranged from 0.0001 to 0.73 in 11 experiments [see the text for details]). In the representative experiment shown, citrate addition to Δfur mutant cultures resulted in a small (16%) but statistically significant (P < 0.05) increase in peak luminescence. Data shown were collected from the same experiment and are representative of at least three independent experiments for each strain.
Iron-mediated regulation of luminescence is independent of RyhB.
In other organisms, many of Fur's effects are mediated by its regulation of the small regulatory RNA RyhB (61–63), and we therefore wanted to determine whether iron limitation influences luminescence indirectly through RyhB. To test this possibility, we assayed the effect of citrate addition on luminescence of a ΔryhB mutant and found that citrate addition increased luminescence similar to the increased brightness observed in wild-type cultures (Fig. 5). This result indicates that Fur influences luminescence in response to citrate independently of RyhB.
Chelator-mediated luminescence induction requires native LuxR-LuxI regulation.
We considered the possibility that citrate-mediated iron limitation and derepression of the Fur regulon might influence luminescence through metabolic changes influencing bioluminescence rather than by affecting expression of the lux operon. To test whether native LuxR-LuxI regulation of luminescence was required for citrate-mediated enhancement of luminescence, we used strain JB22, which has the genes directly responsible for bioluminescence (luxCDABEG) under the control of a constitutive nonnative promoter. Addition of citrate to JB22 cultures did not result in any change in luminescence (Fig. 5), indicating that this effect of citrate is dependent on regulation of the native luxI promoter, which requires LuxR-mediated activation. Although JB22 is brighter than ES114 under these conditions, the luminescence of JB22 is still 2 to 3 orders of magnitude lower than its maximal luminescence capacity (12), suggesting that if citrate mediated a luminescence-enhancing effect independent of native lux transcription, we would still see enhanced luminescence in JB22 despite its higher basal luminescence.
Bioinformatic analysis identifies a putative Fur binding site upstream of litR.
To investigate the mechanism of Fur-mediated regulation of luminescence, we performed a virtual footprint analysis (64) to locate putative Fur binding sites in the V. fischeri genome, searching for matches to a weighted 18-bp Fur box determined in E. coli (Fig. 6A). As a frame of reference, this analysis returned a position weight matrix (PWM) score of 16.22 for a putative Fur box upstream of the heme uptake/utilization cluster (i.e., upstream of VF_1225), which is known to be Fur regulated (e.g., see reporter data in Fig. 3). Among other putative Fur binding sites elsewhere in the V. fischeri genome, we identified a site in the sequence upstream of the litR gene with a PWM score of 19.63 (Fig. 6A), a better match than in the Fur box of our Fur-dependent reporter. Moreover, the putative Fur box upstream of litR appeared embedded between sequences that matched reasonable −10 and −35 transcriptional promoter elements (Fig. 6B). Because LitR is a transcriptional activator of luxR (Fig. 1), we further investigated a possible role for LitR in Fur-mediated regulation of luminescence.
Fig 6.

Virtual footprint analysis of possible Fur binding sites. (A) Comparison of the sequence logo of the E. coli Fur binding site position weight matrix (PWM) used in the virtual footprint analysis of the V. fischeri ES114 genome (http://prodoric.tu-bs.de/vfp/) and putative Fur binding sites identified upstream of the Fur-regulated heme uptake cluster gene VF_1225 and the litR gene. For the weighted matrix, the y axis indicates bit scores for each nucleotide, and the x axis indicates the Fur box nucleotide position. The putative Fur binding site upstream of VF_1225 is located 44 bp upstream of the ATG codon and has a score of 16.22, while the site upstream of litR is located 40 bp upstream of the ATG codon, with a score of 19.63. Nucleotides that are identical between the two putative binding sites are in boldface type, and nucleotides identical to bases in the PWM sequence logo are shaded with the corresponding nucleotide color. (B) Position of the putative Fur binding site upstream of litR. The litR translational start site is in boldface type, the putative Fur binding site is highlighted in yellow, and possible −10 and −35 sequences are underlined.
litR is repressed by Fur and is required for luminescence induction in response to iron limitation.
We hypothesized that Fur represses litR under iron-rich conditions, but when iron is limiting, Fur-mediated repression of litR is relieved, resulting in elevated levels of LitR, increased luxR expression levels, and brighter luminescence. Two pheromone signaling pathways converge at LuxO (Fig. 1), which is upstream of LitR in the regulatory hierarchy. Consistent with our hypothesis, addition of citrate to luxO mutant cultures resulted in an increase in luminescence similar to what was observed for wild-type cultures (Fig. 5), indicating that the effect of citrate on luminescence is downstream of LuxO. Next, we added citrate to litR mutant cultures and found no change in luminescence (Fig. 5), indicating that the effect of citrate requires litR as well as fur.
To test if Fur regulates litR expression, we constructed a lacZ-based litR promoter reporter plasmid (pAS120) and assayed for fur-dependent regulation. We found elevated PlitR-lacZ expression levels in the Δfur mutant relative to the wild type (Fig. 7A), suggesting that Fur represses litR expression under iron-rich conditions. Based on our model of the pheromone-mediated regulatory hierarchy in V. fischeri (Fig. 1), we predicted that Fur's ultimate effect on luminescence is mediated by LitR's activation of luxR. To test this possibility, we assayed luxR promoter activity in the wild type and the Δfur, litR, and Δfur litR mutants. Consistent with our prediction, we observed elevated expression levels of a PluxR-lacZ reporter in the Δfur mutant compared to its expression levels in the wild type, and this increase was dependent on litR (Fig. 7B). These data indicate that the iron-dependent regulator Fur ultimately modulates expression of both key pheromone-dependent transcriptional activators, LitR and LuxR, in V. fischeri.
Fig 7.
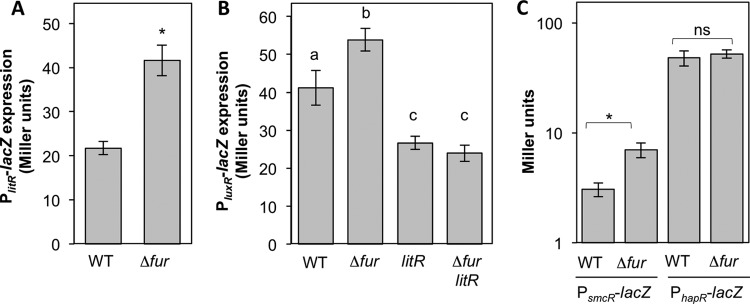
Effects of Fur on litR, luxR, smcR, and hapR transcriptional reporters. In all panels, V. fischeri cultures of ES114 (wild type), YLM111 (Δfur), JB19 (litR::erm), or ANS63 (Δfur litR::erm) were grown in aerobic shake flasks in SWTO medium supplemented with 43 μM FeSO4. In panels A and C, for litR, smcR, and hapR transcriptional reporters, cells harboring pAS120, pAS123, and pAS128 were harvested at an OD595 of ∼0.5. In panel B, for the luxR transcriptional reporter, cells harboring pJLB170 were harvested at an OD595 of ∼1.0. Error bars indicate standard deviations (n = 4 [A and B] and n = 3 [C]). Asterisks in panels A and C indicate a significant difference of the indicated pairwise comparison (P < 0.005) by Student's t test, while the comparison labeled “ns” was not significant (P > 0.05). In panel B, lowercase letters shared between bars indicate no statistically significant difference (P > 0.2), whereas different letters indicate a significant difference (P < 0.001), based on a one-way analysis of variance and Tukey's honestly significant difference test. Data in each panel are representative of at least three independent experiments.
Fur also represses expression of the Vibrio vulnificus litR homolog smcR.
Because Fur and LitR homologs are widespread in the Vibrionaceae, we asked whether this Fur-mediated regulation of a pheromone signaling master regulator is conserved. Interestingly, a previous study examining the Fur regulons in five sequenced Vibrio species identified putative Fur binding sites upstream of genes encoding LitR homologs in V. parahaemolyticus and V. vulnificus but not in Vibrio salmonicida or V. cholerae (65). Based on the presence or absence of predicted Fur binding sites determined previously by Ahmad et al. (65), we hypothesized that Fur would repress expression of V. vulnificus smcR but not influence expression of V. cholerae hapR. To test this, we constructed lacZ-based promoter reporter plasmids for V. vulnificus smcR (pAS123) and V. cholerae hapR (pAS128) and assayed for fur-dependent regulation of these reporters in wild-type V. fischeri and the V. fischeri Δfur mutant. Although the promoters in our constructs were cloned from different strains than those analyzed by Ahmad et al. (65), the presence or absence of putative Fur binding sites was conserved between strains within each species. Consistent with the predictions by Ahmad et al., we found elevated PsmcR-lacZ expression levels in the Δfur mutant relative to those in the wild type (Fig. 7C); however, PhapR-lacZ expression was unaffected by Fur (Fig. 7C). These data suggest the Fur-mediated repression of pheromone signaling master regulators is not limited to the control of LitR in V. fischeri and that the consensus Fur binding site described previously by Ahmad et al. is effective at predicting Fur binding sites in members of the Vibrionaceae.
DISCUSSION
The accumulation of bacterial pheromones may be influenced by high cell density, but pheromone-mediated regulatory circuits in bacteria are also influenced by environmental factors, indicating that they are not simply census-taking systems. For example, in V. fischeri, the LuxR-LuxI pheromone-dependent regulatory system is also controlled by density-independent factors (36–38). Both the pheromone synthase (LuxI) and its cognate pheromone receptor (LuxR) are regulated in response to environmental conditions, as are LuxI and LuxR homologs in other bacteria. Expanding on previous findings (39, 40), we have now shown that iron limitation leads to derepression of Fur-regulated genes (Fig. 3), resulting in a fur- and litR-dependent increase in luminescence (Fig. 2A and 4A). Based on our data, we propose that this effect is due to a Fur-dependent increase in the level of the LitR quorum-sensing regulator (Fig. 7A), which influences luxR expression (Fig. 7B). Because luxI is cotranscribed with the genes directly underlying light production, it is likely that this enhanced luminescence parallels an effect on 3OC6 synthesis as well. Thus, elements of the V. fischeri pheromone (3OC6-mediated) regulatory circuit are modulated by Fur and iron availability.
This connection between the iron-dependent regulator Fur and pheromone-mediated regulation could be relevant in a natural environment for V. fischeri, the host light organ. Previous work studying the Vibrio-squid symbiosis indicated that the squid light organ has low iron availability (41, 42). We speculate that the Fur- and LitR-dependent response described above might contribute to luminescence induction in symbiotic cells. Fidopiastis et al. showed previously that while a litR mutant achieved wild-type levels of colonization and luminescence in juvenile squid at 24 h postinoculation, the litR mutant displayed a 1-h delay in the onset of detectable luminescence compared to the wild type during squid colonization (29). Thus, while LitR-mediated regulation of the LuxR-LuxI regulatory system is not required for luminescence induction in symbiotic cells, given that the light organ appears to be a low-iron environment resulting in derepression of Fur-regulated genes (42), we speculate that Fur-mediated control of litR might contribute to the onset of symbiotic luminescence during initial infection.
This model of the role of Fur in symbiotic luminescence induction would be easier to test if it invoked Fur activating litR rather than relieving repression of litR, because in that case, a fur mutant would be predicted to have a phenotype similar to that of a litR mutant. Because our model proposes a role for Fur in repressing litR under iron-rich culture conditions but not in the host, the symbiotic phenotype of the Δfur mutant is not helpful in testing our model. Future experiments examining the role(s) and levels of LitR in early and late colonization will help elucidate whether the regulatory connection between Fur and LitR has symbiotic significance.
Although LitR regulates bioluminescence through its role as an activator of luxR transcription, LitR clearly regulates additional genes, some of which appear to have symbiotic relevance (29). A litR mutant outcompeted the wild type in a squid coinfection assay, and it also had altered colony morphology (29). In this study, we noticed modest growth effects of the litR mutation, further suggesting that LitR regulates other genes of physiological importance and possibly related to adaptation to low-iron environments.
While the squid light organ has low iron levels, this environmental factor is likely not host specific, because seawater also can be iron limiting. However, while both of these low-iron environments may lead to Fur-mediated derepression of litR in V. fischeri, only conditions leading to sufficiently high concentrations of 3OC6 pheromone would result in LuxR activation and enhanced luminescence. Therefore, we speculate that when V. fischeri is free-living or in the host, these low-iron conditions derepress the Fur regulon, including litR, which regulates other functions in addition to luxR expression. In free-living cells, pheromone diffuses away; however, in the squid light organ, pheromone levels accumulate to stimulatory levels due to high cell density and other host factors promoting pheromone synthesis, resulting in activation of LuxR and bright luminescence. Future work focused on elucidating LitR-regulated genes in V. fischeri may help reveal the connection between iron and LitR and its regulatory role in free-living versus symbiotic cells.
In other vibrios, LitR homologs similarly control a number of functions, and our findings here demonstrate that Fur-mediated regulation of LitR homologs could have implications beyond V. fischeri. Most other species of Vibrio lack the LuxR-LuxI system, and instead, a LitR homolog acts as the master regulator for pheromone-mediated behaviors. As examples, the LitR homologs in V. vulnificus (SmcR), V. harveyi (LuxRVh), V. parahaemolyticus (OpaR), and V. cholerae (HapR) control a range of behaviors and systems, including biofilm formation, type III secretion, toxins, and virulence factors (30, 32, 66–73). Interestingly, previous work by Ahmad et al. identified putative Fur binding sites upstream of litR, smcR, and opaR but not hapR (65), and transcriptional reporter assays shown here using wild-type and Δfur V. fischeri strains demonstrate that Fur represses expression of litR and smcR but not hapR (Fig. 7C). It will be interesting to determine how iron levels and Fur influence the SmcR regulon in V. vulnificus and whether Fur also regulates OpaR in V. parahaemolyticus. In any case, the reach of these regulons and the evidence for V. fischeri suggest that modulation of LitR by Fur could have impacts well beyond the luminescence phenotype described here.
The connection of LitR to Fur begs the question of why the LitR regulon, and possibly the other LitR homologs, would be modulated in response to iron availability in the local environment. Interestingly, for V. parahaemolyticus, a previous microarray analysis of transcripts regulated by OpaR included genes that appear to be involved in iron transport (66); however, these were a small portion of the total regulon. Moreover, an iron transport system in V. vulnificus was identified previously in a genome-wide search using a consensus SmcR binding sequence (74). While it is intriguing to think that LitR and/or homologs like OpaR and SmcR could be involved in modulating a response to low iron, these regulators also control factors involved in host colonization (66, 71). Thus, Fur might modulate these regulons to enhance expression in response to low iron availability, which is a characteristic typical of many host tissues.
There is similar evidence of iron levels regulating pheromone-mediated signaling in non-Vibrio species. For instance, in response to iron limitation, Pseudomonas aeruginosa increased transcription levels of lasR, which encodes an acyl-homoserine lactone-dependent transcriptional activator homologous to V. fischeri LuxR, and LasR-regulated proteins were also significantly modulated by iron limitation (75). A separate study demonstrated a lasI- and lasR-dependent increase in expression levels of the lasI pheromone synthase gene when iron was limited (76). Moreover, work with Streptococcus pneumoniae demonstrated that the autoinducer synthase LuxS mediates iron-dependent regulation of biofilm formation and competence (77). While these bacteria do not have LitR homologs, the connection between iron- and pheromone-mediated regulation is intriguing and suggests that iron levels may be a conserved density-independent regulator of pheromone systems in organisms outside the Vibrionaceae. Further studies of the connection between Fur and pheromone signaling in V. fischeri may elucidate properties that can be generalized to a broader range of bacteria.
ACKNOWLEDGMENTS
We thank Diana Downs, Elizabeth Mann, and Anne K. Dunn for helpful discussions.
This research was supported by the National Science Foundation (NSF) under grants CAREER MCB-0347317, IOS-0841480, OCE-0929081, and IOS-1121106. A.N.S. was supported by funds awarded by the University of Georgia Graduate School and DOD, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate (NDSEG) fellowship 32 CFR 168a.
Footnotes
Published ahead of print 11 January 2013
REFERENCES
- 1. Bassler BL, Losick R. 2006. Bacterially speaking. Cell 125:237–246 [DOI] [PubMed] [Google Scholar]
- 2. Miller MB, Bassler BL. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165–199 [DOI] [PubMed] [Google Scholar]
- 3. Waters CM, Bassler BL. 2005. Quorum sensing: cell-to-cell communication in bacteria. Annu. Rev. Cell Dev. Biol. 21:319–346 [DOI] [PubMed] [Google Scholar]
- 4. Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176:269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gray KM, Garey JR. 2001. The evolution of bacterial LuxI and LuxR quorum sensing regulators. Microbiology 147:2379–2387 [DOI] [PubMed] [Google Scholar]
- 6. Fuqua C, Winans SC, Greenberg EP. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727–751 [DOI] [PubMed] [Google Scholar]
- 7. Dunn AK, Stabb EV. 2007. Beyond quorum sensing: the complexities of prokaryotic parliamentary procedures. Anal. Bioanal. Chem. 387:391–398 [DOI] [PubMed] [Google Scholar]
- 8. Frederix M, Downie AJ. 2011. Quorum sensing: regulating the regulators. Adv. Microb. Physiol. 58:23–80 [DOI] [PubMed] [Google Scholar]
- 9. Platt TG, Fuqua C. 2010. What's in a name? The semantics of quorum sensing. Trends Microbiol. 18:383–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McFall-Ngai MJ, Ruby EG. 1991. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science 254:1491–1493 [DOI] [PubMed] [Google Scholar]
- 11. Wei SL, Young RE. 1989. Development of symbiotic bacterial bioluminescence in a nearshore cephalopod, Euprymna scolopes. Mar. Biol. 103:541–546 [Google Scholar]
- 12. Bose JL, Rosenberg CS, Stabb EV. 2008. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch. Microbiol. 190:169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578–4586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Engebrecht J, Nealson K, Silverman M. 1983. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773–781 [DOI] [PubMed] [Google Scholar]
- 15. Stabb EV. 2006. The Vibrio fischeri-Euprymna scolopes light organ symbiosis, p 204–218 In Thompson FL, Austin B, Swings J. (ed), The biology of vibrios. ASM Press, Washington, DC [Google Scholar]
- 16. Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444–2449 [DOI] [PubMed] [Google Scholar]
- 17. Antunes LCM, Ferreira RBR, Lostroh CP, Greenberg EP. 2008. A mutational analysis defines Vibrio fischeri LuxR binding sites. J. Bacteriol. 190:4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Devine JH, Shadel GS, Baldwin TO. 1989. Identification of the operator of the lux regulon from the Vibrio fischeri strain ATCC7744. Proc. Natl. Acad. Sci. U. S. A. 86:5688–5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nealson KH, Platt T, Hastings JW. 1970. Cellular control of the synthesis and activity of the bacterial luminescent system. J. Bacteriol. 104:313–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gilson L, Kuo A, Dunlap PV. 1995. AinS and a new family of autoinducer synthesis proteins. J. Bacteriol. 177:6946–6951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuo A, Callahan SM, Dunlap PV. 1996. Modulation of luminescence operon expression by N-octanoyl-L-homoserine lactone in ainS mutants of Vibrio fischeri. J. Bacteriol. 178:971–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lupp C, Ruby EG. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 186:3873–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bassler BL, Wright M, Silverman MR. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273–286 [DOI] [PubMed] [Google Scholar]
- 24. Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545–549 [DOI] [PubMed] [Google Scholar]
- 25. Freeman JA, Bassler BL. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 181:899–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lupp C, Urbanowski M, Greenberg EP, Ruby EG. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50:319–331 [DOI] [PubMed] [Google Scholar]
- 27. Miyashiro T, Wollenberg MS, Cao X, Oehlert D, Ruby EG. 2010. A single qrr gene is necessary and sufficient for LuxO-mediated regulation in Vibrio fischeri. Mol. Microbiol. 77:1556–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neiditch MB, Federle MJ, Miller ST, Bassler BL, Hughson FM. 2005. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol. Cell 18:507–518 [DOI] [PubMed] [Google Scholar]
- 29. Fidopiastis PM, Miyamoto CM, Jobling MG, Meighen EA, Ruby EG. 2002. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 45:131–143 [DOI] [PubMed] [Google Scholar]
- 30. Jobling MG, Holmes RK. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023–1034 [DOI] [PubMed] [Google Scholar]
- 31. McCarter LL. 1998. OpaR, a homolog of Vibrio harveyi LuxR, controls opacity of Vibrio parahaemolyticus. J. Bacteriol. 180:3166–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee JH, Rhee JE, Park U, Ju HM, Lee BC, Kim TS, Jeong HS, Choi SH. 2007. Identification and functional analysis of Vibrio vulnificus SmcR, a novel global regulator. J. Microbiol. Biotechnol. 17:325–334 [PubMed] [Google Scholar]
- 33. Showalter RE, Martin MO, Silverman MR. 1990. Cloning and nucleotide sequence of luxR, a regulatory gene controlling bioluminescence in Vibrio harveyi. J. Bacteriol. 172:2946–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boettcher KJ, Ruby EG. 1995. Detection and quantification of Vibrio fischeri autoinducer from symbiotic squid light organs. J. Bacteriol. 177:1053–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dunn AK, Millikan DS, Adin DM, Bose JL, Stabb EV. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72:802–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bose JL, Kim U, Bartkowski W, Gunsalus RP, Overley AM, Lyell NL, Visick KL, Stabb EV. 2007. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol. Microbiol. 65:538–553 [DOI] [PubMed] [Google Scholar]
- 37. Lyell NL, Dunn AK, Bose JL, Stabb EV. 2010. Bright mutants of Vibrio fischeri ES114 reveal conditions and regulators that control bioluminescence and expression of the lux operon. J. Bacteriol. 192:5103–5114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Septer AN, Bose JL, Dunn AK, Stabb EV. 2010. FNR-mediated regulation of bioluminescence and anaerobic respiration in the light-organ symbiont Vibrio fischeri. FEMS Microbiol. Lett. 306:72–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Haygood MG, Nealson KH. 1985. Mechanisms of iron regulation of luminescence in Vibrio fischeri. J. Bacteriol. 162:209–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dunlap PV. 1992. Iron control of the Vibrio fischeri luminescence system in Escherichia coli. Arch. Microbiol. 157:235–241 [DOI] [PubMed] [Google Scholar]
- 41. Graf J, Ruby EG. 2000. Novel effects of a transposon insertion in the Vibrio fischeri glnD gene: defects in iron uptake and symbiotic persistence in addition to nitrogen utilization. Mol. Microbiol. 37:168–179 [DOI] [PubMed] [Google Scholar]
- 42. Septer AN, Wang Y, Ruby EG, Stabb EV, Dunn AK. 2011. The haem-uptake gene cluster in Vibrio fischeri is regulated by Fur and contributes to symbiotic colonization. Environ. Microbiol. 13:2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bose JL, Wollenberg MS, Colton DM, Mandel MJ, Septer AN, Dunn AK, Stabb EV. 2011. Contribution of rapid evolution of the luxR-luxI intergenic region to the diverse bioluminescence outputs of Vibrio fischeri strains isolated from different environments. Appl. Environ. Microbiol. 77:2445–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stabb EV, Schaefer A, Bose JL, Ruby EG. 2008. Quorum signaling and symbiosis in the marine luminous bacterium Vibrio fischeri, p 233–250 In Winans SC, Bassler BL. (ed), Chemical communication among microbes. ASM Press, Washington, DC [Google Scholar]
- 45. Stabb EV, Reich KA, Ruby EG. 2001. Vibrio fischeri genes hvnA and hvnB encode secreted NAD(+)-glycohydrolases. J. Bacteriol. 183:309–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Miller JH. 1992. A short course in bacterial genetics, p 456 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 47. Dunn AK, Martin MO, Stabb EV. 2005. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid 54:114–134 [DOI] [PubMed] [Google Scholar]
- 48. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 49. Herrero M, de Lorenzo V, Timmis KN. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stabb EV, Ruby EG. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413–426 [DOI] [PubMed] [Google Scholar]
- 51. Boettcher KJ, Ruby EG. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reference deleted. [Google Scholar]
- 53. Dunn AK, Stabb EV. 2008. Genetic analysis of trimethylamine N-oxide reductases in the light organ symbiont Vibrio fischeri ES114. J. Bacteriol. 190:5814–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stabb EV, Butler MS, Adin DM. 2004. Correlation between osmolarity and luminescence of symbiotic Vibrio fischeri strain ES114. J. Bacteriol. 186:2906–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Matsukawa T, Ikeda S, Imai H, Yamada M. 2002. Alleviation of the two-cell block of ICR mouse embryos by polyaminocarboxylate metal chelators. Reproduction 124:65–71 [DOI] [PubMed] [Google Scholar]
- 56. Staubli A, Boelsterli UA. 1998. The labile iron pool in hepatocytes: prooxidant-induced increase in free iron precedes oxidative cell injury. Am. J. Physiol. 274:G1031–G1037 [DOI] [PubMed] [Google Scholar]
- 57. Craig SA, Carpenter CD, Mey AR, Wyckoff EE, Payne SM. 2011. Positive regulation of the Vibrio cholerae porin OmpT by iron and fur. J. Bacteriol. 193:6505–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lee HJ, Bang SH, Lee KH, Park SJ. 2007. Positive regulation of fur gene expression via direct interaction of Fur in a pathogenic bacterium, Vibrio vulnificus. J. Bacteriol. 189:2629–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mey AR, Wyckoff EE, Kanukurthy V, Fisher CR, Payne SM. 2005. Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect. Immun. 73:8167–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bagg A, Neilands JB. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471–5477 [DOI] [PubMed] [Google Scholar]
- 61. Davis BM, Quinones M, Pratt J, Ding Y, Waldor MK. 2005. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J. Bacteriol. 187:4005–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Masse E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Oglesby AG, Murphy ER, Iyer VR, Payne SM. 2005. Fur regulates acid resistance in Shigella flexneri via RyhB and ydeP. Mol. Microbiol. 58:1354–1367 [DOI] [PubMed] [Google Scholar]
- 64. Munch R, Hiller K, Grote A, Scheer M, Klein J, Schobert M, Jahn D. 2005. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21:4187–4189 [DOI] [PubMed] [Google Scholar]
- 65. Ahmad R, Hjerde E, Hansen GA, Haugen P, Willassen NP. 2009. Prediction and experimental testing of ferric uptake regulator regulons in vibrios. J. Mol. Microbiol. Biotechnol. 16:159–168 [DOI] [PubMed] [Google Scholar]
- 66. Gode-Potratz CJ, McCarter LL. 2011. Quorum sensing and silencing in Vibrio parahaemolyticus. J. Bacteriol. 193:4224–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hammer BK, Bassler BL. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101–104 [DOI] [PubMed] [Google Scholar]
- 68. Henke JM, Bassler BL. 2004. Quorum sensing regulates type III secretion in Vibrio harveyi and Vibrio parahaemolyticus. J. Bacteriol. 186:3794–3805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Liu Z, Miyashiro T, Tsou A, Hsiao A, Goulian M, Zhu J. 2008. Mucosal penetration primes Vibrio cholerae for host colonization by repressing quorum sensing. Proc. Natl. Acad. Sci. U. S. A. 105:9769–9774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Miller MB, Skorupski K, Lenz DH, Taylor RK, Bassler BL. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303–314 [DOI] [PubMed] [Google Scholar]
- 71. Shao CP, Lo HR, Lin JH, Hor LI. 2011. Regulation of cytotoxicity by quorum-sensing signaling in Vibrio vulnificus is mediated by SmcR, a repressor of hlyU. J. Bacteriol. 193:2557–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhu J, Mekalanos JJ. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647–656 [DOI] [PubMed] [Google Scholar]
- 73. Zhu J, Miller MB, Vance RE, Dziejman M, Bassler BL, Mekalanos JJ. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 99:3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee DH, Jeong HS, Jeong HG, Kim KM, Kim H, Choi SH. 2008. A consensus sequence for binding of SmcR, a Vibrio vulnificus LuxR homologue, and genome-wide identification of the SmcR regulon. J. Biol. Chem. 283:23610–23618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kim EJ, Wang W, Deckwer WD, Zeng AP. 2005. Expression of the quorum-sensing regulatory protein LasR is strongly affected by iron and oxygen concentrations in cultures of Pseudomonas aeruginosa irrespective of cell density. Microbiology 151:1127–1138 [DOI] [PubMed] [Google Scholar]
- 76. Bollinger N, Hassett DJ, Iglewski BH, Costerton JW, McDermott TR. 2001. Gene expression in Pseudomonas aeruginosa: evidence of iron override effects on quorum sensing and biofilm-specific gene regulation. J. Bacteriol. 183:1990–1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Trappetti C, Potter AJ, Paton AW, Oggioni MR, Paton JC. 2011. LuxS mediates iron-dependent biofilm formation, competence, and fratricide in Streptococcus pneumoniae. Infect. Immun. 79:4550–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]