Abstract
Resources from the Sinorhizobium meliloti Rm1021 open reading frame (ORF) plasmid libraries were used in a medium-throughput method to construct a set of 50 overlapping deletion mutants covering all of the Rm1021 pSymA megaplasmid except the replicon region. Each resulting pSymA derivative carried a defined deletion of approximately 25 ORFs. Various phenotypes, including cytochrome c respiration activity, the ability of the mutants to grow on various carbon and nitrogen sources, and the symbiotic effectiveness of the mutants with alfalfa, were analyzed. This approach allowed us to systematically evaluate the potential impact of regions of Rm1021 pSymA for their free-living and symbiotic phenotypes.
INTRODUCTION
The genome of the soil bacterium Sinorhizobium meliloti Rm1021 contains three replicons: the chromosome and two megaplasmids, pSymA and pSymB (1). The smallest megaplasmid, pSymA, is 1.3 Mb long and contains many genes defining the ability of the bacteria to establish a nitrogen-fixing symbiotic association with forage legumes. pSymA also contains open reading frames (ORFs) that have been shown or are predicted to be involved in a range of functions that allow S. meliloti to survive and successfully compete with other soil microbes (2).
The complete genome sequence of S. meliloti strain Rm1021 has been determined (3), and this enabled the development of several tools allowing genome-wide profiling of S. meliloti Rm1021 (4–6). The S. meliloti ORFeome project cloned 6,314 of the S. meliloti Rm1021 ORFs into Gateway entry vectors (7, 8) that allow the subsequent in vivo transfer of cloned ORFs into various destination vectors. This system permits using standardized approaches to manipulate the whole set of genes. For example, 6,290 ORF-specific transcriptional fusions to β-glucuronidase (gusA) and green fluorescent protein (gfp) reporter genes were constructed and used to analyze the expression pattern of the 30 sugar kinase genes annotated in S. meliloti (9). The resources generated in the ORFeome project are suitable for generation of deletion mutations using an FLP-FLP recombination target (FRT) recombination strategy (7); genes predicted to code for a denitrification pathway (7) or to be exported using the twin arginine transport pathway (Tat) (10) have been deleted. This work describes the construction of a set of 50 overlapping deletions that cover all of the pSymA plasmid except the origin of replication and linked replication genes. Free-living and symbiotic phenotypes of the mutants were analyzed to link phenotypic characteristics of S. meliloti Rm1021 to specific gene clusters located on pSymA.
MATERIALS AND METHODS
Bacterial strains and media.
Strains and plasmids used in this study are listed in Table 1. S. meliloti strains were grown at 30°C on yeast extract-mannitol (YMB), minimal NH4 medium (Min-NH4) (17), minimal-mannitol-NH4 (MMNH4) (17), or TY medium (18). Supplements were typically added at 2 g/liter for carbon sources and 0.2 g/liter for nitrogen sources. Escherichia coli strains were grown on LB (19) at 37°C unless otherwise noted. Antibiotics were filter sterilized and added to the medium at the following concentrations (μg/ml): tetracycline (Tet), 10 for replicating plasmids and 1 for plasmids inserted into the S. meliloti chromosome; chloramphenicol (Cam), 50; spectinomycin (Spc), 100; streptomycin (Str), 200; kanamycin (Kan), 40; rifampin (Rif), 100; and gentamicin (Gen), 50 (except as indicated for specific procedures, e.g., Spc200 = 200 μg/ml spectinomycin). To select for plasmid loss, 5% sucrose was added to media.
Table 1.
Plasmids and strains
| Strain or plasmid | Relevant genotype or characteristic | Source or reference |
|---|---|---|
| E. coli strains | ||
| DB3.1 | F− gyrA462 endA1 Δ(sr1-recA) mcrB mrr hsdS20(rB−mB−) supE44 ara-14 galK2 lacY1 proA2 rpsL20(Strr) xyl-5 Δleu mtl-1 | 11 |
| DB3.1 λpir | λpir lysogen of DB3.1 | 7 |
| DH5α | φ80lacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rk−, mk+) supE44 relA1 deoR phoA Δ(lacZYA-argF)U169 | 12 |
| DH5α λpir | λpir lysogen of DH5α | 13 |
| DH5αR λpir | Rifr derivative of DH5α λpir | 7 |
| S17-1 | pro hsdR recA (RP4-2[Tc::Mu][Km::Tn7]) | 14 |
| S. meliloti strains | ||
| Rm1021 | SU47 str-21 | 15 |
| 1021::pRG1SMaxxxx | Rm1021 with pRG1 construct integrated into pSymA | This work |
| 1021::pD2SMaxxxx | Rm1021 with pD2 construct integrated into pSymA | This work |
| 1021::pRG1SMaxxxxΔpD2SMaxxxx | Rm1021 with a deletion of DNA between but not including the numbered pSymA ORFs | This work |
| Rm1021xpK19gntR-ins | Rm1021 with pK19sacBmob integrated into gntR | This work |
| Rm1021xpK19SMa2341 | Rm1021 with pK19sacBmob integrated into SMa2341 | This work |
| Rm1021xpK19SMa2343 | Rm1021 with pK19sacBmob integrated into Ma2343 | This work |
| Rm1021xpK19SMa2349 | Rm1021 with pK19sacBmob integrated into SMa2349 | This work |
| Rm1021xpK19SMa5012 | Rm1021 with pK19sacBmob integrated into SMa5012 | This work |
| Rm1021xpK19SMa2357 | Rm1021 with pK19sacBmob integrated into SMa2357 | This work |
| Rm1021xpK19SMa1288 | Rm1021 with pK19sacBmob integrated into SMa1288 | This work |
| Rm1021xpK19SMa1299 | Rm1021 with pK19sacBmob integrated into SMa1299 | This work |
| Rm1021xpK19SMa1292 | Rm1021 with pK19sacBmob integrated into SMa1292 | This work |
| Rm1021xpK19SMa1296 | Rm1021 with pK19sacBmob integrated into SMa1296 | This work |
| Rm1021xpK19SMa0796 | Rm1021 with pK19sacBmob integrated into SMa0796 | This work |
| Plasmids | ||
| pMK2017 | Ampr; FRT-ccdB-Camr-FRT cassette | 7 |
| pMK2016-2 | Spcr Strr; oriV oriTcolE1 with FRT cassette from pMK2015 | This work |
| pMK2030 | Tetr Camr; gfp and gusA reporter vector | 9 |
| pBH474 | Genr RK2 derivative FLP | 7 |
| pESMaxxxx | Kanr; entry vector with ORF inserted | 8 |
| pRG1SMaxxxx | pMK2030 reporter plasmid with ORF inserted | This work |
| pD2SMaxxxx | pMK2016-2 destination vector with ORF inserted | This work |
| pK19sacBmob | Kanr | 16 |
| pK19gntR-ins | pK19mob (200 bp of SMa0246) | This work |
| pK19SMa2341_ins | pK19mob (200 bp of SMa2341) | This work |
| pK19SMa2343_ins | pK19mob (200 bp of SMa2343) | This work |
| pK19SMa2349_ins | pK19mob (200 bp of SMa2349) | This work |
| pK19SMa5012_ins | pK19mob (200 bp of SMa5012) | This work |
| pK19SMa2357_ins | pK19mob (200 bp of SMa2357) | This work |
| pK19SMa1288_ins | pK19mob (200 bp of SMa1288) | This work |
| pK19SMa1299_ins | pK19mob (200 bp of SMa1299) | This work |
| pK19SMa1292_ins | pK19mob (200 bp of SMa1291) | This work |
| pK19SMa1296_ins | pK19mob (200 bp of SMa1296) | This work |
| pK19SMa0796_ins | pK19mob (200 bp of SMa0796) | This work |
Plasmid constructions.
pMK2016-2 was constructed (Fig. 1) to allow deletions to be generated when paired with strains derived from the transcriptional fusion plasmids described earlier (9). A total of 50 pRG1SMaxxxx reporter constructs that were to be one of the deletion endpoints were selected from the ORF transcriptional fusion plasmid library (9). In vivo recombination via pentaparental mating (7) was used to transfer another set of 50 pSymA ORFs from entry vectors to pMK2016-2 to create the pD2SMaxxxx plasmids in E. coli. This mating used DH5αR as the final recipient and selected pD2SMaxxxx recombinants on LB Spc100 Rif100 agar. Detailed methods for high-throughput plasmid and strain construction are included in SI-1 in the supplemental material.
Fig 1.
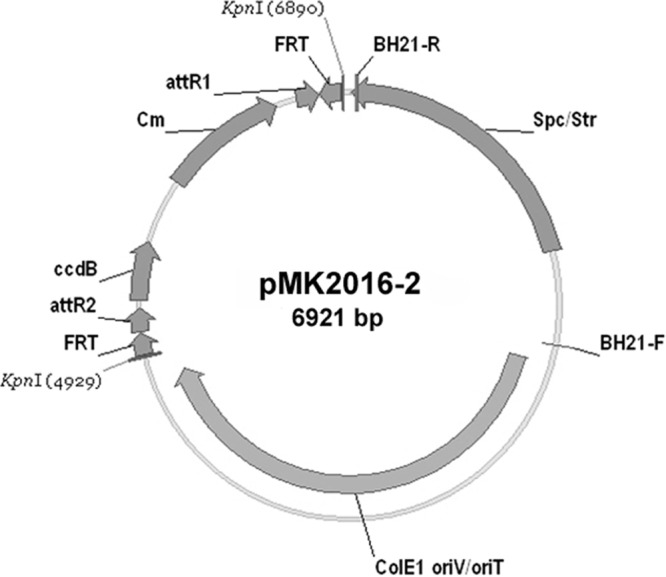
Deletion plasmid pMK2016-2. pMK2016-2 was constructed by using KpnI to excise the attR1-Camr-ccdB-attR2 cassette from pMK2016, in which the FRT elements face outward, and replacing it with a KpnI fragment from pMK2017 containing an attR1-Camr-ccdB-attR2 cassette, in which the FRT elements face inward. Together with the pMK2030 reporter plasmid, pMK2016-2 can be used to construct deletions with defined endpoints.
Construction of deletions in Rm1021 pSymA.
A transcriptional fusion plasmid (pRG1SMaxxxx) and a deletion plasmid (pD2SMaxxxx) were sequentially introduced into S. meliloti Rm1021, and then the intervening DNA was removed by FLP-FRT-mediated recombination (Fig. 2). Triparental matings using DH5αR λpir (pRG1SMaxxxx), DH5α (pRK2013), and Rm1021 were done as described in reference 9 to integrate the reporter fusion plasmids into the chromosome by selecting homologous recombinants on MMNH4 Tet1 Str200 medium. Recombination was confirmed by PCR using two primers, one within the plasmid and one from the adjacent flanking regions. To insert the other endpoint of a desired deletion into these strains, another triparental mating was carried out, using DH5αR (pD2SMaxxxx), DH5α (pRK2013), and the appropriate Rm1021::pRG1SMaxxxx strain. These matings were streaked for isolation on MM NH4 Str200 Tet1 Spc400 agar plates, selecting for strains containing both the pRG1SMa and pD2SMa integrated plasmids and counterselecting against the donors.
Fig 2.
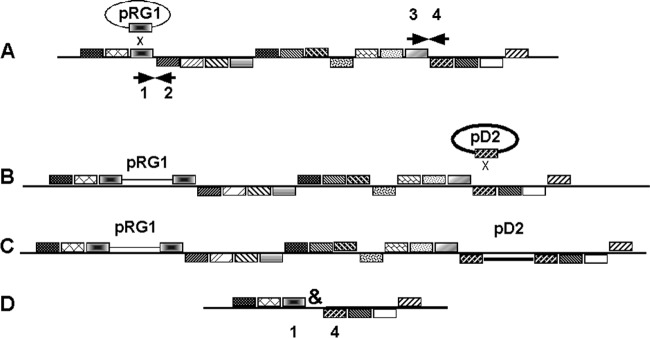
Construction of deletions. To construct a deletion missing the region of pSymA from gene 2 through gene 3, plasmids were constructed that contained ORF1 in a plasmid pRG1 context and ORF4 in a plasmid pD2 context. The pRG1 plasmid was recombined into pSymA first (transition from panel A to panel B, A→B), selecting for Tetr clones, and then the pD2 plasmid was introduced (B→C), selecting for Spcr clones (C). After the FLP plasmid pBH474 was mated into the strain, recombination between FRT sequences at the left of the pRG1 insert and the right of the pD2 insert leads to deletion of the desired segment and generates a scar sequence (&).
Using the Flp recombinase to convert the double integrant strains to the desired pSymA deletion mutants was accomplished as described in reference 7 by introducing the FLP-containing plasmid pBH474. A single Genr colony from each mating plate was streaked to isolate colonies, and about 25 colonies were then picked and tested for growth on MM NH4 Tet1, MM NH4 Spc200, and MM NH4 Str200 agar. Colonies that were Tets and Spcs were streak plated on YMB-5% sucrose to eliminate pBH474. Colonies appearing on the sucrose plates were tested for Gens, and those that had lost all three antibiotic resistances were considered candidate deletions.
To confirm the deletions, paired gene-specific PCR primers used originally to amplify and clone the pSymA ORFs for the entry vector library construction (8) were chosen that would anneal to the distal ends of ORFs flanking each deletion (ORFs 1 and 4 in Fig. 2; see also SI-2 in the supplemental material) in order to amplify the intervening DNA that remains. In some cases, this PCR failed. We hypothesized that this was due to internal hairpin structures (shown as “&” in Fig. 2) generated during the resolution of the FRT sequences. To test this, we paired a flanking primer with one of two other primers, FRT-scar-R or FRT-scar-F (see SI-3 and Table S1 in the supplemental material), each of which anneals to a sequence within the partially symmetrical sequence that would be generated by the FRT recombination, shortening the potential region of symmetry that might be amplified. In all cases where the previous PCR had failed, using the appropriate FRT-scar primer generated a PCR fragment of the expected size.
Genetic techniques.
DNA manipulations were carried out using standard procedures (19). To construct S. meliloti single-crossover mutations in a specific gene, approximately 200 bp in the middle of the gene was amplified by PCR, and this DNA fragment was cloned into pK19mob (14). These plasmids were then recombined into S. meliloti to generate gene-specific mutations. The primers used for these PCRs are listed in Table S1 in the supplemental material.
Nadi test.
The pSymA deletion mutants were tested for cytochrome oxidase activity using the Nadi cytochrome oxidase test (20) in which indophenol blue is produced by a reaction that depends on the level of bacterial cytochrome c and cytochrome c oxidases. Mutants that remain white have decreased respiration via cytochrome c oxidase.
Growth measurements.
To evaluate growth, S. meliloti strains were first grown on YMB plates for 48 h. Cells were suspended in 1 ml of Min-salt solution (minimal medium without either a carbon or nitrogen source) with a final optical density at 600 nm (OD600) of around 0.600. A total of 2 μl of the cell suspension was added to microplate wells containing 200 μl of minimal NH4 medium (Min-NH4) that contained 2 g/liter of the indicated carbon compound or minimal mannitol medium (MM) that contained 0.2 g/liter of the indicated nitrogen compound. Growth at 30°C was monitored for 48 to 72 h by measuring the absorbance at 600 nm every 30 min using a SPECTRAmax 250 microplate spectrophotometer system (Molecular Devices, Sunnyvale, CA).
Alternatively, the cells were suspended in Min-salt solution to an OD600 of 0.5, and successive 10-fold dilutions of the cell suspensions were prepared in a 96-well microplate. Aliquots of these dilutions were then transferred onto solid medium using a sterile bolt replicator. After 3 to 7 days, the sizes of colonies were scored.
Plant tests.
Alfalfa (Medicago sativa cv. Champ) was used for all nodulation studies (21). Four to five weeks after inoculation, plants were harvested and shoot dry mass and root nodule formation were examined.
RESULTS AND DISCUSSION
Construction of an S. meliloti Rm1021 pSymA megaplasmid deletion library.
Fifty pairs of adjacent ORFs were chosen from pSymA as deletion endpoints using the criteria that the genes were transcribed convergently and that they had approximately 25 ORFs between them (Fig. 2). The convergent transcription criterion was used to minimize the possibility that a deletion would disrupt operons and generate null mutations without removing the gene itself.
Procedures for generating deletion mutants are described briefly in Materials and Methods and in more detail in SI-1 in the supplemental material. For each deletion, in vivo recombination mediated by lambda integrase using a pentaparental mating in Escherichia coli was used to generate a pRG1 reporter plasmid to serve as one end of the deletion and a corresponding pD2 deletion plasmid for the other end of the deletion. As indicated in Fig. 2, these were sequentially mated into S. meliloti Rm1021, selecting for homologous recombination by selecting for the antibiotic resistance gene carried on each plasmid. Plasmid pBH474, carrying the yeast FLP recombinase, was then mated into the S. meliloti strain carrying both plasmids to generate the desired deletion. After curing pBH474, the deletions were first confirmed by loss of the antibiotic resistances and then by PCR.
Phenotypic characterization of the mutants.
The strategy described above allowed us to generate a set of deletions covering the entire pSymA (Fig. 3; see Table S2 in the supplemental material) with the exception of the origin of replication. All pSymA deletion strains were viable, indicating that none of the pSymA genes were absolutely required for free-living growth. This is generally consistent with the previously published isolation and analysis of a pSymA-minus derivative of S. meliloti Rm2011, a related strain (2). Rm2011 lacking pSymA did not grow on MMNH4, but none of the Rm1021 deletions we recovered had this property. This could be due to strain differences between Rm1021 and Rm2011 or to the growth defect of Rm2011 lacking pSymA being due to lack of more than one subregion of pSymA.
Fig 3.
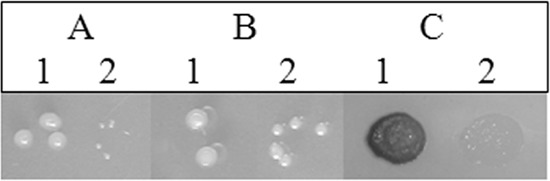
Phenotype complementation with ferric citrate. Strains were grown on Min-succinate-NH4 (A) and Min-succinate-NH4 supplemented with 1 mM ferric citrate (B). (C) TMPD oxidation. 1, Rm1021; 2, Rm1021::SMa2321ΔSMa2359 strain.
Cytochrome c respiration and iron and potassium uptake.
A significant number of the genes comprising the S. meliloti free-living cytochrome c respiratory chains as well as the genes involved in cytochrome c maturation have been identified (22). Some of these genes are located in pSymA, including three reiterations of the fixNOQP genes (1, 23) encoding the symbiotically essential high-affinity terminal oxidase required for microaerobic respiration (21, 24, 25). These operons and the cytochrome-related ORFs (SMa0007, SMa0657, SMa0659, SMa1232, SMa1233, and SMa1487) are not involved in free-living cytochrome c respiration. To identify genes that might be involved in cytochrome c respiration in free-living cells, we screened the deletion mutant library strains for the ability of the strains to oxidize TMPD (N,N,N′,N′-tetramethyl-p-phenylenediamine), which accepts electrons from cytochrome c oxidase and generates a blue color. After exposure to TMPD, the Rm1021::SMa2321ΔSMa2359 strain remained white, suggesting that at least one gene missing in the strain was involved in cytochrome c respiration (see SI-4 in the supplemental material). The Rm1021::SMa2321ΔSMa2359 strain grew poorly on MM-NH4 and Min-succinate-NH4 media (see SI-5A and B), but it grew well on complex media such as YMB (see SI-5C) or LB (see Table S3).
The Rm1021::SMa2321ΔSMa2359 strain was missing 27 annotated ORFs (see Table S4 in the supplemental material), including rhbABCDEF and rthA, genes that are involved in biosynthesis and transport of rhizobactin Rm1021, an iron siderophore produced under iron limitation (26). To test whether deleting the rhb and rth genes caused the iron starvation and consequential restriction in growth and cytochrome c respiration of the mutant, we analyzed the growth and TMPD oxidase activity of the mutant in the presence of 1 mM ferric citrate. Ferric citrate rescued the growth of the mutant on Min-succinate-NH4 (see SI-5D in the supplemental material; Fig. 3), but it did not restore a blue TMPD oxidase phenotype (Fig. 3). Moreover, visualization of c-type cytochromes showed that the deletion strain produces less cytochrome c than Rm1021 even when the strains were grown in the presence of ferric citrate (see SI-6). This suggested that iron deficiency did not directly decrease cytochrome c respiration.
The Rm1021::SMa2321ΔSMa2359 strain also lacks the kdpABC operon (SMa2325 to SMa2333), which codes for a unique pSymA potassium transport system (see Table S4 in the supplemental material). KDP is a P-type ATPase that plays an important role in potassium uptake under low K+ concentration (27, 28). To eliminate the possibility that the decrease of cytochrome c respiration is linked to K+ deficiency, we grew the strain in the presence of 10 mM KCl and evaluated TMPD activity. Supplementation of Min-succinate-HN4 medium with KCl did not reverse the mutant's growth or TMPD phenotypes.
The inability of Fe citrate or KCl to complement the defect in cytochrome c respiration suggests that ORFs SMa2325 to SMa2339 were probably not involved in Rm1021 respiration. Moreover, SMa2323 was also deleted in the Rm1021::pRG1SMa2291ΔpD2SMa2325 strain, which did not exhibit a TMPD̄ phenotype, suggesting that SMa2323 is also not involved in cytochrome c respiration. This left 10 ORFs, SMa2341 to SMa2357, which could potentially play a role in S. meliloti respiration. The predicted functions of the genes do not suggest which ones might be involved in respiration.
Insertion mutations were constructed in three of the remaining single genes (SMa2341 [transcriptional regulator], SMa2357 [adenylate/guanylate cyclase], and SMa2343 [oxidoreductase]) and in SMa5012 and SMa2349, genes at the beginning of two putative operons. The TMPD phenotype of these mutants was tested, but none of the constructed mutants was TMPD̄.
Osmotic stress.
We evaluated the effect of pSymA deletions on Rm1021 osmoadaptation by growing the strains on TY medium with 2%, 2.5%, and 3% NaCl. Only one strain, Rm1021::pRG1SMa0785ΔpD2SMa0814, was more sensitive to NaCl than Rm1021. The strain was missing 18 ORFs (see Table S4 in the supplemental material), four of which were also deleted in another mutant that had normal salt tolerance. Two others were associated with transposase and ISRm23 insertion sequences. One of the remaining 12 ORFs, SMa0796, codes for a putative aldehyde dehydrogenase that is 82% identical to Rhizobium leguminosarum bv. trifolii WSM1325 protein annotated as a betaine aldehyde dehydrogenase (BADH) and 39% identical and 98% similar to S. meliloti Rm1021 BetB BADH. Insertion mutations in SMa0796 did not decrease the ability of Rm1021 to resist high-salt concentrations, but while this suggests that SMa0796 is not critical for Rm1021 osmoadaptation, it is possible that the insertions studied have residual activity.
The annotation of pSymA predicted several genes that could be involved in S. meliloti Rm1021 osmoadaptation. Among them are a putative BADH (betB2/SMa1731); a glycine betaine ABC transporter; a periplasmic solute-binding protein, SMa1729; a transcriptional regulator, SMa1726, which is homologues to the betI transcriptional repressor of bet operon; and lipase, SMa1727. Osmoadaptation was not affected in the Rm1021::pRG1SMa1696ΔpD2SMa1740 strain, which lacks SMa1726, SMa1727, SMa1729, and SMa1731, indicating that these genes might not be involved in the S. meliloti Rm1021 salt stress response.
Growth properties of the mutants.
We tested the ability of the deletion mutants to assimilate a variety of carbon and nitrogen sources (see Table S3 in the supplemental material). The Rm1021::pRG1SMa0273ΔpD2SMa0335 strain grew very slowly on all rich and defined media tested (see Table S3). Growth of the Rm1021::pRG1SMa1005ΔpD2SMa1082 strain was severely affected on TY, YMB, and LB complex media and on several defined media. However, the Rm1021::pRG1SMa1005ΔpD2SMa1082 strain grew relatively well on Min-mannitol medium with glutamine, arginine, asparagine, or glycine as nitrogen sources or on Min-NH4 medium with γ-aminobutyrate (GABA) or succinate as carbon sources. The Rm1021::pRG1SMa0554ΔpD2SMa0599 strain could not utilize mannitol as a carbon source but grew well on Min-NH4 supplemented with succinate, malate, arabinose, galactose, or trehalose, suggesting that it could have specific defects in mannitol assimilation (see Table S3). The Rm1021::pRG1SMa0748ΔpD2SMa0787 strain formed much bigger colonies than Rm1021 on almost all defined media (see Table S3).
Glycine, GABA, serine, gluconate, and inosine utilization.
An S. meliloti Rm2011 mutant lacking pSymA lost the ability to use inosine, GABA, serine, glycine, or gluconate as sole carbon sources (2). We tested the Rm1021 pSymA deletion mutants for their ability to use these compounds.
Glycine.
S. meliloti strain Rm1021, as well as all deletion mutants, did not grow on Min-NH4 medium supplemented with glycine as the carbon source, but only the mutants with general growth defects could not use glycine as the nitrogen source when it was added to Min-mannitol medium.
GABA.
We did not find any mutants with poor growth associated specifically with the inability to assimilate GABA. This suggests that the ability of S. meliloti Rm1021 to assimilate GABA as a carbon source may rely on more than one region of pSymA. Alternatively, the strains Rm1021 and Rm2011 might have some differences in the genetic determinants of GABA assimilation.
Serine, gluconate, and inosine utilization.
S. meliloti Rm1021 and all but one deletion mutant grew slowly on these compounds. The exception, the Rm1021::pRG1SMa1266ΔpD2SMa1302 strain, did not grow on serine, gluconate, or inosine as the sole carbon source (see Table S3 in the supplemental material). This deletion strain was missing 16 genes (see Table S4). The first and last of these genes were also deleted in other mutants that were not affected in serine, gluconate, or inosine utilization.
Previously, it was shown that a putative tripartite ATP-independent periplasmic (TRAP) transporter that is negatively regulated by a GntR family regulator is required for gluconate utilization by S. meliloti Rm1021 (29) and that a mutation in the GntR repressor promoted the ability of Rm1021 to grow on gluconate. It is possible that the Rm1021::pRG1SMa1266ΔpD2SMa1302 deletion removed an activator of the TRAP transporter, causing a complete loss of gluconate uptake by the mutant. In our experiments, introduction of a gntR mutation into the Rm1021 genome increased the growth rate of Rm1021 on gluconate but not on serine or inosine. Adding the gntR mutation to the Rm1021::pRG1SMa1266ΔpD2SMa1302 strain did not change the strain's ability to use any of these compounds.
To identify the genes that might be involved in serine, gluconate, and inosine utilization, we generated insertion mutations in SMa1288 (carboxyl-lyase), SMa1291 (protease), SMa1296 (Zn-dependent AdhA1 alcohol dehydrogenase) (30), and SMa1299 (transcriptional regulator). The Rm1021xpK19SMa1296 mutant failed to grow on inosine but grew like Rm1021 on serine and grew slightly slower on gluconate (Fig. 4). However, it still grew better on gluconate than the Rm1021::pRG1SMa1266ΔpD2SMa1302 strain. This indicated that adhA1 might be critical for inosine catabolism and play some role in gluconate catabolism in Rm1021.
Fig 4.
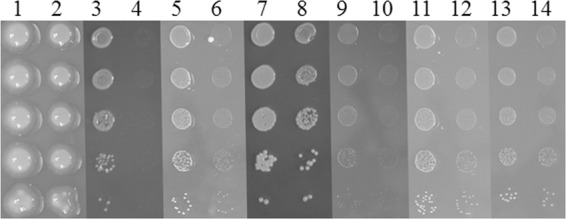
Growth properties of the Rm1021xpK19SMa1296 strain. The Rm1021 (1, 3, 5, 7, 9, 11, 13) and Rm1021xpK19SMa1296 (2, 4, 6, 8, 10, 12, 14) strains were grown on MMNH4 (1, 2) and Min-NH4 supplemented with 0.2% inosine (3, 4), 0.2% gluconate (5, 6), 0.2% serine (7, 8), 0.5 mM riboflavin (9, 10), 0.01% ribose (11, 12), or 0.01% adonitol (13, 14). The rows are sequential 10-fold dilutions of the cultures.
Mutation in adhA1 also affected the ability of Rm1021 to use riboflavin for growth (Fig. 4). One possible step in inosine and riboflavin catabolism is hydrolysis of the glycosidic bond between the ribose ring and the hypoxanthine and flavin moieties, respectively. This hydrolysis would enable further catabolism of the products. The Rm1021::pRG1SMa1266ΔpD2SMa1302 and Rm1021xpK19SMa1296 strains had decreased ability to grow on 0.01% ribose or adonitol (ribitol) (Fig. 4). E. coli can use the carbohydrate moiety of nucleosides through the pentose phosphate pathway in reactions catalyzed by ribokinase, ribose phosphate isomerase, and ribulose-5-phosphate-3-epimerase (31) with ribulose-5P as one of the intermediates. Geddes and Oresnik (32) showed that adonitol enters central metabolism in S. meliloti through a pentose phosphate intermediate that leads to production of ribulose-5P. A chromosomal locus necessary for the conversion of adonitol into ribulose-5P was found on the Rm1021 chromosome (32). Based on the growth properties of the Rm1021::pRG1SMa1266ΔpD2SMa1302 and Rm1021xpK19SMa1296 strains, we speculate that an alcohol dehydrogenase located on pSymA, adhA1, might be required to catabolize carbohydrates derived from nucleoside catabolism through the 5-carbon pentose phosphate pathway.
Nodulation and nitrogen fixation.
As expected, most deletion mutants had normal symbiotic performance, but four of the mutants did not form an effective symbiosis with alfalfa (Table 2). The Rm1021::pRG1SMa0785ΔpD2SMa0814 and Rm1021::pRG1SMa1191ΔpD2SMa1232 strains formed nodules on alfalfa roots (Nod+) but were defective in nitrogen fixation (Fix−) (Table 2). The Rm1021::pRG1SMa0785ΔpD2SMa0814 strain lacks the FdxN ferredoxin, which is essential for S. meliloti nitrogen fixation (33). A large cluster of fix genes (fixGHI1S1, fixN1O1Q1P1, fixLJ, fixT1, fixK1, and fixM) was deleted in the Rm1021::pRG1SMa1191ΔpD2SMa1232 strain. Deletion of fixN1O1Q1P1 (23), fixT (34), fixK1 (35), and fixM (35) does affect nitrogen fixation because of complementation by other regions of the genome (23, 35), but deletion of fixLJ (36) should result in a Fix− phenotype. The Rm1021::pRG1SMa0805ΔpD2SMa0863 and Rm1021::pRG1SMa0861ΔpD2SMa0907 strains were missing extended regions of nod genes which affected their ability to nodulate alfalfa (Nod−) (Table 2).
Table 2.
Symbiotic phenotype of selected pSymA deletion mutants
| Strain | No. of nodules/plant | Nodule color | Shoot dry wt (mg/shoot) |
|---|---|---|---|
| Rm1021 | 4.3 ± 1.2 | Red | 17.52 |
| 1021::pRG1SMa0785ΔpD2SMa0814 | 16.2 ± 5.5 | White | 3.34 |
| 1021::pRG1SMa0805ΔpD2SMa0863 | 0.3 ± 07 | White | 3.07 |
| Rm1021::pRG1SMa0861ΔpD2SMa0907 | 0 | 4.70 | |
| 1021::pRG1SMa1191ΔpD2SMa1232 | 19.3 ± 3.4 | White | 3.80 |
| 1021::pRG1SMa2101ΔpD2SMa2171 | 3.9 ± 1.7 | Red | 20.13 |
| No inoculation | 0.2 ± 0.6 | White | 4.04 |
SMa0719, an ORF deleted in the Rm1021::pRG1SMa0697ΔpD2SMa0750 strain, codes for a short-chain dehydrogenase/reductase. SMa0719 mutants were reported to have decreased utilization of a number of carbon sources and decreased symbiotic performance (37). The Rm1021::pRG1SMa0697ΔpD2SMa0750 mutant grew slower on Min-mannitol medium supplemented with taurine and on Min-NH4 with either arabinose, galactose, or trehalose (see Table S3 in the supplemental material). In contrast to the reported phenotype of the single-gene mutant (37, 38), the deletion strain grew well with GABA as a carbon source. However, the deletion strain formed an effective symbiosis with alfalfa. The difference between this report and the earlier one (37) might be due to different plant growth conditions that masked a marginal symbiotic phenotype of the mutant.
Two GntR-type regulators on pSymA, GtrA (SMa0160) and GtrB (SMa0222), had been shown to affect S. meliloti Rm1021 growth and nodulation (38). These ORFs were deleted in the Rm1021::pRG1SMa0137ΔpD2SMa0191 and Rm1021::pRG1SMa0185ΔpD2SMa0235 strains, respectively. However, neither strain exhibited altered growth properties on any tested media, including rich LB and defined Min-mannitol-glutamate media (data not shown). The GtrA and GtrB proteins were suggested to act as repressors of gene transcription in response to the absence of some metabolites (38). It is possible that the large deletions in the Rm1021::pRG1SMa0137ΔpD2SMa0191 and Rm1021::pRG1SMa0185ΔpD2SMa0235 strains also removed the target gene(s), relieving problems associated with the absence of the repressors. A similar scenario was described for control of gluconate utilization, where a mutation in the repressor promoted the growth of S. meliloti Rm1021 on gluconate, while a second mutation in the target gene negated the effect of the first mutation (29). We could not detect a significant difference in the symbiotic performance of the Rm1021::pRG1SMa0137ΔpD2SMa0191 and Rm1021::pRG1SMa0185ΔpD2SMa0235 strains, but this also could be attributed to simultaneous deletion of both regulator and target genes. It also could result from different plant growth conditions, which might complicate observation of marginal symbiotic phenotypes.
Conclusions.
The publication of the complete S. meliloti Rm1021 genome sequence brought new approaches to the study of its free-living and symbiotic physiology, increasing the importance of the S. meliloti-Medicago symbiosis as a model system for rhizobium-legume associations. However, making functional assignments to over 6,000 genes is still daunting, especially because testing the most interesting aspects of the system involves a lengthy assay for symbiotic function. The block deletion method presented here is an example of a systematic way of identifying regions that contain genes involved in generating a phenotype and then using more targeted methods to identify which genes in the regions are involved. Although it can be used in any context where it would be strategic to construct deletions with defined endpoints, the method is most likely to be useful in mapping plasmids or virulence islands that do not contain essential genes. It is likely that it could be used in any context where there are adequate levels of homologous recombination and the FLP-FRT system is functional.
Constructing deletions within the pSymA plasmid used a library of Gateway-compatible ORFeome plasmids and their derivatives (8, 9) and developed a plasmid, pRK2016-2, that could be used with a previously constructed reporter plasmid to generate deletions by using the yeast FRT recombination sequences. Deletions were confirmed by PCR using primers in the adjacent genes, although it was sometimes necessary to use a PCR primer located within the deletion scar, which appeared to interfere with amplification across the entire sequence. The construction of a series of 50 plasmids missing different defined regions of the pSymA replicon (Fig. 3) is a logical continuation of the ORFeome project and illustrates the flexibility of the system.
Analyzing these deletions allowed us to identify small sets of genes that play a role in S. meliloti Rm1021 cytochrome c respiration, salt stress adaptation, or serine and gluconate catabolism. Additionally, we showed that adhA1 plays a critical role in inosine utilization, probably by affecting the catabolism of the nucleoside moiety through the pentose phosphate pathway. This information could be used as a new starting point for research groups interested in rhizobial stress response, respiration, and carbon assimilation. Additional phenotypic characterization of the pSymA deletion library could help identify more regions involved in additional metabolic functions of S. meliloti Rm1021. They might be especially useful in locating genes that affect host range, both by confirming that some are in previously identified regions or by finding new regions where the strains containing specific deletions have defects in infection or nitrogen fixation.
ACKNOWLEDGMENTS
This work was supported by the Agricultural Research Center at Washington State University, grant DE-FG03-96ER20225 from the Energy Biosciences Program at the United States Department of Energy, and a grant from the Microbial Genetics Program at the U.S. National Science Foundation (MCB-0133258).
We thank Brenda Schroeder for discussions early in the project and the WSU Laboratory for Biotechnology and Bioanalysis for sequencing support.
Footnotes
Published ahead of print 18 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02974-12.
REFERENCES
- 1. Barnett MJ, Fisher RF, Jones T, Komp C, Abola AP, Barloy-Hubler F, Bowser L, Capela D, Galibert F, Gouzy J, Gurjal M, Hong A, Huizar L, Hyman RW, Kahn D, Kahn ML, Kalman S, Keating DH, Palm C, Peck MC, Surzycki R, Wells DH, Yeh KC, Davis RW, Federspiel NA, Long SR. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA megaplasmid. Proc. Natl. Acad. Sci. U. S. A. 98:9883–9888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Oresnik IJ, Liu SL, Yost CK, Hynes MF. 2000. Megaplasmid pRme2011a of Sinorhizobium meliloti is not required for viability. J. Bacteriol. 182:3582–3586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Galibert F, Finan TM, Long SR, Pühler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, Bothe G, Boutry M, Bowser L, Buhrmester J, Cadieu E, Capela D, Chain P, Cowie A, Davis RW, Dreano S, Federspiel NA, Fisher RF, Gloux S, Godrie T, Goffeau A, Golding B, Gouzy J, Gurjal M, Hernández-Lucas I, Hong A, Huizar L, Hyman RW, Jones T, Kahn D, Kahn ML, Kalman S, Keating DH, Kiss E, Komp C, Lelaure V, Masuy D, Palm C, Peck MC, Pohl TM, Portetelle D, Purnelle B, Ramsperger U, Surzycki R, Thébault P, Vandenbol M, Vorhölter FJ, Weidner S, Wells DH, Wong K, Yeh KC, Batut J. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668–672 [DOI] [PubMed] [Google Scholar]
- 4. Becker A, Berges H, Krol E, Bruand C, Rüberg S, Capela D, Lauber E, Meilhoc E, Ampe F, de Bruijn FJ, Fourment J, Francez-Charlot A, Kahn D, Küster H, Liebe C, Pühler A, Weidner S, Batut J. 2004. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol. Plant Microbe Interact. 17:292–303 [DOI] [PubMed] [Google Scholar]
- 5. Cowie A, Cheng J, Sibley CD, Fong Y, Zaheer R, Patten CL, Morton RM, Golding GB, Finan TM. 2006. An integrated approach to functional genomics: construction of a novel reporter gene fusion library for Sinorhizobium meliloti. Appl. Environ. Microbiol. 72:7156–7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Djordjevic MA. 2004. Sinorhizobium meliloti metabolism in the root nodule: a proteomic perspective. Proteomics 4:1859–1872 [DOI] [PubMed] [Google Scholar]
- 7. House BL, Mortimer MW, Kahn ML. 2004. New recombination methods for Sinorhizobium meliloti genetics. Appl. Environ. Microbiol. 70:2806–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schroeder BK, House BL, Mortimer MW, Yurgel SN, Maloney SC, Ward KL, Kahn ML. 2005. Development of a functional genomics platform for Sinorhizobium meliloti: construction of an ORFeome. Appl. Environ. Microbiol. 71:5858–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Humann JL, Schroeder BK, Mortimer MW, House BL, Yurgel SN, Maloney SC, Ward KL, Fallquist HM, Ziemkiewicz HT, Kahn ML. 2008. Construction and expression of sugar kinase transcriptional gene fusions by using the Sinorhizobium meliloti ORFeome. Appl. Environ. Microbiol. 74:6756–6765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pickering BS, Yudistira H, Oresnik IJ. 2012. Characterization of the twin-arginine transport secretome in Sinorhizobium meliloti and evidence for host-dependent phenotypes. Appl. Environ. Microbiol. 78:7141–7144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bernard P, Kezdy KE, Van Melderen L, Steyaert J, Wyns L, Pato ML, Higgins PN, Couturier M. 1993. The F plasmid CcdB protein induces efficient ATP-dependent DNA cleavage by gyrase. J. Mol. Biol. 234:534–541 [DOI] [PubMed] [Google Scholar]
- 12. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 13. Platt R, Drescher C, Park SK, Phillips GJ. 2000. Genetic system for reversible integration of DNA constructs and lacZ gene fusions into the Escherichia coli chromosome. Plasmid 43:12–23 [DOI] [PubMed] [Google Scholar]
- 14. Simon R, Quandt J, Klipp W. 1989. New derivatives of transposon Tn5 suitable for mobilization of replicons, generation of operon fusions and induction of genes in Gram-negative bacteria. Gene 80:161–169 [DOI] [PubMed] [Google Scholar]
- 15. Leigh JA, Signer ER, Walker GC. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. U. S. A. 82:6231–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schäfer A, Tauch A, Jäger Kalinowski WJ, Thierbach G, Pühler A. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73 [DOI] [PubMed] [Google Scholar]
- 17. Somerville JE, Kahn ML. 1983. Cloning of the glutamine synthetase I gene from Rhizobium meliloti. J. Bacteriol. 156:168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Beringer JE. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188–198 [DOI] [PubMed] [Google Scholar]
- 19. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 20. Marrs B, Gest H. 1973. Genetic mutations affecting the respiratory electron-transport system of the photosynthetic bacterium Rhodopseudomonas capsulata. J. Bacteriol. 114:1045–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yurgel SN, Berrocal J, Wilson C, Kahn ML. 2007. Pleiotropic effects of mutations that alter the Sinorhizobium meliloti cytochrome c respiratory system. Microbiology 153:399–410 [DOI] [PubMed] [Google Scholar]
- 22. Delgado MJ, Bedmar EJ, Downie JA. 1998. Genes involved in the formation and assembly of rhizobial cytochromes and their role in symbiotic nitrogen fixation. Adv. Microb. Physiol. 40:191–231 [DOI] [PubMed] [Google Scholar]
- 23. Renalier MH, Batut J, Ghai J, Terzaghi B, Ghérardi M, David M, Garnerone AM, Vasse J, Truchet G, Huguet T. 1987. A new symbiotic cluster on the pSym megaplasmid of Rhizobium meliloti 2011 carries a functional fix gene repeat and a nod locus. J. Bacteriol. 169:2231–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Preisig O, Anthamatten D, Hennecke H. 1993. Genes for a microaerobically induced oxidase complex in Bradyrhizobium japonicum are essential for a nitrogen-fixing endosymbiosis. Proc. Natl. Acad. Sci. U. S. A. 90:3309–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Preisig O, Zufferey R, Thöny-Meyer L, Appleby CA, Hennecke H. 1996. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 178:1532–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lynch D, O'Brien J, Welch T, Clarke P, Cuiv PO, Crosa JH, O'Connell M. 2001. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183:2576–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Epstein W. 2003. The roles and regulation of potassium in bacteria. Prog. Nucleic Acid Res. Mol. Biol. 75:293–320 [DOI] [PubMed] [Google Scholar]
- 28. Domínguez-Ferreras A, Muñoz S, Olivares J, Soto MJ, Sanjuán J. 2009. Role of potassium uptake systems in Sinorhizobium meliloti osmoadaptation and symbiotic performance. J. Bacteriol. 191:2133–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Steele TT, Fowler CW, Griffitts JS. 2009. Control of gluconate utilization in Sinorhizobium meliloti. J. Bacteriol. 191:1355–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Willis LB, Walker GC. 1998. Identification of the Rhizobium meliloti alcohol dehydrogenase gene (adhA) and heterologous expression in Alcaligenes eutrophus. Biochim. Biophys. Acta 1384:197–203 [DOI] [PubMed] [Google Scholar]
- 31. Sprenger GA. 1995. Genetics of pentose-phosphate pathway enzymes of Escherichia coli K-12. Arch. Microbiol. 164:324–330 [DOI] [PubMed] [Google Scholar]
- 32. Geddes BA, Oresnik IJ. 2012. Genetic characterization of a complex locus necessary for the transport and catabolism of erythritol, adonitol and L-arabitol in Sinorhizobium meliloti. Microbiology 158:2180–2191 [DOI] [PubMed] [Google Scholar]
- 33. Klipp W, Reilander H, Schluter A, Krey R, Pühler A. 1989. The Rhizobium meliloti fdxN gene encoding a ferredoxin-like protein is necessary for nitrogen fixation and is cotranscribed with nifA and nifB. Mol. Gen. Genet. 216:293–302 [DOI] [PubMed] [Google Scholar]
- 34. Foussard M, Garnerone AM, Ni F, Soupène E, Boistard P, Batut J. 1997. Negative autoregulation of the Rhizobium meliloti fixK gene is indirect and requires a newly identified regulator, FixT. Mol. Microbiol. 25:27–37 [DOI] [PubMed] [Google Scholar]
- 35. Batut J, Daveran-Mingot ML, David M, Jacobs J, Garnerone AM, Kahn D. 1989. fixK, a gene homologous with fnr and crp from Escherichia coli, regulates nitrogen fixation genes both positively and negatively in Rhizobium meliloti. EMBO J. 8:1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. David M, Daveran ML, Batut J, Dedieu A, Domergue O, Ghai J, Hertig C, Boistard P, Kahn D. 1988. Cascade regulation of nif gene expression in Rhizobium meliloti. Cell 54:671–683 [DOI] [PubMed] [Google Scholar]
- 37. Jacob AI, Adham SA, Capstick DS, Clark SR, Spence T, Charles TC. 2008. Mutational analysis of the Sinorhizobium meliloti short-chain dehydrogenase/reductase family reveals substantial contribution to symbiosis and catabolic diversity. Mol. Plant Microbe Interact. 21:979–987 [DOI] [PubMed] [Google Scholar]
- 38. Wang Y, Chen AM, Yu AY, Luo L, Yu GQ, Zhu JB, Wang YZ. 2008. The GntR-type regulators gtrA and gtrB affect cell growth and nodulation of Sinorhizobium meliloti. J. Microbiol. 46:137–145 [DOI] [PubMed] [Google Scholar]


