Abstract
Lactobacilli (Lactobacillales: Lactobacillaceae) are well known for their roles in food fermentation, as probiotics, and in human health, but they can also be dominant members of the microbiota of some species of Hymenoptera (ants, bees, and wasps). Honey bees and bumble bees associate with host-specific lactobacilli, and some evidence suggests that these lactobacilli are important for bee health. Social transmission helps maintain associations between these bees and their respective microbiota. To determine whether lactobacilli associated with social hymenopteran hosts are generally host specific, we gathered publicly available Lactobacillus 16S rRNA gene sequences, along with Lactobacillus sequences from 454 pyrosequencing surveys of six other hymenopteran species (three sweat bees and three ants). We determined the comparative secondary structural models of 16S rRNA, which allowed us to accurately align the entire 16S rRNA gene, including fast-evolving regions. BLAST searches and maximum-likelihood phylogenetic reconstructions confirmed that honey and bumble bees have host-specific Lactobacillus associates. Regardless of colony size or within-colony oral sharing of food (trophallaxis), sweat bees and ants associate with lactobacilli that are closely related to those found in vertebrate hosts or in diverse environments. Why honey and bumble bees associate with host-specific lactobacilli while other social Hymenoptera do not remains an open question. Lactobacilli are known to inhibit the growth of other microbes and can be beneficial whether they are coevolved with their host or are recruited by the host from environmental sources through mechanisms of partner choice.
INTRODUCTION
Lactobacillus is the largest genus of lactic acid bacteria (LAB), containing species that are well known for their roles in food production and human health (1). Lactobacilli convert sugars to lactic acid and other acids, and some species are used in food fermentations to preserve foods, contribute flavor, or inhibit the growth of other bacteria (2). Other lactobacilli are used as human probiotics; for example, L. reuteri protects children from rotavirus gastroenteritis (3). Lactobacillus is of obvious importance to humans for both economic and health reasons.
Lactobacilli also associate with diverse nonhuman animals, and there is evidence that some lactobacilli can protect these hosts from pathogens (4, 5). For example, Lactobacillus reuteri is found in the gastrointestinal tracts of pigs, rodents, chickens, and humans and aids in protection of the host from pathogens (6). Lactobacillus plantarum is one of the five dominant bacterial phylotypes found in Drosophila melanogaster intestinal tracts (7), promoting larval growth in low-nutrient medium (8). A mixture of LAB, including Lactobacillus species, help protect honey bee larvae from Paenibacillus larva and Melissococcus plutonius, the causative agents of American and European foulbrood, respectively (9, 10). The Lactobacillus phylotypes Firm4 and Firm5, in conjunction with other members of the bumble bee microbiota, protect bumble bee workers from the trypanosome pathogen Crithidia bombi (11, 12). LAB have also been hypothesized to play a role in fermenting pollen stored by honey bees, thereby protecting the stored bee bread from spoilage (13), although firm evidence for this hypothesis is still lacking.
Several authors suggested recently that the relationships between honey and bumble bees (corbiculate apids) and their microbiotae, both of which include related Lactobacillus phylotypes, have been shaped by coevolutionary processes (10, 14, 15). Our recent phylogeny (16), based on a short fragment of the 16S rRNA gene, agreed with suggestions that the Firm3, Firm4, and Firm5 honey bee- and bumble bee-associated Lactobacillus phylotypes are host specific, which suggests that between-species transmission of these phylotypes is uncommon or absent (16). Horizontal transmission has not been excluded, but both honey and bumble bees can acquire microbes through within-colony social contact (11, 17). In contrast, solitary and primitively eusocial sweat bees associate with lactobacilli related to those that occur on flowers (16). It is currently unknown whether sociality per se or some unique aspect of corbiculate apid biology maintains the relationship between honey and bumble bees and their host-specific lactobacilli.
Here we explore the host specificity of lactobacilli that associate with hymenopteran hosts that exhibit a range of social structures. First, we used very accurate comparative secondary structure models as templates to create a larger set of 16S rRNA structure models that represent all of the major forms of structural diversity present in the primary groups of lactobacilli. Second, we used these structure models to construct a comprehensive and highly accurate alignment of full-length or nearly full-length 16S rRNA gene sequences. To determine whether host specificity is common in associates of insects that live in large societies or is limited to associates of corbiculate apids, we constructed two additional alignments that included 16S-amplicon 454-pyrosequencing surveys of several bee and ant species. We then used these structure-based alignments to reconstruct the phylogenetic history of the genus Lactobacillus.
MATERIALS AND METHODS
Full and nearly full-length Lactobacillus 16S rRNA gene sequences.
To obtain a comprehensive representation of this diverse genus, we downloaded publicly available, full-length or nearly full-length sequences of the 16S rRNA gene. First, we searched NCBI's nucleotide database (18) for complete Lactobacillus 16S rRNA gene sequences. Next, we searched the Ribosomal Database Project (RDP [19]) for Lactobacillus 16S sequences longer than 1,200 bases and of good quality, as designated by RDP. To include sequences from bee-associated lactobacilli, we downloaded associated sequences from NCBI PubMed entries of bee-bacterial studies and additionally searched NCBI's nucleotide database for bee-associated lactobacilli. To avoid redundant sequences and obtain a more tractable representation of the genus, we clustered sequences of ≥97% sequence similarity with the program CD-HIT (20). Since this clustering may have eliminated some sequences from type strains that were represented by longer sequences that shared ≥97% sequence identity with the type strain sequence, we obtained the accession numbers of Lactobacillus type specimens from RDP. We then searched our file for the accession numbers of these type sequences and added missing sequences back into the alignment. We additionally searched NCBI taxonomy for Lactobacillus species and added reference sequences (21), when available, for the species that were missing in our alignment. Finally, we searched the list of prokaryotic names with standing in nomenclature (22) to verify that our alignment did indeed contain representatives from all described lactobacilli. This set of 16S rRNA sequences included identified, cultured, unidentified, and uncultured Lactobacillus species. As outgroups, we added sequences from 14 bacterial species from across the Bacilli but with emphasis on the Lactobacillales. To investigate diversity of paralogous gene copies within a genome, we included all 16S rRNA gene copies from whole-genome sequence of five ingroup species and one outgroup species. Our final alignment contained 19 outgroup sequences (from 14 species), 158 sequences from undescribed or unidentified Lactobacillus species, and 224 sequences from 160 named, but not necessarily published, Lactobacillus species for a total of 401 sequences.
Partial and complete 16S rRNA gene sequences, including hymenopteran associates.
To integrate 454 pyrosequencing data into the alignment of complete or nearly complete full-length sequences, we built a second alignment spanning the length of the 16S rRNA gene. We coded the five prime and three prime ends of the 454 sequences as missing data. The effect of missing data on phylogenetic reconstructions is an ongoing research area with no simple consensus (23, 24). We therefore planned to interpret the results of the partial and complete 16S alignment with missing data with caution.
We started with the 401 sequences described above and added sequences from three separate 16S rRNA-amplicon 454-pyrosequencing studies of bacteria associated with Hymenoptera: the sweat bees (Halictidae) Augochlora pura, Halictus ligatus, and Megalopta genalis (16); an attine fungus-growing ant, Mycocepurus smithii (K. Kellner et al., unpublished); and the fire-ants, Solenopsis invicta and Solenopsis geminata (25; R. M. Plowes et al., unpublished). We previously published the halictid data (16), which we obtained from one wild nest of A. pura (Virginia), one laboratory nest of H. ligatus (Virginia), and two wild nests of M. genalis (Barro Colorado Island, Panama). We also sampled one colony of M. smithii in the wild in Gamboa, Panama, and then brought the nest into the laboratory and sampled it again 6 months later. We collected S. invicta and S. geminata-complex samples in their native range in Argentina and in their invasive range in Taiwan, Texas, Florida, and California (Plowes et al., unpublished). These data represent subsets of data sets from previous or forthcoming publications (see, for example, references 16 and 25). The three studies used the same 16S rRNA primers: Gray28F 5′-GAGTTTGATCNTGGCTCAG and Gray519r 5′-GTNTTACNGCGGCKGCTG, which span V1-V3 in the secondary structure of 16S rRNA. V1-V3 are variable regions that have been shown to accurately differentiate closely related lactobacilli (26). Research and Testing Laboratories (Lubbock, TX) generated the 454 sequence for the three studies at different times. The sweat bee- and Mycocepurus-associated reads were 454 sequenced forward from Gray28f, while the Solenopsis-associated sequences were 454 sequenced reverse from Gray519r (these later sequences were reverse complemented for our analysis).
We used previously described pipelines to denoise and otherwise quality check the 454 16S rRNA reads (16, 25). The sweat bee-associated Lactobacillus sequences were depleted of low-quality sequences and chimeras, denoised, and assigned to phylotype as described in McFrederick et al. (16). The Mycocepurus- and Solenopsis-associated bacterial sequences were processed in the program mothur (27). First, we denoised the data with the shhh.flows command. We then removed any sequences with mismatches in the primer binding site or the barcode, or homopolymer runs of over eight bases, and removed primers and barcodes from the sequences. To remove chimeric sequences, we used the chimera.uchime command, and deleted the detected chimeras. Next, we ran BLAST searches against a 16S rRNA database curated by the Medical Biofilm Research Institute (MBRI; Lubbock, TX). We selected sequences with top BLAST hits against the MBRI database of at least 90% sequence identity to Lactobacillus sequences. We chose a minimum of 90% sequence identity in order to include novel Lactobacillus sequences, while keeping in mind that any sequences that do not belong in the genus Lactobacillus should fall outside of our ingroup in our phylogenetic analysis. As a validation of our BLAST searches, we ran additional BLAST searches of the 454 sequences against the entire nucleotide collection at NCBI. Next, to maximize the phylogenetic signal of the short 454 reads, we removed 454 sequences that were <350 bases long. We then clustered sequences of 97% or greater sequence identity with the program CD-HIT (20). After quality control and clustering, we added 84 sweat bee-associated, 79 Mycocepurus-associated, and 56 Solenopsis-associated sequences to our alignment, for a total of 620 sequences in the entire alignment.
The hymenopteran hosts included here vary widely in both geography and natural history. Augochlora pura and H. ligatus both live in North America, but A. pura is a solitary bee that nests in rotten logs (28), whereas H. ligatus is a primitively eusocial halictid that forms small colonies in the soil (29). Megalopta genalis is a socially polymorphic, neotropical sweat bee that builds nests in decaying branches found in the forest understory (30, 31). Mycocepurus smithii is a neotropical non-leaf-cutting fungus-growing ant that forms colonies with an average of 77 workers in reproductive nests in Puerto Rico (32) but can also form much larger single colonies that occupy more than one nest in the Brazilian Amazon (33, 34). In contrast, S. invicta is originally from South America but has become widespread in its invasive range and can reach colony sizes of 220,000 individuals (35, 36). Solenopsis geminata variants have been introduced to many locations, including Taiwan and possibly Florida (H. Axen, unpublished data).
Partial 16S rRNA gene sequences, including hymenopteran associates.
To investigate the phylogenetic utility of the short 454-pyrosequencing data, we created a third alignment spanning positions 30 to 517 in the E. coli 16S rRNA gene sequence. We used the entire length of the 454 data and trimmed the 401 full-length sequences to span only the regions overlapping the 454 pyrosequencing reads. By creating three separate alignments, we were able to compare results and determine differences in the phylogenetic reconstructions based on partial versus full length versus a combination of partial and complete 16S rRNA sequences.
Sequence alignment and RNA secondary structure.
We used covariation analysis to identify RNA secondary structure that is common to a set of sequences that are known to have the same function and higher-order structure (37). Covariation analyses identify all types of canonical and noncanonical base pairs with a set of nucleotides that covary with one another, regardless of their proximity to other structural elements (38, 39). Approximately 97% of the base pairs, including all of the noncanonical base pairs, pseudoknots, and other irregular structural elements in the comparative 16S rRNA secondary structure models are in the crystal structure of the ribosomal subunit (40). The strength of the covariation for each predicted base pair has been quantified and used as a confidence rating for each base pair (http://www.rna.ccbb.utexas.edu/SAE/2A/nt_Frequency/BP/). Given this very high accuracy, as gauged with high-resolution crystal structures, we are most confident that all or nearly all of the base pairs in the comparative structure models for the rRNAs from different organisms are correct, including all of the 16S rRNA secondary structure models for the genus Lactobacillus (for a complete discussion of covariation analysis, see File S1 in the supplemental material).
The accuracy of our predicted secondary structure models are directly associated with the quality of the sequence alignment. Better alignments facilitate better comparative structure models. Moreover, vice versa, very accurate comparative structure models are the basis for the juxtaposition of nucleotides that are components of similar and analogous structural elements, especially in highly variable regions that can have large variances in the number of nucleotides with little or no sequence identity. Thus, our secondary structure-based alignments do not attempt to maximize sequence identity a priori. Instead, they attempt to maximize the alignment of similar structural elements.
After the 16S rRNA sequences were semiautomatically aligned with the template-based alignment program CRWAlign (41), we refined the alignment manually with the alignment editor AE2 (developed by T. Macke, Scripps Research Institute, San Diego, CA [42]). This tool was developed for Sun Microsystems (Santa Clara, CA) workstations running the Solaris operating system. The manual alignment process utilizes the CRWAlign program to identify nucleotides in a column that might not map to the same locations in the secondary and tertiary structure. A visual inspection of the alignment determines if these flagged nucleotides should be realigned. If necessary, this is done manually with the AE2 alignment editor. For regions of the alignment with high sequence similarity and minimal variance in the number of nucleotides, the information in the primary structure is sufficient to align sequences with confidence. In contrast, for more variable regions in closely related sequences or between more distantly related sequences, a high-quality alignment can only be produced when secondary and/or tertiary structure information is included.
Secondary structure diagrams.
Secondary structure diagrams were generated after the first secondary structure model was derived with comparative methods. Although the Comparative RNA Web (CRW) site (http://www.rna.ccbb.utexas.edu/) contains more than 200 16S rRNA secondary structure diagrams that sample the diversity within the Bacteria, we generated 47 secondary structure diagrams for the present study: 38 from taxa representing all of the major phylogenetic groups of Lactobacillus and nine from related organisms. We generated the secondary structure diagrams with the interactive secondary structure program XRNA (written in the C programming language for Sun Microsystems workstations running the Solaris operating system by B. Weiser and H. Noller, University of California, Santa Cruz, CA). All structure diagrams are available as online supplemental files at http://www.rna.ccbb.utexas.edu/.
Phylogenetic analyses.
To reconstruct the phylogenetic history of the lactobacilli, we conducted separate maximum-likelihood analyses on our three alignments. We first determined that GTR+I+Γ was the most appropriate model of sequence evolution for each of our alignments using the Akaike Information Criterion (AIC) in Modeltest 3.7 (43) and PAUP* 4b10 (44). We then ran 20 independent search replicates on each alignment in the program GARLI 2.0 (45), with the genthreshfortopoterm, stopgen, and stoptime settings set to 10,000,000. We allowed the program to estimate all parameter values during the runs. To assess branch support we conducted 100 bootstrap pseudoreplicates on each alignment, using a genthreshfortopoterm setting of 5,000,000. We used Mesquite (46), to calculate patristic distances across the entire tree. To calculate divergence, we used the branch info tool in Mesquite (46) to measure the distance from the base of monophyletic groups of hymenopteran associates to the most recent common ancestor with their closest relatives.
Data availability.
We deposited the 84 sweat bee-associated, 79 Mycocepurus-associated, and 56 Solenopsis-associated sequences in the genetic sequence database at the National Center for Biotechnical Information (NCBI GenBank accession numbers KC354148 to KC354367).
RESULTS
Phylogenetic reconstructions of our three 16S rRNA gene sequence alignments resulted in similar clade structure for all trees (Fig. 1 and 2; see also Fig. S1 and see Table S1 in the supplemental material for simple sequence statistics for all alignments). The phylogeny based only on full-length 16S rRNA sequences (Fig. 1) was in nearly complete agreement with the phylogeny based on a combination of partial and complete 16S rRNA sequences (Fig. 2). The phylogeny based on only partial 16S sequences (see Fig. S1 in the supplemental material), however, differed in the branching patterns between and within clades compared to the full-length phylogeny, but the terminal clade composition remained stable. Sequence alignments and detailed phylogenetic trees are available at TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S13670), and a detailed version of Fig. 2 with bootstrap support values and taxon labels is available in the online supplemental files (see Fig. S2 in the supplemental material).
Fig 1.
16S rRNA gene maximum-likelihood phylogeny of the genus Lactobacillus, based on full or nearly full-length sequences only. Branch widths are proportional to bootstrap support from 100 pseudoreplicates. Highlighted branches represent Lactobacillus associates of honey or bumble bees. Major clades are indicated with curved lines and named according to previous studies (47–50).
Fig 2.
16S rRNA gene maximum-likelihood tree of the genus Lactobacillus, based on partial and complete sequences combined. Branch widths are proportional to bootstrap support from 100 pseudoreplicates. Highlighted branches represent Hymenoptera associated lactobacilli as indicated in the figure legend. Clades containing lactobacilli associated with humans are indicated by a human figure, whereas clades containing lactobacilli used in food fermentation are indicated by a piece of cheese. Major clades are indicated with curved lines and named according to previous studies (47–50).
16S rRNA secondary structural models.
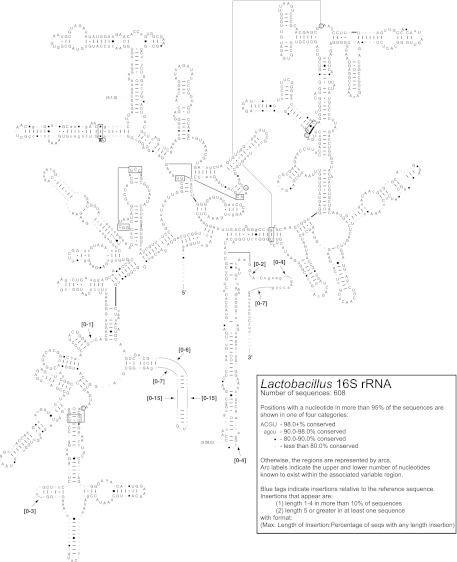
We created secondary structural models from 16S rRNA gene sequences of 38 lactobacilli and nine other Firmicutes (see File S1 in the supplemental material). In addition, the extent of primary and secondary structure conservation for 608 16S rRNA sequences representing the entire lactobacilli genus was mapped onto a L. acidophilus secondary structure diagram (Fig. 3). The first hairpin stem and loop from the five prime end of the molecule (the V1 region) was the most variable region in Lactobacillus 16S structure. Our structural models suggest that next generation sequencing surveys of bacterial communities aimed at elucidating the diversity of Lactobacillus should use primers targeting the V1 region.
Fig 3.
Secondary structure of the 16S rRNA molecule in the genus Lactobacillus. The degree of conservation in the molecule as determined by analysis of 608 sequences is superimposed on the structure of the Lactobacillus acidophilus 16S molecule. Conservation of each nucleotide across the 608 analyzed Lactobacillus species is indicated as follows: uppercase letters (>98% conservation), lowercase letters (90 to 98% conservation), closed circles (80 to 90% conservation), or open circles (<80% conservation). Canonical nucleotide pairs (G-C or A-U) are represented by dashes, wobble nucleotide pairs (G-U) are represented by small closed circles, A-G nucleotide pairs are represented by large open circles, and all other pairs are represented by large closed circles.
Full-length 16S rRNA gene phylogeny.
Our maximum-likelihood analysis of full- or nearly full-length 16S rRNA gene sequences recovers six major monophyletic clades, which we identify in accordance with previous studies: L. salivarius, L. delbrueckii (acidophilus), L. casei, L. buchneri, L. plantarum, and L. reuteri (Fig. 1) (47–50). The maximum-likelihood bootstrap support values for these clades varied greatly: the L. salivarius and L. reuteri clades showed moderate support (82 and 85%, respectively), while other clades showed weak support at their deepest nodes. The deeper branches connecting these major clades uniformly showed little support. The clade containing the greatest number of taxa and the greatest sequence diversity was the L. delbrueckii (acidophilus) clade, which was comprised of 160 taxa and covered a maximum patristic distance of 1.21, out of a global maximum of 1.51 across the entire tree (Table 1).
Table 1.
Maximum patristic distances of the major clades in the most likely full-length 16S phylogenetic tree, out of a maximum patristic distance of 1.51 across the entire tree
| Clade | Patristic distance |
|---|---|
| L. salivarius | 0.5 |
| L. casei | 0.44 |
| L. buchneri | 0.54 |
| L. plantarum | 0.34 |
| L. reuteri | 0.55 |
| L. delbrueckii (acidophilus) | 1.21 |
Partial and complete 16S rRNA gene phylogeny, including hymenopteran associates.
The phylogenetic trees determined from the analysis of the partial and complete 16S rRNA alignment and the full-length 16S rRNA alignment were very similar (Fig. 1 and 2). The main difference between the two phylogenies was the placement of the reuteri clade. The reuteri clade was sister to the plantarum clade in the full-length phylogeny (Fig. 1), whereas in the partial and complete phylogeny the members of the reuteri clade were reconstructed paraphyletic as a series of branching lineages leading to the monophyletic L. delbrueckii (acidophilus) clade (Fig. 2). Clade memberships and within-clade branching patterns were very similar between the full-length and the partial and complete phylogenies, as were the bootstrap support values (Fig. 1 and 2).
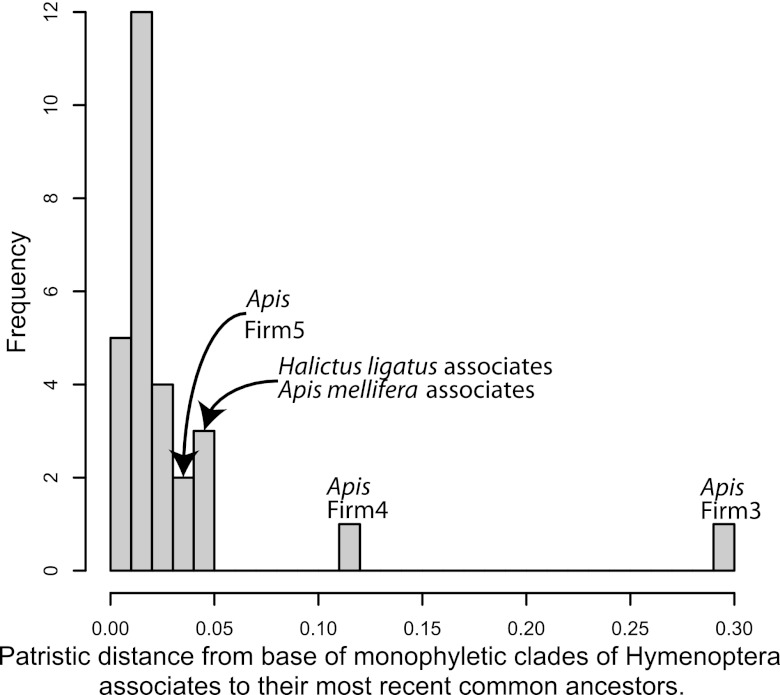
Of the Hymenoptera-associated sequences, only the honey and bumble bee-associated Firm3 and Firm4 (F3 and F4 sensu [51]) clades were host specific and highly diverged from other lactobacilli (Fig. 4). Firm5 also formed a monophyletic group, but was not as diverged from other lactobacilli as Firm3 and Firm4 (Fig. 2 and 4). In addition, two sequences from Solenopsis clustered within the Firm5 clade. In contrast to the Firm3-Firm5 clades, most Hymenoptera-associated sequences were scattered throughout the genus and either clustered with or were closely related to lactobacilli known from vertebrate hosts or from diverse environmental sources (Fig. 2; see also Fig. S2 in the supplemental material). Most reads from the pyrosequencing surveys, however, tended to cluster into several operational taxonomic units (OTUs) per host (Table 2). The most abundant Lactobacillus OTUs associated with Solenopsis, for example, were closely related to L. sakei and L. plantarum, both of which are used in food fermentation (2). The most abundant M. smithii-associated OTUs were closely related to several lactobacilli that are common inhabitants of the intestinal tracts of rodents, birds, and humans (Table 2). These M. smithii-associated OTUs were found in both fungus-garden and worker samples (see Table S2 in the supplemental material). As already reported in McFrederick et al. (16), halictids associated mainly with Lactobacillus OTUs that were related to flower-inhabiting, fructophilic lactobacilli (52) or to a clade that includes flower-inhabiting and fructophilic lactobacilli (53) and lactobacilli used in sourdough fermentation (54). Some of the halictid-associated OTUs formed monophyletic clades, but these clades did not exhibit the same level of divergence as the Firm3 and Firm4 clades (Fig. 2 and 4). Many of the terminal branches that included Hymenoptera-associated sequences exhibited high bootstrap support (see Fig. S2 in the supplemental material).
Fig 4.
Histogram of patristic distances from the base of monophyletic clades of Hymenoptera associates to their most recent common ancestor. The honey bee-associated Firm3 and Firm4 clades exhibited the greatest divergence, while the honey bee-associated Firm5 clade exhibited an amount of divergence similar to that of some other Hymenoptera associates.
Table 2.
Ten most abundant Lactobacillus OTUs (clustered at ≥97% sequence identity) associated with three ant and three bee species (bee species are binned as halictids)a
| Host | Source of read | No. of reads | Closest phylogenetic relative | Best BLAST hit | Sequence identity (%) |
|---|---|---|---|---|---|
| Solenopsis | Brood geminata TX | 5,993 | Undescribed | L. sakei | 92 |
| Brood geminata TW | 2,118 | L. plantarum | L. plantarum | 99 | |
| Brood invicta TX | 935 | L. curvatus | L. curvatus | 99 | |
| Brood invicta TX | 756 | L. brevis | L. brevis | 99 | |
| Pooled invicta TW | 661 | L. plantarum | L. plantarum | 99 | |
| Pooled invicta TW | 324 | L. plantarum | L. plantarum | 99 | |
| Pooled geminata TW | 261 | L. paracasei | L. casei | 99 | |
| Brood geminata TW | 167 | L. kimchicus | L. odoratitofui | 99 | |
| Brood geminata TW | 156 | Undescribed | L. sakei | 93 | |
| Brood geminata TW | 116 | L. vaccinostercus | L. vaccinostercus | 99 | |
| Mycocepurus smithii | Garden | 14,152 | L. johnsonii | L. johnsonii | 99 |
| Worker | 3,748 | L. crispatus | L. crispatus | 99 | |
| Garden | 2,794 | L. salivarius | L. salivarius | 97 | |
| Garden | 1,059 | L. aviarius | L. aviarius | 99 | |
| Garden | 806 | L. crispatus | L. crispatus | 98 | |
| Worker | 598 | L. salivarius | L. salivarius | 99 | |
| Garden | 586 | L. reuteri | L. reuteri | 99 | |
| Worker | 431 | L. salivarius | L. salivarius | 98 | |
| Worker | 420 | L. aviarius/L. johnsonii | L. aviarius | 90 | |
| Garden | 408 | L. johnsonii | L. johnsonii | 95 | |
| Halictids | Larva | 20,669 | L. kunkeei/L. ozensis | L. kunkeei | 93 |
| Pollen | 15,761 | L. fructivorans | L. fructivorans | 91 | |
| Larva | 3,923 | L. kunkeei/L. ozensis | L. kunkeei | 93 | |
| Pollen | 3,631 | L. kunkeei/L. ozensis | L. kunkeei | 94 | |
| Pollen | 1,991 | L. kunkeei/L. ozensis | L. kunkeei | 93 | |
| Pollen | 1,557 | L. fructivorans | L. fructivorans | 90 | |
| Larva | 1,455 | L. kunkeei/L. ozensis | L. kunkeei | 93 | |
| Pollen | 1,285 | L. kunkeei/L. ozensis | L. kunkeei | 94 | |
| Frass | 1,102 | L. paracasei | L. casei | 99 | |
| Pollen | 397 | L. kunkeei/L. ozensis | L. kunkeei | 94 |
The source of read indicates the types of samples from which the OTUs were isolated (TX, Texas; TW, Taiwan; pooled, workers and brood from five colonies). The number of reads represents the number of reads that cluster into each OTU. The closest phylogenetic relative is the closest Lactobacillus species in our phylogenetic reconstruction with which a particular read clustered (Fig. 2). The best BLAST hit is the top hit to a named Lactobacillus species from a BLAST search of NCBI's entire nucleotide collection. The sequence identity is the percent sequence identity of the query to the best BLAST hit. Several OTUs shared top BLAST hits to the same Lactobacillus species but shared <97% sequence identity to each other.
Partial 16S rRNA gene phylogeny, including Hymenoptera associates.
The deeper branching patterns in the partial 16S phylogeny did not agree with the full-length phylogeny. For example, the partial 16S rRNA analysis recovered the L. salivarius clade as a paraphyletic series of branching lineages leading to the monophyletic L. delbrueckii (acidophilus) clade (see Fig. S1 in the supplemental material). The L. plantarum and L. buchneri clades were also no longer monophyletic in the partial phylogeny but instead were both broken into two separate clades. The L. reuteri and L. casei clades remained monophyletic in the partial 16S analysis. Although the partial 16S analysis was unable to resolve deeper branches in the Lactobacillus phylogeny, the membership of the terminal clades was stable across all analyses, and many of the terminal groups received high bootstrap support even in the partial 16S analysis (see Fig. S1 in the supplemental material).
DISCUSSION
Host specificity of Hymenoptera-associated lactobacilli.
Hymenoptera ranging from a solitary species (A. pura) to species that live in colonies numbering in the hundreds of thousands (Solenopsis) associate with lactobacilli that are either found in other hosts or the environment. Social structure by itself does not determine whether hymenopteran hosts associate with lactobacilli that are host-specific or that are more recent acquisitions from the environment. Honey bees (colony sizes up to 60,000 [55]) and bumble bees (colony sizes from 50 to >400 [55]) are the only hymenopteran hosts examined to date that associate with lactobacilli that appear to be host specific and highly diverged from other lactobacilli.
Previous studies already suggested that the corbiculate apids associate with host-specific lactobacilli (see, for example, references 14, 15, and 56), but it remains unclear why honey and bumble bees are special in this regard. The phylotypes associated with Apis mellifera form a more consistent association with their hosts compared to Bombus, and this may be related to how A. mellifera founds new colonies (15, 56). Apis mellifera colonies are founded via swarming, where a colony divides and approximately half of the workers leave with the old queen to form a new colony, whereas most ants, social wasps, and bumble bees are nonswarming (see reference 57 and references therein). The founding of colonies by thousands of individuals may allow for the between-generation maintenance of multiple strains that have been identified within A. mellifera-associated phylotypes (56). Why Bombus species, in which nests are always founded by single queens (58), maintain associations with host-specific bacteria remains to be determined in detailed studies of dispersing Bombus queens, as well as the changes in associated microbial communities during colony founding. Bombus terrestris, the bumble bee whose microbiota has been best studied (11, 12, 59), shares nectar via honeypots inside the colony (60). Bombus species do not engage in oral-oral food exchange (trophallaxis) (61), so shared food stores (e.g., honeypots), social contact (e.g., grooming), feces, or the nest environment may serve as important means for social transmission of bumble bee microbiota. In contrast, trophallaxis between nestmates or other contact within the hive is known to be important for the establishment of the microbiota of A. mellifera (17). Megalopta and Solenopsis also share food within a colony via trophallaxis (31, 62) but do not harbor host-specific microbes. Trophallaxis alone, therefore, does not automatically lead to the maintenance of host-specific microbes.
Our phylogenetic analyses and BLAST searches suggest likely mechanisms regarding how environmental lactobacilli are recruited into association with sweat bees and ants. As we previously reported, sweat bees associate with lactobacilli that are related to lactobacilli isolated from flowers, and may therefore obtain these bacteria from flowers (16). Mycocepurus smithii belongs to the group of attine ants that do not collect leaves to sustain growth of their fungus gardens, but instead collect insect frass, flower parts, seeds, and fruit-flesh (63). Two of the dominant Lactobacillus OTUs associated with M. smithii (L. johnsonii and L. crispatus) have been isolated from A. mellifera guts (64), indicating that these lactobacilli may occur in the guts and frass of other insects. Mycocepurus smithii colonies may therefore recruit L. johnsonii, L. crispatus, and L. salivarius from insect frass that they collect to sustain their fungus gardens. These lactobacilli are found in both M. smithii fungus gardens and workers, indicating that they can occupy broad ecological niches. Alternatively, M. smithii may be collecting fecal material of vertebrates such as mice, birds, or pigs, all of which are hosts to the lactobacilli associated with M. smithii.
Solenopsis, on the other hand, is omnivorous (65) and may be obtaining lactobacilli from plant, insect, or vertebrate food sources. The most abundant Lactobacillus in Solenopsis was most closely related to an undescribed Lactobacillus species from fermented tea. This OTU was found to associate with S. invicta and S. geminata in Argentina, North America, and Taiwan, indicating that it might play an important role in the biology of Solenopsis. Lactobacillus plantarum, whose comparatively large genome may allow it to inhabit a variety of environments (66), was also found to associate with S. invicta and S. geminata in Argentina, North America, and Taiwan. Lactobacillus plantarum is found in dairy, meat, plants, and the human gastrointestinal tract and may be recruited by Solenopsis from any of these sources. It is currently unknown whether these associations derived from an acquisition by a Solenopsis lineage ancestral to the invasive Solenopsis lineages and were then vertically transmitted within these Solenopsis lineages or whether Lactobacillus is recruited continually from the environment by Solenopsis. Notably, we found several Firm5 sequences, which are thought to be specific to honey and bumble bees (15), associated with Solenopsis. These sequences were not abundant, suggesting that they may be transient and perhaps picked up by Solenopsis when scavenging dead A. mellifera or Bombus workers.
The function of lactobacilli in hymenopteran hosts outside of the honey and bumble bees is still relatively unexplored, but the available evidence suggests that lactobacilli facilitate digestion of sugars and inhibit the growth of other microbes through acidification, thereby benefiting their hosts. Although 16S rRNA phylotype does not translate well into functional phenotypes (67), lactobacilli exhibit some general properties which may serve as exaptations for the symbiotic habit. For example, lactic acid bacteria (LAB) digest a variety of sugars, with the main end product being lactic acid, although other acids and ethanol are also common end products (49). By lowering the pH of their environment, LAB inhibit the growth of many other bacteria (68). LAB are also known for the secretion of bacteriocins, which are compounds that inhibit the growth of other bacteria (68). For example, L. sakei produces several bacteriocins that inhibit pathogenic and spoilage organisms (69). These general properties of lactobacilli mean that they are likely to be beneficial to hymenopteran hosts, and further research should determine the relative importance of acidification versus bacteriocin-secretion in the biology of ant- and bee-associated Lactobacillus.
Utility of the 16S rRNA gene for Lactobacillus identification and phylogeny.
Although our analysis of 16S rRNA did not resolve the deeper branches in the Lactobacillus phylogeny, 16S rRNA did place sequences into terminal clades with confidence. Clade membership in all of our analyses was generally well supported and largely agrees with the clade structures suggested in previous studies (47–50). By using alignments based on full-length sequences only, partial and complete sequences combined, and partial sequences only, we were able to assess the influence of missing data or 454 pyrosequencing length data on phylogenetic reconstructions. Although the analysis using only partial sequences performed poorly with regard to the placement of deeper branches, all of our analyses placed sequences into terminal clades with confidence.
Other studies have found that short pyrosequencing data can be placed accurately into an existing full-length phylogeny (70), and our results suggest that if a highly variable region can be accurately aligned, alignments built on short pyrosequencing sized data can accurately place sequences into terminal clades. Our secondary structural models of 16S rRNA indicate that the hairpin stem and loop in V1 is the most variable region in Lactobacillus. Next-generation sequencing surveys targeting V1 should provide the best taxonomic resolution for closely related lactobacilli.
Conclusions.
We created highly accurate comparative models of 16S rRNA secondary structure and used these models to precisely align the entire 16S rRNA gene, including fast-evolving regions. Maximum-likelihood phylogenetic reconstructions using our structurally informed sequence alignments revealed that honey and bumble bees associate with host-specific Lactobacillus. In contrast, regardless of colony size or within-colony oral sharing of food (trophallaxis), sweat bees and ants associate with lactobacilli that are closely related to those found in vertebrate hosts or in diverse environments. Host specificity therefore appears to be the exception rather than the rule for Hymenoptera-associated lactobacilli. Nest founding by colony fission has been proposed as a likely means for long-term transmission of microbiota within honey bee lineages (15). M. smithii may facultatively found nests by colony fissioning (34), and yet we found that M. smithii does not associate with host-specific lactobacilli. This suggests that (i) facultative colony fissioning is not sufficient to maintain long-term associations between a social-insect lineage and all host-specific microbiota; (ii) regular colony founding by single queens may disrupt long-term associations because new microbiota are frequently acquired by a social-insect lineage during that stage; and (iii) obligate colony fissioning is more likely than facultative colony fissioning to maintain long-term host-microbe specificities in a social insect. Studies of lactobacilli associated with other obligate colony-fissioning Hymenoptera such as meliponine bees and related solitary and communal nesters such as euglossine bees (55) will help determine whether colony-fissioning or some other characteristic of apid biology promotes host specificity. Regardless of how they are acquired, lactobacilli are known to inhibit the growth of other microbes and may be beneficial whether they are coevolved with their host or are recruited by the host from environmental sources through mechanisms of partner choice.
ACKNOWLEDGMENTS
This material is based on work supported by National Science Foundation grant PRFB-1003133 awarded to Q.S.M. and grant DEB-0919519 awarded to U.G.M., Texas Higher Education Coordinating Board grant 01923 awarded to L. E. Gilbert, and National Institutes of Health grant GM067317 awarded to R.R.G.
We appreciate technical assistance from D. A. Estrada in the initial processing of the Solenopsis sequences. We thank three anonymous reviewers for their helpful comments.
Footnotes
Published ahead of print 4 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03681-12.
REFERENCES
- 1. Dellaglio F, Felis GE. 2005. Taxonomy of lactobacilli and bifidobacteria, p 25 In Tannock G. (ed), Probiotics and prebiotics: scientific aspects. Caister Aademic Press, Norfolk, United Kingdom [Google Scholar]
- 2. Leroy F, De Vuyst L. 2004. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci. Technol. 15:67–78 [Google Scholar]
- 3. Shornikova AV, Casas IA, Mykkanen H, Salo E, Vesikari T. 1997. Bacteriotherapy with Lactobacillus reuteri in rotavirus gastroenteritis. Pediatr. Infect. Dis. J. 16:1103–1107 [DOI] [PubMed] [Google Scholar]
- 4. Walter J, Britton RA, Roos S. 2011. Host-microbial symbiosis in the vertebrate gastrointestinal tract and the Lactobacillus reuteri paradigm. Proc. Natl. Acad. Sci. U. S. A. 108:4645–4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cross ML. 2002. Microbes versus microbes: immune signals generated by probiotic lactobacilli and their role in protection against microbial pathogens. FEMS Immunol. Med. Microbiol. 34:245–253 [DOI] [PubMed] [Google Scholar]
- 6. Casas IA, Dobrogosz WJ. 2000. Validation of the probiotic concept: Lactobacillus reuteri confers broad-spectrum protection against disease in humans and animals. Microb. Ecol. Health Dis. 12:247–285 [Google Scholar]
- 7. Wong CNA, Ng P, Douglas AE. 2011. Low diversity bacterial community in the gut of the fruitfly Drosophila melanogaster. Environ. Microbiol. 13:1889–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Storelli G, Defaye A, Erkosar B, Hols P, Royet J, Leulier F. 2011. Lactobacillus plantarum promotes Drosophila systemic growth by modulating hormonal signals through TOR-dependent nutrient sensing. Cell Metab. 14:403–414 [DOI] [PubMed] [Google Scholar]
- 9. Forsgren E, Olofsson TC, Vasquez A, Fries I. 2010. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 41:99–108 [Google Scholar]
- 10. Vasquez A, Forsgren E, Fries I, Paxton RJ, Flaberg E, Szekely L, Olofsson TC. 2012. Symbionts as major modulators of insect health: lactic acid bacteria and honeybees. PLoS One 7:e33188 doi:10.1371/journal.pone.0033188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koch H, Schmid-Hempel P. 2011. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc. Natl. Acad. Sci. U. S. A. 108:19288–19292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Koch H, Schmid-Hempel P. 2012. Gut microbiota instead of host genotype drive the specificity in the interaction of a natural host-parasite system. Ecol. Lett. 15:1095–1103 [DOI] [PubMed] [Google Scholar]
- 13. Vasquez A, Olofsson TC. 2009. The lactic acid bacteria involved in the production of bee pollen and bee bread. J. Apicult. Res. 48:189–195 [Google Scholar]
- 14. Koch H, Schmid-Hempel P. 2011. Bacterial communities in Central European bumblebees: low diversity and high specificity. Microb. Ecol. 62:121–133 [DOI] [PubMed] [Google Scholar]
- 15. Martinson VG, Danforth BN, Minckley RL, Rueppell O, Tingek S, Moran NA. 2011. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 20:619–628 [DOI] [PubMed] [Google Scholar]
- 16. McFrederick QS, Wcislo WT, Taylor DR, Ishak HD, Dowd SE, Mueller UG. 2012. Environment or kin: whence do bees obtain acidophilic bacteria? Mol. Ecol. 21:1754–1768 [DOI] [PubMed] [Google Scholar]
- 17. Martinson VG, Moy J, Moran NA. 2012. Establishment of characteristic gut bacteria during development of the honey bee worker. Appl. Environ. Microbiol. 78:2830–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Benson DA, Boguski MS, Lipman DJ, Ostell J. 1997. GenBank. Nucleic Acids Res. 25:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang Y, Niu B, Gao Y, Fu L, Li W. 2010. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics 26:680–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pruitt KD, Tatusova T, Brown GR, Maglott DR. 2012. NCBI Reference Sequences (RefSeq): current status, new features, and genome annotation policy. Nucleic Acids Res. 40:D130–D135 doi:10.1093/nar/gkr1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Euzeby JP. 2012. List of prokaryotic names with standing in nomenclature. http://www.bacterio.cict.fr
- 23. Lemmon AR, Brown JM, Stanger-Hall K, Lemmon EM. 2009. The effect of ambiguous data on phylogenetic estimates obtained by maximum likelihood and Bayesian inference. Syst. Biol. 58:130–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiens JJ, Morrill MC. 2011. Missing data in phylogenetic analysis: reconciling results from simulations and empirical data. Syst. Biol. 60:719–731 [DOI] [PubMed] [Google Scholar]
- 25. Ishak HD, Plowes R, Sen R, Kellner K, Meyer E, Estrada DA, Dowd SE, Mueller UG. 2011. Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb. Ecol. 61:821–831 [DOI] [PubMed] [Google Scholar]
- 26. Kullen MJ, Sanozky-Dawes RB, Crowell DC, Klaenhammer TR. 2000. Use of the DNA sequence of variable regions of the 16S rRNA gene for rapid and accurate identification of bacteria in the Lactobacillus acidophilus complex. J. Appl. Microbiol. 89:511–516 [DOI] [PubMed] [Google Scholar]
- 27. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stockhammer KA. 1966. Nesting habits and life cycle of a sweat bee, Augochlora pura (Hymenoptera: Halictidae). J. Kans. Entomol. Soc. 39:157–192 [Google Scholar]
- 29. Michener CD, Bennett FD. 1977. Geographical variation in nesting biology and social organization of Halictus ligatus. Univ. Kans. Sci. Bull. 51:233–260 [Google Scholar]
- 30. Wcislo WT, Arneson L, Roesch K, Gonzalez V, Smith A, Fernandez H. 2004. The evolution of nocturnal behaviour in sweat bees, Megalopta genalis and M. ecuadoria (Hymenoptera: Halictidae): an escape from competitors and enemies? Biol. J. Linn. Soc. Lond. 83:377–387 [Google Scholar]
- 31. Wcislo WT, Gonzalez VH. 2006. Social and ecological contexts of trophallaxis in facultatively social sweat bees, Megalopta genalis and M. ecuadoria (Hymenoptera, Halictidae). Insectes Sociaux 53:220–225 [Google Scholar]
- 32. Fernandez-Marin H, Zimmerman JK, Wcislo WT, Rehner SA. 2005. Colony foundation, nest architecture, and demography of a basal fungus-growing ant, Mycocepurus smithii (Hymenoptera, Formicidae). J. Nat. Hist. 39:1735–1743 [Google Scholar]
- 33. Rabeling C, Gonzales O, Schultz TR, Bacci M, Garcia MVB, Verhaagh M, Ishak HD, Mueller UG. 2011. Cryptic sexual populations account for genetic diversity and ecological success in a widely distributed, asexual fungus-growing ant. Proc. Natl. Acad. Sci. U. S. A. 108:12366–12371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rabeling C, Lino-Neto J, Cappellari SC, Dos-Santos IA, Mueller UG, Bacci M. 2009. Thelytokous parthenogenesis in the fungus-gardening ant Mycocepurus smithii (Hymenoptera: Formicidae). PLoS One 4:e6781 doi:10.1371/journal.pone.0006781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tschinkel WR. 1988. Colony growth and the ontogeny of worker polymorphism in the fire ant, Solenopsis invicta. Behav. Ecol. Sociobiol. 22:103–115 [Google Scholar]
- 36. Morrison LW, Porter SD, Daniels E, Korzukhin MD. 2004. Potential global range expansion of the invasive fire ant, Solenopsis invicta. Biol. Invasions 6:183–191 [Google Scholar]
- 37. Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius R, Gutell R, Hogan JJ, Noller HF. 1980. Secondary structure model for bacterial 16S rRNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 8:2275–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gutell R, Power A, Hertz G, Putz E, Stormo G. 1992. Identifying constraints on the higher-order structure of RNA: continued development and application of comparative sequence analysis methods. Nucleic Acids Res. 20:5785–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shang L, Xu W, Ozer S, Gutell RR. 2012. Structural constraints identified with covariation analysis in rRNA. PLoS One 7:e39383 doi:10.1371/journal.pone.0039383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gutell RR, Lee JC, Cannone JJ. 2002. The accuracy of rRNA comparative structure models. Curr. Opin. Struct. Biol. 12:301–310 [DOI] [PubMed] [Google Scholar]
- 41. Gardner DP, Xu W, Miranker DP, Ozer S, Cannone JJ, Gutell RR. 2012. An accurate scalable template-based alignment algorithm, p 237–243 In Proceedings of the 2012 IEEE International Conference on Bioinformatics and Biomedicine IEEE Computer Society, Washington, DC: doi:10.1109/BIBM.2012.6392676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Larsen N, Olsen GJ, Maidak BL, McCaughey MJ, Overbeek R, Macke TJ, Marsh TL, Woese CR. 1993. The ribosomal database project. Nucleic Acids Res. 21:3021–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818 [DOI] [PubMed] [Google Scholar]
- 44. Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), v.4b10. Sinauer Associates, Sunderland, MA [Google Scholar]
- 45. Zwickl DJ. 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. University of Texas at Austin, Austin, TX. [Google Scholar]
- 46. Maddison WP, Maddison DR. 2010. Mesquite: a modular system for evolutionary analysis, version 2.74. http://mesuiteproject.org
- 47. Canchaya C, Claesson MJ, Fitzgerald GF, Van Sinderen D, O'Toole PW. 2006. Diversity of the genus Lactobacillus revealed by comparative genomics of five species. Microbiology 152:3185–3196 [DOI] [PubMed] [Google Scholar]
- 48. Felis GE, Dellaglio F. 2007. Taxonomy of lactobacilli and bifidobacteria. Curr. Issues Intest. Microbiol. 8:44–61 [PubMed] [Google Scholar]
- 49. Pot B, Tsakalidou E. 2009. Taxonomy and metabolism of Lactobacillus, p 3–58 In Ljungh A, Wadstrom T. (ed), Lactobacillus molecular biology: from genomics to probiotics. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 50. Salvetti E, Torriani S, Felis GE. 2012. The genus Lactobacillus: a taxonomic update. Probiotics Antimicrob. Proteins 4:217–226 [DOI] [PubMed] [Google Scholar]
- 51. Babendreier D, Joller D, Romeis J, Bigler F, Widmer F. 2007. Bacterial community structures in honeybee intestines and their response to two insecticidal proteins. FEMS Microbiol. Ecol. 59:600–610 [DOI] [PubMed] [Google Scholar]
- 52. Endo A, Irisawa T, Futagawa-Endo Y, Takano K, du Toit M, Okada S, Dicks LMT. 2012. Characterization and emended description of Lactobacillus kunkeei as a fructophilic lactic acid bacterium. Int. J. Syst. Evol. Microbiol. 62:500–504 [DOI] [PubMed] [Google Scholar]
- 53. Endo A, Futagawa-Endo Y, Sakamoto M, Kitahara M, Dicks LMT. 2010. Lactobacillus florum sp. nov., a fructophilic species isolated from flowers. Int. J. Syst. Evol. Microbiol. 60:2478–2482 [DOI] [PubMed] [Google Scholar]
- 54. Corsetti A, Settanni L. 2007. Lactobacilli in sourdough fermentation. Food Res. Int. 40:539–558 [Google Scholar]
- 55. Michener CD. 1974. The social behavior of the bees. Belknap Press, Cambridge, MA [Google Scholar]
- 56. Engel P, Martinson VG, Moran NA. 2012. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. U. S. A. 109:11002–11007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ratnieks FLW, Reeve HK. 1991. The evolution of queen-rearing nepotism in social Hymenoptera: effects of discrimination costs in swarming species. J. Evol. Biol. 4:93–115 [Google Scholar]
- 58. Goulson D. 2003. Bumblebees: their behaviour and ecology. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 59. Koch H, Cisarovsky G, Schmid-Hempel P. 2012. Ecological effects on gut bacterial communities in wild bumblebee colonies. J. Anim. Ecol. 81:1202–1210 [DOI] [PubMed] [Google Scholar]
- 60. Dornhaus A, Chittka L. 2005. Bumble bees (Bombus terrestris) store both food and information in honeypots. Behav. Ecol. 16:661–666 [Google Scholar]
- 61. Schmid-Hempel P. 2001. On the evolutionary ecology of host-parasite interactions: addressing the question with regard to bumblebees and their parasites. Naturwissenschaften 88:147–158 [DOI] [PubMed] [Google Scholar]
- 62. Sorensen A, Busch TM, Vinson SB. 1985. Trophallaxis by temporal subcastes in the fire ant, Solenopsis invicta, in response to honey. Physiol. Entomol. 10:105–111 [Google Scholar]
- 63. Mehdiabadi NJ, Schultz TR. 2010. Natural history and phylogeny of the fungus-farming ants (Hymenoptera: Formicidae: Myrmicinae: Attini). Myrmecol. News 13:37–55 [Google Scholar]
- 64. Carina Audisio M, Torres MJ, Sabate DC, Ibarguren C, Apella MC. 2011. Properties of different lactic acid bacteria isolated from Apis mellifera L. bee-gut. Microbiol. Res. 166:1–13 [DOI] [PubMed] [Google Scholar]
- 65. Lofgren CS, Banks WA, Glancey BM. 1975. Biology and control of imported fire ants. Annu. Rev. Entomol. 20:1–30 [DOI] [PubMed] [Google Scholar]
- 66. Kleerebezem M, Boekhorst J, Van Kranenburg R, Molenaar D, Kuipers OP, Leer R, Tarchini R, Peters SA, Sandbrink HM, Fiers MWEJ. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. U. S. A. 100:1990–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schloss PD, Westcott SL. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl. Environ. Microbiol. 77:3219–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Naidu A, Bidlack W, Clemens R. 1999. Probiotic spectra of lactic acid bacteria (LAB). Crit. Rev. Food Sci. Nutr. 39:13–126 [DOI] [PubMed] [Google Scholar]
- 69. Champomier-Verges MC, Chaillou S, Cornet M, Zagorec M. 2001. Lactobacillus sakei: recent developments and future prospects. Res. Microbiol. 152:839–848 [DOI] [PubMed] [Google Scholar]
- 70. Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. 2007. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 35:e120 doi:10.1093/nar/gkm541 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We deposited the 84 sweat bee-associated, 79 Mycocepurus-associated, and 56 Solenopsis-associated sequences in the genetic sequence database at the National Center for Biotechnical Information (NCBI GenBank accession numbers KC354148 to KC354367).