Abstract
Prophages make up 12% of the enterohemorrhagic Escherichia coli genome and play prominent roles in the evolution and virulence of this food-borne pathogen. Acquisition and loss of and rearrangements within prophage regions are the primary causes of differences in pulsed-field gel electrophoresis (PFGE) patterns among strains of E. coli O157:H7. Sp11 and Sp12 are two tandemly integrated and putatively defective prophages carried by E. coli O157:H7 strain Sakai. In this study, we identified 3 classes of deletions that occur within the Sp11-Sp12 region, at a frequency of ca. 7.74 × 10−4. One deletion resulted in a precise excision of Sp11, and the other two spanned the junction of Sp11 and Sp12. All deletions resulted in shifts in the XbaI fragment pattern observed by PFGE. We sequenced the inducible prophage pool of Sakai but did not identify any mature phage particles corresponding to either Sp11 or Sp12. Deletions containing pchB and psrC, which are Sp11-carried genes encoding proteins known or suspected to regulate type III secretion, did not affect the secretion levels of the EspA or EspB effector. Alignment of the Sp11-Sp12 DNA sequence with its corresponding regions in other E. coli O157:H7 and O55:H7 strains suggested that homologous recombination rather than integrase-mediated excision is the mechanism behind these deletions. Therefore, this study provides a mechanism behind the previously observed genetic instability of this genomic region of E. coli O157:H7.
INTRODUCTION
Enterohemorrhagic Escherichia coli (EHEC) O157:H7 is a devastating food-borne pathogen that causes diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome (HUS) (1). The industrialization of agriculture, the expanding scale of food production, and the complicated food distribution chain have likely contributed to the increased occurrence and scale of E. coli O157:H7 outbreaks (2, 3). One of the best-studied strains, designated Sakai, was responsible for a massive outbreak in 1996 that was linked to contaminated radish sprouts and resulted in approximately 12,680 clinical cases (4, 5). During such outbreaks, molecular subtyping methods, including pulsed-field gel electrophoresis (PFGE), are used to identify sources and routes of transmission of the pathogen to the food supply. Although this method has high discriminatory power (6, 7), one drawback is that the banding patterns can often be unstable due to rearrangements and deletions within the prophage-containing regions of the genome (8–11).
The well-studied E. coli O157:H7 strains carry approximately 18 prophages or prophage remnants (12–14). In the genome of strain Sakai, 11 of its 18 annotated prophages (designated Sp1 through Sp18) are lambdoid and carry numerous genes related to virulence (15). For instance, Sp5 encodes Stx2, a toxin strongly associated with the development of HUS (16, 17). Although in silico analysis suggested that many of these prophages are defective, Asadulghani et al. (15) demonstrated that some can be induced or packaged and may even transduce other E. coli strains. The constant loss and acquisition of prophages are identified as the major mechanisms in the evolution of this pathogen (11, 12, 15, 18–20). In addition to integrase-mediated phage integration and excision, homologous recombination between related prophages can also contribute to the genomic instability of E. coli O157:H7 (9, 11).
E. coli O157:H7 also carries non-prophage-associated virulence genes that were acquired by horizontal gene transfer, such as the locus for enterocyte effacement (LEE) (13). The LEE island is also present in the genomes of enteropathogenic E. coli and a mouse pathogen, Citrobacter rodentium (21–23). The LEE in strain Sakai is integrated at the tRNA gene selC; it encodes an outer membrane protein (intimin), the structural and regulatory proteins for a type III secretion system (T3SS), and the effector proteins secreted by the T3SS (13). When E. coli O157:H7 attaches to enterocytes, the LEE-associated protein Tir is translocated from the bacterial cytoplasm across the epithelial cell membrane, where it serves as a receptor for intimin, an additional LEE-encoded protein expressed on the bacterial surface. This process leads to the intimate attachment of bacteria and the effacement of microvilli (24, 25), designated the attaching and effacing (A/E) phenotype (26, 27).
A variety of virulence functions for LEE-encoded type III secretion effectors have been described. For example, EspA is involved in forming a physical bridge between the bacterium and the eukaryotic cell surface (28), and EspB is essential for the protein translocation process and A/E lesion formation (29). Genes within the LEE are regulated by a number of factors, including those that are carried by prophages (30–33). Note that the positive regulator gene pchB (ECs2182) (32) and the negative regulator gene psrC (ECs2191) (33) are both carried within Sp11.
In this study, we identified three classes of rare deletion events within the tandem prophages Sp11 and Sp12 of Sakai, and we suggest that homologous recombination rather than integrase-mediated excision causes these deletions. These deletions affect the fragment patterns observed by PFGE, which could complicate epidemiologic investigations. Unexpectedly, although the deletions excised pscC and/or pchB from the genome, we did not observe an effect on EspA/B secretion compared to the wild-type (WT) strains. Our observation adds to previous reports of genomic instability of E. coli O157:H7 (8–11). Additionally, the application of high-throughput sequencing to define precise deletion events was demonstrated. Future studies are needed to ascertain the impact of these deletions on host biology.
MATERIALS AND METHODS
Bacterial strains and culture media.
The bacterial strains and plasmids used in this study are described in Table 1. Strains were routinely grown in liquid or solid lysogeny broth (LB) medium (37). All stocks were maintained at −80°C in 10% glycerol. Chloramphenicol (Cam; 25 μg/ml), streptomycin (Str; 100 μg/ml), kanamycin (Kan; 50 μg/ml), cefixime (Cef; 0.05 μg/ml), and/or potassium tellurite (Tel; 2.5 μg/ml) was added to the medium if indicated. In selecting for the loss of pDS132-carried sacB, LB agar with 5% (wt/vol) sucrose and without NaCl (LB-sucrose agar) was used (34). A Str-resistant (Strr) mutant of E. coli O157:H7 Sakai was selected for by plating an overnight culture of the wild-type strain on LB agar supplemented with Str. Putative Strr colonies were streaked on the same medium for purification.
Table 1.
Strains and plasmids used in this study
| E. coli strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| SM10λpir | Host for maintenance and conjugation of pDS132 derivatives | Lab collection |
| Sakai | O157:H7; stx1+ stx2+ | Wei Zhang, Division of Biology, Illinois Institute of Technology |
| Sakai-Strr | Streptomycin-resistant mutant of Sakai | This study |
| SK-18 | Second-crossover derivative of Sakai::(pDS132::ΔtDNA5); ΔSp11; type I deletion mutant | This study |
| SK-11 | Second-crossover derivative of Sakai::(pDS132::ΔtDNA5); type II deletion mutant | This study |
| SK-17 | Second-crossover derivative of Sakai::(pDS132::ΔtDNA5); type III deletion mutant | This study |
| SKΔpchB | Sakai with pchB deletion | This study |
| Plasmids | ||
| pDS132 | Suicide vector; Camr; encodes levansucrase | 34 |
| pDS132::ΔtDNA5 | pDS132 carrying concatenated fragments flanking ileZ5-argO5 | This study |
| pDS132::ΔtDNA1 | pDS132 carrying concatenated fragments flanking ileZ1-argN1-argO1 | This study |
| pDS132::ΔtDNA3 | pDS132 carrying concatenated fragments flanking ileZ3-argO3 | This study |
| pKD4 | Template plasmid for aph gene; Kanr | 35 |
| pKM200 | Red recombinase expression plasmid; Camr | 36 |
Screening for spontaneous deletion mutation within the Sp11-Sp12 region of E. coli O157:H7 Sakai.
A suicide plasmid-based method similar to that previously described (38) was used to detect and quantify the frequency of spontaneous deletions within Sp11 and Sp12. With the Sakai genome as a template, PCRs were designed with primer pairs tDNA5-LL plus tDNA5-LR and tDNA5-RL plus tDNA5-RR (see Table S1 in the supplemental material), which amplified 330 bp upstream (positions 2189181 to 2189510 in the Sakai genome [GenBank accession number BA000007]) and 399 bp downstream (positions 2189759 to 2190157) of the Sp11-carried tRNA genes ileZ5-argO5 (tDNA5), respectively. The two amplicons overlapped by 28 bp, and an NotI site was located within this overlap. Approximately 20 ng of each PCR product and primers tDNA5-LL and tDNA5-RR were used in a second round of PCR. The resulting PCR product was digested with XbaI and cloned into the corresponding site in pDS132 (34). The constructed plasmid (pDS132::ΔtDNA5) was transformed into E. coli SM10λpir, which was permissive for replication of the suicide vector. Conjugation was conducted between E. coli SM10λpir(pDS132::ΔtDNA5) and Sakai in the same manner as previously described (38), and selection for recombinants was performed on LB agar with Cam, Cef, and Tel. Putative Sakai::(pDS132::ΔtDNA5) colonies were restreaked on the same medium for purification.
Selection for the spontaneous loss of pDS132-carried sacB, which encodes levansucrase and results in a sucrose-sensitive phenotype, was performed as previously described (38). Briefly, 0.5 μl of an overnight Sakai::(pDS132::ΔtDNA5) culture was diluted in 10 ml fresh LB broth and grown with shaking at 37°C for 8 h. Selection for plasmid excision was performed by plating aliquots of broth on LB-sucrose agar. Colonies were harvested, and their sensitivity to Cam was confirmed by replica plating on plain LB agar and LB agar supplemented with Cam. For each isolate, PCRs using the primer pairs shown in Table S1 in the supplemental material were used to estimate the left and right boundaries of each deletion event.
Illumina sequencing and data analysis.
Genomic DNAs from SK-11 and SK-17 (Table 1) were extracted using a Wizard genomic DNA purification kit (Promega, Madison, WI) and then submitted to the Institute for Genome Sciences at the School of Medicine, University of Maryland. High-throughput sequencing was performed using a HiSeq2000 sequencing system (Illumina, Hayward, CA). The reads were assembled by SeqMan NGen (DNASTAR, Madison, WI), using the genome sequence of Sakai as the template, and sequencing depth reports were generated using SeqMan Pro (DNASTAR, Madison, WI). Primer pairs SK11-L plus SK11-R and SK17-L plus SK17-R (see Table S1 in the supplemental material) were designed based on the sequencing results.
Quantifying the frequency of spontaneous loss of sacB.
Sakai::(pDS132::ΔtDNA1) and Sakai::(pDS132::ΔtDNA3) (Table 1) were constructed in the same manner as described above. Briefly, primer pairs tDNA1-LL plus tDNA1-LR and tDNA1-RL plus tDNA1-RR were used to generate DNA fragments flanking ileZ1-argN1-argO1 (530 bp and 700 bp, respectively), and tDNA3-LL plus tDNA3-LR and tDNA3-RL plus tDNA3-RR (see Table S1 in the supplemental material) were used to generate DNA fragments flanking ileZ3-argO3 (811 bp and 858 bp, respectively). The respective amplicons were concatenated by overlap PCR and cloned into pDS132 in a manner similar to that described above for pDS132::ΔtDNA5. The resulting plasmids were transformed into E. coli SM10λpir and transferred into Sakai by conjugation.
Frequencies of spontaneous loss of sacB were compared among Sakai::(pDS132::ΔtDNA5), Sakai::(pDS132::ΔtDNA1), and Sakai::(pDS132::ΔtDNA3). Bacteria were grown from frozen stocks overnight in liquid LB at 37°C with shaking. The cultures were diluted 1:20,000 (vol/vol) in fresh LB. After another overnight shaking incubation at 37°C, each culture was serial diluted and plated on LB agar and LB-sucrose agar. CFU were determined on both media, and the frequency of sucrose resistance (Sucr), which indicated the loss of sacB, was calculated as CFULB-sucrose agar/CFULB agar. For each comparison, two experimental replicates with two biological replicates each were performed.
Primer pairs tDNA1out-L plus tDNA1out-R and tDNA3out-L plus tDNA3out-R (see Table S1 in the supplemental material) flanked ileZ1-argN1-argO1 and ileZ3-argN3-argO3, respectively. PCRs using these primers were performed to confirm that Sucr was caused only by homologous recombination-mediated resolution of the plasmids from strains Sakai::(pDS132::ΔtDNA1) and Sakai::(pDS132::ΔtDNA3).
PFGE.
PFGE analyses using XbaI or AvrII (an isoschizomer of BlnI) (NEB, Ipswich, MA) were performed using the standardized protocol from the Centers for Disease Control and Prevention (39).
Phage DNA isolation and sequencing.
Sakai was grown from frozen stocks in liquid LB overnight at 37°C with shaking. The culture was diluted to an optical density at 600 nm (OD600) of 0.05 in LB supplemented with 45 ng/ml ciprofloxacin (40). After 8 h of incubation with shaking at 37°C, lysates were collected from the culture supernatants by centrifugation (4,000 × g, 10 min, 4°C) and filtration (0.2-μm cellulose acetate filter; VWR, Philadelphia, PA). Phage DNA was then isolated from each lysate by a method similar to a previously described method (15). Briefly, the supernatant samples were incubated with 0.6 U/ml DNase I (Promega, Madison, WI) and 1.8 μg/ml RNase (Promega, Madison, WI) at 37°C for 1 h. Polyethylene glycol 8000 (Promega, Madison, WI) and NaCl were added to the samples, to final concentrations of 10% (wt/vol) and 1 M, respectively. After overnight incubation at 4°C, the samples were centrifuged at 5,000 × g for 2 h (4°C). The pellet was resuspended in SM buffer (0.58% NaCl, 0.2% MgSO4 · 7H2O, 1 M Tris-Cl [pH 7.5], and 0.01% gelatin) and was incubated at 65°C for 1 h with 50 μg/ml (final concentration) of proteinase K (EMD, Darmstadt, Germany). Phenol-chloroform extraction was carried out to extract phage DNA. DNA was further purified using a Clean and Concentrator kit (Zymo Research, Irvine, CA).
The extracted total DNA was submitted to the Penn State Genome Core Facility for sequencing using a Roche GS 454 FLX+ system. The raw reads were assembled using SeqMan Pro (DNASTAR, Madison, WI). The contigs generated from each strain were then aligned against the whole genome sequence of Sakai by using the Artemis comparison tool (ACT) (41).
Construction of pchB knockout mutants.
Deletion of pchB (ECs2191) in Sakai was performed using a one-step inactivation method (32, 36). Briefly, the Kanr gene aph was amplified using pKD4 (Table 1) as the template and the primer pair pchBKO-L plus pchBKO-R (see Table S1 in the supplemental material), each containing a 50-bp region homologous to the 5′ or 3′ end of pchB. The PCR product was electroporated into Sakai harboring pKM200, which encodes λ Red, Gam, and Bet (36), and plated onto LB supplemented with Kan. The pchB mutants were screened from the harvested Kan-resistant colonies by PCR using the primer pair pchB-L plus pchB-R (see Table S1). The replacement of pchB with aph increased the size of the amplicon from 772 bp to 1,935 bp.
Analysis of secreted proteins.
Quantification of EspA and EspB secreted from Sakai and its derivatives was performed using a method adapted from those previously described (32, 33). Bacteria were grown from frozen stocks statically overnight at 37°C in Dulbecco's modified Eagle's medium (DMEM) (with 4.5 g/liter glucose and l-glutamine, without sodium pyruvate; Cellgro, Manassas, VA). A total of 2 ml was diluted into 28 ml of fresh DMEM and incubated statically at 37°C until an OD600 of 0.8. The bacteria were removed by centrifugation (4,000 × g, 30 min) and filtration through 0.2-μm cellulose acetate filters (VWR, Philadelphia, PA). Bovine serum albumin (BSA; NEB, Ipswich, MA) was added to the culture supernatant, to a final concentration of 2 μg/ml, as a coprecipitant, and the secreted proteins in each sample were precipitated with 10% (vol/vol) trichloroacetic acid. Supernatants were incubated overnight at 4°C, followed by 30 min of centrifugation at 4,000 × g. Protein pellets were carefully dried with a Kimwipe (Kimberly-Clark, Neenah, WI) and then dissolved in 120 μl of 1 M Tris-Cl (pH 8.0).
Western blot analysis was performed for protein quantification. A mixture of 8 μl of the resuspended sample and 2 μl 5× Laemmli buffer (60 mM Tris-Cl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, and 0.01% bromophenol blue) was loaded onto a 4 to 20% precast linear gradient polyacrylamide gel (Bio-Rad, Hercules, CA). Electrophoresis was conducted in a Mini-PROTEAN system (Bio-Rad, Hercules, CA) at 180 V for 45 min. The same system (40 V, 16 h, 4°C) was also used to transfer protein from the polyacrylamide gel to a 0.45-μm polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA). Rabbit anti-EspB and anti-EspA (kindly provided by James Kaper) and rabbit anti-BSA (Li-Cor, Lincoln, NE) primary antibodies and IRDye 680RD goat anti-rabbit IgG (Li-Cor, Lincoln, NE) secondary antibody were used to probe each membrane. An Odyssey infrared imaging system (Li-Cor, Lincoln, NE) was used for fluorescence detection. The signal intensity from each sample was analyzed using the Odyssey application software, version 3.0, and the data are presented in arbitrary units. The percentage of EspA or EspB secretion (in arbitrary units) from Sakai derivatives compared with the WT strain was calculated as (Espmutant/BSAmutant)/(EspWT/BSAWT).
Statistical analysis.
All protein quantification experiments were performed with three biological repeats. Data were analyzed using one-way analysis of variance (ANOVA) by Minitab 16.2.0.
RESULTS
Identifying spontaneous deletions within tandem prophages Sp11 and Sp12.
As part of a separate study, we attempted to construct chromosomal deletions of the rare codon tRNA genes (ileZ5 and argO5) carried within the prophage previously designated Sp11 from E. coli O157:H7 Sakai (13). The protocol, as described before (34), started with the cloning of DNA fragments that are 5′ and 3′ of the tRNA genes into the suicide vector pDS132. The resulting plasmid, designated pDS132::ΔtDNA5 (Table 1), was integrated into the Sakai chromosome as described in Materials and Methods. Next, a second crossover involving homologous recombination-mediated excision of the plasmid was selected for on LB-sucrose agar. We expected that this protocol would generate either an ileZ5-argO5 deletion mutant or a WT revertant. We harvested 20 second-crossover isolates and screened for mutants and WT revertants by PCR using the primer pair tDNA5-LL plus tDNA5-RR (see Table S1 in the supplemental material). The size of the amplicons was expected to be either 759 bp (ileZ5-argO5 deletion mutant) or 1,001 bp (WT revertant). However, among the 20 isolates, 3 appeared to be WT, and no amplicon was observed with the other 17 isolates. Given the potential instability of the prophage region where ileZ5-argO5 is located, we reasoned that the latter isolates may have resulted from a genomic deletion(s) that included one or both of the primer binding sites. PCRs involving 11 primer pairs (Fig. 1; see Table S1) were performed to define the limits of the deleted region(s).
Fig 1.
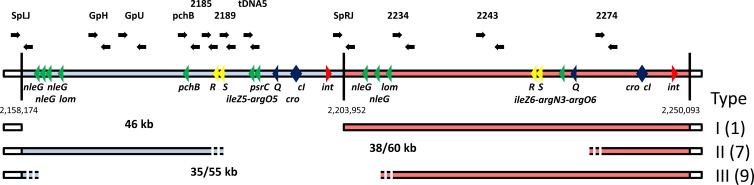
Spontaneous deletions within the Sp11-Sp12 prophage region of Sakai. Genomic loci of Sp11 (light blue) and Sp12 (light red) are shown. Genes related to prophage regulation (blue), to integration/excision (red), to bacterial host lysis (yellow), and putatively to virulence (green) are marked by triangles. Primer pairs (see Table S1 in the supplemental material) used to estimate the range of deletions are indicated using black arrows. Three different classes of deletions were identified. One included the entire Sp11 prophage (46 kb), while the boundaries of the other two deletion types were not definite (shown by dashed lines). The estimated minimal and maximal sizes of each deletion type are listed. The designated type of deletion and the frequency of observation (indicated in parentheses) among 20 second-crossover isolates are shown to the right of each deletion scheme.
We first hypothesized that the initial PCR failures were due to excision of the entire Sp11. However, using the primer pair SpLJ-L plus SpRJ-R (Fig. 1; see Table S1 in the supplemental material), annealing at the 5′ and 3′ ends of the reported Sp11 integration site, an amplicon was generated only using DNA from SK-18 (Table 1) as the template. Sequencing of the amplicon confirmed that this strain had a precise deletion of Sp11 (data not shown), and we designated this event a type I deletion (Fig. 1).
We were also able to identify 2 additional classes of mutants (Fig. 1). For the 7 isolates designated type II, PCR results were positive using primer pairs 2185-F/R and 2274-F/R but negative using primer pairs 2189-F/R and 2243-F/R or any that targeted regions in between (Fig. 1). The distances between primers 2185-R and 2274-F and primers 2189-F and 2243-R are 60 kb and 38 kb, respectively, providing an estimate of the size of the deletion. A third class of deletions (type III) was observed with the remaining 9 isolates, which were PCR positive using primer pairs SpLJ-F/R and 2234-F/R, but for which no amplification was observed using primer pairs GpH-F/R and SpRJ-F/R or any that targeted regions in between (Fig. 1). The size of the deletion in this case was estimated to be between 35 kb (distance between GpH-F and SpRJ-R) and 55 kb (SpLJ-R and 2234-F). Attempts to more precisely define the limits of the DNA deletion by PCR were hindered by the extensive sequence identity between Sp11 and Sp12 (see Fig. S1 in the supplemental material).
This entire experiment was also repeated with Strr Sakai (Table 1). After conjugation and selection for the second crossover, 20 isolates were obtained and screened. Using the PCR-based method described above, we observed that three isolates carried the ileZ5-argO5 deletion, one carried the Sp11 deletion (type I), 12 were categorized as having a type II deletion, and 4 had a type III deletion.
Identifying the boundaries of deletions in Sakai derivatives SK-11 and SK-17.
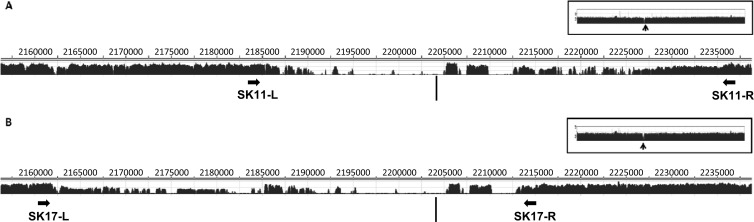
In order to further narrow down the boundaries of the SK-11 (type II deletion) and SK-17 (type III deletion) (Table 1) deletions, we sequenced these two strains by Illumina sequencing and aligned the reads on the Sakai genome (GenBank accession number BA000007) (Fig. 2). We observed average sequencing depths (defined as the average number of sequence reads that aligned for each base) of 235 and 279 by aligning reads obtained from SK-11 and SK-17, respectively. As expected, the sequencing depth was much less within the Sp11-Sp12 chromosomal regions, confirming that chromosomal deletions exist within this locus in the genomes of SK-11 and SK-17.
Fig 2.
Templated assembly of Sakai derivatives within the Sp11-Sp12 region. The reads from whole-genome sequencing of SK-11 (A) and SK-17 (B) were aligned against the genome of WT Sakai, and a sequencing depth report was generated by SeqMan. In each case, the coordinates of the Sakai genome are shown at the top, and a bar graph presenting the detailed sequencing depth is shown at the bottom. The overall sequencing depth for each genome is shown in an inset, with an arrow highlighting the gap in the Sp11-Sp12 region.
To further confirm that deletions occurred within the regions predicted by sequence alignments, PCR was performed using the primer pair SK11-L (binding nucleotides 2184026 to 2184045 in the Sakai genome) plus SK11-R (binding nucleotides 2237331 to 2237312) (see Table S1 in the supplemental material). The PCR generated an amplicon of 2.3 kb using the SK-11 genome as the template; no product was seen when the WT Sakai genome was used as the template (data not shown). An alignment between bp 2184045 to 2186356 (ca. 2.3 kb downstream of the left primer) and bp 2235008 to 2237320 (ca. 2.3 kb upstream of the right primer) showed a 99% identity (2,292/2,314 bp). The two boundaries of the deletion in SK-11 are therefore likely to be located within these two homologous regions. The estimated total deletion size in this case was 51.0 kb.
A similar analysis was conducted to identify the boundaries of deletion in SK-17. The primer pair SK17-L (bp 2160569 to 2160588) plus SK17-R (bp 2213863 to 2213844) (see Table S1 in the supplemental material) amplified a product of 2.5 kb with the SK-17 genome as the template (data not shown). The sequences between bp 2160621 to 2163138 in the Sakai genome (2.5 kb downstream of the left primer) and bp 2211344 to 2213863 (2.5 kb upstream of the right primer) were 98% identical (2,460/2,523 bp). Therefore, the boundaries of the deletion in SK-17 occurred within these two homologous regions; the estimated size of this deletion was 50.8 kb.
Frequency of deletion events involving prophages Sp11 and Sp12.
The combined frequency of deletions within Sp11 and Sp12 was quantified by measuring the loss of the counterselective marker sacB from Sakai::(pDS132::ΔtDNA5). After an overnight incubation of Sakai::(pDS132::ΔtDNA5), the average frequency of reversion to Sucr was 9.1 × 10−4 (Table 2). As 34 of the 40 deletions characterized above were type I, II, or III, we estimated the rate of spontaneous deletion to be 7.74 × 10−4, while that of a plasmid-mediated second crossover would be 1.37 × 10−4. In order to determine whether large deletions also occur surrounding tRNA genes carried within other lambdoid prophages found within the Sakai genome, we performed similar experiments using Sakai::(pDS132::ΔtDNA1) and Sakai::(pDS132::ΔtDNA3) (Table 1). These two strains contained pDS132 derivatives integrated at the Sp4-carried ileZ1-argN1-argO1 operon and the Sp9-carried ileZ3-argO3 operon. Similar to ileZ5-argO5, these genes are localized immediately downstream of the predicted prophage late promoter (13). The frequencies of reversion to Sucr for Sakai::(pDS132::ΔtDNA1) and Sakai::(pDS132::ΔtDNA3) after overnight incubation in LB were 4.9 × 10−5 and 2.1 × 10−4, respectively (Table 2). Ten Sucr colonies derived from Sakai::(pDS132::ΔtDNA1) and 10 derived from Sakai::(pDS132::ΔtDNA3) were picked and PCR amplified with primer pairs tDNA1outL/R and tDNA3outL/R (see Table S1 in the supplemental material), respectively. The PCR results indicated that all 20 Sucr isolates underwent a plasmid-mediated second crossover, resulting in either deletion of the tRNA genes or WT revertants (data not shown).
Table 2.
Frequencies of reversion to Sucr in different merodiploid strains
| Triala | Frequency of reversion to Sucrb |
||
|---|---|---|---|
| Sakai::(pDS132::ΔtDNA5) | Sakai::(pDS132::ΔtDNA1) | Sakai::(pDS132::ΔtDNA3) | |
| 1 | 6.23 × 10−04 | 3.95 × 10−05 | |
| 2 | 8.59 × 10−04 | 5.77 × 10−05 | |
| 3 | 1.30 × 10−03 | 2.96 × 10−04 | |
| 4 | 8.51 × 10−04 | 1.33 × 10−04 | |
Each trial was conducted with two technical repeats.
The frequencies of reversion to Sucr were calculated as the colony count on LB-sucrose agar/total colony count on LB agar.
Type I, II, and III deletions change the XbaI pulsotype of strain Sakai.
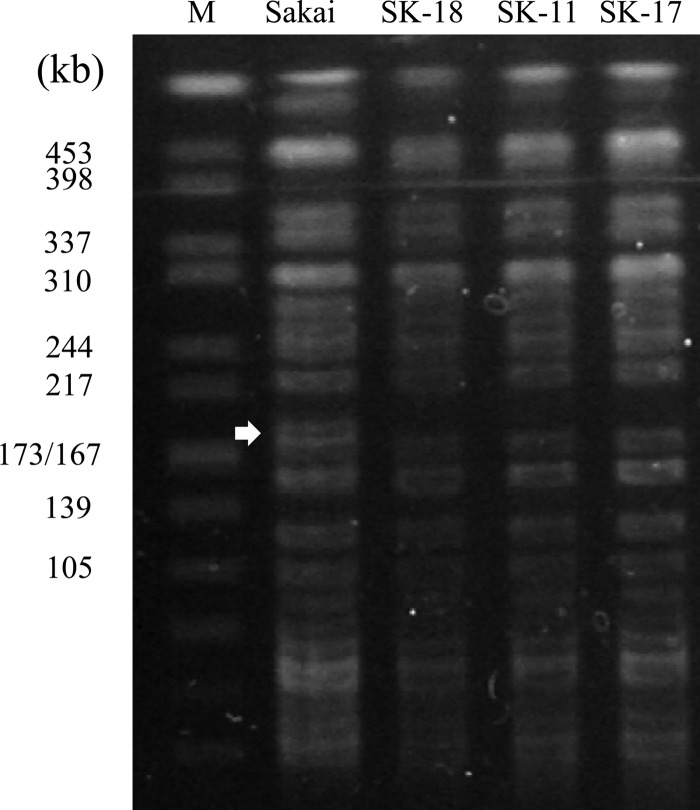
WT Sakai and derivative strains SK-18 (type I), SK-11 (type II), and SK-17 (type III) (Table 1) were analyzed by PFGE using the restriction enzyme XbaI. In the vicinity of Sp11 and Sp12 within the Sakai genome are 3 XbaI sites, positioned at nucleotides 2014272, 2199534, and 2357116 (5′ ends of the restriction sites). The PCR results described above predicted that the middle restriction site would be absent in all deletion mutants (Fig. 1), resulting in the disappearance of the 187-kb and 157-kb bands seen by XbaI-PFGE analysis of the WT and the appearance of a 298-kb (for SK-18) or 293-kb (for SK-11 and SK-17) band for the deletion derivatives. As expected (Fig. 3), the 187-kb band clearly disappeared from all three mutants, though we were unable to identify the disappearance of the 157-kb band or the emergence of new 298- or 293-kb bands. This was probably due to the presence of overlapping bands of similar sizes (Fig. 3).
Fig 3.
Comparison of PFGE profiles of E. coli O157:H7 Sakai and its derivatives. A total of 4 strains were digested using XbaI in this assay. A Salmonella enterica serotype Braenderup reference standard (H9812), also restricted with XbaI, was used as a size standard (M). The approximate size of each marker band is presented in kilobases. The white arrow highlights the 187-kb band observed only with WT Sakai.
Absence of inducible phage matching the Sp11-Sp12 region.
Our next question was whether excised Sp11-Sp12 DNA might be packaged inside phage particles. We performed high-throughout sequencing on the ciprofloxacin-induced prophage pools from Sakai. The assembly of 34,324 reads (average length, 322 bp) from the Sakai lysate resulted in a total of 30 contigs that were longer than 1 kb. However, none of these contigs were aligned within the Sp11-Sp12 tandem prophage region (data not shown).
pchB and psrC deletions have no detectable effect on the secretion of EspA and EspB.
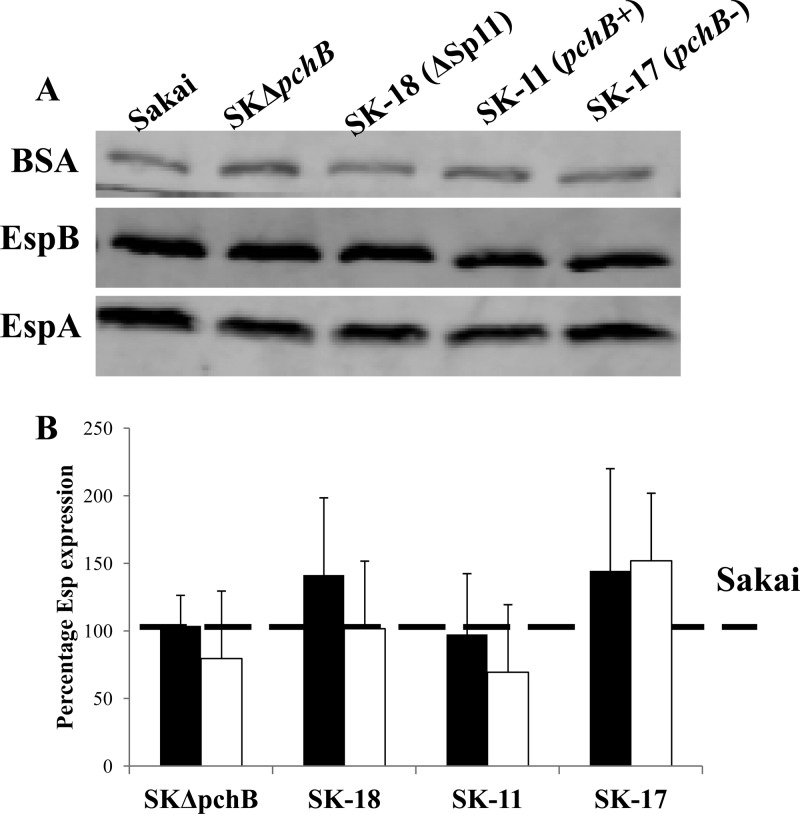
Several studies have indicated that pchB and psrC positively and negatively affect effector secretion, respectively (32, 33, 42). In order to determine whether deletion of these genes affected Esp secretion, we evaluated protein accumulation in supernatants by using a Western blot assay (Fig. 4A). Three strains, namely, SK-18 (type I; pchB psrC), SK-11 (type II; pchB+ psrC), and SK-17 (type III; pchB psrC) (Table 1), were chosen as representatives of each deletion class. WT Sakai and the pchB knockout mutant SKΔpchB (Table 1) were included as well. We normalized the secretion of Esp proteins from each mutant strain against that in WT Sakai (Fig. 4B). Although slight differences in Esp secretion levels were seen among the Sp11-Sp12 deletion mutants, the differences were not statistically significant (P = 0.54 by one-way ANOVA).
Fig 4.
Effect of pchB on secretion of Esp proteins. (A) Representative Western blot results for BSA (top) and for EspB (middle) and EspA (bottom) of Sakai and its derivatives. (B) The intensities of Esp protein bands (closed bars, EspB; open bars, EspA) were analyzed using the Odyssey application software. The relative Esp secretion levels of the derivative strains compared to wild-type Sakai were calculated as (Espderivative/BSAderivative)/(EspWT/BSAWT). All results for Esp quantification are averages for three biological repeats, and error bars represent 1 standard deviation (SD).
DISCUSSION
EHEC genomes contain a significant amount of prophage DNA, which contributes to evolution and virulence (12, 20). Sakai, one of the prototypical strains of E. coli O157:H7, carries 18 prophages and 6 prophage-like elements, amounting to 16% of the chromosome (13). Although in silico analysis suggested that most of these prophages are defective, many are still excised, undergo packaging, and can be transduced into a recipient (15). The high sequence similarity among prophages provides the potential for large-scale recombination within the bacterial chromosome (8, 9, 11), which also contributes to the genetic instability of the bacterial genome. In the current study, we identified and characterized a group of deletion events within the tandem prophages Sp11 and Sp12 in Sakai and evaluated the effects of these mutations on PFGE patterns and on the secretion of two T3SS proteins. These deletions collectively occurred at a frequency of 7.74 × 10−4, similar to that reported for the complete excision of O-island 43 and O-island 48, two other horizontally acquired regions in E. coli O157:H7, in strain EDL933 (43).
Both Sp11 and Sp12 are lambdoid prophages of approximately 46 kb (13). Previous in silico analysis (15) of these prophages identified disruptions in both of their integrase genes, which are essential for phage integration and excision. A similar organization was also observed in two other fully sequenced E. coli O157:H7 strains, i.e., EC4115 (GenBank accession number NC011353) and TW14395 (GenBank accession number NC013008) (see Fig. S2A and B and S3 in the supplemental material). However, in another fully sequenced E. coli O157:H7 strain, EDL933 (GenBank accession number NC002655), we observed a remnant of Sp12, but Sp11 was not found adjacent (see Fig. S2C). We did not observe prophages with high sequence similarity to Sp11 and Sp12 in the corresponding genomic region of E. coli O55:H7 (see Fig. S4), the predecessor of O157:H7 (19, 44–46). These observations collectively highlighted the dynamic changes within this region that have occurred during the course of O157:H7 evolution.
Genomic instability within and surrounding the Sp11-Sp12 prophage region has been reported previously. For example, Iguchi et al. (9) and Kotewicz et al. (47) identified several chromosomal inversions of between 250 kb and 1.4 Mb within regions corresponding to Sp11-Sp12. In the present study, we identified 3 different deletion events within the Sp11-Sp12 prophage region: one led to the deletion of the entire Sp11 prophage (type I), the second occurred approximately between ECs2185 and ECs2274 (type II), and the third occurred approximately between ECs2154 and ECs2234 (type III) (Fig. 1). All three classes of deletions resulted in altered XbaI PFGE patterns compared to the WT pattern (Fig. 3). Another restriction enzyme often used in PFGE assays of E. coli O157:H7 is BlnI (39). According to the published Sakai genome sequence (GenBank accession number BA000007), the two BlnI sites are located at genome positions 1690887 and 2561471 (5′ ends of the restriction sites) and therefore flank the Sp11-Sp12 region. Thus, all deletions characterized here would shift the mobility of an 871-kb WT band to one between 811 kb and 836 kb (depending on the size of the deletion). This difference in mobility would not likely be resolved in our PFGE experiments.
Two mechanisms that could explain the observed deletion events are recombination between homologous regions of Sp11 and Sp12 and integrase-mediated phage excision. Sequences with high similarity can be found in different prophages, explaining why some sequence reads were aligned within the Sp11-Sp12 region in Fig. 2. Homologous recombination between prophages was reported to be one of the primary causes of genomic deletions and rearrangements in E. coli O157:H7 (9–11, 47). Several observations from the current study support this hypothesis. First, the predicted or known boundaries of each class of deletions are located within the homologous DNA regions between Sp11 and Sp12 (Fig. 1; see Fig. S1 in the supplemental material). For example, Sp11 and Sp12 share identical sequences in the first 284 bp of the 5′ end (indicated by the vertical red bar in the ACT alignment on the left side of Fig. S1), which we speculate could promote the formation of type I deletions. Similar arguments can be made to explain type II and type III deletions (see Fig. S1). The 5′ and 3′ ends of the deletions observed in strains SK-11 (type II) and SK-17 (type III) were all internal to regions longer than 2 kb that are homologous between Sp11 and Sp12 (Fig. 2). Second, neither Sp11 nor Sp12 is predicted to encode a functional integrase (15), which is essential for phage excision. Third, we repeatedly observed a lower frequency of type I deletions than deletions of the other two classes (Fig. 1; see Fig. S1), which is in line with the previously reported correlation between the frequency of homologous recombination and the length of homologous sequence (48). Lastly, the homologous recombination hypothesis was also supported by the high-throughput sequencing results. Indeed, no phage particle responsible for Sp11 or Sp12 was identified in the inducible prophage pool of Sakai. This observation was in line with the previous suggestion from Asadulghani et al. (15) that neither Sp11 nor Sp12 is capable of being excised from the genome as an active phage.
E. coli O157:H7 acquired many of its virulence-related genes through phage-mediated horizontal gene transfer (20, 49). Some of these prophage-carried genes are transcriptional regulators of other virulence genes. Examples include the pch genes (32, 42, 50), psr genes (33), and rgdR (30), which were shown to regulate the expression of genes within the LEE island. Interestingly, Sp11 encodes both a positive LEE regulator (pchB) and a putative negative regulator (psrC) (Fig. 1). We analyzed the secretion levels of EspA and EspB from strains SK-18 (pchB psrC), SK-11 (pchB+ psrC), and SK-17 (pchB psrC), as well as a pchB mutant (SKΔpchB) and the wild-type strain. Similar to the work of Honda et al. (42), we did not observe a difference in secretion of the effector protein EspB in strains lacking pchB. The same group also previously reported that a pchB mutant of strain Sakai resulted in a slight decrease in EspA secretion (32), and for reasons that are unclear, our results did not support this observation. As for the psr regulator, Tree et al. (33) identified two homologs, designated psrA and psrB, within the genome of E. coli O157:H7 strain EDL933 and determined that a deletion of psrA but not psrB increased the secretion of EspB/D (not differentiated by the method used) and EspA. These investigators also identified four other psr homologs, designated psrC through psrF, within the genomes of other pathogenic E. coli strains, and they observed that these genes are commonly carried within prophages that also carry genes encoding T3SS effectors and pch homologs. Our data suggest that psrC, which is found within the genomes of several E. coli O157:H7 strains but is missing from EDL933 (33), is phenotypically more similar to psrB in that inactivation does not increase effector secretion. Whether overexpression would negatively affect EspA and EspB secretion and/or affect other regulatory circuits as seen with psrB remains to be determined.
In conclusion, this study provides mechanistic insight into the previously reported genomic instability within the Sp11-Sp12 prophage region. While the observed deletions may excise regulators of the E. coli O157:H7 T3SS, we were unable to demonstrate an effect on secretion, at least with EspA or EspB. The deletions were shown, however, to affect DNA fragment patterns observed by PFGE. This adds to a number of previous reports (8–11) describing mechanisms of genomic rearrangements in E. coli O157:H7 that may limit the reliability of PFGE in epidemiology studies. The genetic mechanisms promoting these deletions, their potential effects on host biology, and the frequencies of these deletions in different environments, including food, cattle, and the human gastrointestinal tract, are subjects for future study.
ACKNOWLEDGMENTS
This work was funded by USDA-NIFA grant 2009-03611, by start-up funds through the Penn State University Department of Food Science and College of Agricultural Sciences, and by a Casida Development Professorship to E.G.D.
We thank the Penn State Genomics Core Facility (University Park, PA) for generating DNA sequence information. We also thank James Kaper and Jane Michalski (University of Maryland, School of Medicine) for providing the antibodies and Mark Eppinger (University of Maryland, School of Medicine, Institute for Genome Sciences) for assistance with generating genome sequence data.
Footnotes
Published ahead of print 11 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03682-12.
REFERENCES
- 1. Griffin PM, Tauxe RV. 1991. The epidemiology of infections caused by Escherichia coli O157:H7, other enterohemorrhagic E. coli, and the associated hemolytic uremic syndrome. Epidemiol. Rev. 13:60–98 [DOI] [PubMed] [Google Scholar]
- 2. Beutin L. 2006. Emerging enterohaemorrhagic Escherichia coli, causes and effects of the rise of a human pathogen. J. Vet. Med. B 53:299–305 [DOI] [PubMed] [Google Scholar]
- 3. Nyachuba DG. 2010. Foodborne illness: is it on the rise? Nutr. Rev. 68:257–269 [DOI] [PubMed] [Google Scholar]
- 4. Fukushima H, Hashizume T, Morita Y, Tanaka J, Azuma K, Mizumoto Y, Kaneno M, Matsu-Ura MO, Konma K, Kitani T. 1999. Clinical experiences in Sakai City Hospital during the massive outbreak of enterohemorrhagic Escherichia coli O157 infections in Sakai City, 1996. Pediatr. Int. 41:213–217 [DOI] [PubMed] [Google Scholar]
- 5. Watanabe H, Wada A, Inagaki Y, Itoh K, Tamura K. 1996. Outbreaks of enterohaemorrhagic Escherichia coli O157:H7 infection by two different genotype strains in Japan, 1996. Lancet 348:831–832 [DOI] [PubMed] [Google Scholar]
- 6. Byrne CM, Erol I, Call JE, Kaspar CW, Buege DR, Hiemke CJ, Fedorka-Cray PJ, Benson AK, Wallace FM, Luchansky JB. 2003. Characterization of Escherichia coli O157:H7 from downer and healthy dairy cattle in the Upper Midwest region of the United States. Appl. Environ. Microbiol. 69:4683–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davis MA, Hancock DD, Besser TE, Call DR. 2003. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7. J. Clin. Microbiol. 41:1843–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Iguchi A, Osawa R, Kawano J, Shimizu A, Terajima J, Watanabe H. 2002. Effects of repeated subculturing and prolonged storage at room temperature of enterohemorrhagic Escherichia coli O157:H7 on pulsed-field gel electrophoresis profiles. J. Clin. Microbiol. 40:3079–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iguchi A, Iyoda S, Terajima J, Watanabe H, Osawa R. 2006. Spontaneous recombination between homologous prophage regions causes large-scale inversions within the Escherichia coli O157:H7 chromosome. Gene 372:199–207 [DOI] [PubMed] [Google Scholar]
- 10. Shima K, Wu Y, Sugimoto N, Asakura M, Nishimura K, Yamasaki S. 2006. Comparison of a PCR-restriction fragment length polymorphism (PCR-RFLP) assay to pulsed-field gel electrophoresis to determine the effect of repeated subculture and prolonged storage on RFLP patterns of Shiga toxin-producing Escherichia coli O157:H7. J. Clin. Microbiol. 44:3963–3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshii N, Ogura Y, Hayashi T, Ajiro T, Sameshima T, Nakazawa M, Kusumoto M, Iwata T, Akiba M. 2009. Pulsed-field gel electrophoresis profile changes resulting from spontaneous chromosomal deletions in enterohemorrhagic Escherichia coli O157:H7 during passage in cattle. Appl. Environ. Microbiol. 75:5719–5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. 2011. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc. Natl. Acad. Sci. U. S. A. 108:20142–20147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hayashi T, Makino K, Ohnishi M, Kurokawa K, Ishii K, Yokoyama K, Han CG, Ohtsubo E, Nakayama K, Murata T. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11–22 [DOI] [PubMed] [Google Scholar]
- 14. Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Pósfai G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA, Blattner FR. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533 [DOI] [PubMed] [Google Scholar]
- 15. Asadulghani M, Ogura Y, Ooka T, Itoh T, Sawaguchi A, Iguchi A, Nakayama K, Hayashi T. 2009. The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 5:e1000408 doi:10.1371/journal.ppat.1000408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Friedrich AW, Bielaszewska M, Zhang W-L, Pulz M, Kuczius T, Ammon A, Karch H. 2002. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J. Infect. Dis. 185:74–84 [DOI] [PubMed] [Google Scholar]
- 17. Werber D, Fruth A, Buchholz U, Prager R, Kramer MH, Ammon A, Tschäpe H. 2003. Strong association between Shiga toxin-producing Escherichia coli O157 and virulence genes stx2 and eae as possible explanation for predominance of serogroup O157 in patients with haemolytic uraemic syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 22:726–730 [DOI] [PubMed] [Google Scholar]
- 18. Kudva IT, Evans PS, Perna NT, Barrett TJ, Ausubel FM, Blattner FR, Calderwood SB. 2002. Strains of Escherichia coli O157:H7 differ primarily by insertions or deletions, not single-nucleotide polymorphisms. J. Bacteriol. 184:1873–1879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kyle JL, Cummings CA, Parker CT, Quiñones B, Vatta P, Newton E, Huynh S, Swimley M, Degoricija L, Barker M, Fontanoz S, Nguyen K, Patel R, Fang R, Tebbs R, Petrauskene O, Furtado M, Mandrell RE. 2012. Escherichia coli serotype O55:H7 diversity supports parallel acquisition of bacteriophage at Shiga toxin phage insertion sites during evolution of the O157:H7 lineage. J. Bacteriol. 194:1885–1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, Kodama T, Abe H, Nakayama K, Kurokawa K. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:17939–17944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Deng W, Li Y, Vallance BA, Finlay BB. 2001. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect. Immun. 69:6323–6335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 92:1664–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McDaniel TK, Kaper JB. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399–407 [DOI] [PubMed] [Google Scholar]
- 24. Creuzburg K, Middendorf B, Mellmann A, Martaler T, Holz C, Fruth A, Karch H, Schmidt H. 2011. Evolutionary analysis and distribution of type III effector genes in pathogenic Escherichia coli from human, animal and food sources. Environ. Microbiol. 13:439–452 [DOI] [PubMed] [Google Scholar]
- 25. Delahay RM, Frankel G, Knutton S. 2001. Intimate interactions of enteropathogenic Escherichia coli at the host cell surface. Curr. Opin. Infect. Dis. 14:559–565 [DOI] [PubMed] [Google Scholar]
- 26. Donnenberg MS, Kaper JB, Finlay BB. 1997. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 5:109–114 [DOI] [PubMed] [Google Scholar]
- 27. Elliott SJ, Wainwright LA, McDaniel TK, Jarvis KG, Deng Y, Lai L-C, McNamara BP, Donnenberg MS, Kaper JB. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1–4 [DOI] [PubMed] [Google Scholar]
- 28. Knutton S, Rosenshine I, Pallen MJ, Nisan I, Neves BC, Bain C, Wolff C, Dougan G, Frankel G. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wolff C, Nisan I, Hanski E, Frankel G, Rosenshine I. 1998. Protein translocation into host epithelial cells by infecting enteropathogenic Escherichia coli. Mol. Microbiol. 28:143–155 [DOI] [PubMed] [Google Scholar]
- 30. Flockhart AF, Tree JJ, Xu X, Karpiyevich M, McAteer SP, Rosenblum R, Shaw DJ, Low CJ, Best A, Gannon V, Laing C, Murphy KC, Leong JM, Schneiders T, La Ragione R, Gally DL. 2012. Identification of a novel prophage regulator in Escherichia coli controlling the expression of type III secretion. Mol. Microbiol. 83:208–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Islam MS, Bingle LEH, Pallen MJ, Busby SJW. 2011. Organization of the LEE1 operon regulatory region of enterohaemorrhagic Escherichia coli O157:H7 and activation by GrlA. Mol. Microbiol. 79:468–483 [DOI] [PubMed] [Google Scholar]
- 32. Iyoda S, Watanabe H. 2004. Positive effects of multiple pch genes on expression of the locus of enterocyte effacement genes and adherence of enterohaemorrhagic Escherichia coli O157:H7 to HEp-2 cells. Microbiology 150:2357–2371 [DOI] [PubMed] [Google Scholar]
- 33. Tree JJ, Roe AJ, Flockhart A, McAteer SP, Xu X, Shaw D, Mahajan A, Beatson SA, Best A, Lotz S, Woodward MJ, La Ragione R, Murphy KC, Leong JM, Gally DL. 2011. Transcriptional regulators of the GAD acid stress island are carried by effector protein-encoding prophages and indirectly control type III secretion in enterohemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 80:1349–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Philippe N, Alcaraz J-P, Coursange E, Geiselmann J, Schneider D. 2004. Improvement of pCVD442, a suicide plasmid for gene allele exchange in bacteria. Plasmid 51:246–255 [DOI] [PubMed] [Google Scholar]
- 35. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murphy KC, Campellone KG. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic E. coli. BMC Mol. Biol. 4:11 doi:10.1186/1471-2199-4-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. 2006. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 61:1267–1282 [DOI] [PubMed] [Google Scholar]
- 39. Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, Wrigley D, Barrett T, Ribot E. 2005. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43:1045–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang X, McDaniel AD, Wolf LE, Keusch GT, Waldor MK, Acheson DW. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664–670 [DOI] [PubMed] [Google Scholar]
- 41. Carver TJ, Rutherford KM, Berriman M, Rajandream M-A, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21:3422–3423 [DOI] [PubMed] [Google Scholar]
- 42. Honda N, Iyoda S, Yamamoto S, Terajima J, Watanabe H. 2009. LrhA positively controls the expression of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli by differential regulation of their master regulators PchA and PchB. Mol. Microbiol. 74:1393–1411 [DOI] [PubMed] [Google Scholar]
- 43. Bielaszewska M, Middendorf B, Tarr PI, Zhang W, Prager R, Aldick T, Dobrindt U, Karch H, Mellmann A. 2011. Chromosomal instability in enterohaemorrhagic Escherichia coli O157:H7: impact on adherence, tellurite resistance and colony phenotype. Mol. Microbiol. 79:1024–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leopold SR, Magrini V, Holt NJ, Shaikh N, Mardis ER, Cagno J, Ogura Y, Iguchi A, Hayashi T, Mellmann A, Karch H, Besser TE, Sawyer SA, Whittam TS, Tarr PI. 2009. A precise reconstruction of the emergence and constrained radiations of Escherichia coli O157 portrayed by backbone concatenomic analysis. Proc. Natl. Acad. Sci. U. S. A. 106:8713–8718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wick LM, Qi W, Lacher DW, Whittam TS. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 187:1783–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhou Z, Li X, Liu B, Beutin L, Xu J, Ren Y, Feng L, Lan R, Reeves PR, Wang L. 2010. Derivation of Escherichia coli O157:H7 from its O55:H7 precursor. PLoS One 5:e8700 doi:10.1371/journal.pone.0008700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kotewicz ML, Jackson SA, LeClerc JE, Cebula TA. 2007. Optical maps distinguish individual strains of Escherichia coli O157:H7. Microbiology (Reading, Engl.) 153:1720–1733 [DOI] [PubMed] [Google Scholar]
- 48. Fujitani Y, Yamamoto K, Kobayashi I. 1995. Dependence of frequency of homologous recombination on the homology length. Genetics 140:797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64–67 [DOI] [PubMed] [Google Scholar]
- 50. Yang Z, Kim J, Zhang C, Zhang M, Nietfeldt J, Southward CM, Surette MG, Kachman SD, Benson AK. 2009. Genomic instability in regions adjacent to a highly conserved pch prophage in Escherichia coli O157:H7 generates diversity in expression patterns of the LEE pathogenicity island. J. Bacteriol. 191:3553–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]