Abstract
Chemolithoautotrophic nitrifying bacteria release soluble organic compounds, which can be substrates for heterotrophic microorganisms. The identities of these heterotrophs and the specificities of their interactions with nitrifiers are largely unknown. In this study, we incubated nitrifying activated sludge with 13C-labeled bicarbonate and used stable isotope probing of 16S rRNA to monitor the flow of carbon from uncultured nitrifiers to heterotrophs. To facilitate the identification of heterotrophs, the abundant 16S rRNA molecules from nitrifiers were depleted by catalytic oligonucleotides containing locked nucleic acids (LNAzymes), which specifically cut the 16S rRNA of defined target organisms. Among the 13C-labeled heterotrophs were organisms remotely related to Micavibrio, a microbial predator of Gram-negative bacteria. Fluorescence in situ hybridization revealed a close spatial association of these organisms with microcolonies of nitrite-oxidizing sublineage I Nitrospira in sludge flocs. The high specificity of this interaction was confirmed by confocal microscopy and a novel image analysis method to quantify the localization patterns of biofilm microorganisms in three-dimensional (3-D) space. Other isotope-labeled bacteria, which were affiliated with Thermomonas, colocalized less frequently with nitrifiers and thus were commensals or saprophytes rather than specific symbionts or predators. These results suggest that Nitrospira spp. are subject to bacterial predation, which may influence the abundance and diversity of these nitrite oxidizers and the stability of nitrification in engineered and natural ecosystems. In silico screening of published next-generation sequencing data sets revealed a broad environmental distribution of the uncultured Micavibrio-like lineage.
INTRODUCTION
Chemolithoautotrophic ammonia- and nitrite-oxidizing microorganisms catalyze nitrification, which is a key process of the biogeochemical nitrogen cycle and important for excess nitrogen elimination from sewage in biological wastewater treatment plants (WWTPs). Key nitrifiers in most domestic and industrial WWTPs are ammonia-oxidizing bacteria (AOB) (1) related to the genera Nitrosomonas and Nitrosospira (2, 3) and nitrite-oxidizing bacteria (NOB) of the genus Nitrospira (3–5). Usually these organisms occur in tight cell clusters, which are embedded in the extracellular matrix of biofilms or activated sludge flocs (5, 6). Aside from the nitrifiers, most nitrifying bioreactors host a great diversity of other organisms, most of which presumably are heterotrophs that feed on organic substrates present in the sewage (7). Interestingly, however, soluble microbial products (SMP) released by the autotrophic AOB and NOB, and decaying nitrifier biomass, also can support the growth of heterotrophs in WWTPs (8) and in other environments such as drinking water treatment facilities (9). Heterotrophic growth supported by nitrifiers as primary producers can be quite extensive, as heterotrophic bacteria represented 50% of the microbial community in a nitrifying biofilm that received ammonia and bicarbonate-CO2 as the sole energy and carbon sources, respectively (10). Accordingly, the growth of nitrifiers and the resulting increase of heterotrophic biomass can pose serious hygienic problems in sensitive applications such as drinking water treatment (11, 12). Dissecting the flow of nutrients from nitrifiers to heterotrophs is a nontrivial task that requires cultivation-independent methods to detect the in situ uptake and assimilation of substrates. Previous studies (10, 13) applied fluorescence in situ hybridization with rRNA-targeted probes (FISH) and microautoradiography (MAR) (14) to monitor the cross-feeding of heterotrophs by nitrifiers in biofilm. This elegant approach revealed a niche differentiation among heterotrophs, which fed on different radiolabeled organics (10) and metabolites or cellular decay products of nitrifiers (13). These results, which were based on the use of FISH probes covering large phylogenetic groups, led to the question of how specific such interactions might be at a higher phylogenetic resolution. It would also be interesting to analyze the in situ spatial distribution of the nitrifiers and heterotrophs, because localization patterns can provide important hints about the specificity and nature of interactions among microbes in biofilms and flocs (15).
In this study, we combined stable isotope probing of RNA (RNA-SIP) (16) with the full-cycle rRNA approach (17) to specifically identify heterotrophic bacteria that received carbon from nitrifiers in activated sludge. Stable isotope probing has successfully been used to monitor the carbon flow in natural environments (18–23). Following the incubation of activated sludge with H13CO3− and NH4+ or NO2−, 13C-labeled RNA was separated from unlabeled RNA by isopycnic centrifugation. To facilitate the identification of heterotrophs, we used novel locked nucleic acid enzymes (LNAzymes) (24) to specifically deplete the 16S rRNA of AOB and NOB in the complex RNA mixture prior to the isopycnic centrifugation. The 16S rRNA in the separated fractions was reverse transcribed and PCR amplified, and the amplicons were characterized by terminal restriction fragment length polymorphism (T-RFLP) analysis, cloning, and sequencing. Once the 16S rRNA sequences of potential cross-feeding heterotrophs had been obtained by RNA-SIP, we designed specific FISH probes to detect the respective organisms in situ. Most interestingly, this approach revealed a previously unknown interaction between Nitrospira-like NOB and an uncultured alphaproteobacterium. The high specificity of this interaction was demonstrated by quantifying the spatial localization patterns of these organisms in the sludge flocs.
MATERIALS AND METHODS
Sludge sampling and stable isotope labeling.
Activated sludge samples were collected from a full-scale nitrifying sequencing batch reactor of the municipal WWTP of Ingolstadt, Germany. The suspended biomass was diluted 1:8 (vol/vol) in sludge supernatant, and 30-ml aliquots of the diluted sludge were incubated in the dark in six 100-ml sterilized crimp-sealed flasks in a water bath set at 23°C (the temperature in the WWTP reactor was 20°C) with horizontal shaking (60 rpm). Ammonium (0.25 mM) was added to two flasks, one containing also NaH13CO3 (2 mM) and the other one NaH12CO3 (2 mM). Two other flasks, also containing either [13C]bicarbonate or [12C]bicarbonate, were amended with 0.25 mM nitrite. During the incubation, ammonium and nitrite were replenished by adding, on average, 50 μl of a NH4Cl or NaNO2 stock solution (final concentration, 0.25 mM) every 3 h with an automated peristaltic pump. A preliminary experiment had shown that this rate of feeding did not cause accumulation of ammonium or nitrite, respectively (data not shown). These incubations were done to label with 13C first the autotrophic AOB or NOB and subsequently any heterotrophs feeding on 13C-labeled exudates or biomass of nitrifiers. The experiments including [12C]bicarbonate were controls for SIP. To distinguish carbon fixation by AOB or NOB from nitrifier-independent anaplerotic CO2 fixation (25), the last two flasks were amended only with NaH13CO3 or NaH12CO3 and contained neither ammonium nor nitrite. The headspace of all flasks was aerated, by using an automated air pump, every 4 h for 2 min. The depletion of ammonium in the respective flasks was regularly confirmed colorimetrically (26), whereas the use of nitrite was monitored by using nitrite test stripes (Merckoquant; Merck, Darmstadt, Germany). The pH was adjusted daily to 7.5 by adding appropriate amounts of 175 mM [12C]NaHCO3 or [13C]NaHCO3 (the final concentration of HCO3− was 5 to 10 mM). Prior to the start of the incubation and at selected time points (12 h, 36 h, 4.5 days, 12 days, 21 days, and 41 days), 1 ml of each culture was fixed in 2% (vol/vol) formaldehyde for 3 h at 4°C, and 4 ml of each culture was subdivided into four equal aliquots. The sludge in these aliquots was shortly allowed to settle by gravitation, the supernatant was removed, and the concentrated sludge was frozen in liquid N2. Fixed aliquots were resuspended in a 1:1 mixture of 1× phosphate-buffered saline and 96% (vol/vol) ethanol and kept at −20°C, whereas frozen aliquots were stored at −80°C until further use.
Nucleic acid extraction.
Frozen activated sludge pellets were resuspended in 1 ml of TRIzol (Invitrogen, Carlsbad, CA). The suspension was transferred into Lysing Matrix A tubes (MP Biomedicals, Solon, OH), and cells were disrupted in a bead beater (Bio 101, Vista, CA) at 6.5 m/s for 45 s. Subsequently, RNA and DNA were extracted according to the recommendations provided with TRIzol. DNA was removed from RNA preparations by DNase I (Sigma-Aldrich, St. Louis, MO) digestion in a modified (slightly lower pH and addition of CaCl2) DNase I buffer (20 mM Tris-HCl [pH 8.0], 2 mM MgCl2, 0.2 mM CaCl2) for 45 min at 37°C. After the digestion, the RNA was extracted again with TRIzol.
Isotope ratio mass spectrometry (IRMS).
The 13C content of total extracted RNA was measured on days 12, 21, and 41 to determine the incorporation of 13C into biomass during the incubations. RNA concentrations were determined using a NanoDrop ND-1000 spetrophotometer (NanoDrop Technologies, Wilmington, DE). An aliquot of 500 ng of RNA was added into a tin capsule already containing 7 μg of unlabeled proline-sucrose solution. This spiking was necessary to increase the total amount of C and to improve the reliability of measuring the 13C content in small amounts of 13C-enriched RNA. Samples were dried at 60°C overnight and analyzed by an elemental analyzer (EA 1110, CE Instruments, Wigan, United Kingdom) coupled via a ConFlo III device to the isotope ratio mass spectrometer (DeltaPLUS; Thermo Fisher Scientific). The 13C content of the RNA was calculated according to the following formula: atoms percent 13CRNA = (atoms percent 13Ctotal × Ctotal − atoms percent 13Cspike × Cspike)/(Ctotal − Cspike).
LNAzyme digestion.
The LNAzymes used in this study were purchased from Exiqon (Vedbaek, Denmark) and are listed in Table S1 in the supplemental material. The LNAzyme-mediated depletion of the native 16S rRNA of AOB and NOB was carried out as described elsewhere (24). We simultaneously applied two LNAzymes with helper probes (see Table S1) to achieve a significant depletion of the target 16S rRNA types of nitrifiers. After cleavage, the LNAzymes were digested with DNase I for 45 min at 37°C. Subsequently, 10 μl of 50 mM EDTA was added, and DNase I was inactivated by heating the mixture to 70°C for 10 min. After this step, the noncleaved rRNA (of microorganisms other than the known nitrifiers) was ready for isopycnic centrifugation.
Isopycnic centrifugation.
Cesium trifluoroacetate (GE Healthcare, Munich, Germany) density gradients were prepared as described by Whiteley et al. (27). A total of 500 ng of RNA was added to the gradient solution, which had a starting density of 1.8 g/ml. OptiSeal tubes (4.9 ml) filled with the gradient solution were sealed and spun in a VTi 90 rotor in an Optima L-100 XP ultracentrifuge (Beckman Coulter, Brea, CA) at 130,000 × g for 72 h. The gradients were then fractionated by displacing the gradient solution with water by using a syringe pump (World Precision Instruments, Sarasota, FL) at a flow rate of 0.75 ml/min. Fractions were collected at intervals of 20 s, and the density of each fraction was determined by using a digital refractometer (AR 200; Reichert Analytical Instruments, Depew, NY). Glycogen (30 μg; Ambion, Austin, TX) and 2 volumes of ice-cold isopropanol were added to each of the fractions, which then were stored at −20°C until further processing. To precipitate RNA, the samples were centrifuged at 14,000 × g at 4°C for 20 min. The supernatant was discarded, and the pellet was washed with 500 μl of ice-cold 70% (vol/vol) ethanol and again centrifuged (14,000 × g and 4°C for 5 min). After brief air-drying, the pellet was resuspended in 50 μl of diethyl pyrocarbonate (DEPC)-treated water and stored at −80°C. To treat water with DEPC, 1 ml of DEPC was added to 1 liter of ultrapure water (Milli-Q; Millipore, Bedford, MA) in a prebaked (180°C for 6 h) glass bottle. This water was then stirred for 24 h on a magnetic stirrer and finally autoclaved (121°C for 15 min) before use.
PCR and RT-PCR.
All PCRs and reverse transcriptase PCRs (RT-PCRs) were performed as described in reference 24. The primers used in this study for PCR and RT-PCR are listed in Table S1 in the supplemental material and were obtained from Thermo Fisher Scientific (Ulm, Germany). For PCRs and for RT-PCRs, the nucleic acid templates were diluted 1:10 in ultrapure water in order to avoid PCR inhibition by highly concentrated template or inhibitory compounds.
T-RFLP.
For T-RFLP analysis, primer 8F (see Table S1 in the supplemental material) labeled with 6-carboxyfluorescein (6-FAM) was used in PCRs or RT-PCRs. Amplicons of the correct size were excised from agarose gels and purified by using the QiaQuick gel extraction kit (Qiagen). BesTRF software (28) was used to determine which restriction enzyme yielded the optimal resolution of T-RFLP for specific phylogenetic groups. Approximately 100 ng of purified amplicons was digested for 3 h at 37°C using either the restriction endonuclease MspI or RsaI (Fermentas). The digestion products were analyzed by using an ABI 3130xl genetic analyzer (Applied Biosystems, Foster City, CA) and PeakScanner 1.0 software (Applied Biosystems). An in-house Perl script was used to remove an artifact peak that was present in all T-RFLP profiles in this and a previous (24) study. Real peaks were distinguished from noise by using the method described by Abdo et al. (29) and by using the Perl and “R” scripts provided on their website (http://www.ibest.uidaho.edu/tools/trflp_stats/index.php).
Cloning, sequencing, and phylogenetic analysis of 16S rRNA genes.
Cloning and sequencing of PCR-amplified 16S rRNA genes were performed as described elsewhere (24). The obtained 16S rRNA gene sequences were aligned using the SINA Webaligner (http://www.arb-silva.de/aligner) and imported into ARB (30) using the SILVA 108 sequence database (31). The sequence alignments were refined manually. Phylogenetic trees were computed by maximum likelihood analysis (RAxML [32]) with a 50% conservation filter for Bacteria. Chimeric sequences were detected by independent phylogenetic analyses of the first, central, and last ≈300 base positions. Sequences showing unstable phylogenetic affiliations in these tests were further inspected for anomalies by using Mallard software (33).
Next-generation sequencing (NGS) reads of 16S rRNA gene amplicons from environmental samples were extracted from the SRA (34) and VAMPS (http://vamps.mbl.edu/resources/databases.php) databases on 4 and 5 June 2012. The sequence data sets (see Table S4 in the supplemental material) were downloaded manually according to the classification of “ecological metagenomes” provided by the NCBI Taxonomy Browser (http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?mode=Root). NGS data sets from the same kind of environment were saved in a common folder on disk for subsequent processing. Raw fasta files were produced from fastq files by using the fastq-dump program of the sratoolkit software suite (version 2.2.0), which is available on the SRA website. Sequences longer than 200 bp were then collected in newly created, specific databases for each kind of environment by using tools of the BLAST+ suite (35). These databases were searched by BLAST (36) using the Micavibrio-like P7H12 clone sequence (JQ815012), representatives of different Nitrospira lineages, and known predatory bacteria as queries (see Table S3 in the supplemental material). To avoid false-positive hits due to partial sequence similarities, the BLAST results were filtered by a self-written Perl script to exclude all NGS reads, whose alignment to the most similar query sequence was shorter than 80% of the NGS read length and whose sequence similarity to the query along this aligned region was below 97%. The 80% alignment length threshold was chosen because the raw NGS reads still contained barcode and primer sequences, and because the sequence read quality was often low toward the 5′ and 3′ ends. The hits which met the minimal alignment length and similarity criteria were counted for each query organism.
Probe design, FISH, and digital image analysis.
16S rRNA-targeted oligonucleotide probes specific for organisms identified in this study were designed by using the “Probe Design” and “Probe Match” tools of ARB (30). To determine the optimal stringent hybridization conditions, formamide concentration series (37) were carried out using the new probes and activated sludge, which was collected during our experiments and contained the respective target organisms according to the T-RFLP data. FISH with rRNA-targeted oligonucleotide probes was performed according to reference 38 with the probes listed in Table S1. Probes labeled with FLUOS or with the dye Cy3 or Cy5 were obtained from Thermo Fisher. Stacked optical sections (z-stacks) of probe-labeled bacteria in sludge flocs were recorded by using a Zeiss LSM 510 Meta confocal laser scanning microscope equipped with two HeNe lasers (543 and 633 nm, respectively) and one argon laser (458, 477, 488, and 514 nm). The xy resolution of the images was 512 by 512 or 1,024 by 1,024 pixels, and the axial distances between the images in the z-stacks were 0.25 to 0.5 μm. Biomass objects of probe-target organisms were detected by three-dimensional (3-D) image segmentation based on voxel intensity thresholding by the RATS-L method (39). The detected 3-D objects (cells and cell aggregates) were counted automatically by using our image analysis software daime (39). The spatial arrangement patterns of probe-target organisms were quantified in z-stacks by a new 3-D implementation of the original 2-D “Inflate Algorithm” (15) that is part of the daime software. In all these analyses, the Micavibrio-like alphaproteobacterium was the “analyzed population.” The “reference population” was either sublineage I Nitrospira or other NOB (sublineage II Nitrospira). In the 3-D version of the algorithm, all biomass objects of the reference population are virtually and iteratively enlarged (“inflated”) in 3-D space by 3-D dilations with a spherical structuring element (see Fig. S1 in the supplemental material). The size of the structuring element determines the increase in size (in μm) of the inflated objects after each dilation. Thus, a given number of dilations also indicates a particular distance in 3-D space from the original, not dilated objects of the “reference” population. After each dilation, the overlapping voxels of the inflated “reference” population and the unmodified “analyzed” population are counted (Fig. S1). From this number, we subtract the number of voxels that overlapped after the previous dilation. The result is divided by the total number of voxels that belong to the analyzed population in the z-stack. This fraction represents the density of the analyzed population at the current distance from the reference population. The current distance (in μm) is inferred from the respective number of dilations (see above). As a null hypothesis, we assume that the populations are randomly distributed. This is tested by repeating the whole procedure with artificial image stacks, which contain the real reference population and an in silico-generated and randomly distributed analyzed population, whose overall density in the image stack is equal to that of the real analyzed population. The fractions of overlapping voxels, which have been obtained with the real images, are divided by the respective fractions obtained with these artificial images. Then a value of 1 indicates random distribution of the real analyzed population at the respective distance. Finally, these normalized values are plotted against the corresponding distances (in μm) between the reference and analyzed populations. In these plots, values of 1 indicate random spatial distribution of the populations, values of >1 indicate coaggregation, and values of <1 indicate mutual avoidance. Further details of the algorithm are provided in reference 15. 3-D visualizations of confocal image stacks were rendered by the daime software.
Nucleotide sequence accession numbers.
The nucleotide sequences obtained in this work have been deposited in the GenBank database (accession numbers JQ814956 to JQ815075).
RESULTS
Stable isotope probing and phylogenetic analyses.
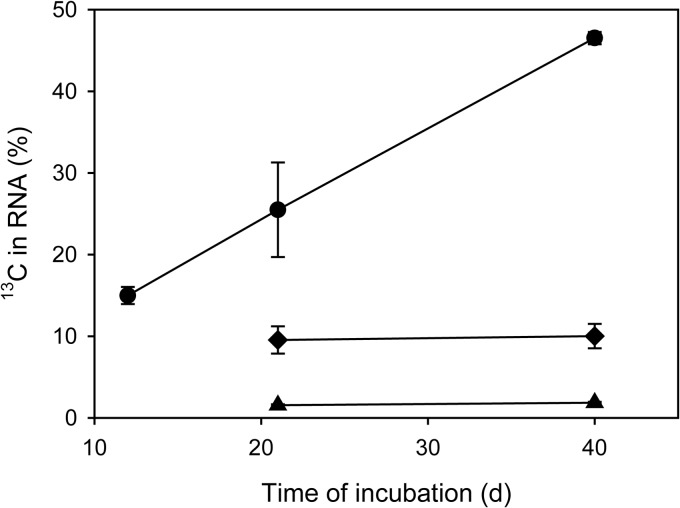
A quick prescreening analysis of the extracted (but not LNAzyme-treated) RNA by IRMS showed a substantial increase of its 13C content during the incubation of activated sludge with ammonium (Fig. 1). This increase is consistent with the expected autotrophic fixation of the added [13C]bicarbonate by the nitrifiers. During the incubations with nitrite, the increase of the 13C content was much weaker (Fig. 1), and as expected, no increase was observed in the absence of ammonia and nitrite (Fig. 1). We assumed that a successful SIP-based identification of heterotrophs that received carbon from nitrifiers would require a considerable incorporation of 13C into their rRNA. Hence, we did not further analyze the samples that were incubated with nitrite due to the relatively weak 13C labeling. Instead, we focused on the incubations with ammonium and on the negative control without any added energy source. RNA extracted from these samples after 21 days of incubation was subjected to isopycnic centrifugation and subsequent bacterial 16S rRNA-specific RT-PCR and T-RFLP analysis. Although the 13C labeling had been even stronger after 41 days (Fig. 1), we did not analyze this time point, because we assumed an increasing amount of rRNA from nonnitrifiers to be labeled due to carbon exchange among heterotrophs instead of direct carbon transfer from the nitrifiers. As expected, after incubation with ammonium and H13CO3−, the 16S rRNA in the heaviest fractions of the density gradient (density > 1.82 g/ml) was dominated by 16S rRNA of AOB and NOB, as the T-RFLP profiles obtained from these fractions contained prominent peaks characteristic for these organisms (data not shown). This result was confirmed by RT-PCR and cloning of 16S rRNA from these fractions and by sequencing of 45 randomly selected clones, which all carried 16S rRNA genes related to Nitrosomonas or Nitrospira (data not shown). The dominance of labeled 16S rRNA from nitrifiers complicated the identification of other organisms that were 13C labeled during the experiment and thus had received labeled carbon from the nitrifiers. A conceptually simple solution would be to sequence large numbers of randomly selected clones, possibly after prescreening the clones by restriction fragment length polymorphism analysis or a similar technique, in order to find cloned 16S rRNA genes of nonnitrifiers. We wanted to avoid such a labor-intensive screening and sequencing of many clones and decided to use a method that would further improve the sensitivity of T-RFLP, where dominant nitrifier peaks might mask changes of peaks belonging to other organisms. Therefore, we applied catalytic oligodeoxyribonucleotides containing locked nucleic acids (LNAzymes) that cleave target RNA molecules with a high specificity at a defined site (40, 41). If specific LNAzymes cleave native rRNA molecules between the primer binding sites, the cleaved rRNA is not amplified during RT-PCR (see Fig. S2 in the supplemental material). Hence, appropriate LNAzymes can be used to deplete selected dominant rRNA types in a complex mixture, and this depletion leads to a relative increase in the abundance of rarer sequence types (24). Consequently, we repeated the isopycnic centrifugation of RNA, but prior to this step, we used previously developed LNAzymes targeting the genera Nitrosomonas and Nitrospira (24) (see Table S1 in the supplemental material) to deplete the 16S rRNA molecules of these nitrifiers. RT-PCR and T-RFLP analysis were performed as described above. Based on a comparison of the resulting T-RFLP profiles to the profiles obtained without LNAzyme treatment, this approach resulted in a 4-fold decrease of the 16S rRNA genes of nitrifiers from the heavy fractions of the density gradient (data not shown). Thus, the identification of other 16S rRNA types was greatly facilitated. Nevertheless, 13C-labeled 16S rRNA from nitrifiers still was abundant in the heavy RNA fractions, showing that the LNAzyme-based digestion of their rRNA was not complete (Fig. 2).
Fig 1.
13C content of RNA, which was extracted from activated sludge incubated with H13CO3−, as measured by IRMS. Circles represent the experiment with NH4+ as the energy source and H13CO2− as the carbon source, diamonds the experiment with NO2− and H13CO2−, and triangles the control experiment without any added energy source and H13CO2−.
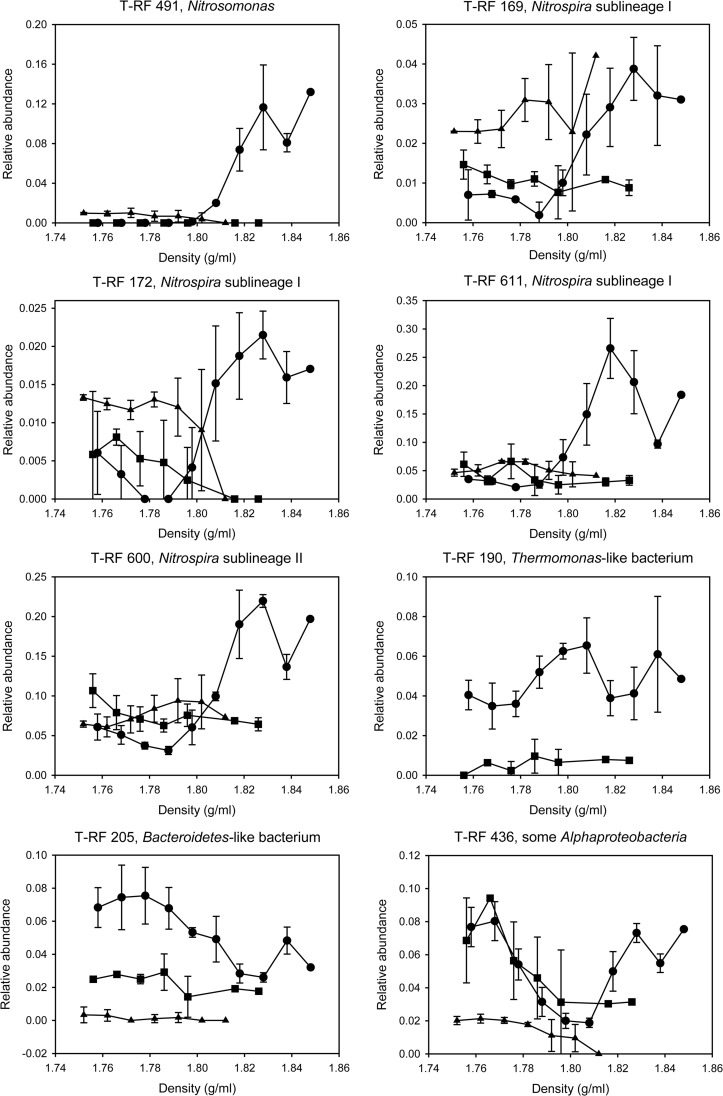
Fig 2.
Relative abundances of T-RFs, after digestion with MspI, which represent key nitrifying and heterotrophic populations in the light and heavy fractions of the density gradients after RNA-SIP. The analysis was conducted after the samples had been incubated for 21 days. Error bars show 1 standard deviation (two replicates). Missing data points indicate that no PCR amplicon could be obtained at these gradient densities in the respective experiment. Circles represent the experiment with NH4+ as the energy source and H13CO2− as the carbon source, squares the control experiment with NH4+ and H12CO2−, and triangles the control experiment without any added energy source and H13CO2− (in this control experiment, Thermomonas T-RF 190 was not detected above the threshold applied for peak calling). Note the different y axis scales in the graphs.
Like for the non LNAzyme-treated sample, a 16S rRNA gene library was established from the heavy fraction (density = 1.823 g/ml) of the LNAzyme-treated RNA sample (incubated for 21 days with H13CO3− and NH4+) after RT-PCR. Approximately 900-bp-long sequences were obtained from 120 randomly selected clones. Based on a 97% sequence similarity threshold, 37 operational taxonomic units (OTUs) were defined, which were distributed across 10 bacterial phyla (see Table S2 in the supplemental material). The most frequent OTUs in the library represented AOB (OTUs 2 to 4; 10.8% abundance), NOB (OTUs 30 to 32; 45% abundance), Micavibrio-related alphaproteobacteria (OTU 9; 5% abundance), Thermomonas-related betaproteobacteria (OTU 5; 5% abundance), and a member of the phylum Bacteroidetes (OTU 17; 5.8% abundance) (see Table S2).
Based on the 16S rRNA sequences, which had been retrieved from the sample incubated with NH4+ and after LNAzyme treatment of the RNA, we could assign additional T-RFLP peaks to organisms in the T-RFLP profiles. As expected, the terminal restriction fragments (T-RFs) belonging to AOB or NOB increased in relative abundance in the heavy fractions after incubation with NH4+ and [13C]bicarbonate (Fig. 2). However, if the RT-PCR products were cleaved by the restriction endonuclease MspI, an additional 436-nucleotide-long T-RF was identified to be abundant in the clearly 13C-labeled fractions and also in the lighter fractions (Fig. 2). According to an in silico analysis, several different alphaproteobacterial sequences from the clone library could have yielded this T-RF. Analyses of the sequences by BesTRF software (28) showed that the restriction enzyme RsaI would allow us to distinguish these alphaproteobacterial sequences in the T-RFLP profiles. Indeed, T-RFLP analyses using RsaI revealed that the aforementioned prominent T-RF from the 13C-labeled (heavy) fractions represented the Micavibrio-like alphaproteobacterial OTU that was also abundant in the 16S rRNA gene library (see Table S2 and Fig. S3 in the supplemental material). Another T-RF representing a gammaproteobacterial, Thermomonas-like OTU (see Table S2) was detected mainly in fractions of intermediate density (Fig. 2; see also Fig. S3 in the supplemental material), indicating that this group was not 13C labeled as strongly as the Micavibrio-like alphaproteobacterial OTU. The T-RFLP analyses of the density fractions showed that for all other detected organisms, the ratio of labeled to unlabeled RNA was lower than for the Micavibrio-like and Thermomonas-like OTUs. For example, although a high number (5.8%) of clones from the heavy RNA fraction belonged to the Bacteroidetes (see Table S2), the majority of this population was detected in the light fractions and thus was mainly unlabeled (Fig. 2; see also Fig. S3 in the supplemental material).
None of the aforementioned T-RFs increased in relative abundance in the heavy density gradient fractions after incubation of the activated sludge with [12C]bicarbonate (Fig. 2). This result confirmed that the increase, which was observed with [13C]bicarbonate, was not an artifact but resulted from the incorporation of 13C into the RNA of the respective organisms. Consistently, in the control experiments with [13C]bicarbonate but without added NH4+ or NO2−, the T-RFs of the nitrifiers and of the Micavibrio-like bacteria did not increase in the heavy compared to the light fractions (Fig. 2). No T-RF of the Thermomonas-like OTU was detected in these experiments. Thus, the labeling of the Thermomonas- or Micavibrio-like organisms was detectable only after incubation of the sludge with ammonium and was most likely not due to heterotrophic CO2 fixation, which could in principle also cause positive signals in SIP experiments (25).
In situ detection of nitrifiers and heterotrophs.
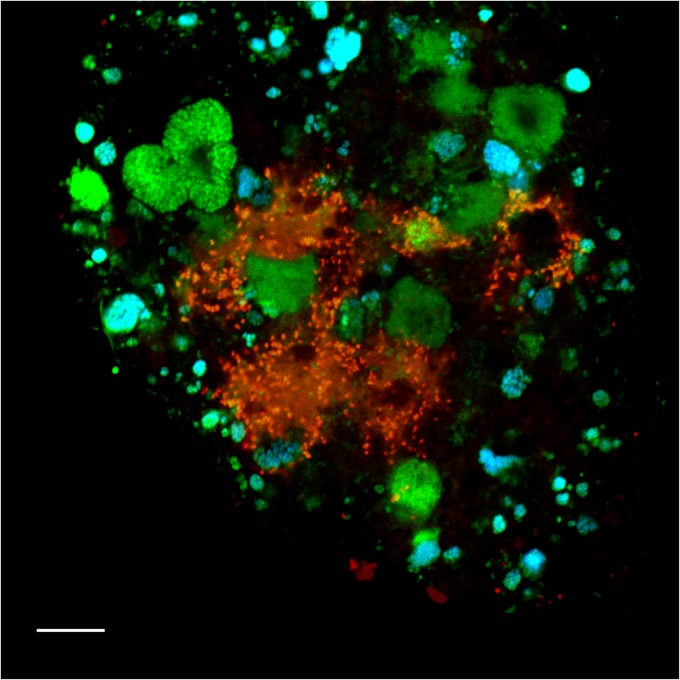
FISH with specific 16S rRNA-targeted oligonucleotide probes (see Table S1 in the supplemental material) was used to confirm that the organisms, which had clearly been labeled by 13C according to SIP, indeed were present in the activated sludge samples. As expected, betaproteobacterial AOB and Nitrospira-like NOB were abundant in the original activated sludge from the WWTP and in the incubated samples. By using suitable probes (42), Nitrospira of the phylogenetic sublineages I and II of this genus (5) could be distinguished and were found to coexist in the sludge. All detected AOB and NOB formed tight microcolonies embedded in the sludge flocs. They frequently occurred in close spatial vicinity, which reflects their mutualistic symbiosis (4, 43). FISH with the newly designed probe Thmm115, which targets the Thermomonas-like betaproteobacterium (see Table S1 in the supplemental material), detected small (1- to 1.3-μm) rod-shaped cells. These organisms did not form aggregates, but they occasionally agglomerated in the neighborhood of nitrifiers (Fig. 3). However, loose aggregations of Thmm115-labeled cells were also found at large distances from any detectable nitrifiers, and no specific spatial coaggregation pattern of these populations was observed. Consistent with the T-RFLP data, the Thermomonas-like organism was most abundant in the samples from day 21 of the incubations supplemented with ammonium. It was not detected by FISH and T-RFLP in the original activated sludge.
Fig 3.
Fluorescence micrograph showing the Thermomonas-like organism (red), Nitrospira (cyan), and other bacteria (green) detected by FISH in activated sludge. Applied probes were Thmm115 (labeled with Cy3), Ntspa662 (labeled with Cy5), and the EUB338 probe mix (labeled with FLUOS). The Thermomonas-like bacterium appears more red than yellow due to a weak EUB338 signal obtained for this organism. Bar = 10 μm.
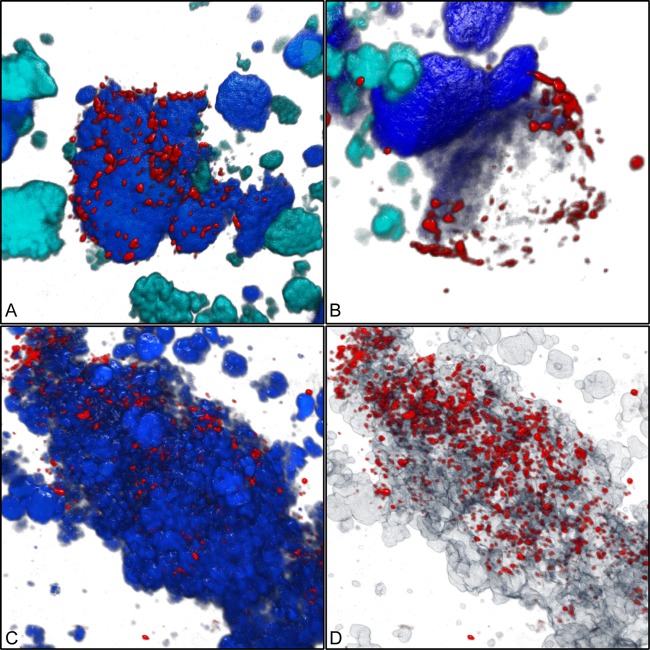
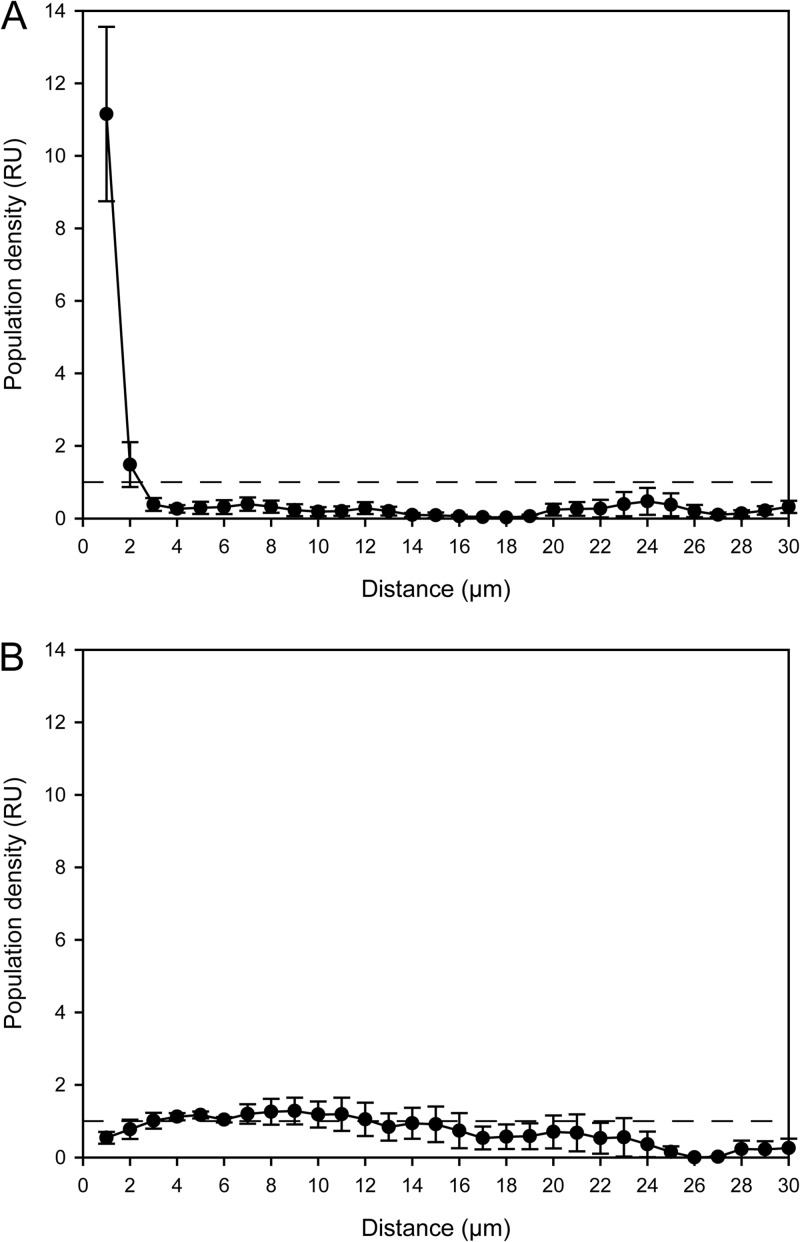
Both newly designed oligonucleotide probes targeting the Micavibrio-like organism (see Table S1 in the supplemental material) detected short (0.7- to 1-μm) cells that were almost exclusively found in the direct vicinity of Nitrospira sublineage I cell colonies (Fig. 4). This very close spatial colocalization was observed in all investigated incubated samples and in original activated sludge collected at different time points. While only few of the probe-labeled Nitrospira cell aggregates colocalized with the Micavibrio-like bacterium, the latter was almost never found at larger distances away from Nitrospira sublineage I. Interestingly, we found several Nitrospira colonies that were surrounded by Micavibrio-like cells, which seemed to be attached to the surface of the Nitrospira clusters and sometimes appeared to have migrated or grown into the Nitrospira aggregates (Fig. 4A, C, and D). Occasionally, Micavibrio-like cells were found close to weakly fluorescent FISH-labeled Nitrospira clusters (Fig. 4B). To verify the qualitative observation that the Micavibrio-like organism and sublineage I Nitrospira specifically coaggregated, we applied digital image analysis to quantify the spatial distribution patterns of the nitrifiers and the Micavibrio-like bacteria. Previously developed algorithms for spatial analyses of microbial populations (15, 39) process batches of 2-D images, which must be taken at random x, y, and z positions within a specimen. This approach works best if the target populations are so abundant that they occur in most of the recorded images. The Micavibrio-like bacteria, however, were rare in the sludge samples. They would have been found only in a very few of the randomly taken 2-D images, which might not contain sufficient information about localization patterns with other organisms. This problem was solved by a 3-D approach that analyzed a larger volume around the few detected Micavibrio-like bacteria and thus lowered the risk of overlooking colocalized other bacteria. We acquired 3-D confocal z-stacks at four positions where the Micavibrio-like organism was detected and recorded the fluorescent signals of the different FISH probes targeting this bacterium, sublineage I Nitrospira, and sublineage II Nitrospira. The recorded volumes ranged from 33,203 to 71,150 μm3. Automated counting of 3-D objects by the daime software revealed that the image stacks together contained at least 538 Micavibrio-like cells and small cell aggregates, 236 cell clusters of sublineage I Nitrospira, and 502 cell clusters of sublineage II Nitrospira. The count of Micavibrio-like bacteria is an underestimation, because the software could not split all adjacent Micavibrio-like cells in the 3-D images prior to counting. Of the 236 sublineage I Nitrospira cell clusters, 9 (3.8%) were surrounded by Micavibrio-like bacteria. The multicolor z-stacks were analyzed by a new 3-D implementation of the 2-D “Inflate Algorithm” (15) as described in Materials and Methods. These analyses confirmed a very pronounced coaggregation of the Micavibrio-like organism with sublineage I Nitrospira within short distances, below 2 μm (Fig. 5A). Beyond this distance range, the density of Micavibrio was lower than expected if the populations were randomly distributed (Fig. 5A). As only a few cell clusters of sublineage I Nitrospira were surrounded by Micavibrio-like cells (see above), this result shows that the tendency to coaggregate was a feature of Micavibrio only and not of Nitrospira. In contrast, the spatial arrangement pattern of the Micavibrio-like bacterium and sublineage II Nitrospira lacked any coaggregation signal (Fig. 5B).
Fig 4.
Three-dimensional visualization showing the coaggregation of the Micavibrio-like bacterium with sublineage I Nitrospira. (A) Micavibrio-like cells (red) attached to a cell aggregate of sublineage I Nitrospira (blue). Other nitrifiers (AOB and sublineage II Nitrospira) are shown in cyan. Applied probes were HSAL723 and HSAL866 (both labeled with Cy3), Ntspa662 (labeled with Cy5, rendered as blue), Ntspa1151 (labeled with FLUOS, rendered as cyan), and NEU, Cluster6a192, and Nso1225 (labeled with FLUOS, rendered as cyan). (B) Micavibrio-like cells surrounding a partly dark (possibly degraded) sublineage I Nitrospira cell aggregate. Colors and probes are like in panel A. (C) Micavibrio-like cells (red) attached to a cell aggregate of Nitrospira (probe Ntspa662; blue). (D) Same position as shown in panel C, but Nitrospira clusters are rendered semitransparent to show that Micavibrio-like cells also penetrated the inner parts of the Nitrospira cell colonies.
Fig 5.
Spatial arrangement pattern analysis of the Micavibrio-like organism and nitrifiying bacteria. In either plot the horizontal dashed line (y = 1) indicates random distribution at the respective distance. Values above this line indicate coaggregation (i.e., the Micavibrio-like cells were more abundant at these distances than expected for randomly distributed populations). Values below this line indicate that the Micavibrio-like cells were less abundant at these distances than expected for randomly distributed populations. Error bars show the standard errors of the means (n = 4). RU, relative units. (A) Coaggregation of Micavibrio-like bacteria with sublineage I Nitrospira; (B) lack of coaggregation between Micavibrio-like bacteria and sublineage II Nitrospira.
Screening of NGS data sets for Micavibrio-like bacteria.
An in silico screening of published NGS data sets detected organisms, which are closely related to the Micavibrio-like bacterium analyzed in this study, in other bioreactors and in freshwater, soil, and extreme (e.g., hydrothermal vents) environments (see Fig. S4 in the supplemental material). Interestingly, the percentages of sequence reads from these organisms were similar to the percentages of reads from some known obligate bacterial predators, which were also detected in the same habitat types. However, the frequency of all considered predators ranged over several orders of magnitude in most data sets (see Fig. S4).
DISCUSSION
Nitrifying microorganisms, which fix inorganic carbon, can support the growth of heterotrophic organisms by excreting organic compounds or by providing substrates to saprophytes after cell lysis (8, 13). Although nitrifiers might have important functions as primary producers, in addition to their roles in nitrogen cycling, little is known about the specificity and nature of their interactions with heterotrophs in engineered or natural ecosystems. It remains largely unexplored which heterotrophs benefit from AOB and/or NOB and whether these organisms are mutualistic symbionts, merely commensals or saprophytes, or even parasites of the nitrifiers. Experiments using radiolabeled substrates and FISH-MAR successfully started to dissect the complex food web in nitrifying activated sludge (10, 13). This approach is efficient if broad group-specific FISH probes are used in the FISH-MAR experiments, but it lacks the high phylogenetic resolution that may be needed to identify specific biological interactions. An alternative would be to establish from the sample a general 16S rRNA clone library and use this sequence information to design many (possibly hundreds) highly specific FISH probes, which could then be used in numerous FISH-MAR experiments to specifically identify the substrate-labeled organisms. This “brute force” approach would be very tedious and time-consuming and may easily overlook rare organisms that are underrepresented in the clone library. Therefore, we followed a different strategy and applied RNA-SIP, which is a true discovery tool that directly identifies also unknown isotope-labeled organisms and provides access to their 16S rRNA sequences for further analysis. Subsequently, we completed the full-cycle rRNA approach (17) by using FISH with newly designed probes to detect two isotope-labeled putative heterotrophs in situ.
Stable isotope probing.
Betaproteobacterial AOB and Nitrospira-like NOB were expected to be active and fix inorganic carbon during the incubations of activated sludge in the presence of NH4+, whereas only NOB should be active nitrifiers during the incubations with NO2−. Consistently, 16S rRNA genes of both AOB and NOB were abundant in a clone library established from 13C-labeled RNA after incubation with ammonium and H13CO3. The weak 13C labeling of RNA in the late phase of the incubation with NO2− indicates an unexpectedly low carbon fixation and/or ribosome biosynthesis by Nitrospira under the applied conditions, although nitrite was removed during this incubation (data not shown) and the supplied nitrite concentration (0.25 mM) should have supported the growth of Nitrospira spp. (42), which are adapted to relatively low nitrite concentrations (44–46). This outcome of the incubations with NO2− did not impair our study, because the experiments with NH4+ allowed us to analyze interactions of heterotrophs with AOB and NOB.
The 16S rRNA sequences of 13C-labeled heterotrophs were difficult to obtain due to the dominance of 16S rRNA from nitrifiers in the 13C-labeled RNA fraction. This problem was solved by applying LNAzymes, which can strongly facilitate the detection of rare organisms in highly uneven microbial communities (24). In this study, the use of LNAzymes helped us to obtain from the heavy rRNA fraction several sequences of organisms not known to be nitrifiers (see Table S2 in the supplemental material). Two of these organisms, a Thermomonas- and a Micavibrio-like bacterium, were 13C labeled in the presence of NH4+ as indicated by the T-RFLP profiles from the light and heavy fractions and the control experiments (Fig. 2; see also Fig. S3 in the supplemental material). The other organisms (see Table S2) may also have obtained 13C from nitrifiers, but the labeling was too weak to be confirmed by the combination of SIP and T-RFLP analysis. In addition, LNAzyme 215 is not perfectly specific and might thus have cleaved the 16S rRNA of some nonnitrifiers (see Table S1). As the original activated sludge samples and the RNA preparations had not been screened for presence of these nonnitrifiers prior to LNAzyme treatment, we cannot exclude the possibility that unspecific action of LNAzyme 215 hampered their detection in our experiments. To confirm uptake of small amounts of carbon from nitrifiers by the other putative heterotrophs (see Table S2), a more sensitive method like FISH-MAR with H14CO3− could be used in future experiments with newly designed FISH probes that target the sequences obtained by SIP.
In situ analysis and lifestyle of the Thermomonas- and Micavibrio-like bacteria.
In this study, we focused on the two potential heterotrophs that were labeled according to SIP. FISH showed that one of these organisms, the Thermomonas-like bacterium, sometimes occurred in the spatial neighborhood of nitrifiers (Fig. 3) but also grew far away from any known AOB and NOB. Together with the relatively weak 13C labeling, this distribution pattern suggests that the Thermomonas-like organism is not a specific symbiont of nitrifiers but a heterotroph that occasionally takes up soluble organic products excreted by nitrifiers, or substrates set free due to the decay of nitrifier biomass, or extracellular polymeric substances (EPS) that enclose the microcolonies of nitrifiers. This lifestyle would be consistent with the properties of the closely related isolate Thermomonas brevis, which was obtained from a denitrifying bioreactor, reduces nitrate and nitrite, and grows on a variety of substrates, including sugars and N-acetylglucosamine (47). Its transiently high abundance in our incubations but not the original sludge indicates that this organism was only temporarily favored, either by the incubation conditions or by biotic factors such as the decline and cell lysis of another population in the sludge.
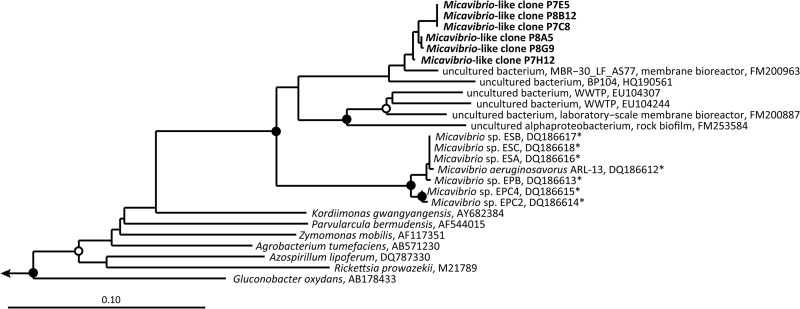
The coaggregation of the Micavibrio-like bacterium and Nitrospira sublineage I, which was observed in the incubated samples and in the source activated sludge, indicates an interaction of these organisms different from the nonobligate saprophytism or commensalism proposed for Thermomonas. The absence of the Micavibrio-like organism at larger distances from Nitrospira and its quantified strong tendency to tightly coaggregate with sublineage I Nitrospira (Fig. 5A), but not with other Nitrospira members (Fig. 5B), suggest that this interaction is highly specific and, at least in the examined sludge, also obligate for Micavibrio. The characterized members of the genus Micavibrio (48) are obligate epibiotic predators of Gram-negative bacteria (49, 50). These motile organisms attach to the outer membrane of their prey, but different from other predators such as Bdellovibrio, they do not penetrate the periplasmic space and divide simply by binary fission. The attacked cells are completely consumed (51). Furthermore, Micavibrio spp. are able to prey on target bacteria that aggregate within biofilms and thus are not limited to attacking planktonic cells (52, 53). This mode of predation strikingly resembles the direct attachment of the Micavibrio-like cells to Nitrospira microcolonies in the sludge flocs (Fig. 4) and would lead to 13C labeling of the Micavibrio-like population. It would also explain the occasional agglomerations of Micavibrio-like bacteria around apparently empty space or weakly probe-labeled sublineage I Nitrospira clusters (Fig. 4B). Completely or partly degraded prey cells would not be detectable by FISH, or they would emit only dim fluorescence signals due to the degradation or leakage of ribosomes. The high specificity for Nitrospira sublineage I is in line with the high prey specificity that has been observed for Micavibrio and other bacterial predators (51, 54, 55). The observation that most sublineage I Nitrospira microcolonies did not coaggregate with Micavibrio-like cells would also be consistent with a predator-prey relationship, where usually only a fraction of the prey population is attacked by the predator at any time. Shemesh and Jurkevitch (56) observed that bacterial prey populations developed a transient resistance to predation by Bdellovibrio, which was a plastic phenotypic response rather than a genetic mutation. As this resistance was not total, both the predator and the prey survived (56). It is tempting to speculate that only few sublineage I Nitrospira colonies were surrounded by Micavibrio-like cells, because the major part of the Nitrospira population had developed resistance. Moreover, different T-RFs representing sublineage I Nitrospira (Fig. 2) indicate a high microdiversity in this sublineage that was not resolved by the applied FISH probes, which had a broader specificity. Most of these sublineage I populations might not have been susceptible to predation by the Micavibrio-like bacteria. Alternatively, many of the Nitrospira clusters may have been inaccessible to the Micavibrio-like organism if they were deeply embedded in EPS or located in the center of dense activated sludge flocs, or they were simply not encountered yet by Micavibrio-like cells seeking prey. Other possible interactions between Nitrospira and the Micavibrio-like organism would be inconsistent with the current notion that Micavibrio spp. are predators but cannot be excluded without direct evidence for predation. For example, the detected transfer of 13C could also be explained by a mutualistic symbiosis that involves the transfer of substrates. Such a mutualistic relationship can also be reflected by a tight spatial coupling of the partners (for example, see reference 57). Moreover, the Micavibrio-like organism might be a previously unrecognized autotrophic nitrifier that directly fixed inorganic 13C and colocalized with other nitrifiers, because AOB and NOB are mutualistic symbionts (43) and often coaggregate (4, 42). A mutualistic relationship of any kind, however, would be beneficial for both partners. It should thus be reflected by mutual coaggregation, which was not observed for Nitrospira relative to the Micavibrio-like bacterium. If the latter indeed was a novel nitrifier, one would also not expect such a highly specific colocalization with only one Nitrospira sublineage. It seems to be more likely that the Micavibrio-like organism, if not a predator, could be a highly specialized heterotrophic commensal or saprophyte feeding on particular organic products or cell components of sublineage I Nitrospira. Further hints could be obtained from chemical and isotopic analyses of sludge pore water and the floc matrix. This could show whether extracellular 13C-labeled compounds are small molecules, which likely have been secreted by living Nitrospira, or proteins and other larger cell components that more probably have been released from lysed Nitrospira cells. Such analyses could be difficult to conduct with the complex microbial community in activated sludge but should be more straightforward once coenrichment cultures of Nitrospira and Micavibrio-like bacteria become available. As outlined in reference 49, bacterial predators such as Micavibrio might have evolved from saprophytic ancestors, which attached to the envelope of dead cells and secreted lytic enzymes. According to this evolutionary model, the common ancestry with the known predatory Micavibrio species (Fig. 6) would be consistent with a saprophytic or a predatory lifestyle of the Micavibrio-like organism.
Fig 6.
Maximum likelihood tree based on 16S rRNA sequences, which shows the phylogenetic affiliation of the Micavibrio-like bacteria detected in this study (in boldface). Strains which are known predators are marked by an asterisk. Sequences retrieved from WWTPs in other studies are marked by “WWTP.” Solid circles on tree nodes indicate >90% and open circles >70% maximum likelihood bootstrap support (1,000 iterations). Representative beta- and gammaproteobacteria were used as outgroups. The scale bar indicates 0.1 estimated change per nucleotide.
Ecological considerations.
This study provides further evidence of a role for autotrophic nitrifiers as primary producers, which supply heterotrophic microbial community members with organic sources of carbon and energy. While such carbon transfer was detectable in the activated sludge samples analyzed in this study, this additional ecological function of nitrifiers likely is more important in oligotrophic environments where the ambient concentrations of organic nutrients are much lower than in wastewater. Examples include drinking water treatment facilities (9, 11, 12) and even cave ecosystems that may be driven primarily by nitrite oxidation (58). As demonstrated for the Micavibrio-like organism, interactions of heterotrophic bacteria with nitrifiers can be more specific than previously recognized. So far, only protozoan grazing (23, 59, 60) and bacteriophage attack (61) have been considered biological controls of nitrifier populations. Our results indicate that bacterial predation may be another regulator of nitrification. Whether its impact on the population densities of nitrifiers is marginal or significant, and whether it may account for nitrification failure in WWTPs, awaits clarification in future research. However, predation could even have positive effects. The specific control of nitrifier populations by predators may prevent any particular nitrifier strain from becoming dominant. Thus, predators could be a selective force toward a high diversity of susceptible and resistant strains within nitrifier communities, which may lead to high functional redundancy and thus improve the overall resistance of a nitrifying WWTP or a natural ecosystem to environmental perturbations (62).
Micavibrio-related 16S rRNA sequences have been retrieved from other bioreactors (7, 10, 63) and from soils (64) (Fig. 6). In addition, our analysis of NGS data sets revealed the presence of these organisms in many kinds of habitats (see Fig. S4 in the supplemental material). Solely based on their phylogenetic affiliation and sequence similarity to known Micavibrio spp., one cannot determine whether all these environmental Micavibrio-like organisms are predators. Here we show that one member of this group (Fig. 6) is involved in a highly specific interaction with other Gram-negative bacteria (Nitrospira). However, the aforementioned screening of NGS data sets did not reveal any correlation between the occurrence of Micavibrio-like bacteria and members of the genus Nitrospira (data not shown). This may indicate that no biological interactions exist between these organisms in the respective habitats, but the lack of a correlation might also be due to possible biases of nucleic acid extraction or PCR amplification in the respective studies. Future research, possibly including genomic comparisons to already sequenced predators such as Micavibrio aeruginosavorus (50) and Bdellovibrio bacteriovorus (65), may illuminate the lifestyle of the Micavibrio-like bacteria and their impact on the ecological functions of their interaction partners or prey.
ACKNOWLEDGMENTS
We thank Christian Baranyi for technical assistance and Norbert Kohler and Konrad Thoma for providing us with sludge samples from the WWTP Ingolstadt. We thank Margarete Watzka for performing the IRMS analyses.
This work was funded by the Graduate School “Symbiotic Interactions” of the University of Vienna and partly by the Vienna Science and Technology Fund (WWTF, grant LS09-040).
Footnotes
Published ahead of print 18 January 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03408-12.
REFERENCES
- 1. Mussmann M, Brito I, Pitcher A, Sinninghe Damste JS, Hatzenpichler R, Richter A, Nielsen JL, Nielsen PH, Muller A, Daims H, Wagner M, Head IM. 2011. Thaumarchaeotes abundant in refinery nitrifying sludges express amoA but are not obligate autotrophic ammonia oxidizers. Proc. Natl. Acad. Sci. U. S. A. 108:16771–16776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koops HP, Purkhold U, Pommerening-Röser A, Timmermann G, Wagner M. 2003. The lithoautotrophic ammonia-oxidizing bacteria, p 778–811 In Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. (ed), The prokaryotes: a handbook on the biology of bacteria, 3rd ed Springer Science+Business Media, New York, NY [Google Scholar]
- 3. Schramm A, de Beer D, Wagner M, Amann R. 1998. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl. Environ. Microbiol. 64:3480–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Juretschko S, Timmermann G, Schmid M, Schleifer K-H, Pommerening-Röser A, Koops H-P, Wagner M. 1998. Combined molecular and conventional analyses of nitrifying bacterium diversity in activated sludge: Nitrosococcus mobilis and Nitrospira-like bacteria as dominant populations. Appl. Environ. Microbiol. 64:3042–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Daims H, Nielsen JL, Nielsen PH, Schleifer KH, Wagner M. 2001. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl. Environ. Microbiol. 67:5273–5284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wagner M, Rath G, Amann R, Koops H-P, Schleifer K-H. 1995. In situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251–264 [Google Scholar]
- 7. Juretschko S, Loy A, Lehner A, Wagner M. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84–99 [DOI] [PubMed] [Google Scholar]
- 8. Rittmann BE, Regan JM, Stahl DA. 1994. Nitrification as a source of soluble organic substrate in biological treatment. Water Sci. Technol. 30:1–8 [Google Scholar]
- 9. Martiny AC, Albrechtsen HJ, Arvin E, Molin S. 2005. Identification of bacteria in biofilm and bulk water samples from a nonchlorinated model drinking water distribution system: detection of a large nitrite-oxidizing population associated with Nitrospira spp. Appl. Environ. Microbiol. 71:8611–8617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kindaichi T, Ito T, Okabe S. 2004. Ecophysiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by microautoradiography-fluorescence in situ hybridization. Appl. Environ. Microbiol. 70:1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Regan JM, Harrington GW, Baribeau H, De Leon R, Noguera DR. 2003. Diversity of nitrifying bacteria in full-scale chloraminated distribution systems. Water Res. 37:197–205 [DOI] [PubMed] [Google Scholar]
- 12. Regan JM, Harrington GW, Noguera DR. 2002. Ammonia- and nitrite-oxidizing bacterial communities in a pilot-scale chloraminated drinking water distribution system. Appl. Environ. Microbiol. 68:73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Okabe S, Kindaichi T, Ito T. 2005. Fate of 14C-labeled microbial products derived from nitrifying bacteria in autotrophic nitrifying biofilms. Appl. Environ. Microbiol. 71:3987–3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee N, Nielsen PH, Andreasen KH, Juretschko S, Nielsen JL, Schleifer K-H, Wagner M. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daims H, Wagner M. 2011. In situ techniques and digital image analysis methods for quantifying spatial localization patterns of nitrifiers and other microorganisms in biofilm and flocs. Methods Enzymol. 496:185–215 [DOI] [PubMed] [Google Scholar]
- 16. Manefield M, Whiteley AS, Griffiths RI, Bailey MJ. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amann RI, Ludwig W, Schleifer K-H. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chauhan A, Cherrier J, Williams HN. 2009. Impact of sideways and bottom-up control factors on bacterial community succession over a tidal cycle. Proc. Natl. Acad. Sci. U. S. A. 106:4301–4306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frias-Lopez J, Thompson A, Waldbauer J, Chisholm SW. 2009. Use of stable isotope-labelled cells to identify active grazers of picocyanobacteria in ocean surface waters. Environ. Microbiol. 11:512–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Glaubitz S, Lueders T, Abraham WR, Jost G, Jurgens K, Labrenz M. 2009. (13)C-isotope analyses reveal that chemolithoautotrophic Gamma- and Epsilonproteobacteria feed a microbial food web in a pelagic redoxcline of the central Baltic Sea. Environ. Microbiol. 11:326–337 [DOI] [PubMed] [Google Scholar]
- 21. Lu YH, Conrad R. 2005. In situ stable isotope probing of methanogenic archaea in the rice rhizosphere. Science 309:1088–1090 [DOI] [PubMed] [Google Scholar]
- 22. Lueders T, Kindler R, Miltner A, Friedrich MW, Kaestner M. 2006. Identification of bacterial micropredators distinctively active in a soil microbial food web. Appl. Environ. Microbiol. 72:5342–5348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moreno AM, Matz C, Kjelleberg S, Manefield M. 2010. Identification of ciliate grazers of autotrophic bacteria in ammonia-oxidizing activated sludge by RNA stable isotope probing. Appl. Environ. Microbiol. 76:2203–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dolinšek J, Dorninger C, Lagkouvardos I, Wagner M, Daims H. Depletion of unwanted nucleic acid templates by selective cleavage: LNAzymes open a new window for detecting rare microbial community members. Appl. Environ. Microbiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Feisthauer S, Wick LY, Kastner M, Kaschabek SR, Schlomann M, Richnow HH. 2008. Differences of heterotrophic (13)CO(2) assimilation by Pseudomonas knackmussii strain B13 and Rhodococcus opacus 1CP and potential impact on biomarker stable isotope probing. Environ. Microbiol. 10:1641–1651 [DOI] [PubMed] [Google Scholar]
- 26. Hood-Nowotny R, Hinko-Najera Umana N, Inselbacher E, Oswald-Lachouani P, Wanek W. 2010. Alternative methods for measuring inorganic, organic, and total dissolved nitrogen in soil. Soil Sci. Soc. Am. J. 74:1018–1027 [Google Scholar]
- 27. Whiteley AS, Thomson B, Lueders T, Manefield M. 2007. RNA stable-isotope probing. Nat. Protoc. 2:838–844 [DOI] [PubMed] [Google Scholar]
- 28. Stres B, Tiedje JM, Murovec B. 2009. BEsTRF: a tool for optimal resolution of terminal-restriction fragment length polymorphism analysis based on user-defined primer-enzyme-sequence databases. Bioinformatics 25:1556–1558 [DOI] [PubMed] [Google Scholar]
- 29. Abdo Z, Schuette UM, Bent SJ, Williams CJ, Forney LJ, Joyce P. 2006. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ. Microbiol. 8:929–938 [DOI] [PubMed] [Google Scholar]
- 30. Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar , Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüßmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glockner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stamatakis A, Ludwig T, Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463 [DOI] [PubMed] [Google Scholar]
- 33. Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734–5741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kodama Y, Shumway M, Leinonen R. 2012. The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 40:D54–D56 doi:10.1093/nar/gkr854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 37. Daims H, Brühl A, Amann R, Schleifer K-H, Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434–444 [DOI] [PubMed] [Google Scholar]
- 38. Daims H, Stoecker K, Wagner M. 2005. Fluorescence in situ hybridisation for the detection of prokaryotes, p 213–239 In Osborn AM, Smith CJ. (ed), Molecular microbial ecology. Bios-Garland, Abingdon, United Kingdom [Google Scholar]
- 39. Daims H, Lücker S, Wagner M. 2006. daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8:200–213 [DOI] [PubMed] [Google Scholar]
- 40. Santoro SW, Joyce GF. 1997. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. U. S. A. 94:4262–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vester B, Lundberg LB, Sorensen MD, Babu BR, Douthwaite S, Wengel J. 2004. Improved RNA cleavage by LNAzyme derivatives of DNAzymes. Biochem. Soc. Trans. 32:37–40 [DOI] [PubMed] [Google Scholar]
- 42. Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, Wagner M, Daims H. 2006. Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ. Microbiol. 8:1487–1495 [DOI] [PubMed] [Google Scholar]
- 43. Stein LY, Arp DJ. 1998. Loss of ammonia monooxygenase activity in Nitrosomonas europaea upon exposure to nitrite. Appl. Environ. Microbiol. 64:4098–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bartosch S, Hartwig C, Spieck E, Bock E. 2002. Immunological detection of Nitrospira-like bacteria in various soils. Microb. Ecol. 43:26–33 [DOI] [PubMed] [Google Scholar]
- 45. Schramm A, de Beer D, van den Heuvel JC, Ottengraf S, Amann R. 1999. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microsensors. Appl. Environ. Microbiol. 65:3690–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, Rattei T, Damsté JS, Spieck E, Le Paslier D, Daims H. 2010. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc. Natl. Acad. Sci. U. S. A. 107:13479–13484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mergaert J, Cnockaert MC, Swings J. 2003. Thermomonas fusca sp. nov. and Thermomonas brevis sp. nov., two mesophilic species isolated from a denitrification reactor with poly(epsilon-caprolactone) plastic granules as fixed bed, and emended description of the genus Thermomonas. Int. J. Syst. Evol. Microbiol. 53:1961–1966 [DOI] [PubMed] [Google Scholar]
- 48. Lambina VA, Afinogenova AV, Lenabad SR, Konovalova SM, Pushkareva AP. 1982. Micavibrio admirandus new-genus new-species. Mikrobiologiia 51:114–117 [PubMed] [Google Scholar]
- 49. Jurkevitch E. 2007. Predatory behaviors in bacteria—diversity and transitions. Microbe 2:67–73 [Google Scholar]
- 50. Wang Z, Kadouri DE, Wu M. 2011. Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics 12:453 doi:10.1186/1471-2164-12-453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Davidov Y, Huchon D, Koval SF, Jurkevitch E. 2006. A new alpha-proteobacterial clade of Bdellovibrio-like predators: implications for the mitochondrial endosymbiotic theory. Environ. Microbiol. 8:2179–2188 [DOI] [PubMed] [Google Scholar]
- 52. Dashiff A, Junka RA, Libera M, Kadouri DE. 2011. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J. Appl. Microbiol. 110:431–444 [DOI] [PubMed] [Google Scholar]
- 53. Kadouri D, Venzon NC, O'Toole GA. 2007. Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl. Environ. Microbiol. 73:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guerrero R, Pedrosalio C, Esteve I, Mas J, Chase D, Margulis L. 1986. Predatory prokaryotes: predation and primary consumption evolved in bacteria. Proc. Natl. Acad. Sci. U. S. A. 83:2138–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rashidan KK, Bird DF. 2001. Role of predatory bacteria in the termination of a cyanobacterial bloom. Microb. Ecol. 41:97–105 [DOI] [PubMed] [Google Scholar]
- 56. Shemesh Y, Jurkevitch E. 2004. Plastic phenotypic resistance to predation by Bdellovibrio and like organisms in bacterial prey. Environ. Microbiol. 6:12–18 [DOI] [PubMed] [Google Scholar]
- 57. Orphan VJ, House CH, Hinrichs KU, McKeegan KD, DeLong EF. 2001. Methane-consuming archaea revealed by directly coupled isotopic and phylogenetic analysis. Science 293:484–487 [DOI] [PubMed] [Google Scholar]
- 58. Holmes AJ, Tujula NA, Holley M, Contos A, James JM, Rogers P, Gillings MR. 2001. Phylogenetic structure of unusual aquatic microbial formations in Nullarbor caves, Australia. Environ. Microbiol. 3:256–264 [DOI] [PubMed] [Google Scholar]
- 59. Graham DW, Knapp CW, Van Vleck ES, Bloor K, Lane TB, Graham CE. 2007. Experimental demonstration of chaotic instability in biological nitrification. ISME J. 1:385–393 [DOI] [PubMed] [Google Scholar]
- 60. Moussa MS, Hooijmans CM, Lubberding HJ, Gijzen HJ, van Loosdrecht MC. 2005. Modelling nitrification, heterotrophic growth and predation in activated sludge. Water Res. 39:5080–5098 [DOI] [PubMed] [Google Scholar]
- 61. Choi JD, Kotay SM, Goel R. 2010. Various physico-chemical stress factors cause prophage induction in Nitrosospira multiformis 25196—an ammonia oxidizing bacteria. Water Res. 44:4550–4558 [DOI] [PubMed] [Google Scholar]
- 62. Daims H, Purkhold U, Bjerrum L, Arnold E, Wilderer PA, Wagner M. 2001. Nitrification in sequencing biofilm batch reactors: lessons from molecular approaches. Water Sci. Technol. 43:9–18 [PubMed] [Google Scholar]
- 63. Huang LN, De Wever H, Diels L. 2008. Diverse and distinct bacterial communities induced biofilm fouling in membrane bioreactors operated under different conditions. Environ. Sci. Technol. 42:8360–8366 [DOI] [PubMed] [Google Scholar]
- 64. Davidov Y, Friedjung A, Jurkevitch E. 2006. Structure analysis of a soil community of predatory bacteria using culture-dependent and culture-independent methods reveals a hitherto undetected diversity of Bdellovibrio-and-like organisms. Environ. Microbiol. 8:1667–1673 [DOI] [PubMed] [Google Scholar]
- 65. Rendulic S, Jagtap P, Rosinus A, Eppinger M, Baar C, Lanz C, Keller H, Lambert C, Evans KJ, Goesmann A, Meyer F, Sockett RE, Schuster SC. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689–692 [DOI] [PubMed] [Google Scholar]